Abstract
Purpose
Supplementing investigator‐specified variables with large numbers of empirically identified features that collectively serve as ‘proxies’ for unspecified or unmeasured factors can often improve confounding control in studies utilizing administrative healthcare databases. Consequently, there has been a recent focus on the development of data‐driven methods for high‐dimensional proxy confounder adjustment in pharmacoepidemiologic research. In this paper, we survey current approaches and recent advancements for high‐dimensional proxy confounder adjustment in healthcare database studies.
Methods
We discuss considerations underpinning three areas for high‐dimensional proxy confounder adjustment: (1) feature generation—transforming raw data into covariates (or features) to be used for proxy adjustment; (2) covariate prioritization, selection, and adjustment; and (3) diagnostic assessment. We discuss challenges and avenues of future development within each area.
Results
There is a large literature on methods for high‐dimensional confounder prioritization/selection, but relatively little has been written on best practices for feature generation and diagnostic assessment. Consequently, these areas have particular limitations and challenges.
Conclusions
There is a growing body of evidence showing that machine‐learning algorithms for high‐dimensional proxy‐confounder adjustment can supplement investigator‐specified variables to improve confounding control compared to adjustment based on investigator‐specified variables alone. However, more research is needed on best practices for feature generation and diagnostic assessment when applying methods for high‐dimensional proxy confounder adjustment in pharmacoepidemiologic studies.
Keywords: causal inference, confounding, machine learning
Key Points
To improve confounding control in healthcare database studies, data‐driven algorithms can be used to leverage the large volume of information in healthcare databases to generate and identify features that indirectly capture information on unmeasured or unspecified confounding factors (proxy confounders).
Three areas to consider for data‐driven high‐dimensional proxy confounder adjustment include: (1) feature generation—transforming raw data into covariates (or features) to be used for proxy adjustment; (2) covariate prioritization, selection and adjustment; and (3) diagnostic assessment.
There is a large literature on methods for high‐dimensional confounder prioritization/selection, but relatively little has been written on best practices for feature generation and diagnostic assessment. Consequently, these areas have particular limitations and challenges when applying machine learning algorithms for high‐dimensional proxy confounder adjustment.
Plain Language Summary
A fundamental obstacle in studies that utilize administrative healthcare databases is unmeasured confounding bias stemming from nonrandomized treatment choices and poorly measured comorbidities. Failing to adjust for important confounding factors can make it difficult to differentiate between outcomes that are due to drug effects or a result of the underlying conditions for which the drug was prescribed. Traditional approaches for confounding adjustment rely on the investigator to specify all factors that may confound a causal treatment‐outcome association. However, adjustment based on investigator‐specified covariates alone is often inadequate because some important confounding factors are often unknown. Furthermore, because routine‐care databases are not collected for research purposes, many important confounding factors are not directly measured in these data sources. To reduce bias caused by unspecified or unmeasured confounders, many studies have proposed using data‐driven algorithms to identify and control for large numbers of variables that are indirectly associated with unmeasured (or unspecified) confounders ('proxy' confounders). Here, discuss various aspects of high‐dimensional proxy confounder adjustment and give an overview of the current literature. We give particular focus on methods that have been impactful in pharmacoepidemiology research.
1. INTRODUCTION
Routinely‐collected healthcare data are increasingly being used to generate real‐world evidence (RWE) to inform decision making in clinical practice, drug development, and health policy. 1 However, unmeasured confounding from non‐randomized treatment allocation and poorly measured information on comorbidities, disease progression, and disease severity remains a fundamental obstacle to effectively utilizing these data sources for RWE generation. 2 Statistical methods should therefore be used to extract the maximum possible information on confounding from the data to minimize the effects of unmeasured confounding so that accurate comparative estimates of treatments' effectiveness and safety can be obtained. Approaches to mitigate confounding bias would ideally be based on causal diagrams and expert knowledge for confounder selection. 3 However, adjustment based on researcher‐specified variables alone is not always adequate because some confounders are either unknown to researchers or not directly measured in these data sources.
To improve confounding control in healthcare database studies, data‐driven algorithms can be used to leverage the large volume of information in these data sources to generate and identify features that indirectly capture information on unmeasured or unspecified confounding factors (proxy confounders). 4 Proxy confounder adjustment is based on the concept that unmeasured confounding can be mitigated by adjusting for large numbers of variables that collectively serve as proxies for unobserved factors. 5 For example, donepezil use (captured in any claims database) could be used as a proxy for cognitive impairment since cognitive impairment and early Alzheimer's disease and related disorders (ADRD) are often unmeasured in administrative data (Figure 1). For more on the concept of proxy confounder adjustment see VanderWeele 3 and Schneeweiss. 4
FIGURE 1.
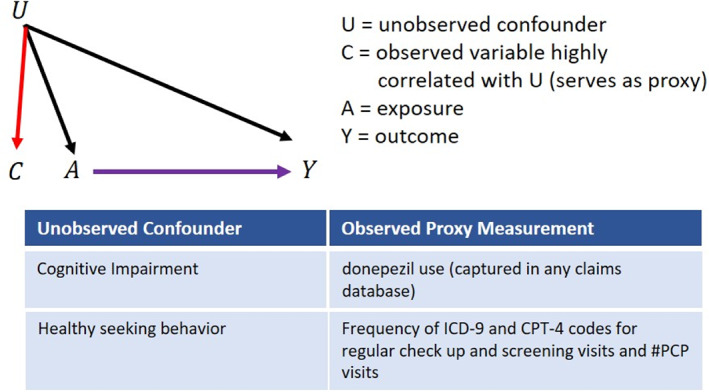
Illustration and examples for ‘proxy confounder’ adjustment.
While researcher‐specified confounders are identified using expert background knowledge, empirical or proxy confounders are identified using empirical associations and coding patterns observed in the data. There is a growing body of evidence showing that complementing researcher‐specified variables with empirically‐identified proxy confounders improves confounding control compared to adjustment based on researcher‐specified confounders alone. 4 , 6 , 7 , 8 , 9 Consequently, there has been a recent focus on the development of data‐driven methods to empirically identify high‐dimensional sets of proxy variables for adjustment in healthcare database studies. 6 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18
In this paper, we discuss the considerations underpinning three areas for data‐driven high‐dimensional proxy confounder adjustment: (1) feature generation—transforming raw data into covariates (or features) to be used for proxy adjustment; (2) covariate prioritization, selection and adjustment; and (3) diagnostic assessment (Figure 2). We review current approaches and recent advancements within each area, including the most widely used approach to proxy confounder adjustment in healthcare database studies (the high‐dimensional propensity score or hdPS). We discuss limitations of the hdPS and survey recent advancements that incorporate the principles of proxy adjustment with machine learning (ML) extensions to improve performance. We further discuss challenges and directions for future development within each area. We give particular focus to diagnostic assessment for causal inference as this has received the least attention when performing high‐dimensional proxy confounder adjustment in the pharmacoepidemiology literature.
FIGURE 2.
