Abstract
The occurrence of neurotrophic tyrosine receptor kinase (NTRK) gene fusions in a wide range of tumor types presents an attractive opportunity for using a tropomyosin receptor kinase (TRK) inhibitor as cancer therapy. Recent clinical studies have demonstrated highly efficacious outcomes associated with the use of TRK inhibitors, such as larotrectinib and entrectinib in NTRK fusion‐bearing cancers, in both adult and pediatric populations. While NTRK gene fusions are commonly found in some uncommon adult and pediatric malignancies, they are also found, albeit rarely, in a wide range of more common malignancies. The potential value of testing for NTRK gene fusions in practically all advanced malignancies is underpinned by the remarkable therapeutic outcomes that TRK inhibitors offer. This requirement presents practical and financial challenges in real‐world oncological practice. Furthermore, different testing platforms exist to detect NTRK gene fusions, each with its advantages and disadvantages. It is, therefore, imperative to develop strategies for NTRK gene fusion testing in an attempt to optimize the use of limited tissue specimen and financial resources, and to minimize the turnaround time. A multidisciplinary task force of Singapore medical experts in both public and private sectors was convened in late 2020 to propose testing algorithms for adult colorectal tumors, sarcomas, non‐small cell lung cancer, and pediatric cancers, with particular adaptation to the Singapore oncological practice. The recommendations presented here highlight the heterogeneity of NTRK‐fusion positive cancers, and emphasize the need to customize the testing methods to each tumor type to optimize the workflow.
Keywords: cancers, next‐generation sequencing, NTRK gene fusion, NTRK testing algorithm, TRK inhibitor
1. BACKGROUND
Neurotrophic tyrosine receptor kinase (NTRK) gene fusions represent actionable genomic alterations of high clinical potential. 1 Physiologically, neurotrophin receptors interact with tropomyosin receptor kinase (TRK) proteins, whose normal biological functions include pain sensation, thermoregulation, memory formation, metabolic homeostasis, and proprioception. 2 Although TRK receptors are predominantly expressed in neuronal tissue, NTRK gene fusions have been found in a wide array of different adult and pediatric tumor types. NTRK gene fusions involve proto‐oncogenes of either NTRK1, NTRK2, or NTRK3. When fused in‐frame with an unrelated gene, the resulting TRK fusion protein, which consists of an intact tyrosine kinase domain, is constitutively active. This chimeric oncoprotein functions as a primary oncogenic driver, activating downstream signaling cascades continuously, via the Ras/mitogen‐activated protein kinase pathway, phospholipase C‐gamma pathway, and phosphatidylinositol‐3 kinase/Akt pathway. This causes the altered transcription of prodifferentiation genes, transcription factors, and prosurvival genes, respectively, thereby promoting tumorigenesis. 3 , 4 , 5
Generally, the occurrence of NTRK gene fusions in cancers can be categorized as low prevalence or high prevalence. Common malignancies typically harbor lower frequencies of NTRK gene fusions, from less than 1% in lung cancers, to 25% in pediatric and young adult differentiated thyroid carcinoma. 6 Conversely, rare cancers, such as secretory breast carcinoma and congenital infantile fibrosarcoma, are enriched for NTRK gene fusions, with frequencies approaching 90%. 7
TRK inhibitors have shown convincing efficacy as tumor‐agnostic drugs across a wide spectrum of adult and pediatric malignancies. Two promising agents 8 that have advanced the furthest in clinical development are larotrectinib 9 and entrectinib, 10 both of which have received accelerated regulatory approvals for the treatment of cancers harboring NTRK gene fusion. In the United States and European Union, larotrectinib is currently the only approved TRK inhibitor for pediatric patients under 12 years of age. 11 Larotrectinib targets TRK fusion proteins specifically, while entrectinib has multikinase activity against ALK, ROS1, and JAK. 12 Integrated clinical data for both larotrectinib and entrectinib have demonstrated objective response rates of 79% 13 and 57%, 14 respectively, with larotrectinib achieving higher than 90% objective response rates in the pediatric population. 15 For larotrectinib, median time to response was as short as 1.8 months, and median duration of response was as long as 35.2 months. 13 Observed responses were independent of histology, age, and the NTRK gene (NTRK1, NTRK2, or NTRK3) involved in the fusion. 16 , 17 Both TRK inhibitors are well‐tolerated, with a favorable safety profile. 13 , 17 In Singapore, both TRK inhibitors have been granted approval by the Health Sciences Authority for the treatment of adult and pediatric patients (12 years of age or older with entrectinib) with solid tumors that have an NTRK gene fusion without a known acquired resistance mutation, are locally advanced or metastatic or where surgical resection is likely to result in severe morbidity, and have progressed following prior therapies or have no satisfactory alternative treatments. 18 , 19
Unsurprisingly, with increasing usage of TRK inhibitors, acquired resistance to first‐generation TRK inhibitors has emerged. 20 , 21 , 22 , 23 Acquired resistance is a typical obstacle that commonly plagues small molecule inhibitors, limiting their clinical utility. 24 , 25 , 26 To overcome this acquired drug resistance, concerted efforts are underway to develop second‐generation TRK inhibitors. 27 , 28
2. CURRENT NTRK GENE FUSION TESTING PLATFORMS
Although clinical studies have clearly demonstrated the utility of TRK inhibitors, the challenge lies in identifying patients with cancers that harbor NTRK gene fusions. 29 Not only do NTRK gene fusions exist across a wide spectrum of cancers, there are also different types of laboratory techniques for the screening and diagnosis of NTRK gene fusions, each with its advantages and limitations. 30 In current clinical practice, the detection of NTRK gene fusions is often made via next‐generation sequencing (NGS). Indeed, the companion diagnostic that is approved by the United States FDA to identify NTRK gene fusions in solid tumors for TRK inhibitors is an NGS‐based assay. 31 RNA‐based NGS is preferable to the DNA‐based platform, particularly to detect novel fusion partners and to overcome the problem of large intronic regions. 32 Besides NGS, fluorescence in situ hybridization (FISH) and reverse‐transcriptase polymerase chain reaction (RT‐PCR) are alternatives that have seen successful deployment clinically. 15 However, FISH testing requires the use of three separate probes and assays, and may not identify intrachromosomal translocations, such as LMNA‐NTRK1, which results from a 1q intrachromosomal deletion. Conventional RT‐PCR lacks multiplex capabilities, and requires specific primers designed to target specific gene fusions. 