Abstract
Scope
Human milk oligosaccharides (HMOs) are complex glycans that are abundant in human milk. The potential impact of a maternal diet on individual HMOs and the association with secretor status is unknown. Thus, this study is aimed to examine the association between maternal diet and HMO profiles.
Methods and results
This is a cross‐sectional study of the MAMI cohort with 101 human milk samples from healthy mothers. HMO profiling is assessed by quantitative HPLC. Maternal dietary information is recorded through an FFQ, and perinatal factors including the mode of delivery, antibiotic exposure, and breastfeeding practices, are collected. A more significant effect of diet on HMO profiles is observed in secretor mothers than in non‐secretor mothers. (Poly)phenols and fibers, both soluble and insoluble, and several insoluble polysaccharides, pectin, and MUFA are associated with the secretor HMO profiles.
Conclusions
Maternal diet is associated with the composition and diversity of HMO in a secretor status‐dependent manner. The relationship between maternal diet and bioactive compounds, including HMOs, which are present in human milk, needs further research due its potential impact on infant development and health outcomes.
Keywords: breast milk, fiber, human milk oligosaccharides, maternal diet, secretor
There is a complex interaction between maternal diet and HMO. Maternal diet is associated with the composition and diversity of HMO human milk in a secretor status‐dependent manner. Dietary fiber and polyphenols are identified as the best dietary predictors of HMO profile in secretor mothers.
1. Introduction
Human milk is the optimal nutrition for infants during early life[ 1 , 2 ] and it contains macro‐ and micronutrients and also, several bioactive components, such as soluble immune factors, peptides, fatty acids, hormones, and stem cells.[ 3 , 4 ] These components, together with milk microbiota work synergistically to promote infant development through by impacting the maturation of the gut and immune system.[ 5 , 6 , 7 ]
Breastfeeding has been associated with a lower prevalence of several diseases, including necrotizing enterocolitis, obesity, and allergies,[ 8 , 9 , 10 ] than formula feeding, although a large variability among studies exists. Breastmilk microbiota and human milk oligosaccharides (HMOs)[ 11 ] have been identified as potential players in the mechanisms behind these observations through the interaction with the immune system during the neonatal period.[ 5 , 12 ] HMOs are complex glycans present in high concentrations in human milk representing the third largest solid component in human milk (5–15 g L−1) after lactose and milk.[ 13 ] More than a hundred of structures have been identified[ 13 , 14 ] and some maternal factors, such as genetics[ 15 , 16 ] and the stage of lactation,[ 17 ] determine HMO concentration and patterns.[ 18 , 19 , 20 , 21 ] However, the effect of other factors has been underexplored and to the best of our knowledge, only a few studies based on dietary interventions have explored the effect of maternal diets[ 11 ] and probiotics supplementation[ 22 ] on the HMO patterns. No information is available on the relationship between the HMO composition and maternal diet in observational studies. Previous data have reported an association between maternal diet and the breast milk microbial communities[ 23 ] as well as with, both maternal[ 24 ] and infant gut microbiota,[ 25 ] with potential impact on health outcomes related to growth trajectories. However, the mechanisms that drive this effect have still not been studied. HMOs and breast milk microbiota have a close relationship since they aid the growth of several beneficial bacteria that could used them to produce bioactive compounds, such as short‐chain fatty acids (SCFAs). The linkage of diets and HMOs is therefore key to understand how maternal diet could affect neonatal microbial colonization and thus, infant and adult health.
The aim of this study was to analyze the relationship between maternal diet and HMO profile in mature breast milk. The exploration of the relationship between maternal diet and HMO patterns could provide valuable knowledge for the development of future strategies targeting the milk composition.
2. Results
2.1. Clinical and Nutritional Profiles and Secretor Status
In this cross‐sectional study, the maternal secretor status phenotype was determined based on the presence or near absence (<100 nmol mL−1) of 2’FL and LNFP‐1 as secretors (n = 76/101, 75%) and nonsecretors (n = 25/101, 25%), respectively. These is in line with the evidence showing that the prevalence of nonsecretor status in a Caucasian population is approximately 20–30%.[ 15 , 26 ] All the gestations were at term (39–40 weeks). The vaginal birth rate was 63.4%, and the exclusive breastfeeding rate up to 1 month after birth was 85% across the population. No significant differences were identified among maternal clinical characteristics according to secretor status phenotype (Table 1 ) neither in macronutrients, dietary fiber nor (poly)phenol intakes.
Table 1.
Clinical and nutritional characteristics of the population
| Total | Secretor | Non‐secretor | p‐value | |
|---|---|---|---|---|
| (n = 101) | (n = 76) | (n = 25) | ||
| Maternal data | ||||
| Maternal age [years] | 34.78 ± 3.90 | 34.8 ± 4# | 34.72 ± 3.5 | 0.996 |
| Pre‐pregnancy BMI [kg m−2] | 22.6 (20.8–25.5) | 22.6 (20.8–25.4) | 22.8 (20.8–26.4) | 0.750 |
| REE [kcal per day] | 1593 (1508–1708) | 1591 (1519–1706) | 1617 (1471–1812) | 0.997 |
| Gestational age [weeks] | 40 (39–40) | 40 (39–40) | 40 (39–40) | 0.763 |
| Gestational weight gain [kg] | 12 (9.5–14.25) | 12 (10–14) | 12 (9.0–15.5) | 0.708 |
