Abstract
Oleandomycin, a macrolide antibiotic produced by Streptomyces antibioticus, contains two sugars attached to the aglycon: l-oleandrose and d-desosamine. oleY codes for a methyltransferase involved in the biosynthesis of l-oleandrose. This gene was overexpressed in Escherichia coli to form inclusion bodies and in Streptomyces lividans, producing a soluble protein. S. lividans overexpressing oleY was used as a biotransformation host, and it converted the precursor l-olivosyl-erythronolide B into its 3-O-methylated derivative, l-oleandrosyl-erythronolide B. Two other monoglycosylated derivatives were also substrates for the OleY methyltransferase: l-rhamnosyl- and l-mycarosyl-erythronolide B. OleY methyltransferase was purified yielding a 43-kDa single band on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The native enzyme showed a molecular mass of 87 kDa by gel filtration chromatography, indicating that the enzyme acts as a dimer. It showed a narrow pH range for optimal activity, and its activity was clearly stimulated by the presence of several divalent cations, being maximal with Co2+. The S. antibioticus OleG2 glycosyltransferase is proposed to transfer l-olivose to the oleandolide aglycon, which is then converted into l-oleandrose by the OleY methyltransferase. This represents an alternative route for l-oleandrose biosynthesis from that in the avermectin producer Streptomyces avermitilis, in which l-oleandrose is transferred to the aglycon by a glycosyltransferase.
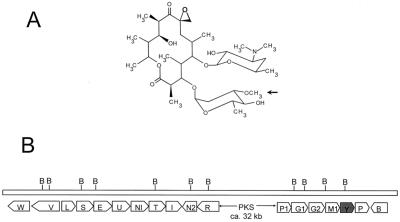
Oleandomycin (Fig. 1A) is a clinically important 14-membered macrolide antibiotic produced by Streptomyces antibioticus. Structurally it contains a macrolactone ring (oleandolide) which is synthesized by the assembly of one starter acetyl-coenzyme A (CoA) and six extender methylmalonyl CoA units. These reactions are catalyzed by a modular type I polyketide synthase comprising three large polypeptides, named OleA1, OleA2, and OleA3 (24, 25). The oleandomycin biosynthetic gene cluster has been fully sequenced and characterized (1, 15, 16, 18, 19, 24, 25) (Fig. 1B). Two DNA regions flanking the genes encoding the three multifunctional polypeptides of the polyketide synthase contain a number of genes that code for other enzymatic functions required for the biosynthesis of the two sugars and their transfer to the aglycon (1, 16, 18), genes encoding an ABC transporter system responsible for secretion (15), genes encoding an inactivating-reactivating enzymatic system that confers self-resistance to the producer organism (18, 26), and a gene encoding a cytochrome P-450 oxygenase involved in epoxidation of the macrolactone ring at carbon 8 (19, 24). The oleandomycin macrolactone ring is glycosylated by two 6-deoxysugars (l-oleandrose and d-desosamine). Two glycosyltransferases (OleG1 and OleG2) are responsible for sugar transfer. Insertional inactivation of oleG1 produced a mutant that accumulates the macrolactone 8,8a-deoxyoleandolide, because both OleG1 and OleG2 glycosyltransferases were affected in this mutant by a polar effect (16). Complementation experiments using eryBV- and eryCIII-minus mutants of the erythromycin producer Saccharopolyspora erythraea showed that oleG1 complemented an eryCIII-minus mutant and oleG2 complemented an eryBV-minus mutant (6). Therefore, it was concluded that d-desosamine and l-oleandrose were the substrates for the OleG1 and OleG2 glycosyltransferases, respectively. Further support for OleG2 as the l-oleandrose transferase came from biotransformation experiments. By feeding erythronolide B (EB) to a Streptomyces albus clone expressing the oleG2 gene and harboring pOLE, a plasmid containing all the genes necessary for the biosynthesis of l-oleandrose, l-oleandrosyl-erythronolide B (OLE-EB) was formed (1). One of the genes in pOLE (oleY) was proposed to encode a 3-O-methyltransferase responsible for the conversion of dTDP-l-olivose into dTDP-l-oleandrose during the biosynthesis of l-oleandrose (1).
FIG. 1.
(A) Chemical structure of the macrolide oleandomycin. The arrow indicates the site of action of the OleY methyltransferase. (B) Gene organization of the oleandomycin biosynthetic gene cluster. oleY is shadowed. For a description of the different genes and their functions, see references 1, 15, 16, 18, 19, 24, and 25.
Here we report the oleY expression, purification of the OleY methyltransferase, and evidence demonstrating that methylation occurs at the C-3-O position of the neutral sugar to generate l-oleandrose during biosynthesis of oleandomycin by S. antibioticus. This methylation event occurs once the sugar is attached to the aglycon. Also we show that this methyltransferase can use as substrates at least two other monoglycosylated derivatives, l-rhamnosyl-erythronolide B (RHA-EB) and l-mycarosyl-erythronolide B (MYC-EB).
MATERIALS AND METHODS
Microorganisms, culture conditions, and plasmids.
