ABSTRACT
Selenium is an important micronutrient for foetal development. MicroRNAs play an important role in the function of the placenta, in communication between the placenta and maternal systems, and their expression can be altered through environmental and nutritional cues. To investigate the associations between placental selenium concentration and microRNA expression in the placenta, our observational study included 393 mother-child pairs from the New Hampshire Birth Cohort Study (NHBCS) and the Rhode Island Child Health Study (RICHS). Placental selenium concentrations were quantified using inductively coupled plasma mass spectrometry, and microRNA transcripts were measured using RNA-seq. We fit negative binomial additive models for assessing the association between selenium and microRNAs. We used the microRNA Data Integration Portal (mirDIP) to predict the target mRNAs of the differentially expressed microRNAs and verified the relationships between miRNA and mRNA targets in a subset of samples using existing whole transcriptome data (N = 199). We identified a non-monotonic association between selenium concentration and the expression of miR-216a-5p/miR-217-5p cluster (effective degrees of freedom, EDF = 2.44 and 2.08; FDR = 3.08 × 10−5) in placenta. Thirty putative target mRNAs of miR-216a-5p and/or miR-217-5p were identified computationally and empirically and were enriched in selenium metabolic pathways (driven by selenoprotein coding genes, TXNRD2 and SELENON). Our findings suggest that selenium influences placental microRNA expression. Further, miR-216a-5p and its putative target mRNAs could be the potential mechanistic targets of the health effect of selenium.
KEYWORDS: Selenium, placenta, hsa-miR-216a-5p, hsa-miR-217-5p, TXNRD2, MACF1
Introduction
As a micronutrient, selenium (Se) is incorporated in selenoproteins that are of importance to human health [1]. During pregnancy, maternal selenium concentrations decrease significantly, likely caused by increased demands for the antioxidative protection imparted by selenoproteins during foetal development [2]. Numerous epidemiologic studies have linked decreased selenium concentration to reproductive and obstetric complications, including recurrent spontaneous abortion, preeclampsia, gestational diabetes, foetal growth restriction, preterm birth, and neurobehavioral health risks in later life of children [3–5]; moreover, the health effect of selenium can be non-monotonic; for example, an inverted U-shaped relationship between prenatal selenium concentration and the neuromotor development of children was detected [6]. Thus, a better understanding of the mechanisms underlying the health effects of selenium will help deepen the understanding of the health effect of selenium and define public health interventions.
MicroRNAs (microRNAs), post-transcriptionally inhibit gene expression through effects on mRNAs stability and translation [7]. Although prior studies are limited, the effect of selenium on microRNA expression was observed in animals and humans [8–10]. It has been reported that selenium supplementation altered microRNA expression in healthy males [11] and in men who have a higher risk of prostate cancer [12]. In-vitro studies of human gastrointestinal cells further demonstrate that a selenium-deficient/supplemented environment could alter microRNA expression, playing a role as feedback control of selenoproteome expression [13–15].
The placenta is a versatile organ that is essential for foetal growth. MicroRNAs play an important role in the placenta, influencing placental function through the regulation of various biological processes, including proliferation, invasion, migration, and apoptosis [16]. Placental microRNAs can be modified by prenatal exposures (reviewed by Maccani et al.) [17], such as maternal cigarette smoking, bisphenol A and alcohol [18]. These exposure-associated microRNA alterations in the placenta may further associate with foetal growth and developmental outcomes. For example, the microRNAs associated with prenatal alcohol exposure interfere with placental development and foetal alcohol spectrum disorder [18]. Our prior study demonstrated that placental microRNA expression is associated with newborn neurobehavioral development [19], but was focused only on a small panel of candidate miRNAs. Therefore, it is of great interest to investigate the impact of prenatal selenium exposures on placental microRNA more comprehensively.
In this study, we aimed to identify Se-associated microRNA expression in the placenta using next-generation sequencing technology; and further computationally predicted the target mRNAs of the differentially expressed microRNAs and verified the associations using our real data.
Methods
Study population
In this observational study, we included 114 and 279 mother-infant pairs who have both selenium and microRNA data available from the Rhode Island Child Health Study (RICHS) and the New Hampshire Birth Cohort Study (NHBCS), respectively (see sample flowchart in Figure S1). The two cohorts have been fully described in our previous studies [20]. Briefly, RICHS recruited mother-child pairs at the Women & Infants Hospital in Providence, Rhode Island, USA, between 2010 and 2013. NHBCS is an ongoing cohort where the mother-child pairs involved in the present study were enrolled from the study’s participating prenatal care clinics in New Hampshire, USA between 2012 and 2013. Only singleton births were recruited into the cohorts. RICHS oversampled small-for-gestational-age (SGA) and large-for-gestational age (LGA) infants, and excluded those with a gestational age less than 37 weeks, and life-threatening conditions or complications. NHBCS recruited women from the prenatal clinics of New Hampshire who used a private and unregulated well for their home drinking water. All the mothers involved in the present study provided written informed consent. The two cohorts were approved by the Institutional Review Board (IRB) at Dartmouth College, Emory University, and RICHS was also approved by the IRB of Women & Infants Hospital of Rhode Island.
Placental specimens collection and selenium (Se) assessment
RICHS and NHBCS shared the same protocols of sample collection, processing, placental selenium assessment, and quality control procedures, of which detailed description can be found in our previous study [21]. Briefly, trained staff collected the tissue samples from the foetal side of the placenta within two hours after birth and avoided including calcium deposits and maternal tissue. Selenium concentration (nanogram (ng) per gram (g), ng/g) in the placental samples (wet tissue) was assessed at the Dartmouth College Trace Element Analysis Core using inductively coupled plasma mass spectrometry and following the EPA method instruction of 3050b and 6020a. The selenium concentration in all the samples passed the detection limit (4.24 ng/g). To ease the biological interpretation, we scaled selenium concentration by dividing the value into the interquartile range (IQR) of selenium.