33 Another complementary approach is immunohistochemistry (IHC). IHC is widely available and reimbursable across different parts of the world, is relatively inexpensive, and has a fast turnaround time of around 2 days. 32 However, with IHC lacking specificity and being a test for TRK protein overexpression, verification of IHC‐positive cases by molecular sequencing (preferably by NGS) is required to confirm the presence of NTRK gene fusions. One important limitation of IHC is its inability to distinguish abnormal TRK fusion proteins from wild‐type TRK proteins that are expressed in tumor tissues with neural and smooth muscle differentiation, such as glioblastoma, neuroblastoma, and gastrointestinal stromal tumors. 30 , 34 Finally, the scoring algorithms for IHC are not standardized. 35 Nonetheless, as IHC is easily deployed, it is reasonable to use it as a screening tool to enrich patient populations for NTRK gene fusions, 36 followed by NGS confirmation of pan‐TRK IHC‐positive cases. 37 In cancers with low prevalences of NTRK gene fusions, this two‐step approach is a pragmatic approach to maximize cost‐effectiveness, which is particularly relevant in nonreimbursable markets, where patients bear the testing costs out‐of‐pocket, as is the case in Singapore. This two‐step approach of IHC screening prior to molecular sequencing is also in line with international guidelines, such as those published by ESMO. 38 , 39 Although FISH and RT‐PCR are feasible tests for determining NTRK gene fusions, NGS, specifically the RNA‐based NGS, remains the preferred and most dependable method whenever it is available. 40
Given the high cost associated with the use of NGS, it is not practical to employ NGS upfront to detect NTRK gene fusions across all tumor types. 41 The treatment of some common cancers, such as non‐small cell lung cancer (NSCLC) and colorectal cancer, is heavily biomarker driven. In such cancers, NGS is already commonly employed in the initial diagnosis or at the time of disease progression in standard clinical practice. The search for NTRK gene fusions in these cancers, which are uncommon, is streamlined and fairly straightforward. On the other hand, there are cancers, such as sarcomas and certain pediatric tumors, where NTRK gene fusions are common. 42 In these cancers, it is imperative to specifically embed NTRK gene fusion testing in the diagnostic and management algorithm to better guide the treatment decisions. 43 , 44
To address these considerations, a multidisciplinary medical expert panel was convened in late 2020 to discuss, debate, and then recommend practice‐oriented guidelines pertaining to the NTRK gene testing and TRK inhibitor usage in the context of Singapore, with a specific focus on the aforementioned cancer types. This task‐force comprised experienced oncologists and pathologists from both the private and public healthcare sectors in Singapore. This review presents the consensus derived from this expert meeting.
3. WHEN TO CONSIDER TRK INHIBITOR IN CLINICAL PRACTICE?
TRK inhibitors offer clinically meaningful efficacy as a highly targeted tumor‐agnostic therapy, with NTRK gene fusion as an actionable target.16,17 TRK inhibitors are indicated for the treatment of adult and pediatric patients (12 years of age or older with entrectinib) with solid tumors that have an NTRK gene fusion without a known acquired resistance mutation, that are locally advanced, metastatic or where surgical resection is likely to result in severe morbidity, and who have no satisfactory alternative treatments or whose cancer has progressed following treatment. 18 , 19
Recommendations on when to consider TRK inhibitors for patients with metastatic/locally advanced cancers harboring NTRK gene fusions
Cancers which are positive for NTRK gene fusion, particularly cancers in which treatment options are limited, should be treated with a TRK inhibitor, whenever accessible. Of note:
► [Non‐small cell lung cancer] TRK inhibitors may be considered as first‐line therapy in specific circumstances, or as subsequent therapy following progression on standard first‐line therapy.
► [Colorectal cancer] TRK inhibitors may be considered as second‐line or third‐line therapy following progression on standard prior therapies.
► [Adult sarcomas] With limited treatment options available for advanced sarcomas, TRK inhibitors may be considered as first‐line therapy. If urgent treatment is clinically indicated and there are no other effective treatments available, based on expert panel opinion, TRK inhibitor may be initiated concurrently with a confirmatory NGS, for IHC‐positive cases in sarcoma subtypes enriched for NTRK gene fusions.
► [Pediatric cancers] In specific pediatric tumors known to harbor NTRK gene fusions, TRK inhibitors may be considered in the first‐line setting if no standard treatment is available, or as subsequent therapy.
4. WHEN AND HOW TO TEST FOR NTRK GENE FUSION IN CLINICAL PRACTICE?
Given the high objective response rates and the durability of such responses following TRK inhibitor treatment, it behoves the clinicians to consider NTRK gene fusion testing in the appropriate clinical setting. 45 Testing methodologies should take into consideration multiple factors, including prevalence of NTRK gene fusions in the specific tumor type, availability of testing platforms, turnaround time, and financial constraints. As recommended by ESMO, tumors in which NTRK gene fusions are highly prevalent are appropriately tested by using FISH and RT‐PCR assays. Otherwise, NGS, in particular RNA‐based sequencing, remains the diagnostic test of choice for confirmation. 38 In clinical practice, the decision to carry out NTRK gene fusion testing is often confounded by the competing availability of other (often less costly) conventional therapeutic options, and the accessibility to TRK inhibitors in that particular locale. The use of tumor‐specific algorithms may provide guidance to the clinicians in deciding when and how to carry out NTRK gene fusion testing, and when to employ TRK inhibitors in the management of cancer patients.
The algorithms presented here are intended to serve as a guide to oncologists and pathologists in Singapore who specialize in the management of NSCLC, colorectal cancer, sarcomas, and pediatric cancers.
Recommendations on when and how to test for NTRK gene fusions for metastatic/locally advanced NTRK‐fusion positive cancers
In general, if NGS is available, it is the preferred testing platform to confirm the presence of absence of NTRK gene fusions, particularly RNA‐based NGS for the detection of novel fusion partners. Alternatively, IHC can be used as a screening test, with NGS used as a confirmation test.
► [Non‐small cell lung cancer] NGS testing is recommended upfront to test for NTRK gene fusions alongside key oncogenic drivers (EGFR, ALK, ROS1, MET, BRAF, and PD‐L1). Alternatively, standard testing for key oncogenic drivers can be performed first. If oncogenic driver is absent, screening by pan‐TRK IHC can be considered, followed by a confirmatory NGS. Upon disease progression, retesting for NTRK gene fusions as well as other actionable molecular alterations may be considered.