S. antibioticus ATCC 11891, an oleandomycin producer, was used as a donor of chromosomal DNA. S. lividans TK21 and S. albus NAG2 (1) were used as hosts for subcloning and for biotransformation experiments. E. coli XL1-Blue (4) was also used as a host for subcloning. pUC18 was used as subcloning vector in E. coli. pQE60 (Qiagen) and pUWL201 (7) were used as expression vectors in E. coli and S. lividans, respectively. pWHM2109 is a plasmid containing all the l-oleandrose genes from the avermectin biosynthetic gene cluster in Streptomyces avermitilis (27). When antibiotic selection of transformants was needed, 100 μg of ampicillin/ml, 25 μg of thiostrepton/ml, or 25 μg of apramycin/ml was used.
DNA manipulation and sequencing.
Plasmid DNA preparations, endonuclease digestions, alkaline phosphatase treatments, ligations, and other DNA manipulations were carried out according to standard procedures for E. coli (21) and for Streptomyces (11).
Constructs for oleY expression in E. coli
A 601-bp fragment containing the 5′ end of oleY was amplified by PCR. The following oligoprimers were used: 5′ GGAATTCTCATGACGTACGACGACCAC 3′ for the 5′ end and 5′ ATCAAGCTTATGCCGATCCCTAGGCTA 3′ for the 3′ end. An EcoRI site was included in the first oligoprimer and a HindIII site was included in the second to facilitate subcloning, and these sites are underlined. In addition, a BspHI site was also included in the 5′ oligoprimer in a region that includes the ATG starting codon to allow subcloning of the gene in expression vectors. PCR conditions were as follows: 100 ng of template DNA was mixed with 30 pmol of each primer and 2 U of Vent DNA polymerase (New England Biolabs) in a total reaction volume of 50 μl containing 2 mM concentration of each deoxynucleoside triphosphate, 10 mM KCl, 10 mM (NH4)2SO4, 20 mM Tris-HCl (pH 8.8), 2 mM MgSO4, 0.1% Triton X-100, and 10% dimethyl sulfoxide (DMSO) (Merck). Polymerization reaction was performed in a thermocycler (MinyCycler; MJ Research) under the following conditions: an initial denaturation of 3 min at 98°C; 30 cycles of 30 s at 98°C, 1 min at 67°C, and 3 min at 72°C; after the 30 cycles, an extra extension step of 5 min at 72°C was added. The PCR product was purified and subcloned into pUC18 as an EcoRI-HindIII fragment, generating pLR14b-1. To add the 3′ end of oleY, this construct was digested with BamHI and HindIII and the released fragment was replaced by a 2.7-kb BamHI-HindIII fragment containing the 3′ end of oleY, the entire oleP, and the 3′ end of oleB, thus reconstituting the entire oleY in pLR14b-2. From this a 1.6-kb BspHI-StuI fragment containing the entire oleY and the 5′ end of oleP was rescued and subcloned into the NcoI-BamHI sites (this last site blunt-ended with Klenow polymerase) of the E. coli expression vector pQE60. This final construct, pLR14b-3, was used for overexpression of oleY in E. coli.
Constructs for oleY expression in Streptomyces
pLR14b-3 was digested with EcoRI and BglII. In this way oleY, preceded by the ribosomal binding site of pQE60, was rescued. This fragment was subcloned into the EcoRI-BamHI sites of the shuttle (E. coli-Streptomyces) expression vector pUWL201. The final construct, pLR14b-4, was used to transform S. lividans TK21 protoplasts. In this construct, expression of oleY is under the control of the strong constitutive erythromycin resistance gene promoter (ermEp) from S. erythraea.
Expression in E. coli and S. lividans
E. coli harboring pLR14b-3 was incubated in 2× TY medium overnight in the presence of 100 μg of ampicillin/ml. Fresh medium was inoculated, and when the culture reached an absorbance at 580 nm of approximately 0.4, induction was initiated by adding a 1 mM final concentration of isopropyl-β-d-thiogalactopyranoside (IPTG). At different time intervals, samples were removed and washed twice by centrifugation and the cells were broken by using an MSE ultrasonic disintegrator at 150 W and 20 kHz (5 pulses of 15 s each with intermittent cooling on ice water). Soluble and insoluble fractions were separated by centrifugation.
For expression in S. lividans, the clone harboring pLR14b-4 was incubated for different time intervals in YEME medium containing 5 μg of thiostrepton/ml. The mycelia were recovered by centrifugation, washed twice in distilled water, and subjected to ultrasonic disruption (5 pulses of 5 s each with intermittent cooling on ice water). Soluble and insoluble fractions were separated by centrifugation.
Purification of the OleY methyltransferase.
