Abstract
During the course of chronic cystic fibrosis (CF) infections, Pseudomonas aeruginosa undergoes a conversion to a mucoid phenotype, which is characterized by overproduction of the exopolysaccharide alginate. Chronic P. aeruginosa infections involve surface-attached, highly antibiotic-resistant communities of microorganisms organized in biofilms. Although biofilm formation and the conversion to mucoidy are both important aspects of CF pathogenesis, the relationship between them is at the present unclear. In this study, we report that the overproduction of alginate affects biofilm development on an abiotic surface. Biofilms formed by an alginate-overproducing strain exhibit a highly structured architecture and are significantly more resistant to the antibiotic tobramycin than a biofilm formed by an isogenic nonmucoid strain. These results suggest that an important consequence of the conversion to mucoidy is an altered biofilm architecture that shows increasing resistance to antimicrobial treatments.
Cystic fibrosis (CF) is the most common inherited lethal genetic disorder in Caucasian populations. Individuals suffering from CF harbor mutations in the cystic fibrosis transmembrane conductance regulator gene, resulting in multiorgan malfunction (50). The most significant manifestation of the disease is in the respiratory tract, which is predisposed to chronic infection (16, 22, 26, 42, 47, 54). The CF patient mounts a massive immune response, which fails to clear these infections but causes a tremendous amount of tissue damage. Ultimately, the patient succumbs to deteriorating lung function caused by these infections and the resulting inflammation.
Infections by the opportunistic pathogen Pseudomonas aeruginosa are the leading cause of morbidity and mortality in CF patients (15, 17). Colonization of CF airways by P. aeruginosa usually occurs early in the lifetime of the host (23). Eventually, the environment in the CF lung causes P. aeruginosa to undergo a switch to a mucoid phenotype (11, 25). The mucoid phenotype is caused by overproduction of alginate, an exopolysaccharide (EPS) consisting of mannuronic acid and guluronic acid monomers. The mucoid conversion is often associated with inactivating mutations in the mucA gene, which encodes an anti-sigma factor of AlgT. AlgT is required for expression of the alginate biosynthetic operon (10, 19, 31, 32). Alginate is thought to have a protective function in a relatively harsh environment in which the bacteria are continually subjected to oxidative stress and attack by the immune system (28, 44, 45). The switch to mucoidy is also thought to promote persistence of P. aeruginosa in the airways and is usually coincident with a downturn in the prognosis of the patient (27).
Another key aspect of P. aeruginosa CF lung infections is the growing body of evidence that they involve biofilm bacteria. Biofilms are surface-attached communities of bacteria embedded in an extracellular matrix of biopolymeric substances and are involved in many types of chronic infections (for recent reviews, see references 5, 6, 8, and 39). Biofilm bacteria are physiologically distinct from free-swimming bacteria of the same species. A hallmark characteristic of biofilm bacteria is that they can be up to 1,000 times more resistant to antibiotics than their free-swimming counterparts (14, 24, 36). In addition, the extrapolymeric matrix produced by biofilm bacteria has been shown to inhibit phagocytosis by cells of the immune system (34). Initial microscopic observations of postmortem lung tissue and the sputa of patients suggested that P. aeruginosa forms biofilms in the CF lung (29, 48). Recent physiological evidence supports this idea, showing a change in quorum-sensing signal profiles comparing free-swimming and biofilm P. aeruginosa organisms (46). These observations are not surprising, since the biofilm mode of growth confers a protective advantage to the bacteria.
Wild-type, nonmucoid P. aeruginosa biofilm formation proceeds through distinct developmental steps based upon observational data (5, 6, 9, 40). After initial attachment of single cells to a surface, the bacteria move on the surface by twitching motility to form clumps of cells or microcolonies (40). The cells continue to proliferate and form a mature biofilm consisting of several layers of cells stacked upon one another. The mature biofilm, depending upon the growth medium, can have a fairly uniform structure or a highly differentiated structure with pillars of cells separated by spaces devoid of bacteria (5, 21). There are a number of studies indicating that the type and amount of EPS produced are important for biofilm formation and structure in different bacterial species (3, 7, 53). However, the effect of alginate overproduction on P. aeruginosa biofilm formation remains undetermined. Some reports have suggested that mucoid P. aeruginosa biofilms are more resistant to antibiotics than nonmucoid biofilms (2, 35). However, these studies were limited in that the strains used for comparison were not isogenic derivatives. Interestingly, a recent report actually demonstrated that laboratory-grown biofilms of nonmucoid P. aeruginosa become mucoid when exposed to hydrogen peroxide, reinforcing the concept that oxidative stress is an important determinant in the conversion to mucoidy (33). Although the conversion to mucoidy and biofilm formation are known to be important components of P. aeruginosa CF lung infections, the relationship between the two is unclear.
In this report, we examine how alginate overproduction influences P. aeruginosa biofilm development and function. We have studied biofilm development of a nonmucoid wild-type P. aeruginosa PAO1 strain and an alginate-overproducing isogenic mucA mutant strain. Our data show that the mucA alginate-overproducing strain exhibits enhanced microcolony formation and a more highly structured mature biofilm. Furthermore, we demonstrate that the mucoid strain produces a biofilm in which the cells are highly resistant to the antibiotic tobramycin. Using an isogenic strain unable to synthesize alginate, an algDmucA double mutant, we verify that the observed effect is due to alginate overproduction, not additional AlgT-regulated gene products. Our observations have led us to propose that the conversion to mucoidy has drastic effects on biofilm structure and may function to promote persistent biofilm populations in the CF lung.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The P. aeruginosa strains were PAO1 and PDO300, a mucA22 derivative of PAO1 constructed by allelic exchange (33). The Escherichia coli strains were HB101 and CC118λpir (18). The plasmids used were pJBA27 (PA1/04/03-gfp-T0-T1 cassette on pUC18Not) (1), pJMT6 (mini-Tn5 transposon delivery vector, tellurite resistance), pRK600 (ColE1 oriV RP4 oriT, helper plasmid in triparental matings, chloramphenicol resistance), and pMH94 (Gfp expression cassette located on a mini-Tn5 transposon element, tellurite and ampicillin resistance).
