Summary
Root‐colonizing bacteria have been intensively investigated for their intimate relationship with plants and their manifold plant‐beneficial activities. They can inhibit growth and activity of pathogens or induce defence responses. In recent years, evidence has emerged that several plant‐beneficial rhizosphere bacteria do not only associate with plants but also with insects. Their relationships with insects range from pathogenic to mutualistic and some rhizobacteria can use insects as vectors for dispersal to new host plants. Thus, the interactions of these bacteria with their environment are even more complex than previously thought and can extend far beyond the rhizosphere. The discovery of this secret life of rhizobacteria represents an exciting new field of research that should link the fields of plant–microbe and insect–microbe interactions. In this review, we provide examples of plant‐beneficial rhizosphere bacteria that use insects as alternative hosts, and of potentially rhizosphere‐competent insect symbionts. We discuss the bacterial traits that may enable a host‐switch between plants and insects and further set the multi‐host lifestyle of rhizobacteria into an evolutionary and ecological context. Finally, we identify important open research questions and discuss perspectives on the use of these rhizobacteria in agriculture.
Introduction
Many higher organisms co‐occur in the same habitat and can interact with each other, providing opportunities for the exchange of microbes between, for example, plants and animals. This exchange can have significant ecological implications but has been little explored.
Over the past decades, it has become clear that the plant rhizosphere microbiome is of great importance for plant health (Berendsen et al., 2012; Mendes et al., 2013; Berg et al., 2017; Trivedi et al., 2020). Rhizosphere microbes can be present in the soil directly around the root, on the root surface, or inside the plant root. They are mostly recruited from bulk soil by plant root exudates (Sasse et al., 2018), and can potentially be pathogenic but also beneficial to the plant. Within the bacterial community of the rhizosphere, many individual members as well as consortia of different species have been identified as plant‐beneficial (Berendsen et al., 2012; Berendsen et al., 2018). They can suppress plant diseases by directly inhibiting pathogens, or indirectly by inducing defence responses in the plant (Pieterse et al., 2014), and some can also promote plant growth in the absence of pathogens (Compant et al., 2010). This makes these bacteria interesting candidates for the biological control of crop diseases or as biological fertilizers.
Plants do not only interact with microbes but also with higher organisms, including insects. Positive interactions with insects such as pollinators and insects that spread plant seeds are essential for plant reproduction. In contrast, herbivorous insects present a constant threat for plant growth and survival. Moreover, many of these insects are important vectors of viral and bacterial plant pathogens (Kluth et al., 2002). Parasitoids and predatory insects may function as bodyguards of the plant by keeping these vectors and pest insects at bay (van Lenteren and Manzaroli, 1999). Like plants, insects harbour microbiomes that include beneficial bacteria and fungi that provide nutrients or protection against pathogens, parasites and predators (Douglas, 2015). They may also contain microorganisms with entomopathogenic activities. These could serve as a source for biological control agents to control pest insects, but they may also threaten crop yields in the case of pollinator pathogens.
The microbiomes of plants and insects have mostly been studied separately from each other, but since plant–insect interactions are so ubiquitous, it is not surprising that interactions between plant‐associated bacteria and insects, and between insect‐associated bacteria and plants have been observed. Recent studies (Allard et al., 2018; Rebolleda Gómez and Ashman, 2019) suggest that pollinators can alter microbial communities of flowers, and it is even suggested that these communities can be used as an indicator of which pollinators visited the plant (Ushio et al., 2015). These studies show that bacteria can be transferred from insects to plants, but they did not examine the function of these bacteria in both insects and plants.
Most examples of interactions between plant‐associated bacteria and insects have been reported for pathogens and pest insects, especially those inhabiting above‐ground parts of the plant. The dispersal of plant pathogens by insect vectors is a common phenomenon (Orlovskis et al., 2015). For example, the bacteria that cause the Huanglongbing disease of citrus are spread from plant to plant by a phloem‐feeding insect (da Graça et al., 2016). A review by Nadarasah and Stavrinides (2011) highlights that insects can also serve as a host for plant pathogenic bacteria. They state that some plant pathogens may originate from entomopathogens, suggesting that plants are not their original habitat. Diminished insect virulence may facilitate the vectoring role of insects in spreading the pathogen to its plant host. For plant pathogenic bacteria, frequent encounters with insects that share their habitat have resulted in the evolution of mechanisms that allow colonization of two different hosts. A loss of virulence or increased virulence towards one or both hosts could change the nature of the symbioses over time, for example from entomopathogenicity to commensalism, or even insect mutualism.
Plant‐beneficial rhizosphere bacteria can influence plant–insect interactions by activating defence responses in plants or by changing volatile compound emissions, and therefore attractiveness, of the plant (Pineda et al., 2013, 2017; Pangesti et al., 2015). In turn, in response to insect herbivory, plants can alter their microbiome to increase resistance to herbivory for the following generation (Kong et al., 2016; Pineda et al., 2017).
Even though many insects live in and on the soil, close to plant roots, direct interactions between insects and rhizosphere bacteria have received little attention. In the late 90s, experiments in controlled microcosms suggested that a Pseudomonas chlororaphis strain could be dispersed by insects (Snyder et al., 1998, 1999). Almost a decade later, the discovery that certain plant‐beneficial Pseudomonas strains produce insecticidal compounds that are highly toxic to a range of pest insects (Péchy‐Tarr et al., 2008) triggered investigations on insects as alternative hosts for these bacteria. Recently, more evidence emerged that plant‐beneficial rhizosphere bacteria can use insects as an alternative host and dispersal vector (Flury et al., 2019; Kim et al., 2019). Perhaps this phenomenon is as common for plant‐beneficial rhizosphere bacteria as it is for foliar plant‐pathogenic bacteria.
In this review, we discuss recently discovered examples of rhizosphere bacteria with an alternative insect‐associated lifestyle, bacterial traits that facilitate this multi‐host lifestyle, how such a lifestyle may evolve, and lastly the ecological and agricultural implications.
Insects as vectors and alternative hosts for rhizosphere bacteria
For plant‐pathogenic bacteria, it is well‐known that they can be vectored by insects (Nadarasah and Stavrinides, 2011; Orlovskis et al., 2015). Like plant pathogens, rhizosphere bacteria frequently encounter insects with which they share their habitat. Lately, evidence is rising that certain plant‐beneficial rhizosphere bacteria can successfully colonize insects and use them as a means of dispersal to the rhizosphere of new host plants. In the following, we discuss two recently published cases of this phenomenon.
Plant‐beneficial Pseudomonas protegens and chlororaphis – opportunistic pathogens of insects?
The genus Pseudomonas is well‐known for its plant‐beneficial rhizosphere species and their role in disease‐suppressive soils and plant growth promotion (Haas and Défago, 2005; Mercado‐Blanco and Bakker, 2007; Kupferschmied et al., 2013; Almario et al., 2014; Thomashow et al., 2019). Among this group of rhizosphere bacteria, Pseudomonas protegens strains such as CHA0 and Pf‐5 can protect plants against various root pathogens (Haas and Défago, 2005; Kupferschmied et al., 2013) (Fig. 1) and can also induce systemic resistance against foliar diseases (Maurhofer et al., 1994; Iavicoli et al., 2003). Interestingly, both P. protegens strains also display an alternative lifestyle in insects. They produce the Fit toxin, an insecticidal protein that is closely related to the Makes caterpillars floppy (Mcf) toxin of Photorhabdus and Xenorhabdus species, both symbionts of entomopathogenic nematodes. Pseudomonas protegens strains kill insect larvae not only when injected into the insect hemocoel (Péchy‐Tarr et al., 2008), but also when fed to larvae of several lepidopteran and dipteran species (Olcott et al., 2010; Ruffner et al., 2013; Loper et al., 2016; Rangel et al., 2016; Ruiu and Mura, 2021) (Fig. 1). Within the P. fluorescens group only the species P. protegens and its close relative Pseudomonas chlororaphis exhibit strong insecticidal activity (Ruffner et al., 2015; Flury et al., 2016; Loper et al., 2016), clearly distinguishing them from other plant‐beneficial pseudomonads.
Fig. 1.
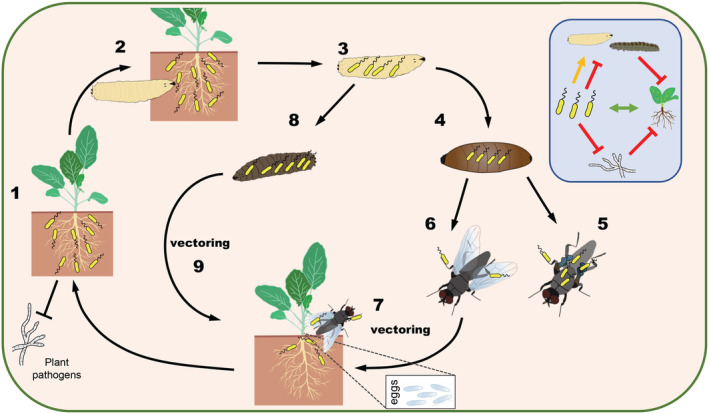
Pseudomonas protegens has a multi‐host lifestyle in plants and insects and can be vectored to new plants by the cabbage root fly Delia radicum. 1. The rhizosphere is colonized by P. protegens, which protects the plant against pathogenic fungi. 2. Insect larvae that feed on the roots of the plant ingest P. protegens cells. 3. Pseudomonas protegens persists in larvae of the cabbage root fly (D. radicum) and other insect larvae. 4. Pseudomonas protegens persists in D. radicum pupae. 5. The pupae develop into deformed adults or 6. into healthy flies. 7. Healthy D. radicum flies lay their eggs at the base of the plant on the soil and can transfer P. protegens cells to the rhizosphere. 8. When P. protegens kills the insect host, it mass replicates inside the larvae. 9. Pseudomonas protegens cells escape from the dead larvae, and migrate into the soil, from where they colonize the rhizosphere. Inlay: Diagram that indicates the nature of the interactions of the bacteria with the insect, the plant and soilborne plant pathogens. Green arrow, beneficial; orange arrow, neutral; red inhibition arrow, negative interaction. The figure has been adapted from Flury et al., 2019 .
Many factors contributing to the insecticidal activity of P. protegens and P. chlororaphis strains have been identified over the last years (Péchy‐Tarr et al., 2008; Flury et al., 2016, 2017; Keel, 2016; Kupferschmied et al., 2016; Loper et al., 2016; Rangel et al., 2016; Vacheron et al., 2019; Vesga et al., 2020). Among them are antimicrobial metabolites, i.e. cyclic lipopeptides, rhizoxin and hydrogen cyanide, which are involved in pathogen suppression and insecticidal activity (Flury et al., 2017), but also factors that are specifically expressed inside insects (Péchy‐Tarr et al., 2013; Kupferschmied et al., 2014; Flury et al., 2017; Vesga et al., 2020), including the Fit toxin, a chitinase, and a two‐partner secretion system. Strain CHA0 has been shown to use these virulence factors and a type VI secretion device with associated toxic effectors to compete with the native insect gut microflora (Vacheron et al., 2019) and to overcome the gut epithelial barrier (Vesga et al., 2020). If the strain reaches the hemocoel it multiplies to very high numbers leading to fatal septicemia. The bacteria then use the insect cadaver as a vessel for mass reproduction, and the billions of resulting bacteria can spread and recolonize plant roots (Fig. 1).
