Abstract
Ovarian cancer is the most lethal gynecological malignancy worldwide with high metastasis and poor prognosis rates. Cancer‐associated fibroblasts (CAFs), a heterogeneous population of cells that constitutes a major component of the tumor microenvironment, secrete extracellular vesicles (EVs) loading with proteins, lipids, and RNAs to promote tumorigenesis. However, the specific roles of CAF‐derived proteins contained in EVs in ovarian cancer remain poorly understood at present. Using the gene expression microarray analysis, we identified a list of dysregulated genes between the α‐SMA+CAF and FAP+CAF subpopulations, from which secretory leukocyte protease inhibitor (SLPI) was chosen for further validation. Quantitative PCR, western blot, immunohistochemistry, and enzyme‐linked immunosorbent assays were used to assess SLPI expression in ovarian cancer cells, tissues, CAFs, and EVs. Additionally, we evaluated the effects of exogenous SLPI on proliferation, migration, invasion, and adhesion of ovarian cancer cells in vitro. Our results showed SLPI protein was upregulated in CAFs, particularly in the FAPhighα‐SMAlowCAF subpopulation, and associated with increased tumor grade and decreased overall survival (OS). Importantly, CAF‐derived SLPI protein could be encapsulated in EVs for delivery to ovarian cancer cells, thus facilitating cell proliferation, migration, invasion, and adhesion via activating the PI3K/AKT and downstream signaling pathways. Moreover, high plasma expression of SLPI encapsulated in EVs was closely correlated with tumor stage in ovarian cancer patients. Our collective results highlight an oncogenic role of plasma EV‐encapsulated SLPI secreted by CAFs in tumor progression for the first time, supporting its potential utility as a prognostic biomarker of ovarian cancer.
Keywords: cancer‐associated fibroblasts, extracellular vesicles, ovarian cancer, SLPI, tumor progression
Abbreviations
- ATCC
American Type Culture Collection
- CAFs
cancer‐associated fibroblasts
- CM
conditioned medium
- DAPI
4′,6‐diamidino‐2′‐phenylindole
- ecDNA
extrachromosomal DNA
- ECM
extracellular matrix
- EMT
epithelial–mesenchymal transition
- EVs
extracellular vesicles
- Fn
fibronectin
- GEPIA
gene expression profiling interactive analysis
- HGSOC
high‐grade serous epithelial ovarian cancer
- ITGBL1
Integrin beta‐like 1
- NTA
nanoparticle tracking analysis
- OS
overall survival
- Prgn
progranulin
- qPCR
quantitative real‐time PCR
- Rb
retinoblastoma tumor suppressor protein
- rhSLPI
recombinant human SLPI protein
- SLPI
secretory leukocyte protease inhibitor
- SNK
Student–Newman–Keuls
- STR
short tandem repeat
- TEM
transmission electron microscope
- WAP
whey acidic protein
1. INTRODUCTION
Ovarian cancer is the most lethal gynecological malignancy worldwide, often diagnosed at advanced stages due to the lack of early diagnostic biomarkers. 1 Despite the rapid development of novel treatment strategies in recent years, such as targeted therapy, antiangiogenesis, and immunotherapy, prognostic improvement remains limited, and the 5‐year survival rate of patients with advanced cancer is still lower than 30%. 2 , 3 Elucidation of the mechanisms underlying ovarian cancer occurrence and progression would provide valuable insights that provide a basis for improving survival rates.
The tumor microenvironment consists of vascular endothelial cells, immune cells, fibroblasts, bone marrow–derived inflammatory cells, and extracellular matrix (ECM). Interactions between the surrounding extracellular microenvironment and cancer cells clearly have significant regulatory roles in tumor development and therapeutic response. 4 , 5 , 6 , 7 Cancer‐associated fibroblasts (CAFs) are major components in the tumor microenvironment that interact with tumor cells and promote tumorigenesis by secreting cytokines, chemokines, and microvesicles in various cancer types. 8 , 9 , 10 , 11 , 12
Extracellular vesicles (EVs) are membrane‐encapsulated nanoparticles secreted from various cells that contain abundant small bioactive molecules, including noncoding RNA, DNA, proteins, and lipids. 13 , 14 EVs have a key role in intercellular communication by delivering bioactive molecules to recipient cells via membrane fusion or receptor–ligand interactions. 15 , 16 , 17 Accumulating evidence strongly suggests that CAF‐derived EVs carrying specific miRNAs promote tumor drug resistance, proliferation, migration, and invasion. 12 , 18 , 19 , 20 In addition to miRNAs, CAF‐derived EV‐mediated protein delivery is reported to contribute importantly to tumor development. Recent studies have demonstrated that CAF‐derived Snail1 promoted epithelial–mesenchymal transition (EMT) progression upon exosomal delivery in lung cancer while CAF‐derived EV‐encapsulated annexin A6 enhanced drug resistance by activating the FAK‐YAP signaling pathway in gastric cancer. 21 , 22 Earlier, Chen and co‐workers 23 reported that exosomal Wnt10b released from CAFs promoted EMT progression in breast cancer epithelial cells and facilitated metastasis. The collective evidence obtained so far indicates that CAF‐derived EV‐encapsulated proteins are significantly involved in the regulation of tumor development. However, limited studies have focused on the effects of EV‐mediated proteins released from stromal fibroblasts in ovarian cancer to date.
Secretory leukocyte protease inhibitor (SLPI) is an 11.7 kDa whey acidic protein (WAP) secreted by mucosal surfaces of the respiratory and gastrointestinal systems. 24 , 25 , 26 Early studies have shown that overexpression of SLPI promoted ovarian cancer cells migration, invasion, and drug resistance. 27 , 28 , 29 However, the issue of whether SLPI‐mediated crosstalk exists between CAFs and tumor cells that can regulate paracrine communication and protumorigenic activity in the tumor microenvironment is yet to be established.
Here, we observed upregulation of SLPI in ovarian cancer cells and CAFs, particularly in the FAPhighα‐SMAlowCAF subpopulation. Importantly, our studies revealed for the first time that CAF‐secreted SLPI was encapsulated in EVs and delivered to tumor cells. Increased expression of SLPI in ovarian cancer cells further promoted cancer cell proliferation, migration, invasion, and adhesion by activating PI3K–AKT–GSK‐3β/NF‐κB/MAPK signaling pathways. Clinically, compared to the plasma EVs from healthy controls, SLPI protein was more highly expressed in plasma EVs of ovarian cancer patients, especially those with advanced disease. Given our finding that CAF‐secreted SLPI stimulates cancer progression via EVs, the expression of SLPI protein in plasma EVs may serve as an effective prognostic biomarker of ovarian cancer.
2. MATERIALS AND METHODS
2.1. Tissue and plasma samples
Ovarian cancer tissue samples were collected from the Third Affiliated Hospital of Harbin Medical University and plasma samples were obtained from Sun Yat‐sen University Cancer Center according to the guidelines of the Ethics Committees of the respective institutions. Informed consent was obtained from the patients or their guardians.
2.2. Cell culture
SKOV3, Caov3, OV‐90, OVCAR3, ES‐2, and OVCAR5 cell lines were purchased from American Type Culture Collection (ATCC). HOSE 6‐3, A2780, UACC‐1598, and HO‐8910 cells were kindly provided by Xin‐Yuan Guan (University of Hong Kong Medical Center). All cell lines were cultured following the protocols described by the ATCC and authenticated using the short tandem repeat (STR) analysis (Microread).