Bacteria were routinely grown in Luria-Bertani (LB) broth or LB agar with antimicrobial agents when necessary. The antimicrobial agents were used at the following concentrations: ampicillin, 100 μg/ml for E. coli; potassium tellurite, 400 μg/ml for E. coli and 150 μg/ml for P. aeruginosa; chloramphenicol, 8 μg/ml for E. coli; tetracycline, 15 μg/ml for E. coli and 60 μg/ml for P. aeruginosa.
Constructions.
DNA manipulations were performed using standard methods. Plasmid isolation was performed by using QIAprep spin miniprep kits (Qiagen, Chatsworth, Calif.), and DNA fragments were excised and purified from agarose gels by GFX PCR kit (Amersham-Pharmacia Biotech, Uppsala, Sweden). pMH94 was constructed by cloning a 2.0-kb NotI-fragment containing the PA1/04/03-gfp-T0-T1 cassette of pJBA27 into the NotI-site of pJMT6. The PA1/04/03-gfp-T0-T1 transposon cassette was inserted into random positions on the chromosomes of P. aeruginosa PAO1 and PDO300 by triparental mating with E. coli CC118λpir(pMH94) and E. coli HB101(pRK600). The transconjugants had a green fluorescent phenotype compatible with fluorescence microscopy and growth characteristics identical to those of the parental strains. The strains had a generation time of 2.3 h when grown in EPRI medium at 30°C.
The PAO1 algDmucA22 double mutant was constructed by double homologous recombination of the plasmid pDJW487 into the PDO300 chromosome. pDJW487 contains the algD operon with most of the algD gene, including the 5′ regulatory sequences, deleted and replaced with a tetracycline marker (D. J. Wozniak, unpublished work). The knockout construct is harbored by pEX100T containing the sacB gene and the bla resistance marker (43). A transformant which was nonmucoid, tetracycline resistant, sucrose tolerant, and carbenicillin sensitive was selected. The mucoid phenotype was complemented by the plasmid pAlgD, which contains a 23-kb BamHI fragment of the algD operon of pAlg2 (38) cloned into the broad-host-range vector pBBR1MCS-5.
Static culture biofilm assay.
The experiments were performed as previously described by O'Toole et al. (40, 41). Overnight cultures of PAO1 and PDO300 were diluted to an optical density at 600 nm (OD600) of 0.1 in fresh EPRI medium; 100-μl culture aliquots were dispensed into the wells of a 96-well polystyrene microtiter dish and incubated for 24 h at 30°C. For the experiments whose results are shown in Fig. 2, comparing initial attachment of the PAO1, mucA22, and the mucA22algD double-mutant strains, the incubation period was 10 h. Biofilm formation was detected by staining with 1% (wt/vol) crystal violet in water. Nonadherent cells and residual dye were removed by a thorough rinsing with water. Cell-associated dye was solubilized with ethanol and measured at OD595.
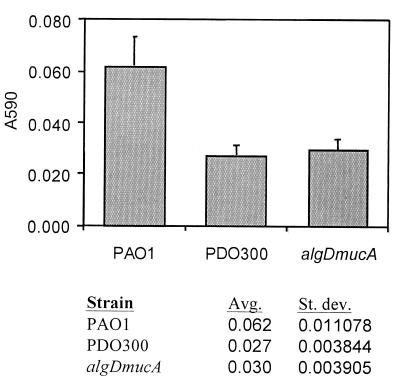
FIG. 2.
Biofilm formation assay comparing the total biofilm biomasses of PAO1, PDO300, and algDmucA strains after 10 h. Biofilms were prepared and stained as described in Materials and Methods. Both PDO300 and the algDmucA double mutant harbored less biomass in their biofilms than did PAO1. Each value was the average of 32 individual replicates. Avg, average; St. dev., standard deviation.
Continuous-culture biofilm reactor system.
Biofilms were cultivated in flow cells with individual channel dimensions of 1 by 4 by 40 mm with a flow of EPRI medium modified to contain 0.005 μg of iron per ml as the growth-limiting substrate (9). The once-through, continuous-culture biofilm reactor system was assembled and prepared as described by Christensen et al. (4). The substratum consisted of a microscope glass coverslip (Knittel 24×50 st1; Knittel Gläser, Braunschweig, Germany). Flow cells were inoculated with exponentially growing cultures in EPRI medium at an OD600 of 0.15. After inoculation, the medium flow was arrested for 1 h. Medium flow was resumed at a constant rate of 0.28 mm/s using a Watson Marlow 205S peristaltic pump (Watson Marlow Ltd., Falmouth, England). The flow cell system was incubated at 30°C. Effluent from the flow cells was collected and plated for determination of viable counts and mucoid phenotype.
Microscopy and image analysis.
Epifluorescence microscopy was performed using a Zeiss AxioPhot epifluorescence microscope (Carl Zeiss, GmbH, Jena, Germany) equipped with fluorescein isothiocyanate and tetramethyl rhodamine isocyanate filter sets and a Zeiss AxioCam camera for image acquisition. Noninvasive monitoring of the biofilm three-dimensional structure was achieved by scanning confocal laser microscopy (SCLM) using a Leica TCS4D system (Leica Lasertechnik, GmbH, Heidelberg, Germany). The 488-nm and the 568-nm laser lines of an ArKr laser were used to excite Gfp/SYTO 9 and propidium iodide (PI), respectively. Stacks of horizontal-plane images captured by SCLM were subjected to quantitative image analysis using the COMSTAT software (21). The program calculates several characteristic biofilm parameters such as biofilm thickness, roughness, and substratum coverage. Simulated fluorescence projections and vertical cross sections through the biofilms were generated by using the IMARIS software package (Bitplane AG, Zürich, Switzerland) running on a Silicon Graphics Indigo2 workstation (Silicon Graphics, Mountain View, Calif.). Images were further processed for display by using the PhotoShop software (Adobe, Mountain View, Calif.).