The ecological relevance of the insect‐associated lifestyle of these pseudomonads is not fully understood yet. For P. protegens lethal oral infections are mainly observed in experiments with laboratory‐reared insect larvae fed on leaves or an artificial diet inoculated with the bacterium. The susceptible insects were either leaf‐feeding lepidopteran species or dipteran species such as Drosophila melanogaster or Musca domestica, which normally feed on fruits or rotting organic material respectively (Péchy‐Tarr et al., 2008, 2013; Olcott et al., 2010; Ruffner et al., 2013; Flury et al., 2016; Loper et al., 2016; Rangel et al., 2016; Ruiu and Mura, 2021). However, P. protegens had no or very little effect on the survival of root‐feeding dipteran and coleopteran pests (Chiriboga et al., 2018; Flury et al., 2019; Jaffuel et al., 2019). This might be due to the different physiology of these insect groups, due to lower bacterial dosages taken up when larvae feed on colonized roots, or due to co‐evolution of root‐feeding insects with rhizobacteria towards an attenuated virulence. The latter would represent an advantage for pseudomonads if the survival of the insects enables the transportation of the bacteria to new root habitats.
Indeed, CHA0 can persist in Delica radicum and Otiorhynchus sulcatus throughout different developmental stages (Flury et al., 2019) until the imago (Fig. 1). This is of significant ecological impact, since adult insects cover larger distances, especially when in search of new host plants for egg deposition. In fact, CHA0 ingested by larvae of D. radicum can be vectored by the resulting adults from the roots of one plant to the roots of another plant under experimental conditions (Flury et al., 2019) (Fig. 1). Already in the 90s a study by Snyder and colleagues showed that P. chlororaphis strain L11, a strong root‐colonizer with the ability to move into the foliage, can persist from the larval to the adult stage in the southern corn rootworm, Diabrotica undecimpunctata subsp. howardi, and can be vectored between corn plants (Snyder et al., 1998). While nothing is known about the biocontrol ability of L11 and the study has received little attention for a long time, this finding now strongly supports the hypothesis that P. protegens and related strains can use insects as means of dispersal.
For many years, interactions with insects had only been studied for plant‐derived P. protegens and P. chlororaphis strains. However, a recent isolation effort from soil organisms revealed that both species are also naturally associated with healthy coleopteran insects and myriapods (Vesga et al., 2021). Like root isolates, the strains isolated from insects have both lepidopteran killing and plant disease suppressive abilities (Vesga et al., 2021). This supports the hypothesis that insects are alternative hosts for these bacteria in natural ecosystems.
In summary, P. protegens and P. chlororaphis can adopt contrasting lifestyles as effective colonizers of plant roots and insects. The same strains possess the tools enabling them to colonize both hosts and likely to switch between them. While the interaction of the bacteria with plants is beneficial for both partners, for the insects it can be commensal or pathogenic (Fig. 1). They probably use the insect as vector for dispersal or as a vessel for survival and multiplication.
Mutualistic interactions between Streptomyces globisporus and pollinators
While P. protegens is either pathogenic or commensal to its insect hosts, a Streptomyces globisporus strain that is almost identical to a plant‐beneficial Streptomyces sp. found in the rhizosphere can engage in a mutualistic interaction with insects (Kim et al., 2019). Streptomyces is a genus of Gram‐positive filamentous bacteria that can be found in a wide variety of habitats ranging from soils (Delgado‐Baquerizo et al., 2018) to insects (Kaltenpoth, 2009) to humans (Kapadia et al., 2007). While some species are plant pathogens (Loria et al., 1997), other species of Streptomyces are associated with disease‐suppressive soils (Cha et al., 2016), and there is evidence that they can improve plant growth (Jog et al., 2014). Additionally, Streptomyces are known to produce volatile organic compounds (VOCs), some of which are used to interact with arthropods (Weisskopf et al., 2021).
Streptomyces globisporus strain SP6C4, isolated from strawberry flowers, protects strawberry flowers and fruits against the fungal pathogen Botrytis cinerea (Kim et al., 2019). Interestingly, the SP6C4 genome is nearly identical (99.99% sequence identity) to the Fusarium‐suppressive strain S4‐7 isolated from strawberry field soil (Cha et al., 2016). Moreover, SP6C4 can endophytically migrate from the root to the stem as well as from the flower to the stem (Kim et al., 2019) (Fig. 2). This indicates that migration from the root to the flower or vice versa might be possible.
Fig. 2.
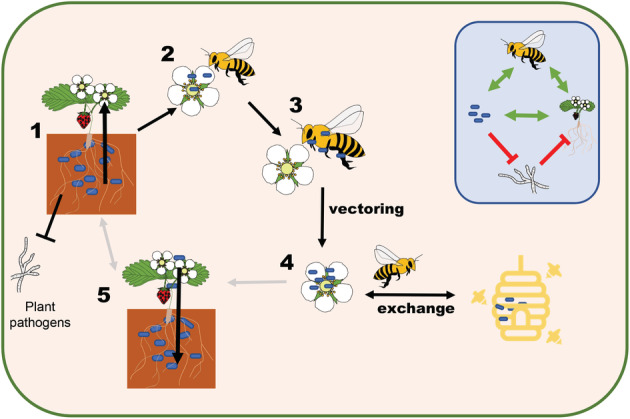
Streptomyces globisporus SP6C4 has a multi‐host lifestyle colonizing strawberry plants and honeybees, which can serve as vectors for dispersal. 1. In the rhizosphere, S. globisporus SP6C4 protects the plant from soil‐borne diseases. 2. SP6C4 can potentially migrate from the rhizosphere to the flower. The bacteria protect the flower from fungal infection. Honeybees pollinate the plant and pick up the bacteria externally and by ingestion. 3. The bacteria protect honeybees from entomopathogens. The honeybee spreads the bacteria to new flowers while pollinating. 4. The honeybee facilitates exchange between the beehive and strawberry flowers. 5. Streptomyces globisporus migrates from the flowers to the rhizosphere. Grey arrows indicate speculative interactions for which the definite evidence is still lacking. Inlay: Diagram that indicates the nature of the interactions of the bacteria with the insect, the plant and soilborne pathogens. Green arrow, positive; red inhibition arrow, negative interactions.
In an experimental setup, honeybees could transmit strain SP6C4 between strawberry flowers and from the beehive to strawberry flowers (Kim et al., 2019) (Fig. 2). Flowers that were ‘inoculated’ by the bees in this experiment showed a decrease of disease symptoms caused by B. cinerea (Kim et al., 2019). Future experiments should point out whether strain SP6C4 can migrate to the roots and if it can provide disease suppression against Fusarium oxysporum. Additionally, it would be interesting to know if strain SP6C4 inoculated in the soil will migrate to strawberry flowers, where it could be vectored by honeybees (Fig. 2).
An earlier 16S rRNA gene‐based microbial community profiling study points towards a similar function of Streptomyces in tomato plants and their bumblebee pollinators (Kwon et al., 2018). The bacterial communities of tomato flowers and bumblebee hives in the same greenhouse became more similar over time, indicating microbe exchange between bumblebees and tomato flowers. Furthermore, S. globisporus consistently showed up in both communities with increasing relative abundance over time. Similar to the results from strawberries, a Streptomyces strain isolated from tomato flowers inhibited the growth of the necrotrophic fungus B. cinerea in an in vitro assay.
These studies show that plants can benefit from insect‐mediated dispersal of beneficial Streptomyces bacteria. In turn, the bacteria, which can colonize below‐ground and above‐ground plant organs as well as insects, presumably profit from the plant host by obtaining nutrients and from the insect host by an increased range of dispersal, and perhaps also nutrient acquisition. But is there also benefit for the insect? An experiment with bumblebees showed that SP6C4 does not only antagonize plant pathogens but also the insect pathogens Serratia marcescens and Paenibacillus larvae, the causative agent of foulbrood disease (Kim et al., 2019). Recently, other Streptomyces strains isolated from pollen in beehives were also shown to protect honeybees against P. larvae (Grubbs et al., 2021). Thus, by dispersing SP6C4, honeybees may be rewarded with healthier flowers and protection against entomopathogens. This means that all three actors profit from the interaction and natural selection might favour this multipartite mutualism (Fig. 2). Interestingly, a recent study implicates that honeybee behaviour is affected by geosmin, a VOC produced by numerous Streptomyces species (Scarano et al., 2021). Low concentrations of geosmin strongly suppress the defensive behaviour of bees. The implications of this behavioural change in bees are unknown, but geosmin produced by Streptomyces coelicolor attracts springtails, which then feed on the bacterial colonies and disperse S. coelicolor spores (Becher et al., 2020). VOCs may also be important for the interaction between S. globisporus and honeybees and could be a topic of future research.
The examples of P. protegens and S. globisporus demonstrate that insects can play an important role as alternative hosts and for the dispersal of plant‐beneficial rhizosphere bacteria and point out that multi‐host lifestyles including plant and insect partners might be much more common than known to date and are thus an intriguing field for future research.
Identifying bacteria with both rhizosphere‐ and insect‐associated lifestyles
Plant‐beneficial bacteria with a multi‐host lifestyle in both plants and insects have neither received broad attention from the field of plant–microbe interactions nor from the field of microbe–insect interactions, but when examining the literature from these fields more closely there are indications that such bacteria may be more common than previously assumed. We will discuss examples of bacterial taxa that potentially switch between rhizosphere‐ and insect‐associated lifestyles, but for which conclusive evidence is not available yet. We focus on bacteria that are mutualistic to the plant and mutualistic or pathogenic to the insect.
Mutualistic interactions between plant‐beneficial Burkholderia spp. and stinkbugs
Members of the genus Burkholderia are typically found in the rhizosphere and often show plant‐beneficial activities (Elliott et al., 2007; de los Santos‐Villalobos et al., 2012; Paungfoo‐Lonhienne et al., 2016). Several stinkbug families, which can be important agricultural pests, harbour obligate symbiotic Burkholderia bacteria in specialized midgut crypts (Kikuchi et al., 2005, 2011). Studies of the stinkbug Riptortus pedestris and its symbiont Burkholderia insecticola have provided important insights into the stinkbug‐Burkholderia symbiosis. Presumably, the bacteria provide a nutritional benefit to their host, resulting in increased growth rates and body mass (Kikuchi et al., 2007). Stinkbugs do not vertically transmit their symbionts, but their nymphs acquire them de novo from the soil every generation (Kikuchi et al., 2007). To overcome the risk of co‐infection with non‐symbiotic and even pathogenic bacteria, stinkbugs possess a tiny organ that serves as a selective filter that only allows their Burkholderia symbionts into the midgut crypts (Ohbayashi et al., 2015). In R. pedestris, colonization by B. insecticola triggers closure of the midgut crypts, so that no other bacteria can enter (Kikuchi et al., 2020). This adaptation allows the stinkbugs to reliably acquire the correct symbiont from the environment.