2.3. Conditioned medium (CM)
Cells were cultured in Dulbecco's modified Eagle's medium with 10% serum. After 24 h, the medium was replaced with Opti‐MEM and cells were cultured for a further 72 h. Subsequently, the CM was recovered, centrifuged at 1000g for 10 min and passed through a 0.22 μm filter (Millipore), and used for experiments as indicated.
2.4. Quantitative real‐time PCR (qPCR)
When cells were in good condition, total RNA was isolated from cells using TRIzol reagent (Invitrogen), followed by reverse transcription of RNA to synthesize the first‐strand cDNA samples using the Transcriptor First Strand cDNA Synthesis Kit (Roche) according to the manufacturer's instructions. The clinical samples (28 normal and 49 ovarian cancer tissues) for qPCR assay were obtained from the Third Affiliated Hospital of Harbin Medical University, and all patients had written approval documents. qPCR analysis was performed as described previously. 30 , 31 , 32
2.5. Western blot analysis
Proteins were isolated using RIPA lysis buffer (Applygen) with 10% protease inhibitor and 10% phosphatase inhibitor. After quantification of the protein concentration using a BCA Protein Assay Kit (Applygen), equal amounts of protein samples were separated via sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to PVDF membranes. After blocking with 5% skimmed milk, membranes were immunoblotted with primary antibodies overnight at 4°C. The next day, membranes were incubated with secondary antibodies for 1h at room temperature and scanned using a Tanon 5200 (Tanon) or Amersham Imager 600 (GE Healthcare Life Sciences) chemiluminescence system. The band intensity was assessed with ImageJ.
2.6. Antibodies and reagents
The following antibodies and reagents were utilized: anti‐SLPI (Novus; 1:1000), anti‐PI3K, Akt, phospho‐Akt (S473), phospho‐Akt (T308), GSK‐3β, phospho‐GSK‐3β, IKKα, phospho‐IKKα/β, IκBα, NF‐κB, phospho‐NF‐κB, MAPK, phospho‐MAPK, EV markers (CD9, HSP70, Alix, Annexin‐V, Flotillin‐1 and GM130), α‐SMA, vimentin, FAP (Cell Signaling Technology; 1:1000), anti‐CD63 and TSG101 (System Biosciences; 1:500), anti‐mouse IgG/HRP and anti‐rabbit IgG/HRP secondary antibodies (Cell Signaling Technology; 1:2500). Recombinant human SLPI protein (rhSLPI) was acquired from R&D Systems and 1 μg/ml rhSLPI was used in the study.
2.7. Cell proliferation assay
The MTS assay (Promega) was utilized to assess cell proliferative ability. Tumor cells (with or without rhSLPI) were seeded in 96‐well plates at a density of 3000 cells/well in quintuplicate. MTS solution (100 μl medium + 20 μl MTS) was added to each well and incubated for 2 h at 37°C. The OD value of each well was measured by absorbance at 490 nm during the coculture period and GraphPad 5 software (GraphPad Software) was applied to calculate growth rates.
2.8. EdU assay
Cells were grown on slides in a 24‐well plate at a density of 1 × 105 cells/well. After 24 h, cells were incubated with EdU (10 μM/well) for 2 h at 37°C. Then, the cells were fixed with 4% paraformaldehyde (Boster) and washed three times with 3% bovine serum albumin (BSA)‐phosphate‐buffered saline (PBS), followed by permeabilization with 0.3% Triton X‐100 for 15 min at room temperature. Next, cells were incubated with click additive solution in the dark for 30 min and stained with 4′,6‐diamidino‐2′‐phenylindole (DAPI). Finally, staining images were captured with a fluorescence microscope (Carl Zeiss). Ten random fields were photographed and the EdU‐positive rates were counted (EdU‐positive rate (%) = the number of positive cells/number of total cells × 100%).
2.9. Transwell migration and invasion assays
For the cell migration assay, 3 × 104 tumor cells were resuspended in 200 µl serum‐free RPMI‐1640 and seeded into an upper Transwell chamber (8 μm pore size; Corning). Next, the 500 µl medium containing 10% FBS (with or without rhSLPI) was added to the lower chamber. After coculture for 30 h (A2780) and 24 h (Caov3), cells on the opposite sides of the chambers were fixed with 4% paraformaldehyde and stained with 0.2% crystal violet. Finally, cells were imaged and counted in five random fields using a light microscope (Nikon). For the invasion assay, cells were cocultured using Corning Matrigel invasion chambers, and the cell numbers were counted after 48 h (A2780) and 30 h (Caov3).
2.10. Adhesion assay
A 96‐well‐plate was coated with fibronectin (Fn) at a concentration of 10 µg/ml overnight at 4°C. The plate was washed twice with PBS and blocked with 0.5% BSA for 2 h at 37°C. Next, 2 × 104 tumor cells (with or without rhSLPI) were seeded in a 96‐well plate and cultured for 1 h at 37°C. Cells were washed twice with PBS to remove nonadherent cells and MTS reagent was added to determine the OD values. Finally, the cell adhesion rate was calculated for statistical analysis.
2.11. Immunofluorescence
Cells grown on slides were washed three times with PBS, fixed with 4% paraformaldehyde for 15 min, and then permeabilized with 0.1% Triton X‐100 and blocked with 4% BSA‐PBS. Next, slides were incubated with α‐SMA antibody (Abcam; 1:100), FAP antibody (Cell Signaling Technology; 1:200), vimentin antibody (Cell Signaling Technology; 1:200), and SLPI antibody (Novus; 1:200) overnight at 4°C. After that, the cells were incubated with fluorescence‐conjugated secondary antibody and subjected to DAPI staining. Images were photographed under a confocal fluorescence microscope. For tissues, multicolor immunofluorescence staining of SLPI, Ki67, FAP, and DAPI was performed using a PANO 4‐plex IHC kit (Panovue, China) according to the manufacturer's instructions. In brief, paraffin‐embedded tissue sections were baked in a dry oven for 60 min at 60°C, followed by deparaffinization and rehydration. After antigen retrieval and blocking, sections were washed three times and incubated with primary antibody, followed by the incubation with biotinylated secondary antibody and tyrosine signal amplification diluent. During each round of staining, the slides were subjected to high‐pressure treatment. After all antigens were labeled, the nucleus was subjected to DAPI staining. Images were obtained using a Vectra Polaris slide scanner (PerkinElmer).
2.12. EV isolation and identification
EVs were isolated from culture supernatant and plasma via differential ultracentrifugation as described previously. 33 , 34 In brief, culture supernatant and plasma were centrifuged at 500g for 10 min and subsequently at 12,000g for 30 min at 4°C. CM and plasma were collected and passed through a 0.22 μm filter (Millipore), followed by ultracentrifugation at 100,000g for 70 min for EV collection. EVs were washed with sterilized PBS and purified via further ultracentrifugation at 100,000g for 70 min. Next, EVs were resuspended in PBS, dropped in glow‐discharged copper grids, and stained with 2% phosphotungstic acid. Samples were observed under a J Tecnai G2 F20 ST transmission electron microscope (TEM) or Talos L120C TEM. The size and concentration of EVs were quantified via nanoparticle tracking analysis (NTA) at VivaCell Biosciences with a ZetaView PMX 110 or NanoSight NS300 instrument (Malvern Instruments Ltd.) equipped with NTA 3.0 analytical software (Malvern Instruments Ltd.).
2.13. EV labeling and tracking
Isolated EVs from the culture medium were stained with PKH26 red fluorescent membrane linker dye (Sigma‐Aldrich) in keeping with the manufacturer's instructions. Tumor cells grown on coverslips were incubated with PKH26‐labeled EVs for 12h and prepared for immunofluorescence as described above. Internalization of EVs was examined via confocal fluorescence microscopy.