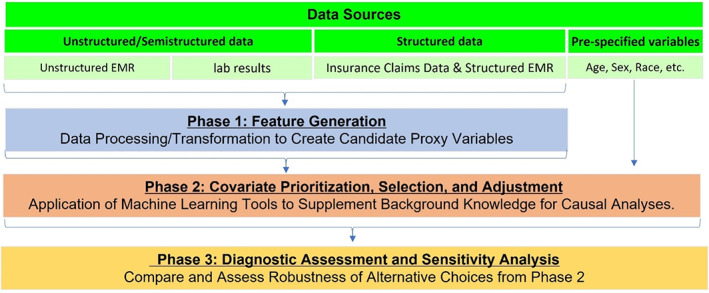
Different phases for high‐dimensional proxy confounder adjustment.
2. GENERATING FEATURES FOR PROXY CONFOUNDER ADJUSTMENT
The first challenge for proxy confounder adjustment is determining how to best leverage the full information content in healthcare databases to generate features (or proxy variables) that best capture confounder information. Several approaches for feature generation of proxy confounders have been applied in the pharmacoepidemiologic literature. These have ranged from very simple approaches that generate binary indicators representing whether or not a given code occurs during a pre‐defined exposure assessment period, 19 to approaches that first process information from healthcare databases into a common data model format with common terminologies and coding schemes representing health concepts. 20 , 21 , 22 Feature engineering can then be applied to the common data model to enable a consistent process across different databases. Examples include the Observational Medical Outcomes Partnership (OMOP) Common Data Model, maintained by the open‐science Observational Health Data Sciences and Informatics (OHDSI) network and also used in the European Health Data and Evidence Network (EHDEN) project, and the National Patient‐Centered Clinical Research Network (PCORnet). 23 , 24 , 25 Generating features consistent with a common data model format can be advantageous for capturing relevant health concepts, but these approaches require more data pre‐processing to extract and transform the original codes into variables representing health concepts.
Instead of generating features based on health concepts, an alternative approach is to generate features based on empirical associations and longitudinal coding patterns observed in the data. Such approaches can be more flexible since they can be independent of the coding system and do not rely on a common data model. 6 The hdPS has become the most widely used tool to generate features based on observed coding patterns in healthcare claims databases. 6 The hdPS generates features by transforming raw medical codes into binary indicator variables based on the frequency of occurrence of each code during a defined pre‐exposure period.
By taking into account the frequency of occurrence of various codes during the covariate assessment period, the hdPS tries to capture information on the intensity of the medical event or drug dispensing. In theory, algorithms could consider more complex longitudinal coding patterns to try and capture additional confounder information. For example, recent work has proposed using neural networks to model a patient's full course of care to consider temporal sequences of a specific course of treatment. 26 The use of neural networks for extracting confounder information by modeling complex coding patterns is promising but examples are limited. 27 , 28
2.1. Challenges in generating features for proxy adjustment from electronic health records
An important limitation of current high‐dimensional proxy confounder adjustment approaches is that they can only use structured electronic healthcare information. However, much of the essential confounder information, such as patient‐reported symptoms, severity, stage and prognosis of the disease, and functional status, is frequently recorded in free‐text notes or reports in electronic health records (EHRs) that are substantially underutilized for confounding adjustment. 29 , 30 Little is known about the impact of incorporating these data for confounding adjustment since unstructured data are not readily analyzable. Natural language processing (NLP) is a subfield of machine learning that can be used to generate variables from unstructured free text. 31 NLP methods are increasingly used to identify health outcomes from EHRs, but the application of NLP algorithms for purposes of identifying high‐dimensional sets of confounding factors is limited. 32 More research is needed on the use of NLP algorithms for generating high‐dimensional sets of proxy confounders and the value of unstructured EHR data in proxy adjustment.
An additional challenge to utilizing EHR data for high‐dimensional confounding control is missing data. While both healthcare claims and EHR data are susceptible to missing information, EHR data is particularly vulnerable due to a lack of continuity and completeness of health records caused by patients seeking care at different delivery systems. 33 , 34 Various approaches for handling missing data have been proposed, including several alternative multiple imputation techniques. Multiple imputation can account for informative missingness under certain untestable assumptions. However, there are many different approaches to handling missing data and no single approach is universally best. 35 Failing to appropriately account for missingness and measurement error when using EHR data can result in analyses that increase rather than reduce bias in estimated treatment effects. 36 , 37 , 38
3. COVARIATE PRIORITIZATION, SELECTION, AND ADJUSTMENT
Once proxy variables have been generated through transformations of the raw data, some degree of dimension reduction is needed to prioritize and select variables for adjustment. Reducing the dimension of covariates is necessary to avoid problems of nonoverlap when adjusting for high‐dimensional sets of covariates. 39 Nonoverlap can result in non‐convergence due to separation when sufficiently many covariates are included in case of logistic regression models. 40 Positivity violations are also a concern for hdPS analyses, as covariate overlap is more difficult to satisfy when controlling for high‐dimensional sets of variables. 39 Even when the sample size may be large enough to effectively preclude problems related to convergence and positivity, it is not practical to consider every possible adjustment set for high‐dimensional data. Machine learning can help researchers reduce the dimension of covariates to avoid issues of nonoverlap and can more flexibly model selected covariates when predicting the treatment and outcome mechanisms.
3.1. hdPS prioritization and its limitations
The hdPS has been the most widely used data‐driven tool in the pharmacoepidemiologic literature for high‐dimensional confounder selection. The hdPS prioritizes or ranks each generated variable based on its potential for bias by assessing the variable's prevalence and univariate, or marginal, association with the treatment and outcome using the Bross bias formula. 6 , 41 , 42 From this ordered list, researchers then specify the number of variables to include for adjustment along with pre‐specified variables. While the hdPS has been shown to often improve confounding control when used to complement investigator‐specified confounders, 6 , 8 , 9 , 43 , 44 there are cases where adjustment for hdPS generated variables had no impact or even harmed the properties of estimators beyond adjustment for researcher‐specified confounders alone. 45 , 46 Limitations of the hdPS prioritization include: (1) the method assesses a variable's potential confounding impact through marginal, or univariate, associations with both treatment and outcome (ideally one would want to consider conditional, or joint, associations among variables); (2) the method requires researchers to subjectively determine how many “proxy” variables to include for adjustment. These limitations can lead to “over adjusting” for variables that can harm the properties of estimators without reducing bias (Figure 3). 47 , 48 , 49 Under adjusting by failing to control for proxy variables that contain important confounder information can also be a concern when implementing the hdPS.
FIGURE 3.
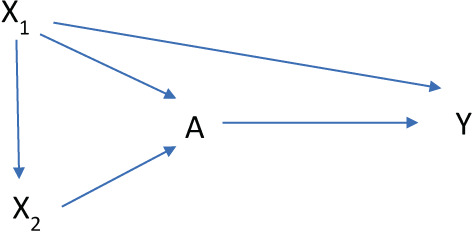
Causal diagram illustrating one scenario where the use of marginal empirical associations for confounder selection can result in over‐adjusting for instrumental variables. In this causal structure, X 2 is marginally associated with both treatment and outcome, but is independent of the outcome after conditioning on X 1.