| Intrapartum antibiotic | 40 (39.6%) | 29 (38.2%) | 11 (44%) | 0.386 |
| Antibiotics during pregnancy | 30 (29.7%) | 22 (28.9%) | 8 (32%) | 0.478 |
| Delivery mode | ||||
| Vaginal | 64 (63.4%) | 49 (64.4%) | 15 (60%) | 0.431 |
| C‐section | 37 (37.6%) | 27 (35.6%) | 10 (40%) | |
| Infant birth weight [g] | 3300 (3022–3570) | 3308 (3021–3565) | 3280 (2990–3670) | 0.953 |
| Gender | ||||
| Female | 55 (54.5%) | 38 (50%) | 17 (68%) | 0.090 |
| Male | 46 (45.5%) | 38 (50%) | 8 (32%) | |
| Exclusive breastfeeding | 86 (85.15%) | 65 (85.5%) | 21 (84%) | >0.999 |
| Dietary dataa) | ||||
| Energy [kcal per day] | 2587 (2207–2988) | 2505 (2204–2951) | 2782 (2318–3105) | 0.294 |
| Total protein [g] | 121.5 (93.3–138.6) | 114.2 (95.6–136.7) | 129.7 (108.5–152.7) | 0.090 |
| Animal source | 66.2 (52.9–85.2) | 63.9 (50.6–81.3) | 76.0 (58.2–91.93) | 0.061 |
| Vegetable source | 45.7 (39.4–56.8) | 45.7 (39.7–55.3) | 48.3 (36.5–58.5) | 0.776 |
| Total lipids [g] | 114.4 (97.8–136.6) | 113.6 (94.4–136.2) | 123.4 (107.8–144.6) | 0.130 |
| SFA | 32.0 (28.0–40.2) | 31.6 (27.8–37.2) | 34.7 (29.6–43.2) | 0.169 |
| MUFA | 54.6 (46.9–64.0) | 54.5 (46.9–63.7) | 55.73 (47.3–66.2) | 0.601 |
| PUFA | 19.0 (15.2–24.1) | 18.4 (15.5–23.5) | 21.0 (16.1–27.5) | 0.227 |
| Total carbohydrates [g] | 258.2 (200.5–296.7) | 257.5 (01.4–295.5) | 270.3 (198.1–327.9) | 0.601 |
| Polysaccharides [g] | 132.1(105.2–158.0) | 132.1 (104.7–150.1) | 131.0 (104.9–172.2) | 0.504 |
| Glucose [g] | 9.1 (6.6–12.2) | 9.1 (6.6–12.8) | 8.5 (6.9–11.7) | 0.701 |
| Lactose [g] | 10.1 (5.8–20.1) | 10.1 (6.5–20.1) | 10.1 (2.9–20.2) | 0.973 |
| Fructose [g] | 9.4 (7.0–12.5) | 9.4 (6.7–13.4) | 9.1 (7.3–12.0) | 0.744 |
| Galactose [g] | 0.25 (0.16–0.39) | 0.26 (0.16–0.4) | 0.2 (0.1–0.34) | 0.165 |
| Dietary fiber [g] | 34.8(28.6–42.7) | 33.7 (27.8–41.7) | 37.3 (30.4–46.4) | 0.173 |
| Insoluble fiber [g] | 21.42 (16.67–27.8) | 20.87 (16.31–26.19) | 23.45 (17.7–32.1) | 0.219 |
| Soluble fiber [g] | 3.92 (3.23–5.34) | 3.63 (3.19–5.34) | 4.53 (3.26–5.45) | 0.334 |
| (Poly)phenols [mg] | 1684.7 (1303.6–2033.6) | 682.6 (1289.5–1981.1) | 1713.0 (1328.2–2283.4) | 0.725 |
Categorical variables are presented as positive cases (percentage of total population) and significant difference between them tested by Fisher's exact test. Differences in quantitative variables between groups were assessed by Mann–Whitney U test and p < 0.05 was considered as significant. #, two samples with missing data; REE, resting energy expenditure.
a) n = 4 participants were removed from the dietary data analysis for over reporting (considered as an energy intake higher than 2.6 time than the average resting energy expenditure [REE] rate of the population calculated according Hronek et al.[ 27 ]
2.2. HMO Profile Is Determined by Maternal Secretor Status Phenotype
As expected, HMO concentrations were dependent on maternal secretor status (Figure 1 , Figure S1, Table S1, Supporting Information). The PCoA showed the distribution of the mothers based on their HMO profiles according to their secretor status (Figure 1A), indicating the variance in the HMO content related to secretor status. Higher total HMO concentrations (p < 0.001) and higher HMO‐bound fucose (p < 0.001) were observed in the milk of secretor mothers compared to nonsecretors mothers (Figure 1B). Specifically, secretor mothers showed a higher presence of 2′FL (p < 0.001), DFL (p < 0.001), LNFP I (p < 0.001), LNFP II (p < 0.001), LNFP III (p < 0.001), LSTc (p < 0.001), DFLNT (p < 0.001), DFLNH (p < 0.001) as well some sialylated HMOs including 3′SL (p = 0.010), 6′SL (p < 0.001), and FDSLNH (p < 0.001) (Figure 1C, Table S1 , Supporting Information). Nonsecretor mothers displayed higher concentrations of 3′FL (p < 0.001). No differences in the amount of HMO‐bound sialic acid were found between secretor and nonsecretor mothers. HMO profiles of secretor mothers showed a higher diversity (p < 0.001) and evenness (p < 0.001) than those found in nonsecretor samples (Figure 1D).
Figure 1.

Secretor phenotypes impact the HMO profile composition and diversity. A) Principal component analysis (PCA) of the mothers according to secretor status based on the HMO content. B) Differences in sialylated (Sia), fucosylated (Fuc), and total HMO (SUM) quantification according to maternal secretor status. C, D) Differences in the quantification of each measured HMO (C) and diversity/evenness richness (D) according to secretor status. Statistical differences are marked as following: * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
2.3. Maternal Nutrient Intakes and HMO Profiles Associations Are Dependent on Secretor Status
A negative association was found between the total amount of secretor HMOs and both, diversity (rho = −0.523, p ≤ 0.001) and evenness (rho = −0.511, p < 0.001) indexes. Specific HMOs in the milk of secretor women were associated with specific nutrient patterns, especially insoluble and soluble fiber, fructose, galactose, hemicellulose, and (poly)phenols, among others (Figure 2A). A higher concentration of total HMO was associated with lower maternal intakes of insoluble fiber, cellulose, hemicellulose, and (poly)phenols. These components were positively associated with some minor HMOs such as FLNH and FDSLNH, among others (Figure 2). Polyphenols were positive correlated to DFLNH (rho = 0.34, p = 0.003) and FLNH (rho = 0.28, p = 0.016), FDSLNH (rho = 0.25, p = 0.034) and DSLNH (rho = 0.24, p = 0.040). In addition, higher intakes of fructose and galactose were associated with higher 2′FL (rho = 0.30, p = 0.010, and rho = 0.24, p = 0.040; respectively) and lower 3′FL (rho = −0.24, p = 0.036, and rho = −0.29, p = 0.015).
Figure 2.
