Abstract
The 1,024-amino-acid acylated hemolysin of Escherichia coli subverts host cell functions and causes cell lysis. Both activities require insertion of the toxin into target mammalian cell membranes. To identify directly the principal toxin sequences dictating membrane binding and insertion, we assayed the lipid bilayer interaction of native protoxin, stably active toxin, and recombinant peptides. Binding was assessed by flotation of protein-liposome mixtures through density gradients, and insertion was assessed by labeling with a photoactivatable probe incorporated into the target lipid bilayer. Both the active acylated hemolysin and the inactive unacylated protoxin were able to bind and also insert. Ca2+ binding, which is required for toxin activity, did not influence the in vitro interaction with liposomes. Three overlapping large peptides were expressed separately. A C-terminal peptide including residues 601 to 1024 did not interact in either assay. An internal peptide spanning residues 496 to 831, including the two acylation sites, bound to phospholipid vesicles and showed a low level of insertion-dependent labeling. In vitro acylation had no effect on the bilayer interaction of either this peptide or the full-length protoxin. An N-terminal peptide comprising residues 1 to 520 also bound to phospholipid vesicles and showed strong insertion-dependent labeling, ca. 5- to 25-fold that of the internal peptide. Generation of five smaller peptides from the N-terminal region identified the principal determinant of lipid insertion as the hydrophobic sequence encompassing residues 177 to 411, which is conserved among hemolysin-related toxins.
The Escherichia coli 110-kDa hemolysin, HlyA, elicits a number of responses from mammalian target cells. HlyA is a potent trigger of G protein-dependent generation of inositol triphosphate and diacylglycerol in granulocytes and endothelial cells, stimulating the respiratory burst and the secretion of vesicular constituents (6, 15). Most recently, it has been shown to contribute to inflammation by inducing Ca2+ oscillations in renal epithelial cells (34). HlyA also alters the membrane permeability of host cells, causing lysis and death (5, 14). Toxin activity is absolutely dependent upon two posttranslational events. The inactive protoxin, proHlyA, is matured intracellularly by HlyC-directed cellular acyl carrier protein (ACP)-dependent fatty acylation of two internal lysine residues, K564 and K690 (16, 17, 31). After export, the acylated toxin binds Ca2+ at a C-terminal domain formed by acidic glycine-rich nonapeptide repeats (4, 9, 21).
The interaction of mature, Ca2+-bound HlyA with eukaryotic membranes appears to be a two-stage interaction: a reversible adsorption that is sensitive to electrostatic forces and an irreversible insertion associated with a change in toxin conformation (1, 24, 27). It is suggested that Ca2+ binding may promote the irreversible insertion, while not directly contributing to a predicted pore-forming structure (8, 21), and it has been shown that binding results in a change in toxin conformation (2, 26). Attempts to establish the role of acylation have provided contradictory results, suggesting either that inactive proHlyA is unable to bind to erythrocyte membranes (9, 21) or that proHlyA and HlyA have equal membrane affinities (3, 24, 29). It seemed possible that some of the contradiction reflected differences in the purification methods and intrinsic differences in samples of extracellular HlyA, these typically being mixtures of unacylated proHlyA and labile active HlyA (30). In addition to the influence of Ca2+ binding and acylation, hydrophobic sequences towards the N terminus of the toxin appear to be important in membrane interaction, as mutations reducing hydrophobicity attenuate pore formation (20, 21, 22).
We have investigated membrane binding and membrane insertion by purified protoxin, acylated toxin, and recombinant protoxin peptides. We used a protein-refolding protocol that achieved extremely stable toxin activity, facilitating the reproducible and direct assay of native (unmutated), chemically unmodified proteins. The effect of maturation on insertion was also established directly by in vitro acylation of protoxin. Two assays were used. Binding to liposomes composed of phospholipids and cholesterol was assayed by flotation through sucrose gradients to ensure separation of membrane-bound and free protein. Integration into liposomes was assessed by insertion-dependent hydrophobic labeling by a photoactivatable radiolabeled probe incorporated into the target lipid bilayer (10, 11). This photo-cross-linking approach has been successfully used to indicate the membrane-inserted regions of several integral membrane proteins and bacterial toxins (12), most notably the botulinum and tetanus neurotoxins (25) and diphtheria toxin (36). The combined results give a direct view of the interaction of the pro(toxin) with lipid bilayers.
MATERIALS AND METHODS
Bacteria and recombinant plasmids.
Recombinant plasmids pEK50 (complete pHly152 hly operon in pBR322), pT7HlyA (hlyA in pAR2529), pT7ApepN (N520, amino acids [aa] 1 to 520 of HlyA in pAR2529), pT7ApepI (I336, aa 496 to 831 of HlyA in pAR3040), and pT7ApepC (C423, aa 601 to 1024 of HlyA in pAR3040) have been described (16, 18, 19, 31). All hly plasmids were carried in E. coli 5KC (recA1 hsdR hsdD), E. coli BL21(DE3) (F− ompT recA rB−), or E. coli MC1061 [F− araD139 Δ(ara-leu)7696 galE15 galK16 Δ(lac)X74 rpsL (Strr) hsdR2, (rK− mK+) mcrA mcrB1]). Bacteria were grown at 37°C with aeration in 2× TY (1.6% Bacto Tryptone, 1% yeast extract, 0.5% NaCl) and 50 μg of ampicillin (Sigma) ml−1. The N-terminal proHlyA peptides were created using pEK50 as a template for the PCR (native Pfu polymerase; Stratagene and Perkin-Elmer GeneAmp PCR System 2400). The primers were engineered to incorporate an NdeI restriction site at the start codon ATG and a BamHI restriction site 3′ of the stop codon, respectively [N1-255, primers 1F (5′-CTGGTTAAGAGGTAATCATATGACAACAATAACCAC-3′) and 1R (5′-CTGCATCTGCATGGATCCGAATTTAGCTTGGTGAAAATTCG C-3′); N1-315, primers 1F and 2R (5′-GAATGAGGGGGATCCATTGCTTATGTCACAGCAGAAGC C-3′); N108-423, primers 2F (5′-GGCCTCACCGAACGGCATATGACTATCTTTGCACC-3′) and 3R (5′-CCATTCAGCAATAACAGGATCCATTTAACTGGCAACATG-3′); N160-423, primers 4F (5′-GGTACTTGCACTTTCCCATATGAAAATAGACGAACTG-3′) and 3R; N255-520, primers 5F (5′-CTGCGATTTCACATATGTTCATTCTGAGCAATG CAG-3′) and 4R (5′-CCTTTCAATGGATCCAAGACTTACTTCTGGAATTCATCC-3′)]. The resulting PCR products were subcloned into the NdeI/BamHI sites of the T7 expression plasmid, pET11C (Novagen).