S. lividans LR14b-4, a recombinant strain that overexpresses oleY (see Results), was used as a source of OleY methyltransferase for enzyme purification. A spore suspension of this strain was used to inoculate a 250-ml Erlenmeyer flask containing 25 ml of TSB (Trypticase soy broth; Oxoid) liquid medium in the presence of 5 μg of thiostrepton/ml. This culture was incubated for 24 h at 30°C on an orbital shaker incubator (200 rpm) and used to inoculate several 2-liter Erlenmeyer flasks containing 500 ml of TSB medium each (also supplemented with 5 μg of thiostrepton/ml) at a 1:100 dilution. After 48 h of incubation at 30°C in an orbital shaker, the mycelia were collected by centrifugation and washed twice with distilled water. The sample was suspended in buffer A (50 mM Tris-HCl buffer, pH 8.0, 1 mM dithiothreitol) and disrupted by two passes through a French press at a pressure of 1,500 lb/in2. DNA was broken by ultrasonic disruption (3 pulses of 15 s each with intermittent cooling on ice water) at 150 W and 20 kHz. Unbroken cells and cellular debris were removed by centrifugation at 30,000 × g for 15 min. Nucleic acids were precipitated with streptomycin sulfate (1% final concentration), and the precipitates were removed by centrifugation. The supernatant was fractionated by precipitation with ammonium sulfate. Fractions precipitating between 25 and 50% saturation were recovered by centrifugation and diluted into buffer A to reach an ammonium sulfate final concentration of 0.8 M. This fraction was applied to a Phenyl-Sepharose 6 Fast Flow (High Sub) column (Pharmacia) previously equilibrated with buffer A plus 0.8 M ammonium sulfate at a flow rate of 3 ml/min. Elution took place with a decreasing linear gradient (0.8 to 0 M), active fractions were concentrated by ammonium sulfate precipitation (95% saturation), and after centrifugation, the precipitates were resuspended in 4 ml of buffer B (50 mM Tris-HCl [pH 8.0], 20% glycerol, 150 mM NaCl). The sample was applied to a Sephacryl S-200 column (2.6 by 90 cm) at a flow rate of 0.3 ml/min. Active fractions were pooled, extensively dialyzed against buffer C (50 mM Tris-HCl, pH 8.0, 20% glycerol), and applied to a Q-Sepharose column (20-ml volume) at a flow rate of 3 ml/min. The column was eluted with a linear gradient of NaCl (0 to 0.6 M followed by a quick salt increase to 1 M) in buffer C. Fractions containing the desired enzymatic activity were pooled and kept in aliquots at −70°C until use.
Enzyme assays.
Methylation of different substrates by OleY methyltransferase was carried out in a reaction mixture containing (for a 50-μl final volume) 1 μl of S-adenosyl-l-[methyl-3H]methionine (0.1 mCi/ml; specific activity, 68 Ci/mmol), 1 μl of different macrolide substrates (100 μM final concentration), and 48 μl of extract or purified protein. After different time incubations at 30°C, the reactions were extracted with 50 μl of ethyl acetate. The radioactivity in 25-μl samples was determined after evaporation, resuspension in 100 μl of methanol, and addition of scintillation cocktail.
Protein determinations and PAGE.
Protein concentration in the different samples was determined by the protein-dye binding assay (3). Expression of oleY in E. coli and in S. lividans was followed by polyacrylamide gel electrophoresis (PAGE) in the presence of sodium dodecyl sulfate (SDS) as described previously (14). Gel staining was carried out with Coomassie blue.
Biotransformation experiments.
Spores of the appropriate recombinant strains were used to inoculate agar plates with solid R5A medium (9), supplemented with 25 μg of thiostrepton/ml and 100-μg/ml concentrations of different macrolides (spiramycin, niddamycin, carbomycin) or glycosylated derivatives (EB, l-olivosyl-erythronolide B[OLV-EB], RHA-EB, and MYC-EB). After 5 days of incubation at 28°C, 2-cm2 pieces of agar were extracted with 2 ml of ethyl acetate and the extracts were evaporated under vacuum and redissolved in 20 μl of methanol.
Chromatographic techniques.
Analysis by thin-layer chromatography (TLC) and high-performance liquid chromatography (HPLC) of the EB-related compounds were performed as described previously (1).
RESULTS
Expression of the OleY methyltransferase gene.
The OleY methyltransferase gene was expressed using different expression systems both in E. coli and in S. lividans. After induction by IPTG of the E. coli strain containing pLR14b-3, a derivative of expression vector pQE60, a protein band of an Mr similar to that expected from the gene sequence was observed in Coomassie blue-stained gels. Analysis of soluble and insoluble fractions by SDS-PAGE showed that most OleY protein was found in the insoluble fraction, probably as inclusion bodies. To attempt its expression as a soluble protein, oleY was also expressed in S. lividans using the expression vector pUWL201. In this case, after incubation of the clone carrying pLR14b-4, a band of a size similar to that expected for OleY was observed, and most of the protein was found in the soluble fraction.
The OleY methyltransferase acts on monoglycosylated macrolactones.