MicroRNAs purification, sequencing, data processing and quality control
RICHS and NHBCS shared the same protocols of microRNAs purification and sequencing, and the same data pre-processing steps, including read trimming, quality control of the trimmed reads, and microRNA quantitation (GRCh38/hg38 assembly). Details have been described in our previous studies [22]. In the quality control of microRNA counts, we excluded the microRNAs with less than one count per million in more than ten percent of samples for each cohort (NRICHS = 230, NNHBCS = 320) using DESeq2 [23]. We retained a total of 802 and 777 out of the mapped 2656 microRNA transcripts in RICHS and NHBCS. One sample of NHBCS was removed in the sample quality control due to an obvious outlier after visual inspection of principal component analysis biplots (Figure S2).
RICHS mRNA purification and sequencing, data processing and quality control
Only RICHS performed mRNA expression measurement using HiSeq 2500 platform (Illumina, San Diego, CA) as described in [24]. FastQC was applied to check the quality of raw reads, and then Spliced Transcripts Alignment to a Reference (STAR) was used to align the qualified reads to the human genome assembly hg19 [25]. In the quality control of mRNA transcripts, we excluded the genes with less than one count per million in more than ten percent of samples using DESeq2 [23]. Thus, we retained a total of 17,482 out of the mapped 50,810 total RNA transcripts in RICHS.
Covariate assessments
We collected the information on potential confounding variables from self-reported questionnaires or medical records. We assessed the association of selenium concentration with maternal age (p = 0.21), pre-pregnancy BMI (p = 0.27), maternal education (p = 0.03, greater than high school vs. high school less), smoking status (p = 0.41, self-reported prenatal smoking vs. prenatal non-smoking), obstetric history (p = 0.51, had previous pregnancy vs. had no previous pregnancy), gestational age (p = 0.59), child sex (p = 0.22), child race/ethnicity (p = 3.18 × 10−16, white vs. non-white), batch (p = 2.00 × 10−16), and RIN (RNA integrity number, p = 7.07 × 10−8), using one-way ANOVA test (categorical variables) or Pearson’s correlation test (continuous variables). We did not include the cohort indicator because the batch effect also captured the variance contributed by the cohorts (Figure S3). Additionally, we observed significant associations between the top two principal components of microRNA expression and some of the potentially confounding factors, including gestational age, maternal education, child race/ethnicity, batch, and RIN (Figure S4). Although with these factors, no significant associations were observed in our data, other studies have implied that maternal age, pre-pregnancy BMI, smoking status, gestational age, and child sex could bias the epigenetic effect of selenium [26–29]. Therefore, we included maternal age, pre-pregnancy BMI, maternal education, smoking status, gestational age, child sex, child race/ethnicity, batch, and RIN (for microRNA analysis only) as covariates in models fit to the microRNA data. To maintain the uniformity of the models, we also adjusted for these covariates in the mRNA analyses.
To capture unknown or unmeasured sources of variations in the pooled microRNA and the RICHS mRNA expression data (e.g., cell-type heterogeneity), we estimated surrogate variables from the normalized counts using the svaseq function in the sva R package [23,30]. We obtained five surrogate variables for microRNA and two for mRNA using the num.sv function in sva with the ‘be’ method and 200 permutations. We included maternal age, pre-pregnancy BMI, maternal education, smoking status, gestational age, child sex, race/ethnicity, batch, RIN for microRNA analysis only) in the null model, and additionally included the selenium concentration in the full model. To avoid over-adjustment, we only included the first two surrogate variables in the models of microRNA/mRNA analysis due to the significant associations of the surrogate variables with selenium as well as the first two principal components of microRNA/mRNA expression (Figure S4).
Statistical analysis
We described the characteristics of the population in RICHS, NHBCS, and the pooled data using mean, standard deviations (SD), median, interquartile range, or percentage. We compared the characteristics between RICHS and NHBCS using Student’s T test for continuous variables or Chi-Squared test for categorical variables. According to the theoretical basis for a nonlinear health effect of selenium, we fit a negative binomial additive model that modelled individual microRNA expression (dependent variable) against selenium concentration (independent variable). Models were fit to the pooled data, adjusting for all the previously mentioned covariates using NBAMSeq R package [31]. The effective degree of freedom (edf) was used to estimate the degree of non-linearity. An edf = 1 indicates a linear relationship; and a higher edf suggests a relationship that deviates from linearity [32]. In our results, there was an overall decreased tendency for the differentially expressed microRNAs as selenium concentration increased, so we further fit a negative binomial generalized linear model using DESeq2 R package [23] to obtain an average effect size of selenium for the differentially expressed microRNA, adjusting for all the covariates. We considered microRNAs with a FDR (false discovery rate) of q < 0.05 to be differentially expressed microRNAs in relation to selenium concentration.
To test the robustness of our findings, we performed three sensitivity analyses, in which we excluded the samples with a) an extreme value of microRNAs expression, b) an extreme value of selenium concentration in the pooled data, and c) smokers because it has been reported that maternal smoking is associated with lower levels of plasma selenium [26]. Extreme values were defined as values lying beyond Q1 – 3 × IQR or Q3 + 3 × IQR.
