Abstract
Intense research efforts over the last two decades have renewed our understanding of plant phylogeography and domestication in the Mediterranean basin. Here we aim to investigate the evolutionary history and the origin of domestication of the carob tree (Ceratonia siliqua), which has been cultivated for millennia for food and fodder. We used >1000 microsatellite genotypes to delimit seven carob evolutionary units (CEUs). We investigated genome‐wide diversity and evolutionary patterns of the CEUs with 3557 single nucleotide polymorphisms generated by restriction‐site associated DNA sequencing (RADseq). To address the complex wild vs. cultivated status of sampled trees, we classified 56 sampled populations across the Mediterranean basin as wild, seminatural or cultivated. Nuclear and cytoplasmic loci were identified from RADseq data and separated for analyses. Phylogenetic analyses of these genomic‐wide data allowed us to resolve west‐to‐east expansions from a single long‐term refugium probably located in the foothills of the High Atlas Mountains near the Atlantic coast. Our findings support multiple origins of domestication with a low impact on the genetic diversity at range‐wide level. The carob was mostly domesticated from locally selected wild genotypes and scattered long‐distance westward dispersals of domesticated varieties by humans, concomitant with major historical migrations by Romans, Greeks and Arabs. Ex situ efforts to preserve carob genetic resources should prioritize accessions from both western and eastern populations, with emphasis on the most differentiated CEUs situated in southwest Morocco, south Spain and eastern Mediterranean. Our study highlights the relevance of wild and seminatural habitats in the conservation of genetic resources for cultivated trees.
Keywords: biogeography, genetic diversity, microsatellite, origin, phylogeography, RADseq
1. INTRODUCTION
Fruit trees played a major role in the development of Mediterranean civilizations during the last millennia (Zohary & Hopf, 2012). Several species survived in refugia during the Pleistocene climatic changes and suffered repeated range expansions and contractions, which shaped their genetic diversity and structure. Their evolutionary histories represent examples of plant evolution under three important drivers (geological, climatic and human), which have been defined as the Mediterranean triptych (Thompson, 2020). Human activities represent the most recent driver of the Mediterranean triptych on shaping global biodiversity (Boivin et al., 2016). Indeed, humans have profoundly modified Mediterranean ecosystems for thousands of years, resulting in a continuum between forest and agrosystems (Quézel & Médail, 2003). As a result, it is difficult to document the evolutionary history of fruit trees, which may have cultivated, feral and wild populations in the same region (Besnard et al., 2018). In exceptional cases, intense dispersal by humans across the Mediterranean has led to the lack of robust genetic structure, for example in the chestnut (Fineschi et al., 2000) or the stone pine (Vendramin et al., 2008). By contrast, recent phylogeographic studies have revealed that imprints of ancestral populations preceding agriculture are still present in the genetic diversity and structure of some Mediterranean cultivated tree species, such as the olive tree and the wild date palm (Besnard et al., 2018; Gros‐Balthazard et al., 2017). Identifying the ancestral genetic legacy is essential to properly conserve genetic resources in the Mediterranean region, and to improve our understanding of the domestication process.
As a general pattern, the domestication of plants in the Mediterranean started in the East, in the Fertile Crescent, and was followed by human‐mediated westward dispersals of crops across the Mediterranean basin (Zeder, 2008; Zohary & Hopf, 2012). However, recent studies suggest that the cultivation of useful plants was not a rare phenomenon throughout the Mediterranean and may have involved local resources from several diversity centres in a protracted process during which genetic admixture, within or between species, played a crucial role (Fuller et al., 2011; Purugganan, 2019; Thompson, 2020).
The carob (Ceratonia siliqua, Fabaceae) is a common tree in traditional Mediterranean orchards which has traditionally been valued, and still is, for its ability to produce food and fodder in marginal lands, notably during unfavourable years. Thus, domestication of the carob tree aimed at increasing pulp in the fruit (Zohary, 2002). New uses have recently emerged, such as producing bioethanol or obtaining galactomannan from seeds as a food stabilizer. A recent review outlined the potential of carob for developing health‐beneficial food products (Brassesco et al., 2021). The carob tree has also been the subject of trials for the ecological restoration and afforestation of degraded lands (Domínguez et al., 2010). Cultivars are propagated clonally by grafting; branches (scions) from selected productive trees are grafted on carob trees (rootstocks) often obtained from seedlings. Then it is assumed that the spread of its cultivation and domestication was linked to the development of grafting methods ca. 3000 years ago (Meyer et al., 2012; Zohary, 2002). As for several crops, the Near East and the Eastern Mediterranean regions were initially proposed as the centre of domestication for the carob tree (de Candolle, 1883; Ramón‐Laca & Mabberley, 2004; Zohary, 2002). Due to the lack of international breeding programmes, each country possesses its own carob varieties and few international exchanges of selected varieties have been reported so far. The countries with the highest numbers of cultivated varieties are Spain, Portugal and Italy (Tous et al., 2013), although a lack of geographical pattern explaining the genetic structure was initially found for Spanish and Italian cultivars (Caruso et al., 2008; La Malfa et al., 2014). A recent study of the world's largest germplasm collection detected genetic structure between cultivars from south Spain and Morocco, and separated from cultivars of northeast Spain, using microsatellite and plastid markers (Di Guardo et al., 2019). This finding was congruent with studies including wild carob populations (Viruel et al., 2018, 2020), which recognized four main genetic groups across the Mediterranean and a strong west–east structure, as documented for several Mediterranean plants (Chen et al., 2014; Désamoré et al., 2011; García‐Verdugo et al., 2021; Migliore et al., 2018; Nieto Feliner, 2014). The scenario resulting from integrating the results from these studies supports the regional use, cultivation and domestication of wild carob trees in several parts of the Mediterranean. The mixed ancestry found in current cultivars was probably the result of the diffusion of selected productive, female or hermaphrodite genotypes via grafting (Di Guardo et al., 2019). Nevertheless, the effects of domestication were not homogeneous across the Mediterranean basin. For example, carob cultivation in orchards was less intensive in Andalusia and Morocco than in the rest of the distribution range (Di Guardo et al., 2019). In these two regions, the carob pods from cultivars have a low pulp content, similar to the wild‐type. By contrast, in the eastern and central Mediterranean areas, especially in Sicily, Crete and Cyprus, carob cultivation is more intensive and supports several traditional uses, suggesting an ancient history of selection and domestication. This geographical variation of domestication efforts is congruent with the habitats and plant communities where carob trees occur in the Mediterranean, being more heterogeneous in the western Mediterranean (Baumel et al., 2018).
In this study, we aim to resolve the complex evolutionary scenario of the carob tree in the Mediterranean and to investigate the footprints of domestication on the overall genetic diversity of this tree. We hypothesize (i) a stronger impact of domestication on the genetic diversity is expected in central and eastern Mediterranean populations, and (ii) human‐mediated gene flow occurred from east to west of domesticated carob trees mainly influenced by the spread of cultivars by Arabs during the Middle Ages (Ramón‐Laca & Mabberley, 2004).
Our first objective was to define evolutionary units based on geographical distribution and genetic structure data obtained from simple sequence repeat (SSR) polymorphisms and single nucleotide polymorphisms (SNPs) obtained by Viruel et al. (2018, 2020). We aimed to delimit geographically homogeneous genetic groups of carob populations (hereafter called CEUs for carob evolutionary units). Then, using these CEUs, we investigated carob genome‐wide diversity and differentiation with data developed using a reduced‐representation genomic approach, restriction‐associated DNA sequencing (RADseq), which has been successfully used to decipher evolutionary history in several tree species (Borrell et al., 2018; Hipp et al., 2020; Hodel et al., 2017; Warschefsky & von Wettberg, 2019). Our second objective was to assess the potential impact of domestication on the genome‐wide diversity of the carob tree. For this, we performed a comparative diversity analysis and search of candidate loci for carob populations in different conditions (wild vs. cultivated). Our third objective was to reconstruct the evolutionary history of the CEUs, including population splits and gene flow estimations.
2. MATERIALS AND METHODS
2.1. Plant material
We used material collected from populations of Ceratonia siliqua across the Mediterranean basin as described in Table S1. Along with the field sampling, it was challenging to distinguish between cultivated and wild trees because carob is currently distributed in a gradient of ecological conditions from natural habitats to cultivated lands. Since leaves for DNA extraction were collected from the canopy, they could belong to the grafted part of the tree. Indeed, clones were reported in Viruel et al. (2020), see also Table S1, but they were very scarce. We classified populations as wild when trees showed several trunks and small curved fruits (wild‐type). By contrast, trees cultivated in orchards with large and noncurved fruits were identified as grafted cultivars. However, in many cases, we could not find any evidence of cultivation, grafted scars or no fruits. Moreover, we often faced abandoned cultivated areas, some of which were burnt, and rootstocks probably replaced the cultivars. Therefore, to address uncertainty regarding wild or cultivated status in our analyses, we assigned each sampled carob tree to one of three types of habitats. Based on field observations, the habitat of each population was recorded as “wild,”, “‘seminatural” or “cultivated.” Examples of these three types of habitats found in our sampling are provided in Figure S1. Habitats where human impact is low and there is no evidence of recent land use were recorded as “‘wild.” These wild habitats correspond mainly to cliffs, rocky slopes or riverbanks. In wild habitats, carob trees are probably of wild or feral origin, but they have an almost null probability of having been planted or grafted. “Semi‐natural” habitats lack evidence of current cultivation but show the clear presence of past human activities such as pasture or old farming where carobs, often consociated with almond or olive trees, are occasionally harvested. In these seminatural habitats, the carob trees may be of wild origin, originating from close thermophilous vegetation that recolonizes the fields and terraces (Baumel et al., 2018), or they can be the descendants of cultivated carob trees (feral trees) or even abandoned cultivated trees. In this case, single plants could be grafted with cultivars and clones were observed (Table S1). Habitats containing carob trees in specialized or consociated orchards (with cereals or other fruit trees) were recorded as “cultivated.” In these cultivated habitats carob trees have a higher probability of having been grafted as verified in some cases (Table S1). When microsatellite makers revealed clones, only one representative genotype was kept.
2.2. Preliminary delimitation of C. siliqua evolutionary units using microsatellite data
We selected localities with at least 10 carob genotypes per population. We used 17 microsatellite markers for genotyping 1019 individuals collected in 56 localities across the Mediterranean basin as described in Table S1. We also scored 15 SNPs present in the flanking regions of the 17 SSR loci following Viruel et al. (2018). We calculated genetic differentiation between localities using the D index (Winter, 2012; mmod R package) and converted it into Euclidean genetic distances based on the coordinates of an NMDS (nonmetric multidimensional scaling; vegan R package). The Euclidean genetic distances and Euclidean geographical distances were then processed by clustgeo (Chavent et al., 2018; clustgeo R package), which relies on Ward's method to minimize intragroup variance for both geographical and genetic distances by calculating K and α parameters: K is the number of clusters and α is a mixing parameter determining the weight of the spatial constraint. Several values of α were tested to optimize the spatial contiguity of populations without deteriorating genetic differentiation structure. Several values of K were also tested. This clustering method provided genetically homogeneous groups of spatially adjacent carob populations, named “CEUs” (carob evolutionary units), which were used in subsequent analyses. A neighbour‐joining (NJ) tree based on pairwise genetic differentiation (Gst, mmod package) among the seven CEUs was built to display the overall differentiation structure.