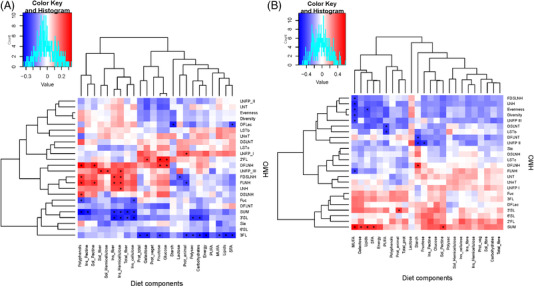
Specific maternal nutrients intakes were related to HMO concentrations in both secretor (A) and nonsecretor mothers (B). Heatmaps of Spearman correlations between HMO and dietary components intake during pregnancy. Significant correlations (p < 0.05) are marked by an asterisk (∗) and q‐values < 0.2. a‐. Blue squares represent negative correlations, whereas red squares show positive correlations.
To explore the effect of nutrient intake in the individual concentrations of each HMO detected in milk samples, multiple linear regressions were used. As Table S2, Supporting Information shows, nutrient intake was related to the concentration of several secretor HMOs in 1‐month milk samples accounting for a considerable variability in HMO concentrations (Table S2, Supporting Information). Generally, fiber and (poly)phenols were the dietary components with significant contributions to secretor HMO concentrations. The regression models thus revealed that each gram of insoluble fiber consumption led to an increase of 0.65 nmol mL−1 of FNLH in mother´s milk.
In nonsecretor women, lower intakes of MUFA were associated with higher concentrations of LNFPIII (rho = −0.41, p = 0.047) LNH (rho = −0.49, p = 0.015), FLNH (rho = −0.42, p = 0.042), and FDSLNH (rho = −0.42, p = 0.042). Furthermore, dietary starch consumption was negative correlated to DFLNT (rho = −0.42, p = 0.043) and LNFPII (rho = −0.49, p = 0.016) (Figure 2B). The multiple linear regressions indicated that fewer of individual HMOs were modulated by maternal nutrients intake in nonsecretor than in secretor mothers (Table S3, Supporting Information).
2.4. Secretor HMO Clusters Were Determined by Maternal Diet
Effect size analysis of each nutrient on the overall structure of HMO content in secretor milk revealed that different types of carbohydrates and (poly)phenols were the main sources driving the HMO profile (Figure 3A). Accordingly, the secretor HMO profiles were associated with (poly)phenols (R 2 = 0.18, p = 0.001) and fibers, both soluble (R 2 = 0.10, p = 0.028) and insoluble fiber (R 2 = 0.15, p = 0.003), and several insoluble polysaccharides, including insoluble cellulose (R 2 = 0.16, p = 0.005), hemicellulose (R 2 = 0.14, p = 0.005), and pectin (R 2 = 0.13, p = 0.015).
Figure 3.
Maternal diet during pregnancy impacts the HMO profile in secretor mothers. A) Polar plots visualizing the amount of variance of HMO profiles that could be explained by the nutrients analyzed using envFit function. The height of the bars reflects the amount of variance (R 2) explained by each covariate. Covariates are colored according to nutrient category. Asterisks indicate significant covariates (p < 0.05). B) Principal component analysis (PCA) showing the differences in the clusters of HMO. C, D) Boxplot indicating the differences in diversity and evenness (C) and the differences in maternal nutrient intake (in percentage of daily energy) according to clusters (D). E) Two dimensional nonmetric multidimensional scaling (NMDS) plot of HMO with the significant variables that impact the HMO profile.

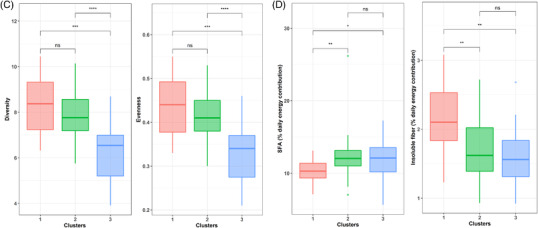
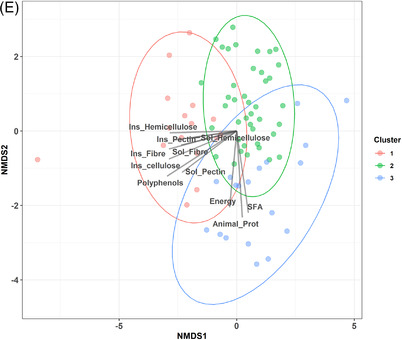
The secretor HMO profile was also grouped into distinct clusters by the k‐means method (Figure 3B). Cluster I was characterized by higher concentrations of LNH, FLNH, DSLNH, and FDSLNH (Figure S2, Supporting Information), Cluster II by higher concentrations of 3′FL and DFLNT, and Cluster III by a higher presence of LNFP I. Significant differences among clusters were identified in terms of HMO diversity (p < 0.001) and evenness (p < 0.001). Cluster I showed higher diversity and evenness than Cluster III (p < 0.001), but it showed no difference in diversity and evenness with Cluster II (p = 0.904 and p = 0.895, diversity and evenness, respectively) (Figure 3C). It was also found that mothers with a Cluster I HMO profile had a higher percentage of insoluble fiber in their daily diets than those in Cluster II (p = 0.007) and Cluster III (p = 0.007) (Figure 3D). Cluster I was characterized by mothers whose diet had a lower percentage of SFA than those in Cluster II (p = 0.021) and Cluster III (p = 0.058). The ordination plot of the mothers based on their HMO production revealed that Cluster III was linked to the consumption of SFA and animal proteins, while Cluster I was linked to (poly)phenols, fibers and hemicellulose, cellulose, and pectin (Figure 3E). These linkages indicate the relationship between the dietary consumption and the mothers’ HMO profiles.
2.5. Maternal Diets Had a Modest Impact on the HMO Profiles of Non‐Secretor Mothers
The effect of maternal diets on the overall structure of the HMO pattern of the nonsecretor mothers was less than that observed in secretor mothers (Figure 4A). Only MUFA intake appeared relevant in the HMO compositions of these mothers, but with no significant effect (R 2 = 0.17, p = 0.116). In nonsecretor mothers, MUFA intake was negatively associated with both diversity (rho = −0.44, p = 0.033) and evenness (rho = −0.47, p = 0.022) in the HMO profiles. In the same line, a negative association was observed between the total HMO with diversity (rho = −0.64, p = 0.001) and evenness (rho = −0.64, p = 0.001) in secretor mothers.
Figure 4.