To further illustrate the function of the identified microRNAs, we explored their target mRNA using the microRNA Data Integration Portal (mirDIP) that integrates 30 computational microRNA-mRNA prediction tools [33], as well as the whole transcriptome mRNA expression data of RICHS. First, we extracted the computationally predicted microRNA-mRNA interactions with the top 5% confidence score from mirDIP. Second, we estimated the correlations between the differentially expressed microRNAs and the predicted mRNA from the first step, using the microRNA and mRNA expression data in RICHS. We removed the batch effect from the normalized expression data in the log2 scale of microRNA and mRNA separately before the correlation test using the removeBatchEffect function in limma R package [34]. First, we conducted Pearson correlation tests in the samples that were involved in the primary analysis of Se-associated microRNA and had mRNA expression data available (N = 85) using psych R package [35]; then we checked the negatively correlated microRNA-mRNA pairs (FDR-adjusted p-value <0.05) in all the samples with microRNA and mRNA data available in RICHS (N = 199). We considered the microRNA-mRNA pairs exhibiting significant negative correlations in both of the tests to be our putative target mRNAs (FDR-adjusted p-value <0.05), which were subsequently used in functional enrichment analysis.
For enrichment analysis, we searched the putative target mRNAs in the pathway-based gene sets and the Gene Ontology (GO)-based sets using the over-representation analysis approach in the ConsensusPathDB [36]. A p-value of the hypergeometric test and an FDR-adjusted p-value were calculated for each pathway and GO category. 17,482 background mRNAs that passed the quality control were used in the analysis.
Research data and code
The raw reads of microRNA and mRNA have been uploaded to the dbGaP database. The accession number is phs001586. Code is available upon request.
Results
Study population
Table 1 displays the characteristics of mother-child pairs enrolled in the present study from RICHS and NHBCS. The mother-child pairs had similar age, pre-pregnancy BMI, gestational age, the prevalence of prenatal smoking and child sex, but some differences were observed in maternal education and infant race/ethnicity. More mothers in NHBCS had an education level greater than high school, and the infants included from NHBCS were all white, reflective of the source population of the parent study. The placental selenium concentration in RICHS and NHBCS was approximately normally distributed in a similar pattern (Figure S5), but the range of the concentrations in NHBCS was wider (RICHS: 195.79–370.79 ng/g, NHBCS: 107.63–527.76 ng/g). Overall, most mothers had a high school education and were non-smokers during pregnancy.
Table 1.
Charateristics of mother-infant pairs included in analyses of RICHS and NHBCS.
| Characteristics | RICHSa | NHBCSb | Pool |
|---|---|---|---|
| (N = 114) | (N = 279) | (N = 393) | |
| Mothers | |||
| Maternal age, years: Mean (SD) | 30.08 (5.47) | 31.11 (4.70) | 30.81 (4.95) |
| Pre-pregnancy BMI, kg/m2: Mean (SD) | 26.43 (6.49) | 26.17 (5.93) | 26.25 (6.09) |
| Education: N (%) | |||
| Greater than high school | 91 (79.8) | 249 (89.2) | 340 (86.5) |
| High school or less | 23 (20.2) | 30 (10.8) | 53 (13.5) |
| Smoking status: N (%) | |||
| Never | 102 (89.5) | 242 (86.7) | 344 (87.5) |
| Any | 12 (10.5) | 37 (13.3) | 49 (12.5) |
| Children | |||
| Sex: N (%) | |||
| Female | 53 (46.5) | 141 (50.5) | 194 (49.4) |
| Male | 61 (53.5) | 138 (49.5) | 199 (50.6) |
| Baby race/ethnicity: N (%) | |||
| White | 74 (64.9) | 279 (100) | 353 (89.8) |
| Other | 40 (35.1) | 0 (0) | 40 (10.2) |
| Gestational age, weeks: Mean (SD) | 39.30 (0.97) | 39.50 (1.39) | 39.50 (1.29) |
| Placental selenium, ng/g: Median (IQR) | 272.00 (39.40) | 277.40 (57.10) | 274.20 (50.50) |
aRICHS, Rhode Island Child Health Study
bNHBCS, New Hampshire Birth Cohort Study
Associations between selenium concentration and individual microRNA expression in placenta
To examine the potential nonlinear relationship between the placental selenium concentration and individual microRNA expression across the genome, we fit negative binomial additive models for selenium concentration on the expression of each individual microRNA, adjusting for maternal age, pre-pregnancy BMI, maternal education, smoking status, gestational age, child sex, race/ethnicity, batch, RIN, and the first two SVA surrogate variables. The Manhattan plot (Figure 1) shows the genome distribution and -log10 p-values of all the filtered microRNA related to selenium. In the nonlinear analysis, we found that the expression of miR-216a-5p (effective degrees of freedom, EDF = 2.44; FDR = 3.08 × 10−5) and miR-217-5p (EDF = 2.08; FDR = 3.08 × 10−5) were significantly associated with selenium in an overall monotonically decreasing trend (Figure 2), but miR-216a-5p has a slightly increased expression at a high level of selenium concentration. The decreasing rate of expression of the two microRNAs tended to be smaller at a higher level of selenium. We further conducted a linear analysis to estimate the average effect size of selenium on the two microRNAs. Overall, for one IQR increase of selenium, there was an estimated 8% and 13% reduction in the expression of miR-216a-5p (log2 fold change = −0.12, log2 standard error = 0.35; FDR = 6.05 × 10−6) and miR-217-5p (log2 fold change = −0.20, log2 standard error = 0.35; FDR = 3.54 × 10−8), respectively, fixing the values of the other covariates in the model. We illustrated the average effect estimates of selenium on all filtered microRNAs in a volcano plot (Figure S6). MiR-216a-5p and miR-217-5p are in the same miRNA genomic cluster. The normalized counts of the two microRNAs are highly correlated in our study (r = 0.81, p = 2.20 × 10−16, Figure S7). We displayed the microRNAs with unadjusted p-values less than 0.05 in nonlinear and linear analyses in Table S1 and Table S2.