The choice of the number of proxy variables to include in an hdPS model to adequately control for confounding without “over adjusting” or “under adjusting” varies according to the properties and structure of a given dataset and cannot be identified by only evaluating marginal associations between variables. Determining how many empirically identified “proxy” confounders to include for adjustment is particularly challenging in studies with rare events — settings relevant to RWE studies. In these settings, previous work has shown unstable effect estimates where results are highly dependent on the number of “proxy” confounders included for adjustment. 9 , 43
3.2. Machine learning extensions for covariate prioritization and selection
To address the limitations outlined above, recent studies have developed extensions for proxy confounder adjustment that combine the principles of proxy confounder adjustment with ML tools for prediction modeling and variable selection. These tools have largely focused on incorporating principles for proxy confounder adjustment with regularized regression and Targeted Learning tools, including Super Learning and Collaborative Targeted variable selection. While other ML tools for variable prioritization and selection are available (e.g., principal components, random forests, feature importance selection with neural networks), here we focus on targeted learning tools and regularized regression as these have been the most widely used approaches in the pharmacoepidemiology literature.
3.2.1. Regularized regression for high‐dimensional proxy confounder adjustment
Regularized regression models use penalized maximum likelihood estimation to shrink imprecise model coefficients toward zero. LASSO is the most commonly used regularized regression model for variable selection in high‐dimensional covariate datasets. 44 , 50 Previous work 20 , 21 , 22 , 51 found that LASSO regression can be used to select a subset of generated “proxy” confounders to supplement researcher‐specified confounders to form the adjustment set for confounding control. To improve the performance of regularized regression for high‐dimensional confounder selection, several studies have developed variations of LASSO that consider covariate associations with both treatment and outcome when penalizing the likelihood function. These recent extensions include: (1) Outcome adaptive LASSO, 17 (2) Group LASSO, 16 (3) Highly Adaptive LASSO, 52 (4) Highly Adaptive Outcome LASSO, 11 and (5) Collaborative Controlled LASSO. 53 Other versions of regularized regression, including ridge regression and elastic net, have also been shown to perform well for confounder selection and can be preferable to the LASSO penalization in certain settings. 51
3.2.2. Combining the hdPS with super learning
Super Learning is an ensemble ML algorithm for prediction modeling that forms a set of predicted values based on the optimal weighted combination of a set of user‐specified prediction models in terms of minimizing cross validated predictive performance. 54 , 55 The flexibility of super learning can be utilized to identify a small number of optimally performing prediction algorithms that generally perform best for a given data structure. Previous work has combined Super Learning with proxy confounder adjustment in high‐dimensional covariate spaces. 18 Super Learning can simplify model selection for propensity score estimation in high dimensions and has been shown to perform well in a number of simulations. 13 , 18
3.2.3. High‐dimensional proxy adjustment with scalable versions of collaborative targeted maximum likelihood estimation
Collaborative targeted maximum likelihood estimation (CTMLE) is an extension of the doubly robust targeted maximum likelihood estimation (TMLE) method. 56 , 57 TMLE consists of fitting an initial outcome model to predict the counterfactual outcomes for each individual, then using the estimated propensity score to fluctuate this initial estimate to optimize a bias/variance tradeoff for a specified causal parameter (i.e., the treatment effect). CTMLE extends TMLE by using an iterative forward selection process to construct a series of TMLE estimators, where each successive TMLE estimator controls for one additional variable to consider how a variable relates to both treatment and outcome after conditioning on a set of previously selected variables. 56 , 57 By taking into account a variable's conditional association with both treatment and outcome, CTMLE avoids “over‐adjustment” to improve the properties of estimators by reducing the likelihood of controlling for variables that are conditionally independent of the outcome after adjusting for a set of previously identified confounders. Recent work has developed adaptations of CTMLE that are computationally scalable to large healthcare databases. 14 These adaptations modify the standard version of CTMLE by including a pre‐ordering of variables to avoid the iterative process of searching through each variable in the selection procedure. Simulations indicate that computational gains are substantial and that combining scalable CTMLE with methods for proxy adjustment work well relative to the standard instantiations of CTMLE, hdPS, and TMLE. 14 , 18
3.3. Adjustment for proxy confounders
Once proxy confounders have been prioritized and selected, researchers must determine a method for adjustment and causal estimation. Propensity score methods (e.g., propensity score matching, inverse probability weighting) using logistic regression for estimation of the propensity score function have become the most common approach for adjustment of selected proxy confounders in the medical literature. 58 , 59 Some evidence suggests that improvements can be gained in both predictive performance and bias reduction when using more flexible ML models for propensity score estimation. 28 , 60 , 61 , 62 Another avenue for improving estimations is to adapt ML algorithms to casual inference. Two important examples are the adaptation of random forest to causal forest and X‐learner, a meta‐algorithm that uses ML methods as an intermediate step in an efficient estimation algorithm. 63 , 64
3.3.1. Machine learning with doubly robust estimation for improved adjustment
Widely used doubly robust methods include TMLE, augmented inverse probability weighting (AIPW), and double ML (e.g., R‐learner). 57 , 65 , 66 , 67 These approaches use a model for both the outcome and the propensity score, requiring only one of the two to be correctly specified for consistent estimation of average treatment effects. Theory and simulations have shown that doubly robust approaches are asymptotically efficient and more robust than conventional singly robust methods like propensity score matching and inverse probability weighting. 68
Recent work has further shown that the use of flexible nonparametric ML models for the estimation of nuisance functions (i.e., the propensity score or outcome model) comes at a cost of slow convergence rates. This slow convergence is particularly problematic within singly robust estimation methods and can yield effect estimates with poor statistical properties with performance deteriorating as the dimension of the data increases (the ‘curse of dimensionality’). 69 This work has further demonstrated that doubly robust methods allow for slower converging nuisance models and, therefore, can mitigate or even resolve such problems. Consequently, recent literature suggests that ML‐based methods for estimation of nuisance functions should be applied within doubly robust frameworks rather than more commonly used singly robust methods. For more on machine learning in causal inference see Kennedy, 69 Naimi et al., 70 , 71 and Zivich et al. 72
4. DIAGNOSTIC VALIDITY ASSESSMENT OF CAUSAL ESTIMATIONS
Evaluating the validity of causal analyses for high‐dimensional proxy adjustment remains challenging but is essential to improving robustness and validity of estimated effects. 73 While held‐out sets and cross‐validation allow a direct comparison of ML predictions to observed target variables, such a straightforward evaluation is infeasible in causal inference and the role of prediction diagnostics for purposes of causal inference is less clear. 47 , 74 , 75 , 76 Below, we survey a list of standard ML diagnostics for model prediction and diagnostics for causal inference with a focus on assessing the performance of models for high‐dimensional proxy adjustment in their ability to reduce bias in estimated treatment effects. We highlight their underlying assumptions and limitations.
4.1. Diagnostics for treatment and outcome model prediction
A process to estimate ML model performance using out‐of‐sample data, such as cross validation, are often recommended to assess model robustness and generalizability and to examine the characteristics of the inferred models to verify the importance of domain‐relevant variables. Below we focus on additional measures with specific importance to causal model diagnostics.
4.1.1. Dichotomous and categorical models
Calibration plots depict the average predicted versus observed (empirical) probability of the studied event in subsets of entities (typically, deciles), to evaluate the accuracy of the predicted probabilities. 77 , 78 Probability estimation accuracy is essential for causal inference, more than it typically is for ML classification tasks, as downstream calculations, for example, inverse probability weighting, may rely on these values as being “true” probabilities. Various metrics can be used to quantitatively measure calibration quality, for example, Hosmer‐Lemeshow goodness of fit test, 79 but these have several drawbacks 80 ; visual inspection of the calibration plots or characterization of its slope and intercept is thus recommended.
