Abstract
The search of antivirals against SARS‐CoV‐2 in available libraries of compounds was initiated as soon as WHO announced that the coronavirus outbreak became a pandemic. That pivotal task has been conducted by both experimental groups in wet‐labs as well as by theoretical chemists in supercomputing centers. The combination of biochemical and clinical intuitions yields first to remdesivir, a broad‐spectrum antiviral that remains as the standard solution for the treatment of severe cases, while paxlovid, molnupiravir and fluvoxamine have been recently proposed as oral alternatives. Unfortunately, the intensive publication of standard virtual screening (VS) simulations might be not the best strategy to increase that short list of antivirals. This contribution joins theory and biological assays to rescore massive VS. Our goal is to critically assess pros and cons of using molecular models for drug repurposing.
Keywords: drug design, antivirals, computational methods – biological assays, main protease, COVID-19
Massive virtual screenings with a focus on SARS‐CoV‐2 are now available in the literature. Unfortunately, standard implementations without further refinements and/or biological assays have shown a limited impact in real clinical applications. To guide next computational efforts, herein we combine theory and experiments to critically assess pros and cons of such methods.
Introduction
There is no precedent for the collective effort for fighting against the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). So far, two lines of research are on the top list for the scientific community: the development of vaccines to stop virus spread across healthy population, and the discover of efficient antivirals to mitigate the effects of the associated disease, COVID‐19. [1] The progress of such strategies is rather dissimilar. Novel vaccines have been rapidly designed by adapting the messenger RNA (mRNA) technology to the SARS‐CoV‐2 architecture. [2] Moderna and Pfizer/BioNTech were granted with the first mRNA‐vaccine licenses, while the viral‐vectored AstraZeneca and Janssen are now approved. [3] A wider market is expected in next months as ca. 200 vaccine candidates have entered different clinical stages. [4] On the contrary, less successful is achieved for antivirals. Remdesivir, a broad‐spectrum antiviral (influenza, coronaviruses, alphaviruses and flaviviruses) originally designed by the Antiviral Drug Discovery and Development Center, was used at an early stage of the pandemic to intravenously treat patients with COVID‐19. [5] However, remdesivir as well as other antivirals (i. e. hydroxychloroquine, lopinavir, and interferon regimens) have shown a moderate action during their clinical applications. [6] Last Pfizer and Merck's contenders – paxlovid and molnupiravir, respectively – seem to offer benefits over that first list of drugs as they are both oral antivirals that can be administrated out of hospitals. [7] Fluvoxamine is also under investigation for its use at the beginning of the infection. [8]
Despite such promising steps, the need of discovering antivirals is still urgent, a challenge that might be addressed by experimental assays in laboratories while being accelerated by using computational methods. The characterization of the first crystal structure of the main protease (Mpro) in April 2020, an enzyme recruited by the virus to complete the replication and transcription steps, was a cornerstone. [9] Indeed, the publication of the crystal in the Protein Data Bank with code 6LU7 opened the door for massive virtual screening (VS). The seminal work by Jiang, Rao and Yang was also an initial attempt of using theory for the search of antiviral drugs. [9] These authors implemented an in‐house database of potential binding compounds in the virtual workflow scheme based on the Schrödinger suite of programs, [10] which integrates Glide as docking engine. [11] Most importantly, the best‐ranked candidates were tested in biological assays, including qRT‐PCR with Vero E6 cells at biosafety level‐3 (BSL‐3) laboratories. Six of the selected compounds were active against Mpro functionality, with half‐maximal inhibitory concentration (IC50) values in the range of 0.67 to 21.4 μM, [9] though initial optimism was tempered down by their low performance in hospitals. Since that work, many VS contributions have suggested other compounds for treating COVID‐19. Collacovid, a free web site that summarizes both peer‐reviewed publications and preprints with a focus on the advanced against the new virus, includes more than 700 entries for works with docking predictions. [12] However, VS without experimental verification is a high‐risk prediction that still carries a high false positive rate. [13] We defined such flaws, e. g., best‐ranked drugs by computational methods but without biological activity, as ‘false theoretical‐friends’. It is therefore critical to confirm VS outcomes with experiments to reach clinical stages.