Figure 1.
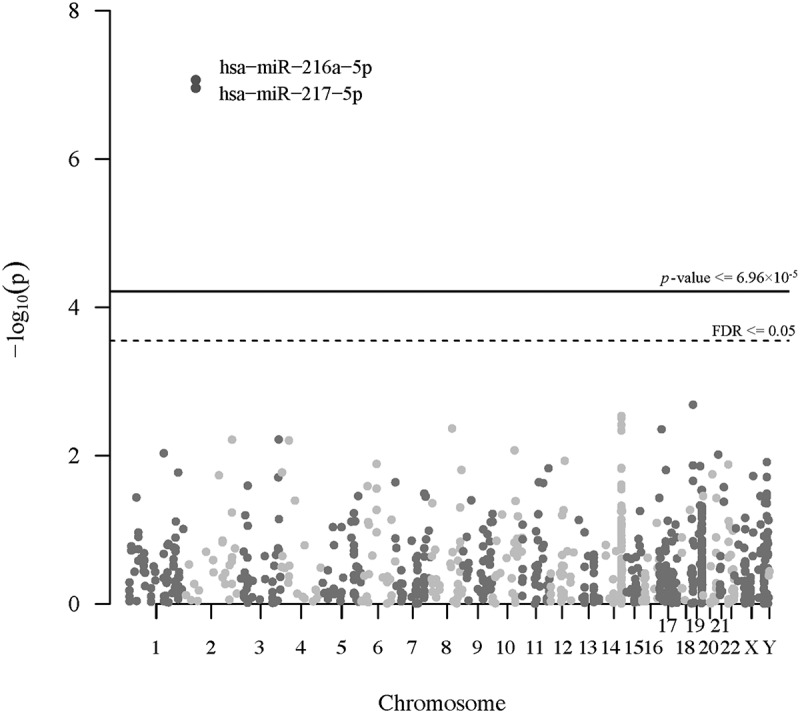
Manhattan plot of the genome distribution and p-values of all the filtered microRNA associations with selenium (N = 393). The solid line and the dashed line represents Bonferroni corrected threshold and the false discovery rate (FDR), respectively.
Figure 2.

The partial effect plots display the component effect of selenium on the differentially expressed microRNAs in the negative binomial additive model, adjusted for maternal age, pre-pregnancy BMI, maternal education, smoking status, gestational age, child sex, child race/ethnicity, batch, RIN, and the first two SVAs. The dashed lines indicate the 95% confidence intervals for the mean of the effect. The distribution of selenium concentration is represented by the rug on the X-axis. Se_IQR: Scaled selenium concentration by dividing the value into the interquartile range of selenium.
In the first sensitivity analysis, we excluded one sample with the extreme value of miR-216a-5p expression; there were no extreme values for miR-217-5p. The association between selenium and miR-216a-5p/miR-217a-5p was robust, and the degree of nonlinearity similar to the primary analysis (EDFmiR-216a-5p = 2.39, pmiR-216a-5p = 1.19 × 10−3, EDFmiR-217-5p = 1.90, pmiR-217-5p = 1.00 × 10−3). In the second sensitivity analysis, we removed five samples with extreme values of selenium concentration, four from NHBCS and one from RICHS. The association between selenium and the miR-216a-5p/miR-217a-5p remained robust, and the degree of nonlinearity was slightly higher in the sensitivity analysis than in the primary analysis (EDFmiR-216a-5p = 2.61, pmiR-216a-5p = 7.89 × 10−7, EDFmiR-217-5p = 2.31, pmiR-217-5p = 7.62 × 10−7). The third sensitivity analysis, which only included non-smokers (N = 344), also indicated robust results (EDFmiR-216a-5p = 2.66, pmiR-216a-5p = 6.38 × 10−8, EDFmiR-217-5p = 2.22, pmiR-217-5p = 3.56 × 10−8).
The target mRNA of the miR-216a-5p and miR-217-5p
We obtained 3443 computationally predicted mRNA for miR-216a-5p from mirDIP. We tested the correlations between the predicted mRNA and the expression of miR-216a-5p. Although there were no predicted mRNA for miR-217-5p in mirDIP, we also tested for correlations between these same mRNA with miR-217-5p because microRNAs in the same cluster evolve to coordinately repress the target genes [37]. A total of 30 out of 3443 predicted mRNAs were negatively correlated with miR-216a-5p and/or miR-217-5p (FDR < 0.05, Table 2), and were considered to be the putative target mRNAs in downstream enrichment analyses. Seven putative target mRNAs, including ATXN7L1, CDK5RAP2, CLSTN1, PTPN14, SELENON, TENM3, TXNRD2, were considered to be the most robust targets as they exhibited statistically significant negative correlations with both miR-216a-5p and miR-217-5p.
Table 2.