Many of the stinkbug‐associated Burkholderia species fall into a distinct ‘stinkbug‐associated and environmental’ (SBE) phylogenetic clade, but symbionts of the stinkbug suborders Largidae and Blissidae fall into the ‘plant‐associated beneficial and environmental (PBE)’ clade of Burkholderia species that also harbours many beneficial rhizosphere bacteria (Takeshita et al., 2015; Gordon et al., 2016). Several PBE species belonging to the genus Burkholderia and the closely related genus Pandoraea that were isolated from the soil or rhizosphere and were fed to symbiont‐free stinkbug nymphs successfully established a symbiosis with the stinkbug. They were only slightly less effective than SBE species (Itoh et al., 2019). It is remarkable that free‐living and plant‐associated bacteria can colonize such a selective and specialized symbiont sorting organ, and it suggests that the mechanisms needed for establishing the symbiosis with stinkbugs were present in the ancestor of the genera Burkholderia and Pandoraea. These mechanisms may also be important for their free‐living and plant‐associated lifestyle. In fact, there is evidence that some SBE species also have a free‐living and possibly plant‐associated lifestyle. The SBE symbiont B. insecticola of the soybean‐feeding stinkbug R. pedestris (syn. R. clavatus) was found on soybean roots, and newly hatched nymphs acquired their symbionts from soybeans potted in field soil (Kikuchi et al., 2007). Future experiments could point out whether the SBE species and the stinkbug symbionts in the PBE clade can also colonize the rhizosphere, and perhaps even have plant‐beneficial properties.
Bacillus thuringiensis: insect pathogen and plant benefactor?
Bacillus thuringiensis is well‐known for its production of insecticidal toxins. Because of this feature, it is used as a biocontrol organism to ward off pest insects in crops. Most research has focused on the biocontrol properties of B. thuringiensis, while its ecology has received less attention. Bacillus thuringiensis has not only been found in insects, but in many different environments such as soils, rhizospheres and plant endospheres, and is possibly vectored between these environments by nematodes (Ruan et al., 2015). Bacillus thuringiensis was found to be naturally present in different plant compartments of cotton in fields that had never been treated with B. thuringiensis products before (Monnerat et al., 2009). Moreover, when added to soil close to cotton roots, B. thuringiensis translocated from the soil to the roots and moved endophytically to the leaves, which then became toxic to two Lepidoptera species (Monnerat et al., 2009). Further research should point out whether B. thuringiensis naturally present in (agro)ecosystems can help plants to ward off insects similar to when it is artificially introduced as a biocontrol agent. Bacillus thuringiensis also has other plant‐beneficial properties. Certain B. thuringiensis strains produce compounds that can promote plant growth or suppress plant pathogens (Azizoglu, 2019). However, it is unknown whether these plant‐beneficial strains produce insecticidal toxins and whether insecticidal B. thuringiensis strains are plant‐beneficial. The identification of B. thuringiensis strains with both insecticidal and plant‐beneficial activities would be highly interesting and could lead to the development of 2‐in‐1 products serving as biopesticides and biofertilizers.
The rhizosphere as alternative habitat for the nematode symbiont and insect pathogen Photorhabdus luminescens
Photorhabdus bacteria live in symbiosis with entomopathogenic nematodes. The nematode enters the insect hemolymph and regurgitates the Photorhabdus bacteria, which produce diverse toxins and cause a lethal septicemia in the insect host (Waterfield et al., 2009). Photorhabdus luminescens exists in two phenotypically different, but genetically identical forms. The so‐called primary cells are living in symbiosis with the nematodes. Upon prolonged cultivation, but also in the insect host, primary cells can convert into secondary cells (Eckstein and Heermann, 2019). These secondary cells do not re‐associate with nematodes, and compared to primary cells they exhibit transcriptional differences, which indicate adaptations to a life outside the insect host (Eckstein et al., 2019). Secondary cells exhibit features similar to known plant‐beneficial rhizosphere bacteria. Unlike primary cells, they exhibit chemotaxis towards root exudates and can colonize Arabidopsis thaliana roots (Regaiolo et al., 2020). On the roots, P. luminescens might exhibit plant‐beneficial traits. In plate inhibition assays, secondary cells inhibited growth of the plant pathogen Fusarium graminearum (Regaiolo et al., 2020). If secondary cells retained insecticidal activity, they could potentially protect the roots against herbivores. In summary, these findings indicate that P. luminescens may exhibit, in addition to its insect and nematode associated lifestyles, adaptations to living in the rhizosphere. Therefore, it would be of great interest to reveal whether secondary Photorhabdus cells are indeed associated with roots in natural contexts.
Is the beewolf symbiont Streptomyces philanthi rhizosphere competent?
Beewolf wasps rear a symbiont, Streptomyces philanthi, in their antennal glands, which they secrete and smear on their honeybee prey and on their brood cells to protect their developing offspring from fungal infections (Strohm and Eduard Linsenmair, 2001; Kaltenpoth et al., 2005, 2006). This form of vertical symbiont transmission (see Box 1) ensures that the larvae ingest the symbiont when hatching, and later incorporate the symbiont into their cocoon, from which the emerging adult reacquires the symbiont. Interestingly, a Streptomyces isolate from the rhizosphere of chili pepper was identified as S. philanthi, based on 100% sequence similarity of a partial sequence of the 16S rRNA gene and it was able to control various plant diseases (Boukaew et al., 2011). Although it is not sure whether this strain is also a beewolf symbiont, the close relatedness to the beewolf‐associated S. philanthi indicates that there might be an overlap of traits that allow S. philanthi to establish a symbiosis with insects and with plants.
Box 1. Horizontal versus vertical symbiont transmission in insects.
Most reported insect symbionts are vertically transmitted obligate mutualists (Salem et al., 2015), whereby the symbionts are directly transmitted from the parent to the offspring. Horizontal transmission, whereby the symbiont is acquired de novo from the environment each generation, is less common in insects. Nevertheless, part of the insect gut microbiome is acquired from the environment through nutrition. The following table shows the characteristics of both types of symbiont transmission.
| Horizontal transmission | Vertical transmission |
|---|---|
|
|
Could beewolf wasps possibly acquire an environmental plant‐beneficial Streptomyces symbiont? Beewolf wasps burrow in the soil where they stash their prey (honeybees) and lay their eggs. The emerging larvae live and pupate in the soil. Beewolves that dig their burrows close to plants may occasionally pick up rhizosphere bacteria in their antennae. However, beewolves have evolved a mechanism that blocks the antennal gland secretions in the antennal segments that contain opportunistic bacteria, which prevents their further spreading (Nechitaylo et al., 2014). This is similar to how stinkbugs close off their symbiont‐housing organs (Kikuchi et al., 2020). Nevertheless, opportunistic environmental Actinobacteria were sometimes found in the antennae in high densities (Nechitaylo et al., 2014). Contact between rhizosphere bacteria and the beewolf's antenna is likely, and one can hypothesize that if the bacterium is very closely related to the beewolf symbiont, and shares traits necessary for survival in the glands, a symbiosis could be established. This remains purely speculative but invites taking a closer look, starting with testing the ability of the rhizosphere isolate to establish a symbiosis with a beewolf.
Bacterial traits that facilitate a multi‐host lifestyle
Many rhizosphere bacteria are known for their metabolic diversity, and they can produce and release a wide range of secondary metabolites (Lucke et al., 2020). This allows them to cope with frequently changing conditions and competition from other microbes. The bacteria discussed in the examples above belong to genera such as Pseudomonas, Burkholderia, Streptomyces or Bacillus. Representatives of these genera can be found in diverse environments and are associated to different hosts ranging from plants to insects to mammals, for which they can be beneficial or pathogenic (Vial et al., 2007; Silby et al., 2011; Seipke et al., 2012; Patiño‐Navarrete and Sanchis, 2017). Many species are free‐living with large genomes, and the wide metabolic range associated with these large genomes is presumably what makes these bacteria so versatile.
We have discussed examples of bacterial species or strains that can successfully switch from an insect host to a plant or vice versa. These bacteria are beneficial to plants, and they can be parasitic, commensal, or mutualistic to insects. They may have undergone host‐specific adaptations, such as the acquirement and/or adaptation of genes enabling the production of the insecticidal Fit toxin of P. protegens and its host‐specific regulation (Péchy‐Tarr et al., 2013; Ruffner et al., 2013; Kupferschmied et al., 2014; Flury et al., 2016; Vesga et al., 2021), but other bacterial traits may be involved in interactions with both plants and insects. Perhaps these bacteria use components of the same genetic toolbox for colonizing very different environments. Indeed, the obligate stinkbug symbiont B. insecticola can be replaced with related rhizosphere species, which provide similar benefits to the stinkbug (Hosokawa et al., 2016). Likewise, multiple bacterial species, including Pseudomonas spp. and Acinetobacter spp., that were isolated from insects were shown to have functions that make them plant‐growth promoting (Indiragandhi et al., 2008; Vallet‐Gely et al., 2010; Vesga et al., 2021). In this section, we describe bacterial traits and mechanisms that facilitate a lifestyle in both the rhizosphere and insects.
Flagella and lipopolysaccharides
Because plants and insects represent very different environments, bacteria may have evolved different colonization strategies for each host. However, there is evidence that some mechanisms are important for colonization of both plants and insects. For example, B. insecticola requires flagellar motility for the passage of the constricted gut region in the stinkbug midgut (Ohbayashi et al., 2015). After passaging through this region and before entering the midgut crypts, motility‐related genes are downregulated under the influence of stress‐related molecules from the insect, causing the bacteria to lose their flagellar motility (Ohbayashi et al., 2019). Rhizosphere bacteria also need flagella to approach and colonize the plant roots (de Weger et al., 1987), after which other proteins such as LapA are necessary for attachment to the root surface (Hinsa et al., 2003). Additionally, research on plant‐beneficial Burkholderia species shows the downregulation of flagellar motility once inside the host plant, similar to what happens in insects (Paungfoo‐Lonhienne et al., 2016). Likewise, in P. protegens, motor activity‐related genes are expressed during the colonization of both the rhizosphere and lepidopteran insects (Vesga et al., 2020).
Lipopolysaccharides (LPS) found on the outer cell membrane of Gram‐negative bacteria play a role in virulence in many different host‐pathogen systems. The O‐antigen polysaccharide is required for the initial colonization of the R. pedestris gut by its Burkholderia symbiont (Kim et al., 2016). Once established in the midgut, the symbionts lack the O‐antigen, but they need the core oligosaccharide for maintenance of the symbiosis (Kim et al., 2017). There is evidence that LPS components, including O‐antigen, are also important for the colonization of roots by rhizosphere bacteria (de Weger et al., 1989; Ormeño‐Orrillo et al., 2008; Li et al., 2021b). In P. protegens CHA0, biosynthetic gene clusters for multiple O‐antigen decorations exist (Kupferschmied et al., 2016), which may reflect an adaptation to colonize different hosts. This is supported by a recent study showing that in CHA0 some O‐antigenic polysaccharide gene clusters are expressed during root and insect colonization, while others are expressed specifically in insect backgrounds (Vesga et al., 2020). CHA0 mutants lacking a specific O‐antigen are no longer resistant to antimicrobial peptides that are a part of the insect immune response and are significantly less virulent to insects (Kupferschmied et al., 2016). These specific O‐antigens are thus likely involved in avoiding recognition by the insect immune response.