2.3. RADseq methodology and sequencing
CEUs were used to select 376 samples representative of the genetic diversity and structure of C. siliqua for RADseq analysis. We sampled eight trees of the only other species in the genus, C. oreothauma native to the Arabian Peninsula, kindly sent by the Oman Botanical Garden, which was used as an outgroup in our analysis. Genomic library preparation and sequencing were conducted by Microsynth ecogenics GmbH. DNA samples (200–400 ng input) were digested with the restriction enzymes EcoRI/MSeI following heat inactivation according to the manufacturer's protocol (New England Biolabs, NEB). Fragments between 500 and 600 bp were selected by automated gel cut, Illumina Y‐shaped adaptors were ligated, and ligation products were bead purified. Each library was then individually barcoded by PCR (polymerase chain reaction) using a dual‐indexing strategy. Individually barcoded libraries were pooled and subsequently purified before sequencing on an Illumina NextSeq platform (300 million of 75‐bp reads per run).
2.4. Bioinformatic pipeline to extract and filter SNPs from RADseq data
The bioinformatic approach used in this study is summarized in Figure 1. fastqc reports (multiqc, Ewels et al., 2016) were used to exclude 26 C. siliqua and four C. oreothauma samples due to low sequencing coverage (i.e., below 0.5 million reads). The remaining 354 samples (350 C. siliqua and four C. oreothauma) had an average of 3 million raw reads, ranging between 0.7 and 15 million. Assembly (Figure 1) was performed using ipyrad (Eaton & Overcast, 2020) in a high‐performance computing cluster (HPC pytheas). Thereafter, a “locus” is an RADseq marker of 65 bp that resulted from the ipyrad workflow, and an “SNP” is a polymorphic position of a specific locus, considering that a locus can contain several SNPs. We conducted an assembly limited to C. siliqua following two steps. First, we selected 36 samples representative of the diversity in C. siliqua with a sequencing coverage ranging from 1.3 to 5.3 million reads to conduct four de novo assemblies with varying thresholds of clustering reads (clust_threshold) between 0.9 and 0.96, the minimum number of samples per locus (min_sample_locus) fixed to 30, the minimum depth (mindepth_statistical) for base calling fixed to 8, the maximum depth fixed to 1000, the minimum size of reads fixed to 50 and the first 5 bp of both ends of a locus trimmed. The maximum number of indels and the maximum percentage of SNPs per locus were set to 1% and 10%, respectively. Default values were used for all other parameters. Considering the number of loci, heterozygosity and error rates, we estimated an optimal clustering threshold of 0.94 (see Table S1). A reference file was generated by extracting the first sequence of each locus with a pyrad2fasta script (available on https://github.com/pimbongaerts). Second, this file was used for a subsequent reference assembly for the 354 samples with the same parameters as previously except for the minimum number of samples per loci, which was fixed to 45. The data set constructed using this reference assembly contained more than 64% missing data. The vcf file obtained from ipyrad was then processed to run a principal components analysis (PCA) using the glPca function of the adegenet R package, and an NJ tree, based on pairwise genetic differentiation (Gst, mmod R package) among the seven CEUs, to check that the structure from RAD markers was congruent with previous analysis based on microsatellite markers.
FIGURE 1.
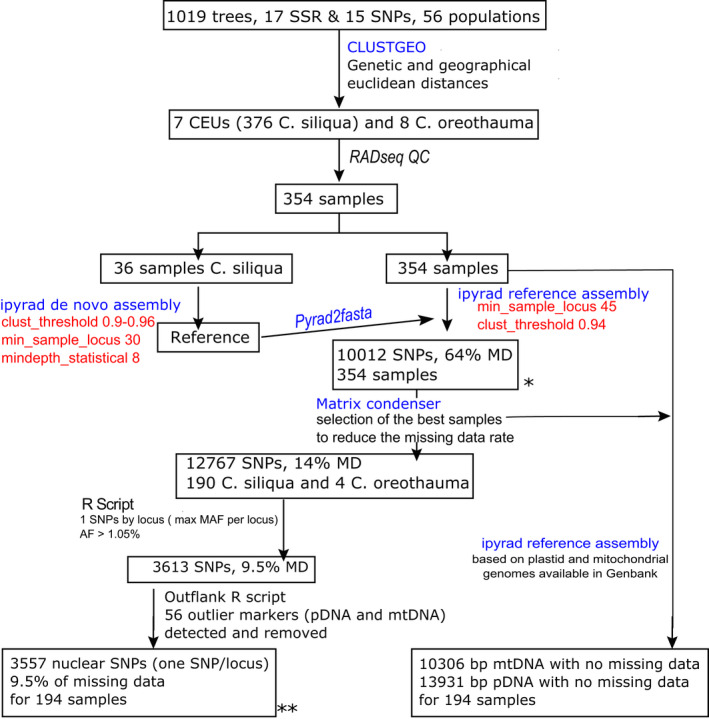
Bioinformatic pipeline used to call and filter SNPs from RADseq data for the carob tree. Asterisks (* and **) correspond to the two data sets of Table 1. Raw data and filtered vcf files (* and **) are available (see Data Availability statement)
We used matrix condenser software (de Medeiros, 2019; de Medeiros & Farrell, 2018) to visualize missing data across samples aiming at maintaining a balance between reducing missing data and not significantly discarding SNPs. We filtered samples to optimize locus coverage and to keep the sampling equilibrium among CEUs and reconducted the reference assembly with ipyrad on a new set of 190 C. siliqua samples. The data were then reduced to one SNP per locus, based on two criteria: SNPs having the maximal minimum allelic frequency per locus and only keeping SNPs with an allelic frequency above 1.05% (i.e., rarest allele present in at least two individuals).
To examine the carob genetic diversity based on neutral processes, such as genetic drift and gene flow, we reduced the effect of outlier loci (i.e., having unexpectedly high differentiation among populations, which could have a great effect on the variance among populations). We used outflank software (Whitlock & Lotterhos, 2015), which produces a lower false‐positive rate compared to other methods (e.g., Silliman, 2019), considering CEUs as populations. The false discovery rate (FDR) (q‐value) was fixed to 0.05. blast searches using the nucleotide NCBI database limited to flowering plant sequences were conducted for outliers, which were mostly assigned to plastid (pDNA) or mitochondrial (mtDNA) genomes. These outlier loci were removed to create a final data set of 3557 SNPs (one SNP per locus) for 190 C. siliqua and four C. oreothauma. Since a subsequent outflank scan failed to detect further outliers (see Results), we considered this set of SNPs as a genome‐wide data set of neutral markers.
2.5. Recovery of plastid and mitochondrial data
To obtain plastid and mitochondrial markers we performed ipyrad reference assembly with default parameters, and changed the maximum alleles per site to one. We used NCBI reference sequence NC‐047061.1, a plastid genome of C. siliqua produced by Zhang et al. (2020), as a reference for plastid assembly. For mitochondrial assembly, we used the GenBank sequence MW448447, a mitochondrial genome of C. siliqua produced by Choi et al. (2021). In both cases, we produced data sets with and without missing data using trimal (Capella‐Gutiérrez et al., 2009).
2.6. Analysis of genetic differentiation and admixture
The final data set of 3,557 SNPs and 190 C. siliqua was converted from genind to genlight, vcf and treemix data formats using the dartr (Gruber et al., 2018) and radiator (Gosselin, 2020) R packages. Population structure analysis was performed using snmf (lea package in R, Frichot & François, 2015), which estimates admixture coefficients from the genotypic matrix assuming K ancestral populations. snmf runs fast even on large data sets and without loss of accuracy compared to other Bayesian modelling software such as admixture (Alexander et al., 2009; Frichot et al., 2014). snmf was run for K = 2–15, with 100 repetitions, regularization parameter set to 250 and 25% of the genotypes masked to compute the cross‐entropy criterion. Barplots showing ancestry coefficients were obtained with the compoplot function in the adegenet R package (Jombart & Ahmed, 2011), with genotypes sorted according to CEUs. To visualize the genetic diversity structure, we performed a PCA using the glPca function of the adegenet R package. Pairwise differentiation among CEUs (Gst) was also computed with the mmod package.
2.7. Phylogenetic relationships between CEUs
Plastid and mitochondrial DNA alignments were used to reconstruct maximum‐likelihood phylogenetic trees with and without missing data as implemented in iq‐tree (Minh et al., 2020). All parameters were set to default and node robustness was estimated by ultrafast bootstrap analysis (1000 iterations). The cophylo function from phytools (Revell, 2012) was used to build a figure comparing pDNA vs. mtDNA phylogenetic trees.
The nuclear data set, which includes four C. oreothauma samples as outgroup, was used to estimate a coalescent‐based tree with the SVDquartets method (Chifman & Kubatko, 2014) as implemented in paup*4.0a (Swofford, 2018). Comparative studies using coalescent inferences have demonstrated that the SVDquartets approach is statistically consistent for different data types (Wascher & Kubatko, 2021). Ten million randomly selected quartets were analysed and node support was assessed based on 1,000 bootstrap replicates. We used the “distribute” option for heterozygous sites. To infer population splits and mixtures across the evolutionary history of the carob tree, we performed a treemix analysis (Pickrell & Pritchard, 2012). A pilot analysis including C. oreothauma genotypes was first run to check for the position of the root followed by a second one without C. oreothauma to obtain more accurate branch lengths within C. siliqua. treemix builds a backbone tree based on population allelic frequency without gene flow and then adds reticulate branches, representing gene flow, aiming at improving the fit of the data. For this analysis, we used CEUs as populations. In the final analysis, one of the CEUs was fixed as an outgroup in agreement with the results obtained in previous analyses.
2.8. Screening for footprints of domestication in RADseq data
Footprints of domestication were investigated by estimating genetic diversity and/or the presence of candidate loci under selection (outliers) for the CEUs. A stronger effect of domestication is expected in cultivated habitats compared to wild and seminatural habitats, and in the eastern CEUs compared to the western ones (Zohary, 2002). Therefore, the analyses were organized according to combinations of these two factors. Observed heterozygosity (H O), Nei's genetic diversity (H E), inbreeding coefficient (f) and the standardized association index , which indicates a lack of genetic mixing when it increases, were estimated using the diversity and poppr R packages (Kamvar et al., 2014; Keenan et al., 2013). Outliers were searched with the outflank method with an FDR threshold of 0.05 (as described previously) considering combinations of habitats and CEUs as populations.