Significant (FDR < 0.05) correlations between miR-216a-5p/miR-217-5p and target mRNAs predicted by miRDIP (N = 199).
| MicroRNA | Target mRNA | Correlation coefficient | raw p-value | FDR |
|---|---|---|---|---|
| hsa-miR-217-5p | ARHGAP35 | −0.42 | 7.47 × 10−5 | 0.034 |
| hsa-miR-217-5p | ATAT1 | −0.38 | 2.77 × 10−4 | 0.044 |
| hsa-miR-216a-5p | ATXN7L1 | −0.39 | 2.14 × 10−4 | 0.043 |
| hsa-miR-217-5p | ATXN7L1 | −0.38 | 3.89 × 10−4 | 0.049 |
| hsa-miR-217-5p | C3orf70 | −0.44 | 3.02 × 10−5 | 0.027 |
| hsa-miR-217-5p | CCDC149 | −0.39 | 2.58 × 10−4 | 0.044 |
| hsa-miR-216a-5p | CDK5RAP2 | −0.41 | 1.13 × 10−4 | 0.035 |
| hsa-miR-217-5p | CDK5RAP2 | −0.38 | 3.84 × 10−4 | 0.049 |
| hsa-miR-217-5p | CLSTN1 | −0.46 | 1.14 × 10−5 | 0.024 |
| hsa-miR-216a-5p | CLSTN1 | −0.40 | 1.26 × 10−4 | 0.037 |
| hsa-miR-217-5p | CNNM3 | −0.39 | 2.32 × 10−4 | 0.044 |
| hsa-miR-217-5p | FOXK1 | −0.38 | 3.49 × 10−4 | 0.047 |
| hsa-miR-217-5p | HTT | −0.39 | 1.94 × 10−4 | 0.043 |
| hsa-miR-217-5p | LPP | −0.39 | 2.30 × 10−4 | 0.044 |
| hsa-miR-216a-5p | LRTOMT | −0.38 | 3.19 × 10−4 | 0.046 |
| hsa-miR-216a-5p | MACF1 | −0.37 | 4.36 × 10−4 | 0.050 |
| hsa-miR-217-5p | PARD3B | −0.38 | 3.35 × 10−4 | 0.047 |
| hsa-miR-217-5p | PITPNB | −0.40 | 1.39 × 10−4 | 0.038 |
| hsa-miR-217-5p | PLXNA1 | −0.43 | 4.10 × 10−5 | 0.028 |
| hsa-miR-217-5p | PTPN14 | −0.48 | 2.60 × 10−6 | 0.021 |
| hsa-miR-216a-5p | PTPN14 | −0.47 | 7.03 × 10−6 | 0.021 |
| hsa-miR-217-5p | RAPH1 | −0.37 | 4.27 × 10−4 | 0.049 |
| hsa-miR-216a-5p | RBM19 | −0.38 | 4.00 × 10−4 | 0.049 |
| hsa-miR-217-5p | RBPMS | −0.43 | 3.49 × 10−5 | 0.028 |
| hsa-miR-217-5p | RERE | −0.40 | 1.75 × 10−4 | 0.042 |
| hsa-miR-217-5p | SELENON | −0.43 | 3.74 × 10−5 | 0.028 |
| hsa-miR-216a-5p | SELENON | −0.37 | 4.39 × 10−4 | 0.050 |
| hsa-miR-217-5p | TENM3 | −0.43 | 4.55 × 10−5 | 0.028 |
| hsa-miR-216a-5p | TENM3 | −0.38 | 2.78 × 10−4 | 0.044 |
| hsa-miR-216a-5p | TNPO2 | −0.40 | 1.45 × 10−4 | 0.039 |
| hsa-miR-217-5p | TNRC6B | −0.41 | 1.08 × 10−4 | 0.035 |
| hsa-miR-217-5p | TTC28 | −0.39 | 1.97 × 10−4 | 0.043 |
| hsa-miR-216a-5p | TXNRD2 | −0.47 | 6.64 × 10−6 | 0.021 |
| hsa-miR-217-5p | TXNRD2 | −0.45 | 1.71 × 10−5 | 0.024 |
| hsa-miR-217-5p | UTRN | −0.39 | 2.52 × 10−4 | 0.044 |
| hsa-miR-217-5p | ZNF445 | −0.43 | 4.70 × 10−5 | 0.028 |
| hsa-miR-216a-5p | ZNF587 | −0.41 | 1.17 × 10−4 | 0.035 |
Enrichment analysis
Enrichment analysis suggested that thirty putative target mRNAs were enriched in two Wikipathways: selenium metabolism and selenoproteins (Wikipathways), and selenium micronutrient network (FDR < 0.05), which were driven by putative target mRNAs of two selenoprotein genes: SELENON (selenoprotein N) and TXNRD2 (thioredoxin reductase 2). Moreover, the putative target mRNAs of PLXNA1 (plexin A1) and ARHGAP35 (Rho GRPase activating protein 35) genes were enriched in the semaphorin interactions pathway (Reactome database) (FDR < 0.05). The enrichment analysis also suggested that the 30 putative target mRNAs were enriched in genes involved in 15 GO biological progresses, of which five GO categories were related to neurological processes (FDR < 0.05) (Figure 3).
Figure 3.

Bar plot displays the significant Gene Ontology (GO) biological processes of the target genes of miR-216a-5p and miR-217-5p. Neurological processes are shown in bold.
Discussion
In the present study, we investigated the association between selenium concentration and microRNAs expression in the placenta on a genome-wide scale and linked the selenium-associated microRNAs and their putative target mRNAs. We found that an increased placental selenium concentration was associated with decreased expression of miR-217-5p and miR-216a-5p. Thirty putative target mRNAs of miR-216a-5p/miR-217-5p were enriched in the selenium metabolism pathways and neurobiological processes.