Siderophores
Siderophores are a class of compounds that chelate iron for uptake from the environment and are commonly produced by fluorescent pseudomonads in the rhizosphere (Höfte and Bakker, 2007; Loper et al., 2012; Zboralski and Filion, 2020). Siderophores play an important ecological role in the rhizosphere. Several rhizosphere bacteria can cross‐utilize siderophores produced by others, providing a competitive advantage (Joshi et al., 2006). This cross‐utilization may directly inhibit pathogen growth (de los Santos‐Villalobos et al., 2012), but siderophores are also known to induce systemic resistance in plants (De Vleesschauwer et al., 2008). Siderophore cross‐utilization is also reported for siderophore‐producing insect gut bacteria, which protect the diamondback moth against entomopathogens (Indiragandhi et al., 2008). Additionally, 79% of the gut bacteria of the grasshopper Sathrophyllia femorata produced siderophores in vitro (Sonawane et al., 2018). Siderophores are also important in pathogenic interactions with insects. For example, pyoverdine expression in P. protegens is highly upregulated once the bacterium has overcome the insect gut epithelium and is proliferating under iron‐limiting conditions in the hemolymph (Vesga et al., 2020).
Antimicrobial compounds
Antimicrobial compounds are a common secondary metabolite class and directly inhibit competing microbes in microbial communities. They can also provide an advantage to hosts when they target important host pathogens. For example, Actinobacteria such as Streptomyces are known for their production of antimicrobial secondary metabolites and are commonly found in the insect gut microbiome (Chevrette et al., 2019) and in the rhizosphere (Boukaew and Prasertsan, 2014; Adegboye and Babalola, 2013; Chen et al., 2018). In the genome of the plant‐beneficial S. globisporus SP6C4 27 biosynthetic gene clusters encoding secondary metabolites were predicted (L. Pronk, unpublished results). This is in line with the finding that defence against pathogenic fungi is the main characteristic shared by the symbioses of S. globisporus with both strawberry plants and honeybees (Kim et al., 2019). The S. philanthi – beewolf symbiosis is also based on antifungal compounds (Koehler et al., 2013) and could explain why the symbiont was found in chili rhizospheres as a plant‐beneficial species (Boukaew et al., 2011). A study showing that S. philanthi culture filtrates were effective in controlling rice sheath blight disease (Boukaew and Prasertsan, 2014) confirms the plant‐beneficial capacities of this insect symbiont. Similarly, P. protegens produces toxic compounds that are important in competition with other microbes in the rhizosphere. Cyclic lipopeptides, rhizoxin and hydrogen cyanide, which contribute to P. protegens' biocontrol activity against soilborne diseases, were found to also play a role during insect infection (Haas and Défago, 2005; Gross and Loper, 2009). Inside the insect these toxic metabolites may provide a competitive advantage against the natural gut microflora, but also directly harm the insect host (Kupferschmied et al., 2013; Loper et al., 2016; Ma et al., 2016; Flury et al., 2017; Vesga et al., 2020).
Evolutionary implications of a multi‐host lifestyle
The previous sections show that some rhizosphere bacteria are capable of a multi‐host lifestyle in both the rhizosphere and insects. Could this be a sign of ongoing evolution towards association with one or the other host, depending on which host provides the most benefits? Or does the adaptation to two hosts represent an evolutionarily stable lifestyle that provides more benefits than costs? As discussed above, some bacterial traits needed in rhizosphere and insect interactions are similar. Insects may frequently encounter rhizosphere bacteria and accidentally ingest them. If these bacteria are already capable of further colonizing the insect, there is a lot of potential for rhizosphere bacteria to develop more insect‐specific adaptations. This makes the rhizosphere a likely source of insect symbionts, be they mutualistic, commensal, or pathogenic. In this section, we discuss examples that show how rhizosphere bacteria may evolve into specialized insect symbionts, and alternatively, how a multi‐host lifestyle may be favoured by evolution.
Obligate insect symbionts may evolve from rhizosphere bacteria
The stinkbug species that were discussed in the previous sections acquire their symbiotic bacteria from the environment. In contrast, the pentatomid stinkbug species Plautia stali vertically transmits its symbionts from the Enterobacteriaceae family, mainly of the genus Pantoea, to its offspring via egg‐smearing (Duron and Noël, 2016). Accordingly, most populations of P. stali harbour uncultivable symbionts with small genomes, a sign of coevolution (Hosokawa et al., 2016). However, some populations associate with cultivable bacteria that have remarkably larger genomes and are also present in the soil habitats of their host populations (Hosokawa et al., 2016). Furthermore, P. stali nymphs hatching from sterilized eggs could establish a symbiosis with free‐living Pantoea strains. Additionally, Pantoea symbionts of different insect species and even of different populations of the same species are only distantly related and do not form a pentatomid‐specific clade (Duron and Noël, 2016; Hosokawa et al., 2016). Since vertical transmission is not perfect and sometimes the original symbionts are replaced with environmental species, symbionts from different stinkbug populations and species show varying degrees of coevolution, ranging from generalist bacteria that can live outside their host to highly specialized bacteria having lost their free‐living ability (Hosokawa et al., 2016; Otero‐Bravo and Sabree, 2021).
It would be interesting to conduct an experimental evolution study to see if plant‐associated Burkholderia and Pantoea species which were able to replace the symbiotic functions of Burkholderia insecticola in the stinkbug R. pedestris (Itoh et al., 2019) could evolve into more specialized insect symbionts that show signs of genome reduction similar to what is observed in other stinkbug symbionts (Takeshita and Kikuchi, 2020), and if horizontal or vertical transmission will become dominant.
A multi‐host lifestyle can be a stable evolutionary state for bacteria
Does coevolution of an environmentally acquired bacterium with its host always direct towards obligate symbiosis with a single host and lead to genome erosion? Or could a multi‐host lifestyle have an adaptive advantage and therefore present an evolutionarily stable alternative?
The direction of symbiont evolution along a parasite‐mutualist continuum cannot be easily predicted and depends on many complex factors such as microbial community complexity, host control mechanisms and mode of transmission (see Box 1) (Drew et al., 2021). A multi‐host lifestyle in both plants and insects implicates a horizontal symbiont transmission mode, which is generally considered to select for parasitism, but it can also facilitate the evolution of defensive traits in mutualists (Drew et al., 2021). Research on S. philanthi, a symbiont of beewolf wasps, shows that a horizontal transmission mode in North American beewolf populations is paired with bacterial traits that are normally associated with free‐living Streptomyces species, such as a large and functionally diverse genome and the capability to grow in standard culture medium (Nechitaylo et al., 2014). In contrast, symbionts in other, geographically separated, beewolf populations showed traits typical for strictly vertically transmitted and highly coevolved species, such as highly reduced genomes and the inability to grow in culture medium. Environmental conditions may determine the dominant mode of transmission of insect symbionts. If the insect environment is consistently nutrient‐rich, and if the symbiont is consistently transmitted to new offspring, there may be no selection pressure for the symbiont to retain its free‐living capacities. This may favour an evolutionary trajectory towards high host‐specificity and vertical transmission. Alternatively, if the host provides a less costly, but relatively nutrient‐poor environment for its symbiont, selection may favour a high metabolic diversity that enables it to live in the environment (Nechitaylo et al., 2014). This generalist lifestyle may make the switch from one host to another easier and could possibly allow for dynamic switching between multiple hosts, depending on the situation. In an often‐changing environment or with an unreliable host, a multi‐host lifestyle may be an adaptive trait.
In this light, acquiring a new host does not necessarily lead to a change along the parasite‐mutualist continuum for the symbiont in the original host, even though there will likely be adaptations to the new host. In a recent study very closely related strains of P. chlororaphis, isolated from roots and from different insect species, exhibited differences in pathogenicity towards Plutella xylostella larvae, but not in their plant‐protection abilities. A genomic analysis revealed single nucleotide polymorphisms and more complex variations (e.g. deletions, insertions) in loci related to insecticidal activity such as the Fit toxin gene cluster, a chitinase gene and a two‐partner secretion system gene (Vesga et al., 2021). These variations may be responsible for the differences in insecticidal activity and in the ability to colonize insects observed for the examined P. chlororaphis strains. An experimental evolution approach recently revealed that in the rhizosphere, P. protegens CHA0 can rapidly genetically adapt to a host plant. Adaptive mutations were found in genes encoding global regulators and cell surface components (LPS O‐antigen) (Li et al., 2021a; Li et al., 2021b). Thus, one could hypothesize that P. protegens may also rapidly adapt to an insect host. However, P. protegens can live in the rhizosphere and in insects and does not show signs of a transition from a plant‐associated lifestyle to an insect‐associated lifestyle. When comparing P. protegens isolates from roots and from insects, no difference was found in their ability to colonize roots, suppress disease, and infect and kill insect larvae (Vesga et al., 2021). Together with the fact that insecticidal factors such as the Fit toxin are upregulated in an insect background, while other pathways are upregulated in plants, this indicates that P. protegens adapted to a lifestyle in both insects and plants (Péchy‐Tarr et al., 2013; Vesga et al., 2020). A well‐equipped large genome and tight host‐adapted regulatory mechanisms likely allow P. protegens and P. chlororaphis to engage in interactions with such contrasting hosts as insects and plants while limiting the energy costs for the diverse lifestyles.
For the S. globisporus – honeybee symbiosis, too little is known about the interaction to tell whether coevolution between the two has occurred. The interaction was ‘set up’ in an artificial environment and it is not clear whether this phenomenon also occurs in natural bee populations. Comparative genomics or transcriptomics of bee‐associated and root‐associated Streptomyces strains in both natural and experimental populations, for example, could provide more insights. Additionally, future research should point out whether the S. globisporus strains found on strawberry flowers and in honeybees still have plant‐beneficial properties in the rhizosphere.
The evolutionary trajectory of symbiont–host interactions is not easy to predict. A multi‐host lifestyle with adaptations towards both hosts appears to be a viable strategy for some plant‐beneficial rhizosphere bacteria. The partly free‐living lifestyle of rhizosphere bacteria in complex microbial communities may contribute to their apparent flexibility. Experimental evolution studies may provide useful insights into the mechanisms of the evolution of multi‐host lifestyles and will help us understand the bigger picture of complex ecosystems.
Ecological and agricultural implications
Plant‐beneficial rhizosphere bacteria and insect symbionts share many features that may allow them to switch between hosts. The ecological implications of this finding are not well understood yet and it may affect the agricultural use of rhizosphere bacteria, since the bacteria discussed above are common members of the rhizosphere microbiome of crop plants, and some are already exploited for their plant beneficial activities (Velivelli et al., 2014). Thus, it is important to study not only their interactions with plants but also with insects, to decipher the ecological roles and the benefits and costs for each actor in these three‐way interactions.
Insect hosts might provide nutrients to the bacteria and possibly serve as shelter, for example during overwintering. However, of particular interest is the fact that insects can transfer the bacteria from plant to plant (Nadarasah and Stavrinides, 2011; Frank et al., 2017; Lòpez‐Fernàndez et al., 2017; Flury et al., 2019). For plant‐pathogenic bacteria it is known that they can even manipulate their host plant into attracting insect vectors (Orlovskis et al., 2015), and their insect vectors into showing dispersal‐related behaviour (Martini et al., 2015). Similar mechanisms may be used by plant‐beneficial rhizosphere bacteria, although this remains to be shown.
