Abstract
The RimM protein in Escherichia coli is associated with free 30S ribosomal subunits but not with 70S ribosomes and is important for efficient maturation of the 30S subunits. A mutant lacking RimM shows a sevenfold-reduced growth rate and a reduced translational efficiency. Here we show that a double alanine-for-tyrosine substitution in RimM prevents it from associating with the 30S subunits and reduces the growth rate of E. coli approximately threefold. Several faster-growing derivatives of the rimM amino acid substitution mutant were found that contain suppressor mutations which increased the amount of the RimM protein by two different mechanisms. Most of the suppressor mutations destabilized a secondary structure in the rimM mRNA, which previously was shown to decrease the synthesis of RimM by preventing the access of the ribosomes to the translation initiation region on the rimM mRNA. Three other independently isolated suppressor mutations created a fusion between rpsP, encoding the ribosomal protein S16, and rimM on the chromosome as a result of mutations in the rpsP stop codon preceding rimM. A severalfold-higher amount of the produced hybrid S16-RimM protein in the suppressor strains than of the native-sized RimM in the original substitution mutant seems to explain the suppression. The S16-RimM protein but not any native-size ribosomal protein S16 was found both in free 30S ribosomal subunits and in translationally active 70S ribosomes of the suppressor strains. This suggests that the hybrid protein can substitute for S16, which is an essential protein probably because of its role in ribosome assembly. Thus, the S16-RimM hybrid protein seems capable of carrying out the important functions that native S16 and RimM have in ribosome biogenesis.
The RimM protein is encoded by the second gene of the trmD operon, which also encodes the ribosomal proteins (r-proteins) S16 and L19 and the tRNA(m1G37)methyltransferase (TrmD) (5). The RimM protein is important for maturation of the 30S ribosomal subunits (4), and it is associated with free 30S subunits but not with those in the 70S ribosomes (3). The amount of RimM in the cell is approximately 12-fold smaller than that of r-proteins (16). These findings suggest that RimM interacts transiently with the 30S ribosomal subunits during the maturation process. Mutants lacking the RimM protein show a sevenfold-decreased growth rate and a reduced translational efficiency (3), probably due to incorrectly matured 30S subunits. The slow growth and translational deficiency of a ΔrimM mutant can be partially suppressed by alterations in the C-terminal part of r-protein S13, which binds 16S rRNA (3). Increased expression of the ribosome binding factor RbfA, which also is important for the maturation of the 30S ribosomal subunits, also suppresses the translational deficiency of a ΔrimM mutant (4). The r-protein S16 is essential for the viability of Escherichia coli (12) and plays an important role in the assembly of the 30S ribosomal subunits (7). However, in vitro-assembled 30S subunits lacking S16 show functional properties similar to those of 30S subunits that contain S16, suggesting that S16 is not directly involved in translation (7). Further, S16 also binds to cruciform DNA and shows DNA-nicking activity (2, 11). The observation of 12-fold-higher levels of S16 than of RimM is explained by translational level regulation (6, 16) through a large mRNA hairpin structure, which involves base paring between the translation initiation region of rimM and sequences approximately 100 nucleotides downstream from the rimM start codon (17, 19). This mRNA hairpin structure prevents access of the ribosomes to the translation initiation region and thereby reduces the translational initiation of rimM.
Here we have found that a RimM protein that contains alanine substitutions for two adjacent conserved tyrosines has a dramatically reduced ability to associate with the 30S ribosomal subunits. Suppressor mutations that increased the growth rate of this rimM mutant either increased the synthesis of the mutant RimM protein up to 10-fold or fused the rpsP gene to rimM. The amount of the mutant RimM protein associated with the 30S ribosomal subunits in the RimM-overproducing strains was similar to that of RimM in wild-type strains. The hybrid S16-RimM protein in the other suppressor strains was found both in free 30S ribosomal subunits and in translationally active 70S ribosomes. Since no native-sized S16 was detected in the ribosomes, this suggests that the hybrid protein can substitute for both RimM and S16.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used are listed in Table 1.
TABLE 1.