► [Colorectal cancer] NGS is recommended upfront to test for NTRK gene fusions alongside key oncogenic drivers (KRAS, NRAS, and BRAF), together with the assessment of MSI status. Alternatively, standard testing for key oncogenic drivers can be performed first. At the time of disease progression after first‐ or second‐line therapies, NGS testing may be considered to seek out additional molecular alterations, including NTRK gene fusions, especially in MSI‐high patients with an absence of key oncogenic drivers.
► [Adult sarcomas] Pan‐TRK IHC is recommended as a screening test, followed by NGS confirmation for NTRK gene fusions. Interpretation of IHC results may be influenced by the sarcoma subtypes. If IHC results are negative for sarcoma subtypes enriched for NTRK gene fusions, NGS confirmation would be still recommended for cases with histological features suspicious for NTRK gene fusions. NTRK testing is currently optional for adult sarcomas with actionable genomic aberrations; however, further research is warranted.
► [Pediatric cancers] Apart from pediatric malignancies in which there is effective treatment resulting in good outcome, NGS testing may be considered upfront for molecular profiling of pediatric malignancies, with NTRK gene fusions as part of the routine diagnostic workup. If NGS is not accessible, FISH, RT‐PCR, and pan‐TRK IHC are reasonable testing alternatives, along with confirmation by NGS depending on the index of suspicion for the underlying NTRK gene fusions. NTRK testing is currently optional for tumors with a clear diagnosis of a tumor type not known to have NTRK gene fusions (e.g., Wilms tumor and neuroblastoma).
5. NTRK DIAGNOSTIC ALGORITHMS ON SELECTED CANCER TYPES
5.1. Non‐small cell lung cancer
In the case of NSCLC, there are a handful of key molecular oncogenic alterations which are targetable, including EGFR, ALK, and ROS1. 46 NTRK gene fusion joins the list of actionable oncogenic drivers for lung cancer. 47 As a bona‐fide target, results from clinical trials support the use of TRK inhibitors in NTRK gene fusion‐positive NSCLC 48 and is endorsed by the National Comprehensive Cancer Network guidelines, which recommend its use in both upfront and subsequent settings. In the presence of a positive NTRK1/2/3 gene fusion, including the situation where NTRK gene fusions are discovered prior to the initiation of first‐line systemic therapy, larotrectinib and entrectinib may be considered as a first‐line therapy in specific clinical circumstances. 49 Both larotrectinib and entrectinib are biologically active in the central nervous system (CNS), and their clinical activity in primary and metastatic CNS tumors is well documented. 8 , 13 , 50 , 51 , 52 , 53 , 54
The presence of numerous potential oncogenic drivers in NSCLC supports the use of NGS testing upfront to optimize tissue utilization. 55 At the time of disease progression, repeat molecular testing for genomic alterations, including NTRK gene fusions, may be valuable for at least two reasons. First, retesting allows a second opportunity to cast a wider net that could take into account technical limitations, such as different primer setups in different gene panels. In this case, a different testing platform could be used to reconfirm that the result is truly negative. Second, retesting may shed light on whether there has been a true biological switch or an emergence of new driver mutations. Refractory lung cancers often harbor alternate driver mutations, as compared to mutually exclusive drivers at the time of diagnosis when the tumor is treatment‐naive. 56 However, given the financial burden associated with repeat NGS testing, such decisions should be made on a case‐by‐case basis.
Although NGS testing upfront will be ideal, a major limitation is the long turnaround time of about 2–3 weeks to obtain results from the molecular analysis, which could delay the initiation of appropriate treatment. 57 Moreover, the utility of deploying NGS assays in NSCLC where there are only a few common actionable targets should be viewed from a cost‐effectiveness perspective. Indeed, most of the NGS results may end up being underutilized. A rational framework that deploys different tests at different phases of the lung cancer management would ensure more efficient use of the limited health resources. Single‐gene testing is still a valid approach to the molecular profiling of lung cancers, especially when NGS is inaccessible or unaffordable. When standard single‐gene testing fails to reveal any oncogenic driver, a screening pan‐TRK IHC may be considered to detect any possibility of NTRK gene fusion. A positive pan‐TRK IHC result should be confirmed before acted upon, preferably via an NGS.
FIGURE 1.
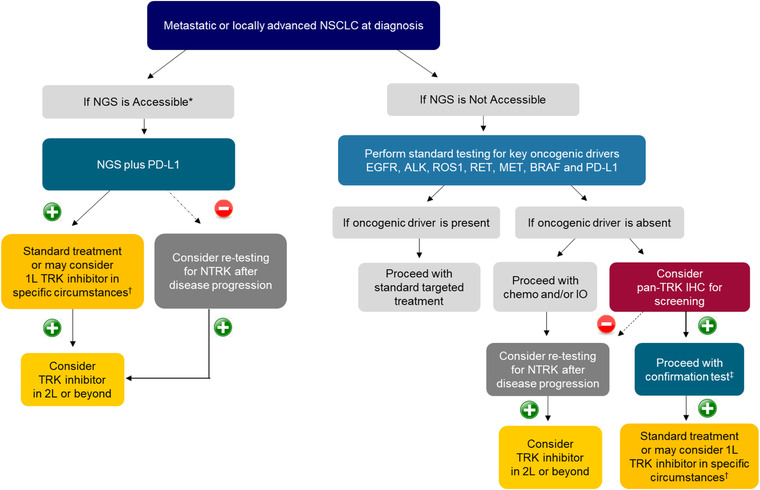
Recommended NTRK gene fusion testing algorithm for NSCLC. (+)/(–): Presence/absence of NTRK gene fusions. *Accessible: Testing platform is available and affordable. †In the 2021 NCCN Guidelines, 49 larotrectinib and entrectinib are recommended as first‐line therapy when NTRK gene fusion is positive. ‡For confirmation test, NGS is preferred, but FISH or RT‐PCR may be used if NGS is not accessible. Abbreviations: ALK, anaplastic lymphoma kinase; BRAF, v‐raf murine sarcoma viral oncogene homolog B; EGFR, epidermal growth factor receptor; IHC, immunohistochemistry; 1L/2L, first/second line; IO, immunotherapy; MET, MET proto‐oncogene, receptor tyrosine kinase; NGS, next‐generation sequencing; NSCLC, non‐small cell lung cancer; NTRK, neurotrophic tyrosine receptor kinase; PD‐L1, programmed death‐ligand 1; RET, proto‐oncogene tyrosine‐protein kinase receptor Ret; ROS1, ROS proto‐oncogene 1, receptor tyrosine kinase
5.2. Colorectal cancer
Microsatellite or mismatch repair protein status has important upfront therapeutic implications in the management of advanced colorectal cancers. 58 Current standard algorithm for colorectal cancer tests for microsatellite status upfront, regardless of the availability of NGS. This is aligned with international clinical practice guidelines. 59
In addition to testing for microsatellite instability (MSI), it is also standard to test for the major predictive molecular markers in colorectal cancers, namely, KRAS, NRAS, and BRAF. An alternative to this single‐gene testing is NGS testing, which includes the testing for NTRK gene fusion. Such NGS testing may be deployed at the time of diagnosis of advanced colorectal cancer, or deferred to the time when the patient experiences disease progression after standard first‐line treatment.