Massive virtual screening of large chemical spaces
There are two excellent examples of ultra‐large virtual screenings with a focus on repurposing drug against the virus. Gorgulla, Arthanari and co‐workers developed what the authors defined as a virtual campaign (VirtualFlow) to search for SARS‐CoV‐2 inhibitors. [14] This was a huge computational effort; ca. 1 billion of molecules were docked into 17 different viral and host targets, with a total of 45 VS simulations. Drug candidates were retrieved from Enamine REAL database and the in‐stock subgroup of molecules deposited in the ZINC15 library of compounds.[ 15 , 16 ] VirtualFlow web site allows for the access to the top 1 million hits (∼0.1 % of the virtual hits), [17] which can be downloaded in DataWarrior format. [18] Of course, the screening of such large number of molecules required a cost‐effective computational method. Accordingly, data deposited in VirtualFlow is based on a rigid target protein while docking score is computed with QuickVina W and QuickVina 2.[ 19 , 20 , 21 ] Both versions use the score function of AutoDock Vina.[ 20 , 21 ] The conducted calculations with the active and dimerization sites for Mpro (labeled in VirtualFlow platform as VS16 and VS18, respectively) yields to a docking score ranging from −11.5 to −8.9 kcal/mol for the top 1 M, a narrow energetic window of 2.6 kcal/mol only. In a parallel contribution, Cherkasov and co‐workers at University of British Columbia screened the whole ZINC15 library rather than a subset, which contains more than 1 billion of compounds. [22] Docked poses were generated by Glide[ 10 , 11 ] within a deep learning platform. First 1000 hits were provided in the supporting material of that work, with docking score ranging from −11.3 to −9.0 kcal/mol. Both Gorgulla and Cherkasov screens used the same target, namely, the PDB structure with code 6LU7. [9]
Ranking hits
Theory reduces libraries from billions to thousands of molecules. This is a critical step, though still not enough for deciding the final candidates to be tested on a wet lab. A more recent study by Halazonetis and co‐workers further refines these abovementioned ultra‐large screens by imposing general requisites such as drug‐likeness and chemical diversity. [23] These additional criteria lead to 207 compounds from Gorgulla collection. [14] However, that screening results on a very weak activity, and all of them were discarded as Mpro inhibitors. On the contrary, the analysis of the 8 selected hits from Cherkasov series reveals that ZINC636416501 and ZINC373659060 compounds exhibit a moderate activity against the Mpro target, with IC50 values of 47 mM and 29 mM, respectively. The limited success of that first selection from massive screens motivated the present contribution, which aims for retrieving novel hits from these libraries.
Molecular Models and Experimental Methods
Computational details
From a theoretical point of view, there is a major dissimilarity between Gorgulla [14] and Cherkasov [22] approaches compared to the seminal work by Yang and co‐workers. [9] Although all these contributions performed docking calculations with the same X‐ray structure (PDB code 6LU7), only the contribution by Yang includes a second level of theory by applying the Molecular Mechanics/Generalized Born Surface Area (MMGBSA) method as implemented in Prime module, [24] which was used as a refinement of the raw VS outcomes. It is well‐known that docking score functions allow for an efficient cost/performance ratio when dealing with large molecular database. However, top‐ranked poses are usually close in energy to provide a clean selection criterium. A subsequent refinement (rescoring) is a required step if meaningful biological activities are looked for. [25] MMGBSA combines molecular mechanics calculations and continuum solvation models while allowing for receptor (protein) relaxation upon ligand binding. Consequently, MMGBSA has been proposed as a method to delivery more refined binding free energies and might be included in the screening workflow to increase the ‘ranking power’ of docking codes. [26]
This contribution aims to assess whether a rescoring strategy recovers other compounds from earlier massive screenings. We first compute the MMGBSA binding energies for the 1000 compounds listed in the publication by Cherkasov. [22] For the sake of comparison, we used the codes integrated in the Schrödinger suite as Yang [9] and Cherkasov, [22] including the force field version (OPLS3). [27] Accordingly, Protein Preparation Wizard module [28] was implemented to cure the Mpro structure deposited in the PDB entry 6LU7. The resulting model system were used to generate the grid at the binding site, e. g., centered into the catalytic dyad residues (His41 and Cys145). A similar procedure is implemented for rescoring the series by Gorgulla. [14] However, structures (poses) were not accessible at VirtualFlow portal. In that case, we used these set to perform the whole virtual screening workflow, including the generation of 3D structures with DataWarrior, following by a sequence that includes high through virtual screening (HTVS) and docking steps with Glide. To enlarge the conformation space of search, ligands are allowed to adopt up to 10 poses into the binding site. Best poses are picked up and used for computing MMGBSA binding energies. For the records, a larger selection was used herein. As discussed in Introduction, first 1 M of the compounds posted on VirtualFlow lies very close in the Vina score scale. If restricted to top‐1000 hits, docking score ranges from −11.50 to −10.30 kcal/mol. However, there are other hits with the same score value. Aiming to provide a wider view, we decided to set a Vina score threshold of −10.30 kcal/mol; 6411 candidates satisfy that requirement and are therefore incorporated into our rescoring strategy.
Experimental methods
The binding to Mpro was determined by using the protocol for protein expression, purification, and activity assay described elsewhere. [23] In that approach, the interaction of the selected molecules was assessed by using differential scanning fluorimetry. Thermal shift assays were conducted in duplicate tests with a LightCycler 480 multi‐well plates 96 (volume=20 μL, Mpro concentration=1 μM), which were subsequently preincubated at room temperature for 20 minutes. An acoustic liquid dispenser (Gen5‐Acoustic Transfer System; EDC Biosystems) is used for dispensing both selected molecules and SYPRO Orange binding dye (Sigma). Fluorescence measures used 465 nm as excitation wavelengths and 580 nm for emission in the temperature ranging from 20 up to 95 °C (step=0.05 °C/s; 11 measures/°C). After addition of the FRET substrate, fluorescence was acquired at 10 minutes intervals over 60 min. Compounds were purchased from Enamine, Molport and Vitas‐M.