Bacterial strains
| Strain | Genotype (mutation[s]) | Origin or referencea |
|---|---|---|
| JML002 | MW136 rimM125 (AGC to AAC in codon 2) | |
| JML020 | MW136 rpsP877 (TAA to CAA) | |
| JML021 | MW136 rpsP878 (TAA to GAA) | |
| JML022 | MW136 rimM128 (AGC to AAC in codon 2) | |
| JML023 | MW136 rimM129 (ATG to ATT in codon 1) | |
| JML024 | MW136 rimM130 (CTC to CTG in codon 29) | |
| JML027 | MW136 rpsP879 (TAA to TTA) | |
| JML028 | MW136 rimM132 (CTC to CTT in codon 29) | |
| JML029 | MW136 rimM133 (ATG to ATT in codon 1) | |
| MW100 | Hfr P4X | 18 |
| MW136 | Hfr P4X rimM120 (TAC to GCT in codon 106 and TAC to GCG in codon 107) | Lövgren et al.b |
| MW144 | MW136 rimM124 (GGG to GGA in codon 27) | |
| PW109 | Hfr P4X ΔrimM102 yfiB::nptI sdr-43 = DUP(′infB to yhbM′) | 4 |
Unless otherwise noted, the origin was this study.
Unpublished results.
Isolation of suppressor mutations.
To isolate faster-growing derivatives of the rimM120 mutant MW136 (Y106A plus Y107A), several independent colonies were grown overnight in Luria broth (1). An aliquot of each overnight culture was streaked out on rich medium plates, in order to detect revertants, simultaneously with its reinoculation into fresh medium. This process was repeated until revertants appeared on the plates. Only one revertant was saved from each original culture for further analyses.
PCR amplification of chromosomal DNA and DNA sequencing.
Regions of the E. coli chromosome were amplified by PCR (9, 15) from colonies resuspended in H2O using Taq DNA polymerase from Roche Diagnostics Scandinavia AB (Bromma, Sweden). Obtained fragments were purified using Quantum Prep PCR Kleen Spin Columns from Bio-Rad Laboratories (Hercules, Calif.). DNA sequencing of PCR fragments was carried out with a Thermo Sequenase II dye terminator cycle sequencing premix kit from Amersham Pharmacia Biotech (Buckinghamshire, England), using an ABI 377 XL DNA Sequencer from PE Applied Biosystems (Stockholm, Sweden).
Western blot analysis with an anti-RimM antiserum after sucrose gradient centrifugation of cellular extracts.
Extracts containing dissociated 70S ribosomes were prepared by disrupting log phase cells in a solution containing 10 mM Tris-HCl (pH 7.5), 50 mM KCl, 6 mM β-mercaptoethanol, and 1 mM MgCl2 using a French press set at 15 MPa. The S30 extracts obtained after centrifugation were fractionated by sucrose gradient centrifugation as described previously (8). Polysome extracts were prepared by freeze-thawing log phase cells in the presence of lysozyme according to the method of Ron et al. (14) and fractionated by sucrose gradient centrifugation mainly as described by Powers and Noller (13). Aliquots from the obtained fractions were analyzed spectrophotometrically at 260 nm. Selected fractions were subjected to Western blot analysis using the ECL kit from Amersham Pharmacia Biotech with antisera against the RimM protein raised in rabbits by Agri Sera AB (Vännäs, Sweden) using the RimM part of a thrombin-cleaved glutathione S-transferase–RimM hybrid protein (data not shown).
Immunoprecipitation of the r-protein S16 and the S16-RimM hybrid protein using an anti-S16 antiserum.
Strains JML020 (rimM120 rpsP877) and JML024 (rimM120 rimM130) were grown in morpholinepropanesulfonic acid minimal medium (10) containing 0.4% glucose, and at an optical density at 600 nm of 0.03; 15 ml of each culture was labeled with 325 μCi of [35S]methionine for four generations. Cells were harvested by centrifugation, washed once in a solution containing 10 mM Tris-HCl (pH 7.5), 50 mM KCl, 10 mM MgCl2, and 6 mM β-mercaptoethanol, and disrupted by sonication. 70S ribosomes and free 30S subunits were purified by sucrose gradient centrifugation, and selected fractions were pooled. The sucrose was removed using Centricon YM-3 centrifugal filter devices from Millipore Corporation (Bedford, Mass.). r-proteins were extracted using acetic acid, precipitated with acetone, solubilized in 8 M urea, and immunoprecipitated with the immunoglobulin G fraction of an anti-S16 antiserum coupled to Sepharose as described previously (18).