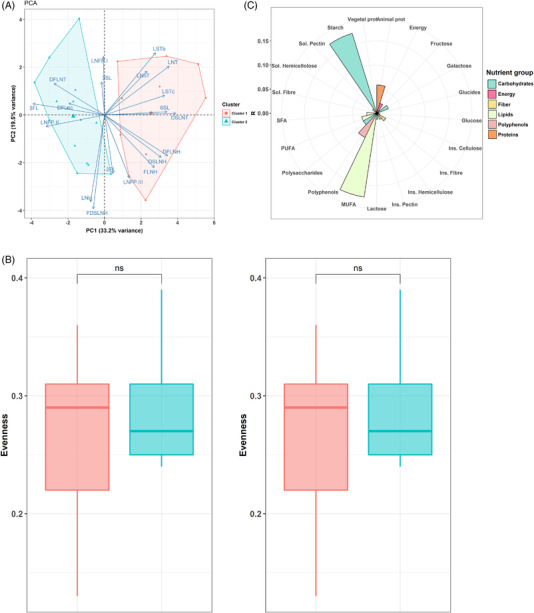
Maternal diet during pregnancy showed a narrow impact in the HMO profile of nonsecretor mothers. A) Principal component analysis (PCA) showing the differences in the clusters of HMO. B) Polar plots visualizing the amount of variance of HMO profiles that could be explained by the as nutrients analyzed using envFit function. The height of the bars reflects the amount of variance (R 2) explained by each covariate. Covariates are colored to according to nutrients category. C) Boxplot indicating the differences in diversity and evenness according to HMO clusters.
Then, the different patterns of HMO profiles in nonsecretor mothers were investigated, and two clusters were identified (Figure 4B). The Cluster I was defined by samples with higher concentrations of sialylated HMO, including 6’SL (p = 0.007), DSLNT (p < 0.001), LSTc (p = 0.001), and LNT (p < 0.001). Cluster II showed higher concentrations of 3′FL (p < 0.001) and LNFP II (p = 0.003) (Figure S3, Supporting Information). No differences in terms of HMO diversity or evenness were found among clusters in nonsecretor mothers. Nonsecretor HMO clusters were not associated with maternal nutrient intakes. Only MUFA intake showed a tendency to differ between both clusters (p = 0.074).
3. Discussion
Limited evidence on the impact of maternal diets on HMOs is available. This study reported the impact of maternal diet on the HMO profile which is dependent on the secretor phenotype. Differences in HMO profile were dependent on maternal secretor status and the specific nutrients intakes, such us fructose/galactose, fiber, and (poly)phenols. This study suggests that HMO profile may be modulated through maternal dietary interventions being this relevant for maternal‐infant health outcomes related to infant microbiota development and immune system maturation.
Human milk is an extremely complex fluid consisting of a wide range of bioactive components, such as nutrients, hormones, cytokines, immunoglobulins, and microbes.[ 3 , 28 , 29 , 30 ] These elements contribute to infant growth during the first months of life through several routes. They not only serve as a nutrient sources but play a role in other physiological processes, including immune system maturation[ 31 ] and gut microbial colonization[ 32 ] with potential impact for infant health.[ 31 , 33 , 34 ]
HMOs are a diverse family of unconjugated glycans that are the third most abundant solid component in the human milk.[ 14 ] They have been considered as a prebiotic source for beneficial bacteria in the infant gut, and could promote infant development.[ 35 ] Beyond the effect of some maternal genetic features, such as FUT2 and FUT3 gene, little is known about the potential factors that modulate the HMO profile. The results of this study, which are in agreement with previous studies,[ 36 , 37 ] showed that clustering the human milk samples according to maternal secretor status greatly explains the variability in HMO profiles.[ 20 , 21 ] Even though, the results regarding the differences in each of the HMO showed some discrepancies probably due to the differences in study designs and intrinsic characteristics of the populations.[ 15 , 20 ] Longitudinal studies have also shown that HMO content varies during the lactation period. Most studies have found that HMOs concentrations are higher at early stages of lactation and decrease later.[ 37 , 38 ] HMOs, along with other milk components, such as lipids and proteins, would be dependent on the lactation period.[ 39 , 40 ]
Other studies have reported minor effects of maternal age, pregestational BMI, delivery mode, and parity[ 15 , 37 , 38 , 41 ] on both, human milk microbiota and HMO content. It has been hypothesized that hormonal, metabolic and immune signals related to these conditions could influence the mother's ability to produce the specific HMOs.[ 41 ] This study did not find any association between maternal BMI and HMO profiles or HMO clusters. Some authors have suggested that obesity and hyperglycemia are linked to HMO biosynthesis[ 42 , 43 ] through the hexosamine pathway.[ 43 ] However, due to our limited number of obesity cases (n = 4 with BMI >30 kg m−2), we could not test these observations.
Differences in HMO profile have been described according to season and geographical location,[ 15 , 44 , 45 ] which likely indicate differences in dietary patterns. Davis et al[ 45 ] found that in the dry season, which is characterized by higher energy intake, a higher total amount of HMO was produced in the African Gambia population. Similarly, Azad et al[ 15 ] also found geographically and seasonal patterns in HMO profiles, but they related these observations to changes in environmental factors more than diet. Although data on the effect of maternal diets on HMO profiles are very scarce, previous studies have shown that the nutritional content of the human milk varied depending on the maternal intake.[ 46 , 47 , 48 ] There is evidence of a link between probiotic consumption and the variations of some HMO concentrations in the colostrum[ 22 ] as well as of the presence of diet‐derived monosaccharides on HMO structure.[ 49 , 50 ] Quin et al.’s found[ 50 ] that diet had a greater impact on HMO content in secretor mothers than in nonsecretor mothers which is in agreement with this study. They also reported that ingested monosaccharides and fruit‐derived fibers were positively correlated with galactose and fucose present in HMOs. Additionally, they positively related several sulfonated/phosphorylated HMOs to monounsaturated and polyunsaturated fats and observed the opposite trend in SFA intake.[ 50 ] Lower sialylated HMO concentrations have been consistently linked to high fat diets (contributing >40% total energy) in the scarce number of studies that has been performed.[ 11 ] Higher concentrations of 3′SL and 6′SL have been observed in obese women than in lean women.[ 37 ] Even this study did not have a case‐control design, and thus, there was not a high‐fat diet group in the analysis; a negative relationship between 3’SL and MUFA in secretor mothers and between MUFA and total sialylated HMO (data not shown) was found, which could support the hypothesis of the association between lipids and sialylated HMO. As mentioned, in agreement with Quin et al., we found that the associations between maternal diets and the HMO profiles were found more relevant in secretor mothers compared to nonsecretor mothers. However, this result needs to be confirmed in other populations since it could be influenced by the lower sample size in the nonsecretor group.