Results and Discussion
Contrary to the selection criteria used by Cherkasov and Gorgulla, which were guided by docking score, we decided to move the focus towards the MMGBSA binding energies. A novel list of compounds is thus generated and tested in the lab. As discussed, our goal is to show whether such refinement helps to recover other active ligands from the massive screenings available in the literature.
Rescoring Cherkasov's series
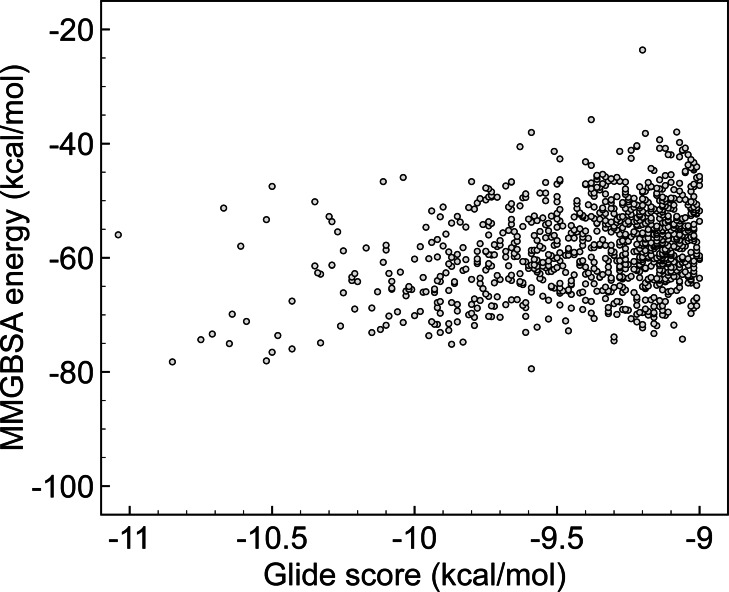
The reported series of ligands by Cherkasov and co‐workers was first assessed. [22] Chemical structures and computed Glide scores were retrieved from the provided supporting material in Ref. [22] and subsequently used for rescoring. Figure 1 illustrates the correlation between these two energetic parameters. It should be noted that the final list of 1000 hits published by Cherkasov only included best‐ranked candidates, e. g., molecules with a Glide score of at least −9.00 kcal/mol, which eventually limits the X‐axis scale. A close inspection of Figure 1 reveals that it is not possible to establish a simple linear dependence. First‐ranked compounds in the docking scale (with Glide in the range of −11.00/−10.50 kcal/mol, located on the left of the graphics) do not systematically correspond with the highest binding energies in the MMGBSA list. If the latter value is used as a selection parameter, novel molecules might be prioritized.
Figure 1.
Computed MMGBSA energies vs. Glide score using results from deep docking by Cherkasov Ref. [22]. All energies are given in kcal/mol.
Table 1 lists top‐compounds on the MMGBSA scale. Computed docking score values by Cherkasov are also included as a reference. It is remarkable that our selection, which is exclusively based on the more refined MMGBSA energies, mostly matches to Halazonetis choice. However, there are two novel compounds in the MMGBSA list that were missing in the earlier conducted experiments, e. g., ZINC1485985070 and ZINC856751528, which correspond to PV‐001017415655 and Z2654860553 entries in Enamine catalogue, respectively. It is worth stressing that these compounds were not at the top hit of deep learning as they correlate with medium docking score values (i. e., the associated Glide scores are less than −10.00 kcal/mol).
Table 1.
Comparison of the predicted energies in the Glide‐based Deep Docking screening as implemented by Cherkasov [a] and the refined MMGBSA binding energies counterparts (both in kcal/mol).
|
Compound |
Glide [kcal/mol][a] |
MMGBSA [kcal/mol] |
|---|---|---|
|
ZINC1485985070 |
−9.59 |
−79.45 |
|
ZINC636416501[b] |
−10.85 |
−78.24 |
|
ZINC67726685[b] |
−10.52 |
−78.06 |
|
ZINC544491491[b] |
−10.50 |
−76.55 |
|
ZINC543523837[c] |
−10.43 |
−75.97 |
|
ZINC856751528 |
−9.87 |
−75.15 |
|
ZINC544491494[d] |
−10.65 |
−75.04 |
[a] Energies reported by Cherkasov and co‐workers, Ref. [22]. [b] Included in the experimental assays by Halazonetis, Ref. [23]. [c] Same scaffold than ZINC544491491. [d] ZINC544491491 enantiomeric form.