Purification of active HlyA from culture supernatant.
E. coli MC1061 transformants carrying plasmid pEK50, which encodes the entire hly operon, were grown to late-exponential-growth phase (A600, ≈1.0). Bacteria were removed by centrifugation (twice at 9,000 × g, 10 min, 4°C), and the supernatant was adjusted to near the isoelectric point of HlyA, pH 4.5, with 1 M malonic acid to aid in precipitation. The solution was gradually brought to a final concentration of 20% ethanol. After 16 h at 4°C, the precipitate was collected by centrifugation (18,500 × g, 30 min, 4°C) and dissolved in 6 M guanidine hydrochloride (GnHCl; Sigma). Protein was purified by repeated solubilization in GnHCl and precipitation by ethanol, followed by high-speed centrifugation (350,000 × g, 10 min, 4°C), removing lipopolysaccharides and contaminating proteins, until the suspension of HlyA was >90% homogenous. The HlyA preparation had an activity of ca. 20,000 hemolytic units per μg, which was maintained undiminished over 72 h at 4°C.
Purification of inactive cytosolic proHlyA and derived peptides.
E. coli BL21(DE3) organisms transformed with recombinant T7 plasmids were grown at 37°C with aeration to mid-exponential phase (A600, ≈0.6). T7 polymerase-directed gene expression was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (Alexis), and cultures were grown for a further 2 h at 37°C. ProHlyA protein was harvested as previously detailed (16) and stored in HED (25 mM HEPES, 5 mM EGTA, 1 mM dithiothreitol, pH 8.0) with 6 M urea. ProHlyA peptides were pelleted (10,000 × g, 10 min, 4°C) and extracted from bacterial debris with 6 M urea. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting with HlyA-specific antibodies verified their integrity. SDS-PAGE gels were calibrated with prestained protein molecular weight markers (New England Biolabs).
In vitro acylation of proHlyA and peptide derivatives.
Proteins (1 μg) in HED with 6 M urea were prediluted in 150 μl of HED plus 100 mM KCl. Palmitoyl ACP or [3H]palmitoyl ACP (100 ng), depending on whether the proteins were to be applied to the insertion assay or the binding assay, respectively, and acyltransferase HlyC (20 ng) were added, and the reaction mixture was incubated for 20 min at 37°C. Palmitoyl ACP (ACP, palmitic acid; Sigma) and [3H]palmitoyl ACP ([3H]palmitic acid; Amersham) were prepared as detailed previously (33). To confirm the acylation reaction, a small aliquot from each [3H]-labeled reaction was analyzed by SDS-10% PAGE; gels were incubated in a solution of Amplify (Amersham) fluorographic reagent for 15 min before drying onto Whatman 3MM filter paper under vacuum at 80°C. The dried gels were exposed at −80°C to preflashed Kodak X-Omat AR film and developed after 48 h. For the unlabeled reaction, a small aliquot of the proHlyA mixture was assayed for hemolytic activity (hemoglobin [A543] released after incubation for 30 min at 42°C with 2% equine erythrocytes in 150 mM NaCl and 20 mM CaCl2; 1 hemolytic unit releases 1 nmol of hemoglobin in 1 h).
Liposome flotation assays.
Phospholipid vesicles (PLV) (liposomes) were prepared fresh from stock solutions (10 mg/ml, dried down from chloroform under vacuum and by nitrogen flushing) of cholesterol, phosphatidyl-choline, phosphatidyl-serine, phosphatidyl-ethenolamine, and sphingomyelin (Sigma), mixed in equimolar proportions. Suspensions (10 mg/ml) were obtained by sonication into buffer A (10 mM Tris, pH 7.4, 150 mM NaCl). Protein samples (2 μg) were renatured by fast dilution into 0.4 ml of buffer A in the presence of 5 mM EGTA or 0.5 mM Ca2+ and centrifuged (50,000 × g, 10 min, 4°C) to eliminate aggregates. Buffer A or buffer A liposome suspension (10 μl) was added, and the mixture was incubated at 37°C for 30 min. Phospholipid-associated and phospholipid-free protein were separated by flotation through sucrose gradients. A sucrose-buffer A solution was added to the mixture to a final concentration of 50% sucrose in 1 ml of total volume. The samples were overlaid with 3.5 ml of 40% sucrose and 0.5 ml of buffer A. Centrifugation was carried out overnight at 75,000 × g for 16 h at 16°C. Ten 1-ml fractions were collected from the gradient, and after trichloroacetic acid precipitation and resuspension in SDS sample buffer, they were analyzed by SDS-PAGE and either Coomassie brilliant blue R250 (Sigma) staining or immunoblotting.