Interestingly, NTRK gene fusions appear to be more prevalent in MSI‐high colorectal cancers, compared to tumors which are microsatellite stable. 60 , 61 , 62 This is particularly true if the tumors are shown to be wild‐type for KRAS, NRAS, and BRAF, or are MLH1 promoter hypermethylated. 63 Therefore, it is recommended that MSI‐high colorectal cancers be tested for NTRK gene fusions via NGS testing, especially if the tumors are not found to have mutation in the KRAS, NRAS, and BRAF genes, nor are MLH1 promoter hypermethylated. Nonetheless, as there have been rare reports of KRAS mutation being identified in MSI‐high colorectal cancers that harbor NTRK gene fusions, 62 NTRK gene fusions should ideally be tested in all MSI‐high colorectal cancers, regardless of the RAS/RAF mutation status.
5.3. Adult sarcomas
Sarcomas are rare but heterogeneous cancers, making up only 1% of adult malignancies, but comprising more than 70 histologic subtypes. Systemic therapy of sarcomas is an area of unmet clinical need, with the vast majority of subtypes having a paucity of actionable driver genetic aberrations, in addition to being relatively resistant to cytotoxic chemotherapy. 64 In this landscape of relatively limited therapeutic choices, NTRK gene fusion is a particularly attractive actionable target in the management of patients with sarcomas, including potentially as first‐line therapy for selected cases with no satisfactory alternative therapy. 44 ,
FIGURE 2.
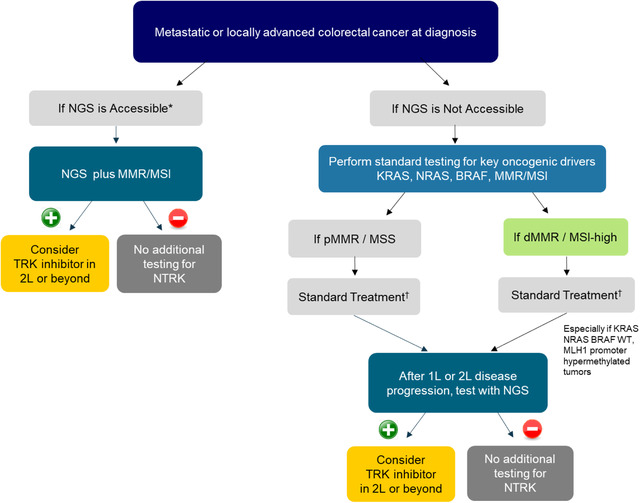
Recommended NTRK gene fusion testing algorithm for colorectal cancer. (+)/(–): Presence/absence of NTRK gene fusions. *Accessible: Testing platform is available and affordable. †Standard treatment: If pMMR/MSS, 1L/2L‐chemotherapy and/or monoclonal antibody, 3L/4L: regorafenib or TAS‐102; if dMMR/MSI‐high: 1L‐pembrolizumab or chemotherapy with or without monoclonal antibody, 2L/3L: chemotherapy and/or monoclonal antibody or pembrolizumab. Abbreviations: BRAF, v‐raf murine sarcoma viral oncogene homolog B; 1L/2L/3L/4L, first/second/third/fourth line; KRAS, Kirsten rat sarcoma viral oncogene homolog; MLH1, MutL Homolog 1; MSI, microsatellite instability; MSS, microsatellite stable; NGS, next‐generation sequencing; NRAS, neuroblastoma RAS viral oncogene homolog; NTRK, neurotrophic tyrosine receptor kinase; p/d/MMR, proficient‐/deficient‐/mismatch repair; WT, wild type
In line with the recommendations from the World Sarcoma Network, 44 a targeted approach to diagnostic testing in sarcomas is recommended based on the prior probability of harboring NTRK gene fusions. For sarcoma subtypes enriched for NTRK gene fusions (e.g., inflammatory myofibroblastic tumor negative for ALK and ROS1 fusions), screening with pan‐TRK IHC is recommended upfront, followed by a confirmatory NGS. If IHC results are positive, treatment with a TRK inhibitor may be initiated concurrently with a confirmatory NGS, if such urgent treatment is clinically indicated and there are no other effective alternative treatments available. If IHC results are negative, NGS confirmation is still recommended to exclude false‐negative IHC, given the relatively high proportion of these tumors harboring NTRK gene fusions.
For sarcoma subtypes associated with low frequency of NTRK gene fusions (e.g., KIT and PDGFRA wild‐type gastrointestinal stromal tumor or sarcomas with complex genomics not associated with characteristic translocations), IHC screening is the recommended initial test. However, positive IHC results in such subtypes should be interpreted with caution in case of myogenic and neural differentiation (e.g., gastrointestinal stromal tumor), due to the high rate of false positivity arising from the detection of wild‐type TRK proteins. 65 If IHC results are negative, routine confirmatory NTRK testing is not recommended, though it can be considered on a case‐by‐case basis following informed discussion regarding specific test characteristics taken in the context of prognosis and therapeutic options for individual cases. The sensitivity of pan‐TRK IHC has been reported to be relatively limited for NTRK3 fusions (55–79%) compared with NTRK2 (89–100%) and NTRK1 (88–96%). 44
For sarcomas that harbor known driver genomic aberrations, routine NTRK testing is not recommended. This is because cancers with known recurrent genetic drivers harboring concurrent driver NTRK fusions are vanishingly rare and have only been reported sporadically. 66 ,
FIGURE 3.