The results of this study revealed a potential link between dietary (poly)phenols and HMO profiles. (Poly)phenols are ubiquitous plant‐derived bioactive compounds with the ability to drive the composition and activity of the intestinal microbiota. Knowledge of the connection between the consumption of these compounds and HMO production in human milk is poor. Nonetheless, some hypotheses could be developed to explain this association. Considering that fruit consumption, one of the main sources of dietary (poly)phenols and fiber, has been associated with higher concentrations of galactose and fucose,[ 50 ] it seems plausible that (poly)phenols may be indirectly related to HMOs. This association is in line with the high direct correlations reported between fucose‐containing sulfated polysaccharides and (poly)phenols in vegetable products.[ 51 ] It has been also reported that dietary (poly)phenol is associated with higher intestinal levels of certain microorganisms belonging to the Bacteroides genus with the capacity to increase fucose levels in the intestinal lumen.[ 52 ]
The potential clinical relevance of these results has yet to be determined. A previous study demonstrated that the shifts in HMO composition may shape the functionality of milk microbiome which could have important consequences for infant colonization.[ 11 , 53 , 54 , 55 ] Indeed, we previously described the impact of maternal diets on the maternal[ 24 ] and infant gut microbiota[ 25 ] and on human milk microbiota,[ 23 ] but the mechanisms behind these observations were unclear. In contrast, the results of the current study indicate a possible link between the maternal nutrition and the mother‐infant microbiome. While HMOs have been shown to modulate the infant microbial communities,[ 56 ] act as substrates for specific bacterial populations,[ 57 , 58 ] and reduce pathogen adhesion,[ 59 ] they may indirectly act by other routes due to their immunomodulatory properties and their capacity to participate in gut barrier maturation.[ 60 ] The results of this study highlight the close link between maternal diet–HMO and suggest a potential mechanism by which maternal diet could impact the infant gut microbiome by shaping HMO in breast milk.
4. Conclusions
Maternal diet is associated with the composition and diversity of HMO human milk in a secretor status‐dependent manner. Dietary fiber, (poly)phenols, and MUFA were the most relevant nutrients for affecting concentrations of HMOs. These findings suggest a complex interaction among different maternal factors and HMOs. This knowledge could support the design of specific dietary interventions that modulate HMO composition to impact infant development and health.
5. Experimental Section
Study Participants and Study Design
A total of 101 healthy mothers from the MAternal MIcrobes (MAMI) cohort were included in this cross‐sectional study at 15–30 days postpartum according to the full availability of biological samples and clinical and dietary data ( Figure S0 , Supporting Information). As described elsewhere,[ 61 ] the MAMI cohort was a prospective and observational mother‐infant cohort in the Spanish Mediterranean area.
Maternal clinical and anthropometric data were collected from the clinical records including maternal age, body mass index (BMI), and pregnancy‐related data including weight gain during pregnancy, antibiotic intake, delivery mode, and intrapartum antibiotic. The project was approved by the Ethical Committee of the Hospital Clínico Universitario Valencia, Spain. The study was registered on the ClinicalTrials.gov platform, with registration number NCT03552939.
Human Milk Sample Collection
Human milk samples were collected from participants at 1 month after delivery according to a previously described protocol.[ 61 ] Briefly, the women were asked to clean their nipples and the surrounding breast skin. The first drops were discarded to avoid possible skin contamination, and the milk sample was collected in a sterile tube using a sterile pump and stored in sterile bottles to normalize the milk collection protocol. Morning collection was recommended. Human milk samples were sent to a biobank and then aliquoted and stored at −80 °C for further analysis.
Maternal Nutritional Assessment and Dietary Intake
Dietary records were collected during the first week after birth using a 140‐item food frequency questionnaire (FFQ) that was completed through personal interviews by trained personnel.[ 23 ] The FFQ questionnaire covered usual foods during pregnancy and their frequency of consumption (daily, weekly, monthly), the number of times the participant consumed a particular food item, the median portion (in household measures‐grams and milliliters), and the size of each participant's portion. The global intake of energy and macro‐ and micronutrients were estimated from the food composition tables developed by the Centro de Enseñanza Superior de Nutrición Humana y Dietética (CESNID),[ 62 ] and the different series of fatty acids were obtained through the information compiled by the United States Department of Agriculture (USDA).[ 63 ] Also, data for the estimation of the different types of fiber was completed from the research work developed by Marlett et al.,[ 63 ] and the phenolic content from Phenol Explorer database.
The FFQ data was validated previously in the dataset[ 23 ] by use of a randomly selected subsample (n = 20) through the use of a 3‐d recall food record questionnaire for the intake of dietary nutrients.