RESULTS
The weak association of a mutant RimM protein with the 30S ribosomal subunits can be compensated for by increased levels of the mutant protein.
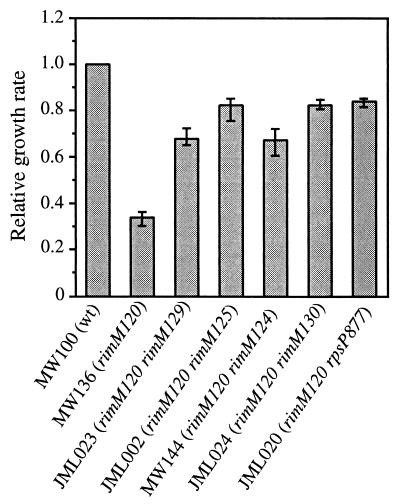
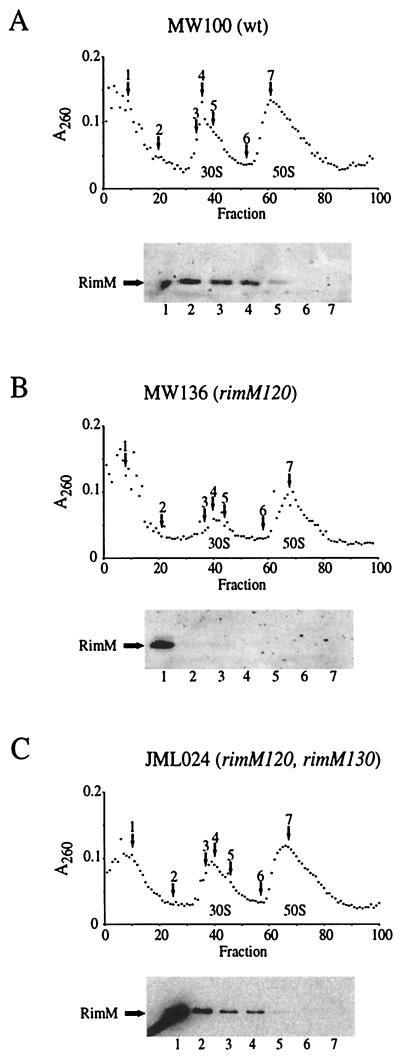
The rimM120 mutant MW136, which contains alanine substitutions in two of the most conserved positions (Y106A and Y107A) of RimM, shows a threefold-lower growth rate than a rimM+ strain (Fig. 1) and a deficiency in the maturation of the 30S subunits evidenced by a reduced processing rate of pre-16S to 16S rRNA (J. M. Lövgren, G. O. Bylund, L. A. C. Lundberg, O. P. Persson, and P. M. Wikström, unpublished results). Since the wild-type RimM protein is associated with free 30S ribosomal subunits (3), we examined whether the two amino acid substitutions in the mutant RimM protein affected its ability to associate with the 30S subunits. The ribosomal subunits in cell extracts from mutant and wild-type strains were separated by sucrose gradient centrifugation under conditions that dissociated the 70S ribosomes into 50S and 30S subunits, and different fractions were screened for the presence of the RimM protein by using an anti-RimM antiserum. The wild-type RimM protein molecules were found associated with free 30S subunits and also in the cytoplasmic fractions, whereas the mutant RimM protein was found mainly in the cytoplasmic fractions (cf. Fig. 2A and B). Thus, the reduced growth rate of the rimM120 mutant and the deficiency in the maturation of the 30S subunits correlate with an inability of the mutant RimM protein to interact with the 30S ribosomal subunits.
FIG. 1.
Relative growth rates of wild-type and different mutant strains. The strains were grown in Luria broth (1), and their growth rates were normalized to that for the wild-type strain, MW100, which had a specific growth rate k (ln2/g, where g is the mass doubling time in hours) of 1.3 to 1.6 in three independent experiments. The variation between the experiments is shown as error bars.
FIG. 2.