C‐statistic (or area under the receiver operating characteristic, ROC, curve), a measure of classification accuracy, is commonly used in standard ML applications. For outcome models, it can be used to assess prediction accuracy over the observed treatment assignment (and assuming, but not verifying, that the causal assumptions hold). For propensity models its utility is less straightforward: an extreme (close to 0 or 1) value, corresponding to a highly discriminative model, may indicate a potential violation of positivity; and, conversely, a value around 0.5, suggesting the model cannot discriminate between treatment groups, is not necessarily a sign for inaccurate model, but potentially good covariate overlap. As a result, some researchers recommended to avoid using C‐statistic in propensity model diagnostics. 75 We note that post‐matching C‐statistic may be used to evaluate covariate balance; see below.
4.1.2. Continuous models
The performance of continuous outcome models can be assessed in each observed treatment group (and observed outcomes) and assuming causal assumptions are met, using standard measures such as the coefficient of determination (R 2) or mean squared error. 78 A poorly performing model for a specific treatment group, for example, over or underestimating outcomes, may subsequently lead to biased effect estimation. As with binary outcome models, poor performance may suggest an inadequate prediction model and guide its improvement.
4.2. Diagnostics for causal inference
Previous work has shown that the use of prediction model diagnostics alone to guide model selection and validity assessment can lead to suboptimal performance for causal inference. 47 , 48 , 49 , 75 , 81 We next survey diagnostic methods to more directly assess assumptions and model validity for purposes of causal inference.
4.2.1. Positivity
An important usage for propensity models for high‐dimensional proxy adjustment is to examine the positivity assumption. This assumption states that every individual has a non‐zero probability to be assigned to any treatment conditional on a sufficient set of covariates. A comparison of propensity score distributions can help in identifying (and potentially excluding) sub‐populations where violations or near violations of the positivity assumption occur. 82 , 83 , 84 While high‐dimensional proxy adjustment assumes that unconfounded treatment effects are more plausible when controlling for large numbers of variables, covariate overlap can be more difficult when adjusting for high‐dimensional sets of variables. 39 Therefore, positivity should be tested at the initial stages of analyses for high‐dimensional proxy adjustment.
4.2.2. Balancing
Propensity score modeling aims to facilitate matching, reweighting or stratification to emulate a random assignment of individuals to treatment groups. Therefore, several studies explored methods to directly evaluate balancing of covariates among these groups. 77 , 78 , 85 In a simulation study, Franklin et al. 85 compared several metrics to assess covariate balance and observed that two had consistently strong associations with bias in estimated treatment effects. The first metric, post‐matching C‐statistic, re‐trains a treatment model on the propensity score matched (similarly, stratified or weighted) sample and assesses its (preferably, lack of) ability to discriminate between patients in different treatment groups using C‐statistic. The second recommended metric, general weighted difference, computes a weighted sum of absolute difference in all individual covariates, all covariate squares, and all pairwise interactions. Other papers have also recommended assessing the standardized mean difference in covariates for PS matching and weighting. 86 , 87
The application of balance diagnostics for high‐dimensional propensity scores is more challenging as it is unclear on which set of variables balance should be assessed. A large literature has shown that balancing variables that are independent of the outcome except through treatment (instrumental variables) harms the properties of estimators. 47 , 49 , 81 In high‐dimensional settings, however, identifying instrumental variables is difficult and previous work has argued that priority should be given to controlling for all confounders at the expense of balancing instruments. 20 , 21 , 48 This has led to some researchers assessing balance on all variables in the database when using propensity scores for high‐dimensional proxy adjustment. 20 , 21 , 22 More research is needed on the best use of balance diagnostics for high‐dimensional propensity score adjustment.
4.2.3. Estimand diagnostics (simulation‐based approaches and negative controls)
Recent studies have suggested methods to assess the overall accuracy of effect estimation using control and synthetic control studies. 20 , 21 , 88 , 89 , 90 These frameworks have largely been based on the use of simulation methods to generate synthetic datasets under constraints where certain relations among variables are known (e.g., the simulated treatment effect) while maintaining much of the complexity and statistical properties of the observed data structure.
Parametric bootstrap (‘Plasmode’ simulation)
Simulation frameworks for model validation in causal inference have largely been based on use of the parametric bootstrap. Such approaches bootstrap subjects from the observed data structure, then use modeled relationships from the original data to inject causal relations between a subset of variables while leaving all other associations among variables unchanged. With treatment‐outcome associations known by design and patterns of confounding that mimic the observed data structure, synthetic datasets have become increasingly popular to provide a benchmark for comparing statistical methods for causal inference.
Franklin et al. 89 proposed using a parametric bootstrap approach, termed ‘plasmode simulation’, to compare causal inference methods in settings specific to healthcare database studies and high‐dimensional propensity scores. Schuler et al. 90 and others 88 , 91 , 92 have proposed variations and extensions of plasmode simulation for model validation in healthcare database studies. Schuemie et al. 20 , 21 use a plasmode simulation‐based approach for generating positive control outcomes to quantify bias due to measured confounders when calibrating effect estimates and confidence intervals. Peterson et al. 93 apply a similar parametric bootstrap method as a diagnostic to assess bias due to violations of positivity. Alaa and van der Schaar 88 developed a validation method that uses the parametric bootstrap and influence functions, which are a key technique in robust statistics.
While simulations can be useful for tailoring analytic choices for causal inference, they also have limitations that deserve attention. Schuler et al. 90 explain that validation frameworks based on the parametric bootstrap are more limited since they are not ‘model free’; they require partial simulation of the data structure. This creates two fundamental challenges when generating synthetic datasets to evaluate causal inference methods: (1) Advani et al. 94 showed that if the simulation framework does not closely approximate the true data generating distribution, then the use of synthetically generated data as a diagnostic tool in causal inference can be misleading; (2) even when the simulation framework closely approximates the true data generating process, Schuler et al. 90 warn that the use of synthetic datasets for model validation could still be biased towards favoring causal inference methods that mimic the modeling choices made when generating the synthetic datasets. These challenges can restrict the usefulness of synthetic datasets for model validation in causal inference. Still, studies have demonstrated that in specific cases, the use of synthetic data to tailor analyses to the study at hand can often improve confounding control relative to the consistent use of any single causal inference method. 88 , 90
Wasserstein Generative Adversarial Networks (WGANs) is an alternative approach to generating synthetic data for simulation‐based model validation in causal inference. 95 GANs estimate the distribution of a particular dataset using a ‘generator’ and a ‘discriminator’. 96 The generator is a flexible neural network to create synthetic data while the discriminator is a competing neural network model that attempts to distinguish between the synthetic and real data. The process is repeated in an iterative fashion until the discriminator is no longer able to distinguish between the synthetic and real data. This technique has become very powerful for supervised and unsupervised ML. 96 WGANS have recently been shown to be useful for generating synthetic datasets that closely approximate the joint correlation structure of an actual dataset for purposes of model validation in causal inference. 95
Negative and positive controls
Another approach that has become increasingly popular for evaluating models for confounder adjustment is the use of real negative controls—exposure‐outcome pairs that are not, as far as we know, causally related. 77 , 97 Such controls can be used to detect residual biases, for example, confounding, in the estimation process. Replicating a known association through use of positive controls can also increase confidence in primary estimates' validity. However, some researchers have argued that identifying positive controls is difficult since the magnitude of known effects is rarely known. 20 , 21 , 22
4.2.4. Sensitivity analyses
Quantitative bias analysis
Estimating an effect from observational data involves multiple, at times somewhat arbitrary, modeling decisions and assumptions, for example, with respect to the definition of confounders, exposures, and outcomes or the statistical analysis. 98 Sensitivity analysis re‐computes the estimated effect under various sets of such decisions 99 or using multiple data sources to verify its robustness. 83 , 100 Sensitivity analyses can also quantify the change that an unmeasured confounder would have on the studied estimand and thus assess its sensitivity to violations of the assumption of no unmeasured confounding. 99 , 101 This can be particularly useful when applying methods for high‐dimensional proxy adjustment as researchers can never be certain how well a set of features captures information on unmeasured factors. The E‐value in particular has become widely used for assessing sensitivity of an estimand to unmeasured confounding in the medical literature. 102 The popularity of the E‐value has largely been due to its simplicity, making its implementation and communication straightforward. However, its simplicity has also been a point of criticism of the method. 103 , 104 , 105 , 106 , 107 Several more comprehensive bias analysis methods have been developed to quantify the impact of various systematic errors to increase confidence that the estimated effects are robust to violations of various assumptions. Lash et al. provide a detailed discussion on methods for quantitative bias analysis. 108 An overview of a subset of diagnostics for causal inference is shown in Table 1.