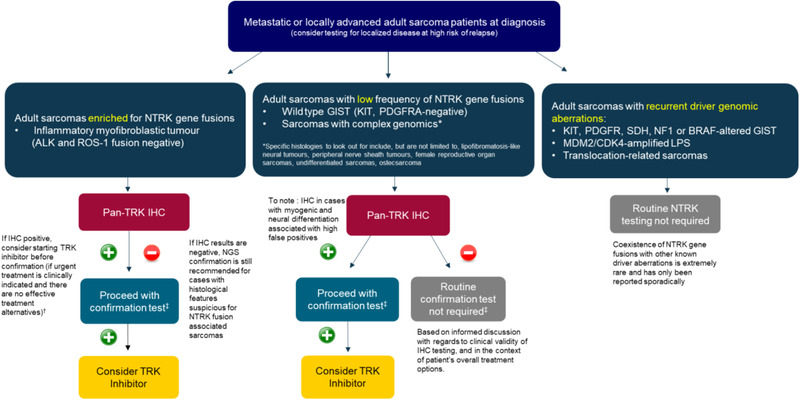
Recommended NTRK gene fusion testing algorithm for adult sarcomas, adapted from expert recommendations from the World Sarcoma Network. 44 (+)/(–): Presence/absence of NTRK gene fusions. †Please note that this is the opinion of the expert panel. Confirmation of NTRK gene fusions is required for TRK inhibitors. ‡For confirmation test, NGS is preferred, but FISH or RT‐PCR may be used if NGS is not accessible. Abbreviations: ALK, anaplastic lymphoma kinase; BRAF, v‐raf murine sarcoma viral oncogene homolog B; CDK4, cyclin‐dependent kinase 4; GIST, gastrointestinal stromal tumor; IHC, immunohistochemistry; KIT, v‐kit Hardy‐Zuckerman 4 feline sarcoma viral oncogene homolog; LPS, liposarcoma; MDM2, mouse double minute 2 homolog; NF1, neurofibromatosis type 1; NGS, next‐generation sequencing; NTRK, neurotrophic tyrosine receptor kinase; PDGFRA, platelet‐derived growth factor receptor alpha; ROS1, ROS proto‐oncogene 1, receptor tyrosine kinase; SDH, succinate dehydrogenase. *Specific histologies to look out for include, but are not limited to, lipofibromatosis‐like neural tumours, peripheral nerve sheath tumours, female reproductive organ sarcomas, undifferentiated sarcomas, osteosarcoma
5.4. Pediatric cancers
In pediatric cancers, the epidemiology of NTRK gene fusions follows the general trend observed in adult cancers, where NTRK gene fusions occur with higher frequencies in specific and rarer tumors. 67 The prevalence of NTRK gene fusions is low in certain pediatric tumors, such as neuroblastoma, Wilms tumor, and hepatoblastoma. In contrast, NTRK gene fusions are more commonly identified in certain rare childhood cancers, namely, infantile fibrosarcomas, gliomas, and thyroid cancers. 42 Pediatric cancers that harbor NTRK gene fusions tend to respond well to TRK inhibitors, with objective response rates reported to be as high as 90%. 68 In pediatric cancers where standard efficacious treatment is lacking, TRK inhibitors may be considered, particularly in aggressive pediatric sarcomas and gliomas where outcomes with conventional therapies are more modest.
Given that pediatric tumors are a highly heterogeneous group of undifferentiated malignancies, molecular profiling upfront may be considered to improve the detection rate of rare genomic alterations, including NTRK gene fusions. The use of NGS as the initial test is relevant in cases (e.g., core biopsies) where the amount of tumor tissue available for testing is limited.
Apart from NGS, the alternative testing platforms of FISH, RT‐PCR, and pan‐TRK IHC are reasonable if NGS is unavailable or unaffordable. Depending on the index of suspicion for the presence of an underlying NTRK gene fusion, a confirmation NGS can be performed after the screening test. It is critical to note that pediatric tumors are extremely varied, and every tumor type is handled differently. On top of that, the patient population is also different because molecular testing is done at different stages of disease and treatment plan. This algorithm described herein is a suggested decision‐making pathway, and will not necessarily apply to all tumor types and patients equally.
5.5. Future directions
While NGS is the endorsed confirmatory test for NTRK gene fusions locally and internationally, its accessibility presents a huge barrier to doctors and patients. Within the Association of Southeast Asian Nations (ASEAN) region, the level of access to NGS is widely varied, reflecting the different levels of technical expertise and financial affordability. Patient access to available treatments, such as TRK inhibitors, is also a vital factor that has a major impact on the real‐world diagnostic and clinical pathways. In addition, government support and third‐party reimbursement for precision medicine are also widely discrepant among the different countries, further complicating the eventual access to highly effective, but costly, new therapeutics.
FIGURE 4.
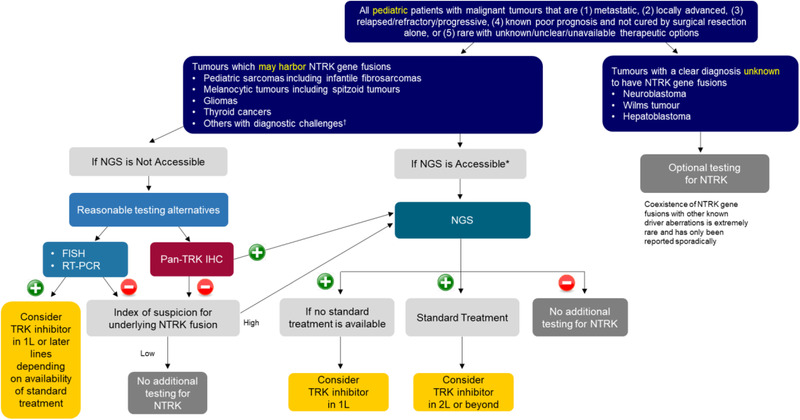
Recommended NTRK gene fusion testing algorithm for pediatric tumors. (+)/(–): Presence/absence of NTRK gene fusions. *Accessible: Testing platform is available and affordable. †Tumors that are challenging to diagnose on the basis of morphology and standard techniques alone. Note: The index of suspicion is high if there is a possibility that the tumor may harbor an NTRK gene fusion. The index of suspicion is low if the possibility that the tumor may harbor an NTRK gene fusion is low, for example, some other key oncogenic driver is identified. Abbreviations: FISH, fluorescence in situ hybridization; IHC, immunohistochemistry; 1L/2L, first/second line; NGS, next‐generation sequencing; NTRK, neurotrophic tyrosine receptor kinase; RT‐PCR, reverse transcription polymerase chain reaction
In this paper, different algorithms are devised for the four different tumor types. Hopefully, in the future, with greater and more uniform access to NGS testing, a more universal testing algorithm based on NGS may become a reality.