Liposome-TID photolabeling.
Just prior to photolabeling, protein samples were renatured by fast dilution from stocks into 1 ml of buffer B (10 mM HEPES, pH 7.4, 150 mM NaCl), in the presence of 5 mM EGTA or 0.5 mM Ca2+ and centrifuged (50,000 × g, 10 min, 4°C) to eliminate aggregates. Under photographic safety lighting, 6.5 μCi of 3-(trifluoromethyl)-3-(m-[125I]iodophenyl) diazirine ([125I]TID) (5-μCi/μl solution in 3:1 ethanol/water; Amersham Pharmacia Biotech) was added to 100 μl of liposome suspension in buffer B and incubated at 37°C for 10 min. This represented an excess of liposomes to ensure complete incorporation of [125I]TID into the hydrophobic phase of the lipids. Ovalbumin, like a small number of other soluble proteins, can be labeled with [125I]TID in solution (13) and was introduced as a strict control for any remaining free [125I]TID. The [125I]TID-liposome mix (20 μl) was added to 1 ml of protein sample and then was incubated for 30 min at 37°C. Samples were illuminated with a long-wave UV fluorescent strip (calibrated separately for each protein), precipitated by trichloroacetic acid, washed with ethanol and acetone, and then separated by SDS-PAGE followed by phosphorimager detection (Packard Cyclone Storage Phosphor System).
RESULTS
Isolation and refolding of prohemolysin and stably active hemolysin.
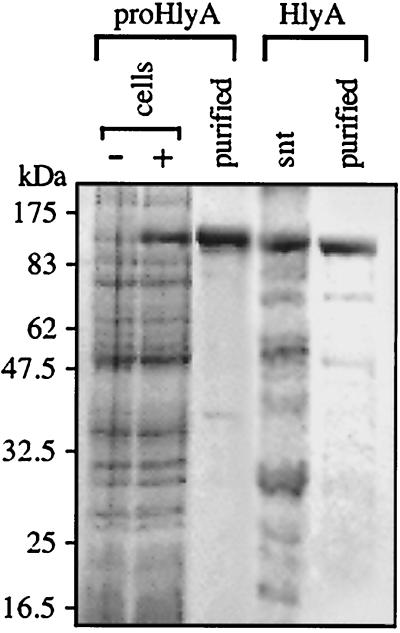
ProHlyA was produced from T7 polymerase-directed gene expression in E. coli as protein aggregates allowing rapid purification by pelleting, repeated washing, and solubilization in urea (Fig. 1). The protoxin was soluble on dilution from stocks and after in vitro acylation. The hemolytically active HlyA protein was isolated from culture supernatants of E. coli expressing the complete hlyCABD operon by repeated washing in 6 M guanidine hydrochloride and ethanol (Fig. 1), which efficiently removed contaminating membrane fragments and prevented the typical formation of hemolytically inactive aggregates by HlyA. This purification method reproducibly allowed reconstitution of stable toxin activity; after refolding and extensive dialysis, the sample was centrifuged at high speed to remove any aggregates and provide homogeneity of the sample. The protein remained soluble, and hemolytic activity was always undiminished over 72 h, ensuring that the results were reproducible and consistent. Proteins were quickly diluted from stocks into assay buffer before use.
FIG. 1.
Isolation of proHlyA and HlyA. Lysates of uninduced (−) and induced (+) cell cultures of E. coli BL21(DE3)(pT7HlyA) and the resulting purified proHlyA protein (2 μg loaded). Secreted HlyA (2 μg) purified from the culture supernatant (snt) of E. coli MC1061 (pEK50). Samples were analyzed by SDS-10% PAGE and Coomassie staining and size-calibrated using standard molecular weight markers.
Interaction of hemolysin and prohemolysin with liposomes.
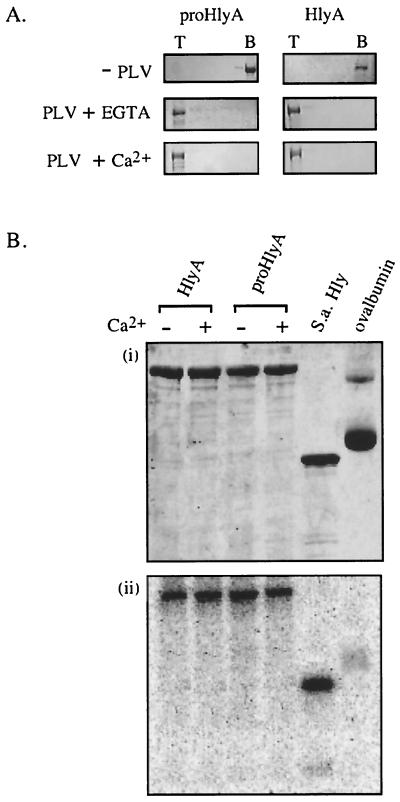
Binding of the protoxin and active toxin to membranes was assessed using PLV (liposomes) composed of phospholipids and cholesterol. Inactive unacylated proHlyA and active acylated HlyA were incubated with liposomes at 37°C for 30 min and mixed with 55% sucrose, overlaid with 40% sucrose, and allowed to float up through a sucrose density gradient during centrifugation. In the absence of PLV, both the toxin and the protoxin remained at the bottom of the gradient (Fig. 2A), but when incubated with liposomes they were both recovered from the top of the gradient, irrespective of whether Ca2+ was present during the incubation. This suggests that neither Ca2+ nor acylation influences the binding of liposomes by (pro)HlyA.
FIG. 2.