LIVE/DEAD BacLight bacterial viability staining.
Bacterial viability in biofilms was assayed by using the LIVE/DEAD BacLight bacterial viability staining kit (Molecular Probes Inc., Eugene, Oreg.). The stain stock solutions of SYTO 9 and PI were diluted 2,000-fold in EPRI medium and injected into the flow channels. The staining was allowed to progress for 15 min with the medium flow arrested. Live SYTO 9-stained cells and dead PI-stained cells were visualized by SCLM using fluorescein isothiocyanate filter and tetramethyl rhodamine isocyanate optical filters, respectively.
Rotating-disk biofilm reactor.
A rotating-disk biofilm reactor system was used for generating quantitative data on biofilm susceptibility to antibiotics. The system consisted of a reactor vessel containing 250 ml of EPRI medium. The reactor was initially operated in a batch mode for 24 h before being switched to chemostat mode at a dilution rate of 0.10 h−1. When the culture reached steady-state condition indicated by a steady optical density of the culture (OD600, ≃0.180), the magnetic stir bar in the reactor was replaced by a stir disk with 18 removable plastic polycarbonate chips. Biofilm formation was allowed to proceed for 24 h before the disk and aliquots of the culture were aseptically retrieved. The chips were removed from the disk wheel and washed in phosphate-buffered saline to remove nonadherent cells. The chips were exposed for 5 h to tobramycin in concentrations in the range of 100 to 0 μg/ml in EPRI medium. After antibiotic exposure, the chips were transferred to phosphate-buffered saline and sonicated for 10 min to resuspend adherent cells. The cell suspension was diluted and plated on LB agar for determination of viable counts (CFU/chip). All experiments were performed at least in triplicate.
RESULTS
Effect of alginate overproduction on biofilm structure.
To investigate the effect of mucoidy on biofilm formation, we compared nonmucoid and mucoid P. aeruginosa strains. The prototrophic wild-type P. aeruginosa PAO1 strain is nonmucoid and produces only trace amounts of alginate. P. aeruginosa PDO300 is a defined isogenic derivative of PAO1 which has been engineered by allelic exchange to contain the mucA22 allele. This mutation leads to constitutive overproduction of the exopolysaccharide alginate. Liquid-culture growth rates of both PAO1 and PDO300 were 2.3 h at 30°C in EPRI growth medium. Initially, we examined and compared biofilm formation by these two strains using epifluorescence and SCLM. To allow fluorescence microscopy, we used the green fluorescent protein (GFP) as a nontoxic biological fluorophore to tag these two strains. A constitutively-expressed allele of GFP was introduced onto the chromosome of PAO1 and PDO300 using a mini-Tn5 transposon cassette. We also verified that the insertion of mini-Tn5 into a critical region of the PAO1 or PDO300 chromosome was not responsible for the observed biofilm phenotypes. To do this we examined both untagged PAO1 and PDO300 biofilms with phase-contrast microscopy and tagged PAO1 and PDO300 bearing a plasmid constitutively expressing GFP. The biofilm architectures formed by these strains were identical to those formed by the mini-Tn5-tagged strains described below (data not shown).
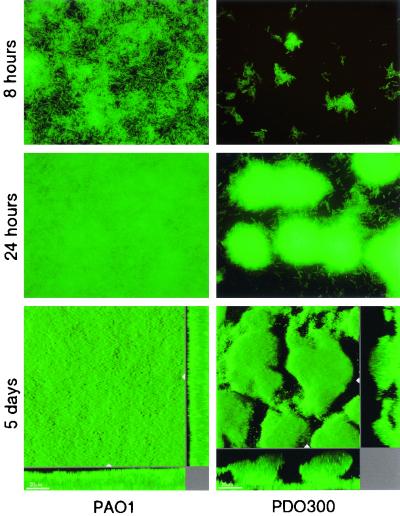
PDO300 appeared to be moderately defective in attachment to the glass surface (Fig. 1) compared to PAO1, which formed a uniform monolayer of cells after 8 h of incubation (Fig. 1). A 96-well microtiter dish assay was used to verify this observation. This assay involves quantitation of biofilm biomass that attaches to the polystyrene walls of the microtiter plate. A comparison of PAO1 and PDO300 using this assay verified that after a 10-h period, PAO1 biofilms contained more biomass than PDO300 biofilms (Fig. 2). Past the initial attachment phase (>20 h), the PAO1 and PDO300 biofilms exhibited significantly different patterns of biofilm development. The PAO1 biofilm developed from a uniform monolayer of attached cells into a biofilm characterized by an almost complete substratum coverage and even biomass distribution (Fig. 1, middle and bottom left panels). PDO300 formed a significantly different biofilm architecture, with attached cells growing exclusively in discrete microcolonies resulting in a low substratum coverage and high structural heterogeneity. The PAO1 and PDO300 biofilms reached a steady state, in which the biofilm architecture did not appreciably change, within 5 to 10 days. The mature PAO1 biofilm exhibited moderate heterogeneity and a high degree of substratum coverage. The average thickness of the steady-state PAO1 biofilm was observed to be about one-half that of the PDO300 biofilm. The steady state PDO300 biofilm was characterized by large microcolonies separated by water channels (Fig. 1, bottom right panel). The mucoid phenotype is unstable, occasionally reverting to a nonmucoid phenotype. Following incubation in biofilm flow cells, PDO300 cells were harvested and plated out to ensure that the bacteria retained a mucoid phenotype. Another concern was that a mutation in an anti-sigma factor gene, mucA, would show pleiotropic effects on genes other than the genes involved in alginate synthesis, effects which would affect biofilm architecture. To test this, we constructed an algDmucA double mutant, which is unable to synthesize alginate. This double mutant formed biofilms with wild-type, nonmucoid architecture, verifying that alginate overproduction is likely to be responsible for the PDO300 biofilm phenotype (data not shown). We also tested the algDmucA double mutant for initial biofilm formation using the 96-well microtiter plate experiment described above. After 10 h, the double mutant had levels of biofilm biomass comparable to that of PDO300 (Fig. 2), suggesting that alginate overproduction is not responsible for this observation.