Moving forward, the development of a regional or international registry for gene fusions would promote data building and sharing, which is especially important for uncommon tumor types. Such a transnational registry would also encourage cooperation and research collaborations between different academic groups. In the realm of NTRK gene fusions and TRK inhibitors, such knowledge sharing should lead to better identification of NTRK gene fusion partners, and more precise clinical and histological characterization of TRK fusion cancers.
Education is an important enabling tool in any new therapeutic domain. Oncologists and pathologists should be made aware of the appropriate tests to screen for and confirm the presence of NTRK gene fusion, with a conscious effort to avoid wastage of precious tissue samples and financial resources. Interdisciplinary learning between oncologists and pathologists is to be encouraged, in order to streamline the diagnosis, molecular profiling, and the treatment of such cancers.
No official guidelines pertaining to NTRK gene fusion testing currently exist in Singapore. This paper represents a concerted interdisciplinary effort to provide a primer on NTRK gene testing in the context of existing therapeutic algorithms, not only for the Singapore medical practitioners, but also providing a template for similar efforts in other parts of the region.
CONFLICT OF INTERESTS
K.H.T.L., H.L.K., K.T.E.C., D.S.W.T., I.B.H.T., F.M., S.Y.S., B.N.K.P., R.A.S., W.S.H., L.A., and S.P.C. attended an advisory board meeting for Bayer about TRK inhibitors clinical use and NTRK gene fusion testing. K.H.T.L., H.L.K., K.T.E.C., D.S.W.T., I.B.H.T., F.M., and S.Y.S. received speakers fees from Bayer. B.N.K.P., R.A.S., W.S.H., L.A., and S.P.C. received consultancy fees from Bayer.
ACKNOWLEDGMENTS
Funding for the development of this manuscript was provided by Bayer (South East Asia) Pte Ltd. Support for medical writing services was provided by Teo Wan Ching Jocelyn, PhD from Alcimed Pte Ltd.
Lim KHT, Kong HL, Chang KTE, et al. Recommended testing algorithms for NTRK gene fusions in pediatric and selected adult cancers: Consensus of a Singapore Task Force. Asia-Pac J Clin Oncol. 2022;18:394–403. 10.1111/ajco.13727
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this publication.
REFERENCES
- 1. Amatu A, Sartore‐Bianchi A, Siena S. NTRK gene fusions as novel targets of cancer therapy across multiple tumour types. ESMO Open. 2016;1(2):e000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amatu A, Sartore‐Bianchi A, Bencardino K, Pizzutilo EG, Tosi F, Siena S. Tropomyosin receptor kinase (TRK) biology and the role of NTRK gene fusions in cancer. Ann Oncol. 2019;30(Suppl 8):viii5‐viii15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nakagawara A. TRK receptor tyrosine kinases: a bridge between cancer and neural development. Cancer Lett. 2001;169(2):107‐114. [DOI] [PubMed] [Google Scholar]
- 4. Khotskaya YB, Holla VR, Farago AF, Mills Shaw KR, Meric‐Bernstam F, Hong DS. Targeting TRK family proteins in cancer. Pharmacol Ther. 2017;173:58‐66. [DOI] [PubMed] [Google Scholar]
- 5. Tan F, Thiele CJ, Li Z. Neurotrophin signaling in cancer. Handbook of Neurotoxicity. New York: Springer New York; 2014. [Google Scholar]
- 6. Albert CM, Davis JL, Federman N, Casanova M, Laetsch TW. TRK fusion cancers in children: a clinical review and recommendations for screening. J Clin Oncol. 2019;37(6):513‐524. [DOI] [PubMed] [Google Scholar]
- 7. Cocco E, Scaltriti M, Drilon A. NTRK fusion‐positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol. 2018;15(12):731‐747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Laetsch TW, Hong DS. Tropomyosin receptor kinase inhibitors for the treatment of TRK fusion cancer. Clin Cancer Res. 2021;27(18):4974‐4982. [DOI] [PubMed] [Google Scholar]
- 9. Mullard A. FDA approves landmark tissue‐agnostic cancer drug. Nat Rev Drug Discov. 2019;18(1):7. [DOI] [PubMed] [Google Scholar]
- 10. Marcus L, Donoghue M, Aungst S, et al. FDA approval summary: entrectinib for the treatment of NTRK‐gene fusion solid tumors. Clin Cancer Res. 2021;27(4):928‐932. [DOI] [PubMed] [Google Scholar]
- 11. Scott LJ. Larotrectinib: first global approval. Drugs. 2019;79(2):201‐206. [DOI] [PubMed] [Google Scholar]
- 12. Ardini E, Menichincheri M, Banfi P, et al. Entrectinib, a pan‐TRK, ROS1, and ALK inhibitor with activity in multiple molecularly defined cancer indications. Mol Cancer Ther. 2016;15(4):628‐639. [DOI] [PubMed] [Google Scholar]
- 13. Hong DS, DuBois SG, Kummar S, et al. Larotrectinib in patients with TRK fusion‐positive solid tumours: a pooled analysis of three phase 1/2 clinical trials. Lancet Oncol. 2020;21(4):531‐540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rolfo C, Dziadziuszko R, Doebele RC, et al. Updated efficacy and safety of entrectinib in patients with NTRK fusion‐positive tumors: integrated analysis of STARTRK‐2, STARTRK‐1 and ALKA‐372‐001. Ann Oncol. 2019;30: Suppl 5: v180. [Google Scholar]
- 15. Laetsch TW, DuBois SG, Mascarenhas L, et al. Larotrectinib for paediatric solid tumours harbouring NTRK gene fusions: phase 1 results from a multicentre, open‐label, phase 1/2 study. Lancet Oncol. 2018;19(5):705‐714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Drilon A, Laetsch TW, Kummar S, et al. Efficacy of larotrectinib in TRK fusion–positive cancers in adults and children. N Engl J Med. 2018;378(8):731‐739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Drilon A, Siena S, Ou S‐HI, et al. Safety and antitumor activity of the multitargeted pan‐TRK, ROS1, and ALK inhibitor entrectinib: combined results from two phase I trials (ALKA‐372‐001 and STARTRK‐1). Cancer Discov. 2017;7(4):400‐409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Health Sciences Authority . New Drug Approvals ‐ Aug 2020. 2020. Available from: https://www.hsa.gov.sg/announcements/new‐drug‐approval/new‐drug‐approvals‐‐‐aug‐2020. Accessed November 12, 2021.