Interaction of proHlyA and HlyA with liposomes. (A) Purified proHlyA and HlyA were incubated without (−PLV) or with (+ PLV) phospholipid vesicles in the presence (+ Ca2+) or absence (+ EGTA) of Ca2+ and analyzed by centrifugation (flotation) through a sucrose gradient. Top (T) and bottom (B) fractions were analyzed by SDS-10% PAGE and Coomassie staining. (B) Insertion-dependent labeling. ProHlyA, HlyA, S. aureus α-hemolysin (S.a.Hly) as a positive control and ovalbumin as a negative control were incubated (2 μg of each) with liposomes in which [125I]TID was incorporated. Ca2+ was present (+) or absent (−). Samples were analyzed by SDS-10% PAGE and developed by Coomassie staining (i) and phosphorimage detection (ii).
The possible influence of acylation and Ca2+ on insertion of the toxin into lipid bilayers was assessed by insertion-dependent labeling by the hydrophobic photoactivatable radiolabeled-probe [125I]TID. The [125I]TID partitions to the hydrophobic core of membranes, from where it reacts specifically with the transmembrane segments of inserted proteins. Ovalbumin was adopted as a strict negative control to ensure that [125I]TID was incorporated into phospholipid vesicles and that only membrane-inserted proteins became efficiently labeled. [125I]TID was incubated with a suspension of liposomes at 37°C for 10 min, to allow incorporation into the hydrophobic phase. An aliquot of the [125I]TID-liposome mixture was then added to refolded HlyA or proHlyA protein samples and incubated for 30 min at 37°C. After SDS-PAGE separation of samples (Fig. 2Bi), labeling was detected by phosphorimage analysis of gels (Fig. 2Bii). Within the concentration range chosen for these experiments, the degree of labeling was directly proportional to the protein concentration in the reaction mixture (data not shown). Labeling of ovalbumin was barely detectable only after extended exposure (Fig. 2B), whereas Staphylococcus aureus alpha-hemolysin, a well-characterized membrane-inserting protein (7), was labeled strongly. The labeling of both proHlyA and HlyA was comparable to that of the S. aureus alpha-hemolysin. Again neither Ca2+ nor acylation influenced the level of insertion.
Interaction of (pro)HlyA peptides with liposomes.
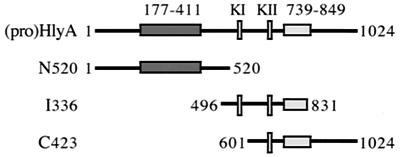
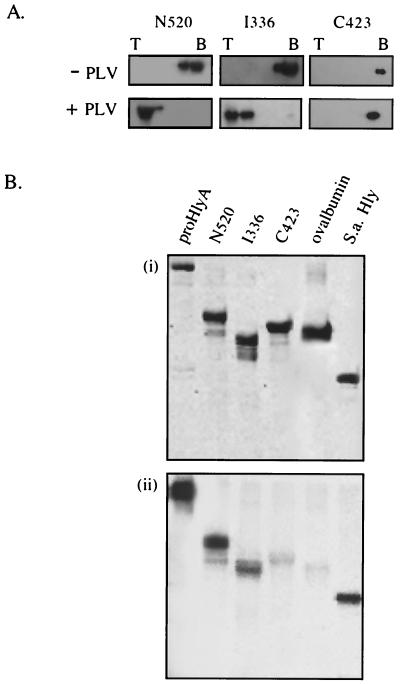
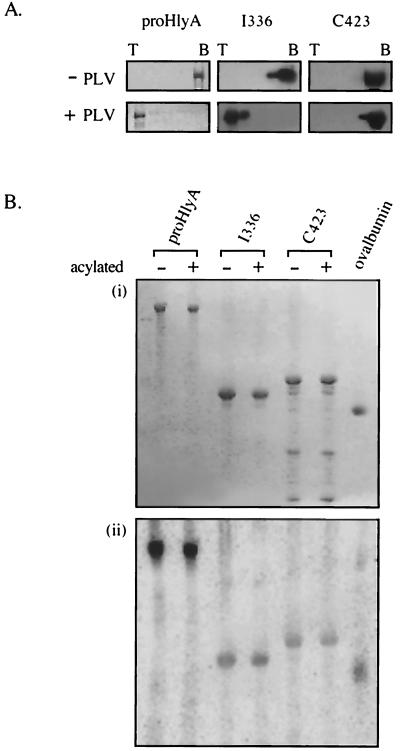
To establish which regions of the toxin molecule are central to membrane interaction, three large overlapping peptides spanning the entire toxin were purified and applied to the phospholipid-binding and insertion assays (Fig. 3). Peptide N520 (aa 1 to 520) encompasses the N-terminal hydrophobic region, and I336 (aa 496 to 831) includes both the KI and KII acylation sites and most of the Ca2+-binding domain, while C423 (aa 601 to 1024) contains the KII acylation site, the entire Ca2+-binding domain, and the secretion signal (19, 31). Sucrose gradient fractions from the resulting flotation assay (Fig. 4A) were immunoblotted with anti-HlyA polyclonal antiserum (the antiserum reacts with all three peptides as well as it does with the HlyA). In the absence of phospholipid vesicles, the three peptides were recovered from the bottom of the gradient. When the peptides were incubated with liposomes, N520 and I336 floated to the top of the sucrose gradient, while C423 remained at the bottom. As with flotation of the full-length (pro)toxin molecules, this was equally true whether Ca2+ was present or not.
FIG. 3.
Representation of proHlyA peptide derivatives N520, I336, and C423 (aa 1 to 520, 496 to 831, and 601 to 1024, respectively), indicating the N-terminal hydrophobic region (dark, residues 177 to 411), acylation sites (KI and KII, lysine residues 564 and 690), and Ca2+-binding domain (light, residues 739 to 849).
FIG. 4.