2.14. Enzyme‐linked immunosorbent assay (ELISA)
CM and EVs were obtained as described above. SLPI protein expression in CM and EVs was quantified using a Human SLPI ELISA Kit (R&D Systems) according to the manufacturer's instructions.
2.15. Gene expression profiling analysis
The Affymetrix GeneChip® Mouse Genome 430 2.0 Array contained 39,000 transcripts representing 34,000 distinct mouse genes, with sequence information based on GeneBank, dbEST, RefSeq, and UniGene databases. Total RNA was extracted and converted to cDNA, which was hybridized with an Affymetrix GeneChip® Mouse Genome 430 2.0 Array for the detection of differentially expressed genes between FAP+CAFs and α‐SMA+CAFs.
2.16. Statistical analysis
Statistical analyses were performed using GraphPad 5 software. Two‐tailed Student's t‐test and one‐way ANOVA with the Student–Newman–Keuls (SNK) test were applied for analysis of relative mRNA expression and cell function experiments, including MTS proliferation, Transwell migration, invasion, and adhesion assays. All data were presented as mean ± SD and p < 0.05 was considered significant difference.
3. RESULTS
3.1. SLPI is overexpressed in CAFs of ovarian cancer, specifically, the FAPhighα‐SMAlow CAF subpopulation
Earlier studies by our group showed that the oncogene Sei‐1 promotes genomic instability of fibroblasts in the tumor microenvironment, forming extrachromosomal DNA (ecDNA), an important genomic feature in cancer. 31 , 32 EcDNA has been shown to regulate tumor progression by driving genome remodeling and altering the shape of its chromatin. 35 , 36 , 37 In addition, Sei‐1‐transfected A2780 cells generated high levels of FAPhighα‐SMAlow CAF subpopulation (FAP+CAFs) in the tumor microenvironment, which displayed ecDNA formation with increasing passage number in vivo, while the vector‐transfected group generated α‐SMAhighFAPlow CAF subpopulation (α‐SMA+CAFs) that failed to form ecDNA. Compared to α‐SMA+CAFs, FAP+CAFs significantly promoted the malignant phenotypes of tumor cells, including migration, invasion, angiogenesis in vitro, and tumorigenesis in vivo (data under publication).
Gene expression profiling was performed to evaluate differentially expressed genes in the α‐SMA+CAF and FAP+CAF subpopulations (Supporting Information: Figure 1A,B). Among the differentially expressed genes, Slpi, Eps8, Il2rg, and Nrn1 levels were significantly increased in FAP+CAFs (Table 1). SLPI is a secretory protein upregulated in various cancer types that promotes the malignant phenotypes. 27 , 38 , 39 However, to our knowledge, no studies to date have focused on the role of CAF‐derived SLPI protein in ovarian cancer development. In the present study, we first confirmed overexpression of SLPI in tumor tissues of ovarian cancer via Gene Expression Profiling Interactive Analysis (GEPIA; Figure 1A). RNA was isolated from 49 high‐grade serous epithelial ovarian cancer (HGSOC) and 28 normal tissue samples, and qPCR was employed to quantify SLPI mRNA expression. The SLPI level was distinctly higher in tumors than those in normal tissues, consistent with GEPIA analysis (Figure 1B). Data obtained from GEPIA additionally disclosed significantly shorter overall survival (OS) of patients with high SLPI expression (Figure 1C). Moreover, SLPI expression was strongly associated with tumor stage in UALCAN database analysis (Figure 1D). More importantly, the analysis of ovarian profile GSE126132 showed that compared to tumor epithelium, SLPI expression was specifically elevated in the tumor CAF (Figure 1E). These data indicated a key role of stromal fibroblasts‐derived SLPI in the occurrence and development of ovarian cancer. Next, we explored the expression and function of SLPI in fibroblasts of ovarian cancer. Based on analysis of gene expression profiles, qPCR results showed significantly higher Slpi mRNA expression in FAP+CAFs relative to α‐SMA+CAFs (Figure 1F) and immunofluorescence experiments revealed that FAP+CAFs had lower α‐SMA but higher FAP and SLPI expression compared to α‐SMA+CAFs (Figure 1G). On western blots, compared to α‐SMA+CAFs, SLPI protein expression was significantly increased in cells and CM of FAP+CAFs (Figure 1H). Furthermore, multicolor immunofluorescence staining showed that Ki67 was expressed in normal ovarian tissues and ovarian cancer tissues, and it seemed that Ki67 was more intensely expressed in tumor tissues. SLPI and FAP were rarely observed in normal ovarian tissues while FAP was mainly expressed in the stromal cells of tumor tissues. SLPI‐positive cells were commonly detected in the epithelial and stromal cells of tumor tissues. Importantly, SLPI, Ki67, and FAP were colocalized in CAFs of tumor tissues (white arrowhead) (Figure 1I). The collective findings support a critical role of SLPI, a secreted protein overexpressed in CAFs, particularly in the FAPhighα‐SMAlowCAF subpopulation, in tumor progression.
Table 1.
Relative expression of elevated genes in FAP+CAFs
| Probe_Set_ID | Gene_Symbol | Fold change | ||
|---|---|---|---|---|
| FAP+CAF1 versus α‐SMA+CAF | FAP+CAF2 versus α‐SMA+CAF | FAP+CAF3 versus α‐SMA+CAF | ||
| 1448377_at | Slpi | 11.293815 | 23.017688 | 22.035244 |
| 1422823_at | Eps8 | 17.572383 | 19.41982 | 21.883900 |
| 1416295_a_at | Il2rg | 26.5812 | 11.21256 | 18.042708 |
| 1428393_at | Nrn1 | 16.9473 | 10.19137 | 14.840687 |
Note: Bold values denote a statistically significant difference at p < 0.05.
Figure 1.