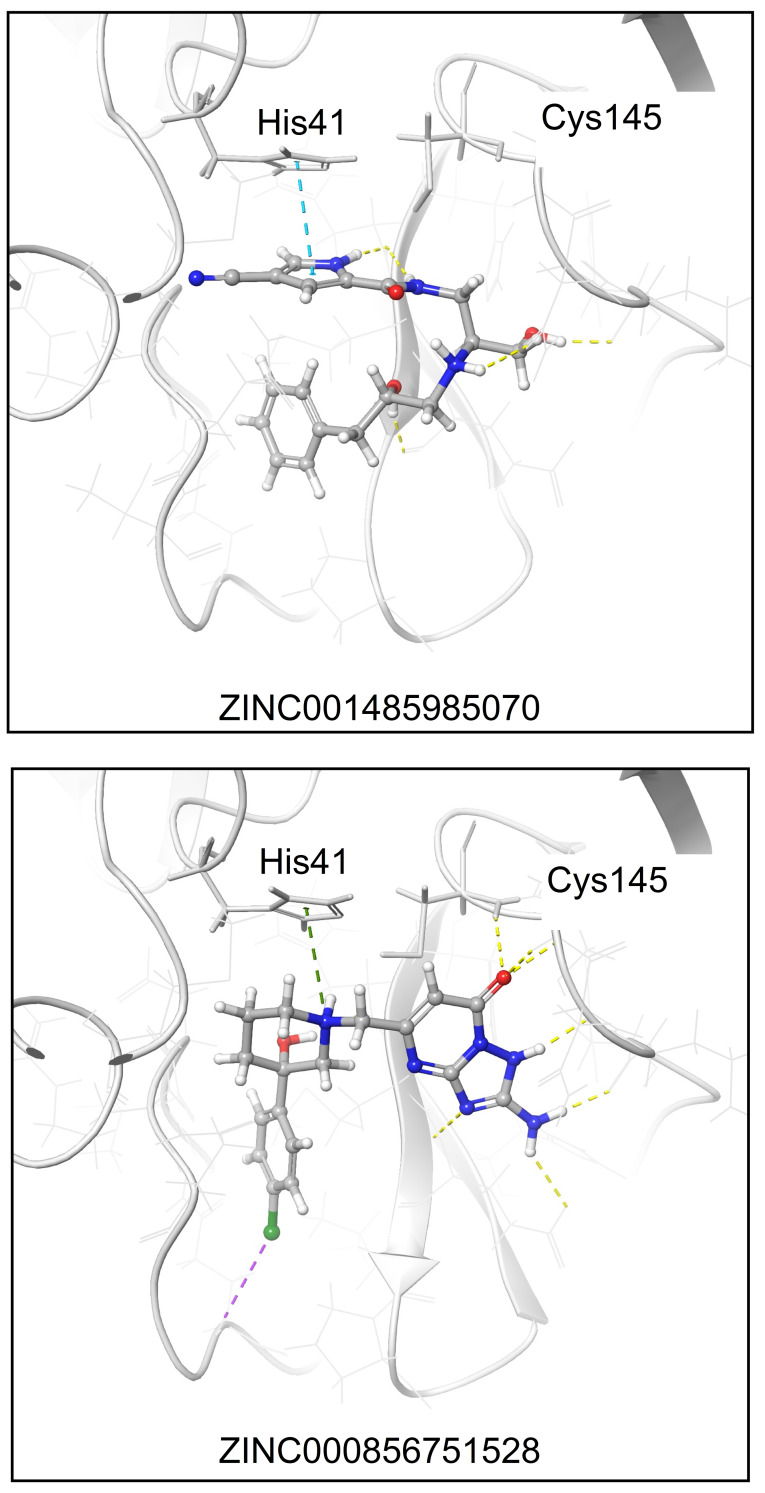
To further investigate the interaction of these two hits with the Mpro target, Figure 2 shows the relaxed structures resulting from MMGBSA. As displayed, the pyrrole motif in ZINC1485985070 allows for a face‐to‐face π‐π stacking (indicated with a blue dashed line) with the catalytic His41 with additional contacts in the binding site by hydrogen‐bonds (H‐bonds, marked with yellow dashed lines). ZINC856751528 is anchored by a π‐cation interaction (green dashed line) with His41 and several H‐bond connections to the region of the Cys145 residue (yellow dashed lines). As ligands seem to be compatible with the Mpro catalytic pocket, both are retained herein for further experimental tests.
Figure 2.
Representation of the ZINC001485985070 and ZINC000856751528 binding modes resolved by MMGBSA. Ligands and Mpro are displayed as balls & sticks and grey cartoons, respectively. Non‐covalent interactions are represented with dashed lines. Color code: blue, π‐π stacking; green, π‐cation stacking; yellow, H‐bonds; purple, halogen‐bonds.
Rescoring Gorgulla's series
A similar procedure is next implemented for rescoring the series by Gorgulla, which is freely available at the website VirtualFlow. [14] As noted above, that alternative series used the Enamine REAL library as the main source of compounds, though also includes a subgroup from ZINC15, namely, the in‐stock set. However, one might expect a minimal overlap with Cherkasov's list as most of the top hits by deep screening (ca. 99 %) are not included in the ZINC15 in‐stock library. [22] Since structural data were not accessible in the VirtualFlow sever, the listed names of compounds are used to generate all 3D structures in DataWarrior. [18] The resulting model systems are subsequently ported to the VS workflow implemented in the Schrödinger suite of programs, [10] including an initial HTVS followed by docking with Glide, and a MMGBSA calculation for the final refinement.