Human Milk Oligosaccharide (HMO) Profile
HMO analysis was performed by HPLC after fluorescent derivatization (Vanquish Quaternary HPLC–fluorescent detection, Thermo Fisher Scientific) as previously outlined in detail by Seferovic et al.[ 11 ] To ensure that analyte loss during the extraction procedure was accounted for, raffinose was added to the human milk samples at the beginning of sample processing. Human milk containing the internal standard was directly applied to solid phase extraction. Oligosaccharides were extracted by high‐throughput solid phase extraction over C18 (Hypercarb‐96, 25 mg bed weight, thermo scientific) and Carbograph microcolumns (Hypersep‐96 C18, 25 mg bed weight, thermos scientific) using a controlled vacuum manifold. Use of high‐throughput microcolumns was validated in multiple different ways: i) establishing parallelism in serial dilutions, ii) spiking milk with individual HMO standards to determine recovery, and iii) comparison with direct in‐sample derivatization. Oligosaccharides were fluorescently labeled with 2‐aminobenzamide (2AB, Sigma) in a 96‐well thermocycler at 65 °C for exactly 2 h. The reaction was stopped abruptly by reducing the thermocycler temperature to 4 °C. The amount of 2AB was titrated to be in excess to account for the high and variable amount of lactose and other glycans in milk samples. Unreacted 2AB was removed by high‐throughput solid phase extraction over silica microcolumns [(Hypersep silica, 25 mg bed weight, thermos scientific). Labeled oligosaccharides were analyzed by HPLC (Dionex Ultimate 3000, Dionex, now Thermo) on an amide‐80 column (15 cm length, 2 mm inner diameter, 3 µm particle size; Tosoh Bioscience)] with a 50‐mmol L−1 ammonium formate–acetonitrile buffer system. Separation was performed at 25 °C and monitored with a fluorescence detector at 360 nm excitation and 425 nm emissions. Peak annotation was based on standard retention times of commercially available HMO standards and a synthetic HMO library and offline mass spectrometric analysis on a Thermo LCQ Duo Ion trap mass spectrometer equipped with a Nano‐ESI‐source. Absolute concentrations were calculated based on HMO standard response curves for each of the annotated HMO and corrected for internal standard recovery. (Oligosaccharide detection limit: ≈20 pmol, dynamic range between 20 and 5000 pmol; milk samples were diluted accordingly). The identified HMOs were: 2′‐fucosyllactose (2′FL), 3‐fucosyllactose (3FL), 3′‐sialyllactose (3′SL), 6′‐sialyllactose (6′SL), difucosyllactose (DFL), difucosyl‐lacto‐N‐hexaose (DFLNH), difucosyl‐lacto‐N‐tetraose (DFLNT), disialyl‐lacto‐N‐hexaose (DSLNH), disialyl‐lacto‐N‐tetraose (DSLNT), fucosyl‐disialyl‐lacto‐N‐hexaose (FDSLNH), fucosyl‐lacto‐N‐hexaose (FLNH), lacto‐N‐fucopentaose (LNFP)I, LNFPII, LNFPIII, lactose‐N‐hexaose (LNH), lacto‐N‐neotetraose (LNnT), lacto‐N‐tetraose (LNT), sialyl‐lacto‐N‐tetraose b (LSTb), and sialyl‐lacto‐N‐tetraose c (LSTc). HMOs were also, classified in sialylated and fucosylated according to their chemical composition. HMO Simpson's diversity and evenness indexes were calculated based on relative HMO abundances.
Statistical Analysis
All statistical analysis was performed using RStudio v.4.1.0[ 64 ] and SPSS Statistics for Windows v. 27.0.[ 65 ] Depending on the data normality, either the Mann–Whitney U test or a t‐test was used for parametric and nonparametric data depending on the data normality to compare the characteristics of the population according to secretor status.
Spearman correlations between specific HMOs and dietary components were performed in RStudio through a ggplots package[ 66 ] and adjustment for multiple testing by FDR method (referred as q‐value in the text) were also calculated. Linear regressions were performed independently for each HMO adjusting by total energy intake in SPPS v.27.0. The following explanatory variables were included: carbohydrates (galactose, lactose, glucose, starch, insoluble, and soluble fiber), lipids (saturated (SFA), monounsaturated (MUFA) and polyunsaturated (PUFA) fatty acids), proteins (animal and vegetable), and (poly)phenols). The variables were included in the model using a forward stepwise mode.
For clustering analysis, mothers were grouped based on their HMO profiles using auto‐scaled abundance of HMOs present in each sample. Optimal number of clusters was estimated with the NbClust package[ 67 ] using the Euclidean distance and the K‐means method. Clusters were visualized using principal component analysis (PCA) with Factoextra package.[ 68 ] The comparison of the HMO concentrations among clusters was assessed by the Kruskal Wallis test with a Dunn's posthoc test and false discovery rate correction for multiple comparisons. The results were plotted using a ggplots2 package.[ 69 ]
The effect of the size and significance of each individual nutrient on the overall HMO pattern was determined using the envit function of the vegan package[ 70 ] on the ordination performed using the nonmetric multidimensional scaling (NMDS) analysis based on the Euclidean distance on the standardized HMO concentrations.
Conflict of Interest
The authors declare no conflict of interest.
Author Contributions
The authors’ responsibilities were as follows—M.C.C.: designed the study; C.M.‐C.: responsible of clinical study, recruitment, and collection of the biological samples; A.F., M.C., and L.B. analyzed milk samples for HMO composition; S.G., M.G.: responsible for the dietary information and analysis; M.S.‐R.: analyzed data and drafted the first draft; and all authors have read and approved the final manuscript.
Supporting information
Supporting Information
Acknowledgements
The authors thank the families involved in the MAMI study as well as all the members of the MAMI cohort study. This work was supported by the European Research Council under the European Union's Horizon 2020 research and innovation program (ERC starting grant, no. 639 226). They would also like to thank the support from LaMaratò‐TV3 (DIM‐2‐ELI, ref. 2018–27/31).
Selma‐Royo M., González S., Gueimonde M., Chang M., Fürst A., Martínez‐Costa C., Bode L., Collado M. C., Maternal Diet Is Associated with Human Milk Oligosaccharide Profile. Mol. Nutr. Food Res. 2022, 66, 2200058. 10.1002/mnfr.202200058
Data Availability Statement
Data available in article supplementary material – individual data available on request from the authors.