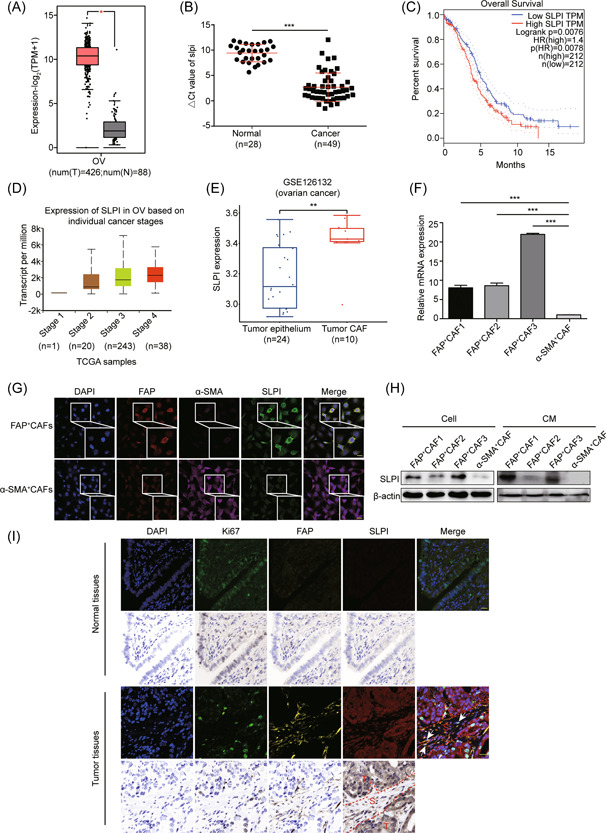
SLPI is specifically upregulated in CAFs of ovarian cancer. (A) Bioinformatics analysis of SLPI expression in ovarian cancer and normal tissues based on the GEPIA database. (B) Quantitative real‐time PCR analysis of relative SLPI mRNA expression in human ovarian cancer and normal tissues. The △CT values (SLPI normalized to β‐actin) were subjected to the Wilcoxon signed‐rank test. Larger △CT values indicate lower expression. (C) GEPIA analysis shows that SLPI expression is related to OS in ovarian cancer. (D) Bioinformatics analysis of the relationship between SLPI and tumor grade in ovarian cancer using UALCAN database. (E) Box plots showing the expression of SLPI in tumor epithelium and CAF of the ovarian profile GSE126132. (F) Quantitative real‐time PCR of relative Slpi mRNAs that are differentially expressed according to gene expression profiling analysis (n = 3). (G) Representative immunofluorescence images of FAP, α‐SMA, and SLPI in FAP+CAFs and α‐SMA+CAFs (scale bar, 20 μm). (H) Western blot of SLPI from cells and CM samples using β‐actin as a loading control. (I) Representative images of multicolor immunofluorescence staining of SLPI, Ki67, FAP, and DAPI in normal and ovarian cancer tissues. The white arrowhead signifies the colocalization of SLPI, Ki67, FAP, and DAPI in tumor stromal tissues. Scale bar, 50 μm.*p < 0.05, **p < 0.01, and ***p < 0.001. CAF, cancer‐associated fibroblast; SLPI, secretory leukocyte protease inhibitor; OS, overall survival; T, tumor; S, stroma. [Color figure can be viewed at wileyonlinelibrary.com]
3.2. Exogenous SLPI promotes tumor cell proliferation, migration, invasion, and adhesion
Because SLPI functions as a secretory protein, the function of exogenous SLPI in ovarian cancer cells was investigated. To exclude interference of endogenous SLPI, we initially examined the SLPI level in a human normal ovarian epithelial cell line (HOSE 6‐3) and ovarian cancer cell lines (A2780, SKOV3, OVCAR3, OV‐90, Caov3, ES‐2, UACC‐1598, and HO‐8910). Our results disclosed markedly lower SLPI expression in Caov3 and HO‐8910 cells (Supporting Information: Figure 1C). Accordingly, A2780, used in previous research, and Caov3 cells were selected for further experiments. To establish the biological function of SLPI, in vitro experiments were performed using rhSLPI. Data from MTS proliferation and EdU assays demonstrated that rhSLPI protein significantly promoted the proliferation of A2780 and Caov3 cells compared with the control group (Figure 2A–C), supporting the theory that CAF‐derived SLPI protein plays a critical role in the progression of ovarian cancer cell growth. Subsequently, we investigated whether exogenous SLPI protein affected the migration and invasion of A2780 and Caov3 cells with the aid of Transwell migration and invasion assays (without or with Matrigel). Notably, compared to the control group, exogenous SLPI protein markedly increased migration (Figure 2D) and invasion abilities (Figure 2E) and promoted adhesion of A2780 and Caov3 cells (Figure 2F). These results suggest that CAF‐secreted SLPI promotes the malignant phenotypes of ovarian cancer cells, including proliferation, migration, invasion, and adhesion.
Figure 2.
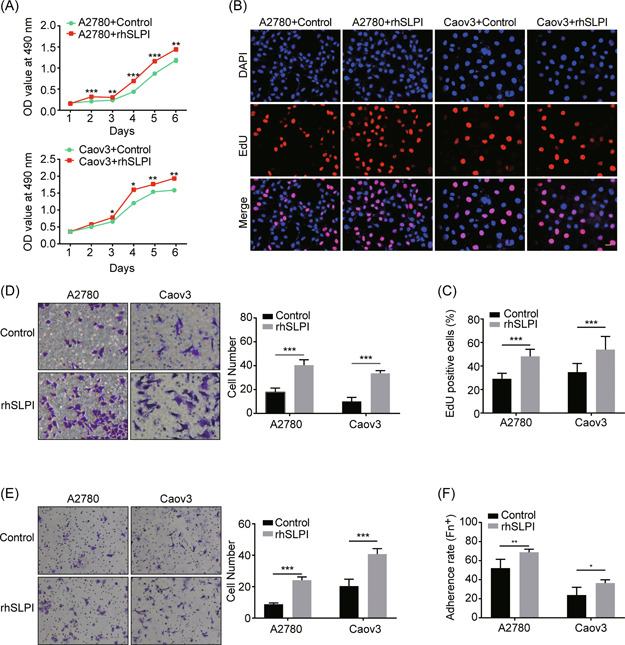
SLPI promotes tumor cell proliferation, migration, invasion, and adhesion. (A, B) Representative images of the proliferation of A2780 and Caov3 cells in the presence (+) and absence (−) of rhSLPI, determined with MTS and EdU assays (scale bar, 20 μm). (C) Quantification of EdU‐positive cells in (B). (D, E) Transwell assay results show that rhSLPI markedly enhances migration and invasion of A2780 and Caov3 cells. Quantitative analysis of migrated and invaded cells is presented in the corresponding right panel. (F) Representative images of A2780 and Caov3 cell adherence rates in the presence (+) and absence (−) of rhSLPI. *p < 0.05, **p < 0.01, and ***p < 0.001. rhSLPI, recombinant human secretory leukocyte protease inhibitor. [Color figure can be viewed at wileyonlinelibrary.com]
3.3. Exogenous SLPI promotes the activation of PI3K–AKT and its downstream signaling pathways in ovarian cancer cells
To further clarify the mechanisms underlying the tumor‐promoting effects of exogenous SLPI in ovarian cancer cells, the PI3K‐AKT pathway associated with tumor growth 40 was examined using rhSLPI in A2780 and Caov3 cells. Expression of PI3K, p‐Akt (S473), and p‐Akt (T308) was significantly increased in A2780 and Caov3 cells treated with rhSLPI, compared with the corresponding control cells (Figure 3A,B). Considering the strong association between GSK‐3β and AKT pathways, 41 we further evaluated GSK‐3β and p‐GSK‐3β expression in A2780 and Caov3 cells (Figure 3A,B). Our data showed a significant increase in the p‐GSK‐3β level after rhSLPI treatment, suggesting that CAF‐secreted SLPI exerts substantial effects on the PI3K–AKT–GSK‐3β signaling pathway in tumor cells. Previous studies have shown that SLPI regulates NF‐κB signaling in tumor cells, 42 one of the major downstream activation pathways of Akt. 41 Accordingly, we further examined the effects of SLPI on critical mediators of the NF‐κB signaling pathway. Levels of p‐NF‐κB and p‐IKKα/β were markedly elevated in rhSLPI‐treated A2780 and Caov3 cells (Figure 3C,D), indicative of rhSLPI‐mediated activation of the NF‐κB pathway under tumor conditions. Previous experiments have additionally shown that SLPI could activate ERK signaling. 29 Consistent with this finding, phosphorylation of MAPK protein was markedly increased after treatment with rhSLPI in A2780 and Caov3 cells in our experiments (Figure 3E,F). These findings collectively indicate that CAF‐derived SLPI promotes ovarian cancer progression through regulatory effects on multiple signaling pathways in tumor cells.
Figure 3.