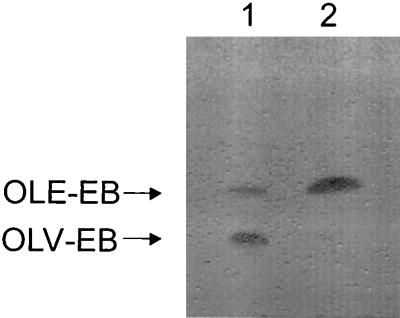
We carried out a series of experiments in order to identify the substrate of the OleY methyltransferase. Initially we tested the possibility that the OleY methyltransferase acts on a dTDP-activated sugar intermediate. Cell extracts of S. lividans LR14b-4 overexpressing oleY were incubated in the presence of an enzymatic system containing dTDP-4-keto-2,6-dideoxyglucose, extracts of two E. coli clones (expressing RmlC 3,5-epimerase and RmlD 4-ketoreductase, respectively), S-adenosyl-methionine, NADPH, and MgCl2 in order to assess action of OleY on a sugar biosynthetic intermediate through its conversion to l-oleandrose. In spite of multiple attempts with different modifications of the system, all the results were unsuccessful (Leticia Rodríguez and Steffan Amman, unpublished results). Subsequent efforts assessed whether OleY methyltransferase acts on the sugar after transfer to the aglycon. Cosmid cosAB35 contains part of the oleandomycin polyketide synthase, a gene involved in resistance to oleandomycin and its secretion, two glycosyltransferases responsible for sugar transfer, cytochrome P-450, and some sugar biosynthesis genes (15). The latter genes include two d-desosamine-specific genes (oleP1 and oleM1) and one gene specific for synthesis of l-oleandrose, oleY. When an S. lividans clone harboring cosAB35 was fed with OLV-EB, a unique biotransformation product was obtained (approximately 40% conversion) as analyzed by TLC (Fig. 2, lane 1). The material in this spot was characterized by HPLC and found to be identical in retention time and absorption spectrum to pure OLE-EB, a previously isolated and characterized compound (1). Since the only difference between these two monoglycosylated macrolactones is the presence of a methyl group at the 3-hydroxy group of l-olivose, this result prompted us to anticipate that methylation of l-olivose was carried out by OleY and that this methylation event occurred once the sugar was attached to the aglycon. To confirm this assumption, a second biotransformation was carried out. Plasmid pLR14b-4 was introduced in S. albus NAG2, a recombinant strain that contains the erythromycin resistance gene integrated into the chromosome and that is therefore resistant to all macrolides. The resultant recombinant strain was fed with OLV-EB, and after incubation the products were analyzed by TLC. Again, a new spot corresponding to OLE-EB was produced with high efficiency (Fig. 2, lane 2).
FIG. 2.
TLC analysis of the reaction products of biotransformation of OLV-EB by S. lividans containing cosAB35 (lane 1) or pLR14b-4 (lane 2). The arrows indicate the mobility of OLV-EB and OLE-EB.
To assess the substrate specificity of the OleY methyltransferase, two other monoglycosylated derivatives were tested: RHA-EB and MYC-EB. Incubation of S. albus NAG2 (pLR14b-4) with these two compounds yielded two new TLC spots, the efficiency being much higher in the case of RHA-EB than in the case of MYC-EB (data not shown). No bioconversion was found when other glycosylated macrolide antibiotics (spiramycin, niddamycin, carbomycin) were added to cultures of S. albus NAG2(pLR14b-4). These three macrolides contain l-mycarose moieties with a free 3-hydroxy group but were not methylated by OleY.
The OleG2 glycosyltransferase does not transfer l-oleandrose.
S. albus NAG2 harboring pOLV or pOLE was able to convert EB into OLV-EB or OLE-EB by biotransformation (1). Based on the above shown results, OleG2 glycosyltransferase transfers l-olivose, which is further converted into l-oleandrose by the OleY methyltransferase. We wondered if OleG2 was also able to recognize and transfer l-oleandrose. In the course of the present work, an l-oleandrose-synthesizing plasmid (pWHM2109) was made available to us. This plasmid contains seven genes (avrCDEFGHI) of the l-oleandrose gene cluster from the avermectin producer S. avermitilis; it directs the biosynthesis of l-oleandrose as shown by the incorporation of this neutral sugar into the avermectin aglycon (27). Protoplasts of S. albus NAG2 were transformed with pOLV, pOLE, and pWHM2109, and selected transformants from each transformation were fed with either EB or OLV-EB for bioconversion. In the presence of either pOLV or pOLE, EB was converted into OLV-EB and OLE-EB, respectively, according to experiments previously described (1). However, bioconversion of neither EB nor OLV-EB was observed in the recombinant strains containing pWHM2109. Since this plasmid directs the biosynthesis of l-oleandrose, the conclusion can be drawn that the formed l-oleandrose sugar is not recognized by the OleG2 glycosyltransferase.
Purification and characterization of the OleY methyltransferase.