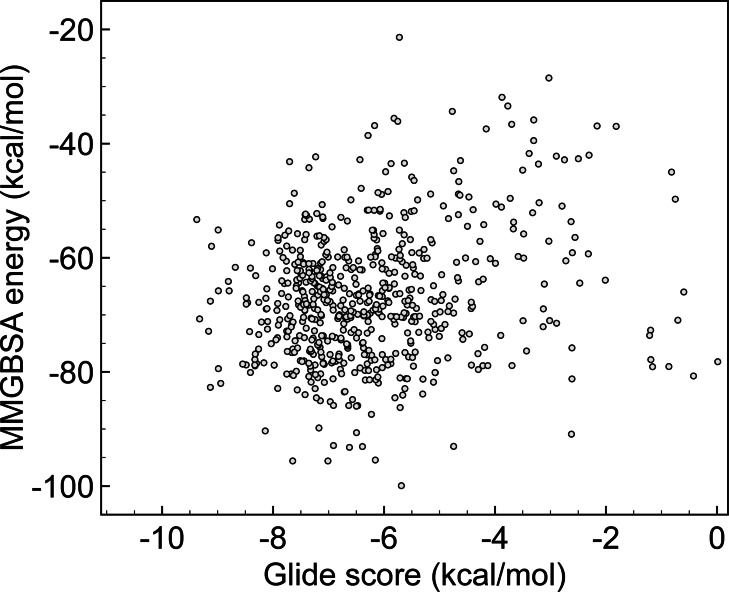
To draw parallels between massive screens, Figure 3 compares MMGBSA energies vs. Glide after rescoring the selected compounds by Gorgulla. [14] We identify several poses with large negative MMGBSA binding energies, e. g., located in the −80.00/−100.00 kcal/mol window, a region that was clean in Cherkasov series (Figure 1). That qualitative picture is completed with the numeric outputs provided in Table 2, which ranks molecules according to MMGBSA values. Original Vina scores are also included. One may note that Glide scores are now more disperse. Indeed, Vina function yields to very similar scores (in the range of −10.00/−10.60 kcal/mol), while that homogeneity disappears with Glide (−4.69/−9.13 kcal/mol). We select molecules with the higher (more negative) MMGBSA energies up to reach the first compound tested by Halazonetis (PV‐0019557066). [23]
Figure 3.
Computed MMGBSA energies vs. Glide score (both in kcal/mol) using results from Gorgulla Ref. [14].
Table 2.
Comparison of the predicted Vina scores as implemented in the VirtualFlow by Gorgulla,[a] computed Glide scores and the refinement at the MMGBSA level of theory (all energies in kcal/mol).
|
Compound[a] |
Vina [kcal/mol][b] |
Glide [kcal/mol] |
MMGBSA [kcal/mol] |
|---|---|---|---|
|
Z1081913254 |
−10.00 |
−5.59 |
−99.93 |
|
Z299581070 |
−10.00 |
−6.39 |
−93.07 |
|
Z1884544179 |
−10.00 |
−4.69 |
−93.04 |
|
ZINC408533192 |
−10.10 |
−6.50 |
−90.64 |
|
ZINC13598149 |
−10.00 |
−7.17 |
−89.81 |
|
PV‐001803361903 |
−10.10 |
−6.23 |
−87.43 |
|
ZINC408526152 |
−10.60 |
−6.57 |
−86.00 |
|
ZINC408528197 |
−10.00 |
−6.48 |
−85.90 |
|
Z1158074909 |
−10.00 |
−6.38 |
−85.88 |
|
ZINC6500491 |
−10.30 |
−6.97 |
−85.22 |
|
ZINC13756054 |
−10.50 |
−7.14 |
−85.05 |
|
ZINC31829105 |
−10.40 |
−5.80 |
−84.52 |
|
ZINC32609526 |
−10.60 |
−7.42 |
−83.98 |
|
PV‐001839244402 |
−10.10 |
−5.03 |
−83.87 |
|
ZINC408529658 |
−10.40 |
−6.70 |
−83.44 |
|
ZINC13509896 |
−10.60 |
−7.57 |
−83.15 |
|
PV‐001863000372 |
−10.00 |
−5.63 |
−82.93 |
|
ZINC32609522 |
−10.30 |
−7.92 |
−82.89 |
|
ZINC13756065 |
−10.30 |
−6.10 |
−81.95 |
|
PV‐001803361903 |
−10.10 |
−9.13 |
−82.70 |
|
Z2232450399 |
−10.20 |
−5.11 |
−81.79 |
|
ZINC101470498 |
−10.00 |
−6.26 |
−81.37 |
|
ZINC64790056 |
−10.00 |
−5.58 |
−81.18 |
|
ZINC4725799 |
−10.20 |
−6.11 |
−80.89 |
|
PV‐001955706661[c] |
−10.00 |
−8.40 |
−80.09 |
[a] Compounds with Z and PV codes are from Enamine library. [b] Energies reported by Gorgulla and co‐workers, Ref. [14]. [c] Included in the experimental assays by Halazonetis, Ref. [23].