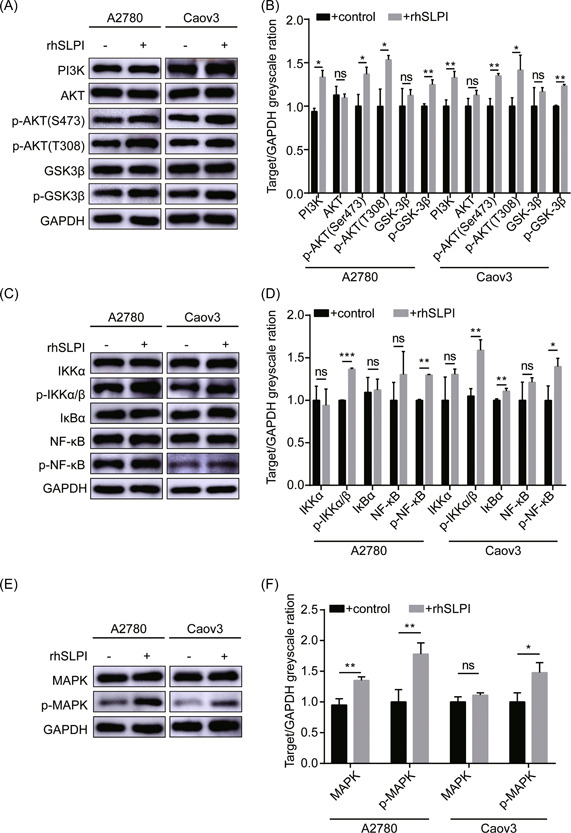
SLPI protein regulates multiple signaling pathways in tumor cells. (A, B) Western blot shows activation of the PI3K–AKT–GSK‐3β pathway in A2780 and Caov3 cells in the presence (+) of rhSLPI. (C, D) Western blot analysis of members of the NF‐κB signaling pathway, including IKKα, p‐IKKα/β, IκBα, NF‐κB, and p‐NF‐κB. (E, F) Western blot analysis of MAPK and p‐MAPK protein levels in A2780 and Caov3 cells with and without rhSLPI. The band intensity was assessed with ImageJ. *p < 0.05, **p < 0.01, and ***p < 0.001. NF‐κB, nuclear factor κB; SLPI, secretory leukocyte protease inhibitor. [Color figure can be viewed at wileyonlinelibrary.com]
3.4. SLPI protein secreted from CAFs is encapsulated in EVs and delivered to tumor cells
EVs are composed of various functional biomolecules, including membrane proteins, intracellular proteins, RNA, and DNA, that are horizontally transferred to recipient cells and participate in intercellular communication. 16 To further ascertain whether EVs mediate the delivery of CAF‐derived SLPI protein to cancer cells, we isolated EVs from FAP+CAF‐CM (FAP+CAF‐EVs) and α‐SMA+CAF‐CM (α‐SMA+CAF‐EVs) via differential ultracentrifugation. TEM revealed that isolated EVs had a double‐layer membrane structure and exhibits typical cup‐shaped morphology (Figure 4A). NTA analysis disclosed a mean vesicle size of 30–150 nm as a typical feature of EVs (Figure 4B). On western blots, the EV markers CD63, TSG101, and HSP70 proteins were positively expressed in these vesicles while the cis‐Golgi compartment‐specific marker GM130 was absent (Figure 4C). Next, to determine whether FAP+CAF and α‐SMA+CAF‐derived EVs are internalized by ovarian cancer cells, EVs were labeled with PKH26 and cocultured with cancer cells for 12 h. PKH26‐labeled EVs were detected in cancer cells using immunofluorescence, indicative of internalization (Figure 4D,E). Notably, SLPI mRNA and protein were highly expressed in FAP+CAF‐EVs but not α‐SMA+CAF‐EVs, consistent with the expression patterns described above (Figure 4F,G). Based on these findings, we propose that CAF‐secreted SLPI protein is encapsulated in EVs and transferred to tumor cells, consequently activating multiple pathways to promote tumor development.
Figure 4.
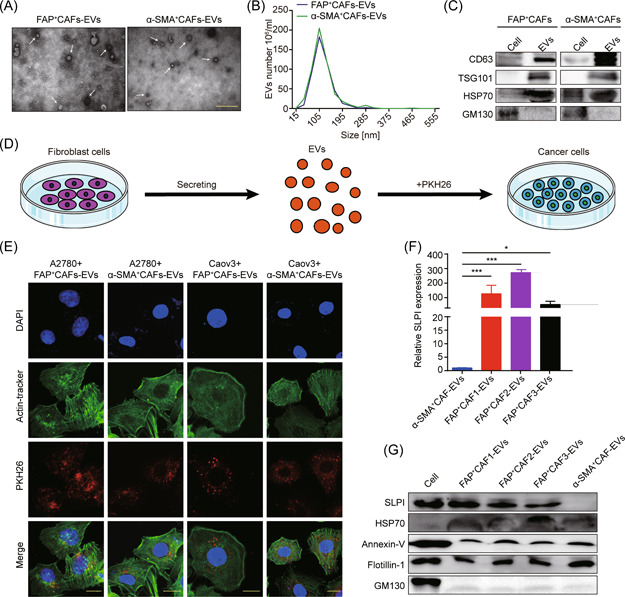
CAF‐secreted SLPI protein is encapsulated in EVs and transferred to tumor cells. (A) Representative TEM micrographs of EVs derived from FAP+CAFs and α‐SMA+CAFs. The white arrowhead signifies EVs (scale bar, 500 nm). (B) NTA analysis of EVs derived from FAP+CAFs and α‐SMA+CAFs. (C) Western blot analysis of CD63, TSG101, HSP70, and GM130 protein levels. (D, E) Internalization of PKH26‐labeled EVs via A2780 and Caov3 cells detected via immunofluorescence (scale bar, 20 μm). (F, G) Quantitative real‐time polymerase chain reaction and western blot results indicate the upregulation of SLPI in FAP+CAFs‐EVs. *p < 0.05 and ***p < 0.001. CAF, cancer‐associated fibroblast; EV, extracellular vesicle; NTA, nanoparticle tracking analysis; SLPI, secretory leukocyte protease inhibitor. [Color figure can be viewed at wileyonlinelibrary.com]
3.5. The SLPI level in EVs in plasma is correlated with tumor stage of ovarian cancer patients
To further elucidate SLPI expression patterns of fibroblasts in ovarian cancer, we examined SLPI levels in human ovarian NFs and primary ovarian CAFs isolated from patient samples using ELISA. CAF marker proteins, such as α‐SMA, vimentin, and FAP, were markedly upregulated, along with SLPI, in CAFs compared to NFs (Figure 5A–D). Surprisingly, CAF1 displaying the highest FAP expression secreted more SLPI protein than other fibroblasts (Figure 5D). These results suggest that SLPI protein is specifically overexpressed in FAPhigh α‐SMAlow CAFs, consistent with the above findings.
Figure 5.
