ABSTRACT
Night shift work is associated with increased breast cancer risk, but the molecular mechanisms are not well-understood. The objective of this study was to explore the relationship between night shift work parameters (current status, duration/years, and intensity) and methylation in circadian genes as a potential mechanism underlying the carcinogenic effects of night shift work. A cross-sectional study was conducted among 74 female healthcare employees (n = 38 day workers, n = 36 night shift workers). The Illumina Infinium MethylationEPIC beadchip was applied to DNA extracted from blood samples to measure methylation using a candidate gene approach at 1150 CpG loci across 22 circadian genes. Linear regression models were used to examine the association between night shift work parameters and continuous methylation measurements (β-values) for each CpG site. The false-discovery rate (q = 0.2) was used to account for multiple comparisons. Compared to day workers, current night shift workers demonstrated hypermethylation in the 5ʹUTR region of CSNK1E (q = 0.15). Individuals that worked night shifts for ≥10 years exhibited hypomethylation in the gene body of NR1D1 (q = 0.08) compared to those that worked <10 years. Hypermethylation in the gene body of ARNTL was also apparent in those who worked ≥3 consecutive night shifts a week (q = 0.18). These findings suggest that night shift work is associated with differential methylation in core circadian genes, including CSNK1E, NR1D1 and ARNTL. Future, larger-scale studies with long-term follow-up and detailed night shift work assessment are needed to confirm and expand on these findings.
KEYWORDS: Night shift work, DNA methylation, circadian genes, circadian rhythm
Introduction
Night shift work is defined as exposure to regular night-time work between the hours of 00:00 and 05:00 for at least three hours, and can include both rotating (i.e., alternating day and night shifts) and permanent night shift schedules [1]. An estimated 1.8 million Canadians work in a schedule with exposure to night shift work, the equivalent of 12% of the working population [2]. In European and North American countries, it is estimated that 13–15% of workers are night shift workers [3]. The proportion of female workers exposed to night shift work is increasing [2], and given that night shift work has been associated with a higher risk of breast cancer [4,5], there is a pressing need to understand the mechanisms by which night shift work may increase cancer risk so as to inform the development of effective intervention/prevention strategies.
It is hypothesized that exposure to artificial light at night and changes in sleep-wake cycles due to night shift work schedules could be responsible for an increase in the risk of cancer [6,7]. This may happen through a number of inter-related biological mechanisms, including suppression of melatonin production, adverse metabolic changes, changes in sex hormone levels, and alteration of the expression of circadian genes [6,7]. Disruption of DNA methylation, which is known to influence gene expression, is implicated in the aetiology of breast cancer, including changes to the methylation of circadian genes [8–11]. However, few studies have investigated the impact of night shift work on circadian gene methylation, and results among studies have differed with respects to specific genes impacted and direction of effects [12–16]. In fact, most studies examined very few circadian genes [12–14].
The objective of this study was to conduct an exploration of the relationship between night shift work and circadian gene methylation by examining well-defined night shift work parameters (current status, duration/years of night shift work, and night shift intensity) and DNA methylation levels at multiple CpG loci and gene regions across 22 circadian genes.
Materials and methods
Female hospital employees from Kingston Health Sciences Center (KHSC) enrolled in a previous study [17] were re-contacted to participate in a new cross-sectional study conducted from July 2019 to March 2020. Participants were eligible if they were 1) still employed at the hospital, 2) working the same schedule as previously recorded: either a shift schedule including a night component, or day-only schedule, and 3) not pregnant. In total, 74 female employees were included who work a fixed day schedule (n = 38) or a rotating schedule including nights (n = 36). Most participants were registered nurses from inpatient units (62%), but staff from laboratory, diagnostic, and support services were also included. This cross-sectional study was approved by the Health Sciences Research Ethics Board at Queen’s University, and all participants provided written informed consent.