3. RESULTS
3.1. Delimitation of CEUs using microsatellite data
Based on genotypes from 17 microsatellite loci (including SSRs and SNPs) and geographical coordinates, we grouped 1,019 carob trees into CEUs. The ClustGeo method, which considers geographical and genetic distances, found seven genetic groups of genotypes which are geographically nonoverlapping (Figure 2). Four of these CEUs are in the Western Mediterranean (W1–W4), C1 in the Central Mediterranean, and E1 and E2 in the Eastern Mediterranean. These CEUs are grouped into two clusters by ClustGeo. W1, W3 and W4 formed the first cluster whereas W2 grouped in the second cluster with C1, E1 and E2. Based on microsatellite SSR markers, the overall genetic differentiation among these seven CEUs is 16% (Gst). Pairwise Gst values visualized by an NJ tree (Figure 2) confirmed the ClustGeo clustering scheme but placed W1 and E2 in intermediate positions along the NJ tree instead of grouping these two CEUs. The sampling and data composition of the seven CEUs in shown in Table 1.
FIGURE 2.
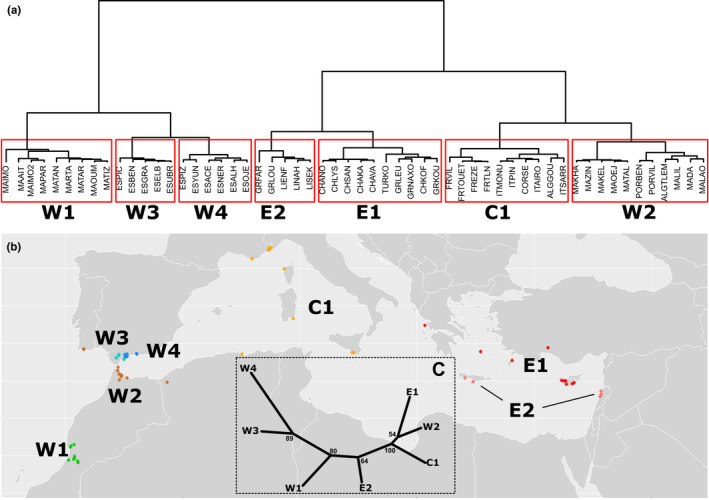
Identification of carob evolutionary units (CEUs) using the ClustGeo method, which considers Euclidean genetic and geographical distances. The analysis used 1,019 genotypes from 56 localities based on 17 SSRs and 15 SNP markers from microsatellite loci. The Ward dendrogram of 56 carob populations with a partition into K = 7 clusters (a) was obtained with a normalized proportion α of explained inertia of 0.2 for the geographical distance and 0.8 for the genetic distance. The seven clusters, or CEUs, are mapped (b) and a neighbour‐joining tree (c) shows pairwise genetic differentiation (Gst SSR markers) among the seven clusters. W1 = southwest Morocco, W2 = mainly north Morocco but also west Algeria and Portugal, W3 = west Andalusia, W4 = East part of Andalusia, C1 = France, Italy and Algeria, E1 = Greece, Turkey and Cyprus, E2 = Lebanon and Crete (Greece); see details in Table 1
TABLE 1.
Sampling and data summarized for seven carob evolutionary units (CEUs) identified by ClustGeo analysis based on genetic differentiation and geographical isolation of 1019 genotypes sampled from 56 locations
| CEU | Observations | Sampling for SSR | Sampling for RAD first selectiona | Sampling for RAD second selectionb |
|---|---|---|---|---|
| W1 | Southwest Morocco, carob trees often exploited, mainly in seminatural habitats but also in wild riparian vegetation and rocky slopes | 9 sites, 169 genotypes | 7 sites, 62 genotypes | 7 sites, 30 genotypes |
| W2 | Mainly north Morocco but also West Algeria and Portugal, carob trees, almost all exploited, are found in cultivated and seminatural habitats, no carob in wild habitats | 11 sites, 173 genotypes | 6 sites, 30 genotypes | 6 sites, 23 genotypes |
| W3 | West Andalusia, carob trees rarely exploited, carob trees frequent in wild habitats mostly on rock outcrops and cliffs | 5 sites, 104 genotypes | 5 sites, 35 genotypes | 5 sites, 18 genotypes |
| W4 | East part of Andalusia, carob trees more exploited than in the rest of the west but sill frequently found in wild habitats | 6 sites, 101 genotypes | 5 sites, 35 genotypes | 5 sites, 21 genotypes |
| C1 | France, Italy and Algeria, high carob crop intensity in Sicily, mostly in seminatural habitats otherwise | 10 sites, 193 genotypes | 7 sites, 71 genotypes | 7 sites, 42 genotypes |
| E1 | Greece, Turkey and Cyprus, high frequency of carob trees in landscape, in seminatural or cultivated habitats and almost all exploited. Only carobs found in riparian vegetation are expected to be wild | 10 sites, 173 genotypes | 8 sites, 53 genotypes | 7 sites, 23 genotypes |
| E2 | Lebanon and Crete (Greece), high frequency of carob trees in the landscape but mostly in abandoned field or orchards in Lebanon but cultivated and exploited in Crete | 5 sites, 106 genotypes | 5 sites, 64 genotypes | 5 sites, 33 genotypes |
Note: a,bSee Materials and methods and Figure 1 to see how these data sets were obtained.
3.2. Assembly of RADseq loci and SNP filtering
The de novo assembly conducted on 36 samples, with a clustering threshold of 0.94 and a minimum number of samples by loci of 30, produced a mean number of 48,828 loci by sample (Table S1), which were reduced to 13,371 loci retained after ipyrad filtering. The sequences of these loci were used as a reference for the assembly of 354 genotypes for 10,012 loci, which revealed a pattern of genetic structure roughly congruent with microsatellite pattern of differentiation (Figure S2). However, these data presented an overall missing data rate of 64%.
To overcome the limitations of the 64% of missing data could have in our results, we searched for genotypes and loci having the highest missing rate as implemented in matrix condenser, and their subsequent removal produced a matrix with 190 samples and 12,767 loci, with 14% missing data rate. After filtering to keep one SNP per locus, the data set included 190 samples for 3,613 loci (or SNPs), with 9.5% overall missing data and a 3.5% median missing rate by genotype. The outflank method detected 56 outlier markers within the data set of 190 samples by 3613 SNPs (Figure S3). The global differentiation computed for these outliers was six times higher than for other SNPs (Gst 0.74 vs. 0.12). Nucleotide blast searches revealed that 27 outliers were assigned to the plastid genome, 14 to the mitochondrial genome, 11 to nuclear genomes and four were not assigned. Excluding the outliers detected by outflank, the new data set included 3,557 unlinked SNPs. For svdquartets and treemix analyses, we filtered out samples with missing data rates above 20%, which generated a matrix containing 171 Ceratonia siliqua and four C. oreothauma genotypes for 3,284 unlinked SNPs and 8% missing data rate.
The plastid and mitochondrial data sets, assembled for 190 C. siliqua and four C. oreothauma samples, held 135,525 and 191,840 bp with 47% and 52% of missing data, respectively. After missing data removal, the pDNA alignment had 13,931 bp with 61 parsimony‐informative sites whereas the mtDNA had 10,306 bp with 19 parsimony‐informative sites.
3.3. Phylogenetic and genetic structure of carob trees across the Mediterranean based on RADseq data
The maximum‐likelihood phylogenetic trees reconstructed with plastid and mitochondrial DNA data obtained without (Figure S5) and with (Figure 3) missing data had a similar topology, but the bootstrap values were higher when missing data were included (Figure S4). Using the alignments without missing data, iq‐tree selected the F81 + F + I substitution model for both pDNA and mtDNA. Haplotypes from the west CEUs are almost all grouped in a clade whereas central and eastern haplotypes (the red dotted square in Figure 3) were nested in one (pDNA) or two (mtDNA) clades. The overall phylogenetic topology shown by cytoplasmic lineages supports a west–east split except for haplotypes from W1 (green), which appear in both west and central‐east lineages. All these haplotypes are from the northern part of W1, situated north of Agadir in the Imouzzer Ida Ou Tanane area. Therefore, the cytoplasmic phylogenetic diversity is almost entirely present in southwest Morocco (W1).
FIGURE 3.
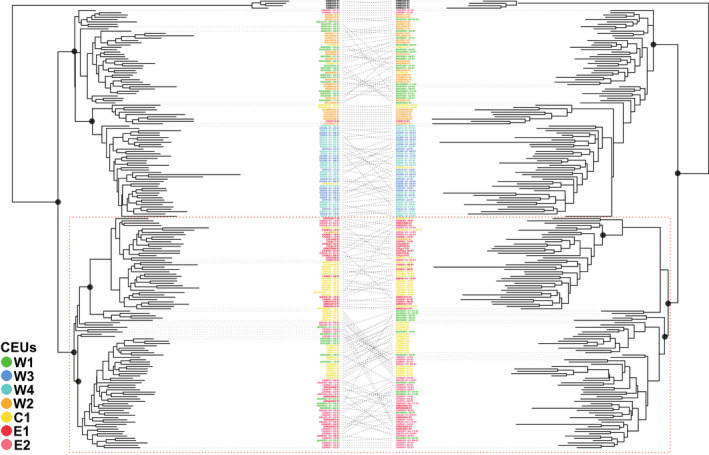
Plastid (left) versus mitochondrial (right) iq‐tree maximum‐likelihood trees obtained for 190 Ceratonia siliqua and four C. oreothauma samples. Black dots indicate bootstrap support above 95% for the main nodes. The dotted red square corresponds to central and eastern CEUs (in yellow and red) except for haplotypes from the northern area of W1 (in green). pDNA and mtDNA have 135,525 and 191,840 bp for 47% and 52% of missing data, respectively. Colour codes correspond to carob evolutionary units (CEUs) and C. oreothauma, the outgroup, is indicated in black
By reducing the rate of missing data and filtering to obtain unlinked SNPs, the genetic structure was more evident as shown in the PCA plot (Figure 4 compared to Figure S2). The phylogenetic tree reconstructed with nuclear data using svdquartets, which included 171 samples of C. siliqua and four samples of C. oreothauma as the outgroup, confirmed the monophyly of C. siliqua and resolved a strongly supported first split of the W1 clade from the remaining C. siliqua lineages (Figure 4a). A subsequent split separated the remaining western CEUs (W2, W3 and W4) from Central and Eastern CEUs (C1, E1 and E2). This phylogenetic topology based on 3557 SNPs is congruent with plastid and mitochondrial data in supporting Southwest Morocco (W1) as sister to all other lineages in the carob evolutionary history. The svdquartets phylogenetic topology is also highly congruent with the K = 4 genetic structure obtained with SSR data (Figure 4a), and these four genetic groups match the following clades: W1, W3 + W4, E1 + E2, C1. The North Moroccan W2 clade is sister to the other three western CEUs in the svdquartets tree, which is congruent with its mixed assignation to different genetic clusters for RADseq data (Figure 4a,c). The genetic clustering estimated with snmf based on RADseq data was repeated from K = 2 to K = 7 ancestral populations. The cross‐entropy criterion suggested two optimal solutions for five and seven groups (Figure S6A), of which K = 7 had the highest probability. The most likely genetic clustering, K = 7 solution (Figure 4a,c), resolved the differentiation of four ancestral populations mostly present in Southwest Morocco (W1), South Spain (W3 and W4) and the eastern CEUs (E1 and E2). However, departures from a good match between microsatellite‐based CEUs and RADseq genetic groups are substantial too. C1 includes individuals with admixture from three ancestral groups, two of them (in orange and yellow, Figure 4) almost restricted to this CEU and the third one, coloured in turquoise (Figure 4), present in all CEUs although with higher proportions in W2, W3 and C1. W2 was resolved as admixed with the contribution of several ancestral populations. Although admixture of W2 was already seen with microsatellite data, it was closer to central and eastern CEUs than with RADseq. The PCA results are consistent with the main genetic groups found with snmf, showing a clear delimitation of three groups, W1, W3 + W4 and E1 + E2, but partial overlap between W2 and C1 (Figure 4b,c). RADseq genetic differentiation among the seven CEUs (Table S2) was moderate with an overall Gst of 11%; all values above the mean involved a west–east differentiation, the highest differentiation being between W4 and E2 (Gst = 19%).