FIG. 1.
Epifluorescence and scanning confocal photomicrographs of the surface-attached communities formed by P. aeruginosa wild type and an isogenic alginate-overproducing mutant. The strains are engineered to contain a gfp expression cassette inserted into the chromosome. The biofilms are grown in once flow-through continuous-culture reaction vessels. (Top) Epifluorescence photomicrographs of the wild type (PAO1) and the mucA22 mutant (PDO300); images were acquired 8 h postinoculation of the biofilm reactor. (Middle) Epifluorescence photomicrographs acquired 24 h postinoculation. (Bottom) Scanning confocal photomicrographs of 5-day-old wild-type and mucA22 mutant biofilms. The larger central plots are simulated fluorescent projections, in which a long shadow indicates a large, high microcolony. Shown in the right and lower frames are vertical sections through the biofilms collected at the positions indicated by the white triangles. Bar, 20 μm.
Software analysis of PAO1 and PDO300 biofilm architecture.
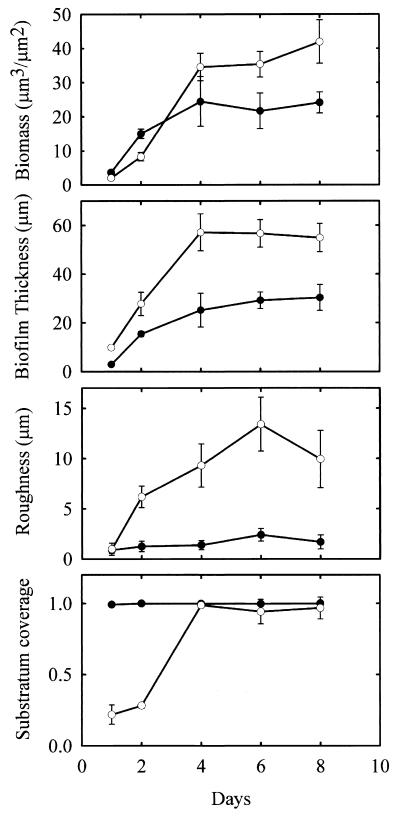
Computer software analysis of SCLM-generated images of PAO1 and PDO300 biofilms allowed quantification of observed differences in biofilm architecture. The COMSTAT software package was used for this analysis as described in Materials and Methods and in a previous report (20). Verifying microscopic observations, COMSTAT analysis indicated that PAO1 and PDO300 formed significantly different biofilms (Fig. 3). During the course of biofilm development, PDO300 consistently formed a thicker and rougher biofilm than PAO1. Thickness is the average depth of the biofilm, while roughness is a measurement of structural heterogeneity. For example, a rough biofilm has many pillars and towers of cells separated by areas devoid of cells, whereas a smooth biofilm consists of a more homogenous layer of cells. PDO300 also formed biofilms that harbored more biomass than PAO1 biofilms (Fig. 3). Another parameter of biofilm architecture that differed between the two strains was substratum coverage. Substratum coverage measures the area of the attachment surface covered by biofilm bacteria. PAO1 quickly covered the entire attachment surface after 24 h of biofilm development, whereas PDO300 covered only 20% of the attachment surface. By the fourth day, both PAO1 and PDO300 had covered most of the attachment surface.
FIG. 3.
Characteristics of P. aeruginosa wild-type (filled circles) and mucA22 mutant (open circles) biofilms. The biomass content, biofilm thickness, roughness coefficient, and substratum coverage were calculated by the COMSTAT image analysis software from scanning confocal image data. The values are averages of six image stacks acquired in four separate experiments. The biomass content is calculated as the biomass volume (μm3) per substratum surface area (μm2). The roughness coefficient describes the variation in biofilm thickness and is a measure of biofilm heterogeneity. The substratum coverage is the fraction of the substratum area covered by biomass. The times at which the biofilms were assayed were 1, 2, 4, 6, and 8 days (as indicated on the x axis).
Antibiotic resistance of P. aeruginosa biofilms.