- 19. Health Sciences Authority . New Drug Approvals ‐ Jan 2021. 2021. Available from: https://www.hsa.gov.sg/announcements/new‐drug‐approval/new‐drug‐approvals‐‐‐jan‐2021. Accessed November 12, 2021.
- 20. Russo M, Misale S, Wei G, et al. Acquired resistance to the TRK inhibitor entrectinib in colorectal cancer. Cancer Discov. 2016;6(1):36‐44. [DOI] [PubMed] [Google Scholar]
- 21. Drilon A, Li G, Dogan S, et al. What hides behind the MASC: clinical response and acquired resistance to entrectinib after ETV6‐NTRK3 identification in a mammary analogue secretory carcinoma (MASC). Ann Oncol. 2016;27(5):920‐926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Estrada‐Bernal A, Le AT, Kutateladze T, Doebele RC. TRK Kinase Domain Mutations that Induce Resistance to a Pan‐TRK Inhibitor. Boston: AACR; 2015. [Google Scholar]
- 23. Hemming ML, Nathenson MJ, Lin J‐R, et al. Response and mechanisms of resistance to larotrectinib and selitrectinib in metastatic undifferentiated sarcoma harboring oncogenic fusion of NTRK1 . JCO Precis Oncol. 2020;4:79‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hamid AB, Petreaca RC. Secondary resistant mutations to small molecule inhibitors in cancer cells. Cancers. 2020;12(4):927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gadgeel SM, Gandhi L, Riely GJ, et al. Safety and activity of alectinib against systemic disease and brain metastases in patients with crizotinib‐resistant ALK‐rearranged non‐small‐cell lung cancer (AF‐002JG): results from the dose‐finding portion of a phase 1/2 study. Lancet Oncol. 2014;15(10):1119‐1128. [DOI] [PubMed] [Google Scholar]
- 26. Mok TS, Wu Y‐L, Ahn M‐J, et al. Osimertinib or platinum–pemetrexed in EGFR T790M–positive lung cancer. N Engl J Med. 2017;376(7):629‐640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Drilon A, Nagasubramanian R, Blake JF, et al. A next‐generation TRK kinase inhibitor overcomes acquired resistance to prior TRK kinase inhibition in patients with TRK fusion–positive solid tumors. Cancer Discov. 2017;7(9):963‐972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Drilon A, Ou S‐HI, Cho BC, et al. Repotrectinib (TPX‐0005) is a next‐generation ROS1/TRK/ALK inhibitor that potently inhibits ROS1/TRK/ALK solvent‐front mutations. Cancer Discov. 2018;8(10):1227‐1236. [DOI] [PubMed] [Google Scholar]
- 29. Solomon JP, Linkov I, Rosado A, et al. NTRK fusion detection across multiple assays and 33,997 cases: diagnostic implications and pitfalls. Mod Pathol. 2020;33(1):38‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Solomon JP, Hechtman JF. Detection of NTRK fusions: merits and limitations of current diagnostic platforms. Cancer Res. 2019;79(13):3163‐3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. U.S. Food and Drug Administration . FDA Approves Companion Diagnostic to Identify NTRK Fusions in Solid Tumors for Vitrakvi. 2020. Available from: https://www.fda.gov/drugs/fda‐approves‐companion‐diagnostic‐identify‐ntrk‐fusions‐solid‐tumors‐vitrakvi. Accessed November 12, 2021.
- 32. Wong D, Yip S, Sorensen PH. Methods for identifying patients with tropomyosin receptor kinase (TRK) fusion cancer. Pathol Oncol Res. 2020;26(3):1385‐1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hsiao SJ, Zehir A, Sireci AN, Aisner DL. Detection of tumor NTRK gene fusions to identify patients who may benefit from tyrosine kinase (TRK) inhibitor therapy. J Mol Diagn. 2019;21(4):553‐571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brčić I, Godschachner TM, Bergovec M, et al. Broadening the spectrum of NTRK rearranged mesenchymal tumors and usefulness of pan‐TRK immunohistochemistry for identification of NTRK fusions. Mod Pathol. 2021;34(2):396‐407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. de Winne K, Sorber L, Lambin S, et al. Immunohistochemistry as a screening tool for NTRK gene fusions: results of a first Belgian ring trial. Virchows Arch. 2021;478(2):283‐291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hechtman JF, Benayed R, Hyman DM, et al. Pan‐TRK immunohistochemistry is an efficient and reliable screen for the detection of NTRK fusions. Am J Surg Pathol. 2017;41(11):1547‐1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Murphy DA, Ely HA, Shoemaker R, et al. Detecting gene rearrangements in patient populations through a 2‐step diagnostic test comprised of rapid IHC enrichment followed by sensitive next‐generation sequencing. Appl Immunohistochem Mol Morphol. 2017;25(7):513‐523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yoshino T, Pentheroudakis G, Mishima S, et al. JSCO—ESMO—ASCO—JSMO—TOS: international expert consensus recommendations for tumour‐agnostic treatments in patients with solid tumours with microsatellite instability or NTRK fusions. Ann Oncol. 2020;31(7):861‐872. [DOI] [PubMed] [Google Scholar]
- 39. Marchiò C, Scaltriti M, Ladanyi M, et al. ESMO recommendations on the standard methods to detect NTRK fusions in daily practice and clinical research. Ann Oncol. 2019;30(9):1417‐1427. [DOI] [PubMed] [Google Scholar]
- 40. Bormann Chung C, Lee J, Barritault M, et al. 13P Evaluating targeted next‐generation sequencing (NGS) assays and reference materials for NTRK fusion detection. Ann Oncol. 2020;31: Suppl 5, S1221. [DOI] [PubMed] [Google Scholar]
- 41. Zito Marino F, Pagliuca F, Ronchi A, et al. NTRK fusions, from the diagnostic algorithm to innovative treatment in the era of precision medicine. Int J Mol Sci. 2020;21(10):3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shulman DS, DuBois SG. The evolving diagnostic and treatment landscape of NTRK‐fusion‐driven pediatric cancers. Pediatr Drugs. 2020;22(2):189‐197. [DOI] [PubMed] [Google Scholar]
- 43. Banerji N, Kanjilal S. Increased intrinsic chemo‐resistance during progression of malignant soft tissue sarcoma. Cancer Res. 2005;65(9 Supplement):1199. [Google Scholar]
- 44. Demetri GD, Antonescu CR, Bjerkehagen B, et al. Diagnosis and management of tropomyosin receptor kinase (TRK) fusion sarcomas: expert recommendations from the World Sarcoma Network. Ann Oncol. 2020;31(11):1506‐1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Solomon JP, Benayed R, Hechtman JF, Ladanyi M. Identifying patients with NTRK fusion cancer. Ann Oncol. 2019;30:viii16‐viii22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Masters GA, Temin S, Azzoli CG, et al. Systemic therapy for stage IV non–small‐cell lung cancer: American Society of Clinical Oncology Clinical Practice Guideline Update. J Clin Oncol. 2015;33(30):3488‐3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Farago AF, Azzoli CG. Beyond ALK and ROS1: RET, NTRK, EGFR and BRAF gene rearrangements in non‐small cell lung cancer. Transl Lung Cancer Res. 2017;6(5):550‐559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Farago A, Kummar S, Moreno V, et al. MA09.07 Activity of larotrectinib in TRK fusion lung cancer. J Thorac Oncol. 2019;14(10):S283‐S284. [Google Scholar]
- 49. NCC Network . NCCN Guidelines. Nonsmall Cell Lung Cancer, Version 2.2021. 2021.