Interaction of HlyA peptides with liposomes. (A) Flotation of peptides after incubation either without (−PLV) or with (+PLV) phospholipid vesicles in the presence of Ca2+. As before, sucrose gradient top (T) and bottom (B) fractions were analyzed by SDS-10% PAGE and immunoblotting with anti-HlyA antisera. (B) Insertion-dependent labeling. ProHlyA and peptide derivatives were incubated with liposomes in which TID was incorporated. Samples were analyzed by SDS-10% PAGE and developed by Coomassie staining (i) and phosphorimage detection (ii).
These results correlated with those of the parallel insertion-dependent photolabeling assay (Fig. 4B). Peptide C423 showed only the background labeling of the ovalbumin negative control, while peptides N520 and I336 were both labeled with [125I]TID. In repeated assays N520 was reproducibly strongly labeled at ca. 40 to 50% of the level evident in the entire (pro)HlyA, while I336 was consistently labeled at ca. 2 to 10% of (pro)HlyA. The data indicate that although part of the internal peptide spanning the acylation region is lipid inserted, the principal membrane-inserting domain lies towards the N terminus. As I336 contains both acylation sites, we sought to establish whether acylation of these was critical. I336 was therefore acylated in vitro by purified acyltransferase HlyC and either palmitoyl ACP or [3H]palmitoyl ACP, depending on whether the proteins were to be applied to the liposome insertion or phospholipid-binding assays, respectively. Peptide C423 and proHlyA were also in vitro acylated. Acylation was confirmed either by detection of 3H signal or by assaying a small aliquot of the proHlyA reaction mixture for hemolytic activity. The results show that in vitro acylation had no effect on the phospholipid-binding ability of I336, C423, or proHlyA (Fig. 5A) or the extent to which they became photolabeled after incubation with liposomes incorporating [125I]TID (Fig. 5B).
FIG. 5.
Effect of in vitro acylation on interaction with liposomes. (A) Flotation of in vitro-acylated proHlyA, I336, and C423 after incubation without (−PLV) or with (+PLV) phospholipid vesicles in the presence of Ca2+. Sucrose gradient top (T) and bottom (B) fractions were analyzed by SDS-10% PAGE and immunoblotting with anti-HlyA antisera. (B) Insertion-dependent labeling. ProHlyA and derivatives before (−) and after (+) in vitro acylation were incubated with liposomes in which TID was incorporated. Samples were analyzed by SDS-10% PAGE and developed by Coomassie staining (i) and phosphorimage detection (ii).
Mapping of the N-terminal toxin sequences required for binding and insertion.
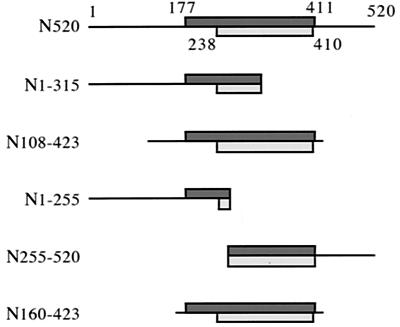
Since toxin peptide N520 was indicated to be the principal inserting domain in the liposome insertion assay, five smaller peptides spanning this region were generated (Fig. 6). They were designed to assess in particular the importance of the extreme N terminus, the conserved hydrophobic region determined by residues 177 to 411, and the stretch of residues 238 to 410, which is predicted to generate amphipathic α-helices. The five peptides, 255 to 315 amino acids long, were purified from inclusion bodies and solubilized in urea. They were diluted immediately before application to both the flotation and photolabeling assays. All of the peptides were nonhemolytic.
FIG. 6.
Representation of peptides spanning the (pro)HlyA N terminus, indicating the hydrophobic region (dark, residues 177 to 411), which includes the amphipathic α-helical sequence (light, residues 238 to 410).
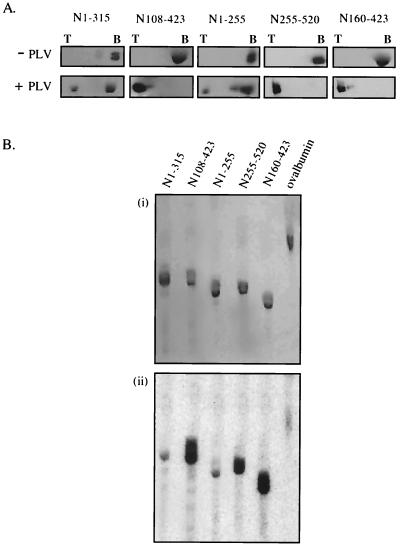
Immunoblotting of sucrose gradient fractions showed that as expected, in the absence of phospholipid vesicles all five of the peptides remained at the bottom of the gradient. However, when the peptides were incubated with liposomes, three peptides, N108-423, N160-423, and N255-520, shared the ability of the parental peptide N520 to float. In each case, apparently 100% was found at the top of the gradient (Fig. 7A). In contrast, repeated assays showed that 90 to 95% of peptides N1-315 and N1-255 remained unbound at the bottom of the gradient. In the photolabeling insertion assay (Fig. 7B), the peptides that had floated were strongly labeled. In repeated assays, the signals from peptides N108-423 and N160-423 were indistinguishable from each other and comparable to that seen in the parental peptide N520. N255-520, lacking part of the hydrophobic sequence, also inserted, but it was consistently labeled to approximately one-half the level of the first two. The two nonfloating peptides N1-315 and N1-255 gave only a very weak insertion signal after prolonged exposure; this was at most marginally stronger than the ovalbumin background. We conclude that they cannot insert.
FIG. 7.