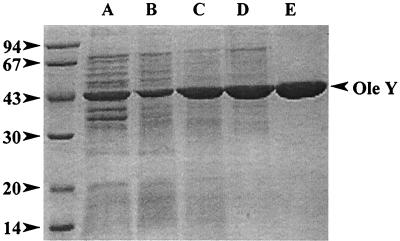
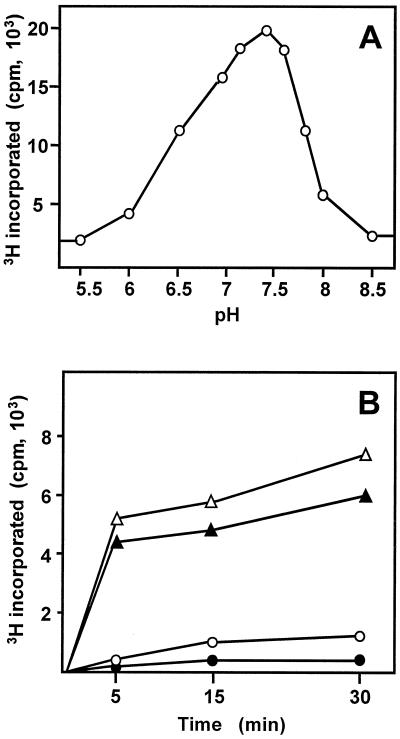
Using as starting material the S. lividans(pLR14b-4) recombinant strain (overproducing OleY), OleY methyltransferase was purified using a four-step purification procedure: ammonium sulfate precipitation and three chromatographic steps using hydrophobic (Phenyl-Sepharose), gel filtration (Sephacryl S-200), and anionic exchange (Q-Sepharose) resins (Table 1). The purified enzyme showed a single band of about 43 kDa on SDS-PAGE and Coomassie staining, as expected, for the deduced product of oleY (Fig. 3). The enzyme was quite stable (in the presence of 20% glycerol) when kept at low temperature, keeping most of the activity after 30 days at −70°C. The native molecular mass of the enzyme was approximately 87 kDa, indicating that the native enzyme acts as a dimer. Activity of the enzyme occurred within a narrow range of pH, with optimal pH between 7.2 to 7.6 and the activity drastically decreasing below pH 6.5 and over pH 7.8 (Fig. 4A). Methylation activity was stimulated by the presence of different divalent cations (100 μM final concentrations), such as Co2+ (11-fold), Mn2+ (7-fold), Zn2+ (5-fold), Mg2+ (3.5-fold), and Fe2+ (1.5-fold), whereas the enzyme activity was strongly inhibited by the presence of Hg2+ and Cu2+ or 1 mM EDTA. Different glycosylated substrates were used for methylation. OLV-EB and RHA-EB were good substrates for the enzyme, but MYC-EB was a poor substrate (Fig. 4B). No methylation was found when niddamycin, carbomycin, or spiramycin was used as substrate.
TABLE 1.
Purification of OleY methyltransferase activity
| Purification step | Sp act (pmol · min−1 · mg of protein−1) | Purification (fold) |
|---|---|---|
| Cell extraction | 0.03 | 1 |
| (NH4)2SO4 precipitation | 0.04 | 1.52 |
| Phenyl-Sepharose elution | 0.05 | 1.52 |
| Sephacryl S-200 elution | 0.99 | 29.77 |
| Q-Sepharose elution | 2.56 | 76.89 |
FIG. 3.
SDS-PAGE analysis of the different steps in the purification of the OleY methyltransferase. Lane A, cell extract; lane B, ammonium sulfate precipitation; lane C, active fractions eluted from the phenyl-Sepharose column; lane D, active fractions eluted from the Sephacryl S-200 column; lane E, active fractions eluted from the Q-Sepharose column.
FIG. 4.
Characterization of OleY methyltransferase. (A) Influence of pH on the OleY activity. The enzyme activity was measured using OLV-EB as substrate (100 μM) at different pHs in 50 mM phosphate buffer. (B) Methylating activity of OleY on different glycosylated macrolide derivatives. Assays were carried out in the presence of 100 μM OLV-EB (Δ), 100 μM RHA-EB (▴), 100 μM MYC-EB (○), or no substrate (control [●]).