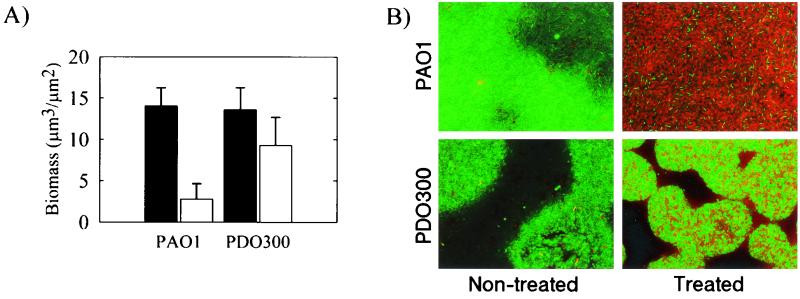
The effects of the markedly different biofilm architectures of PAO1 and PDO300 on biofilm function are unknown. One important function of a biofilm is the protection it confers to members of the biofilm community. Therefore, PAO1 and PDO300 biofilms were assayed for their resistance to the antibiotic tobramycin. Two separate techniques were used to examine antibiotic resistance. The first technique involved the biofilm flow cells used in the experiments described above. Established 24-h-old biofilms grown on EPRI medium were subjected to another 24 h of continuous flow of either tobramycin (2.0 μg/ml) in EPRI medium or EPRI medium alone. The PAO1 biofilm was much more sensitive to tobramycin treatment than was the PDO300 biofilm (Fig. 4A). In the absence of treatment, both PAO1 and PDO300 biofilms had comparable biomasses. Viable biomass was determined by SCLM analysis of GFP-tagged PAO1 and PDO300 biofilms (30). GFP expression has previously been used as a viability marker, and control experiments verified that tobramycin-killed bacteria do not fluoresce (data not shown). However, when PAO1 and PDO300 biofilms were subjected to tobramycin treatment the PAO1 biofilms showed a significant drop in viable biomass (Fig. 4A). These biofilms were also subjected to viability staining (Fig. 4B). Viability was measured by cell membrane integrity, with dead cells having membranes that are permeable to the nucleic acid stain PI indicated by red fluorescence (Fig. 4B). Both viable and dead cells are stained with the nucleic acid stain SYTO 9, indicated by green fluorescence (Fig. 4B). Control experiments comparing CFU and viability staining of liquid and biofilm cultures of P. aeruginosa demonstrated that our staining technique accurately represented the proportion of live and dead cells (data not shown). Almost all of the cells in the PAO1 biofilm were dead; however, many cells were alive in the PDO300 biofilm.
FIG. 4.
Increased tobramycin resistance of a P. aeruginosa PDO300 biofilm. (A) Viable biomass content of gfp-expressing P. aeruginosa wild-type and PDO300 mutant biofilms after 24 h of exposure to 2.0 μg of tobramycin per ml (open bars) and nontreated controls (filled bars). gfp fluorescence is a marker of cell viability and allows quantification of the viable biomass by COMSTAT image analysis of SCLM image data. (B) Visualization of live (green fluorescence) and dead (red fluorescence) cells by LIVE/DEAD BacLight bacterial viability staining kit. The treated biofilms were exposed to tobramycin as described above. Viability was measured by GFP fluorescence (A) and by SYTO 9 viability staining (B).
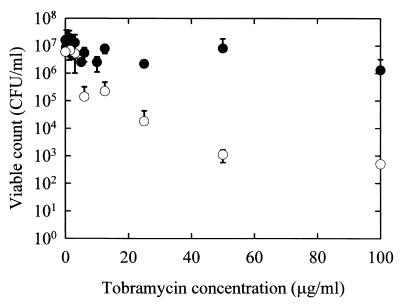
A rotating-disk reactor system (see Materials and Methods), which allows quantification of cells in a biofilm by direct viable count, was also used to assay the susceptibility of PAO1 and PDO300 biofilms to tobramycin. This system utilizes a lexan disk affixed to a stir bar and 18 polycarbonate chips that can be inserted into the lexan disk. When the disk is placed into a liquid culture, it is spun using the stir bar. Only biofilm bacteria accumulate on the polycarbonate chips. Following culturing, the chips can be removed and then exposed to antibiotics in 96-well microtiter plates. Following antibiotic treatment the biofilm bacteria can be removed by sonication and assessed for viability by plate counts. Biofilms were grown for 24 h. Within a range of antibiotic concentrations of 2 to 100 μg/ml, the PDO300 biofilms grown in the rotating disk reactor were up to 1,000 times more resistant to tobramycin than were the PAO1 biofilms (Fig. 5). For cells obtained from liquid cultures of PAO1 and PDO300 (planktonic cells), the MICs of tobramycin were comparable (∼1 μg/ml) (data not shown).
FIG. 5.
Assay of tobramycin sensitivity in a rotating-disk biofilm reactor system. ○, P. aeruginosa wild-type biofilm; ●, PDO300 mutant biofilm. The values represent averages of three separate experiments. Biofilms were treated for 5 h. The planktonic MIC of tobramycin for both these strains is 1 μg/ml.
DISCUSSION
In this study we report a relationship between the conversion to mucoidy by P. aeruginosa and biofilm development and function. A mucoid, i.e., alginate-overproducing, strain developed a highly differentiated biofilm, resulting in a more structurally heterogeneous biofilm than that produced by a comparable nonmucoid strain. A qualitative and quantitative analysis of biofilm architecture shows that PDO300 forms microcolonies very early in biofilm development, although fewer cells are attached to the substratum at the initial attachment stage. We further characterized this defect in initial attachment using 96-well microtiter plate biofilm assays (Fig. 2). Since both PDO300 and the algDmucA double mutant exhibited the same level of biofilm biomass, we believe that alginate overproduction is not responsible for reduced levels of biofilm biomass early on in biofilm development (Fig. 2). This observation may be explained by the absence of flagella in a mucA background. Flagellum expression has been shown to be negatively regulated by AlgT (13). The flagellum has also been shown to play an important role in the attachment of P. aeruginosa during the initial steps of biofilm formation (40). However, past the initial attachment stage, an analysis of older biofilms (beyond 24 h) shows that the architectures of PAO1 and the algDmucA double mutant are the same. This suggests that alginate overproduction is the primary cause of observed differences in biofilm architecture. The mucoid biofilms also proved to be more resistant to the antibiotic tobramycin than biofilms formed by the nonmucoid strain. This difference in antibiotic resistance to tobramycin is not seen in planktonic cells of PAO1 and PDO300. This suggests that the alginate overproduction in the CF lung may play a role in forming biofilms that are more resistant to antimicrobial stresses. A point that should be clarified concerns the structure of the wild-type biofilm. Some reports have demonstrated that PAO1 forms highly structured biofilms (9). However, biofilm architecture is significantly influenced by growth medium (51), and the particular growth medium we selected for PAO1 results in fairly flat, undifferentiated biofilms that are several layers of cells thick (about 30 μm [Fig. 1 and 3]). The growth medium used in this study was chosen because it maintains a stable mucoid phenotype in the biofilm population.