Plants may benefit twice if insect‐vectored rhizosphere bacteria have plant‐beneficial properties as well as insecticidal activity. Insect‐pathogenic rhizobacteria might be dispersed by infected larvae before they die, spread from cadavers, or in the case of non‐lethal infections, might be vectored by adults emerging from colonized larvae still carrying the bacteria. Such bacteria could act as bodyguards that defend the plant against diseases and pest insects (Elliot et al., 2000) and one could imagine such a function for P. protegens, P. chlororaphis or B. thuringiensis. Strains with this dual plant‐beneficial activity could be used for biocontrol of plant diseases and pest insects. Microbe‐based control of agricultural insect pests may be an environmentally friendly alternative to chemical insecticides, which are in part responsible for the worldwide decline of insect populations (Seibold et al., 2019). However, the risk of accidentally infecting plant‐beneficial insects (e.g. pollinators) should first be thoroughly assessed and the host specificity of the bacteria should be determined. Their level of pathogenicity may vary across insect groups, and even developmental stages. For example, CHA0 is particularly active against lepidopteran insects, and more specifically their larval stage, but is not pathogenic towards bumblebees (Péchy‐Tarr et al., 2008, 2013; Kupferschmied et al., 2013; Ruffner, 2013; Ruffner et al., 2013).
It will also be interesting to further study the ecological role of insect‐mediated dispersal for plant‐beneficial rhizosphere strains that show no or very weak activity against insects, such as species from the P. fluorescens group other than P. protegens and P. chlororaphis (Péchy‐Tarr et al., 2008; Flury et al., 2016). Strains of the species P. brassicacearum, P. thivervalensis and P. kilonensis, all with potent antifungal activity, were found to colonize Spodoptera littoralis larvae upon oral uptake, reaching levels of up to 106 cfu/larva without killing the insect (Flury et al., 2016). Thus, these bacteria might enter commensal interactions with the insect. If an insect vector that is only mildly damaging to a plant disperses highly effective plant‐beneficial bacteria, the net effect may be positive for plant health.
From a plant perspective, plant‐beneficial rhizosphere bacteria are ideally dispersed by plant‐beneficial insects such as pollinators. Pollinators are already being used as ‘flying doctors’ to spread biocontrol products to crop plants (Biobest Group, 2021). Having pollinators spread bacteria that are both plant‐beneficial and insect‐beneficial would be ideal and may reduce the use of chemical pesticides and increase the health of pollinator hives while improving crop yield. For example, in the strawberry‐honeybee‐Streptomyces symbiosis (Kim et al., 2019), both the plant and the insect benefit from the bacteria, which has anti‐microbial properties that protect both hosts against pathogens. The dispersal of S. globisporus by honeybees goes from flower to flower, and it is yet unknown whether the bacteria can re‐colonize the rhizosphere from there (Kim et al., 2019). However, honeybees that die and fall on the ground may also be a viable dispersal route. Additionally, beewolf wasps that hunt honeybees bury their prey in the ground as food for their larvae. From there, S. globisporus may recolonize the rhizosphere. Interestingly, beewolf wasps also harbour Streptomyces symbionts, which they may acquire from the environment. It would be interesting to investigate whether S. globisporus can also colonize beewolf wasps, and how this affects the ecology of its other hosts.
Similar to S. globisporus, the Burkholderia symbionts of stinkbugs are both plant‐beneficial and insect‐beneficial. However, the insect‐beneficial lifestyle of the bacterium might negatively impact the plant because stinkbugs are herbivorous. Additionally, Burkholderia capable of degrading the pesticide fenitrothion were found in soybean and sugarcane rhizospheres in fields that were sprayed with this chemical for pest management (Tago et al., 2015). Part of the stinkbug population in these fields harboured the pesticide‐degrading strains in their symbiont organ, and were therefore more resistant to the pesticide (Kikuchi et al., 2012). Chemical control of stinkbugs is difficult, and symbiont‐mediated pesticide resistance may enhance this problem. Given that many soil‐dwelling and rhizosphere bacteria can degrade toxic compounds and are capable of horizontal gene transfer, a potential insect‐associated lifestyle of such bacteria may thus have great ecological and agronomical consequences. The added possibility of insect‐mediated dispersal of pesticide‐degrading bacteria adds to the complexity and should be taken seriously. When introducing rhizobacteria in the field for biocontrol of plant diseases, it should be considered that those bacteria might not stay with their plant host. These bacteria potentially also associate with the insects or other fauna that are interacting with the plant, and which might disperse the bacteria into new habitats.
In this context, it is important to understand with which hosts the bacteria associate under natural conditions. The experiments with P. protegens CHA0 revealed that the outcome of an interaction with insects substantially depends on the insect species and can range from commensalism to pathogenicity. Despite decades of research on plant‐beneficial rhizobacteria, still little is known about their specificity for certain host plants. Microbiome research revealed that different plant species and even cultivars assemble specific microbiomes (Pérez‐Jaramillo et al., 2016; Fitzpatrick et al., 2018; Wei et al., 2019). Rhizosphere bacteria with a multi‐host lifestyle might have preferences for certain plant hosts and on top of that possess adaptations to certain insect host species. Future research should point out whether bacteria adapted to a certain host‐plant species (e.g. cabbage) are also specifically adapted to the associated insects (e.g. herbivores feeding on cabbage). The adaptation and restriction to certain plant and insect host species might vary between different plant‐beneficial rhizosphere bacteria as it does between different plant‐pathogenic bacteria (Shikano et al., 2017). Investigating these host specificities and adaptations will help to understand the complex tritrophic interactions between rhizobacteria, plants and insects. This knowledge may then be utilized to develop more sustainable and safe agricultural practices.
Concluding remarks
Recent discoveries provide experimental evidence that rhizosphere bacteria can exploit insects both as an alternative host and as dispersal vector. Additionally, there is evidence that the reported cases are not unique. Where insects and rhizosphere bacteria frequently encounter each other, interactions may be established. This is supported by the fact that many insect symbionts are closely related to rhizosphere bacteria and may deliver the same symbiotic services to their host, such as nutrient provision or disease resistance. Furthermore, whereas environmental acquisition of insect symbionts is considered rare, it does occur and may lead to relatively stable symbioses. The genomic equipment of rhizosphere bacteria and their metabolic versatility may allow them to deal with a frequently changing environment. Even insects, which have developed mechanisms to selectively acquire their desired symbiont, can be successfully colonized by environmental bacteria. This multi‐host lifestyle of rhizosphere bacteria implicates that interactions in the rhizosphere are even more complex than previously thought. In Fig. 3 we try to illustrate what these interactions may look like based on the discussed examples. Besides plant‐mediated effects of rhizosphere bacteria on above‐ground insects (Pineda et al., 2017), direct interactions of rhizosphere bacteria with both above‐ground and below‐ground insects should also be considered. Future research should focus on the capacity of rhizosphere bacteria to colonize important pest insects, pollinators and other insects present in agroecosystems, and on the specificity of these interactions. Furthermore, experimental evidence of dispersal of rhizosphere bacteria by insects is so far limited to laboratory conditions with either axenic soil or only from spray‐inoculated flowers to flowers. Experiments using natural soils should verify whether bacteria established in the rhizosphere can indeed be acquired by insects and persist when they face competition of other bacteria. Another focus should lie on possible agricultural exploitation. Pathogenic rhizobacteria–insect interactions could lead to new biological pest control methods and spreading of bacteria with antifungal activity by insects might be interesting for plant protection as well.
Fig. 3.
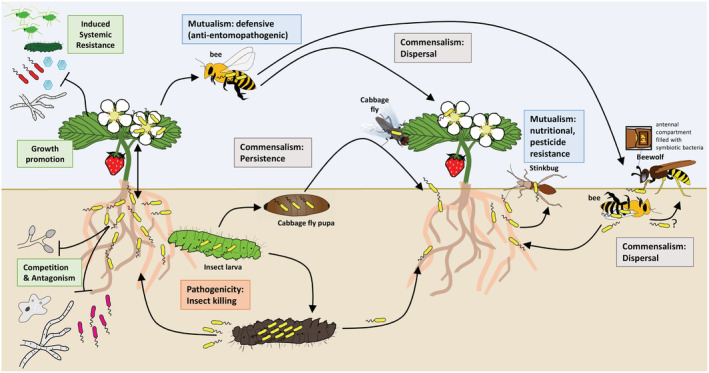
Associations of rhizosphere bacteria with insect hosts might be diverse and range from mutualism to pathogenicity: Rhizosphere bacteria have multiple plant‐beneficial effects. They protect the plants against pathogens by competition, antagonism and induction of plant defences. However, they can also associate with plant‐interacting insects. Pseudomonas protegens CHA0 infects and kills lepidopteran insect larvae upon ingestion. Inside the insect body the bacteria multiply to very high numbers. From the cadaver the bacteria can recolonize plant roots. In nature, CHA0 might be rather an opportunistic pathogen, which often only persists inside insects without killing the host. In the case of the cabbage fly, CHA0 is able to persist transstadially and is dispersed by emerging adults to new host plants. Rhizosphere bacteria may move into aerial plant parts. Streptomyces globisporus colonizes strawberry flowers from where it is vectored by bees to new strawberry plants. It is yet unknown whether S. globisporus can move from the flower to the roots. Beewolves hunt honeybees and bury them in the soil. Thereby bee‐associated bacteria may get access to colonize plant roots. Moreover, beewolf wasps harbour Streptomyces pilanthi which they acquire from the environment. The streptomycete may be capable of colonizing plant roots as well. Stinkbugs harbour Burkholderia symbionts that provide a nutritional benefit to the insect. The bacteria are acquired from the environment and selected by a special symbiont sorting organ in the insect.
In conclusion, plant‐beneficial rhizosphere bacteria have kept their insect‐associated lifestyle well‐hidden for a long time. Over the last years, we have started to lift the veil covering their secrets, opening a thrilling new research field. This will lead to a greater understanding of complex ecological interactions, and it may stimulate sustainable agriculture by developing cleverer uses of biocontrol bacteria.
ACKNOWLEDGMENT
Open Access Funding provided by Forschungsinstitut fur biologischen Landbau. [Correction added on 3 June 2022, after first online publication: CSAL funding statement has been added.]
Contributor Information
Lotte J. U. Pronk, Email: lotte.pronk@wur.nl.
Pascale Flury, Email: pascale.flury@fibl.org.
References
- Adegboye, M.F. , and Babalola, O.O. (2013) Phylogenetic characterization of culturable antibiotic producing Streptomyces from rhizospheric soils. Mol Biol 03: 1–7. [Google Scholar]
- Allard, S.M. , Ottesen, A.R. , Brown, E.W. , and Micallef, S.A. (2018) Insect exclusion limits variation in bacterial microbiomes of tomato flowers and fruit. J Appl Microbiol 125: 1749–1760. [DOI] [PubMed] [Google Scholar]
- Almario, J. , Muller, D. , Défago, G. , and Moënne‐Loccoz, Y. (2014) Rhizosphere ecology and phytoprotection in soils naturally suppressive to Thielaviopsis black root rot of tobacco. Environ Microbiol 16: 1949–1960. [DOI] [PubMed] [Google Scholar]
- Azizoglu, U. (2019) Bacillus thuringiensis as a biofertilizer and biostimulator: a mini‐review of the little‐known plant growth‐promoting properties of Bt . Curr Microbiol 76: 1379–1385. [DOI] [PubMed] [Google Scholar]
- Becher, P.G. , Verschut, V. , Bibb, M.J. , Bush, M.J. , Molnár, B.P. , Barane, E. , et al. (2020) Developmentally regulated volatiles geosmin and 2‐methylisoborneol attract a soil arthropod to Streptomyces bacteria promoting spore dispersal. Nat Microbiol 5: 821–829. [DOI] [PubMed] [Google Scholar]
- Berendsen, R.L. , Pieterse, C.M.J. , and Bakker, P.A.H.M. (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17: 478–486. [DOI] [PubMed] [Google Scholar]
- Berendsen, R.L. , Vismans, G. , Yu, K. , Song, Y. , De Jonge, R. , Burgman, W.P. , et al. (2018) Disease‐induced assemblage of a plant‐beneficial bacterial consortium. ISME J 12: 1496–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg, G. , Köberl, M. , Rybakova, D. , Müller, H. , Grosch, R. , and Smalla, K. (2017) Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol Ecol 93: 1–9. [DOI] [PubMed] [Google Scholar]
- Biobest Group . (2021) Flying Doctors® Hive. URL biobestgroup.com.