DISCUSSION
Seven genes have been identified to be involved in the biosynthesis of the sugar l-oleandrose in the oleandomycin gene cluster in S. antibioticus (1, 16). Five of these genes (oleV, oleW, oleL, oleU, and oleY) code for l-oleandrose-specific enzymes, and two of them (oleS and oleE) code for enzymes catalyzing early common steps in the biosynthesis of the two deoxysugars in oleandomycin, l-oleandrose and d-desosamine. l-Oleandrose is a 2,6-dideoxysugar that possesses a 3-O-methyl group. Comparison of oleY-deduced product with proteins in databases showed similarities with two other proteins proposed to encode sugar O-methyltransferases: TylE from Streptomyces fradiae, a tylosin producer (10), and SnogY from Streptomyces nogalater, a nogalamycin producer (28). OleY was suggested to be responsible for catalyzing the 3-O-methylation step during l-oleandrose biosynthesis (1, 16). It was assumed that this methylation, converting l-olivose into l-oleandrose, would occur during the biosynthesis of the sugar, with l-oleandrose being transferred to the oleandomycin macrolactone ring. This assumption was supported by the isolation of this neutral sugar in its activated form (dTDP-l-oleandrose) from S. avermitilis, a producer of the antihelmintic avermectin that also contains l-oleandrose (as a disaccharide) attached to the aglycon (22). Evidence reported in this paper indicates that the OleG2 glycosyltransferase transfers the unmethylated derivative, l-olivose, which is then converted into l-oleandrose once attached to the oleandomycin aglycon. In vivo biotransformation experiments and in vitro enzymatic assays show that oleY codes for a methylating enzyme able to convert the l-olivose-containing monoglycosylated aglycon (OLV-EB) into its 3-O-methylated derivative, OLE-EB. Only a few other examples of sugar methylation taking place once the sugars have been attached to the aglycon have been reported. TylE and TylF are two O-methyltransferases catalyzing final steps in tylosin biosynthesis in S. fradiae by successively methylating the hydroxy groups at C-2 and C-3 positions of the 6-deoxyallose sugar moiety, thus converting demethylmacrocin into macrocin and then into tylosin (10, 23). These two enzymes act once the three sugars in tylosin have been incorporated into the tylactone. In erythromycin biosynthesis by S. erythraea, eryG codes for a 3-O-methyltransferase that converts l-mycarose into l-cladinose (and therefore erythromycin C into erythromycin A). This is the final step in erythromycin biosynthesis and occurs once both sugars, l-mycarose and d-desosamine, are attached to the aglycon (17). The action of the OleY methyltransferase differs from that of these three methyltransferases. Thus, TylE, TylF, and EryG methylate fully glycosylated intermediates catalyzing final steps in tylosin and erythromycin biosynthesis, respectively. In contrast, OleY acts on a monoglycosylated intermediate that is further converted into a diglycosylated derivative by the transfer of d-desosamine by the OleG1 glycosyltransferase. EryG and OleY methylate sugars (l-mycarose and l-olivose, respectively) attached to the same position of 14-membered aglycons, and they act on a hydroxy group attached to the same carbon atom of the sugar. However, their similarity at the protein level is quite low (18% identical amino acids). This can be due to the fact that the two enzymes act on different sugar substrates and at different steps in the biosynthesis of the corresponding macrolide antibiotic. However, EryG is also able to methylate OLV-EB as OleY does. When a S. erythraea mutant lacking the polyketide synthase genes (JC2 mutant) (20) was grown in the presence of either OLV-EB or OLE-EB, an identical diglycosylated erythronolide B derivative was found in both cases after the biotransformation experiment (A. F. Braña, unpublished results). This should be the result of the incorporation of the d-desosamine moiety by the EryCIII glycosyltransferase (in both OLV-EB and OLE-EB experiments) followed by a 3-O-methylation event (mediated by the EryG enzyme in the case of the OLV-EB experiment). It is noticeable that methylation of l-olivose by EryG occurs in a hydroxy group that shows different stereochemistry than the hydroxy group at l-mycarose (usual substrate for EryG).
OleY methyltransferase was purified to near homogeneity in a four-step procedure and quite efficiently converted OLV-EB and also RHA-EB into 3′-O-methylated derivatives. However, methylation of MYC-EB was less efficient. Some structural features that differentiate l-mycarose from l-olivose and l-rhamnose are the presence of a methyl group at C-3 of l-mycarose and the different stereochemistry of the hydroxy group at C-3. These differences could account for the low methylating activity of OleY on MYC-EB. The native OleY enzyme was found to be a dimer, which is in agreement with two other macrolide sugar O-methyltransferases previously characterized, TylE and TylF, which were found to be a trimer and a dimer, respectively (2, 13).
l-Oleandrose is not a sugar frequently found in nature. As far as we know it is present in only two macrolide bioactive compounds: the antibacterial oleandomycin and the antihelmintic avermectin. Interestingly, the producer organisms of these two compounds seem to have evolved two different pathways for synthesis and incorporation of the same sugar into the aglycon. In S. avermitilis, l-oleandrose is synthesized as a nucleotide-activated sugar that is transferred to the aglycon by a glycosyltransferase which incorporates two successive l-oleandrose moieties (12, 27). In contrast, the oleandomycin producer S. antibioticus synthesizes the unmethylated derivative, l-olivose, which is transferred to the aglycon by the OleG2 glycosyltransferase and then converted into l-oleandrose by the OleY methyltransferase (Fig. 5). This intermediate is subsequently glycosylated with d-desosamine by the OleG1 glycosyltransferase, thus generating the final compound, oleandomycin. Data shown in this paper demonstrate that OleG2 cannot transfer l-oleandrose if it is present in the cytoplasm in its nucleotide-activated form, thus excluding a role for OleG2 as an l-oleandrose glycosyltransferase. Another difference between the two sugar pathways resides in the 2-deoxygenation step. Two enzymes are required to catalyze this deoxygenation, a dehydratase and a reductase. Experimental biochemical evidence for the activity of these two enzymes has been reported for the biosynthesis of granaticin and oleandomycin in Streptomyces violaceoruber Tü22 and in S. antibioticus Tü99, respectively (8), and for the biosynthesis of tylosin in S. fradiae (5). 2,3-Dehydration is first catalyzed by a small family of Zn2+-dependent dehydratases (5) that share a high degree of similarity at the protein level. Two mechanisms exist for catalyzing the C-3 reduction step. In one way, exemplified by the Gra-orf26 or the Tü99-orf11, the reduction forms a sugar in which the C-3 hydroxy group is equatorial (8). In the other approach, exemplified by TylC2, the resulting hydroxy group is axial (5). The reduction step in the biosynthesis of l-olivose in S. antibioticus is carried out by OleW. This protein is identical at the protein level to the Tü99-orf11 reductase, and therefore the hydroxy group at C-3 should be equatorial. On the other hand, in l-oleandrose biosynthesis in S. avermitilis the enzyme responsible for the reduction step, AveBVIII, shows a high similarity to TylC2 (60% identical amino acids), and therefore it should produce a sugar intermediate with an axial hydroxy group at C-3.