The role of EPS in determining biofilm architecture has been reported for a number of different bacterial strains. A change in primary EPS production has been shown to be responsible for the conversion of the Vibrio cholerae smooth-colony phenotype to the rough-colony phenotype (53). This study also demonstrated that the rough-colony phenotype of V. cholerae forms thick, robust biofilms with distinct architecture compared to the smooth-colony biofilms. A report by Danese et al. showed that E. coli mutants unable to synthesize the EPS colanic acid were deficient in their ability to attach to abiotic surfaces compared to the isogenic wild-type strain (7). These two examples list only a few cases where EPS production has a significant influence on biofilm formation and architecture. We show that an alginate-overproducing strain of P. aeruginosa has significant effects on biofilm architecture and development. The PDO300 biofilm was thicker and rougher and exhibited enhanced microcolony formation (Fig. 1, compare top and middle right micrographs). The significance of PDO300's tendency to form microcolonies is unclear. After initial attachment of a cell, enhanced microcolony formation may result from daughter cells originating from the initial attached bacterium being ensnared in the alginate matrix and held in close proximity to the parent cell. However, in the case of the nonmucoid strain, daughter cells may be released into the bulk liquid. Our data are consistent with those of Nivens et al., who recently demonstrated that a mucoid, clinical CF isolate of P. aeruginosa produced a biofilm consisting of microcolonies up to 40 μm in depth, whereas nonmucoid revertants produced a flat and dense biofilm about 6 μm thick (37). The study further established that the structural difference was due to alginate, with the O acetylation of alginate appearing to be pivotal for the biofilm structure.
A consequence of enhanced microcolony formation and thicker, alginate-containing biofilms would be an increased resistance of these bacterial cells to antimicrobial agents. Two of the contributing factors to this resistance are that the EPS matrix represents a physical and chemical barrier and that due to nutritional gradients, cells buried in a biofilm are reduced in metabolic activity, rendering them less susceptible to antibiotics which primarily target the metabolically active cells (12, 14, 49, 52). Therefore, in a harsh environment such as the CF lung, enhanced microcolony formation would be a selective advantage. Using laboratory-grown biofilms, Mathee et al. demonstrated that nonmucoid biofilms subjected to oxidative stress actually induced a conversion to the mucoid phenotype (33). Previous studies have also indicated that mucoid strains of P. aeruginosa are more resistant to antibiotics than nonmucoid strains. However, these studies were limited in that the strains tested were not isogenic.
Using a wild-type strain of P. aeruginosa and an isogenic mucA strain, we show that alginate overproduction does indeed increase the resistance of biofilm bacteria to the antibiotic tobramycin. We demonstrated this using two independent biofilm-culturing techniques. The time points at which antibiotic resistance was assayed for mucoid and nonmucoid strains for both biofilm-culturing techniques were selected so that the same amounts of cells were present in the two biofilms. The flow cell culturing technique combined with viability staining allowed visualization of treated and untreated biofilms and the relative distribution of live and dead cells (Fig. 4B). The nonmucoid biofilm was almost entirely killed, while the mucoid biofilm remained viable. Most of the dead biomass remained attached to the substratum for the nonmucoid biofilms. The fact that most of the cells in the nonmucoid biofilm were killed may be due to the prolonged exposure (24 h) to a dose of tobramycin greater than the MIC. The small number of dead cells in the mucoid biofilm appeared to be concentrated towards the periphery of the microcolony; however, dead cells could be seen throughout the microcolony. Tobramycin appears to kill more biofilm bacteria in the flow cells when the staining method is used than when GFP fluorescence is used as a viability indicator (compare relative drops in viability after treatment in Fig. 4A and B). However, it should be kept in mind that the data in Fig. 4A represent a large number of image stacks which have been analyzed by COMSTAT, while Fig. 4B shows a single snapshot of a biofilm cross section. It is also possible that the viability staining is overestimating the degree of killing, or conversely, that measuring GFP fluorescence as a viability marker is underestimating the degree of killing. The rotating-disk reactor biofilms allowed quantification of viable biomass using traditional plate counts (Fig. 5). These experiments demonstrated that mucoid biofilms were up to 1,000 times more resistant to tobramycin than were the nonmucoid biofilms; however, planktonically the MICs of tobramycin for PAO1 and PDO300 were the same (1 μg/ml). The combination of the two assays for antibiotic resistance strongly supports the conclusion that alginate overproduction protects cells in a biofilm from tobramycin. Furthermore, due to the distribution of live and dead cells within the biofilm, we believe that enhanced microcolony formation creates an antimicrobial-resistant zone in the interior of the microcolony and is an important element of the increased resistance of mucoid biofilms.
The data presented here support a model indicating that a primary role of alginate overproduction by mucoid P. aeruginosa is to alter biofilm architecture, which results in more antimicrobial-resistant biofilms. These results strongly support previous studies, which have indicated that the conversion to mucoidy plays a protective role for the bacterial population. These results also reemphasize the importance of EPS production for biofilm architecture. Future studies will address questions arising from this work. For example, does alginate overproduction influence recruitment of free-swimming cells from the environment into the biofilm? Is the biofilm architecture associated with alginate overproduction a general characteristic of cells overproducing an exopolysaccharide? Does alginate overproduction affect detachment rates? These questions are central to understanding the ecology of P. aeruginosa CF lung infections. Such an understanding may assist in designing therapeutic strategies for treating chronic P. aeruginosa infections.
ACKNOWLEDGMENTS
We thank Kalai Mathee for invaluable input and discussion. We also thank Dan Wozniak for supplying us with pDJW487 and Pradeep Singh for helpful discussions.
G.T. is supported by NSF CH9810378. The present work was supported by grants from the Danish Medical Research Council and the Danish Plasmid Foundation to M.G.