FIGURE 4.
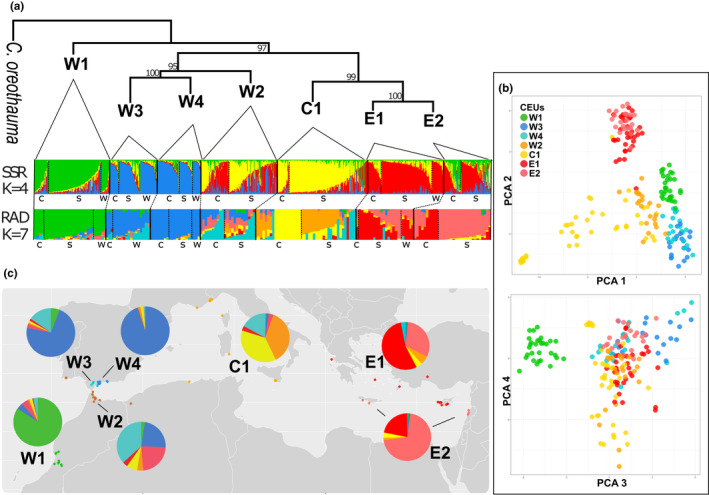
Population genetic structure of the carob tree. (a) svdquartets tree of seven genetically and geographically homogeneous groups (CEUs) based on RADseq markers. Genetic admixture plots are based on four ancestral populations for SSR markers (1019 genotypes, 17 loci) and on seven ancestral populations for RADseq markers (190 genotypes, 3557 neutral unlinked SNPs). Within CEUs, genotypes were organized by habitats and according to their admixture coefficient (c = cultivated, s = seminatural and w = wild). (b) PCA scatterplots of RADseq genotypes (accumulated variance of the first four components = 15.2%). (c) Map of genetic admixture based on RADseq markers
3.4. Impact of dispersals on the genetic diversity and structure of C. siliqua
For the treemix analysis, we rooted the C. siliqua phylogenetic tree with W1, following svdquartets reconstructions which included C. oreothauma as outgroup. The tree topology reconstructed by treemix (Figure 5) is similar to that of svdquartets, showing the same west–east differentiation. The main differences concerned the position of W2. The svdquartets phylogenetic tree reconstructed W2 sister to other western CEUs, in congruence with cytoplasmic lineages, whereas treemix resolved W2 as intermediate between western (W1, W3, W4) and central and eastern CEUs (C1, E1, E2). treemix topology agrees with the highly admixed pattern in W2 inferred by snmf (Figure 2a) and with its low to moderate differentiation with respect to other CEUs (Table S2). The treemix model without gene flow explained 96% of the covariance (Figure 3a), and the addition of four dispersals resulted in 99.8% of the total covariance explained (Figure 3b). Three of the four dispersals identified E2 as the source of introgression, into W1, W3 and W2, whereas the fourth dispersal connected the central‐eastern CEUs to W4, again indicating a westward dispersal.
FIGURE 5.
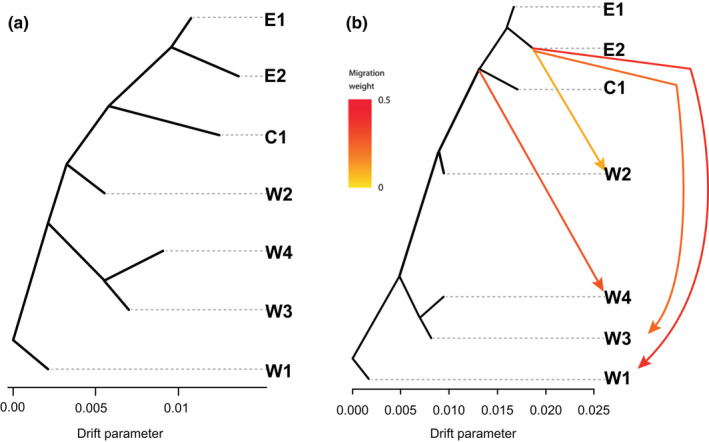
Evolutionary history of the carob tree reconstructed with treemix. Maximum‐likelihood trees obtained without (a) and with gene flow (b) events explaining 96% and 99% of the variance, respectively. The colour of the arrows indicates the gene flow weight, which is the fraction of ancestry derived from the gene flow edge
In summary, the phylogeographic history of the carob tree is mainly derived from west‐to‐east expansions. Southwest Morocco (W1) was reconstructed as sister to all other lineages of C. siliqua, and the populations in the northern part of this area are phylogenetically related to the eastern lineages. On the one hand, genomic information from cytoplasmic lineages, as well as from multispecies coalescent analyses from nuclear data (svdquartets), support two dispersal routes, one giving origin to the western CEUs and the second to the eastern CEUs. On the other hand, treemix, admixture and pairwise F ST analyses, all based on allelic frequencies, support a simplified west‐to‐east differentiation through northern Morocco (W2). The secondary east–west dispersals detected by treemix were in agreement with microsatellite data, for example the shared co‐ancestry between W1 and E2 genotypes (Figure 4a, SSR) or the high mixing rate of W2.
3.5. Impact of carob cultivation effort on carob genetic diversity
Overall, for microsatellite markers, the admixture plot (Figure 4a) revealed the same pattern within each of the CEUs, that is, a similar gradient of ancestries from nonadmixed to highly admixed regardless of the type of habitat. A slightly higher admixture can be observed in seminatural populations. However, considering the RADseq markers, a striking difference is observed for C1 where 12 genotypes collected in cultivated habitats constitute a genetically pure (yellow, Figure 4) group. These genotypes correspond to grafted branches of monumental carob trees living near Raguza in Sicily. Although not forming clones, they are genetically very similar, a pattern that was not detected with microsatellites. This genetic cluster is rarely found outside Sicily, although it only scarcely occurs in France (C1) and north Morocco. In C1, the other genotypes sampled in cultivated habitats, coming from other monumental trees or known cultivars, are more genetically mixed and are closer to the genotypes found in seminatural habitats, which are mainly composed of a cluster that is almost exclusive to this area (orange, Figure 4). Higher admixture was detected in western Spain (W3) with north Morocco (W2) than in eastern Spain (W2). In the eastern Mediterranean, a genetic group (light red, mostly in E2) was mainly detected in seminatural populations, and it constituted the predominant genetic group in Lebanon (Figure 4a).
Genetic diversity values are reported in Table 2. For microsatellite data, Nei's diversity (H E) values decrease from western CEUs and wild or seminatural habitats to eastern and/or cultivated habitats, regardless of the markers. This is less pronounced for RADseq data. Interestingly, the association index , which increases as mixing decreases, shows a very clear effect of decreased genetic mixing in cultivated habitats for microsatellite data. The reduced sampling for RADseq does not allow such a comparison but supports that central (C1) and eastern CEUs experienced less mixing than western ones. The inbreeding index (f) suggests both deficit and excess of heterozygosity. The deficit is significant for both RADseq and SSR data for W1, W2, E1 and E2 whereas an overall excess is observed for C1 for RADseq and a few other cases. The highest values of genetic diversity are observed in the western CEUs, whereas in the east, but also with its cultivation, the genetic diversity of carob decreases as does the genetic mixture (increased , Table 2).
TABLE 2.
Estimates of genetic diversity based on microsatellites (17 SSR loci) and RADseq markers (3557 SNPs) for seven Ceratonia siliqua units (CEUs). Within each CEU, samples were split into groups according to three habitat types (cultivated, seminatural or wild)
| SSR | RAD | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | H O | H E | f |
|
N | H O | H E | f |
|
||
| W1_cultivated | 30 | 0.496 | 0.564 | 0.088* | 22* | 4 | — | — | — | — | |
| W1_seminatural | 119 | 0.508 | 0.578 | 0.106* | 18* | 21 | 0.223 | 0.257 | 0.088* | 6* | |
| W1_wild | 20 | 0.532 | 0.576 | 0.052 ns | 5 ns | 5 | — | — | — | — | |
| W1 | 169 | 0.509 | 0.579 | 0.109* | 17* | 30 | 0.222 | 0.260 | 0.109* | 6* | |
| W3_cultivated | 20 | 0.513 | 0.5 | −0.062 ns | 68* | 3 | — | — | — | — | |
| W3_seminatural | 48 | 0.533 | 0.559 | 0.04 ns | 8 ns | 0 | — | — | — | — | |
| W3_wild | 36 | 0.549 | 0.532 | −0.038 ns | 8 ns | 15 | 0.220 | 0.241 | 0.037 ns | 3* | |
| W3 | 104 | 0.535 | 0.544 | 0.016 ns | 17* | 18 | 0.221 | 0.239 | 0.037_ns | 3* | |
| W4_cultivated | 50 | 0.505 | 0.516 | 0.018 ns | 42* | 8 | 0.222 | 0.207 | −0.135* | 28* | |
| W4_seminatural | 31 | 0.502 | 0.56 | 0.084 ns | 15* | 9 | 0.227 | 0.237 | −0.026 ns | 4* | |
| W4_wild | 20 | 0.594 | 0.543 | −0.128* | 5 ns | 4 | — | — | — | — | |
| W4 | 101 | 0.522 | 0.545 | 0.040 ns | 24* | 21 | 0.224 | 0.229 | −0.010 ns | 9* | |
| W2_cultivated | 62 | 0.456 | 0.48 | 0.011 ns | 36* | 10 | 0.208 | 0.228 | −0.075* | 19* | |
| W2_seminatural | 111 | 0.477 | 0.524 | 0.069* | 16* | 13 | 0.183 | 0.233 | 0.085 ns | 195 ns | |
| W2 | 173 | 0.47 | 0.513 | 0.06* | 25* | 23 | 0.194 | 0.236 | 0.114* | 167_ns | |
| C1_cultivated | 22 | 0.412 | 0.473 | 0.074 ns | 25* | 19 | 0.217 | 0.189 | −0.059* | 188* | |
| C1_seminatural | 171 | 0.417 | 0.459 | 0.071* | 18* | 23 | 0.243 | 0.238 | −0.029* | 23* | |
| C1 | 193 | 0.416 | 0.461 | 0.076* | 17* | 42 | 0.230 | 0.221 | −0.016* | 54* | |
| E1_cultivated | 33 | 0.473 | 0.471 | −0.019 ns | 122* | 1 | — | — | — | — | |
| E1_seminatural | 113 | 0.448 | 0.506 | 0.096* | 18* | 18 | 0.219 | 0.237 | 0.032 ns | 60* | |
| E1_wild | 27 | 0.459 | 0.479 | 0.013 ns | 20* | 5 | — | — | — | — | |
| E1 | 173 | 0.454 | 0.501 | 0.076* | 20* | 24 | 0.217 | 0.240 | 0.060* | 43* | |
| E2_cultivated | 39 | 0.425 | 0.477 | 0.106 ns | 34* | 10 | 0.224 | 0.241 | 0.010 ns | 11* | |
| E2_seminatural | 67 | 0.475 | 0.517 | 0.067 * | 16* | 22 | 0.200 | 0.223 | 0.067 ns | 11* | |
| E2 | 106 | 0.456 | 0.517 | 0.106* | 23* | 32 | 0.208 | 0.235 | 0.091* | 14* | |
Note: An asterisk f or significantly different from zero based upon 500 and 100 bootstrap iterations, respectively.