In vitro assays
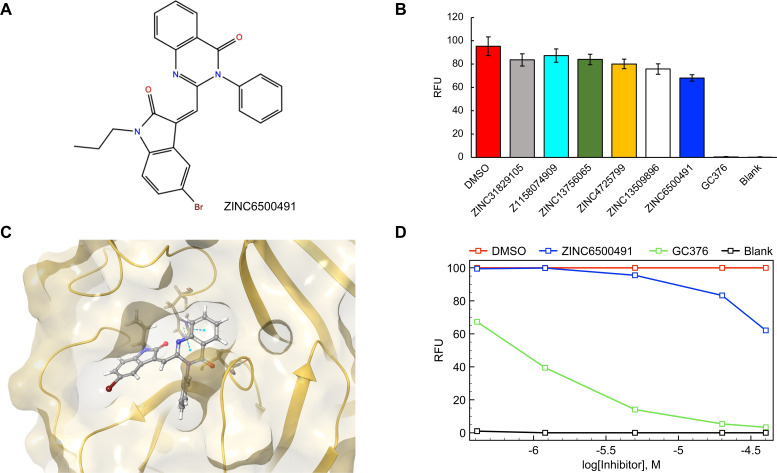
As described, duplicated experiments are performed for all compounds listed in Tables 1 and 2. DMSO and blank were used as controls. GC376, which was used as a positive Mpro inhibitor control by Halazonetis and co‐workers, [23] was also tested. Only ZINC6500491 (MolPort‐000‐706‐737), which ranks 10th in the rescored VirtualFlow list (Table 2) is found to have a measurable inhibition, with a main protease activity of 62 % at 40 μM [all other in vitro activities at a final concentration of 40 μM for their ability to inhibit Mpro are provided in the Supporting Information]. Dose‐response curve for compound ZINC6500491 was assessed at 0.4, 1.2, 5, 20 and 40 μM as final concentration. Unfortunately, Figure 4 illustrates that this compound inhibits Mpro with a IC50 value of about 0.8 mM, which is far from the expected activity for a drug with applicability in the treatments of COVID‐19. Our finding contrasts with the measured IC50 for GC376 (positive control), which is about 1 μM, with a Mpro activity of 3 % at 40 μM.
Figure 4.
(A) Chemical structure of the only active drug, ZINC6500491. (B) Inhibitory activity of the top hits tested at a concentration of 40 μM in an in vitro protease‐activity assay. The increase in fluorescence intensity was normalized to the DMSO control, where RFU states for relative fluorescence units. (C) Predicted binding mode of ZINC6500491 in the active site. Dashed blue lines represent the π‐π interaction with the catalytic His41 residue. (D) Dose‐response curves for ZINC6500491 at 0.4, 1.2, 5, 20 and 40 μM. DMSO, blank and the positive Mpro inhibitor GC376 are used as controls.
Conclusions
Virtual screening (VS) of ultra‐large libraries of compounds must use methods light enough to deal with a huge number of possible candidates. This is the logical prerequisite to deliver quick results at an affordable computational cost. However, docking codes (the core engine of VS approaches) are based on a series of simplifications that prevent a direct extrapolation to biological scenarios. In addition, there are other physical‐chemical properties that might affect to the route that the ligand must overcome prior reaching its target, including solubility, pK a, and logP, to cite a few. All in all, it becomes essential to increase both accuracy and criticism of the produced outputs.
One can envisage several alternatives to further enhance VS predictions if experiments are not accessible/available. More than one score function might be combined to produce a consensus top‐ranked list,[ 29 , 30 ] while X‐ray screening have recently identified allosteric sites that might provide a wider description of virus target.[ 31 , 32 ] In addition, pockets are not static but prone to modifications in in vivo conditions, and a preliminary molecular dynamics (MD) stage could be implemented to produce trajectories of the targets. [33] A series of representative structures (or clusters) can be subsequently used for conducting VS simulations. MD might be also used to confirm the stability of the binding pose.[ 34 , 35 ] Finally, the simulation of a reduced number of drugs allows to use of more advanced levels of theory, e. g., ab initio or multiscale QM/MM.[ 36 , 37 ] Such refinements, which are extensive to other viral proteins, [38] pave the way for improving predictions, though all are concomitant with the raise of the computational cost.
Of course, VS methods are still welcomed to help into the design of the missing antivirals, including COVID‐19. Our purpose is to advise that the overproduction of VS works that simulate a single crystal and a few randomly selected molecules should be reduced. Otherwise, standard VS works risks repurposing most of approved drugs as plausible treatments for COVID‐19. The increasing number of ‘false theoretical‐friends’ in the literature will not help to rationalize the search of an efficient antiviral. Indeed, our work is a clear example. Without the performed in vitro assays, we might have presented ZINC6500491 as a promising Mpro inhibitor as it is predicted to bind the catalytic site by anchoring His41. However, our experiments revealed that it fails to block the enzymatic activity. VS is a valuable tool, but post‐refinement and/or experimental data should be always reported if a real clinical impact is sought. That conclusion is equally valid for the current pandemic as well as for the next one.
Conflict of interest
The authors declare no conflict of interest.
1.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Supporting Information
Acknowledgements
The author is in debt with Giacomo Rossetti and Marianna Ossorio, Department of Molecular Biology, University of Geneva, for providing the access to all experimental facilities and biological data. This work used the computational resources of the Centro de Supercomputación de Galicia (CESGA) supported by the Partnership for Advanced Computing in Europe (PRACE) COVID‐19 Fast Track Call for Proposals – Allocation Decision – Proposal COVID19‐85.