Night shift work
Information on history of night shift work was self-reported through a questionnaire. Night shift work parameters included: a) current night shift work status; b) years of night shift work exposure (duration); and c) average number of consecutive night shifts per week over the past two years. A typical night shift schedule consisted of a rotating shift schedule of two consecutive 12-hour day shifts, followed by two consecutive 12-hour night shifts with at least three working hours between midnight and 05:00, and five consecutive free days. This schedule was variable, with some participants working rotating weeks of only night shifts or only day shifts, or a mix of day and night shifts with no set schedule (for part-time workers). However, due to hospital policy, all night shifts were 12-hour shifts from 19:00 to 07:00. The typical day worker schedule included five consecutive 8-hour shifts starting at 08:00 or 09:00.
DNA methylation
Fasting blood samples (4–5 mL) were collected by the study nurse. In order to minimize diurnal variation in lymphocyte cell counts, all blood samples were collected within the same three-hour time window (06:30 to 09:30). Blood was collected in PAXGene DNA tubes and stored in a − 20°C freezer. To isolate buffy coats, genomic DNA was directly extracted from thawed buffy coats using the QIASymphony SP (Qiagen, Crawley, UK) instrument and the QIAsymphony DNA Midi Kit (Qiagen, Crawley, UK). DNA was quantified using the Quant-iTTM PicoGreen dsDNA kit (Invitrogen), and was bisulphite converted according to manufacturer specifications. DNA was whole-genome amplified, enzymatically fragmented, purified, and applied to the Illumina Infinium MethylationEPIC beadchip (Illumina, San Diego, CA). The EPIC beadchips were analysed using the Illumina iScan system (Illumina, San Diego, CA). Quality control measures included verifying the integrity of the DNA using agarose gel electrophoresis and randomization of samples across chips and plates.
The minfi R package was used to perform quantile normalization of the methylation data. Multi-dimensional scaling plots and principal component analyses were performed to identify sources of variation among samples; no strong sources of variation were identified, and no probes were removed (see Supplementary Figure S1). Filtering was done using the methods described by Maksimovic et al [18]. We excluded probes that had detection p-values greater than or equal to 0.05 in at least one sample (n = 6,831), SNP-related probes (n = 28,767), and cross-reactive probes (n = 40,775) [19]. Since all participants were female, no probes located on the sex chromosomes were removed. β-values were also calculated using the minfi package.
The array included 1150 CpG loci distributed across the 22 circadian genes, as sourced from the UCSC Genome Browser, hg19 assembly [20] (padding of ± 5000 base pairs): CLOCK, ARNTL, ARNTL2, NPAS2, PER1, PER2, PER3, CRY1, CRY2, NR1D1, NR1D2, RORA, DEC1, BHLE41, TIMELESS, CSNK1A1, CSNK1D, CSNK1E, CSNK2A1, CSNK2A2, MTNR1A and MTNR1B. These genes were chosen a priori because many are considered core clock genes whose protein products are essential for the generation and regulation of circadian rhythms [21].
Estimation of white blood cell type distribution
Proportions of six white blood cell types (neutrophils, monocytes, B-cells, Natural Killer (NK) cells, CD4 + T-cells, and CD8 + T-cells) were estimated from methylation data using Houseman’s reference-based approach implemented in the ‘estimatecellcounts2ʹ function from the FlowSorted.Blood.EPIC package in R22, [22].
Statistical analysis
We applied multiple linear regression techniques to examine the association between night shift work parameters and continuous β-values for each CpG site. Confounders in the relationship between night shift work and circadian gene methylation were chosen a priori using a directed acyclic graph (DAG) (see Supplementary Figure S2). All analyses were adjusted for age (<40 years/40-54 years/55+ years), household income (<$75,000/$75,000–99,999/$100,000+), part/full-time status, occupational exposure to radiation (very often/often vs. not often), and occupational exposure to disinfectants (very often/often vs. not often). Proportion of leukocytes cell types (excluding neutrophils to avoid multicollinearity) were also included in all models to adjust for leukocyte cell profile. To account for multiple testing, q-values were calculated based on the false discovery rate (FDR) [23]. In this exploratory study, associations with a q-value less than or equal to 0.20 were deemed noteworthy.