Cellular localization of RimM proteins in the wild type (A), rimM120 mutant (B), and rimM120 mutant overexpressing RimM (C). Cell extracts were fractionated by sucrose gradient centrifugation during conditions that dissociated the 70S ribosomes into 50S and 30S subunits. Selected fractions, indicated by arrows above the A260 curve, were screened for the presence of the RimM proteins in Western blotting experiments using a polyclonal anti-RimM antiserum. The locations of the 50S and 30S ribosomal subunits are indicated below the A260 curve.
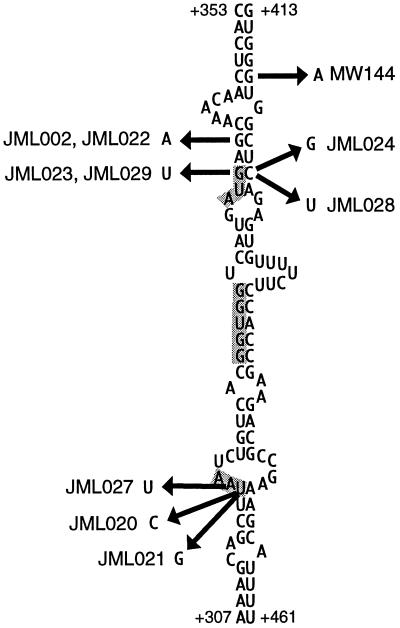
In the process of isolating second-site suppressor mutations that potentially would restore the binding of the mutant RimM protein to the 30S subunits, faster-growing derivatives of strain MW136 (rimM120) were isolated (Fig. 1) that contained mutations which were tightly linked to the rimM120 mutation, as demonstrated by the ability to transfer these mutations together with the rimM120 mutation to the wild-type strain MW100 by the phage P1 (data not shown). Several of these suppressor mutations were in the first 29 codons of rimM, some of which did not alter the amino acid sequence of RimM. All of these mutations destabilized a hairpin structure in the mRNA (Fig. 3) that previously has been shown to decrease translation initiation of rimM, probably by preventing the access of the ribosomes to the Shine-Dalgarno (SD) sequence and start codon (17, 19). Accordingly, four out of four of these mutations that were tested for their effect on the expression of rimM120 were found to increase the levels of the mutant RimM protein (Fig. 4). These results suggest that overexpression of the mutant protein compensates for a reduced function of the protein. This interpretation is supported by the finding that the amount of the mutant protein associated with the 30S ribosomal subunits is higher in the suppressor mutant JML024 (rimM120 rimM130) overexpressing RimM than in the rimM120 mutant MW136 (cf. Fig. 2C and B).
FIG. 3.
Suppressor mutations in the lower part of the mRNA secondary structure that prevents access of the ribosomes to the translational initiation region of rimM (19). The numbering is relative to the transcriptional start site of the trmD operon mRNA (6). The rpsP stop codon, SD sequence, and start codon for rimM are shaded.
FIG. 4.
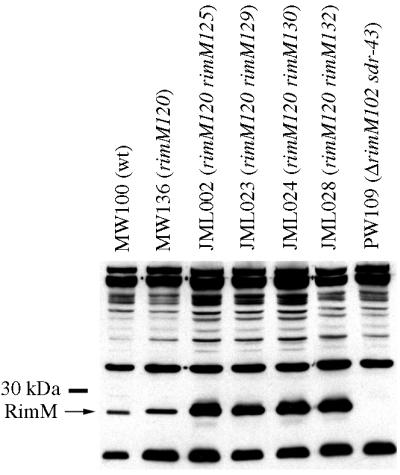
Amounts of RimM proteins in different strains. The amounts of the wild-type RimM and rimM120 mutant proteins in total cell extracts of strains MW100 (wt), MW136 (rimM120), JML002 (rimM120 rimM125), JML023 (rimM120 rimM129), JML024 (rimM120 rimM130), JML028 (rimM120 rimM132), and PW109 (ΔrimM102 sdr-43) were determined by using an anti-RimM antiserum in a Western blotting experiment.
An S16-RimM hybrid protein suppresses the slow growth of the rimM120 mutant.