Abbreviations: f, Wright's inbreeding coefficient; H E, expected heterozygosity or Nei's genetic diversity; H O, observed heterozygosity; N, number of genotypes analysed; , standardized form of the index of association accounting for multilocus linkage (×1000).
Based on the differentiation between genetic groups formed by the combination of CEUs and habitats (Table 2), outflank did not detect candidate loci with an FDR (q‐value) below 0.05, which indicates that the hypothesis of neutrality cannot be rejected (Figure S7).
4. DISCUSSION
Intense research on plant phylogenetics and phylogeography over the last two decades has allowed the discovery of several major biogeographic trends in the Mediterranean basin (Comes, 2004; García‐Verdugo et al., 2021; Nieto Feliner, 2014) and renewed our understanding of plant domestication (Purugganan, 2019). Following an initial focus on biogeographic refugia, recent studies have revealed the genetic imprints of past environmental changes and dispersal processes, some involving the entire Mediterranean basin (see reviews in Fady & Conord, 2010; García‐Verdugo et al., 2021; Médail & Diadema, 2009; Migliore et al., 2018; Nieto Feliner, 2011; Nieto Feliner, 2014; Thompson, 2020; Vargas et al., 2018). Our study reveals a new phylogeographic scenario for a Mediterranean tree species: the ancestral gene pools of carob (i.e., CEUs) originated from a biogeographic refugium probably located in Southwest Morocco, and a subsequent west‐to‐east expansion. Our results also highlight that carob domestication has mainly relied on the use of locally selected varieties, albeit punctuated by long‐distance westward dispersal events by humans, which match major cultural migration waves by Greeks, Romans and Arabs.
4.1. Evolutionary history of the carob tree
Our phylogeographical reconstruction conclusively rejects a long‐standing hypothesis that proposes an introduced origin of the carob tree in most of the Mediterranean. An eastern Mediterranean or southern Arabian origin, followed by human‐mediated expansions, were proposed by several authors partly based on linguistic evidence from vernacular names, Ceratonia siliqua and C. oreothauma occurrences in western Asia, and carob agricultural practices (reviewed in Ramón‐Laca & Mabberley, 2004). However, genetic data from SSR and plastid markers based on a thorough population sampling across the Mediterranean (Viruel et al., 2020) revealed a strong west vs. central‐east pattern, suggesting a low human influence on the main current patterns of genetic diversity and structure of the carob tree across the Mediterranean. The comprehensive review of carob fossil data presented in Viruel et al. (2020) did not provide support for an eastern origin of C. siliqua. Instead, fossil records show a mostly continuous presence of Ceratonia around the palaeo‐Mediterranean Sea since the Oligocene with a progressive decline starting ~20 million years ago.
Compared to SSR data, the increased resolution of the RADseq results presented here has allowed us to bridge phylogenetic and population genetic inferences (Parchman et al., 2018). In addition, the inclusion of Ceratonia oreothauma, the sister species of C. siliqua—which diverged ~6.4 millon years ago (Viruel et al., 2020)—allows us to root the carob tree history, thus providing a relative time frame for the successive splits that occurred along with the evolutionary history of the carob tree. Coalescent‐based models based on SSR data could not disprove the existence of smaller refugia in the eastern Mediterranean, and two disjunct refugia after the Last Interglacial (LIG) were suggested as the most likely evolutionary scenario for the current genetic structure of C. siliqua (Viruel et al., 2020). The RADseq data generated in our study provide a substantially higher resolution for the evolutionary history of the carob tree. The fact that the southwest Morocco CEU (W1) exhibits a high genetic diversity and is sister to all other Mediterranean carob lineages in the phylogenies (Table 2, Figures 3 and 4) supports the existence of a long‐term refugium in the foothills of the High Atlas Mountains near the Atlantic coast, from where the ancestral population of C. siliqua has possibly emerged. From this ancestral population, the carob tree probably followed two dispersal routes: one northward that reached northwest Africa and south Spain (W2, W3 and W4 CEUs), and another towards the east that gave rise to the central‐eastern CEUs. Mitochondrial and plastid evidence extracted from RADseq data also support the existence of an ancestral pool in southwest Morocco. Specifically, mtDNA and pDNA haplotypes present in the northern part of southwest Morocco (Imouzzer Ida Ou Tanane area) are closely related to eastern haplotypes, thus indicating this latter area as close to the source of the eastern populations. This pattern was already apparent in 1472‐bp‐long plastid markers exhibiting only three substitutions (Viruel et al., 2020). This new evolutionary scenario explains the west–east split in the carob genetic diversity by dispersal from an ancestral population. Footprints of this ancestral population were found in the genetic profiles of the populations located in southern Morocco. However, the number of dispersal routes estimated from this ancestral population varied in different analyses. Two dispersal routes were less supported by analyses based on allelic frequencies (pairwise F ST, admixture plots or treemix). Indeed, they positioned north Morocco (W2) at the crossroad of west, central and east CEUs, suggesting that more than one genepool from the west contributed to central and eastern populations. This is represented by the “turquoise” genetic cluster (Figure 4a) inferred with RADseq data, which is mostly present in north Morocco (W2), Southwest Spain (W3) and the Central Mediterranean (C1), with a slight presence in the east (E1). However, it is noteworthy that all cytoplasmic haplotypes from north Morocco belong to the western group and are not related to eastern haplotypes, when using either RADseq (this study) or Sanger sequencing (Viruel et al., 2020). This strong nuclear–cytoplasmic incongruence supports that north Morocco mostly played a receiver rather than a donor role (Currat et al., 2008) during carob history, thus supporting two dispersal routes as the most likely scenario.
Viruel et al. (2020) estimated that the divergence between the west and east part of the carob range could be as old as 1,815 generations ago (95% confidence interval: 400–4640 generations). This age estimate therefore favour a west‐to‐east expansion before the Last Glacial (~21,000 years ago). Species distribution modelling (SDM) indicates that both the LIG (~116,000 years ago) and the Last Glacial Maximum (LGM) were periods of range contraction for the carob tree during the Pleistocene (Viruel et al., 2020). Moreover, SDM predicted that some areas in the north African and south European Atlantic coasts could have been continuously suitable over the last 130,000 years. Southwest Morocco has been identified as a biogeographic refugium and even as a diversification cradle for several taxonomic groups, such as Dracaena draco and several elements of the Macaronesian flora (Médail & Quézel, 1999), Olea europaea L. ssp. maroccana (Médail et al., 2001), Hypochaeris sp. (Ortiz et al., 2009), Daboiea sp. vipers (Martínez‐Freiría et al., 2017), Astragalus edulis (Bobo‐Pinilla et al., 2018), Lavatera maritima (Villa‐Machío et al., 2018) and Buthus sp. scorpions (Klesser et al., 2021). Although Mediterranean phylogeographic studies have focused mostly on glacial refugia, recent studies have highlighted South and West Morocco as a long‐term stable refugium for plant populations during the LIG and LGM (Bobo‐Pinilla et al., 2018; García‐Castaño et al., 2014; Migliore et al., 2018; Villa‐Machío et al., 2018; Viruel et al., 2020). This area is characterized by a highly complex topography with the foothills of the Atlas Mountains near the Atlantic Ocean that may have buffered environmental conditions (i.e., climatic stability, frequent sea fogs) favouring higher species persistence during unfavourable periods of pronounced climate continentality.
4.2. Footprints of domestication in the current genetic structure of the carob tree across the Mediterranean
Although disentangling the history of cultivated plants is complex, our phylogeographic investigation in the carob tree sheds light on its domestication history. Our previous study based on SSR data (Viruel et al., 2020) suggested that local domestication events from wild populations were the most likely scenario. Consistent with this, the RADSeq data, based on a more intense genome‐wide sampling, cannot explain domestication solely based on historical translocations. Agricultural practices in the carob tree are based on propagation by grafting (Zohary, 2002), although seeds could have also been transported for propagation or their use as a unit of weight for gemstones (Turnbull et al., 2006). In either case, if domestication was based only on westward propagations of cultivars from the east, maternal (eastern) haplotypes would remain today in the western Mediterranean. Instead, our results are conclusive in supporting that although the dispersal of selected varieties (vegetatively propagated) between remote geographical areas may have contributed to the local gene pools, this was not the main force of domestication in carob trees.
The use of genomic data at the intraspecific level has allowed us to identify footprints of domestication in crop models where PCR‐based molecular markers had previously failed. In the date palm, genomic data revealed that human‐mediated dispersal imprints were superimposed on a previous phylogeographic structure (Flowers et al., 2019; Gros‐Balthazard et al., 2017). In the carob tree, translocation of eastern domesticated varieties into differentiated western populations does fit with the patterns found in geographically intermediate groups (W2, C1 and E1). These are less differentiated, which is explained by high rates of admixture (Figure 2). To untangle the role of human‐based dispersals in these strong genetic admixtures, we used allelic frequency‐based models aiming at estimating the intensity and origin of dispersal events throughout the carob evolutionary tree (Figure 5): the results of treemix recover westwards dispersals that were mostly originated from E2 (Lebanon and Crete), or central‐eastern CEUs (E2, E1 and C1). These translocations match with the beginning of carob agriculture in the eastern Mediterranean, and its dispersal by Greeks, Romans and later by Arabs in historical times (Ramón‐Laca & Mabberley, 2004; Viruel et al., 2020). They may have contributed to the genetic admixed pool used locally for cultivation as observed in north Morocco (W2).