J. P. Cerón-Carrasco, ChemMedChem 2022, 17, e202200278.
Data Availability Statement
The data that support the findings of this study are available in the Supporting Information for this article.: (i) in vitro inhibitory activity of the top hits tested at a concentration of 40 μM in an in vitro protease‐activity assay, with fluorescence data acquired at 10 minutes intervals over 60 min; (ii) dose‐response curves for the only active drug and the positive control, ZINC6500491 and GC376, respectively; (iii) PDB files with the generated poses.
References
- 1. Pardi N., Weissman D., Nat. Biomed. Eng. 2020, 4, 1128–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Krammer F., Nature 2020, 586, 516–527. [DOI] [PubMed] [Google Scholar]
- 3. Gupta R. K., Nat. Rev. Immunol. 2021, 21, 340–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Draft Landscape of COVID-19 Candidate Vaccines. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (last accessed, 20th February 2021).
- 5. Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G., Cell Res. 2020, 30, 269–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO Solidarity Trial Consortium, N. Engl. J. Med. 2021, 384, 497–511. [DOI] [PMC free article] [PubMed]
- 7. Cully M., Nat. Rev. Drug Discovery 2022, 21, 3–5. [DOI] [PubMed] [Google Scholar]
- 8. Reis G., dos Santos Moreira-Silva E. A., Medeiros Silva D. C., Thabane L., Cruz Milagres A., Ferreira T. S., Quirino dos Santos C. V., de Souza Campos V. H., Ribeiro Nogueira A. M., Figueiredo Guimaraes de Almeida A. P., Diniz Callegari E., Dias de Figueiredo Neto A., Monteiro Savassi L. C., Campos Simplicio M. I., Barra Ribeiro L., Oliveira R., Harari O., Forrest J. I., Ruton H., Sprague S., McKay P., Glushchenko A. V., Rayner C. R., Lenze E. J., Reiersen A. M., Guyatt G. H., Mills E. J., Lancet Glob Health 2022, 10, e42–51.34717820 [Google Scholar]
- 9. Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L. W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H., Nature 2020, 582, 289–293. [DOI] [PubMed] [Google Scholar]
- 10.Schrödinger Release 2019–1. Schrödinger, LCC, New York, 2019.
- 11. Friesner R. A., Banks J. L., Murphy R. B., Halgren T. A., Klicic J. J., Mainz D. T., Repasky M. P., Knoll E. H., Shelley M., Perry J. K., Shaw D. E., Francis P., Shenkin P. S., J. Med. Chem. 2004, 47, 1739–1749. [DOI] [PubMed] [Google Scholar]
- 12.https://www.collabovid.org (last accessed, 20th February 2022).
- 13. Scior T., Bender A., Tresadern G., Medina-Franco J. L., Martínez-Mayorga K., Langer T., Cuanalo-Contreras K., Agrafiotis D. K., J. Chem. Inf. Model. 2012, 52, 867–881. [DOI] [PubMed] [Google Scholar]
- 14. Gorgulla C., Padmanabha Das K. M., Leigh K. E., Cespugli M., Fischer P. D., Wang Z.-F., Tesseyre G., Pandita S., Shnapir A., Calderaio A., Gechev M., Rose A., Lewis N., Hutcheson C., Yaffe E., Luxenburg R., Herce H. D., Durmaz V., Halazonetis T. D., Fackeldey K., Patten J. J., Chuprina A., Dziuba I., Plekhova A., Moroz Y., Radchenko D., Tarkhanova O., Yavnyuk I., Gruber C., Yust R., Payne D., Näär A. M., Namchuk M. N., Davey R. A., Wagner G., Kinney J., Arthanari H. A., iScience 2021, 24, 102021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.https://zinc15.docking.org (last accessed, 20th February 2022).
- 16.https://enamine.net/compound-collections (last accessed, 20th February 2022).
- 17.https://vf4covid19.hms.harvard.edu (last accessed, 20th February 2022).
- 18.https://openmolecules.org (last accessed, 20th February 2022).
- 19. Alhossary A., Handoko S. D., Mu Y., Kwoh C.-K., Bioinformatics 2015, 31, 2214–2216. [DOI] [PubMed] [Google Scholar]
- 20. Hassan N. M., Alhossary A. A., Mu Y., Kwoh Y. C.-K., Sci. Rep. 2017, 7, 15451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Trott O., Olson A. J., J. Comput. Chem. 2020, 31, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ton A.-T., Gentile F., Hsing M., Ban F., Cherkasov A., Mol. Inf. 2020, 39, 2000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rossetti G. G., Ossorio M. A., Rempel S., Kratzel A., Dionellis V. S., Barriot S., Tropia L., Gorgulla C., Arthanari H., Thiel V., Mohr P., Gamboni R., Halazonetis T. D., Sci. Rep. 2022, 12, 2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schrödinger Release 2019–1: Prime, Schrödinger, LLC, New York, NY, 2021.