TABLE 1.
Examples of diagnostic metrics for causal inference.
| Condition being tested | Possible diagnostic checks | Limitations and comments |
|---|---|---|
| Positivity | Overlap of estimated PS across treatment groups |
Impact of limited overlap can depend on the adjustment approach Including more covariates for adjustment can decrease overlap. Consequently, it can be difficult to determine the optimal adjustment set in terms of maximizing confounding control vs bias due to nonoverlap |
| Conditional Exchangeability on Measured Covariates | Covariate balance across treatment groups after PS adjustment |
Primarily used for PS analyses. Less useful for causal inference approaches that model that outcome directly, including doubly robust methods. Can be difficult to quantify the impact of residual imbalance on bias in estimated treatment effects Can be difficult to determine on which variables balance should be assessed (e.g., do not want to balance instrumental variables). |
| Prediction diagnostics to assess correct model specification |
Can reward PS models that include instruments More useful for causal inference approaches that model the outcome, including doubly robust methods |
|
| Simulation‐based approaches for generating synthetic datasets to evaluate bias in estimated treatment effects |
A very general approach that is applicable to any causal inference method Requires advanced simulation techniques to closely approximate the confounding structure of the study population |
|
| Violation of conditional exchangeability due to unmeasured confounding | Real negative and positive control exposures and/or outcomes |
Can be useful to identifying bias caused by unmeasured confounders Can be difficult to identify good negative and/or positive controls |
| Sensitivity to hidden biases (e.g., unmeasured confounding, misclassification) | E‐value |
Implementation and communication is simple and straightforward Recent critiques have argued that the E‐value can be misleading due to its simplicity |
| Formal quantitative bias analysis | Several approaches have been proposed to conduct in‐depth sensitivity analyses for hidden biases. These can provide more detailed assessment of robustness of causal analyses, but are subject to underlying assumptions and can be tedious to implement |
5. DISCUSSION
In this paper, we have provided an overview of high‐dimensional proxy confounder adjustment in studies utilizing electronic healthcare databases. We have focused on three areas for proxy adjustment: (1) feature generation, (2) covariate prioritization, selection, and adjustment, and (3) validity assessment. We have discussed recent ML extensions and paths for future research within each area. Much attention has been given to the development of ML tools for confounder selection and adjustment for high‐dimensional proxy adjustment. These tools have great potential to improve confounding control in healthcare database studies. However, less attention has been given to advancing methods for feature generation and validity assessment for proxy confounder adjustment. Future research is warranted to investigate the optimal methods that extract the relevant confounding information to generate features for proxy adjustment while preserving scalability and data‐adaptability to large healthcare databases. Future research is also needed in the development of diagnostic methods to evaluate and compare the validity of alternative approaches to high‐dimensional proxy adjustment in healthcare database studies.
Finally, although ML tools can be beneficial in identifying empirical associations among large numbers of covariates, empirical associations by themselves are not sufficient to determine causal relations. 109 , 110 , 111 We emphasize the importance of using substantive knowledge to obtain an understanding of the data and the underlying causal structure before applying ML procedures for confounding control. 109 , 110 , 111 ML procedures should not replace background knowledge, but should be used to complement investigator input when controlling for confounding.
AUTHOR CONTRIBUTIONS
Richard Wyss, Chen Yanover, and Tal El‐Hay drafted the manuscript. All authors revised the manuscript for important intellectual content and approved the final manuscript to be submitted for publication.
FUNDING INFORMATION
Funding to support this manuscript development was provided by the International Society for Pharmacoepidemiology (ISPE).
CONFLICT OF INTEREST
Robert W. Platt has consulted for Amgen, Biogen, Merck, Nant Pharma, and Pfizer. Dimitri Bennett is an employee of Takeda. Grammati Sari is employed by Visible Analytics Ltd. Hongbo Yuan is an employee of CADTH. Andrew R. Zullo receives research grant funding from Sanofi Pasteur to support research on infections and vaccinations in nursing homes unrelated to this manuscript. Mugdha Gokhale is a full‐time employee of Merck and owns stocks in Merck. Elisabetta Patorno is supported by a career development grant K08AG055670 from the National Institute on Aging. She is researcher of a researcher‐initiated grant to the Brigham and Women's Hospital from Boehringer Ingelheim, not directly related to the topic of the submitted work.
ACKNOWLEDGMENTS
This manuscript is endorsed by the International Society for Pharmacoepidemiology (ISPE). In addition, we would like to thank Ehud Karavani and Itay Manes, IBM Research – Haifa, for insightful discussions and comments. In addition, we also thank the ISPE members for reviewing and providing comments on our manuscript.
Wyss R, Yanover C, El‐Hay T, et al. Machine learning for improving high‐dimensional proxy confounder adjustment in healthcare database studies: An overview of the current literature. Pharmacoepidemiol Drug Saf. 2022;31(9):932‐943. doi: 10.1002/pds.5500
Richard Wyss and Chen Yanover are the co‐first authors.