- 50. Doebele R, Paz‐Ares L, Farago AF, et al. Abstract CT131: Entrectinib in NTRK‐Fusion Positive (NTRK‐FP) Non‐Small Cell Lung Cancer (NSCLC): Integrated Analysis of Patients Enrolled in Three Trials (STARTRK‐2, STARTRK‐1 and ALKA‐372‐001). Clinical Trials. American Association for Cancer Research; 2019. [Google Scholar]
- 51. Rosen EY, Schram AM, Young RJ, et al. Larotrectinib demonstrates CNS efficacy in TRK fusion‐positive solid tumors. JCO Precis Oncol. 2019;3: PO.19.00009. 10.1200/PO.19.00009. eCollection 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li J, Zhu H, Sun L, Xu W, Wang X. Prognostic value of site‐specific metastases in lung cancer: a population based study. J Cancer. 2019;10(14):3079‐3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Perreault S, van Tilburg CM, Geoerger B, et al. Efficacy and safety of larotrectinib in adult and pediatric patients with tropomyosin receptor kinase (TRK) fusion‐positive primary central nervous system tumors. J Clin Oncol. 2021;39(15_suppl) 2002‐2002. [Google Scholar]
- 54. McDermott R, van Tilburg CM, Farago AF, et al. 1955P Survival benefits of larotrectinib in an integrated dataset of patients with TRK fusion cancer. Ann Oncol. 2020;31:S1101. [Google Scholar]
- 55. Tan AC, Lai GGY, Tan GS, et al. Utility of incorporating next‐generation sequencing (NGS) in an Asian non‐small cell lung cancer (NSCLC) population: incremental yield of actionable alterations and cost‐effectiveness analysis. Lung Cancer. 2020;139:207‐215. [DOI] [PubMed] [Google Scholar]
- 56. Lin JJ, Shaw AT. Resisting resistance: targeted therapies in lung cancer. Trends Cancer. 2016;2(7):350‐364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Uguen A. Reconsidering the turnaround times for BRAF V600 mutation analysis in non‐small‐cell lung cancer: a molecular diagnosis in one day is achievable for rapid treatment choices. Curr Oncol. 2019;26(4):e595‐e596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bittoni A, Sotte V, Meletani T, Cantini L, Giampieri R, Berardi R. Immunotherapy in colorectal cancer treatment: actual landscape and future perspectives. J Cancer Metastasis Treat. 2018;4:55. 10.20517/2394-4722.2018.37 [DOI] [Google Scholar]
- 59. Shinagawa T, Tanaka T, Nozawa H, et al. Comparison of the guidelines for colorectal cancer in Japan, the USA and Europe. Ann Gastroenterol Surg. 2017;2(1):6‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Deihimi S, Lev A, Slifker M, et al. BRCA2, EGFR, and NTRK mutations in mismatch repair‐deficient colorectal cancers with MSH2 or MLH1 mutations. Oncotarget. 2017;8(25):39945‐39962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pietrantonio F, di Nicolantonio F, Schrock AB, et al. ALK, ROS1, and NTRK rearrangements in metastatic colorectal cancer. J Natl Cancer Inst. 2017;109(12). 10.1093/jnci/djx089 [DOI] [PubMed] [Google Scholar]
- 62. Bridgewater J, Jiao X, Parimi M, et al. Abstract 394: Prognosis and Molecular Characteristics of Patients with TRK Fusion Cancer in the 100,000 Genomes Project. Clinical Research (Excluding Clinical Trials). American Association for Cancer Research; 2021. [Google Scholar]
- 63. García‐Alfonso P, García‐Carbonero R, García‐Foncillas J, et al. Update of the recommendations for the determination of biomarkers in colorectal carcinoma: National Consensus of the Spanish Society of Medical Oncology and the Spanish Society of Pathology. Clin Transl Oncol. 2020;22(11):1976‐1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gamboa AC, Gronchi A, Cardona K. Soft‐tissue sarcoma in adults: an update on the current state of histiotype‐specific management in an era of personalized medicine. CA Cancer J Clin. 2020;70(3):200‐229. [DOI] [PubMed] [Google Scholar]
- 65. Gatalica Z, Xiu J, Swensen J, Vranic S. Molecular characterization of cancers with NTRK gene fusions. Mod Pathol. 2019;32(1):147‐153. [DOI] [PubMed] [Google Scholar]
- 66. Rosen EY, Goldman DA, Hechtman JF, et al. TRK fusions are enriched in cancers with uncommon histologies and the absence of canonical driver mutations. Clin Cancer Res. 2020;26(7):1624‐1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Okamura R, Boichard A, Kato S, Sicklick JK, Bazhenova L, Kurzrock R. Analysis of NTRK alterations in pan‐cancer adult and pediatric malignancies: implications for NTRK‐targeted therapeutics. JCO Precis Oncol. 2018(2). 10.1200/PO.18.00183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. van Tilburg CM, DuBois SG, Albert CM, et al. Larotrectinib efficacy and safety in pediatric TRK fusion cancer patients. J Clin Oncol. 2019;37(15_suppl) 10010‐10010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this publication.