- 25. Hou T., Wang J., Li Y., Wang W., J. Chem. Inf. Model. 2011, 51, 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sun H., Li Y., Shen M., Tian S., Xu L., Pan P., Guan Y., Hou T., Phys. Chem. Chem. Phys. 2014, 28, 22035–22045. [DOI] [PubMed] [Google Scholar]
- 27. Harder E., Damm W., Maple J., Wu C., Reboul M., Xiang J. Y., Wang L., Lupyan D., Dahlgren M. K., Knight J. L., Kaus J. W., Cerutti D. S., Krilov G., Jorgensen W. L., Abel R., Friesner R. A., J. Chem. Theory Comput. 2016, 12, 281–296. [DOI] [PubMed] [Google Scholar]
- 28.Schrödinger Release 2019–1: Protein Preparation Wizard; Epik, Schrödinger, LLC, New York, NY, 2019.
- 29. Muñoz Robles V., Ortega-Carrasco E., Alonso-Cotchico L., Rodriguez-Guerra J., Lledós A., Maréchal J. D., ACS Catal. 2015, 5, 2469–2480. [Google Scholar]
- 30. Francés-Monerris A., García-Iriepa C., Iriepa I., Hognon C., Miclot T., Barone G., Monari A., Marazzi M., Phys. Chem. Chem. Phys. 2021, 23, 22957–2297. [DOI] [PubMed] [Google Scholar]
- 31. Günther S., Reinke P. Y. A., Fernández-García Y., Lieske J., Lane T. J., Ginn H. M., Koua F. H. M., Ehrt C., Ewert W., Oberthuer D., Yefanov O., Meier S., Lorenzen K., Krichel B., Kopicki J. D., Gelisio L., Brehm W., Dunkel I., Seychell B., Gieseler H., Norton-Baker B., Escudero-Pérez B., Domaracky M., Saouane S., Tolstikova A., White T. A., Hänle A., Groessler M., Fleckenstein H., Trost F., Galchenkova M., Gevorkov Y., Li C., Awel S., Peck A., Barthelmess M., Schlünzen F., Lourdu Xavier P., Werner N., Andaleeb H., Ullah N., Falke S., Srinivasan V., França B. A., Schwinzer M., Brognaro H., Rogers C., Melo D., Zaitseva-Doyle J. J., Knoska J., Peña-Murillo G. E., Mashhour A. R., Hennicke V., Fischer P., Hakanpää J., Meyer J., Gribbon P., Ellinger B., Kuzikov M., Wolf M., Beccari A. R., Bourenkov G., von Stetten D., Pompidor G., Bento I., Panneerselvam S., Karpics I., Schneider T. R., Garcia-Alai M. M., Niebling S., Günther C., Schmidt C., Schubert R., Han H., Boger J., Monteiro D. C. F., Zhang L., Sun X., Pletzer-Zelgert J., Wollenhaupt J., Feiler C. G., Weiss M. S., Schulz E. C., Mehrabi P., Karničar K., Usenik A., Loboda J., Tidow H., Chari A., Hilgenfeld R., Uetrecht C., Cox R., Zaliani A., Beck T., Rarey M., Günther S., Turk D., Hinrichs W., Chapman H. N., Pearson A. R., Betzel C., Meents A., Science 2021, 372, 642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. El Ahdab D., Lagardère L., Inizan T. J., Célerse F., Liu C., Adjoua O., Jolly L.-H., Gresh N., Hobaika Z., Ren P., Maroun R. G., Piquemal J.-P., J. Phys. Chem. Lett. 2021, 12, 6218–6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schmidtke P., Bidon-Chanal A., Luque F. J., Barril X., Bioinformatics 2011, 27, 3276–3285. [DOI] [PubMed] [Google Scholar]
- 34. Sun C., Zhu L., Zhang C., Song C., Wang C., Zhang M., Xie Y., H. F. Schaefer III , J. Comput. Chem. 2017, 39, 889–900. [DOI] [PubMed] [Google Scholar]
- 35. Hari Narayana Moorthy N. S., Brás N. F., Ramos M. J., Fernandes P. A., In Silico Pharmacol. 2021, 9, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ramos-Guzmán C. A., Ruiz-Pernía J. J., Tuñón I., ACS Catal. 2020, 10, 12544–12554. [DOI] [PubMed] [Google Scholar]
- 37. Ramos-Guzmán C. A., Ruiz-Pernía J. J., Tuñón I., ACS Catal. 2021, 11, 4157–4168. [DOI] [PubMed] [Google Scholar]
- 38. Francés-Monerris A., Hognon C., Miclot T., García-Iriepa C., Iriepa I., Terenzi A., Grandemange S., Barone G., Marazzi M., Monari A., J. Proteome Res. 2020, 19, 4291–4315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Supporting Information
Data Availability Statement
The data that support the findings of this study are available in the Supporting Information for this article.: (i) in vitro inhibitory activity of the top hits tested at a concentration of 40 μM in an in vitro protease‐activity assay, with fluorescence data acquired at 10 minutes intervals over 60 min; (ii) dose‐response curves for the only active drug and the positive control, ZINC6500491 and GC376, respectively; (iii) PDB files with the generated poses.