Funding information International Society of Pharmacoepidemiology
REFERENCES
- 1. Corrigan‐Curay J, Sacks L, Woodcock J. Real‐world evidence and real‐world data for evaluating drug safety and effectiveness. JAMA. 2018;320:867‐868. [DOI] [PubMed] [Google Scholar]
- 2. Streeter AJ, Lin NX, Crathorne L, et al. Adjusting for unmeasured confounding in nonrandomized longitudinal studies: a methodological review. J Clin Epidemiol. 2017;87:23‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. VanderWeele TJ. Principles of confounder selection. Eur J Epidemiol. 2019;34:211‐219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schneeweiss S. Automated data‐adaptive analytics for electronic healthcare data to study causal treatment effects. Clin Epidemiol. 2018;10:771‐788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schneeweiss S, Avorn J. A review of uses of health care utilization databases for epidemiologic research on therapeutics. J Clin Epidemiol. 2005;58:323‐337. [DOI] [PubMed] [Google Scholar]
- 6. Schneeweiss S, Rassen JA, Glynn RJ, Avorn J, Mogun H, Brookhart MA. High‐dimensional propensity score adjustment in studies of treatment effects using health care claims data. Epidemiology. 2009;20:512‐522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guertin JR, Rahme E, Dormuth CR, LeLorier J. Head to head comparison of the propensity score and the high‐dimensional propensity score matching methods. BMC Med Res Methodol. 2016;16:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guertin JR, Rahme E, LeLorier J. Performance of the high‐dimensional propensity score in adjusting for unmeasured confounders. Eur J Clin Pharmacol. 2016;72:1497‐1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Patorno E, Glynn RJ, Hernandez‐Diaz S, Liu J, Schneeweiss S. Studies with many covariates and few outcomes: selecting covariates and implementing propensity‐score‐based confounding adjustments. Epidemiology. 2014;25:268‐278. [DOI] [PubMed] [Google Scholar]
- 10. Ertefaie A, Asgharian M, Stephens DA. Variable selection in causal inference using a simultaneous penalization method. J Causal Inference. 2018;6(1). 10.1515/jci-2017-0010 [DOI] [Google Scholar]
- 11. Ju C, Benkeser D, van der Laan M. Flexible collaborative estimation of the average causal effect of a treatment using the outcome‐highly‐adaptive Lasso. arXiv:1806.06784 [stat.ME]; 2018.
- 12. Ju C, Benkeser D, van der Laan MJ. Robust inference on the average treatment effect using the outcome highly adaptive lasso. arXiv preprint; 2019. [DOI] [PubMed]
- 13. Ju C, Combs M, Lendle SD, Franklin JM, Wyss R, Schneeweiss S, van der Laan MJ. Propensity score prediction for electronic healthcare databases using Super Learner and High‐Dimensional Propensity Score methods. arXiv preprint; 2017. [DOI] [PMC free article] [PubMed]
- 14. Ju C, Gruber S, Lendle SD, et al. Scalable collaborative targeted learning for high‐dimensional data. Stat Methods Med Res. 2017;28:532‐554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ju C, Wyss R, Franklin JM, Schneeweiss S, Häggström J, van der Laan MJ. Collaborative‐controlled LASSO for constructing propensity score‐based estimators in high‐dimensional data. Stat Methods Med Res. 2017;28:1044‐1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koch B, Vock DM, Wolfson J. Covariate selection with group lasso and doubly robust estimation of causal effects. Biometrics. 2018;74:8‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shortreed SM, Ertefaie A. Outcome‐adaptive lasso: variable selection for causal inference. Biometrics. 2017;73:1111‐1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wyss R, Schneeweiss S, van der Laan M, Lendle SD, Ju C, Franklin JM. Using super learner prediction modeling to improve high‐dimensional propensity score estimation. Epidemiology. 2018;29:96‐106. [DOI] [PubMed] [Google Scholar]
- 19. Wyss R, Ellis AR, Brookhart MA, et al. Matching on the disease risk score in comparative effectiveness research of new treatments. Pharmacoepidemiol Drug Saf. 2015;24:951‐961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schuemie MJ, Hripcsak G, Ryan PB, Madigan D, Suchard MA. Empirical confidence interval calibration for population‐level effect estimation studies in observational healthcare data. Proc Natl Acad Sci U S A. 2018;115:2571‐2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schuemie MJ, Ryan PB, Hripcsak G, Madigan D, Suchard MA. Improving reproducibility by using high‐throughput observational studies with empirical calibration. Philos Trans A: Math Phys Eng Sci. 2018;376:20170356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tian Y, Schuemie MJ, Suchard MA. Evaluating large‐scale propensity score performance through real‐world and synthetic data experiments. Int J Epidemiol. 2018;47:2005‐2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Overhage JM, Ryan PB, Reich CG, Hartzema AG, Stang PE. Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc. 2012;19:54‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kent S, Burn E, Dawoud D, et al. Common problems, common data model solutions: evidence generation for health technology assessment. PharmacoEconomics. 2021;39:275‐285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fleurence RL, Curtis LH, Califf RM, Platt R, Selby JV, Brown JS. Launching PCORnet, a national patient‐centered clinical research network. J Am Med Inform Assoc. 2014;21:578‐582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rassen J. A neural‐network driven longitudinal propensity score for evaluation of drug treatment effects. Proceedings of the International Conference on Pharmacoepidemiology & Therapeutic Risk Management, Berlin, Germany; 2020.
- 27. Weberpals J, Becker T, Davies J, et al. Deep learning‐based propensity scores for confounding control in comparative effectiveness research: a large‐scale, real‐world data study. Epidemiology. 2021;32:378‐388. [DOI] [PubMed] [Google Scholar]
- 28. Louizos C, Shalit U, Mooij JM, Sontag D, Zemel R, Welling M. Causal effect inference with deep latent‐variable models. Proceedings of teh 31st International Conference on Neural Information Processing Systems of NeurIPS'17; 2017.
- 29. Dong YH, Alcusky M, Maio V, et al. Evidence of potential bias in a comparison of beta blockers and calcium channel blockers in patients with chronic obstructive pulmonary disease and acute coronary syndrome: results of a multinational study. BMJ Open. 2017;7:e012997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Setoguchi S, Warner Stevenson L, Stewart GC, et al. Influence of healthy candidate bias in assessing clinical effectiveness for implantable cardioverter‐defibrillators: cohort study of older patients with heart failure. BMJ. 2014;348:g2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nadkarni PM, Ohno‐Machado L, Chapman WW. Natural language processing: an introduction. J Am Med Inform Assoc. 2011;18:544‐551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Afzal Z, Masclee GMC, Sturkenboom M, Kors JA, Schuemie MJ. Generating and evaluating a propensity model using textual features from electronic medical records. PLoS One. 2019;14:e0212999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin KJ, Schneeweiss S. Considerations for the analysis of longitudinal electronic health records linked to claims data to study the effectiveness and safety of drugs. Clin Pharmacol Ther. 2016;100:147‐159. [DOI] [PubMed] [Google Scholar]
- 34. Lin KJ, Singer DE, Glynn RJ, Murphy SN, Lii J, Schneeweiss S. Identifying patients with high data completeness to improve validity of comparative effectiveness research in electronic health records data. Clin Pharmacol Ther. 2018;103:899‐905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. de Vries P, Bas BL, van Smeden M, RHH G. Propensity score estimation using classification and regression trees in the presence of missing covariate data. Epidemiol Methods. 2018;7(1). 10.1515/em-2017-0020 [DOI] [Google Scholar]