References
- 1. Eidelman A. I., Schanler R. J., Pediatrics 2012, 129, e827.22371471 [Google Scholar]
- 2. WHO 2017. [Google Scholar]
- 3. Mazzocchi A., Giannì M. L., Morniroli D., Leone L., Roggero P., Agostoni C., De Cosmi V., Mosca F., Nutrients 2019, 11, 1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hamosh M., Pediatr. Clin. North Am. 2001, 48, 69. [DOI] [PubMed] [Google Scholar]
- 5. Gao Y., Davis B., Zhu W., Zheng N., Meng D., Walker W. A., Am. J. Physiol. Gastrointest. Liver Physiol. 2021, 320, G521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garwolińska D., Namieśnik J., Kot‐Wasik A., Hewelt‐Belka W., J. Agric. Food Chem. 2018, 66, 11881. [DOI] [PubMed] [Google Scholar]
- 7. Toscano M., De Grandi R., Grossi E., Drago L., Front. Microbiol. 2017, 8, 2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Herrmann K., Carroll K., Breastfeed. Med. 2014, 9, 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kelishadi R., Farajian S., Adv. Biomed. Res. 2014, 3, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oddy W. H., Breastfeed. Rev. 2012, 20, 7. [PubMed] [Google Scholar]
- 11. Seferovic M. D., Mohammad M., Pace R. M., Engevik M., Versalovic J., Bode L., Haymond M., Aagaard K. M., Sci. Rep. 2020, 10, 22092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Walker W. A., Iyengar R. S., Pediatr. Res. 2015, 77, 220. [DOI] [PubMed] [Google Scholar]
- 13. Thurl S., Munzert M., Boehm G., Matthews C., Stahl B., Nutr. Rev. 2017, 75, 920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bode L., Glycobiology 2012, 22, 1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Azad M. B., Robertson B., Atakora F., Becker A. B., Subbarao P., Moraes T. J., Mandhane P. J., Turvey S. E., Lefebvre D. L., Sears M. R., Bode L., J. Nutr. 2018, 148, 1733. [DOI] [PubMed] [Google Scholar]
- 16. Cabrera‐Rubio R., Kunz C., Rudloff S., García‐Mantrana I., Crehuá‐Gaudiza E., Martínez‐Costa C., Collado M. C., J. Pediatr. Gastroenterol. Nutr. 2019, 68, 256. [DOI] [PubMed] [Google Scholar]
- 17. Austin S., de Castro C. A., Bénet T., Hou Y., Sun H., Thakkar S. K., Vinyes‐Pares G., Zhang Y., Wang P., Nutrients 2016, 8, 346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Urashima T., Fukuda K., Hirabayashi J., Comprehensive Glycoscience, 2nd ed, vol. 5. Elsevier, 2021, pp. 389–439. [Google Scholar]
- 19. Urashima T., Hirabayashi J., Sato S., Kobata A., Trends Glycosci. Glycotechnol. 2018, 30, SE51. [Google Scholar]
- 20. Thurl S., Munzert M., Henker J., Boehm G., Mller‐Werner B., Jelinek J., Stahl B., Br. J. Nutr. 2010, 104, 1261. [DOI] [PubMed] [Google Scholar]
- 21. Chaturvedi P., Warren C. D., Altaye M., Morrow A. L., Ruiz‐Palacios G., Pickering L. K., Newburg D. S., Glycobiology 2001, 11, 365. [DOI] [PubMed] [Google Scholar]
- 22. Seppo A. E., Kukkonen A. K., Kuitunen M., Savilahti E., Yonemitsu C., Bode L., Järvinen K. M., JAMA Pediatr. 2019, 173, 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cortes‐Macías E., Selma‐Royo M., García‐Mantrana I., Calatayud M., González S., Martínez‐Costa C., Collado M. C., J. Nutr. 2021, 151, 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Selma‐Royo M., García‐Mantrana I., Calatayud M., Parra‐Llorca A., Martínez‐Costa C., Collado M. C., Eur. J. Nutr. 2020, 60, 1429. [DOI] [PubMed] [Google Scholar]
- 25. García‐Mantrana I., Selma‐Royo M., González S., Parra‐Llorca A., Martínez‐Costa C., Collado M. C., Gut Microbes 2020, 11, 962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Metgud R., Khajuria Mamta N., Ramesh G., J. Clin. Diagn. Res. 2016, 10, ZC01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hronek M., Zadak Z., Hrnciarikova D., Hyspler R., Ticha A., Nutrition 2009, 25, 947. [DOI] [PubMed] [Google Scholar]
- 28. Munblit D., Treneva M., Peroni D. G., Colicino S., Chow L. Y., Dissanayeke S., Pampura A., Boner A. L., Geddes D. T., Boyle R. J., Warner J. O., Nutrients 532, 2017, 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boix‐Amorós A., Puente‐Sánchez F., du Toit E., Linderborg K. M., Zhang Y., Yang B., Salminen S., Isolauri E., Tamames J., Mira A., Collado M. C., Appl. Environ. Microbiol. 2019, 85, e02994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hunt K. M., Foster J. A., Forney L. J., Schütte U. M. E., Beck D. L., Abdo Z., Fox L. K., Williams J. E., McGuire M. K., McGuire M. A., PLoS One 2011, 6, e21313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. van den Elsen L. W. J. J., Garssen J., Burcelin R., Verhasselt V., Front. Pediatr. 2019, 7, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ho N. T., Li F., Lee‐Sarwar K. A., Tun H. M., Brown B. P., Pannaraj P. S., Bender J. M., Azad M. B., Thompson A. L., Weiss S. T., Azcarate‐Peril M. A., Litonjua A. A., Kozyrskyj A. L., Jaspan H. B., Aldrovandi G. M., Kuhn L., Nat. Commun. 2018, 9, 4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Azad M. B., Vehling L., Chan D., Klopp A., Nickel N. C., McGavock J. M., Becker A. B., Mandhane P. J., Turvey S. E., Moraes T. J., Taylor M. S., Lefebvre D. L., Sears M. R., Subbarao P., Pediatrics 2018, 142, 20181092. [DOI] [PubMed] [Google Scholar]
- 34. Munblit D., Peroni D. G., Boix‐Amorós A., Hsu P. S., Van't Land B., Gay M. C. L., Kolotilina A., Skevaki C., Boyle R. J., Collado M. C., Garssen J., Geddes D. T., Nanan R., Slupsky C., Wegienka G., Kozyrskyj A. L., Warner J. O., Nutrients 2017, 9, 894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lagström H., Rautava S., Ollila H., Kaljonen A., Turta O., Mäkelä J., Yonemitsu C., Gupta J., Bode L., Am. J. Clin. Nutr. 2020, 111, 769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Totten S. M., Zivkovic A. M., Wu S., Ngyuen U., Freeman S. L., Ruhaak L. R., Darboe M. K., German J. B., Prentice A. M., Lebrilla C. B., J. Proteome Res. 2012, 11, 6124. [DOI] [PubMed] [Google Scholar]
