To the Editor,
For the canonical antibody isotypes, antigen recognition is driven by the variable regions of both heavy and light chains. In contrast, single‐domain antibodies or nanobodies (nb) are the antigen‐binding moiety of heavy chain only antibodies occurring in camelid species and cartilaginous fish. 1 , 2 Their small‐size, high‐yield production, and stability render nanobodies versatile building blocks for the development of binders and unorthodox activatory or inhibitory multi‐domain derivatives. 3 In the allergological context, single nanobodies were used as anti‐IgE molecules and as binding moiety for allergens. 4 , 5
Our aim was to generate allergen‐specific nanobodies and establish a nanobody‐based IgE format in the context of hymenoptera venom allergy (Figure 1A,B). In venom‐allergic patients, the IgE response is typically directed to a set of major and minor allergens. 6 The best‐characterized and most abundant honeybee venom (HBV) allergens are phospholipase A2 (Api m 1) and hyaluronidase (Api m 2) (Figure S1A). Therefore, native Api m 1 and recombinant Api m 2 were used for llama immunization (Figure S1B,C). Immune libraries were generated from peripheral blood mononuclear cells. Enrichment by phage display and immunoreactivity of individual phage clones was shown by ELISA (Figure S2). After expression and purification from bacterial supernatant, the nanobodies showed the expected molecular masses in SDS‐PAGE and immunoreactivity to their target in ELISA (Figure S3A–C).
FIGURE 1.
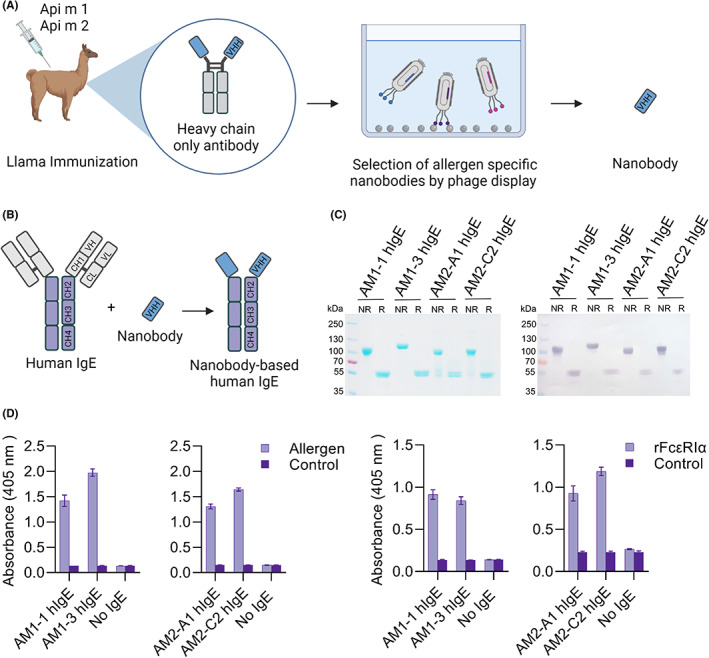
Generation of nanobody‐based human IgE against nApi m 1 and rApi m 2 (A, B) Schematic representation of the nanobody generation. Upon immunization of llamas with nApi m 1 and rApi m 2, immune repertoire libraries were generated and selected by phage display. Nanobody‐based human IgEs (nb‐IgEs) were then generated by fusion of obtained allergen‐specific nanobodies to the Fc domains of human IgE. (C) SDS‐PAGE and immunoblotting analysis of purified nb‐IgEs under non‐reducing (NR) and reducing (R) conditions. The recombinant nb‐IgE were detected using an anti‐IgE antibody conjugated to alkaline phosphatase. (D) The immunoreactivity of purified nb‐IgEs was further assessed in ELISA. Binding to immobilized allergen or FcεRIα was detected using an anti‐IgE antibody conjugated to alkaline phosphatase. Data are mean ± s.d. of triplicates
Exemplary nanobodies against Api m 1 (AM1‐1 and AM1‐3) and against Api m 2 (AM2‐A1 and AM2‐C2) were then converted into homodimeric IgE formats by fusion to IgE CH2‐4 domains. After purification from supernatant of stably transfected HEK293 cells, SDS‐PAGE and immunoblotting analyses corroborated proper dimerization of the nb‐IgE (Figure 1C, Figure S4). Expression yields of 10–20 mg/L pointed at a favorable performance of the nb‐IgE in mammalian hosts as compared to the limited yields often observed for entire IgE antibodies. 7 The nb‐IgEs remained reactive to both their particular target allergen and the FcεRIα in ELISA (Figure 1D).
The antibodies exhibited sIgE reactivity in Euroline assays to HBV and individual allergens without cross‐reactivity to other HBV or yellow jacket venom allergens (Figure 2A,B). Furthermore, individual nb‐IgE clones were applied on the ImmunoCAP test system. Concentration‐dependent sIgE reactivity was detected for Api m 1, Api m 2, and HBV (Figure 2C) corresponding well to the total IgE levels. The sIgE reactivity against Api m 1 was found highly comparable to that of HBV. In contrast, a clearly reduced IgE reactivity to HBV compared with the component Api m 2 pointed toward a lower sensitivity of HBV for patients with sensitization to Api m 2 possibly due to a limited accessibility of immunoreactive Api m 2 in the test. Furthermore, an oligoclonal artificial human serum was established by combining the 4 nb‐IgE against Api m 1 and Api m 2 in equimolar concentration. Assessment of sIgE reactivity verified the added sIgE levels for HBV, Api m 1, and Api m 2 (Figure 2D). Notably, the reduced sIgE level for HBV reflected the reduced reactivity of the individual Api m 2‐specific nb‐IgE to HBV. Hence, artificial sera comprising molecularly defined IgE surrogates of adjustable concentration might replace human sera in applications such as assessment of diagnostic test systems, round‐robin tests, and diagnostic and therapeutic extracts, but also mechanistical analyses in basic research.