The placental selenium concentration of the two cohorts had similar distributions, but the range of selenium in NHBCS is wider. Similar to our previous work [5], the samples involved in the present study had a relatively high level of selenium concentration, compared to other populations in European or Asian countries [38–40], mostly because of a higher selenium intake than the recommended dietary allowance in the United States population [41,42]. Therefore, our population was unlikely to be selenium deficient, although we could not compare the placenta-specific selenium levels due to the lack of a recommended range in the placenta.
Few studies have investigated the microRNA effects of selenium exposure, particularly in placenta tissue and pregnancy. Our study linked the placental selenium concentration to the expression of the miR-216a-5p and miR-217-5p cluster, which are new observations. Alehagen et al. reported seventy differentially expressed microRNAs in plasma of healthy people who were given selenium and coenzyme Q10 supplementation [11]. Although neither miR-216a-5p or miR-217-5p were detected in their study, six microRNAs (miR-139-5p, miR-326, miR-28-5p, miR-151a-5p, miR-126-5p, miR-301a-3p) found in their study were verified in both nonlinear and linear analyses in our study (p< 0.05). Another study of human colon adenocarcinoma cells showed altered expression of twelve microRNAs in response to selenium depletion [13], and two of the microRNAs (miR-373 and miR-28-5p) were also detected in our study (p < 0.05). The consistency between the two prior studies and the present study suggested the robustness of our findings. The inconsistent results between our study and others could be attributed to the tissue-specific expression of microRNAs [43], different measurements, microRNAs coverage, or the selenium concentration range. Although no study has reported the modifications of miR-216a-5p or miR-217-5p in the placenta, the two microRNAs have been well known as tumour suppressors involved in the proliferative, invasive, and apoptotic processes that are also important for placental development [44–46]. In fact, tumour tissue and placental tissue have strikingly comparable microenvironments and mechanisms to achieve proliferation, invasion, and immunosuppression [47], so we speculate a comparable role of miR-216a-5p and miR-217-5p in the placenta. Two studies of gastric cancer have demonstrated that overexpressed miR-217 suppressed cancer cell metastasis and invasion by targeting Protein Tyrosine Phosphatase Non-Receptor Type 14 (PTPN14) [45,48] that was also detected to be a putative target mRNA of both miR-216a-5p and miR-217-5p in the present study.
As expected, the putative target mRNAs of miR-216a-5p and miR-217-5p were enriched in two selenium-related pathways, driven by SELENON and TXNRD2. The two genes encode selenoprotein N and thioredoxin reductase 2, which are selenoproteins containing selenocysteine and regulating redox-related calcium homoeostasis [49]. The effects of selenium on regulating the expression and activity of SELENON and TXNRD2 have been well demonstrated in both human and animal studies [50,51]. For placental tissue, Watson et al. reported an upregulated level of thioredoxin reductase in trophoblast cells in responding to selenium supplementation [51].
Selenium can change the expression of selenoproteins via regulating microRNAs expression in direct or indirect ways (reviewed by Lu et al.) [52]. Hence, the finding of miR-216a-5p/miR-217-5p in our study might provide a candidate mechanism to explain the link between selenium supply and the expression of selenoprotein genes, but this speculation needs to be further examined in a larger sample size and placental tissue.
Selenium also has important neurological functions. Our previous work has revealed that placental selenium was associated with neuromotor development at birth [5]. In the present study, the putative target mRNAs of the selenium-associated microRNAs were enriched in a relatively high number of neurological processes in GO biological process. However, we did not further explore the neurological effects in newborns due to the data availability. Future studies are warranted to investigate the health effects of selenium-associated microRNA modifications and their target mRNAs on neurobehavioral development.
To our best knowledge, this is the first study to explore the microRNA effects of placental selenium on an epigenome-scale, and the first to integrate analysis of selenium exposure, microRNA, and target mRNA. The parallel study design of RICHS and NHBCS makes the two cohorts comparable and allows us to pool their samples to increase the statistical power. However, we should interpret and generalize our findings within the context of the limitations of the present study. First, our work is limited by the relatively small sample size. Second, this is an observational study where we collected the placental samples at birth, so we cannot imply causation or detect the microRNA effects of selenium throughout pregnancy. Third, we cannot thoroughly eliminate the residual confounding caused by some unknown or unmeasured factors, such as the complex population of placental cells, although we adjusted the surrogate variables in the models and our results were robust. Fourth, we measured selenium instead of selenium-containing proteins that have direct functions on the biological processes. Fifth, we verified the predicted targets in our real mRNA data to reduce the false positives as much as possible, but we did not consider the tissue information and mechanism of miRNA-mRNA interaction. Future studies will need to identify the miRNA targets by reliable experimental strategies in placenta tissue. Sixth, we should be cautious about the interpretation of the filtered miRNA tagets. The mRNA expression levels do not necessarily correlate with the downstream protein expression, especially for the selenoproteins that might be directly influenced by the selenium supply. Finally, future studies will need to evaluate the neurobehavioral outcomes in relation to selenium, microRNAs and mRNA as we did not access this question directly in this study.
Conclusion
This study elucidates microRNA expression modifications in relation to prenatal selenium, adding to evidence of the cellular effects of selenium exposure as a potential mechanism for its protective role. Our findings suggest that selenium-associated miR-216a-5p putatively targets mRNA, TXNRD2 and MACF1, which are involved in selenoprotein regulation. This study enhances our understanding of the influence of early-life environmental exposures on foetal development by modifying microRNA and downstream mRNA expression.
Supplementary Material
Acknowledgments
We acknowledge the cooperation of the participants enrolled in the RICHS and NHBCS, and the contribution of the research staff who worked for the two cohorts.