Surprisingly, three of the chromosomal suppressor mutations (carried by strains JML020, JML021, and JML027) changed the stop codon of rpsP preceding rimM to different sense codons (Fig. 3). These three strains were among the fastest-growing suppressor strains isolated, having growth rates that corresponded to approximately 85% of that of a rimM+ strain (exemplified by strain JML020 in Fig. 1). Since rpsP and rimM are in the same reading frame and there are no additional stop codons between the two genes, the two genes had been fused in the three suppressor strains. As shown in Fig. 5, a hybrid S16-RimM protein of the expected size was produced in large amounts with strain JML020, as demonstrated by using an anti-RimM antiserum in Western blotting experiments. Similar results were obtained for the two other strains, JML021 and JML027, that contain mutations in the rpsP stop codon (data not shown). Also, a protein of the expected size for native RimM protein reacted with the antibodies. At this point we cannot distinguish whether this protein was the result of initiation at the rimM start codon or was a degradation product of the hybrid protein. We would like to emphasize that the amount of native-size RimM protein in the suppressor strain JML020 was not larger than that in the rimM120 mutant MW136 (Fig. 5), demonstrating that the mechanism behind the suppression was not an increased level of the native-sized RimM protein. In the total extract of strain JML020, there seemed to be degradation products of the S16-RimM hybrid protein (Fig. 5). To investigate whether the hybrid protein or any of the proposed degradation products were responsible for the observed suppression, a cellular extract of strain JML020 was fractionated by sucrose gradient centrifugation and different fractions were screened with an anti-RimM antiserum in a Western blotting experiment. The fractions corresponding to the 30S subunits contained large amounts of the S16-RimM protein but very small amounts of native-sized RimM and no detectable amounts of any degradation products of the hybrid protein (Fig. 6). Thus, these findings suggest that the hybrid S16-RimM protein is responsible for the observed suppression.
FIG. 5.
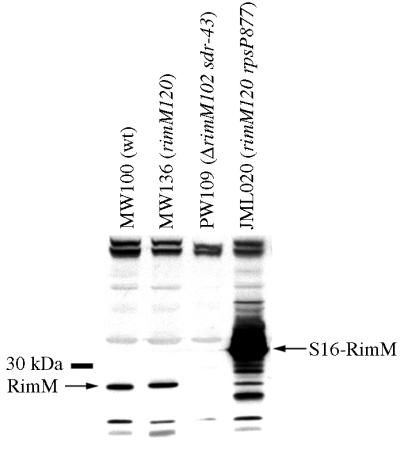
Production of an S16-RimM hybrid protein in the suppressor strain JML020 containing a mutation in the rpsP stop codon. The presence of the S16-RimM hybrid protein and native-sized RimM proteins was screened for in total cell extracts of strains MW100 (wt), MW136 (rimM120), PW109 (ΔrimM102 sdr-43), and JML020 (rimM120 rpsP877) by using an anti-RimM antiserum in a Western blotting experiment.
FIG. 6.
Cellular localization of the S16-RimM hybrid protein. A polysome extract of strain JML020 (rimM120 rpsP877) was fractionated by sucrose gradient centrifugation, and the indicated fractions were screened for the presence of the S16-RimM hybrid protein by using an anti-RimM antiserum in a Western blotting experiment. The locations of the 50S and 30S ribosomal subunits, 70S ribosomes, and polysomes are indicated below the A260 curve. The identity of native-sized RimM protein was determined by running a total extract of the wild-type strain MW100 and a molecular weight marker on the protein gel.
The S16-RimM hybrid protein replaces S16 in mature ribosomes.