- Boukaew, S. , Chuenchit, S. , and Petcharat, V. (2011) Evaluation of Streptomyces spp. for biological control of Sclerotium root and stem rot and Ralstonia wilt of chili pepper. BioControl 56: 365–374. [Google Scholar]
- Boukaew, S. , and Prasertsan, P. (2014) Suppression of rice sheath blight disease using a heat stable culture filtrate from Streptomyces philanthi RM‐1‐138. Crop Prot 61: 1–10. [Google Scholar]
- Cha, J.Y. , Han, S. , Hong, H.J. , Cho, H. , Kim, D. , Kwon, Y. , et al. (2016) Microbial and biochemical basis of a Fusarium wilt‐suppressive soil. ISME J 10: 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Zhou, D. , Qi, D. , Gao, Z. , Xie, J. , and Luo, Y. (2018) Growth promotion and disease suppression ability of a Streptomyces sp. CB‐75 from banana rhizosphere soil. Front Microbiol 8: 2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrette, M.G. , Carlson, C.M. , Ortega, H.E. , Thomas, C. , Ananiev, G.E. , Barns, K.J. , et al. (2019) The antimicrobial potential of Streptomyces from insect microbiomes. Nat Commun 10: 516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiriboga, X. , Guo, H. , Campos‐Herrera, R. , Röder, G. , Imperiali, N. , Keel, C. , et al. (2018) Root‐colonizing bacteria enhance the levels of (E)‐β‐caryophyllene produced by maize roots in response to rootworm feeding. Oecologia 187: 459–468. [DOI] [PubMed] [Google Scholar]
- Compant, S. , Clément, C. , and Sessitsch, A. (2010) Plant growth‐promoting bacteria in the rhizo‐ and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem 42: 669–678. [Google Scholar]
- da Graça, J.V. , Douhan, G.W. , Halbert, S.E. , Keremane, M.L. , Lee, R.F. , Vidalakis, G. , and Zhao, H. (2016) Huanglongbing: an overview of a complex pathosystem ravaging the world's citrus. J Integr Plant Biol 58: 373–387. [DOI] [PubMed] [Google Scholar]
- de los Santos‐Villalobos, S. , Barrera‐Galicia, G.C. , Miranda‐Salcedo, M.A. , and Peña‐Cabriales, J.J. (2012) Burkholderia cepacia XXVI siderophore with biocontrol capacity against Colletotrichum gloeosporioides . World J Microbiol Biotechnol 28: 2615–2623. [DOI] [PubMed] [Google Scholar]
- De Vleesschauwer, D. , Djavaheri, M. , Bakker, P.A.H.M. , and Höfte, M. (2008) Pseudomonas fluorescens WCS374r‐induced systemic resistance in rice against Magnaporthe oryzae is based on pseudobactin‐mediated priming for a salicylic acid‐repressible multifaceted defense response. Plant Physiol 148: 1996–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Weger, L.A. , Bakker, P.A.H.M. , Schippers, B. , van Loosdrecht, M.C.M. , and Lugtenberg, B.J.J. (1989) Pseudomonas spp. with mutational changes in the O‐antigenic side chain of their lipopolysaccharide are affected in their ability to colonize potato roots. In Signal Molecules in Plants and Plant‐Microbe Interactions, Lugtenberg, B.J.J. (ed). Berlin, Heidelberg: Springer, pp. 197–202. [Google Scholar]
- de Weger, L.A. , van Der Vlugt, C.I.M. , Wijfjes, A.H.M. , Bakker, P.A. , Schippers, B. , and Lugtenberg, B. (1987) Flagella of a plant‐growth‐stimulating Pseudomonas fluorescens strain are required for colonization of potato roots. J Bacteriol 169: 2769–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado‐Baquerizo, M. , Oliverio, A.M. , Brewer, T.E. , Benavent‐gonzález, A. , Eldridge, D.J. , Bardgett, R.D. , et al. (2018) Bacteria found in soil. Science 325: 320–325. [DOI] [PubMed] [Google Scholar]
- Douglas, A.E. (2015) Multiorganismal insects: diversity and function of resident microorganisms. Annu Rev Entomol 60: 17–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew, G.C. , Stevens, E.J. , and King, K.C. (2021) Microbial evolution and transitions along the parasite–mutualist continuum. Nat Rev Microbiol 19: 623–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duron, O. , and Noël, V. (2016) A wide diversity of Pantoea lineages are engaged in mutualistic symbiosis and cospeciation processes with stinkbugs. Environ Microbiol Rep 8: 715–727. [DOI] [PubMed] [Google Scholar]
- Eckstein, S. , Dominelli, N. , Brachmann, A. , and Heermann, R. (2019) Phenotypic heterogeneity of the insect pathogen Photorhabdus luminescens: insights into the fate of secondary cells. Appl Environ Microbiol 85: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckstein, S. , and Heermann, R. (2019) Regulation of phenotypic switching and heterogeneity in Photorhabdus luminescens cell populations. J Mol Biol 431: 4559–4568. [DOI] [PubMed] [Google Scholar]
- Elliot, S.L. , Sabelis, M.W. , Janssen, A. , Van der Geest, L.P.S. , Beerling, E.A.M. , and Fransen, J. (2000) Can plants use entomopathogens as bodyguards? Ecol Lett 3: 228–235. [Google Scholar]
- Elliott, G.N. , Chen, W.‐M. , Chou, J.‐H. , Wang, H.‐C. , Sheu, S.‐Y. , Perin, L. , et al. (2007) Burkholderia phymatum is a highly effective nitrogen‐fixing symbiont of Mimosa spp. and fixes nitrogen ex planta. New Phytol 173: 168–180. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick, C.R. , Copeland, J. , Wang, P.W. , Guttman, D.S. , Kotanen, P.M. , and Johnson, M.T.J. (2018) Assembly and ecological function of the root microbiome across angiosperm plant species. Proc Natl Acad Sci U S A 115: E1157–E1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flury, P. , Aellen, N. , Ruffner, B. , Péchy‐Tarr, M. , Fataar, S. , Metla, Z. , et al. (2016) Insect pathogenicity in plant‐beneficial pseudomonads: phylogenetic distribution and comparative genomics. ISME J 10: 2527–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flury, P. , Vesga, P. , Dominguez‐Ferreras, A. , Tinguely, C. , Ullrich, C.I. , Kleespies, R.G. , et al. (2019) Persistence of root‐colonizing Pseudomonas protegens in herbivorous insects throughout different developmental stages and dispersal to new host plants. ISME J 13: 860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flury, P. , Vesga, P. , Péchy‐Tarr, M. , Aellen, N. , Dennert, F. , Hofer, N. , et al. (2017) Antimicrobial and insecticidal: cyclic lipopeptides and hydrogen cyanide produced by plant‐beneficial Pseudomonas strains CHA0, CMR12a, and PCL1391 contribute to insect killing. Front Microbiol 8: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, A. , Saldierna Guzmán, J. , and Shay, J. (2017) Transmission of bacterial endophytes. Microorganisms 5: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, E.R.L. , McFrederick, Q. , and Weirauch, C. (2016) Phylogenetic evidence for ancient and persistent environmental symbiont reacquisition in Largidae (Hemiptera: Heteroptera). Appl Environ Microbiol 82: 7123–7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, H. , and Loper, J.E. (2009) Genomics of secondary metabolite production by Pseudomonas spp. Nat Prod Rep 26: 1408. [DOI] [PubMed] [Google Scholar]
- Grubbs, K.J. , May, D.S. , Sardina, J.A. , Dermenjian, R.K. , Wyche, T.P. , Pinto‐Tomás, A.A. , et al. (2021) Pollen Streptomyces produce antibiotic that inhibits the honey bee pathogen Paenibacillus larvae . Front Microbiol 12: 632637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, D. , and Défago, G. (2005) Biological control of soil‐borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol 3: 307–319. [DOI] [PubMed] [Google Scholar]
- Hinsa, S.M. , Espinosa‐Urgel, M. , Ramos, J.L. , and O'Toole, G.A. (2003) Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol Microbiol 49: 905–918. [DOI] [PubMed] [Google Scholar]
- Höfte, M. , and Bakker, P.A.H.M. (2007) Competition for iron and induced systemic resistance by siderophores of plant growth promoting rhizobacteria. Microb Siderophores 12: 121–133. [Google Scholar]
- Hosokawa, T. , Ishii, Y. , Nikoh, N. , Fujie, M. , Satoh, N. , and Fukatsu, T. (2016) Obligate bacterial mutualists evolving from environmental bacteria in natural insect populations. Nat Microbiol 1: 1–7. [DOI] [PubMed] [Google Scholar]
- Iavicoli, A. , Boutet, E. , Buchala, A. , and Metraux, J.P. (2003) Induced systemic resistance in Arabidopsis thaliana in response to root inoculation with Pseudomonas fluorescens CHA0. Mol Plant Microbe Interact 16: 851–858. [DOI] [PubMed] [Google Scholar]
- Indiragandhi, P. , Anandham, R. , Madhaiyan, M. , and Sa, T.M. (2008) Characterization of plant growth‐promoting traits of bacteria isolated from larval guts of Diamondback moth Plutella xylostella (Lepidoptera: Plutellidae). Curr Microbiol 56: 327–333. [DOI] [PubMed] [Google Scholar]
- Itoh, H. , Jang, S. , Takeshita, K. , Ohbayashi, T. , Ohnishi, N. , Meng, X.‐Y. , et al. (2019) Host–symbiont specificity determined by microbe–microbe competition in an insect gut. Proc Natl Acad Sci U S A 116: 22673–22682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffuel, G. , Imperiali, N. , Shelby, K. , Campos‐Herrera, R. , Geisert, R. , Maurhofer, M. , et al. (2019) Protecting maize from rootworm damage with the combined application of arbuscular mycorrhizal fungi, Pseudomonas bacteria and entomopathogenic nematodes. Sci Rep 9: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jog, R. , Pandya, M. , Nareshkumar, G. , and Rajkumar, S. (2014) Mechanism of phosphate solubilization and antifungal activity of Streptomyces spp. isolated from wheat roots and rhizosphere and their application in improving plant growth. Microbiol (United Kingdom) 160: 778–788. [DOI] [PubMed] [Google Scholar]
- Joshi, F. , Archana, G. , and Desai, A. (2006) Siderophore cross‐utilization amongst rhizospheric bacteria and the role of their differential affinities for Fe3+ on growth stimulation under iron‐limited conditions. Curr Microbiol 53: 141–147. [DOI] [PubMed] [Google Scholar]
- Kaltenpoth, M. (2009) Actinobacteria as mutualists: general healthcare for insects? Trends Microbiol 17: 529–535. [DOI] [PubMed] [Google Scholar]
- Kaltenpoth, M. , Goettler, W. , Dale, C. , Stubblefield, J.W. , Herzner, G. , Roeser‐Mueller, K. , and Strohm, E. (2006) “ Candidatus Streptomyces philanthi”, an endosymbiotic streptomycete in the antennae of Philanthus digger wasps. Int J Syst Evol Microbiol 56: 1403–1411. [DOI] [PubMed] [Google Scholar]