Since core circadian genes are known to regulate downstream expression of genes related to inflammation, it is possible that inflammation could be a mediator in the relationship [24,25]. A sensitivity analysis was also conducted that did not adjust for white blood cell composition (a potential proxy for inflammation). In addition, smoking status and alcohol use are strong predictors of DNA methylation [26,27]. A sensitivity analysis was also conducted that additionally adjusted for smoking status (current or recently quit/past smoker/never smoker) and alcohol use (never or <1 drink a month/1-4 drinks a month/2+ drinks a week). We also examined the association using continuous M-values, rather than β-values. However, results were similar to our original findings and are not presented. All data analyses were conducted using R (version 4.0.4).
Results
Characteristics of study population
Current night shift workers were more likely to be younger and postmenopausal than day workers and were more likely to report working 10 or more years of night shifts (Table 1). Day and night workers were similar in terms smoking status, part-time status, and white blood cell composition. Current night shift workers were also more likely to report being regularly exposed to radiation and disinfectants, compared to day workers, more likely to have a higher household income over $100,000 CDN, and slightly more likely to report a family history of cancer (Table 1).
Table 1.
Demographic data among current day and night shift study participants.
| Current day worker (n = 38) | Current night shift worker (n = 36) | |
|---|---|---|
| Age (years)(n, %) | ||
| <40 | 7 (18.4) | 16 (44.4) |
| 40–54 | 24 (63.2) | 11 (30.6) |
| ≥55 | 7 (18.4) | 9 (25.0) |
| Menopausal status(n, %) | ||
| Postmenopausal | 12 (31.6) | 14 (38.9) |
| Premenopausal | 26 (68.4) | 22 (61.1) |
| Education(n, %) | ||
| High school | 2 (5.3) | 0 (0.0) |
| Post-secondary diploma | 19 (50.0) | 19 (52.8) |
| University undergraduate degree or higher | 17 (44.8) | 17 (47.2) |
| Household Income (in Canadian $)(n, %) | ||
| <$75,000 | 10 (26.3) | 3 (8.3) |
| $75,000-$99,999 | 9 (23.7) | 8 (22.2) |
| ≥$100,000 | 19 (50.0) | 25 (69.4) |
| Smoking status(n, %) | ||
| Never smoked | 24 (63.2) | 25 (69.4) |
| Past smoker | 10 (26.3) | 9 (25.0) |
| Currently smoke/recently quit | 4 (10.5) | 2 (5.6) |
| Family history of cancer(n, %) | ||
| Yes | 17 (44.7) | 19 (52.7) |
| No | 21 (55.3) | 16 (44.4) |
| Missing | 0 (0.0) | 1 (2.8) |
| Alcohol consumption(n, %) | ||
| ≥2+ drinks/week | 10 (26.3) | 10 (27.7) |
| 1–4 drinks/month | 18 (47.4) | 15 (41.7) |
| Never or <1 drink/month | 10 (26.3) | 11 (30.6) |
| Occupational exposure to radiation(n, %) | ||
| Not often | 32 (84.2) | 19 (52.8) |
| Very often/often | 6 (15.8) | 17 (47.2) |
| Occupational exposure to disinfectants(n, %) | ||
| Not often | 15 (39.5) | 1 (2.8) |
| Very often/often | 23 (60.5) | 35 (97.2) |
| White blood cell-type composition(mean, SD) | ||
| B cells (%) | 2.73 (1.40) | 3.59 (1.98) |
| CD8 T cells (%) | 6.91 (2.95) | 6.95 (3.92) |
| CD4 T cells (%) | 18.47 (4.95) | 19.51 (6.40) |
| Natural killer cells (%) | 3.14 (2.24) | 2.59 (2.41) |
| Monocytes (%) | 6.92 (2.50) | 6.63 (2.16) |
| Neutrophils (%) | 61.31 (7.40) | 59.96 (7.56) |
| Characteristics of work schedule | ||
| Status(n, %) | ||
| Part-time | 5 (13.2) | 5 (13.9) |
| Full-time | 33 (86.8) | 31 (86.1) |
| Night shift work duration(n, %) | ||
| <10 years | 26 (68.4) | 10 (27.8) |
| ≥10 years | 12 (31.6) | 26 (72.2) |
| Night shift work intensity(n, %) | ||
| <3 consecutive night shifts/week | - | 25 (69.4) |
| ≥3 consecutive night shifts/week | - | 11 (30.6) |
Night shift work and circadian gene methylation
Results for the 10 circadian CpG sites with the lowest q-values in association with current night shift status, night shift duration, and night shift intensity are summarized in Tables 2–4, respectively. Results for all 1150 CpG loci are provided in Supplementary Tables S1–3. These results are also summarized as a volcano plot in Figure 1, where loci with the strongest associations (q ≤ 0.20) are highlighted for each analysis.