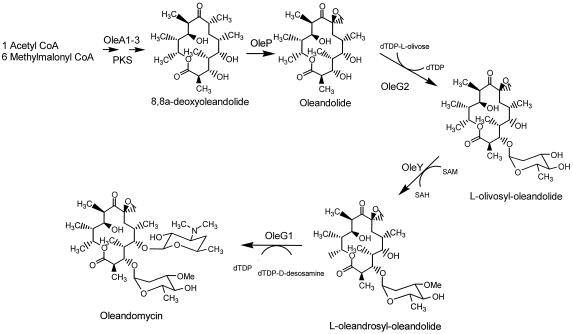
FIG. 5.
Proposed pathway for the post-polyketide steps in oleandomycin biosynthesis.
ACKNOWLEDGMENTS
This research was supported by grants from the European Union (BIO4-CT96-0080 and QLK3-CT-1999-00095).
We thank Glaxo Smithkline Beecham (Stevenage, United Kingdom) for providing EB, MYC-EB, and RHA-EB, Peter F. Leadlay for providing the S. erythraea JC2 mutant, and S.-E. Wohlert and C. R. Hutchinson for providing plasmid pWHM2109.
REFERENCES
- 1.Aguirrezabalaga I, Olano C, Allende N, Rodriguez L, Braña A F, Méndez C, Salas J A. Identification and expression of genes involved in l-oleandrose biosynthesis and its intermediate l-olivose in the oleandomycin producer Streptomyces antibioticus. Antimicrob Agents Chemother. 2000;44:1266–1275. doi: 10.1128/aac.44.5.1266-1275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer N J, Kreuzman A J, Dotzlaf J E, Yeh W K. Purification, characterization, and kinetic mechanism of S-adenosyl-l-methionine:macrocin O-methyltransferase from Streptomyces fradiae. J Biol Chem. 1988;263:15619–15625. [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Bullock W O, Fernández J M, Short J M. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with β-galactosidase selection. BioTechniques. 1987;5:376. [Google Scholar]
- 5.Chen H, Agnihotri G, Guo Z, Que Chen N L S X H, Liu H-W. Biosynthesis of mycarose: isolation and characterization of enzymes involved in the C-2 deoxygenation. J Am Chem Soc. 1999;121:8124–8125. [Google Scholar]
- 6.Doumith M, Legrand R, Lang C, Salas J A, Raynal M C. Interspecies complementation in Saccharopolyspora erythraea: elucidation of the function of oleP1, oleG1 and oleG2 from the oleandomycin biosynthetic gene cluster of Streptomyces antibioticus and generation of new erythromycin derivatives. Mol Microbiol. 1999;34:1039–1048. doi: 10.1046/j.1365-2958.1999.01666.x. [DOI] [PubMed] [Google Scholar]
- 7.Doumith M, Weingarten P, Wehmeier U F, Salah-Bey K, Benhamou B, Capdevila C, Michel J M, Piepersberg W, Raynal M C. Analysis of genes involved in 6-deoxyhexose biosynthesis and transfer in Saccharopolyspora erythraea. Mol Gen Genet. 2000;264:477–485. doi: 10.1007/s004380000329. [DOI] [PubMed] [Google Scholar]
- 8.Draeger G, Park S-H, Floss H G. Mechanism of the 2-deoxygenation step in the biosynthesis of the deoxyhexose moieties of the antibiotics granaticin and oleandomycin. J Am Chem Soc. 1999;121:2611–2612. [Google Scholar]
- 9.Fernández E, Weiβbach U, Sánchez Reillo C, Braña A F, Méndez C, Rohr J, Salas J A. Identification of two genes from Streptomyces argillaceus encoding two glycosyltransferases involved in the transfer of a disaccharide during the biosynthesis of the antitumor drug mithramycin. J Bacteriol. 1998;180:4929–4937. doi: 10.1128/jb.180.18.4929-4937.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fouces R, Mellado E, Diez B, Barredo J L. The tylosin biosynthetic cluster from Streptomyces fradiae: genetic organization of the left region. Microbiology. 1999;145:855–868. doi: 10.1099/13500872-145-4-855. [DOI] [PubMed] [Google Scholar]
- 11.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces. A laboratory manual. Norwich, England: The John Innes Foundation; 1985. [Google Scholar]
- 12.Ikeda H, Nonomiya T, Usami M, Ohta T, Omura S. Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc Natl Acad Sci USA. 1999;96:9509–9514. doi: 10.1073/pnas.96.17.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kreuzman A J, Turner J R, Yeh W K. Two distinctive O-methyltransferases catalyzing penultimate and terminal reactions of macrolide antibiotic (tylosin) biosynthesis. Substrate specificity, enzyme inhibition, and kinetic mechanism. J Biol Chem. 1988;263:15626–15633. [PubMed] [Google Scholar]