- Kaltenpoth, M. , Göttler, W. , Herzner, G. , and Strohm, E. (2005) Symbiotic bacteria protect wasp larvae from fungal infestation. Curr Biol 15: 475–479. [DOI] [PubMed] [Google Scholar]
- Kapadia, M. , Rolston, K.V.I. , and Han, X.Y. (2007) Invasive Streptomyces infections: six cases and literature review. Am J Clin Pathol 127: 619–624. [DOI] [PubMed] [Google Scholar]
- Keel, C. (2016) A look into the toolbox of multi‐talents: insect pathogenicity determinants of plant‐beneficial pseudomonads. Environ Microbiol 18: 3207–3209. [DOI] [PubMed] [Google Scholar]
- Kikuchi, Y. , Hayatsu, M. , Hosokawa, T. , Nagayama, A. , Tago, K. , and Fukatsu, T. (2012) Symbiont‐mediated insecticide resistance. Proc Natl Acad Sci U S A 109: 8618–8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi, Y. , Hosokawa, T. , and Fukatsu, T. (2007) Insect‐microbe mutualism without vertical transmission: a stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl Environ Microbiol 73: 4308–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi, Y. , Hosokawa, T. , and Fukatsu, T. (2011) An ancient but promiscuous host‐symbiont association between Burkholderia gut symbionts and their heteropteran hosts. ISME J 5: 446–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi, Y. , Meng, X.‐Y. , and Fukatsu, T. (2005) Gut symbiotic bacteria of the genus Burkholderia in the broad‐headed bugs Riptortus clavatus and Leptocorisa chinensis (Heteroptera: Alydidae). Appl Environ Microbiol 71: 4035–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi, Y. , Ohbayashi, T. , Jang, S. , and Mergaert, P. (2020) Burkholderia insecticola triggers midgut closure in the bean bug Riptortus pedestris to prevent secondary bacterial infections of midgut crypts. ISME J 14: 1627–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.R. , Cho, G. , Jeon, C.W. , Weller, D.M. , Thomashow, L.S. , Paulitz, T.C. , and Kwak, Y.S. (2019) A mutualistic interaction between Streptomyces bacteria, strawberry plants and pollinating bees. Nat Commun 10: 4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.K. , Jang, H.A. , Kim, M.S. , Cho, J.H. , Lee, J. , Di Lorenzo, F. , et al. (2017) The lipopolysaccharide core oligosaccharide of Burkholderia plays a critical role in maintaining a proper gut symbiosis with the bean bug Riptortus pedestris . J Biol Chem 292: 19226–19237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.K. , Park, H.Y. , and Lee, B.L. (2016) The symbiotic role of O‐antigen of Burkholderia symbiont in association with host Riptortus pedestris . Dev Comp Immunol 60: 202–208. [DOI] [PubMed] [Google Scholar]
- Kluth, S. , Kruess, A. , and Tscharntke, T. (2002) Insects as vectors of plant pathogens: mutualistic and antagonistic interactions. Oecologia 133: 193–199. [DOI] [PubMed] [Google Scholar]
- Koehler, S. , Doubský, J. , and Kaltenpoth, M. (2013) Dynamics of symbiont‐mediated antibiotic production reveal efficient long‐term protection for beewolf offspring. Front Zool 10: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, H.G. , Kim, B.K. , Song, G.C. , Lee, S. , and Ryu, C.M. (2016) Aboveground whitefly infestation‐mediated reshaping of the root microbiota. Front Microbiol 7: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupferschmied, P. , Chai, T. , Flury, P. , Blom, J. , Smits, T.H.M. , Maurhofer, M. , and Keel, C. (2016) Specific surface glycan decorations enable antimicrobial peptide resistance in plant‐beneficial pseudomonads with insect‐pathogenic properties. Environ Microbiol 18: 4265–4281. [DOI] [PubMed] [Google Scholar]
- Kupferschmied, P. , Maurhofer, M. , and Keel, C. (2013) Promise for plant pest control: root‐associated pseudomonads with insecticidal activities. Front Plant Sci 4: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupferschmied, P. , Péchy‐Tarr, M. , Imperiali, N. , Maurhofer, M. , and Keel, C. (2014) Domain shuffling in a sensor protein contributed to the evolution of insect pathogenicity in plant‐beneficial Pseudomonas protegens . PLoS Pathog 10: e1003964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, Y. , Lee, J.T. , Kim, H.S. , Jeon, C. , and Kwak, Y.S. (2018) Comparative tomato flower and pollinator hive microbial communities. J Plant Dis Prot 125: 115–119. [Google Scholar]
- Li, E. , de Jonge, R. , Liu, C. , Jiang, H. , Friman, V.P. , Pieterse, C.M.J. , et al. (2021a) Rapid evolution of bacterial mutualism in the plant rhizosphere. Nat Commun 12: 3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, E. , Zhang, H. , Jiang, H. , Pieterse, C.M.J. , Jousset, A. , Bakker, P.A.H.M. , and de Jonge, R. (2021b) Experimental‐evolution‐driven identification of Arabidopsis rhizosphere competence genes in Pseudomonas protegens . MBio 12: e0092721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loper, J.E. , Hassan, K.A. , Mavrodi, D.V. , Davis, E.W., 2nd , Lim, C.K. , Shaffer, B.T. , et al. (2012) Comparative genomics of plant‐associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet 8: e1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loper, J.E. , Henkels, M.D. , Rangel, L.I. , Olcott, M.H. , Walker, F.L. , Bond, K.L. , et al. (2016) Rhizoxin, orfamide A, and chitinase production contribute to the toxicity of Pseudomonas protegens strain Pf‐5 to Drosophila melanogaster . Environ Microbiol 18: 3509–3521. [DOI] [PubMed] [Google Scholar]
- Lòpez‐Fernàndez, S. , Mazzoni, V. , Pedrazzoli, F. , Pertot, I. , and Campisano, A. (2017) A phloem‐feeding insect transfers bacterial endophytic communities between grapevine plants. Front Microbiol 8: 834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loria, R. , Bukhalid, R.A. , Fry, B.A. , and King, R.R. (1997) Plant pathogenicity in the genus Streptomyces . Plant Dis 81: 836–846. [DOI] [PubMed] [Google Scholar]
- Lucke, M. , Correa, M.G. , and Levy, A. (2020) The role of secretion systems, effectors, and secondary metabolites of beneficial rhizobacteria in interactions with plants and microbes. Front Plant Sci 11: 589416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Z. , Geudens, N. , Kieu, N.P. , Sinnaeve, D. , Ongena, M. , Martins, J.C. , and Höfte, M. (2016) Biosynthesis, chemical structure, and structure‐activity relationship of orfamide lipopeptides produced by Pseudomonas protegens and related species. Front Microbiol 7: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini, X. , Hoffmann, M. , Coy, M.R. , Stelinski, L.L. , and Pelz‐Stelinski, K.S. (2015) Infection of an insect vector with a bacterial plant pathogen increases its propensity for dispersal. PLoS One 10: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurhofer, M. , Keel, C. , Haas, D. , and Defago, G. (1994) Pyoluteorin production by Pseudomonas fluorescens strain CHA0 is involved in the suppression of Pythium damping‐off of cress but not of cucumber. Eur J Plant Pathol 100: 221–232. [Google Scholar]
- Mendes, R. , Garbeva, P. , and Raaijmakers, J.M. (2013) The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev 37: 634–663. [DOI] [PubMed] [Google Scholar]
- Mercado‐Blanco, J. , and Bakker, P.A.H.M. (2007) Interactions between plants and beneficial Pseudomonas spp.: exploiting bacterial traits for crop protection. Antonie van Leeuwenhoek 92: 367–389. [DOI] [PubMed] [Google Scholar]
- Monnerat, R.G. , Soares, C.M. , Capdeville, G. , Jones, G. , Martins, É.S. , Praça, L. , et al. (2009) Translocation and insecticidal activity of Bacillus thuringiensis living inside of plants. J Microbial Biotechnol 2: 512–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarasah, G. , and Stavrinides, J. (2011) Insects as alternative hosts for phytopathogenic bacteria. FEMS Microbiol Rev 35: 555–575. [DOI] [PubMed] [Google Scholar]
- Nechitaylo, T.Y. , Westermann, M. , and Kaltenpoth, M. (2014) Cultivation reveals physiological diversity among defensive ‘Streptomyces philanthi’ symbionts of beewolf digger wasps (Hymenoptera, Crabronidae). BMC Microbiol 14: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi, T. , Futahashi, R. , Terashima, M. , Barrière, Q. , Lamouche, F. , Takeshita, K. , et al. (2019) Comparative cytology, physiology and transcriptomics of Burkholderia insecticola in symbiosis with the bean bug Riptortus pedestris and in culture. ISME J 13: 1469–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi, T. , Takeshita, K. , Kitagawa, W. , Nikohc, N. , Koga, R. , Meng, X.Y. , et al. (2015) Insect's intestinal organ for symbiont sorting. Proc Natl Acad Sci U S A 112: E5179–E5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olcott, M.H. , Henkels, M.D. , Rosen, K.L. , Walker, F.L. , Sneh, B. , Loper, J.E. , and Taylor, B.J. (2010) Lethality and developmental delay in Drosophila melanogaster larvae after ingestion of selected Pseudomonas fluorescens strains. PLoS One 5: e12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlovskis, Z. , Canale, M.C. , Thole, V. , Pecher, P. , Lopes, J.R.S. , and Hogenhout, S.A. (2015) Insect‐borne plant pathogenic bacteria: getting a ride goes beyond physical contact. Curr Opin Insect Sci 9: 16–23. [DOI] [PubMed] [Google Scholar]
- Ormeño‐Orrillo, E. , Rosenblueth, M. , Luyten, E. , Vanderleyden, J. , and Martínez‐Romero, E. (2008) Mutations in lipopolysaccharide biosynthetic genes impair maize rhizosphere and root colonization of Rhizobium tropici CIAT899. Environ Microbiol 10: 1271–1284. [DOI] [PubMed] [Google Scholar]
- Otero‐Bravo, A. , and Sabree, Z.L. (2021) Multiple concurrent and convergent stages of genome reduction in bacterial symbionts across a stink bug family. Sci Rep 11: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangesti, N. , Pineda, A. , Dicke, M. , and van Loon, J.J.A. (2015) Variation in plant‐mediated interactions between rhizobacteria and caterpillars: potential role of soil composition. Plant Biol 17: 474–483. [DOI] [PubMed] [Google Scholar]
- Patiño‐Navarrete, R. , and Sanchis, V. (2017) Evolutionary processes and environmental factors underlying the genetic diversity and lifestyles of Bacillus cereus group bacteria. Res Microbiol 168: 309–318. [DOI] [PubMed] [Google Scholar]