Table 2.
Circadian gene methylation differences between current night shift and day shift workers.
| Illumina ID | Chromosome | Position | Gene | Gene Groupa | Relation to CpG Islandb | Meanc methylation in day workers (SD) | Meanc methylation in night shift workers (SD) | Adjustedd mean difference (95% CI) | q-valuee |
|---|---|---|---|---|---|---|---|---|---|
| cg14718583 | 22 | 38,725,060 | CSNK1E | 5ʹUTR | Open sea | 87.72 (1.84) | 88.57 (1.57) | 1.69 (0.87, 2.52) | 0.18 |
| cg20667664 | 17 | 38,254,448 | NR1D1 | Body | North Shore | 78.97 (1.65) | 78.29 (1.51) | −1.53 (−2.38, −0.67) | 0.52 |
| cg12596843 | 11 | 45,864,825 | CRY2 | - | North Shelf | 12.73 (1.28) | 12.41 (0.84) | −0.89 (−1.43, −0.35) | 0.67 |
| cg23929615 | 22 | 38,794,688 | CSNK1E | TSS200 | Island | 52.41 (2.28) | 51.06 (2.09) | −2.07 (−3.34, −0.79) | 0.67 |
| cg10472395 | 15 | 61,070,548 | RORA | Body | Open Sea | 82.50 (1.38) | 81.96 (1.45) | −1.09 (−1.80, −0.39) | 0.69 |
| cg10976861 | 2 | 239,149,937 | PER2 | TSS1500 | South Shore | 33.86 (2.11) | 32.64 (1.61) | −1.58 (−2.60, −0.55) | 0.69 |
| cg26113056 | 15 | 61,521,643 | RORA | TSS200 | Island | 8.97 (1.56) | 8.57 (1.21) | −1.03 (−1.71, −0.35) | 0.69 |
| cg23965982 | 15 | 61,055,848 | RORA | Body | Open Sea | 83.96 (1.91) | 83.44 (1.41) | −1.23 (−2.07, −0.38) | 0.78 |
| cg16774421 | 12 | 27,481,875 | ARNTL2 | - | North Shelf | 92.20 (1.33) | 91.85 (1.14) | −0.91 (−1.54, −0.27) | 0.78 |
| cg13184823 | 4 | 187,476,599 | MTNR1A | TSS200 | Island | 38.96 (8.07) | 41.24 (6.26) | 5.42 (1.59, 9.25) | 0.78 |
a. Functional region of gene as indicated in Illumina annotation.
b. Position relative to CpG Island as indicated in Illumina annotation.
c. Unadjusted mean.
d. All analyses adjusted for age, income, part/full-time status, occupational exposures to radiation and disinfectants, and leukocyte cell profile. Day workers are the referent category.
e. Q-values represent p-values adjusted for multiple testing using the FDR method.
Table 3.