REFERENCES
- 1.Andersen J B, Sternberg C, Poulsen L K, Bjorn S P, Givskov M, Molin S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol. 1998;64:2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anwar H, Strap J L, Chen K, Costerton J W. Dynamic interactions of biofilms of mucoid Pseudomonas aeruginosa with tobramycin and piperacillin. Antimicrob Agents Chemother. 1992;36:1208–1214. doi: 10.1128/aac.36.6.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baselga R, Albizu I, De La Cruz M, Del Cacho E, Barberan M, Amorena B. Phase variation of slime production in Staphylococcus aureus: implications in colonization and virulence. Infect Immun. 1993;61:4857–4862. doi: 10.1128/iai.61.11.4857-4862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen B B, Sternberg C, Andersen J B, Palmer R J, Jr, Nielsen A T, Givskov M, Molin S. Molecular tools for study of biofilm physiology. Methods Enzymol. 1999;310:20–42. doi: 10.1016/s0076-6879(99)10004-1. [DOI] [PubMed] [Google Scholar]
- 5.Costerton J W, Lewandowski Z, Caldwell D E, Korber D R, Lappin-Scott H M. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 6.Costerton J W, Stewart P S, Greenberg E P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 7.Danese P N, Pratt L A, Kolter R. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J Bacteriol. 2000;182:3593–3596. doi: 10.1128/jb.182.12.3593-3596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davey M E, O'Toole G A. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 10.DeVries C A, Ohman D E. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternate sigma factor, and shows evidence for autoregulation. J Bacteriol. 1994;176:6677–6687. doi: 10.1128/jb.176.21.6677-6687.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doggett R G. Incidence of mucoid Pseudomonas aeruginosa from clinical sources. Appl Microbiol. 1969;18:936–937. doi: 10.1128/am.18.5.936-937.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans D J, Brown M R, Allison D G, Gilbert P. Susceptibility of bacterial biofilms to tobramycin: role of specific growth rate and phase in the division cycle. J Antimicrob Chemother. 1990;25:585–591. doi: 10.1093/jac/25.4.585. [DOI] [PubMed] [Google Scholar]
- 13.Garrett E S, Perlegas D, Wozniak D J. Negative control of flagellum synthesis in Pseudomonas aeruginosa is modulated by the alternative sigma factor AlgT (AlgU) J Bacteriol. 1999;181:7401–7404. doi: 10.1128/jb.181.23.7401-7404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert P, Das J, Foley I. Biofilm susceptibility to antimicrobials. Adv Dent Res. 1997;11:160–167. doi: 10.1177/08959374970110010701. [DOI] [PubMed] [Google Scholar]
- 15.Gilligan P H. Microbiology of airway disease in patients with cystic fibrosis. Clin Microbiol Rev. 1991;4:35–51. doi: 10.1128/cmr.4.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldman M J, Anderson G M, Stolzenberg E D, Kari U P, Zasloff M, Wilson J M. Human beta-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell. 1997;88:553–560. doi: 10.1016/s0092-8674(00)81895-4. [DOI] [PubMed] [Google Scholar]
- 17.Govan J R, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hershberger C D, Ye R W, Parsek M R, Xie Z D, Chakrabarty A M. The algT (algU) gene of Pseudomonas aeruginosa, a key regulator involved in alginate biosynthesis, encodes an alternative sigma factor (sigma E) Proc Natl Acad Sci USA. 1995;92:7941–7945. doi: 10.1073/pnas.92.17.7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heydorn A, Ersboll B K, Hentzer M, Parsek M R, Givskov M, Molin S. Experimental reproducibility in flow-chamber biofilms. Microbiology. 2000;146:2409–2415. doi: 10.1099/00221287-146-10-2409. [DOI] [PubMed] [Google Scholar]
- 21.Heydorn A, Nielsen A T, Hentzer M, Sternberg C, Givskov M, Ersboll B K, Molin S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 22.Hoiby H N. Microbiology of lung infections in cystic fibrosis patients. Acta Paediatr Scand Suppl. 1982;301:33–54. [PubMed] [Google Scholar]
- 23.Hoiby N. Epidemiological investigations of the respiratory tract bacteriology in patients with cystic fibrosis. Acta Pathol Microbiol Immunol Scand Sect B. 1974;82:541–550. [PubMed] [Google Scholar]
- 24.Høiby N, Espersen F, Fromsgaard A, Giwercman B, Jensen E T, Johansen H K, Koch C, Kronborg G, Pedersen S S, Pressler T, Kharazmi A. Biofilm, foreign bodies and chronic infections. Ugeskr Laeger. 1994;156:5998–6005. . (In Danish.) [PubMed] [Google Scholar]
- 25.Iacocca V F, Sibinga M S, Barbero G J. Respiratory tract microbiology in cystic fibrosis. Am J Dis Child. 1963;106:316–324. doi: 10.1001/archpedi.1963.02080050317012. [DOI] [PubMed] [Google Scholar]
- 26.Joris L, Dab I, Quinton P M. Elemental composition of human airway surface fluid in healthy and diseased airways. Am Rev Respir Dis. 1993;148:1633–1637. doi: 10.1164/ajrccm/148.6_Pt_1.1633. [DOI] [PubMed] [Google Scholar]
- 27.Koch C, Hoiby N. Pathogenesis of cystic fibrosis. Lancet. 1993;341:1065–1069. doi: 10.1016/0140-6736(93)92422-p. [DOI] [PubMed] [Google Scholar]