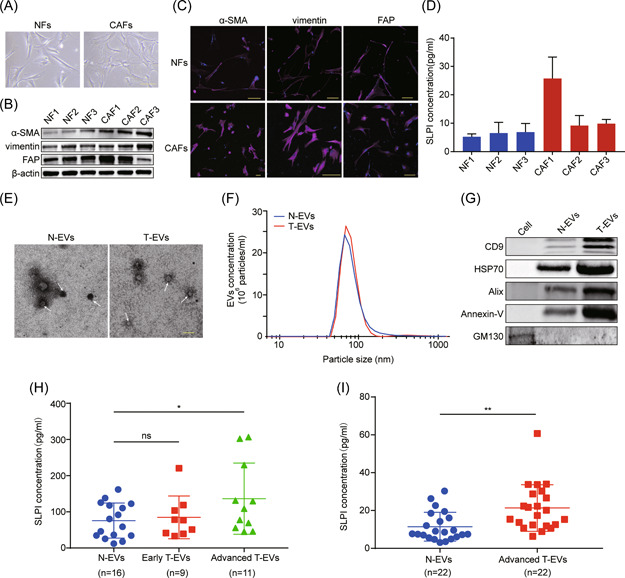
SLPI protein is overexpressed in CAFs and enriched in plasma EVs of ovarian cancer patients. (A) Representative cell morphology images of fibroblasts isolated from human normal ovary and ovarian cancer tissues (scale bar, 100 μm). (B, C) The expression of α‐SMA, vimentin, and FAP was determined via western blot and immunofluorescence staining (scale bar, 100 μm). (D) Enzyme‐linked immunosorbent assay (ELISA) analysis of SLPI expression in CM of fibroblasts. (E) Representative TEM images of EVs isolated from plasma of ovarian cancer patients and healthy controls. The white arrowhead signifies EVs (scale bar, 200nm). (F) NanoSight particle tracking analysis of size distribution and number of EVs from plasma of ovarian cancer patients and healthy women. (G) Western blot results show the increased CD9, HSP70, Alix, and Annexin‐V and decreased GM130 levels in isolated EVs. (H) ELISA analysis of SLPI protein levels in plasma EVs from 16 healthy women, 9 early ovarian cancer, and 11 advanced ovarian cancer patients. (I) ELISA results show the elevated expression of SLPI in plasma EVs of patients with advanced ovarian cancer. *p < 0.05; **p < 0.01. CAF, cancer‐associated fibroblast; EV, extracellular vesicle; SLPI, secretory leukocyte protease inhibitor. [Color figure can be viewed at wileyonlinelibrary.com]
Additionally, we analyzed SLPI protein expression within circulating EVs in plasma from HGSOC patients and healthy women. The double‐layer membrane structure and size of EVs were confirmed via TEM and NanoSight analyses (Figure 5E,F). EV markers, such as CD9, HSP70, Alix, and Annexin‐V proteins, were upregulated as expected, along with the absence of GM130 expression, further verifying the enrichment of EVs (Figure 5G). Next, EVs of plasma were purified via differential ultracentrifugation and SLPI protein expression in plasma EVs was examined via ELISA. Compared to the control group, SLPI expression was significantly higher in plasma EVs of ovarian cancer patients, especially those with advanced‐stage disease (Figure 5H). We additionally collected plasma samples of 22 healthy controls and advanced cancer patients and isolated plasma EVs for detection of SLPI expression. The results consistently showed high expression of SLPI in plasma EVs of patients with advanced cancer (Figure 5I), suggesting that the SLPI protein level was associated with poor outcomes and may thus effectively serve as a prognostic biomarker for ovarian cancer.
4. DISCUSSION
The tumor microenvironment comprises heterogeneous numbers and types of cells. CAFs constitute the major stromal cell type in the tumor microenvironment and are strongly associated with tumor initiation, invasion, metastasis, and drug resistance. 8 However, the potential roles of CAFs and underlying mechanisms in ovarian cancer are not well understood at present. In this study, we demonstrated specific upregulation of SLPI in the FAPhighα‐SMAlowCAF subtype in ovarian cancer. Notably, CAF‐secreted SLPI protein could be encapsulated in EVs and transferred to tumor cells, facilitating tumor progression through regulatory effects on multiple signaling mechanisms, including PI3K–AKT–GSK‐3β, NF‐κB, and MAPK pathways (Figure 6).
Figure 6.
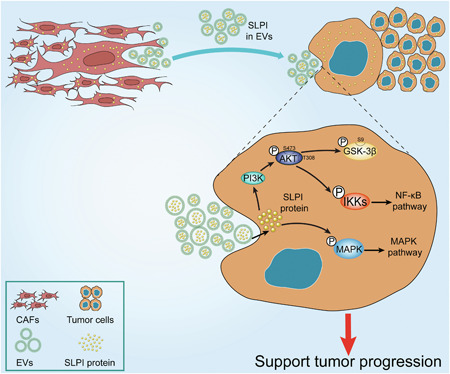
A proposed model illustrating the role of CAF‐derived SLPI protein encapsulated in EVs in regulating tumor progression in ovarian cancer cells. CAF, cancer‐associated fibroblast; EV, extracellular vesicle; SLPI, secretory leukocyte protease inhibitor. [Color figure can be viewed at wileyonlinelibrary.com]
SLPI, an 11.7 kDa secreted WAP, is upregulated in various cancer types (ovarian, breast, lung, and gastric cancer) and implicated in malignant phenotypes of tumors. 28 , 38 , 39 , 43 Previous studies indicated SLPI‐bound progranulin (Prgn) in ovarian cancer cells. SLPI is reported to protect Prgn from elastase‐mediated degradation, enhance cyclin D1 levels, and stimulate cell proliferation. 27 , 28 In addition, SLPI physically interacts with retinoblastoma tumor suppressor protein (Rb), facilitating the release of FoxM1 from the Rb–FoxM1 complex, and activating downstream target genes that promote breast cancer metastasis. 38 Conversely, other studies have reached the opposite conclusion that SLPI inhibits cell growth by inducing an increase in TNF and decrease in E‐cadherin expression, leading to activation of apoptotic pathways. 44 , 45 To date, the majority of studies have focused on the role of SLPI in tumor cells, and no reports on the crosstalk between CAF‐secreted SLPI and tumor cells in the tumor microenvironment are available in the literature. Moreover, no data have been published on the expression and function of CAF‐derived SLPI in ovarian cancer. Our study showed that SLPI was overexpressed in CAFs relative to NFs. Furthermore, stromal SLPI modulated the malignant phenotypes of cancer cells through regulation of various signaling pathways. PI3K/AKT is an important signal transduction pathway associated with cell proliferation and apoptosis. 40 Mechanistically, exogenous SLPI protein promoted the activation of the PI3K/AKT pathway, stimulating downstream molecules, including GSK‐3β and major mediators of NF‐κB signaling. Other studies have similarly demonstrated that SLPI promotes paclitaxel resistance in tumor cells via activation of the ERK pathway. 29 Consistent with this finding, exogenous SLPI protein induced an increase in p‐MAPK in our experiments, indicating activation of MAPK signaling. We additionally explored the role of CAF‐derived SLPI in tumorigenesis in vivo but failed to observe any significant effects (data not shown). Further studies are therefore required to validate these observations.
Increasing evidence suggests that CAFs are heterogeneous populations that vary in terms of molecular, genomic, and functional contexts. 46 Recently, four CAF subpopulations (CAF‐S1–S4) have been described in HGSOC, each with different molecular characteristics and functions. Mesenchymal HGSOC has a high content of CAF‐S1 fibroblasts, which promote tumor immunosuppression via enhancement of attraction, survival, and differentiation of CD25+FOXP3+ T lymphocytes. 47 Moreover, with the development of single‐cell RNA sequencing technology, accumulating CAF subpopulations have been identified, further indicating diversity within the fibroblast population. In breast cancer, Bartoschek et al. 48 identified four spatially and functionally distinct CAF subsets (vCAFs, mCAFs, cCAFs, and dCAFs), which were associated with vascular development, ECM signature, cell cycle, and cell differentiation, respectively. Another recent report revealed three distinct CAF subpopulations (myCAFs, iCAFs, and apCAFs) in pancreatic ductal adenocarcinoma, among which apCAFs activated CD4+T cells and decreased the CD8+T cell:Treg ratio, leading to immune suppression in the tumor microenvironment. Interestingly, SLPI was specifically overexpressed in apCAFs, supporting the association of SLPI+CAFs with tumor immune regulation. 49 Furthermore, FAP+CAFs are reported to promote tumor immunosuppression via activating STAT3‐CCL2 signaling in the tumor microenvironment. 50 In the present study, SLPI was significantly upregulated in FAP+CAFs expressing high levels of FAP. In view of these observations, we proposed that the SLPI+FAP+CAF subtype is tightly associated with immune regulation in ovarian cancer. Further experimental validation of these results is in progress.