The second footprint of domestication was observed in genetic diversity. Our analyses revealed an association between lower genetic diversity (and lower genetic admixture) and a higher carob cultivation intensity. This pattern was well observed in the central CEU (C1), where cultivated varieties are most diffused compared to other CEUs (Di Guardo et al., 2019). In C1, we detected a genetic group of individuals lacking admixture, corresponding to the monumental carobs of the Ragusa district (Sicily, Italy). These monumental carobs have been harvested without interruption for centuries, and they are genetically very close to each other, although not clones. Other cultivars collected near Raguza, either from monumental trees or recently grafted, were closely related to genotypes found in seminatural habitats of C1 and differentiated from other CEUs (orange in Figure 4a). These genetic patterns of cultivated C1 individuals constitute additional support for diffusion of selected genotypes at the local scale, rather than long‐distance dispersal. This local process played a major role in the domestication of carob. Despite this pattern, we did not detect any candidate loci under selection due to domestication pressures, which could be explained by the limitations of our method and sampling or by a low effect of domestication on the carob genome, or even by the suitability of carob wild traits for human needs, which might have been good enough for their use as fodder.
Compared to other perennial crop species for which candidate and adaptive loci have been found by whole genome sequencing as well as RADseq (Alves‐Pereira et al., 2020; Cornejo et al., 2018; Groppi et al., 2021), a relatively lower impact of selection is likely in carob. Moreover, domestication leading to fine‐tuning of gene expression patterns rather than genome‐wide evolution, as observed in olive (Gros‐Balthazard et al., 2019), may be almost undetectable by a reduced‐representation genomics approach such as RADseq. In north Morocco, forest–agrosystems continuity coupled with close wild–domesticated relationships characterize the local domestication of olive and fig trees (e.g., Achtak et al., 2010; Aumeeruddy‐Thomas et al., 2017). However, a low impact of domestication on genetic diversity at the range‐wide level, as shown here for the carob tree, is unique by comparison to other Mediterranean fruit trees. This is probably the reason why the footprints of demographic history are still present and detectable.
4.3. Conservation of genetic diversity within CEUs
Defining the structure of genome‐wide diversity is essential for preserving the genetic resources of cultivated species and for future breeding practices (Purugganan, 2019). We used an integrative approach that combines geographical and genetic differentiation to characterize evolutionary units for C. siliqua across the Mediterranean. In a survey including 1,019 samples, seven nonoverlapping CEUs were identified as the best solution to minimize intragroup variance and obtain homogenous groups without geographical overlap. A thorough sampling across the Mediterranean using nuclear SSR and SNP data allows grouping the seven CEUs into four genetic clusters (Figure 4): Southwest Morocco (W1), Iberian Peninsula (W3, W4), central Mediterranean, and north Morocco (C1, W2) and eastern Mediterranean (E1, E2). RADseq data further resolved this genetic structuring across the Mediterranean by identifying seven genetic clusters (Figure 4a,c), which in some cases fully matched with a CEU (e.g., W1) or two CEUs (W3, W4), whereas, in other cases, a mixture of more than one genetic cluster was observed (e.g., C1). These data permit a better interpretation of the genetic diversity patterns between CEUs and are thus important for future designs of ex situ conservation actions. Our results suggest that a moderate genetic diversity is uniformly distributed across CEUs (Table 2). Only a slightly higher genetic diversity was estimated in western CEUs mostly based on SSR loci. Although W2 and C1 CEUs are highly admixed, these factors did not entail an increase in genetic diversity compared to nonadmixed clusters. Conservation of genetic resources for the carob tree should recover genetic diversity found across the Mediterranean by preserving populations from western and eastern CEUs, and prioritizing the most differentiated ones (Table S3). C1, which contains three genetic groups and for which carob cultivars have been well characterized, specifically in Italy, should benefit from more investigations on carob evolution under domestication.
AUTHOR CONTRIBUTIONS
H.S., A.B., F.M., S.L.M., M.B.K., L.O., G.N.F. and J.V. conceived, planned the study and collected samples. F.L.M. performed the DNA extraction and quality assessment. J.V., A.B. and V.F. performed curation and analysis of microsatellite data. A.B. performed RADseq data curation and SNP filtering. A.B. and J.V. provided the analysis, tables and Figures. A.B., J.V., G.N.F. and F.M. interpreted the results. A.B. and J.V. drafted the manuscript. J.V., G.N.F., S.L.M. and M.D.G. edited the manuscript. J.V., G.N.F. and A.B. wrote the final manuscript. H.S. was in charge of funding acquisition and project administration. All authors read and approved the final version.
CONFLICT OF INTEREST
The authors declared no conflict of interest for this article.
OPEN RESEARCH BADGES
This article has earned an Open Data Badge for making publicly available the digitally‐shareable data necessary to reproduce the reported results. The data is available at https://bigd.big.ac.cn/bioproject.
Supporting information
Appendix S1
Appendix S2
ACKNOWLEDGEMENTS
This study is part of the DYNAMIC project supported by the French national agency of research (ANR‐14‐CE02‐0016) and benefited from equipment and services from the molecular biology facility (SCBM) at IMBE (Marseille, France). All bioinformatics and simulations were done on the High‐Performance Computing Cluster from the Pytheas informatic facility (OSU Institut Pytheas Aix Marseille Univ, INSU‐CNRS UMS 3470). J.V. benefited from a Postdoc Fellowship funded by DYNAMIC and a Marie Skłodowska‐Curie Individual Fellowship (704464 – YAMNOMICS – MSCA‐IF‐EF‐ST). We thank the following for their help to complete our sampling: Annette Patzelt (Oman Botanic Garden), Minas Papadopulos (Department of Forests of the Republic of Cyprus), Zahra Djabeur (Oran University), Nabil Benghanem (Tizi‐Ouzou University), Gianluigi Bacchetta (Cagliari University), Sonja Yakovlev (Paris‐Sud University), Errol Vela (Montpellier University), Maria Panitsa (Patras University) and the services of Junta de Andalucia. We thank the three anonymous reviewers for their helpful comments.
Baumel, A. , Nieto Feliner, G. , Médail, F. , La Malfa, S. , Di Guardo, M. , Bou Dagher Kharrat, M. , Lakhal‐Mirleau, F. , Frelon, V. , Ouahmane, L. , Diadema, K. , Sanguin, H. , & Viruel, J. (2022). Genome‐wide footprints in the carob tree (Ceratonia siliqua) unveil a new domestication pattern of a fruit tree in the Mediterranean. Molecular Ecology, 31, 4095–4111. 10.1111/mec.16563
Handling Editor: Ana Caicedo
DATA AVAILABILITY STATEMENT
Full information on population sampling and microsatellite data are available in Viruel et al. (2020) and deposited in DRYAD (https://doi.org/10.5061/dryad.k7m020r). Raw RADseq reads are deposited at NCBI SRA database under Bioproject accession PRJNA793764. Assemblies from ipyRAD (vcf files), filtered R data files and R scripts of analyses are available at https://osf.io/t5m8y/.
REFERENCES
- Achtak, H. , Ater, M. , Oukabli, A. , Santoni, S. , Kjellberg, F. , & Khadari, B. (2010). Traditional agroecosystems as conservatories and incubators of cultivated plant varietal diversity: The case of fig (Ficus carica L.) in Morocco. BMC Plant Biology, 10(1), 1–12. 10.1186/1471-2229-10-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, D. H. , Novembre, J. , & Lange, K. (2009). Fast model‐based estimation of ancestry in unrelated individuals. Genome Research, 19(9), 1655–1664. 10.1101/gr.094052.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves‐Pereira, A. , Clement, C. R. , Picanço‐Rodrigues, D. , Veasey, E. A. , Dequigiovanni, G. , Ramos, S. L. F. , Baldin Pinheiro, J. , Pereira de Souza, A. , & Zucchi, M. I. (2020). A population genomics appraisal suggests independent dispersals for bitter and sweet manioc in Brazilian Amazonia. Evolutionary Applications, 13, 342–361. 10.1111/eva.12873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumeeruddy‐Thomas, Y. , Moukhli, A. , Haouane, H. , & Khadari, B. (2017). Ongoing domestication and diversification in grafted olive–oleaster agroecosystems in Northern Morocco. Regional Environmental Change, 17(5), 1315–1328. 10.1007/s10113-017-1143-3 [DOI] [Google Scholar]
- Baumel, A. , Mirleau, P. , Viruel, J. , Bou Dagher Kharrat, M. , La Malfa, S. , Ouahmane, L. , Diadema, K. , Moakhar, M. , Sanguin, H. , & Médail, F. (2018). Assessment of plant species diversity associated with the carob tree (Ceratonia siliqua, Fabaceae) at the Mediterranean scale. Plant Ecology and Evolution, 151, 185–193. 10.5091/plecevo.2018.1423 [DOI] [Google Scholar]
- Besnard, G. , Terral, J. F. , & Cornille, A. (2018). On the origins and domestication of the olive: A review and perspectives. Annals of Botany, 121(3), 385–403. 10.1093/aob/mcx145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobo‐Pinilla, J. , Peñas de Giles, J. , López‐González, N. , Mediavilla, S. , & Martínez‐Ortega, M. M. (2018). Phylogeography of an endangered disjunct herb: Long‐distance dispersal, refugia and colonization routes. AoB Plants, 10, ply047. 10.1093/aobpla/ply047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin, N. L. , Zeder, M. A. , Fuller, D. Q. , Crowther, A. , Larson, G. , Erlandson, J. M. , Denham, T. , & Petraglia, M. D. (2016). Ecological consequences of human niche construction: Examining long‐term anthropogenic shaping of global species distributions. Proceedings of the National Academy of Sciences, 113(23), 6388–6396. 10.1073/pnas.1525200113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell, J. S. , Wang, N. , Nichols, R. A. , & Buggs, R. J. (2018). Genetic diversity maintained among fragmented populations of a tree undergoing range contraction. Heredity, 121, 304–318. 10.1038/s41437-018-0132-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brassesco, M. E. , Brandão, T. R. , Silva, C. L. , & Pintado, M. (2021). Carob bean (Ceratonia siliqua L.): A new perspective for functional food. Trends in Food Science & Technology, 114, 310–322. 10.1016/j.tifs.2021.05.037 [DOI] [Google Scholar]