- Paungfoo‐Lonhienne, C. , Lonhienne, T.G.A. , Yeoh, Y.K. , Donose, B.C. , Webb, R.I. , Parsons, J. , et al. (2016) Crosstalk between sugarcane and a plant‐growth promoting Burkholderia species. Sci Rep 6: 37389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péchy‐Tarr, M. , Borel, N. , Kupferschmied, P. , Turner, V. , Binggeli, O. , Radovanovic, D. , et al. (2013) Control and host‐dependent activation of insect toxin expression in a root‐associated biocontrol pseudomonad. Environ Microbiol 15: 736–750. [DOI] [PubMed] [Google Scholar]
- Péchy‐Tarr, M. , Bruck, D.J. , Maurhofer, M. , Fischer, E. , Vogne, C. , Henkels, M.D. , et al. (2008) Molecular analysis of a novel gene cluster encoding an insect toxin in plant‐associated strains of Pseudomonas fluorescens . Environ Microbiol 10: 2368–2386. [DOI] [PubMed] [Google Scholar]
- Pérez‐Jaramillo, J.E. , Mendes, R. , and Raaijmakers, J.M. (2016) Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol Biol 90: 635–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M.J. , Zamioudis, C. , Berendsen, R.L. , Weller, D.M. , Van Wees, S.C.M. , and Bakker, P.A.H.M. (2014) Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol 52: 347–375. [DOI] [PubMed] [Google Scholar]
- Pineda, A. , Kaplan, I. , and Bezemer, T.M. (2017) Steering soil microbiomes to suppress aboveground insect pests. Trends Plant Sci 22: 770–778. [DOI] [PubMed] [Google Scholar]
- Pineda, A. , Soler, R. , Weldegergis, B.T. , Shimwela, M.M. , Van Loon, J.J.A. , and Dicke, M. (2013) Non‐pathogenic rhizobacteria interfere with the attraction of parasitoids to aphid‐induced plant volatiles via jasmonic acid signalling. Plant Cell Environ 36: 393–404. [DOI] [PubMed] [Google Scholar]
- Rangel, L.I. , Henkels, M.D. , Shaffer, B.T. , Walker, F.L. , Davis, E.W. , Stockwell, V.O. , et al. (2016) Characterization of toxin complex gene clusters and insect toxicity of bacteria representing four subgroups of Pseudomonas fluorescens . PLoS One 11: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebolleda Gómez, M. , and Ashman, T.L. (2019) Floral organs act as environmental filters and interact with pollinators to structure the yellow monkeyflower (Mimulus guttatus) floral microbiome. Mol Ecol 28: 5155–5171. [DOI] [PubMed] [Google Scholar]
- Regaiolo, A. , Dominelli, N. , Andresen, K. , and Heermann, R. (2020) The biocontrol agent and insect pathogen Photorhabdus luminescens interacts with plant roots. Appl Environ Microbiol 86: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan, L. , Crickmore, N. , Peng, D. , and Sun, M. (2015) Are nematodes a missing link in the confounded ecology of the entomopathogen Bacillus thuringiensis? Trends Microbiol 23: 341–346. [DOI] [PubMed] [Google Scholar]
- Ruffner, B. (2013) Insecticidal activity in plant‐associated pseudomonads. Molecular basis and ecological relevance. PhD Thesis. [DOI] [PubMed]
- Ruffner, B. , Péchy‐Tarr, M. , Höfte, M. , Bloemberg, G. , Grunder, J. , Keel, C. , and Maurhofer, M. (2015) Evolutionary patchwork of an insecticidal toxin shared between plant‐associated pseudomonads and the insect pathogens Photorhabdus and Xenorhabdus . BMC Genomics 16: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffner, B. , Péchy‐Tarr, M. , Ryffel, F. , Hoegger, P. , Obrist, C. , Rindlisbacher, A. , et al. (2013) Oral insecticidal activity of plant‐associated pseudomonads. Environ Microbiol 15: 751–763. [DOI] [PubMed] [Google Scholar]
- Ruiu, L. , and Mura, M.E. (2021) Oral toxicity of Pseudomonas protegens against muscoid flies. Toxins (Basel) 13: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem, H. , Florez, L. , Gerardo, N. , and Kaltenpoth, M. (2015). An out‐of‐body experience: the extracellular dimension for the transmission of mutualistic bacteria in insects. Proceedings of the Royal Society B: Biological Sciences, 282: 20142957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse, J. , Martinoia, E. , and Northen, T. (2018) Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci 23: 25–41. [DOI] [PubMed] [Google Scholar]
- Scarano, F. , Deivarajan Suresh, M. , Tiraboschi, E. , Cabirol, A. , Nouvian, M. , Nowotny, T. , and Haase, A. (2021) Geosmin suppresses defensive behaviour and elicits unusual neural responses in honey bees. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibold, S. , Gossner, M.M. , Simons, N.K. , Blüthgen, N. , Ambarl, D. , Ammer, C. , et al. (2019) Arthropod decline in grasslands and forests is associated with drivers at landscape level. Nature 574: 1–34. [DOI] [PubMed] [Google Scholar]
- Seipke, R.F. , Kaltenpoth, M. , and Hutchings, M.I. (2012) Streptomyces as symbionts: an emerging and widespread theme? FEMS Microbiol Rev 36: 862–876. [DOI] [PubMed] [Google Scholar]
- Shikano, I. , Rosa, C. , Tan, C.‐W. , and Felton, G.W. (2017) Tritrophic interactions: microbe‐mediated plant effects on insect herbivores. Annu Rev Phytopathol 55: 313–331. [DOI] [PubMed] [Google Scholar]
- Silby, M.W. , Winstanley, C. , Godfrey, S.A.C. , Levy, S.B. , and Jackson, R.W. (2011) Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev 35: 652–680. [DOI] [PubMed] [Google Scholar]
- Snyder, W.E. , Tonkyn, D.W. , and Kluepfel, D.A. (1998) Insect‐mediated dispersal of the rhizobacterium Pseudomonas chlororaphis . Phytopathology 88: 1248–1254. [DOI] [PubMed] [Google Scholar]
- Snyder, W.E. , Tonkyn, D.W. , and Kluepfel, D.A. (1999) Transmission of a genetically engineered rhizobacterium by grasshoppers in the laboratory and field. Ecol Appl 9: 245–253. [Google Scholar]
- Sonawane, M.S. , Chaudhary, R.D. , Shouche, Y.S. , and Sayyed, R.Z. (2018) Insect gut bacteria: a novel source for siderophore production. Proc Natl Acad Sci India Sect B – Biol Sci 88: 567–572. [Google Scholar]
- Strohm, E. , and Eduard Linsenmair, K. (2001) Females of the European beewolf preserve their honeybee prey against competing fungi. Ecol Entomol 26: 198–203. [Google Scholar]
- Tago, K. , Okubo, T. , Itoh, H. , Kikuchi, Y. , Hori, T. , Sato, Y. , et al. (2015) Insecticide‐degrading Burkholderia symbionts of the stinkbug naturally occupy various environments of sugarcane fields in a southeast Island of Japan. Microbes Environ 30: 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita, K. , and Kikuchi, Y. (2020) Genomic comparison of insect gut symbionts from divergent Burkholderia subclades. Genes (Basel) 11: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita, K. , Matsuura, Y. , Itoh, H. , Navarro, R. , Hori, T. , Sone, T. , et al. (2015) Burkholderia of plant‐beneficial group are symbiotically associated with bordered plant bugs (Heteroptera: Pyrrhocoroidea: Largidae). Microbes Environ 30: 321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow, L.S. , Kwak, Y.S. , and Weller, D.M. (2019) Root‐associated microbes in sustainable agriculture: models, metabolites and mechanisms. Pest Manag Sci 75: 2360–2367. [DOI] [PubMed] [Google Scholar]
- Trivedi, P. , Leach, J.E. , Tringe, S.G. , Sa, T. , and Singh, B.K. (2020) Plant–microbiome interactions: from community assembly to plant health. Nat Rev Microbiol 18: 607–621. [DOI] [PubMed] [Google Scholar]
- Ushio, M. , Yamasaki, E. , Takasu, H. , Nagano, A.J. , Fujinaga, S. , Honjo, M.N. , et al. (2015) Microbial communities on flower surfaces act as signatures of pollinator visitation. Sci Rep 5: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacheron, J. , Péchy‐Tarr, M. , Brochet, S. , Heiman, C.M. , Stojiljkovic, M. , Maurhofer, M. , and Keel, C. (2019) T6SS contributes to gut microbiome invasion and killing of an herbivorous pest insect by plant‐beneficial Pseudomonas protegens . ISME J 13: 1318–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet‐Gely, I. , Novikov, A. , Augusto, L. , Liehl, P. , Bolbach, G. , Péchy‐Tarr, M. , et al. (2010) Association of hemolytic activity of Pseudomonas entomophila, a versatile soil bacterium, with cyclic lipopeptide production. Appl Environ Microbiol 76: 910–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lenteren, J.C. , and Manzaroli, G. (1999) Evaluation and use of predators and parasitoids for biological control of pests in greenhouses. In Integrated Pest and Disease Management in Greenhouse Crops. Dordrecht: Springer, pp. 183–201. [Google Scholar]
- Velivelli, S.L.S. , De Vos, P. , Kromann, P. , Declerck, S. , and Prestwich, B.D. (2014) Biological control agents: from field to market, problems, and challenges. Trends Biotechnol 32: 493–496. [DOI] [PubMed] [Google Scholar]
- Vesga, P. , Augustiny, E. , Keel, C. , Maurhofer, M. , and Vacheron, J. (2021) Phylogenetically closely related pseudomonads isolated from arthropods exhibit differential insect‐killing abilities and genetic variations in insecticidal factors. Environ Microbiol 23: 5378–5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesga, P. , Flury, P. , Vacheron, J. , Keel, C. , Croll, D. , and Maurhofer, M. (2020) Transcriptome plasticity underlying plant root colonization and insect invasion by Pseudomonas protegens . ISME J 14: 2766–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vial, L. , Groleau, M.C. , Dekimpe, V. , and Déziel, E. (2007) Burkholderia diversity and versatility: an inventory of the extracellular products. J Microbiol Biotechnol 17: 1407–1429. [PubMed] [Google Scholar]
- Waterfield, N.R. , Ciche, T. , and Clarke, D. (2009) Photorhabdus and a host of hosts. Annu Rev Microbiol 63: 557–574. [DOI] [PubMed] [Google Scholar]
- Wei, F. , Zhao, L. , Xu, X. , Feng, H. , Shi, Y. , Deakin, G. , et al. (2019) Cultivar‐dependent variation of the cotton rhizosphere and endosphere microbiome under field conditions. Front Plant Sci 10: 1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisskopf, L. , Schulz, S. , and Garbeva, P. (2021) Microbial volatile organic compounds in intra‐kingdom and inter‐kingdom interactions. Nat Rev Microbiol 19: 391–404. [DOI] [PubMed] [Google Scholar]
- Zboralski, A. , and Filion, M. (2020) Genetic factors involved in rhizosphere colonization by phytobeneficial Pseudomonas spp. Comput Struct Biotechnol J 18: 3539–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]