- 36. Granger E, Sergeant JC, Lunt M. Avoiding pitfalls when combining multiple imputation and propensity scores. Stat Med. 2019;38:5120‐5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Leyrat C, Seaman SR, White IR, et al. Propensity score analysis with partially observed covariates: how should multiple imputation be used? Stat Methods Med Res. 2019;28:3‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Perkins NJ, Cole SR, Harel O, et al. Principled approaches to missing data in epidemiologic studies. Am J Epidemiol. 2018;187:568‐575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. D'Amour A, Ding P, Feller A, Lei L, Sekhon J. Overlap in observational studies with high‐dimensional covariates. J Econ. 2021;221:644‐654. [Google Scholar]
- 40. Albert A, Anderson JA. On the existence of maximum likelihood estimates in logistic regression models. Biometrika. 1984;71:1‐10. [Google Scholar]
- 41. Bross ID. Spurious effects from an extraneous variable. J Chronic Dis. 1966;19:637‐647. [DOI] [PubMed] [Google Scholar]
- 42. Wyss R, Fireman B, Rassen JA, Schneeweiss S. Erratum: high‐dimensional propensity score adjustment in studies of treatment effects using health care claims data. Epidemiology. 2018;29:e63‐e64. [DOI] [PubMed] [Google Scholar]
- 43. Rassen JA, Glynn RJ, Brookhart MA, Schneeweiss S. Covariate selection in high‐dimensional propensity score analyses of treatment effects in small samples. Am J Epidemiol. 2011;173:1404‐1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Franklin JM, Eddings W, Glynn RJ, Schneeweiss S. Regularized regression versus the high‐dimensional propensity score for confounding adjustment in secondary database analyses. Am J Epidemiol. 2015;182:651‐659. [DOI] [PubMed] [Google Scholar]
- 45. Toh S, Garcia Rodriguez LA, Hernan MA. Confounding adjustment via a semi‐automated high‐dimensional propensity score algorithm: an application to electronic medical records. Pharmacoepidemiol Drug Saf. 2011;20:849‐857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Austin PC, Wu CF, Lee DS, Tu JV. Comparing the high‐dimensional propensity score for use with administrative data with propensity scores derived from high‐quality clinical data. Stat Methods Med Res. 2019;29:568‐588. [DOI] [PubMed] [Google Scholar]
- 47. Brookhart MA, Schneeweiss S, Rothman KJ, Glynn RJ, Avorn J, Stürmer T. Variable selection for propensity score models. Am J Epidemiol. 2006;163:1149‐1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Myers JA, Rassen JA, Gagne JJ, et al. Effects of adjusting for instrumental variables on bias and precision of effect estimates. Am J Epidemiol. 2011;174:1213‐1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bhattacharya J, Vogt WB. Do instrumental variables belong in propensity scores? NBER Technical Working Paper no. 343. National Bureau of Economic Research, 2007.
- 50. Low YS, Gallego B, Shah NH. Comparing high‐dimensional confounder control methods for rapid cohort studies from electronic health records. J Comp Eff Res. 2016;5:179‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Karim ME, Pang M, Platt RW. Can we train machine learning methods to outperform the high‐dimensional propensity score algorithm? Epidemiology. 2018;29:191‐198. [DOI] [PubMed] [Google Scholar]
- 52. Benkeser D, van der Laan M. The highly adaptive lasso estimator. Proceedings of the IEEE International Conference on Data Science and Advanced Analytics, pp. 689–696; 2016. [DOI] [PMC free article] [PubMed]
- 53. Ju C, Wyss R, Franklin JM, Schneeweiss S, Häggström J, van der Laan MJ. Collaborative‐controlled LASSO for constructing proopensity score‐based estimators in high‐dimensional data. Stat Methods Med Res. 2019;28:1044‐1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. van der Laan MJ, Polley EC, Hubbard AE. Super learner. Stat Appl Genet Mol Biol. 2007;6:25. [DOI] [PubMed] [Google Scholar]
- 55. Polley EC, Rose S, van der Laan MJ. Super learning. Targeted Learning. Springer; 2011:43‐66. [Google Scholar]
- 56. van der Laan MJ, Gruber S. Collaborative double robust targeted maximum likelihood estimation. Int J Biostat. 2010;6(1). 10.2202/1557-4679.1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. der Laan MJV, Rose S. Targeted Learning: Causal Inference for Observational and Experimental Data. Springer; 2011. [Google Scholar]
- 58. Brookhart MA, Wyss R, Layton JB, Sturmer T. Propensity score methods for confounding control in nonexperimental research. Circ Cardiovasc Qual Outcomes. 2013;6:604‐611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sturmer T, Wyss R, Glynn RJ, Brookhart MA. Propensity scores for confounder adjustment when assessing the effects of medical interventions using nonexperimental study designs. J Intern Med. 2014;275:570‐580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee BK, Lessler J, Stuart EA. Improving propensity score weighting using machine learning. Stat Med. 2010;29:337‐346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pirracchio R, Petersen ML, van der Laan M. Improving propensity score estimators' robustness to model misspecification using super learner. Am J Epidemiol. 2015;181:108‐119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Westreich D, Lessler J, Funk MJ. Propensity score estimation: neural networks, support vector machines, decision trees (CART), and meta‐classifiers as alternatives to logistic regression. J Clin Epidemiol. 2010;63:826‐833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kunzel SR, Sekhon JS, Bickel PJ, Yu B. Metalearners for estimating heterogeneous treatment effects using machine learning. Proc Natl Acad Sci. 2019;116:4156‐4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wager S, Athey S. Estimation and inference of heterogeneous treatment effects using random forests. J Am Stat Assoc. 2018;113:1228‐1242. [Google Scholar]
- 65. Chernozhukov V, Chetverikov D, Demirer M, et al. Double/debiased machine learning for treatment and structural parameters. Econ J. 2018;21:C1‐C68. [Google Scholar]
- 66. van der Laan MJ, Gruber S. Targeted minimum loss based estimation of causal effects of multiple time point interventions. Int J Biostat. 2012;8(1). 10.1515/1557-4679.1370 [DOI] [PubMed] [Google Scholar]
- 67. Nie X, Wager S. Quasi‐oracle estimation of heterogeneous treatment effects. Biometrika. 2021;108:299‐319. [Google Scholar]
- 68. Funk MJ, Westreich D, Wiesen C, Stürmer T, Brookhart MA, Davidian M. Doubly robust estimation of causal effects. Am J Epidemiol. 2011;173:761‐767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kennedy EH. Semiparametric theory and empirical processes in causal inference. Statistical Causal Inferences and Their Applications in Public Health Research. Springer; 2016:141‐167. [Google Scholar]
- 70. Naimi AI, Kennedy EH. Nonparametric double robustness. arXiv preprint arXiv:1711.7137; 2017.
- 71. Naimi AI, Mishler AE, Kennedy EH. Challenges in obtaining valid causal effect estimates with machine learning algorithms. arXiv preprint arXiv:1711.07137; 2017. [DOI] [PubMed]
- 72. Zivich PN, Breskin A. Machine learning for causal inference: on the use of cross‐fit estimators. Epidemiology. 2021;32:393‐401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hernan MA, Hsu J, Healy BA. A second chance to get causal inference right: a classification of data science tasks. CHANCE. 2019;32:42‐49. [Google Scholar]
- 74. Holland PW. Statistics and causal inference. J Am Stat Assoc. 1986;81:945‐960. [Google Scholar]
- 75. Westreich D, Cole SR, Funk MJ, Brookhart MA, Sturmer T. The role of the c‐statistic in variable selection for propensity score models. Pharmacoepidemiol Drug Saf. 2011;20:317‐320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wyss R, Ellis AR, Brookhart MA, et al. The role of prediction modeling in propensity score estimation: an evaluation of logistic regression, bCART, and the covariate‐balancing propensity score. Am J Epidemiol. 2014;180:645‐655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. OHDSI . The Book of OHDSI: Observational Health Data Sciences and Informatics. OHDSI; 2019. [Google Scholar]
- 78. Shimoni Y, Karavani E, Ravid S, et al. An evaluation toolkit to guide model selection and cohort definition in causal inference. arXiv preprint; 2019.