Liposome interaction of N-terminal (pro)HlyA peptides. (A) Flotation of N-terminal peptides, after incubation with (+ PLV) or without (− PLV) phospholipid vesicles. Sucrose gradient top (T) and bottom (B) fractions were analyzed by SDS-10% PAGE and immunoblotting with anti-HlyA antisera. (B) Insertion-dependent labeling of N-terminal peptides during incubation with liposomes in which TID was incorporated. Samples were analyzed by SDS-10% PAGE and developed by Coomassie staining (i) and phosphorimage detection (ii).
DISCUSSION
The HlyA (RTX) family of membrane-targeted toxins produces a variety of effects in mammalian host cells and causes lysis by inserting into and disrupting target membranes. In this work, we investigated the binding of proHlyA and stably active HlyA to lipid bilayers by flotation of liposome-native (pro)toxin mixtures. Both inactive unacylated proHlyA and active acylated HlyA bound phospholipid vesicles with an efficiency approaching 100%. This is in agreement with earlier assays of toxin binding to erythrocytes, assessed by cosedimentation (24) and by flow cytometry (3), and to lipid vesicles, assessed by flotation or fluorescence (29). Our studies also showed that both the protoxin and toxin were strongly labeled by photoactivatable [125I]TID incorporated into the hydrophobic phase of the lipid bilayer, where it reacts with transmembrane regions of inserted proteins (10, 11).
These data indicated that acylation had no influence on liposome interaction. To provide additional, direct data to resolve this contentious and important aspect of hemolysin behavior, we performed in vitro acylation of proHlyA (with palmitate), which efficiently matures the majority of the protoxin (the exported hemolysin is a heterogeneous mixture of acylated and unacylated protein, [30]). The in vitro acylation confirmed that the fatty acids do not influence liposome binding or insertion. Our results also indicated that the ability of (pro)HlyA to bind and insert into phospholipid vesicles was not dependent on Ca2+ being bound by the glycine-rich repeat domain. This conclusion agrees with that reached in work performed in parallel using mutant HlyA variants chemically labeled with cysteine-specific fluorescent probes (28). Insertion into liposomes in the presence and absence of Ca2+ was detected by spectroscopic assay of emission shifts (28), but the presence of 10 mM Ca2+ in such spectrophotometric analyses is potentially problematic, as it can cause fusion of liposomes and distort the signal (35). Indeed, in this particular study most of the derivatized residues did generate apparently nonspecific shifts in emission spectra (28). Our biochemical assessment of binding and insertion by the native unmodified proteins provides direct evidence of Ca2+-independent membrane insertion.
If neither Ca2+ binding nor acylation is essential for interaction with liposomes yet both are absolute requirements for toxin activity, what roles do they play in toxin action? It has been shown that the toxin lytic action requires the binding of Ca2+ in solution prior to its interaction with membranes (1) and that this binding induces a change in tertiary structure (2, 26). Ca2+-dependent conformation change could ensure productive insertion of HlyA into target cell membranes. There are several possible roles for acylation (32). The acyl groups may also guarantee proper insertion of the toxin into lipid bilayers. They could provide anchorage points in the membrane, preventing essential domains looping away from the membrane surface, or they might ensure that the toxin contacts the membrane in a correctly folded conformation. Alternatively, after insertion, acylation might promote protein-protein interactions that are involved in potential oligomerization or between HlyA and components of a signal transduction pathway.
To establish which regions of the 1,024-residue toxin molecule are principally responsible for the binding and insertion, three large overlapping peptide derivatives of the native protoxin were assessed by the same two assays. A peptide (I336) spanning the internal region of the toxin bound to liposomes efficiently but displayed 10- to 50-fold less insertion-dependent labeling than the full-length toxin, a level that remained unchanged after in vitro acylation. This indicates that some residues within the internal region of (pro)HlyA can penetrate the lipid bilayer, in agreement with a report (24) stating that monoclonal antibodies directed against epitopes surrounding the acylation sites do not react with the toxin after binding to red blood cells. It is plausible that regions surrounding the acyl groups, which are likely to interact with the target cell membrane, also penetrate the lipid bilayer. The C-terminal peptide C423 showed no membrane affinity in either assay, irrespective of Ca2+ binding and in vitro acylation, indicating that the C-terminal region of the toxin does not become inserted upon contact with liposomes. A previous study (3) with a slightly shorter C-terminal peptide (HlyA626-1023) saw some cosedimentation with erythrocytes. In our study, assessment by the less contentious flotation assay showed that the C-terminal region of the toxin does not bind to liposomes and exhibits no insertion-dependent labeling in the [125I]TID assay. Our data show that the principal region inserting into the target membrane lies within the N-terminal half of (pro)HlyA. Peptide N520 bound to phospholipid vesicles and was insertion labeled to about 50% of the level seen in the entire (pro)toxin. Smaller peptides were created to determine which sequences within the N-terminal 520 residues interacted with the membrane, in particular to determine the importance of the only pronounced hydrophobic sequence, residues 177 to 411, in the otherwise hydrophilic HlyA. Within this sequence lies a predicted α-helical region (aa 238 to 410), which is highly conserved within the RTX toxin family. Mutations altering the hydrophobicity of this region reduce or abolish the pore-forming activity of the protein (19, 20, 21), and it has been hypothesized to play a role in membrane disruption (22, 23). The results of the two assays in our study correlated closely. The two peptides spanning the entire hydrophobic region (N108-423 and N160-423) both bound and inserted. In contrast, there was no efficient binding or insertion by two others (N1-255 and N1-315) that include the (pro)HlyA N terminus but not the intact hydrophobic domain. The fifth peptide (N255-520), incorporating most of the hydrophobic region but not the (pro)HlyA N terminus, also bound, but insertion-dependent labeling was one-half that of N108-423 and N160-423.