MK, CM, TP, BJ, KH, JC, DK designed research; KH and AB conducted research; FYT analyzed data; FYT wrote the paper; EK and TE provided essential analysis suggestions; CM had primary responsibility for final content. All authors read and approved the final manuscript.
Funding Statement
This work was supported by the National Institutes of Health (NIH-NIEHS P01ES022832, NIH-NIEHS National Institute of Environmental Health Sciences R24ES028507, NIH-NIEHS R01ES025145, NIH-NIEHS P30ES019776; NIH-NIMHD National Institute on Minority Health and Health Disparities R01MD011698; NIH-NIGMS National Institute of General Medical Sciences P20GM104416); the U.S. Environmental Protection Agency (EPA) (U.S. EPA grant RD83544201).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Rayman MP. Selenium and human health. Lancet. 2012;379:1256–1268. [DOI] [PubMed] [Google Scholar]
- [2].Pieczyńska J, Grajeta H. The role of selenium in human conception and pregnancy. J Trace Elem Med Biol. 2015;29:31–38. [DOI] [PubMed] [Google Scholar]
- [3].Mistry HD, Pipkin FB, Redman CW, et al. Selenium in reproductive health. [Internet] Am J Obstet Gynecol; 2012;206. Available from: https://pubmed.ncbi.nlm.nih.gov/21963101/ [DOI] [PubMed] [Google Scholar]
- [4].Skröder HM, Hamadani JD, Tofail F, et al. Selenium status in pregnancy influences children’s cognitive function at 1.5 years of age. Clin Nutr. 2015;34:923–930. [DOI] [PubMed] [Google Scholar]
- [5].Tian F-Y, Everson TM, Lester B, et al. Selenium-associated DNA methylation modifications in placenta and neurobehavioral development of newborns: an epigenome-wide study of two U.S. birth cohorts. Environ Int. 2020;137:105508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Amorós R, Murcia M, González L, et al. Maternal selenium status and neuropsychological development in Spanish preschool children. Environ Res. 2018;166:215–222. [DOI] [PubMed] [Google Scholar]
- [7].Catalanotto C, Cogoni C, Zardo G. MicroRNA in control of gene expression: an overview of nuclear functions. Int J Mol Sci. 2016;17:1712. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5085744/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yang T, Cao C, Yang J, et al. miR-200a-5p regulates myocardial necroptosis induced by Se deficiency via targeting RNF11. Redox Biol. 2017;15:159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wu RT, Cao L, Mattson E, et al. Opposing impacts on healthspan and longevity by limiting dietary selenium in telomere dysfunctional mice. Aging Cell. 2017;16:125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Guo X, Yang Q, Zhang W, et al. Associations of blood levels of trace elements and heavy metals with metabolic syndrome in Chinese male adults with microRNA as mediators involved. Environ Pollut. 2019;248:66–73. [DOI] [PubMed] [Google Scholar]
- [11].Alehagen U, Johansson P, Aaseth J, et al. Significant changes in circulating microRNA by dietary supplementation of selenium and coenzyme Q10 in healthy elderly males. A subgroup analysis of a prospective randomized double-blind placebo-controlled trial among elderly Swedish citizens. PLoS ONE. 2017;12:e0174880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gontero P, Marra G, Soria F, et al. A randomized double-blind placebo controlled phase I–II study on clinical and molecular effects of dietary supplements in men with precancerous prostatic lesions. Chemoprevention or “chemopromotion”? Prostate. 2015;75:1177–1186. [DOI] [PubMed] [Google Scholar]
- [13].Maciel-Dominguez A, Swan D, Ford D, et al. Selenium alters miRNA profile in an intestinal cell line: evidence that miR-185 regulates expression of GPX2 and SEPSH2. Mol Nutr Food Res. 2013;57:2195–2205. [DOI] [PubMed] [Google Scholar]
- [14].Potenza N, Castiello F, Panella M, et al. Human MiR-544a modulates SELK expression in hepatocarcinoma cell lines. PLOS ONE Public Library Sci. 2016;11:e0156908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tarek M, Louka ML, Khairy E, et al. Role of microRNA-7 and selenoprotein P in hepatocellular carcinoma. Tumour Biol SAGE Publications Ltd STM. 2017;39:1010428317698372. [DOI] [PubMed] [Google Scholar]
- [16].Miska EA. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev. 2005;15:563–568. [DOI] [PubMed] [Google Scholar]
- [17].Maccani MA, Marsit CJ. Exposure and fetal growth-associated miRNA alterations in the human placenta. Clin Epigenet BioMed Central. 2011;2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tseng AM, Mahnke AH, Wells AB, et al. Maternal circulating miRNAs that predict infant FASD outcomes influence placental maturation. Life Sci Alliance. 2019;2. Available from: https://www.life-science-alliance.org/content/2/2/e201800252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Maccani MA, Padbury JF, Lester BM, et al. Placental miRNA expression profiles associated with measures of infant neurobehavioral outcomes. Pediatr Res. 2013;74:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gilbert-Diamond D, Cottingham KL, Gruber JF, et al. Rice consumption contributes to arsenic exposure in US women. Proc Natl Acad Sci U S A. 2011;108:20656–20660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Punshon T, Li Z, Marsit CJ, et al. Placental metal concentrations in relation to maternal and infant toenails in a US cohort. Environ Sci Technol. 2016;50:1587–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kennedy EM, Hermetz K, Burt A, et al. Placental microRNA expression associates with birthweight through control of adipokines: results from two independent cohorts. Epigenetics. 2021;16:770–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Deyssenroth MA, Peng S, Hao K, et al. Whole-transcriptome analysis delineates the human placenta gene network and its associations with fetal growth. BMC Genomics. 2017;18:520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics Oxford Acad. 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bashar SK, Mitra AK. Effect of smoking on vitamin A, vitamin E, and other trace elements in patients with cardiovascular disease in Bangladesh: a cross-sectional study. Nutr J. 2004;3:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bleys J, Navas-Acien A, Guallar E. Serum selenium and diabetes in U.S. adults. Diabetes Care. 2007;30:829–834. [DOI] [PubMed] [Google Scholar]