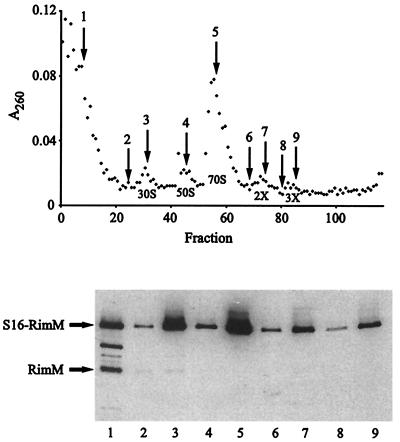
In the screening of the cell extract of strain JML020 fractionated by sucrose gradient centrifugation, the S16-RimM protein was found not only in the 30S subunits but also in 70S ribosomes and polysomes (Fig. 6). These results suggested that the S16-RimM hybrid protein had replaced the r-protein S16 in translationally active 70S ribosomes; however, it was formally possible that native-size S16 was present in the majority of the ribosomes due to proteolytic cleavage of the hybrid protein. Therefore, we labeled strains JML020 and JML024 in vivo with [35S]methionine and probed for the r-protein S16 and the S16-RimM hybrid protein by immunoprecipitation with an anti-S16 antiserum in fractions corresponding to 30S ribosomal subunits and 70S ribosomes after separation of cellular extracts by sucrose gradient centrifugation. No native-size S16 was found in the 30S subunits or in the 70S ribosomes from strain JML020 when they were immunoprecipitated with the anti-S16 antiserum; however, in agreement with the results from the Western blotting experiments with the anti-RimM antiserum, the S16-RimM hybrid protein was found in both the 30S subunits and the 70S ribosomes (Fig. 7). In the control strain JML024, native S16 was found in both the 30S subunits and the 70S ribosomes. The lower intensity of the band corresponding to S16 in the control strain JML024 than that of the S16-RimM hybrid protein in strain JML020 results from the fact that S16 contains only one methionine, whereas the hybrid protein is supposed to contain eight. Taken together, these results suggest that in addition to being able to substitute for RimM, the hybrid S16-RimM protein also can substitute for S16 in translationally active 70S ribosomes.
FIG. 7.
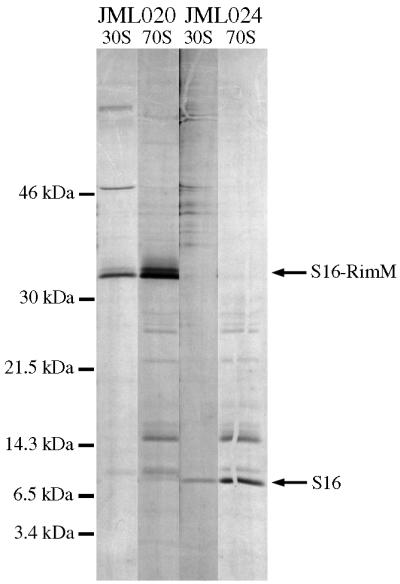
Analysis of ribosomes for the presence of the r-protein S16. Acid-soluble r-proteins were subjected to immunoprecipitation with an anti-S16 antiserum after purification of 70S ribosomes and free 30S subunits from total cell extracts of strains JML020 (rimM120 rpsP877) and JML024 (rimM120 rimM130) labeled in vivo with [35S]methionine (see Materials and Methods). The immunoprecipitated proteins were separated on a 10-to-17.5% gradient polyacrylamide gel containing sodium dodecyl sulfate.
DISCUSSION
Here we show that the RimM protein in the rimM120 mutant, in which alanines have been substituted for the tyrosines in positions 106 and 107, is only weakly associated with the 30S ribosomal subunits. The reduced amount of RimM associated with the 30S subunits most likely explains the slow growth rate of the rimM120 mutant, since RimM is important for the maturation of the 30S subunits (4). Several suppressor mutations to the rimM120 mutation increased the amount of the mutant protein, and for at least one of the suppressor strains, the amount of the mutant RimM protein associated with the 30S subunits was comparable to that of the native RimM protein in wild-type cells. Conceivably, an increased concentration of the mutant RimM protein in the cell can compensate for its weak association with the 30S ribosomal subunits.
An mRNA secondary structure that covers the translation initiation region for rimM seems to involve base pairing of the SD sequence and start codon with sequences around 100 nucleotides downstream from the start codon (19). Previously, it was demonstrated that mutations that destabilized the postulated hairpin structure increased translation initiation of rimM-lacZ fusions (17, 19). Similarly, the suppressor mutations to rimM120 that increased the amount of the mutant RimM protein decreased the calculated stability of this hairpin structure (Fig. 3). Thus, the effect of these suppressor mutations on the expression of RimM corroborates the previously proposed model in which the mRNA secondary structure prevents the access of the ribosomes to the SD sequence and start codon of rimM. Further, the suppressor mutations of strains JML023 and JML029 changed the AUG start codon of rimM to AUU, encoding isoleucine. Probably, the AUG codon just upstream from this AUU codon is used as a start codon in these two strains.