- 79. Hosmer DW, Hosmer T, Le Cessie S, Lemeshow S. A comparison of goodness‐of‐fit tests for the logistic regression model. Stat Med. 1997;16:965‐980. [DOI] [PubMed] [Google Scholar]
- 80. Steyerberg EW, Vickers AJ, Cook NR, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology. 2010;21:128‐138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wooldridge J. Should Instrumental Variables be Used as Matching Variables?. Michigan State University; 2009. [Google Scholar]
- 82. Beaudoin FL, Gutman R, Merchant RC, et al. Persistent pain after motor vehicle collision: comparative effectiveness of opioids vs nonsteroidal antiinflammatory drugs prescribed from the emergency department‐a propensity matched analysis. Pain. 2017;158:289‐295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ozery‐Flato M, Goldschmidt Y, Shaham O, et al., Framework for identifying drug repurposing candidates from observational healthcare data medRxiv, 20018366; 2020. [DOI] [PMC free article] [PubMed]
- 84. Ozery‐Flato M, Goldschmidt Y, Shaham O, Ravid S, Yanover C. Framework for identifying drug repurposing candidates from observational healthcare data. JAMIA Open. 2020;3:536‐544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Franklin JM, Rassen JA, Ackermann D, Bartels DB, Schneeweiss S. Metrics for covariate balance in cohort studies of causal effects. Stat Med. 2014;33:1685‐1699. [DOI] [PubMed] [Google Scholar]
- 86. Ali MS, Groenwold RH, Pestman WR, et al. Propensity score balance measures in pharmacoepidemiology: a simulation study. Pharmacoepidemiol Drug Saf. 2014;23:802‐811. [DOI] [PubMed] [Google Scholar]
- 87. Austin PC. Balance diagnostics for comparing the distribution of baseline covariates between treatment groups in propensity‐score matched samples. Stat Med. 2009;28:3083‐3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Alaa A, Van Der Schaar M. Validating causal inference models via influence functions. Proceedings of the International Conference on Machine Learning, Vol. 97, pp. 191–201; 2019. [Google Scholar]
- 89. Franklin JM, Schneeweiss S, Polinski JM, Rassen JA. Plasmode simulation for the evaluation of pharmacoepidemiologic methods in complex healthcare databases. Comput Stat Data Anal. 2014;72:219‐226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Schuler A, Jung K, Tibshirani R, Hastie T, Shah N. Synth‐Validation: Selecting the best causal inference method for a given dataset. arXiv:1711.00083; 2017.
- 91. Bahamyirou A, Blais L, Forget A, Schnitzer ME. Understanding and diagnosing the potential for bias when using machine learning methods with doubly robust causal estimators. Stat Methods Med Res. 2018;28:1637‐1650. [DOI] [PubMed] [Google Scholar]
- 92. Lenis D, Ackerman B, Stuart EA. Measuring model misspecification: application to propensity score methods with complex survey data. Comput Stat Data Anal. 2018;128:48‐57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Petersen ML, Porter KE, Gruber S, Wang Y, van der Laan MJ. Diagnosing and responding to violations in the positivity assumption. Stat Methods Med Res. 2012;21:31‐54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Advani A, Kitagawa T, Slocznski T. Mostly harmless simulations? Using Monte Carlo studies for estimator selection. J Appl Econometrics. 2019;34:893‐910. [Google Scholar]
- 95. Athey S, Imbens GW, Metzger J, Munro EM. Using Wasserstein generative adversarial networks for the design of Monte‐Carlo simulations. NBER Working Paper no. 26566; 2019.
- 96. Goodfellow IJ, Pouget‐Abadie J, Mirza M, et al. Generative adversarial networks. arXiv preprint arXiv:1406.2661; 2014.
- 97. Lipsitch M, Tchetgen Tchetgen E, Cohen T. Negative controls: a tool for detecting confounding and bias in observational studies. Epidemiology. 2010;21:383‐388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sarri G, Patorno E, Yuan H, et al. Framework for the synthesis of non‐randomised studies and randomised controlled trials: a guidance on conducting a systematic review and meta‐analysis for healthcare decision making. BMJ Evid Based Med. 2020;27:109‐119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Delaney JAC, Seeger JD. Sensitivity Analysis. Developing a Protocol for Observational Comparative Effectiveness Research: A user's Guide. Agency for Healthcare Research and Quality; 2013:145‐159. [PubMed] [Google Scholar]
- 100. Suchard MA, Schuemie MJ, Krumholz HM, et al. Comprehensive comparative effectiveness and safety of first‐line antihypertensive drug classes: a systematic, multinational, large‐scale analysis. Lancet. 2019;394:1816‐1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Vanderweele TJ, Arah OA. Bias formulas for sensitivity analysis of unmeasured confounding for general outcomes, treatments, and confounders. Epidemiology. 2011;22:42‐52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. VanderWeele TJ, Ding P. Sensitivity analysis in observational research: introducing the E‐value. Ann Intern Med. 2017;167:268‐274. [DOI] [PubMed] [Google Scholar]
- 103. Greenland S. Commentary: an argument against E‐values for assessing the plausibility that an association could be explained away by residual confounding. Int J Epidemiol. 2020;49:1501‐1503. [DOI] [PubMed] [Google Scholar]
- 104. Ioannidis JPA, Tan YJ, Blum MR. Limitations and misinterpretations of E‐values for sensitivity analyses of observational studies. Ann Intern Med. 2019;170:108‐111. [DOI] [PubMed] [Google Scholar]
- 105. Sjolander A, Greenland S. Are E‐values too optimistic or too pessimistic? Both and neither! Int J Epidemiol. 2022;51:355‐363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. VanderWeele TJ. Are Greenland, Ioannidis and Poole opposed to the cornfield conditions? A defence of the E‐value. Int J Epidemiol. 2022;51:364‐371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. VanderWeele TJ, Mathur MB, Ding P. Correcting misinterpretations of the E‐value. Ann Intern Med. 2019;170:131‐132. [DOI] [PubMed] [Google Scholar]
- 108. Lash TL, Fox MP, MacLehose RF, Maldonado G, McCandless LC, Greenland S. Good practices for quantitative bias analysis. Int J Epidemiol. 2014;43:1969‐1985. [DOI] [PubMed] [Google Scholar]
- 109. Hernan MA, Hernandez‐Diaz S, Werler MM, Mitchell AA. Causal knowledge as a prerequisite for confounding evaluation: an application to birth defects epidemiology. Am J Epidemiol. 2002;155:176‐184. [DOI] [PubMed] [Google Scholar]
- 110. Robins JM. Data, design, and background knowledge in etiologic inference. Epidemiology. 2001;12:313‐320. [DOI] [PubMed] [Google Scholar]
- 111. Wyss R, Sturmer T. Commentary: balancing automated procedures for confounding control with background knowledge. Epidemiology. 2014;25:279‐281. [DOI] [PMC free article] [PubMed] [Google Scholar]