EVs encapsulated within a lipid bilayer comprise multiple bioactive molecules and are released from several cell types. Accumulating evidence suggests that communication between cancer cells and stroma is facilitated by EVs. 51 , 52 Moreover, recent studies have demonstrated that proteins in tumor cell‐derived EVs function in the promotion of tumorigenesis. In colorectal cancer, tumor cells release integrin beta‐like 1 (ITGBL1)‐rich EVs to promote distal metastatic tumor growth through the fibroblast‐niche formation. 53 Furthermore, tumor cell‐derived EV‐encapsulated MET oncoprotein promotes the malignant phenotype of melanoma. 54 Other than tumor cell‐derived EVs carrying proteins, proteins from CAF‐derived EVs also facilitate tumor development. For instance, CAF‐derived EVs carrying Snail1 and annexin A6 proteins enhance EMT and drug resistance, respectively. 21 , 22 Consistently, Li et al. 55 (2017) reported that compared to normal samples, TGF‐β levels were significantly increased in CAF‐derived exosomes from ovarian cancer patients, promoting migration, invasion, and EMT in tumor cells via activation of SMAD signaling. However, to our knowledge, no studies have focused on the role of SLPI protein from CAF‐derived EVs in ovarian cancer. Here, we showed for the first time that CAF‐secreted SLPI protein was encapsulated in EVs and transferred to ovarian cancer cells. Importantly, expression of SLPI protein in circulating plasma CAF‐derived EVs was strongly associated with tumor stage, suggesting a key role in tumor development and supporting the utility of SLPI as a candidate prognostic marker of ovarian cancer.
In summary, our findings provide preliminary evidence that CAF‐derived EVs encapsulating SLPI protein is implicated in the occurrence and progression of ovarian cancer, which requires validation in further studies. Moreover, the mechanisms underlying the stimulatory effects of CAF‐secreted SLPI on target ovarian cancer cells require comprehensive investigation.
AUTHOR CONTRIBUTIONS
Chunyu Zhang and Luyao Sun designed this study. Luyao Sun, Mengyuan Yin, Junni Wei, and He Zhang performed the research. Miaola Ke and Xin Wang assisted in tissue and blood samplings and clinical information collection. Lu Xu and Xing Tian analyzed the data. Luyao Sun and Fei Wang cowrote the manuscript. Chunyu Zhang and Songbin Fu helped revision of the manuscript, funding acquisition, and project administration. All authors have read and approved the final manuscript
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Supplementary information.
ACKNOWLEDGMENTS
The authors would like to thank Xin‐Yuan Guan (University of Hong Kong Medical Center) for his great assistance with the cell lines. This study was supported by the Science and Technology Program of Guangzhou, China (No. 2060206), the National Natural Science Foundation of China (No. 81672572), and the Fundamental Research Funds for the Central Universities of SCUT (No. 2019PY25). The authors thank all the members listed in the article.
Sun L, Ke M, Wang X, et al. FAPhigh α‐SMAlow cancer‐associated fibroblast‐derived SLPI protein encapsulated in extracellular vesicles promotes ovarian cancer development via activation of PI3K/AKT and downstream signaling pathways. Molecular Carcinogenesis. 2022;61:910‐923. 10.1002/mc.23445
Luyao Sun and Miaola Ke contributed equally to this study.
DATA AVAILABILITY STATEMENT
Please contact the corresponding author for all data requests.
REFERENCES
- 1. Ebell MH, Culp MB, Radke TJ. A systematic review of symptoms for the diagnosis of ovarian cancer. Am J Prev Med. 2016;50(3):384‐394. [DOI] [PubMed] [Google Scholar]
- 2. Lheureux S, Braunstein M, Oza AM. Epithelial ovarian cancer: evolution of management in the era of precision medicine. CA Cancer J Clin. 2019;69(4):280‐304. [DOI] [PubMed] [Google Scholar]
- 3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7‐30. [DOI] [PubMed] [Google Scholar]
- 4. Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423‐1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pietras K, Ostman A. Hallmarks of cancer: interactions with the tumor stroma. Exp Cell Res. 2010;316(8):1324‐1331. [DOI] [PubMed] [Google Scholar]
- 6. Turley SJ, Cremasco V, Astarita JL. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat Rev Immunol. 2015;15(11):669‐682. [DOI] [PubMed] [Google Scholar]
- 7. Fane M, Weeraratna AT. How the ageing microenvironment influences tumour progression. Nat Rev Cancer. 2020;20(2):89‐106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sahai E, Astsaturov I, Cukierman E, et al. A framework for advancing our understanding of cancer‐associated fibroblasts. Nat Rev Cancer. 2020;20(3):174‐186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yue H, Li W, Chen R, Wang J, Lu X, Li J. Stromal POSTN induced by TGF‐β1 facilitates the migration and invasion of ovarian cancer. Gynecol Oncol. 2021;160(2):530‐538. [DOI] [PubMed] [Google Scholar]
- 10. Wen S, Hou Y, Fu L, et al. Cancer‐associated fibroblast (CAF)‐derived IL32 promotes breast cancer cell invasion and metastasis via integrin β3‐p38 MAPK signalling. Cancer Lett. 2019;442:320‐332. [DOI] [PubMed] [Google Scholar]
- 11. Feig C, Jones JO, Kraman M, et al. Targeting CXCL12 from FAP‐expressing carcinoma‐associated fibroblasts synergizes with anti‐PD‐L1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci USA. 2013;110(50):20212‐20217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu JL, Wang W, Lan XL, et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial‐mesenchymal transition in colorectal cancer. Mol Cancer. 2019;18(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. György B, Szabó TG, Pásztói M, et al. Membrane vesicles, current state‐of‐the‐art: emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68(16):2667‐2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255‐289. [DOI] [PubMed] [Google Scholar]
- 15. Tkach M, Théry C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164(6):1226‐1232. [DOI] [PubMed] [Google Scholar]
- 16. Fujita Y, Yoshioka Y, Ochiya T. Extracellular vesicle transfer of cancer pathogenic components. Cancer Sci. 2016;107(4):385‐390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kosaka N, Yoshioka Y, Fujita Y, Ochiya T. Versatile roles of extracellular vesicles in cancer. J Clin Invest. 2016;126(4):1163‐1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo H, Ha C, Dong H, Yang Z, Ma Y, Ding Y. Cancer‐associated fibroblast‐derived exosomal microRNA‐98‐5p promotes cisplatin resistance in ovarian cancer by targeting CDKN1A. Cancer Cell Int. 2019;19:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang HW, Shi Y, Liu JB, et al. Cancer‐associated fibroblast‐derived exosomal microRNA‐24‐3p enhances colon cancer cell resistance to MTX by down‐regulating CDX2/HEPH axis. J Cell Mol Med. 2021;25(8):3699‐3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Richards KE, Zeleniak AE, Fishel ML, Wu J, Littlepage LE, Hill R. Cancer‐associated fibroblast exosomes regulate survival and proliferation of pancreatic cancer cells. Oncogene. 2017;36(13):1770‐1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. You J, Li M, Cao LM, et al. Snail1‐dependent cancer‐associated fibroblasts induce epithelial‐mesenchymal transition in lung cancer cells via exosomes. QJM. 2019;112(8):581‐590. [DOI] [PubMed] [Google Scholar]