Interestingly, three of the isolated suppressor mutations changed the stop codon of rpsP (for S16) to sense codons, which resulted in the production of a hybrid S16-RimM protein. This protein, which was found in large amounts in the cell, seems to be responsible for the suppression of the slow growth of the cells, implied to be caused by the weak association of the mutant RimM protein with the 30S ribosomal subunits. The hybrid S16-RimM protein was present in large amounts both in free 30S subunits and in translationally active 70S ribosomes, probably because of the affinity of the S16 moiety of the hybrid protein for the 30S subunits, since RimM is not normally present in 70S ribosomes (3). Thus, the ability of the S16-RimM protein to substitute for RimM in ribosome maturation might depend on the ability of the S16 moiety to guide the hybrid protein to the 30S subunits. Alternatively, the suppression is dependent on RimM-mediated binding of the S16-RimM protein to the 30S subunits and is explained by the high levels in the cell of the hybrid protein, since other suppressor mutations that increased the amount of the mutant RimM protein restored the level of RimM associated with the 30S subunits to that seen in a wild-type strain (Fig. 2). We favor the latter model, since alterations in r-protein S13 suppress the slow growth of a ΔrimM mutant (3), indicating that RimM associates with the 30S subunits in the area where S13 is found, which is on the opposite side from where S16 is located in the 30S subunits (20).
E. coli cells lacking the r-protein S16 are not viable (12), probably because of the important role the S16 protein plays in the assembly of the 30S subunits (7). Thus, the presence of the S16-RimM protein in translationally active ribosomes of cells lacking native S16 protein and the ability of the hybrid protein to support fast growth of the cells suggest that it must be able to substitute for the native S16 protein in ribosome assembly. Considering that r-proteins to a large extent are embedded in the rRNA structure of the ribosome and that the fusion of RimM to S16 adds 189 extra amino acids to the 82 normally present in S16 of E. coli, it was surprising to find the S16-RimM hybrid protein in the 70S ribosomes. However, we note that in the 3-Å resolution structure of the 30S ribosomal subunit from Thermus thermophilus (20), the most COOH-terminal amino acid (E83) of those localized in the structure of S16 protrudes from the 30S subunits (Fig. 8). Assuming that the structure of the 30S subunits in E. coli is similar to that in T. thermophilus, the RimM part of the hybrid protein seems therefore to be present on the surface of the 30S subunits and might not interfere with the assembly of the 30S subunits. The in vitro assembly of the 30S subunits is highly inefficient in the absence of S16; however, once assembled, the 30S subunits lacking S16 are translationally competent (7), suggesting that S16 has no role or little role in translation as such. Thus, the addition of 189 extra amino acids to the COOH-terminal end of S16 might not have any effect on the translational capacity of the ribosomes as long as the assembly function of S16 is not affected.
FIG. 8.
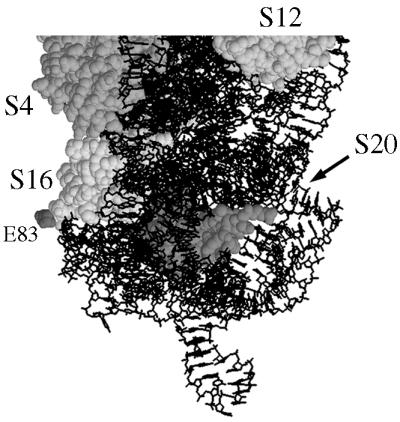
Location of the COOH-terminal end of the r-protein S16 in the 30S subunit of T. thermophilus. The structure of the 30S subunit is from reference 20 and was retrieved from the Protein Data Bank (PDB no. 1FJF). Only a part of the structure is shown, in which the 16S rRNA is presented as a stick model while the indicated r-proteins are presented as space-fill models. The T. thermophilus S16 protein is 88 residues in length, but the most COOH-terminal amino acid in the determined structure is E83.
ACKNOWLEDGMENTS
Olof P. Persson and Glenn R. Björk are acknowledged for stimulating discussions and for their helpful comments on the manuscript.
This work was supported by the Swedish Natural Science Research Council (B-BU 9911), the Carl Trygger Foundation, the Magnus Bergvall Foundation, and the Kempe Foundations.