FIGURE 2.
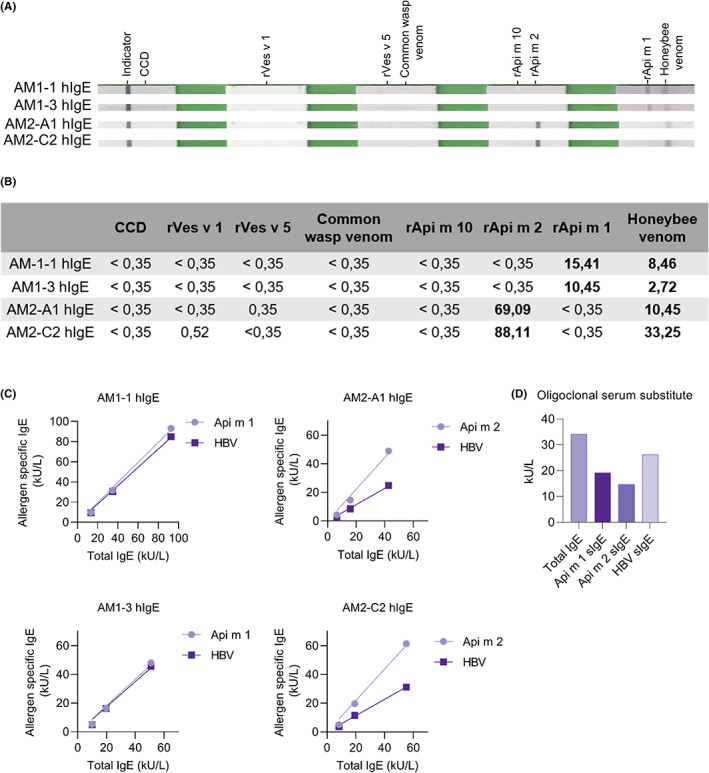
IgE measurements on diagnostic platforms. The immunoreactivity of purified nb‐IgE was further assessed in commercial test platforms. (A and B) Euroline assay. (C) ImmunoCAP measurements of tIgE and sIgE were performed for dilution series of the four different nb‐IgEs. (D) ImmunoCAP measurements of an oligoclonal serum substitute comprising the four different nb‐IgEs. Values are single measurements
In summary, we have established nanobodies as a toolbox for the generation of important downstream formats like the novel nb‐IgE. This platform also paves the way for the generation of other formats including blocking IgG formats, and hence, has the potential to provide advanced tools for diagnostics, functional analyses, and interventional studies.
CONFLICT OF INTEREST
A patent application covering part of the manuscript has been filed by JBA, CS, MM, and ES. The other authors have no financial conflict of interest and nothing to disclose.
Supporting information
Appendix S1
ACKNOWLEDGEMENTS
Gratefully acknowledged is the excellent technical assistance by Nanna Breum Nielsen, Britta Dorn, and Manuel Schulze‐Dasbeck. Figure 1 A and B was created with BioRender.com. This study was supported by the Novo Nordisc Foundation, grant NNF19OC0058484, and the Independent Research Foundation Denmark, grant 9041‐00291A.
REFERENCES
- 1. Hamers‐Casterman C, Atarhouch T, Muyldermans S, et al. Naturally occurring antibodies devoid of light chains. Nature. 1993;363(6428):446‐448. [DOI] [PubMed] [Google Scholar]
- 2. Ward ES, Güssow D, Griffiths AD, Jones PT, Winter G. Binding activities of a repertoire of single immunoglobulin variable domains secreted from Escherichia coli . Nature. 1989;341(6242):544‐546. [DOI] [PubMed] [Google Scholar]
- 3. Konning D, Zielonka S, Grzeschik J, et al. Camelid and shark single domain antibodies: structural features and therapeutic potential. Curr Opin Struct Biol. 2016;45:10‐16. [DOI] [PubMed] [Google Scholar]
- 4. Jabs F, Plum M, Laursen NS, et al. Trapping IgE in a closed conformation by mimicking CD23 binding prevents and disrupts FcepsilonRI interaction. Nat Commun. 2018;9(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zettl I, Ivanova T, Strobl MR, et al. Isolation of nanobodies with potential to reduce patients IgE binding to Bet v 1. Allergy. 2021;77(6):1751‐1760. doi: 10.1111/all.15191 [DOI] [PubMed] [Google Scholar]
- 6. Spillner E, Blank S, Jakob T. Hymenoptera allergens: from venom to “venome”. Front Immunol. 2014;5(77):1‐7. doi: 10.3389/fimmu.2014.00077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Braren I, Blank S, Seismann H, et al. Generation of human monoclonal allergen‐specific IgE and IgG antibodies from synthetic antibody libraries. Clin Chem. 2007;53(5):837‐844. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1