- [28].Kocyigit A, Erel O, Gur S. Effects of tobacco smoking on plasma selenium, zinc, copper and iron concentrations and related antioxidative enzyme activities. Clin Biochem. 2001;34:629–633. [DOI] [PubMed] [Google Scholar]
- [29].Muntau AC, Streiter M, Kappler M, et al. Age-related reference values for serum selenium concentrations in infants and children. Clin Chem. 2002;48:555–560. [PubMed] [Google Scholar]
- [30].Leek JT. svaseq: removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res. 2014;42:e161–e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ren X, Kuan P-F. Negative binomial additive model for RNA-Seq data analysis. BMC Bioinformatics. 2020;21:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zuur AF, Ieno EN, Walker N, et al. Mixed effects models and extensions in ecology with R. New York/NY: Springer New York; 2009, 53. Available from: http://link.springer.com/10.1007/978-0-387-87458-6 [Google Scholar]
- [33].Tokar T, Pastrello C, Rossos AEM, et al. mirDIP 4.1—integrative database of human microRNA target predictions. Nucleic Acids Res. 2018;46:D360–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Revelle WR. psych: procedures for personality and psychological research. 2017. Available from: https://www.scholars.northwestern.edu/en/publications/psych-procedures-for-personality-and-psychological-research
- [36].Kamburov A, Pentchev K, Galicka H, et al. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res. 2011;39:D712–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wang Y, Luo J, Zhang H, et al. microRNAs in the same clusters evolve to coordinately regulate functionally related genes. Mol Biol Evol. 2016;33:2232–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kosik-Bogacka D, Łanocha-Arendarczyk N, Kot K, et al. Concentrations of mercury (Hg) and selenium (Se) in afterbirth and their relations with various factors. Environ Geochem Health. 2018;40:1683–1695. [DOI] [PubMed] [Google Scholar]
- [39].Osman K, Åkesson A, Berglund M, et al. Toxic and essential elements in placentas of Swedish women. Clin Biochem. 2000;33:131–138. [DOI] [PubMed] [Google Scholar]
- [40].Sakamoto M, Yasutake A, Domingo JL, et al. Relationships between trace element concentrations in chorionic tissue of placenta and umbilical cord tissue: potential use as indicators for prenatal exposure. Environ Int. 2013;60:106–111. [DOI] [PubMed] [Google Scholar]
- [41].Institute of Medicine . Dietary reference intakes for vitamin C, vitamin E, selenium, and carotenoids. Washington/D.C.: National Academies Press; 2000. Available from: http://www.nap.edu/catalog/9810. [PubMed] [Google Scholar]
- [42].National Institutes of Health . Office of dietary supplements - selenium. 2019. Available from: https://ods.od.nih.gov/factsheets/Selenium-Consumer/
- [43].Ludwig N, Leidinger P, Becker K, et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res Oxford Academic. 2016;44:3865–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Flum M, Kleemann M, Schneider H, et al. miR-217-5p induces apoptosis by directly targeting PRKCI, BAG3, ITGAV and MAPK1 in colorectal cancer cells. J Cell Commun Signal. 2018;12:451–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Liu Y-P, Sun X-H, Cao X-L, et al. MicroRNA-217 suppressed epithelial-to-mesenchymal transition in gastric cancer metastasis through targeting PTPN14. Eur Rev Med Pharmacol Sci. 2017;21:1759–1767. [PubMed] [Google Scholar]
- [46].Chai L, Yang G. MiR-216a-5p targets TCTN1 to inhibit cell proliferation and induce apoptosis in esophageal squamous cell carcinoma. Cell Mol Biol Lett. 2019;24:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Costanzo V, Bardelli A, Siena S, et al. Exploring the links between cancer and placenta development. Open Biol. 2018;8:180081. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6030113/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wang H, Qin R, Guan A, et al. HOTAIR enhanced paclitaxel and doxorubicin resistance in gastric cancer cells partly through inhibiting miR-217 expression. J Cell Biochem. 2018;119:7226–7234. [DOI] [PubMed] [Google Scholar]
- [49].Stelzer G, Rosen N, Plaschkes I, et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 2016;54:1.30.1–1.30.33. [DOI] [PubMed] [Google Scholar]
- [50].Cao L, Zhang L, Zeng H, et al. Analyses of selenotranscriptomes and selenium concentrations in response to dietary selenium deficiency and age reveal common and distinct patterns by tissue and sex in telomere-dysfunctional mice. J Nutr Oxford Acad. 2017;147:1858–1866. [DOI] [PubMed] [Google Scholar]
- [51].Watson M, van Leer L, Vanderlelie JJ, et al. Selenium supplementation protects trophoblast cells from oxidative stress. Placenta. 2012;33:1012–1019. [DOI] [PubMed] [Google Scholar]
- [52].Lu H-Y, Somuncu B, Zhu J, et al. Selenoproteins and epigenetic regulation in mammals. In Patel VB, Preedy VR, editors. Handbook of nutrition, diet, and epigenetics. Cham: Springer International Publishing; 2019, p. 1803–1817. Available from: http://link.springer.com/10.1007/978-3-319-55530-0_31 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