- 37. Samuel T. M., Binia A., de Castro C. A., Thakkar S. K., Billeaud C., Agosti M., Al‐Jashi I., Costeira M. J., Marchini G., Martínez‐Costa C., Picaud J. C., Stiris T., Stoicescu S. M., Vanpeé M., Domellöf M., Austin S., Sprenger N., Sci. Rep. 2019, 9, 11767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ferreira A. L., Alves R., Figueiredo A., Alves‐Santos N., Freitas‐Costa N., Batalha M., Yonemitsu C., Manivong N., Furst A., Bode L., Kac G., Nutrients 2020, 12, 790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Siziba L. P., Lorenz L., Stahl B., Mank M., Marosvölgyi T., Decsi T., Rothenbacher D., Genuneit J., Nutrients 2019, 11, 2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hartmann P. E., Kulski J. K., J. Physiol. 1978, 275, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Han S. M., Derraik J. G. B., Binia A., Sprenger N., Vickers M. H., Cutfield W. S., J. Nutr. 2021, 151, 1383. [DOI] [PubMed] [Google Scholar]
- 42. Jantscher‐Krenn E., Treichler C., Brandl W., Schönbacher L., Köfeler H., Van Poppel M. N. M., Am. J. Clin. Nutr. 2019, 110, 1335. [DOI] [PubMed] [Google Scholar]
- 43. Hebert L. F., Daniels M. C., Zhou J., Crook E. D., Turner R. L., Simmons S. T., Neidigh J. L., Zhu J. S., Baron A. D., McClain D. A., J. Clin. Invest. 1996, 98, 930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McGuire M. K., Meehan C. L., McGuire M. A., Williams J. E., Foster J., Sellen D. W., Kamau‐Mbuthia E. W., Kamundia E. W., Mbugua S., Moore S. E., Prentice A. M., Kvist L. J., Otoo G. E., Brooker S. L., Price W. J., Shafii B., Placek C., Lackey K. A., Robertson B., Manzano S., Ruíz L., Rodríguez J. M., Pareja R. G., Bode L., Am. J. Clin. Nutr. 2017, 105, 1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Davis J. C. C., Lewis Z. T., Krishnan S., Bernstein R. M., Moore S. E., Prentice A. M., Mills D. A., Lebrilla C. B., Zivkovic A. M., Sci. Rep. 2017, 7, 40466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bzikowska‐Jura A., Sobieraj P., Szostak‐Węgierek D., Wesołowska A., Nutrients 2020, 12, 2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mohammad M. A., Sunehag A. L., Haymond M. W., Am. J. Clin. Nutr. 2009, 89, 1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Innis S. M., Am. J. Clin. Nutr. 2014, 99, 734S. [DOI] [PubMed] [Google Scholar]
- 49. Rudloff S., Obermeier S., Borsch C., Pohlentz G., Hartmann R., Brösicke H., Lentze M. J., Kunz C., Glycobiology 2006, 16, 477. [DOI] [PubMed] [Google Scholar]
- 50. Quin C., Vicaretti S. D., Mohtarudin N. A., Garner A. M., Vollman D. M., Gibson D. L., Zandberg W. F., Hart G. W., J. Biol. Chem. 2020, 295, 4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Skriptsova A., Artic. J. Appl. Phycol. 2014, 27, 545. [Google Scholar]
- 52. Xu J., Bjursell M. K., Himrod J., Deng S., Carmichael L. K., Chiang H. C., Hooper L. V., Gordon J. I., Science (80‐.) 2003, 299, 2074. [DOI] [PubMed] [Google Scholar]
- 53. Smith‐Brown P., Morrison M., Krause L., Davies P. P. S. W., PLoS One 2016, 11, e0161211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lewis Z. Z. T., Totten S. S. M., Smilowitz J. T. J. J. T., Popovic M., Parker E., Lemay D. G., Van Tassell M. L., Miller M. J. M. M. J., Jin Y.‐S. S., German J. B. J. B., Lebrilla C. C. B., Mills D. A., Microbiome 2015, 3, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bai Y., Tao J., Zhou J., Fan Q., Liu M., Hu Y., Xu Y., Zhang L., Yuan J., Li W., Ze X., Malard P., Guo Z., Yan J., Li M., mSystems 2018, 3, e00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sakanaka M., Hansen M. E., Gotoh A., Katoh T., Yoshida K., Odamaki T., Yachi H., Sugiyama Y., Kurihara S., Hirose J., Urashima T., zhong Xiao J., Kitaoka M., Fukiya S., Yokota A., Lo Leggio L., Hachem M. A., Katayama T., Sci. Adv. 2019, 5, 10.1126/SCIADV.AAW7696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sela D. A., Int. J. Food Microbiol. 2011, 149, 58. [DOI] [PubMed] [Google Scholar]
- 58. Matsuki T., Yahagi K., Mori H., Matsumoto H., Hara T., Tajima S., Ogawa E., Kodama H., Yamamoto K., Yamada T., Matsumoto S., Kurokawa K., Nat. Commun. 2016, 7, 11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang Y., Zou Y., Wang J., Ma H., Zhang B., Wang S., Nutrients 2020, 12, 1284. [Google Scholar]
- 60. Plaza‐Díaz J., Fontana L., Gil A., Nutrients 2018, 10, 1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. García‐Mantrana I., Alcántara C., Selma‐Royo M., Boix‐Amorós A., Dzidic M., Gimeno‐Alcañiz J., Úbeda‐Sansano I., Sorribes‐Monrabal I., Escuriet R., Gil‐Raga F., Parra‐Llorca A., Martínez‐Costa C., Collado M. C. M. C., team M., Collado M. C. M. C., García‐Mantrana I., Alcántara C., Gimeno‐Alcañiz J., Selma‐Royo M., Boix‐Amorós A., Dzidic M., Baüerl C., Villoldo E., Zafra C., Olivares L., Pérez‐Martínez G., Mira A., Ferrer M. D., Martínez Santamaria J., Ahicart A., et al., BMC Pediatr. 2019, 19, 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cervera P., Farran A., Zamora‐Ros R., Rev. Esp. Salud Publica 2004, 78, 407. [Google Scholar]
- 63. Marlett J. A. J., Cheung T. T. F., J. Am. Diet. Assoc. 1997, 97, 1139. [DOI] [PubMed] [Google Scholar]
- 64. R. Studio Team (2020) . RStudio: Integrated Development for R. RStudio, PBC, Boston, MA http://www.rstudio.com/.
- 65. IBM Corp. Released 2020 . IBM SPSS Statistics for Windows, Version 27.0. Armonk, NY: IBM Corp.
- 66. Warners G. R., Bolker B., Bonebakker L., Gentleman W., Liaw H. A., Lumley T., Maechler M., Magrusson A., Moeller S., Schwartz M., Venables B., 2019.
- 67. Malika C., Nadia G., Veronique B., Azam N., J. Stat. Softw. 2014, 61, 1. [Google Scholar]
- 68. Kassambara A., Mundt F., 2017.
- 69. Wickham H., 2016.
- 70. Dixon P., J. Veg. Sci. 2003, 14, 927. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
Data available in article supplementary material – individual data available on request from the authors.