Circadian gene methylation differences between workers with ≥10 years of night shift work duration and workers with <10 years.
| Illumina ID | Chromosome | Position | Gene | Gene Groupa | Relation to CpG Islandb | Meanc methylation in <10 years workers (SD) | Meanc methylation in ≥10 years workers (SD) | Adjustedd mean difference (95% CI) | q-valuee |
|---|---|---|---|---|---|---|---|---|---|
| cg20667664 | 17 | 38,254,448 | NR1D1 | Body | North Shore | 79.13 (1.73) | 78.15 (1.33) | −1.60 (−2.33 – 0.87) | 0.08 |
| cg22405816 | 9 | 118,135,937 | DEC1 | 5ʹUTR | Open Sea | 84.68 (2.47) | 83.25 (2.65) | −1.84 (−2.87,0.81) | 0.32 |
| cg00436663 | 22 | 38,793,933 | CSNK1E | Body | Island | 7.54 (0.93) | 7.07 (0.85) | −0.74 (−1.18, – 0.31) | 0.32 |
| cg24219929 | 15 | 60,884,748 | RORA | Body | Island | 9.83 (7.03) | 6.37 (3.38) | −4.81 (−7.63, – 1.98) | 0.32 |
| cg18204040 | 12 | 26,279,190 | BHLHE41 | TSS1500 | Island | 10.69 (0.72) | 10.20 (0.69) | −0.56 (−0.90, −0.22) | 0.32 |
| cg06233947 | 16 | 58,230,251 | CSNK2A2 | Body | North Shore | 92.20 (1.17) | 91.32 (1.49) | −1.13 (−1.81, – 0.44) | 0.32 |
| cg26724232 | 15 | 60,885,912 | RORA | Body | South Shore | 81.46 (3.30) | 83.61 (2.65) | 2.24 (0.85, 3.63) | 0.32 |
| cg09473510 | 4 | 187,476,573 | MTNR1A | TSS200 | Island | 19.92 (8.21) | 23.99 (7.45) | 5.19 (1.97, 8.41) | 0.32 |
| cg13154331 | 22 | 38,795,868 | CSNK1E | TSS1500 | South Shore | 70.52 (2.44) | 71.55 (2.17) | 1.52 (0.57, 2.48) | 0.32 |
| cg07250429 | 15 | 61,051,202 | RORA | Body | Open Sea | 84.76 (1.22) | 83.98 (1.57) | −1.03 (−1.68, –0.38) | 0.32 |
a. Functional region of gene as indicated in Illumina annotation.
b. Position relative to CpG Island as indicated in Illumina annotation.
c. Unadjusted mean.
d. All analyses adjusted for age, income, part/full-time status, occupational exposures to radiation and disinfectants, and leukocyte cell profile. Workers with <10 years of night shift work duration are the referent category.
e. Q-values represent p-values adjusted for multiple testing using the FDR method.
Table 4.
Circadian gene methylation differences between workers with ≥3 consecutive night shifts/week and <3 consecutive night shifts/week.