- 14.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1971;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 15.Olano C, Rodriguez A M, Méndez C, Salas J A. A second ABC transporter is involved in oleandomycin resistance and its secretion by Streptomyces antibioticus. Mol Microbiol. 1995;16:333–343. doi: 10.1111/j.1365-2958.1995.tb02305.x. [DOI] [PubMed] [Google Scholar]
- 16.Olano C, Rodriguez A M, Michel J M, Méndez C, Raynal M C, Salas J A. Analysis of a Streptomyces antibioticus chromosomal region involved in oleandomycin biosynthesis that contains two glycosyltransferases responsible for glycosylation of the macrolactone ring. Mol Gen Genet. 1998;259:299–308. doi: 10.1007/s004380050816. [DOI] [PubMed] [Google Scholar]
- 17.Paulus T J, Tuan J S, Luebke V E, Maine G T, DeWitt J P, Katz L. Mutation and cloning of eryG, the structural gene for erythromycin O-methyltransferase from Saccharopolyspora erythraea, and expression of eryG in Escherichia coli. J Bacteriol. 1990;172:2541–2546. doi: 10.1128/jb.172.5.2541-2546.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quirós L M, Aguirrezabalaga I, Olano C, Méndez C, Salas J A. Two glycosyltransferases and a glycosidase are involved in oleandomycin modification during its biosynthesis by Streptomyces antibioticus. Mol Microbiol. 1998;28:1177–1186. doi: 10.1046/j.1365-2958.1998.00880.x. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez A M, Olano C, Méndez C, Hutchinson C R, Salas J A. A cytochrome P450-like gene possibly involved in oleandomycin biosynthesis by Streptomyces antibioticus. FEMS Microbiol Lett. 1995;127:117–120. doi: 10.1111/j.1574-6968.1995.tb07459.x. [DOI] [PubMed] [Google Scholar]
- 20.Rowe C J, Cortés J, Gaisser S, Staunton J, Leadlay P F. Construction of new vectors for high-level expression in actinomycetes. Gene. 1998;216:215–223. doi: 10.1016/s0378-1119(98)00327-8. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Schulman M D, Acton S L, Valentino D L, Arison B H. Purification and identification of dTDP-oleandrose, the precursor of the oleandrose units of the avermectins. J Biol Chem. 1990;265:16965–16970. [PubMed] [Google Scholar]
- 23.Seno E T, Balz R. Properties of S-adenosyl-l-methionine: macrocin O-methyltransferase in extracts of Streptomyces fradiae strains which produce normal or elevated levels of tylosin and in mutants blocked in specific O-methylations. Antimicrob Agents Chemother. 1981;20:370–377. doi: 10.1128/aac.20.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shah S, Xue Q, Tang L, Carney J R, Betlach M, McDaniel R. Cloning, characterization and heterologous expression of a polyketide synthase and P-450 oxidase involved in the biosynthesis of the antibiotic oleandomycin. J Antibiot. 2000;53:502–508. doi: 10.7164/antibiotics.53.502. [DOI] [PubMed] [Google Scholar]
- 25.Swan D G, Rodriguez A M, Vilches C, Méndez C, Salas J A. Characterisation of a Streptomyces antibioticus gene encoding a type I polyketide synthase which has an unusual coding sequence. Mol Gen Genet. 1994;242:358–362. doi: 10.1007/BF00280426. [DOI] [PubMed] [Google Scholar]
- 26.Vilches C, Hernández C, Méndez C, Salas J A. Role of glycosylation and deglycosylation in the biosynthesis of and resistance to oleandomycin in the producer organism Streptomyces antibioticus. J Bacteriol. 1992;174:161–165. doi: 10.1128/jb.174.1.161-165.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wohlert S-E, Lomovskaya N, Kulowski K, Fonstein L, Occi J L, Gewain K M, MacNeil D J, Hutchinson C R. Insights about the biosynthesis of the avermectin deoxysugar l-oleandrose through heterologous expression of Streptomyces avermitilis deoxysugar genes in Streptomyces lividans. Chem Biol. 2001;8:681–700. doi: 10.1016/s1074-5521(01)00043-6. [DOI] [PubMed] [Google Scholar]
- 28.Ylihonko K, Tuikkanen J, Jussila S, Cong L, Mäntsälä P. A gene cluster involved in nogalamycin biosynthesis from Streptomyces nogalater: sequence analysis and complementation of early-block mutations in the anthracycline pathway. Mol Gen Genet. 1996;251:113–120. doi: 10.1007/BF02172908. [DOI] [PubMed] [Google Scholar]