- Capella‐Gutiérrez, S. , Silla‐Martínez, J. M. , & Gabaldón, T. (2009). trimAl: A tool for automated alignment trimming in large‐scale phylogenetic analyses. Bioinformatics, 25(15), 1972–1973. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso, M. , La Malfa, S. , Pavlíček, T. , Frutos Tomñs, D. , Gentile, A. , & Tribulato, E. (2008). Characterisation and assessment of genetic diversity in cultivated and wild carob (Ceratonia siliqua L.) genotypes using AFLP markers. The Journal of Horticultural Science and Biotechnology, 83(2), 177–182. 10.1080/14620316.2008.11512367 [DOI] [Google Scholar]
- Chavent, M. , Kuentz‐Simonet, V. , Labenne, A. , & Saracco, J. (2018). ClustGeo: An R package for hierarchical clustering with spatial constraints. Computational Statistics, 33(4), 1799–1822. 10.1007/s00180-018-0791-1 [DOI] [Google Scholar]
- Chen, C. , Qi, Z. C. , Xu, X. H. , Comes, H. P. , Koch, M. A. , Jin, X. J. , Fu, C. X. , & Qiu, Y. X. (2014). Understanding the formation of Mediterranean–African–Asian disjunctions: Evidence for Miocene climate‐driven vicariance and recent long‐distance dispersal in the Tertiary relict Smilax aspera (Smilacaceae). New Phytologist, 204, 243–255. 10.1111/nph.12910 [DOI] [PubMed] [Google Scholar]
- Chifman, J. , & Kubatko, L. (2014). Quartet inference from SNP data under the coalescent model. Bioinformatics, 30(23), 3317–3324. 10.1093/bioinformatics/btu530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, I. S. , Wojciechowski, M. F. , Ruhlman, T. A. , & Jansen, R. K. (2021). In and out: Evolution of viral sequences in the mitochondrial genomes of legumes (Fabaceae). Molecular Phylogenetics and Evolution, 163, 107236. 10.1016/j.ympev.2021.107236 [DOI] [PubMed] [Google Scholar]
- Comes, H. P. (2004). The Mediterranean region: A hotspot for plant biogeographic research. New Phytologist, 164, 11–14. http://www.jstor.org/stable/1514398 [DOI] [PubMed] [Google Scholar]
- Cornejo, O. E. , Yee, M. C. , Dominguez, V. , Andrews, M. , Sockell, A. , Strandberg, E. , Livingstone, D., III , Stack, C. , Romero, A. , Umaharan, P. , Royaert, S. , Tawari, N. R. , Ng, P. , Gutierrez, O. , Phillips, W. , Mockaitis, K. , Bustamante, C. D. , & Motamayor, J. C. (2018). Population genomic analyses of the chocolate tree, Theobroma cacao L., provide insights into its domestication process. Communications Biology, 1, 167. 10.1038/s42003-018-0168-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currat, M. , Ruedi, M. , Petit, R. J. , & Excoffier, L. (2008). The hidden side of invasions: Massive introgression by local genes. Evolution, 62, 1908–1920. 10.1111/j.1558-5646.2008.00413.x [DOI] [PubMed] [Google Scholar]
- de Candolle, A. (1883). Origine des plantes cultivées. Germer Baillière et Cie. [Google Scholar]
- de Medeiros, B.A.S. , (2019). Matrix Condenser v.1.0. https://github.com/brunoasm/matrix_condenser/
- de Medeiros, B. A. S. , & Farrell, B. D. (2018). Whole‐genome amplification in double‐digest RADseq results in adequate libraries but fewer sequenced loci. PeerJ, 6, e5089. 10.7717/peerj.5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Désamoré, A. , Laenen, B. , Devos, N. , Popp, M. , González‐Mancebo, J. M. , Carine, M. A. , & Vanderpoorten, A. (2011). Out of Africa: North‐westwards Pleistocene expansions of the heather Erica arborea . Journal of Biogeography, 38(1), 164–176. 10.1111/j.1365-2699.2010.02387.x [DOI] [Google Scholar]
- Di Guardo, M. , Scollo, F. , Ninot, A. , Rovira, M. , Hermoso, J. F. , Distefano, G. , La Malfa, S. , & Batlle, I. (2019). Genetic structure analysis and selection of a core collection for carob tree germplasm conservation and management. Tree Genetics & Genomes, 15(3), 1–14. 10.1007/s11295-019-1345-6 30546292 [DOI] [Google Scholar]
- Domínguez, M. T. , Madejón, P. , Marañón, T. , & Murillo, J. M. (2010). Afforestation of a trace‐element polluted area in SW Spain: Woody plant performance and trace element accumulation. European Journal of Forest Research, 129(1), 47–59. 10.1007/s10342-008-0253-3 [DOI] [Google Scholar]
- Eaton, D. A. , & Overcast, I. (2020). ipyrad: Interactive assembly and analysis of RADseq datasets. Bioinformatics, 36(8), 2592–2594. 10.1093/bioinformatics/btz966 [DOI] [PubMed] [Google Scholar]
- Ewels, P. , Magnusson, M. , Lundin, S. , & Käller, M. (2016). MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics, 32(19), 3047–3048. 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fady, B. , & Conord, C. (2010). Macroecological patterns of species and genetic diversity in vascular plants of the Mediterranean basin. Diversity and Distributions, 16(1), 53–64. 10.1111/j.1472-4642.2009.00621.x [DOI] [Google Scholar]
- Fineschi, S. , Taurchini, D. , Villani, F. , & Vendramin, G. G. (2000). Chloroplast DNA polymorphism reveals little geographical structure in Castanea sativa Mill.(Fagaceae) throughout southern European countries. Molecular Ecology, 9, 1495–1503. 10.1046/j.1365-294x.2000.01029.x [DOI] [PubMed] [Google Scholar]
- Flowers, J. M. , Hazzouri, K. M. , Gros‐Balthazard, M. , Mo, Z. , Koutrumpa, K. , Perrakis, A. , Ferrand, S. , Khierralah, H. S. M. , Fuller, D. Q. , Aberlenc, F. , Fournaraki, C. , & Purugganan, M. D. (2019). Cross‐species hybridization and the origin of North African date palms. Proceedings of the National Academy of Sciences., 116, 1651–1658. 10.1073/pnas.1817453116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frichot, E. , & François, O. (2015). LEA: An R package for landscape and ecological association studies. Methods in Ecology and Evolution, 6(8), 925–929. 10.1111/2041-210X.12382 [DOI] [Google Scholar]
- Frichot, E. , Mathieu, F. , Trouillon, T. , Bouchard, G. , & François, O. (2014). Fast and efficient estimation of individual ancestry coefficients. Genetics, 196(4), 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller, D. Q. , Willcox, G. , & Allaby, R. G. (2011). Cultivation and domestication had multiple origins: Arguments against the core area hypothesis for the origins of agriculture in the Near East. World Archaeology, 43(4), 628–652. 10.1080/00438243.2011.624747 [DOI] [Google Scholar]
- García‐Castaño, J. L. , Terrab, A. , Ortiz, M. A. , Stuessy, T. F. , & Talavera, S. (2014). Patterns of phylogeography and vicariance of Chamaerops humilis L. (Palmae). Turkish Journal of Botany, 38(6), 1132–1146. 10.3906/bot-1404-38 [DOI] [Google Scholar]
- García‐Verdugo, C. , Mairal, M. , Tamaki, I. , & Msanda, F. (2021). Phylogeography at the crossroad: Pleistocene range expansion throughout the Mediterranean and back‐colonization from the Canary Islands in the legume Bituminaria bituminosa . Journal of Biogeography, 48(7), 1622–1634. 10.1111/jbi.14100 [DOI] [Google Scholar]
- Gosselin, T. (2020). radiator: RADseq Data Exploration, Manipulation and Visualization using R. R package version 1.1.9 https://thierrygosselin.github.io/radiator/. 10.5281/zenodo.3687060 [DOI]
- Groppi, A. , Liu, S. , Cornille, A. , Decroocq, S. , Bui, Q. T. , Tricon, D. , & Decroocq, V. (2021). Population genomics of apricots unravels domestication history and adaptive events. Nature Communications, 12(1), 1–16. 10.1038/s41467-021-24283-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros‐Balthazard, M. , Besnard, G. , Sarah, G. , Holtz, Y. , Leclercq, J. , Santoni, S. , Wegmann, D. , Glémin, S. , & Khadari, B. (2019). Evolutionary transcriptomics reveals the origins of olives and the genomic changes associated with their domestication. The Plant Journal, 100(1), 143–157. 10.1111/tpj.14435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros‐Balthazard, M. , Galimberti, M. , Kousathanas, A. , Newton, C. , Ivorra, S. , Paradis, L. , Vigouroux, Y. , Carter, R. , Tengberg, M. , Battesti, V. , Santoni, S. , Falquet, L. , Pintaud, J.‐C. C. , Terral, J.‐F. F. , & Wegmann, D. (2017). The discovery of wild date palms in Oman reveals a complex domestication history involving centers in the Middle East and Africa. Current Biology, 27, 2211–2218. 10.1016/j.cub.2017.06.045 [DOI] [PubMed] [Google Scholar]
- Gruber, B. , Unmack, P. J. , Berry, O. F. , & Georges, A. (2018). dartr: An r package to facilitate analysis of SNP data generated from reduced representation genome sequencing. Molecular Ecology Resources, 18(3), 691–699. 10.1111/1755-0998.12745 [DOI] [PubMed] [Google Scholar]
- Hipp, A. L. , Manos, P. S. , Hahn, M. , Avishai, M. , Bodénès, C. , Cavender‐Bares, J. , Crowl, A. A. , Deng, M. , Denk, T. , Fitz‐Gibbon, S. , Gailing, O. , González‐Elizondo, M. S. , González‐Rodríguez, A. , Grimm, G. W. , Jiang, X.‐L. , Kremer, A. , Lesur, I. , McVay, J. D. , Plomion, C. , … Valencia‐Avalos, S. (2020). Genomic landscape of the global oak phylogeny. New Phytologist, 226, 1198–1212. 10.1111/nph.16162 [DOI] [PubMed] [Google Scholar]
- Hodel, R. G. J. , Chen, S. , Payton, A. C. , McDaniel, S. F. , Soltis, P. , & Soltis, D. E. (2017). Adding loci improves phylogeographic resolution in red mangroves despite increased missing data: Comparing microsatellites and RAD‐Seq and investigating loci filtering. Scientific Reports, 7, 17598. 10.1038/s41598-017-16810-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart, T. , & Ahmed, I. (2011). adegenet 1.3‐1: New tools for the analysis of genome‐wide SNP data. Bioinformatics, 27(21), 3070–3307. 10.1093/bioinformatics/btr521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamvar, Z. N. , Tabima, J. F. , & Grünwald, N. J. (2014). Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ, 2, e281. 10.7717/peerj.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan, K. , McGinnity, P. , Cross, T. F. , Crozier, W. W. , & Prodöhl, P. A. (2013). diveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods in Ecology and Evolution, 4(8), 782–788. 10.1111/2041-210X.12067 [DOI] [Google Scholar]