Our data indicate that the hydrophobic stretch is critical and that HlyA177-411 is most likely the principal region that inserts. The results of our direct insertion assay are strongly complementary to those from a parallel spectroscopic study of the same toxin (28). In the latter case, 13 HlyA derivatives were coupled to fluorescent probes via single cysteine residues introduced throughout the first 739 amino acids of the toxin, and their fluorescence emission was followed in solution and after mixing with liposomes. This approach indicated that the only cysteines that were inserted were located within the same HlyA N-terminal hydrophobic stretch. The identical results obtained from the two parallel independent approaches provide strong evidence that the HlyA177-411 sequence is the principal inserting domain. Such synergistic experimental approaches can now be extended to assess the depth of (pro)HlyA insertion (this requires a substantial scale-up in the case of the cross-linking approach) and investigate the multimeric state of the inserted toxin and the precise role of the fatty acids.
ACKNOWLEDGMENTS
We thank E. Koronakis and J. Eswaran for critical reading of the manuscript.
Work was funded by a Medical Research Council (MRC) Programme Grant (C. Hughes and V. Koronakis) and an MRC studentship (C. Hyland).
REFERENCES
- 1.Bakas L, Ostolaza H, Vaz W L C, Goni F M. Reversible adsorption and nonreversible insertion of Escherichia coli α-hemolysin into lipid bilayers. Biophys J. 1996;71:1869–1876. doi: 10.1016/S0006-3495(96)79386-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakas L, Veiga M P, Soloaga A, Ostolaza H, Goni F M. Calcium-dependent conformation of E. coli alpha-haemolysin. Implications for the mechanism of membrane insertion and lysis. Biochim Biophys Acta. 1998;1368:225–234. doi: 10.1016/s0005-2736(97)00181-8. [DOI] [PubMed] [Google Scholar]
- 3.Bauer M E, Welch R A. Association of RTX toxins with erythrocytes. Infect Immun. 1996;64:4665–4672. doi: 10.1128/iai.64.11.4665-4672.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumann U, Wu S, Flaherty K M, McKay D B. Three-dimensional structure of the alkaline protease of Pseudomonas aeruginosa: a two-domain protein with a calcium binding parallel beta roll motif. EMBO J. 1993;12:3357–3364. doi: 10.1002/j.1460-2075.1993.tb06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhakdi S, Mackman N, Nicaud J M, Holland I B. Escherichia coli hemolysin may damage target cell membranes by generating transmembrane pores. Infect Immun. 1986;52:63–69. doi: 10.1128/iai.52.1.63-69.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhakdi S, Martin E. Superoxide generation by human neutrophils induced by low doses of Escherichia coli hemolysin. Infect Immun. 1991;59:2955–2962. doi: 10.1128/iai.59.9.2955-2962.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhakdi S, Tranum-Jensen J. Alpha-toxin of Staphylococcus aureus. Microbiol Rev. 1991;55:733–751. doi: 10.1128/mr.55.4.733-751.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boehm D F, Welch R A, Snyder I S. Calcium is required for binding of Escherichia coli hemolysin (HlyA) to erythrocyte membranes. Infect Immun. 1990;58:1951–1958. doi: 10.1128/iai.58.6.1951-1958.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boehm D F, Welch R A, Snyder I S. Domains of Escherichia coli hemolysin (HlyA) involved in binding of calcium and erythrocyte membranes. Infect Immun. 1990;58:1959–1964. doi: 10.1128/iai.58.6.1959-1964.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunner J, Semenza G. Selective labeling of the hydrophobic core of membranes with 3-(trifluoromethyl)-3-(m-[125I]iodophenyl)diazirine, a carbene-generating reagent. Biochemistry. 1981;20:7174–7182. doi: 10.1021/bi00528a019. [DOI] [PubMed] [Google Scholar]
- 11.Brunner J. Photochemical labeling of apolar phase of membranes. Methods Enzymol. 1989;172:628–687. doi: 10.1016/s0076-6879(89)72037-1. [DOI] [PubMed] [Google Scholar]
- 12.Brunner J. New photolabeling and crosslinking methods. Annu Rev Biochem. 1993;62:483–514. doi: 10.1146/annurev.bi.62.070193.002411. [DOI] [PubMed] [Google Scholar]
- 13.Brunner J. Use of photocrosslinkers in cell biology. Trends Cell Biol. 1996;6:154–157. doi: 10.1016/0962-8924(96)40001-0. [DOI] [PubMed] [Google Scholar]
- 14.Gadeberg O V, Orskov I. In vitro cytotoxic effect of α-hemolytic Escherichia coli on human blood granulocytes. Infect Immun. 1984;45:255–260. doi: 10.1128/iai.45.1.255-260.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimminger F, Rose F, Sibelius U, Meinhardt M, Potzsch B, Spriestersbach R, Bhakdi S, Suttorp N, Seeger W. Human endothelial cell activation and mediator release in response to the bacterial exotoxins Escherichia coli hemolysin and Staphylococcal α-toxin. J Immunol. 1997;159:1909–1916. [PubMed] [Google Scholar]
- 16.Hardie K R, Issartel J P, Koronakis E, Hughes C, Koronakis V. In vitro activation of Escherichia coli prohaemolysin to the mature membrane-targeted toxin requires HlyC and a low molecular weight cytosolic polypeptide. Mol Microbiol. 1991;5:1669–1679. doi: 10.1111/j.1365-2958.1991.tb01914.x. [DOI] [PubMed] [Google Scholar]