- 28.Krieg D P, Helmke R J, German V F, Mangos J A. Resistance of mucoid Pseudomonas aeruginosa to nonopsonic phagocytosis by alveolar macrophages in vitro. Infect Immun. 1988;56:3173–3179. doi: 10.1128/iai.56.12.3173-3179.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lam J, Chan R, Lam K, Costerton J W. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect Immun. 1980;28:546–556. doi: 10.1128/iai.28.2.546-556.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lowder M, Unge A, Maraha N, Jansson J K, Swiggett J, Oliver J D. Effect of starvation and the viable-but-nonculturable state on green fluorescent protein (GFP) fluorescence in GFP-tagged Pseudomonas fluorescens A506. Appl Environ Microbiol. 2000;66:3160–3165. doi: 10.1128/aem.66.8.3160-3165.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin D W, Holloway B W, Deretic V. Characterization of a locus determining the mucoid status of Pseudomonas aeruginosa: AlgU shows sequence similarities with a Bacillus sigma factor. J Bacteriol. 1993;175:1153–1164. doi: 10.1128/jb.175.4.1153-1164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin D W, Schurr M J, Mudd M H, Govan J R, Holloway B W, Deretic V. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci USA. 1993;90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathee K, Ciofu O, Sternberg C, Lindum P W, Campbell J I, Jensen P, Johnsen A H, Givskov M, Ohman D E, Molin S, Hoiby N, Kharazmi A. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology. 1999;145:1349–1357. doi: 10.1099/13500872-145-6-1349. [DOI] [PubMed] [Google Scholar]
- 34.Meluleni G J, Grout M, Evans D J, Pier G B. Mucoid Pseudomonas aeruginosa growing in a biofilm in vitro are killed by opsonic antibodies to the mucoid exopolysaccharide capsule but not by antibodies produced during chronic lung infection in cystic fibrosis patients. J Immunol. 1995;155:2029–2038. [PubMed] [Google Scholar]
- 35.Nichols W W, Evans M J, Slack M P, Walmsley H L. The penetration of antibiotics into aggregates of mucoid and non-mucoid Pseudomonas aeruginosa. J Gen Microbiol. 1989;135:1291–1303. doi: 10.1099/00221287-135-5-1291. [DOI] [PubMed] [Google Scholar]
- 36.Nickel J C, Wright J B, Ruseska I, Marrie T J, Whitfield C, Costerton J W. Antibiotic resistance of Pseudomonas aeruginosa colonizing a urinary catheter in vitro. Eur J Clin Microbiol. 1985;4:213–218. doi: 10.1007/BF02013600. [DOI] [PubMed] [Google Scholar]
- 37.Nivens D E, Ohman D E, Williams J, Franklin M J. Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J Bacteriol. 2001;183:1047–1057. doi: 10.1128/JB.183.3.1047-1057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohman D E, Chitnis C E. Genetic regulation of alginate structure in Pseudomonas aeruginosa. Antibiot Chemother. 1989;42:56–61. doi: 10.1159/000417604. [DOI] [PubMed] [Google Scholar]
- 39.O'Toole G A, Kaplan H B, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 40.O'Toole G A, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 41.O'Toole G A, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 42.Pier G B, Grout M, Zaidi T S. Cystic fibrosis transmembrane conductance regulator is an epithelial cell receptor for clearance of Pseudomonas aeruginosa from the lung. Proc Natl Acad Sci USA. 1997;94:12088–12093. doi: 10.1073/pnas.94.22.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schweizer H P. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol Microbiol. 1992;6:1195–1204. doi: 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 44.Simpson J A, Smith S E, Dean R T. Alginate inhibition of the uptake of Pseudomonas aeruginosa by macrophages. J Gen Microbiol. 1988;134:29–36. doi: 10.1099/00221287-134-1-29. [DOI] [PubMed] [Google Scholar]
- 45.Simpson J A, Smith S E, Dean R T. Scavenging by alginate of free radicals released by macrophages. Free Radic Biol Med. 1989;6:347–353. doi: 10.1016/0891-5849(89)90078-6. [DOI] [PubMed] [Google Scholar]
- 46.Singh P K, Schaefer A L, Parsek M R, Moninger T O, Welsh M J, Greenberg E P. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 47.Smith J J, Travis S M, Greenberg E P, Welsh M J. Cystic fibrosis airway epithelia fail to kill bacteria because of abnormal airway surface fluid. Cell. 1996;85:229–236. doi: 10.1016/s0092-8674(00)81099-5. [DOI] [PubMed] [Google Scholar]
- 48.Speert D P, Dimmick J E, Pier G B, Saunders J M, Hancock R E, Kelly N. An immunohistological evaluation of Pseudomonas aeruginosa pulmonary infection in two patients with cystic fibrosis. Pediatr Res. 1987;22:743–747. doi: 10.1203/00006450-198712000-00026. [DOI] [PubMed] [Google Scholar]
- 49.Stewart P S. Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrob Agents Chemother. 1996;40:2517–2522. doi: 10.1128/aac.40.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welsh M J, Smith A E. Molecular mechanisms of CFTR chloride channel dysfunction in cystic fibrosis. Cell. 1993;73:1251–1254. doi: 10.1016/0092-8674(93)90353-r. [DOI] [PubMed] [Google Scholar]
- 51.Wolfaardt G M, Lawrence J R, Robarts R D, Caldwell S J, Caldwell D E. Multicellular organization in a degradative biofilm community. Appl Environ Microbiol. 1994;60:434–446. doi: 10.1128/aem.60.2.434-446.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu K D, McFeters G A, Stewart P S. Biofilm resistance to antimicrobial agents. Microbiology. 2000;146:547–549. doi: 10.1099/00221287-146-3-547. [DOI] [PubMed] [Google Scholar]
- 53.Yildiz F H, Schoolnik G K. Vibrio cholerae O1 E1 Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci USA. 1999;96:4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zabner J, Smith J J, Karp P H, Widdicombe J H, Welsh M J. Loss of CFTR chloride channels alters salt absorption by cystic fibrosis airway epithelia in vitro. Mol Cell. 1998;2:397–403. doi: 10.1016/s1097-2765(00)80284-1. [DOI] [PubMed] [Google Scholar]