- Klesser, R. , Husemann, M. , Schmitt, T. , Sousa, P. , Moussi, A. , & Habel, J. C. (2021). Molecular biogeography of the Mediterranean Buthus species complex (Scorpiones: Buthidae) at its southern Palaearctic margin. Biological Journal of the Linnean Society, 133, 166–178. 10.1093/biolinnean/blab014 [DOI] [Google Scholar]
- La Malfa, S. , Currò, S. , Douglas, A. B. , Brugaletta, M. , Caruso, M. , & Gentile, A. (2014). Genetic diversity revealed by EST‐SSR markers in carob tree (Ceratonia siliqua L.). Biochemical Systematics and Ecology, 55, 205–211. 10.1016/j.bse.2014.03.022 [DOI] [Google Scholar]
- Martínez‐Freiría, F. , Crochet, P. A. , Fahd, S. , Geniez, P. , Brito, J. C. , & Velo‐Antón, G. (2017). Integrative phylogeographical and ecological analysis reveals multiple Pleistocene refugia for Mediterranean Daboia vipers in north‐west Africa. Biological Journal of the Linnean Society, 122, 366–384. 10.1093/biolinnean/blx038 [DOI] [Google Scholar]
- Médail, F. , & Diadema, K. (2009). Glacial refugia influence plant diversity patterns in the Mediterranean Basin. Journal of Biogeography, 36(7), 1333–1345. 10.1111/j.1365-2699.2008.02051.x [DOI] [Google Scholar]
- Médail, F. , & Quézel, P. (1999). The phytogeographical significance of SW Morocco compared to the Canary Islands. Plant Ecology, 140(2), 221–244. 10.1023/A:1009775327616 [DOI] [Google Scholar]
- Médail, F. , Quezel, P. , Besnard, G. , & Khadari, B. (2001). Systematics, ecology and phylogeographic significance of Olea europaea L. ssp. maroccana (Greuter & Burdet) P. Vargas et al., a relictual olive tree in south‐west Morocco. Botanical Journal of the Linnean Society, 137(3), 249–266. 10.1111/j.1095-8339.2001.tb01121.x [DOI] [Google Scholar]
- Meyer, R. S. , Duval, A. E. , & Jensen, H. R. (2012). Patterns and processes in crop domestication: An historical review and quantitative analysis of 203 global food crops. New Phytologist, 196, 29–48. 10.1111/j.1469-8137.2012.04253.x [DOI] [PubMed] [Google Scholar]
- Migliore, J. , Baumel, A. , Leriche, A. , Juin, M. , & Médail, F. (2018). Surviving glaciations in the Mediterranean region: An alternative to the long‐term refugia hypothesis. Botanical Journal of the Linnean Society, 187, 537–549. 10.1093/botlinnean/boy032 [DOI] [Google Scholar]
- Minh, B. Q. , Schmidt, H. A. , Chernomor, O. , Schrempf, D. , Woodhams, M. D. , von Haeseler, A. , & Lanfear, R. (2020). IQ‐TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution, 37, 1530–1534. 10.1093/molbev/msaa015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto Feliner, G. (2011). Southern European glacial refugia: A tale of tales. Taxon, 60, 365–372. 10.1002/tax.602007 [DOI] [Google Scholar]
- Nieto Feliner, G. (2014). Patterns and processes in plant phylogeography in the Mediterranean Basin. A review. Perspectives in Plant Ecology, Evolution and Systematics, 16, 265–278. 10.1016/j.ppees.2014.07.002 [DOI] [Google Scholar]
- Ortiz, M. A. , Tremetsberger, K. , Stuessy, T. , Terrab, A. , García‐Castaño, J. L. , & Talavera, S. (2009). Phylogeographic patterns in Hypochaeris sect. Hypochaeris (Asteraceae, Lactuceae) of the western Mediterranean. Journal of Biogeography, 36, 1384–1397. 10.1111/j.1365-2699.2008.02079.x [DOI] [Google Scholar]
- Parchman, T. L. , Jahner, J. P. , Uckele, K. A. , Galland, L. M. , & Eckert, A. J. (2018). RADseq approaches and applications for forest tree genetics. Tree Genetics & Genomes, 14(3), 1–25. 10.1007/s11295-018-1251-3 [DOI] [Google Scholar]
- Pickrell, J. , & Pritchard, J. (2012). Inference of population splits and mixtures from genome‐wide allele frequency data. PLoS Genet, 8(11), e1002967. 10.1038/npre.2012.6956.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan, M. D. (2019). Evolutionary insights into the nature of plant domestication. Current Biology, 29(14), R705–R714. 10.1016/j.cub.2019.05.053 [DOI] [PubMed] [Google Scholar]
- Quézel, P. , & Médail, F. (2003). Écologie et biogéographie des forêts du bassin Méditerranéen. Elsevier Editions. [Google Scholar]
- Ramón‐Laca, L. , & Mabberley, D. J. (2004). The ecological status of the carob‐tree (Ceratonia siliqua, Leguminosae) in the Mediterranean. Botanical Journal of the Linnean Society, 144, 431–436. 10.1111/j.1095-8339.2003.00254.x [DOI] [Google Scholar]
- Revell, L. J. (2012). phytools: An R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution, 3, 217–223. 10.1111/j.2041-210X.2011.00169.x [DOI] [Google Scholar]
- Silliman, K. (2019). Population structure, genetic connectivity, and adaptation in the Olympia oyster (Ostrea lurida) along the west coast of North America. Evolutionary Applications, 12(5), 923–939. 10.1111/eva.12766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D. L. (2018). PAUP*(* Phylogenetic Analysis Using PAUP). Version 4a161.
- Thompson, J. D. (2020). Plant evolution in the Mediterranean: Insights for conservation. Oxford University Press. [Google Scholar]
- Tous, J. , Romero, A. , & Batlle, I. (2013). The Carob tree: Botany, horticulture, and genetic resources. Horticultural Reviews, 41, 385–456. 10.1002/9781118707418.ch08 [DOI] [Google Scholar]
- Turnbull, L. A. , Santamaria, L. , Martorell, T. , Rallo, J. , & Hector, A. (2006). Seed size variability: From carob to carats. Biology Letters, 2(3), 397–400. 10.1098/rsbl.2006.0476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas, P. , Fernández‐Mazuecos, M. , & Heleno, R. (2018). Phylogenetic evidence for a Miocene origin of Mediterranean lineages: Species diversity, reproductive traits and geographical isolation. Plant Biology, 20, 157–165. 10.1111/plb.12626 [DOI] [PubMed] [Google Scholar]
- Vendramin, G. G. , Fady, B. , González‐Martínez, S. C. , Hu, F. S. , Scotti, I. , Sebastiani, F. , Soto, A. , & Petit, R. J. (2008). Genetically depauperate but widespread: The case of an emblematic Mediterranean pine. Evolution: International Journal of Organic. Evolution, 62(3), 680–688. 10.1111/j.1558-5646.2007.00294.x [DOI] [PubMed] [Google Scholar]
- Villa‐Machío, I. , Fernández de Castro, A. G. , Fuertes‐Aguilar, J. , & Nieto Feliner, G. (2018). Out of North Africa by different routes: Phylogeography and species distribution model of the western Mediterranean Lavatera maritima (Malvaceae). Botanical Journal of the Linnean Society, 187, 441–455. 10.1093/botlinnean/boy025 [DOI] [Google Scholar]
- Viruel, J. , Haguenauer, A. , Juin, M. , Mirleau, F. , Bouteiller, D. , Boudagher‐Kharrat, M. , Ouahmane, L. , La Malfa, S. , Médail, F. , Sanguin, H. , Nieto Feliner, G. , & Baumel, A. (2018). Advances in genotyping microsatellite markers through sequencing and consequences of scoring methods for Ceratonia siliqua (Leguminosae). Applications in Plant Sciences, 6, e01201. 10.1002/aps3.1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viruel, J. , Le Galliot, N. , Pironon, S. , Nieto Feliner, G. , Suc, J. P. , Lakhal‐Mirleau, F. , Juin, M. , Selva, M. , Bou Dagher Kharrat, M. , Ouahmane, L. , Malfa, S. , Diadema, K. , Sanguin, H. , Médail, F. , & Baumel, A. (2020). A strong east–west Mediterranean divergence supports a new phylogeographic history of the carob tree (Ceratonia siliqua, Leguminosae) and multiple domestications from native populations. Journal of Biogeography, 47, 460–471. 10.1111/jbi.13726 [DOI] [Google Scholar]
- Warschefsky, E. J. , & von Wettberg, E. J. (2019). Population genomic analysis of mango (Mangifera indica) suggests a complex history of domestication. New Phytologist, 222, 2023–2037. 10.1111/nph.15731 [DOI] [PubMed] [Google Scholar]
- Wascher, M. , & Kubatko, L. (2021). Consistency of SVDQuartets and maximum likelihood for coalescent‐based species tree estimation. Systematic Biology, 70(1), 33–48. 10.1093/sysbio/syaa039 [DOI] [PubMed] [Google Scholar]
- Whitlock, M. C. , & Lotterhos, K. E. (2015). Reliable detection of loci responsible for local adaptation: Inference of a null model through trimming the distribution of FST. The American Naturalist, 186(S1), S24–S36. 10.1086/682949 [DOI] [PubMed] [Google Scholar]
- Winter, D. J. (2012). MMOD: An R library for the calculation of population differentiation statistics. Molecular Ecology Resources, 12(6), 1158–1160. 10.1111/j.1755-0998.2012.03174.x [DOI] [PubMed] [Google Scholar]
- Zeder, M. A. (2008). Domestication and early agriculture in the Mediterranean Basin: Origins, diffusion, and impact. Proceedings of the National Academy of Sciences, 105(33), 11597–11604. 10.1073/pnas.0801317105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, R. , Wang, Y. H. , Jin, J. J. , Stull, G. W. , Bruneau, A. , Cardoso, D. , De Queiroz, L. P. , Moore, J. M. , Zhang, S.‐D. , Chen, S.‐Y. , Wang, J. , Li, D.‐Z. , & Yi, T. S. (2020). Exploration of plastid phylogenomic conflict yields new insights into the deep relationships of Leguminosae. Systematic Biology, 69(4), 613–622. 10.1093/sysbio/syaa013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohary, D. (2002). Domestication of the carob (Ceratonia siliqua L.). Israel Journal of Plant Sciences, 50, 141–115. 10.1560/BW6B-4M9P-U2UA-C6NN [DOI] [Google Scholar]
- Zohary, D. , & Hopf, M. (2012). Domestication of plants in the Old World: The origin and spread of cultivated plants in West Asia, Europe and the Nile Valley. Oxford University Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1
Appendix S2
Data Availability Statement
Full information on population sampling and microsatellite data are available in Viruel et al. (2020) and deposited in DRYAD (https://doi.org/10.5061/dryad.k7m020r). Raw RADseq reads are deposited at NCBI SRA database under Bioproject accession PRJNA793764. Assemblies from ipyRAD (vcf files), filtered R data files and R scripts of analyses are available at https://osf.io/t5m8y/.