- 17.Issartel J P, Koronakis V, Hughes C. Activation of Escherichia coli prohaemolysin to the mature toxin by acyl carrier protein-dependent fatty acylation. Nature. 1991;351:759–761. doi: 10.1038/351759a0. [DOI] [PubMed] [Google Scholar]
- 18.Koronakis V, Hughes C. Identification of the promoters directing in vivo expression of hemolysin genes in Proteus vulgaris and Escherichia coli. Mol Gen Genet. 1988;213:99–104. doi: 10.1007/BF00333404. [DOI] [PubMed] [Google Scholar]
- 19.Koronakis V, Koronakis E, Hughes C. Isolation and analysis of the C-terminal signal directing export of Escherichia coli hemolysin protein across both bacterial membranes. EMBO J. 1989;8:595–605. doi: 10.1002/j.1460-2075.1989.tb03414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ludwig A, Vogel M, Goebel W. Mutations affecting activity and transport of hemolysin in Escherichia coli. Mol Gen Genet. 1987;206:238–245. doi: 10.1007/BF00333579. [DOI] [PubMed] [Google Scholar]
- 21.Ludwig A, Jarchau T, Benz R, Goebel W. The repeat domain of Escherichia coli hemolysin (HlyA) is responsible for its Ca2+-dependent binding to erythrocytes. Mol Gen Genet. 1988;214:553–561. doi: 10.1007/BF00330494. [DOI] [PubMed] [Google Scholar]
- 22.Ludwig A, Schmid A, Benz R, Goebel W. Mutations affecting pore formation by hemolysin from Escherichia coli. Mol Gen Genet. 1991;226:198–208. doi: 10.1007/BF00273604. [DOI] [PubMed] [Google Scholar]
- 23.Menestrina G, Moser C, Pellett S, Welch R. Pore formation by Escherichia coli hemolysin (HlyA) and other members of the RTX toxins family. Toxicology. 1994;87:249–267. doi: 10.1016/0300-483x(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 24.Moayeri M, Welch R A. Prelytic and lytic conformations of erythrocyte-associated Escherichia coli hemolysin. Infect Immun. 1997;65:2233–2239. doi: 10.1128/iai.65.6.2233-2239.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montecucco C, Schiavo G, Bauerlein E, Boquet P, DasGupta B R. Interaction of botulinum and tetanus toxins with the lipid bilayer surface. Biochem J. 1988;251:379–383. doi: 10.1042/bj2510379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ostolaza H, Soloaga A, Goni F M. The binding of divalent cations to Escherichia coli α-hemolysin. Eur J Biochem. 1995;228:39–44. [PubMed] [Google Scholar]
- 27.Ostolaza H, Bakas L, Goni F M. Balance of electrostatic and hydrophobic interactions in the lysis of model membranes by E. coli α-haemolysin. J Membr Biol. 1997;158:137–145. doi: 10.1007/s002329900251. [DOI] [PubMed] [Google Scholar]
- 28.Schindel C, Zitzer A, Schulte B, Gerhards A, Stanley P, Hughes C, Koronakis V, Bhakdi S, Palmer M. Interaction of E. coli hemolysin with biological membranes. A study using cysteine scanning mutagenesis. Eur J Biochem. 2001;268:800–808. doi: 10.1046/j.1432-1327.2001.01937.x. [DOI] [PubMed] [Google Scholar]
- 29.Soloaga A, Ostolaza H, Goni F M, DelaCruz F. Purification of Escherichia coli prohemolysin, and a comparison with the properties of mature α-hemolysin. Eur J Biochem. 1996;238:418–422. doi: 10.1111/j.1432-1033.1996.0418z.x. [DOI] [PubMed] [Google Scholar]
- 30.Stanley P L D, Diaz P, Bailey M J A, Gygi D, Juarez A, Hughes C. Loss of activity in the secreted form of Escherichia coli hemolysin caused by an rfaP lesion in core lipopolysaccharide assembly. Mol Microbiol. 1993;10:781–787. doi: 10.1111/j.1365-2958.1993.tb00948.x. [DOI] [PubMed] [Google Scholar]
- 31.Stanley P, Packman L C, Koronakis V, Hughes C. Fatty acylation of two internal lysine residues required for the toxic activity of Escherichia coli hemolysin. Science. 1994;266:1992–1996. doi: 10.1126/science.7801126. [DOI] [PubMed] [Google Scholar]
- 32.Stanley P, Koronakis V, Hughes C. Acylation of E. coli hemolysin: a unique protein lipidation mechanism underlying toxin function. Microbiol Mol Biol Rev. 1998;62:309–333. doi: 10.1128/mmbr.62.2.309-333.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanley P, Hyland C, Koronakis V, Hughes C. An ordered reaction mechanism for bacterial toxin acylation by the specialised acyltransferase HlyC: formation of a ternary complex with acylACP and protoxin substrates. Mol Microbiol. 1999;34:887–901. doi: 10.1046/j.1365-2958.1999.01648.x. [DOI] [PubMed] [Google Scholar]
- 34.Uhlén P, Laestadius A, Jahnukainen T, Soderblom T, Backhed F, Celsi G, Brismar H, Normark S, Aperia A, Richter-Dahlfors A. Alpha-haemolysin of uropathogenic E. coli induces Ca2+ oscillations in renal epithelial cells. Nature. 2000;405:694–697. doi: 10.1038/35015091. [DOI] [PubMed] [Google Scholar]
- 35.Wilshcut J, Papahadjopoulos D. Ca2+-induced fusion of phospholipid vesicles monitored by mixing of aqueous contents. Nature. 1979;281:690–692. doi: 10.1038/281690a0. [DOI] [PubMed] [Google Scholar]
- 36.Zalman L S, Wisnieski B J. Mechanism of insertion of diptheria toxin: peptide entry and pore size determinations. Proc Natl Acad Sci USA. 1984;81:3341–3345. doi: 10.1073/pnas.81.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]