- 22. Uchihara T, Miyake K, Yonemura A, et al. Extracellular vesicles from cancer‐associated fibroblasts containing Annexin A6 induces FAK‐YAP activation by stabilizing β1 Integrin, enhancing drug resistance. Cancer Res. 2020;80(16):3222‐3235. [DOI] [PubMed] [Google Scholar]
- 23. Chen Y, Zeng C, Zhan Y, Wang H, Jiang X, Li W. Aberrant low expression of p85α in stromal fibroblasts promotes breast cancer cell metastasis through exosome‐mediated paracrine Wnt10b. Oncogene. 2017;36(33):4692‐4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nukiwa T, Suzuki T, Fukuhara T, Kikuchi T. Secretory leukocyte peptidase inhibitor and lung cancer. Cancer Sci. 2008;99(5):849‐855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bouchard D, Morisset D, Bourbonnais Y, Tremblay GM. Proteins with whey‐acidic‐protein motifs and cancer. Lancet Oncol. 2006;7(2):167‐174. [DOI] [PubMed] [Google Scholar]
- 26. Abe T, Kobayashi N, Yoshimura K, et al. Expression of the secretory leukoprotease inhibitor gene in epithelial cells. J Clin Invest. 1991;87(6):2207‐2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Simpkins FA, Devoogdt NM, Rasool N, et al. The alarm anti‐protease, secretory leukocyte protease inhibitor, is a proliferation and survival factor for ovarian cancer cells. Carcinogenesis. 2008;29(3):466‐472. [DOI] [PubMed] [Google Scholar]
- 28. Devoogdt N, Rasool N, Hoskins E, Simpkins F, Tchabo N, Kohn EC. Overexpression of protease inhibitor‐dead secretory leukocyte protease inhibitor causes more aggressive ovarian cancer in vitro and in vivo. Cancer Sci. 2009;100(3):434‐440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rasool N, LaRochelle W, Zhong H, Ara G, Cohen J, Kohn EC. Secretory leukocyte protease inhibitor antagonizes paclitaxel in ovarian cancer cells. Clin Cancer Res. 2010;16(2):600‐609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhu J, Yu Y, Meng X, et al. De novo‐generated small palindromes are characteristic of amplicon boundary junction of double minutes. Int J Cancer. 2013;133(4):797‐806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bao Y, Liu J, You J, et al. Met promotes the formation of double minute chromosomes induced by Sei‐1 in NIH‐3T3 murine fibroblasts. Oncotarget. 2016;7(35):56664‐56675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tian X, Liu C, Wang X, et al. Sei‐1 promotes double minute chromosomes formation through activation of the PI3K/Akt/BRCA1‐Abraxas pathway and induces double‐strand breaks in NIH‐3T3 fibroblasts. Cell Death Dis. 2018;9(3):341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. In: Bonifacino JS, ed. Current Protocols in Cell Biology. Chapter 3: Unit 3.22.2600. John Wiley; 2006. [DOI] [PubMed] [Google Scholar]
- 34. Hoshino A, Kim HS, Bojmar L, et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell. 2020;182(4):1044‐1061, e1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koche RP, Rodriguez‐Fos E, Helmsauer K, et al. Extrachromosomal circular DNA drives oncogenic genome remodeling in neuroblastoma. Nat Genet. 2020;52(1):29‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu S, Turner KM, Nguyen N, et al. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature. 2019;575(7784):699‐703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Turner KM, Deshpande V, Beyter D, et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature. 2017;543(7643):122‐125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kozin SV, Maimon N, Wang R, et al. Secretory leukocyte protease inhibitor (SLPI) as a potential target for inhibiting metastasis of triple‐negative breast cancers. Oncotarget. 2017;8(65):108292‐108302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Du XY, Liu X, Wang ZJ, Wang YY. SLPI promotes the gastric cancer growth and metastasis by regulating the expression of P53, Bcl‐2 and Caspase‐8. Eur Rev Med Pharmacol Sci. 2017;21(7):1495‐1501. [PubMed] [Google Scholar]
- 40. Noorolyai S, Shajari N, Baghbani E, Sadreddini S, Baradaran B. The relation between PI3K/AKT signalling pathway and cancer. Gene. 2019;698:120‐128. [DOI] [PubMed] [Google Scholar]
- 41. Fresno Vara JA, Casado E, de Castro J, Cejas P, Belda‐Iniesta C, González‐Barón M. PI3K/Akt signalling pathway and cancer. Cancer Treat Rev. 2004;30(2):193‐204. [DOI] [PubMed] [Google Scholar]
- 42. Jan Treda C, Fukuhara T, Suzuki T, et al. Secretory leukocyte protease inhibitor modulates urethane‐induced lung carcinogenesis. Carcinogenesis. 2014;35(4):896‐904. [DOI] [PubMed] [Google Scholar]
- 43. Ameshima S, Ishizaki T, Demura Y, Imamura Y, Miyamori I, Mitsuhashi H. Increased secretory leukoprotease inhibitor in patients with nonsmall cell lung carcinoma. Cancer. 2000;89(7):1448‐1456. [DOI] [PubMed] [Google Scholar]
- 44. Nakamura K, Takamoto N, Hongo A, et al. Secretory leukoprotease inhibitor inhibits cell growth through apoptotic pathway on ovarian cancer. Oncol Rep. 2008;19(5):1085‐1091. [PubMed] [Google Scholar]
- 45. Rosso M, Lapyckyj L, Amiano N, et al. Secretory leukocyte protease inhibitor (SLPI) expression downregulates E‐cadherin, induces β‐catenin re‐localisation and triggers apoptosis‐related events in breast cancer cells. Biol Cell. 2014;106(9):308‐322. [DOI] [PubMed] [Google Scholar]
- 46. Louault K, Li RR, DeClerck YA. Cancer‐associated fibroblasts: understanding their heterogeneity. Cancers (Basel). 2020;12:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Givel AM, Kieffer Y, Scholer‐Dahirel A, et al. miR200‐regulated CXCL12β promotes fibroblast heterogeneity and immunosuppression in ovarian cancers. Nat Commun. 2018;9(1):1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bartoschek M, Oskolkov N, Bocci M, et al. Spatially and functionally distinct subclasses of breast cancer‐associated fibroblasts revealed by single cell RNA sequencing. Nat Commun. 2018;9(1):5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Elyada E, Bolisetty M, Laise P, et al. Cross‐species single‐cell analysis of pancreatic ductal adenocarcinoma reveals antigen‐presenting cancer‐associated fibroblasts. Cancer Discov. 2019;9(8):1102‐1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yang X, Lin Y, Shi Y, et al. FAP promotes immunosuppression by cancer‐associated fibroblasts in the tumor microenvironment via STAT3‐CCL2 signaling. Cancer Res. 2016;76(14):4124‐4135. [DOI] [PubMed] [Google Scholar]
- 51. Shoucair I, Weber Mello F, Jabalee J, Maleki S, Garnis C. The role of Cancer‐Associated fibroblasts and extracellular vesicles in tumorigenesis. Int J Mol Sci. 2020;21:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang X, Li Y, Zou L, Zhu Z. Role of exosomes in crosstalk between cancer‐associated fibroblasts and cancer cells. Front Oncol. 2019;9:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ji Q, Zhou L, Sui H, et al. Primary tumors release ITGBL1‐rich extracellular vesicles to promote distal metastatic tumor growth through fibroblast‐niche formation. Nat Commun. 2020;11(1):1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Peinado H, Alečković M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro‐metastatic phenotype through MET. Nat Med. 2012;18(6):883‐891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li W, Zhang X, Wang J, et al. TGFβ1 in fibroblasts‐derived exosomes promotes epithelial‐mesenchymal transition of ovarian cancer cells. Oncotarget. 2017;8(56):96035‐96047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information.
Data Availability Statement
Please contact the corresponding author for all data requests.