| Illumina ID | Chromo-some | Position | Gene | Gene Groupa | Relation to CpG Islandb | Meanc methylation in <3 shifts/week workers (SD) | Meanc methylation in ≥3 shifts/week workers (SD) | Adjustedd mean difference (95% CI) | q-valuee |
|---|---|---|---|---|---|---|---|---|---|
| cg21078679 | 11 | 13,377,829 | ARNTL | Body | Open sea | 85.79 (1.36) | 87.71 (2.14) | 2.21 (1.15, 3.27) | 0.15 |
| cg26449680 | 22 | 38,714,272 | CSNK1E | TSS200 | South Shore | 37.84 (2.92) | 40.81 (2.69) | 1.71 (0.65, 2.77) | 0.97 |
| cg15603424 | 11 | 13,300,592 | ARNTL | 5ʹUTR | Island | 8.10 (4.32) | 12.34 (5.28) | 4.72 (1.65, 7.79) | 0.97 |
| cg14718583 | 22 | 38,725,060 | CSNK1E | 5ʹUTR | Open Sea | 87.97 (1.76) | 89.07 (1.48) | 1.65 (0.56, 2.75) | 0.97 |
| cg27004243 | 17 | 8,055,360 | PER1 | 5ʹUTR | Island | 6.00 (0.64) | 5.45 (0.87) | −0.75 (−1.24, −0.25) | 0.97 |
| cg03092603 | 22 | 38,712,210 | CSNK1E | 5ʹUTR | North Shore | 7.10 (0.72) | 6.52 (0.60) | −0.72 (−1.22, −0.22) | 0.97 |
| cg23633210 | 2 | 101,600,602 | NPAS2 | Body | Open Sea | 86.66 (1.58) | 87.94 (1.36) | 1.47 (0.43, 2.51) | 0.97 |
| cg17367616 | 12 | 27,485,428 | ARNTL2 | TSS1500 | North Shore | 56.72 (2.93) | 55.81 (3.87) | −2.56 (−4.48, −0.65) | 0.97 |
| cg04324336 | 15 | 60,941,269 | RORA | Body | Open Sea | 86.00 (1.45) | 84.89 (1.48) | −1.14 (−2.11, −0.18) | 0.97 |
| cg08924113 | 12 | 27,497,913 | ARNTL2 | Body | Open Sea | 79.10 (4.68) | 76.00 (7.06) | −4.30 (−7.94, −0.66) | 0.97 |
a. Functional region of gene as indicated in Illumina annotation.
b. Position relative to CpG Island as indicated in Illumina annotation.
c. Unadjusted mean.
d. All analyses adjusted for age, income, part/full-time status, occupational exposures to radiation and disinfectants, and leukocyte cell profile. Workers with <3 consecutive night shifts/week are the referent category.
e. Q-values represent p-values adjusted for multiple testing using the FDR method.
Figure 1.
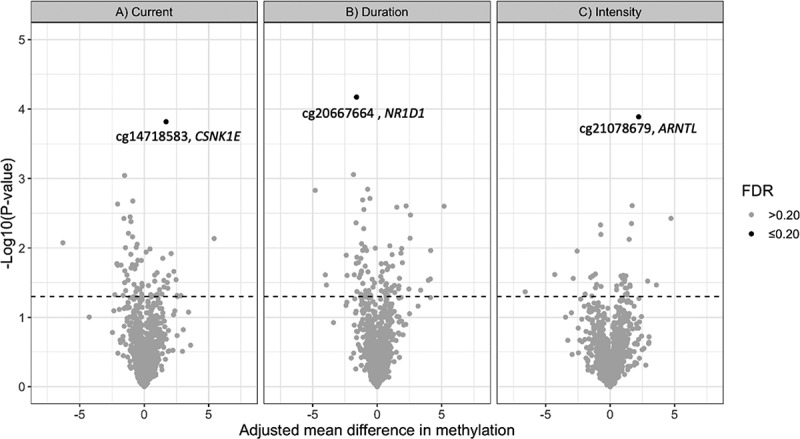
Volcano plot of results from the analysis of 1150 CpG loci across 22 circadian genes in association with A) current night shift work vs. day work, B) night shift work duration (≥10 years vs. <10 years) and C) night shift intensity (≥3 consecutive night shifts/week vs. <3 night shifts/week). The figure plots p-values versus the effect size (adjusted mean difference in methylation β-values). Dotted line represents p = 0.05. Loci with FDR ≤0.20 are highlighted.
Current shift workers had 1.7% higher mean methylation levels at cg14718583, compared to day workers (95% CI: 0.87, 2.52; q-value: 0.18). Cg14718583 is located in the 5ʹUTR of the CSNK1E gene (Table 2). Individuals that worked night shifts for ≥10 years had 1.6% lower mean methylation levels at cg20667664 (95% CI: −2.33, −0.87; q-value: 0.08) compared to those that worked night shifts <10 years. Cg20667664 is located in the gene body of NR1D1 (Table 3). Those who worked ≥3 consecutive night shifts per week had 2.2% higher mean methylation levels at cg21078679 (95%: 1.15, 3.27; q-value: 0.15) compared to those who worked <3 consecutive night shifts per week. Cg21078679 is located in the gene body of ARNTL.