REFERENCES
- 1.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnefoy E. The ribosomal S16 protein of Escherichia coli displaying a DNA-nicking activity binds to cruciform DNA. Eur J Biochem. 1997;247:852–859. doi: 10.1111/j.1432-1033.1997.t01-1-00852.x. [DOI] [PubMed] [Google Scholar]
- 3.Bylund G O, Persson B C, Lundberg L A C, Wikström P M. A novel ribosome-associated protein is important for efficient translation in Escherichia coli. J Bacteriol. 1997;179:4567–4574. doi: 10.1128/jb.179.14.4567-4574.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bylund G O, Wipemo L C, Lundberg L A C, Wikström P M. RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli. J Bacteriol. 1998;180:73–82. doi: 10.1128/jb.180.1.73-82.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byström A S, Hjalmarsson K J, Wikström P M, Björk G R. The nucleotide sequence of an Escherichia coli operon containing genes for the tRNA(m1G)methyltransferase, the ribosomal proteins S16 and L19 and a 21-K polypeptide. EMBO J. 1983;2:899–905. doi: 10.1002/j.1460-2075.1983.tb01519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byström A S, von Gabain A, Björk G R. Differentially expressed trmD ribosomal protein operon of Escherichia coli is transcribed as a single polycistronic mRNA species. J Mol Biol. 1989;208:575–586. doi: 10.1016/0022-2836(89)90149-6. [DOI] [PubMed] [Google Scholar]
- 7.Held W A, Nomura M. Escherichia coli 30 S ribosomal proteins uniquely required for assembly. J Biol Chem. 1975;250:3179–3184. [PubMed] [Google Scholar]
- 8.Lindahl L. Intermediates and time kinetics of the in vivo assembly of Escherichia coli ribosomes. J Mol Biol. 1975;92:15–37. doi: 10.1016/0022-2836(75)90089-3. [DOI] [PubMed] [Google Scholar]
- 9.Mullis K B, Faloona F A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 10.Neidhardt F C, Bloch P L, Smith D F. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oberto J, Bonnefoy E, Mouray E, Pellegrini O, Wikström P M, Rouvière-Yaniv J. The Escherichia coli ribosomal protein S16 is an endonuclease. Mol Microbiol. 1996;19:1319–1330. doi: 10.1111/j.1365-2958.1996.tb02476.x. [DOI] [PubMed] [Google Scholar]
- 12.Persson B C, Bylund G O, Berg D E, Wikström P M. Functional analysis of the ffh-trmD region of the Escherichia coli chromosome by using reverse genetics. J Bacteriol. 1995;177:5554–5560. doi: 10.1128/jb.177.19.5554-5560.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Powers T, Noller H F. Dominant lethal mutations in a conserved loop in 16S rRNA. Proc Natl Acad Sci USA. 1990;87:1042–1046. doi: 10.1073/pnas.87.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ron E Z, Kohler R E, Davis B D. Polysomes extracted from Escherichia coli by freeze-thaw-lysozyme lysis. Science. 1966;153:1119–1120. doi: 10.1126/science.153.3740.1119. [DOI] [PubMed] [Google Scholar]
- 15.Saiki R K, Scharf S, Faloona F, Mullis K B, Horn G T, Erlich H A, Arnheim N. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 16.Wikström P M, Björk G R. Noncoordinate translation-level regulation of ribosomal and nonribosomal protein genes in the Escherichia coli trmD operon. J Bacteriol. 1988;170:3025–3031. doi: 10.1128/jb.170.7.3025-3031.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wikström P M, Björk G R. A regulatory element within a gene of a ribosomal protein operon of Escherichia coli negatively controls expression by decreasing the translational efficiency. Mol Gen Genet. 1989;219:381–389. doi: 10.1007/BF00259610. [DOI] [PubMed] [Google Scholar]
- 18.Wikström P M, Byström A S, Björk G R. Non-autogenous control of ribosomal protein synthesis from the trmD operon in Escherichia coli. J Mol Biol. 1988;203:141–152. doi: 10.1016/0022-2836(88)90098-8. [DOI] [PubMed] [Google Scholar]
- 19.Wikström P M, Lind L K, Berg D E, Björk G R. Importance of mRNA folding and start codon accessibility in the expression of genes in a ribosomal protein operon of Escherichia coli. J Mol Biol. 1992;224:949–966. doi: 10.1016/0022-2836(92)90462-s. [DOI] [PubMed] [Google Scholar]
- 20.Wimberly B T, Brodersen D E, Clemons W M, Morgan-Warren R J, Carter A P, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]