Analyses that did not include adjustment for white blood cell composition and analyses that additionally adjusted for smoking and alcohol intake did not produce materially different results (results not shown).
Discussion
In this exploratory study, three differentially methylated loci in core circadian genes were identified among women working night shifts. We found evidence of CSNK1E hypermethylation in current shift workers, hypomethylation of NR1D1 in workers with ≥10 years of night shift work duration, and hypermethylation of ARNTL in those who worked ≥3 consecutive night shifts per week. Our exploratory analysis suggests that different parameters of night shift work are associated with differential methylation in core circadian genes, including CSNK1E, NR1D1 and ARNTL, and highlights the need for further evaluation of these genes (and others) in future studies.
CSNK1E is a member of the casein kinase I protein family, and is known to encode the kinase CK1ε, which phosphorylates a number of genes including the PERIOD genes PER1 and PER2 [28]. Phosphorylation of PER2 is one of the key circadian pacemakers of the mammalian circadian clock, and is known to be involved in controlling the timing and structure of sleep patterns [29,30]. It is unclear how hypermethylation at the 5ʹUTR region is related to gene expression, as it is likely to be gene dependent [31]. Both CSNK1E and PER2 have been implicated in cancer, including breast cancer. Studies have found that CSNK1E is overexpressed in cancer tissues, while PER2 is under-expressed, which supports the notion that CSNK1E promotes cancer development by downregulation of the PERIOD genes [10,32–35].
We found evidence of hypomethylation in the body of NR1D1 (also known as REV-ERBα) in workers with longer duration of night shift work. NR1D1 is involved in circadian feedback loops as a transcriptional repressor and is thought to play an essential role in circadian clock regulation [36]. Once activated, NR1D1 controls rhythmic oscillations of ARNTL by suppressing its transcription [36]. Although the role of DNA methylation within the gene body is poorly understood, there is evidence that hypomethylation in the gene body affects gene splicing, transcription factors, and chromatin formation [31,37]. It has also been observed that many gene bodies become hypomethylated in cancer [38]. The role of NR1D1 in cancer is less clear, but has been implicated in the development and progression of gastric cancer [39], and activation of NR1D1 suppress proliferation of breast cancer cells [40]. NR1D1 is more commonly implicated in glucose regulation, lipid metabolism, and regulation of inflammatory functions [36].
We found evidence of hypermethylation in the body of ARNTL (also known as BMAL1) among night shift workers working three or more consecutive night shifts a week. ARNTL is a known transcriptional activator which forms a core component of the circadian clock [41]. ARNTL forms a heterodimer with the CLOCK gene that initiates transcription of the PERIOD genes, CRY genes, and NR1D142. ARNTL is also shown to have cancer promoting effects in breast cancer, although the specific molecular mechanism is not well understood [42]. Overexpression of ARNTL is shown to promote the invasion and metastasis of breast cancer cells, and hypomethylation at the promoter region has been observed in breast cancer tissue [10,42,43]. Conversely, hypermethylation at the promoter region (associated with gene silencing) is linked with the development of neoplasia, such as lymphocytic leukaemia, and ovarian cancer [44,45].
Studies that have examined the relationship between night shift work and methylation of circadian genes have produced mixed results with regards to specific circadian genes identified and direction of effects. Similar to our study, Bhatti et al. (2015) found differential methylation in the 5'UTR gene of CSNK1E gene [15]. However, contrary to our findings, they found hypomethylation in the CSNK1E gene, hypomethylation in ARNTL gene near the transcription start site, and found no differences in NR1D115. Samulin Erdem et al. (2017) examined promoter methylation in female nurses with breast cancer (n = 278 cases) or without breast cancer (n = 280 controls) [12]. Similar to our study, among cases they found hypermethylation in ARNTL in shift workers with 3 or more consecutive night shifts and <5 years of duration [12].Reszka et al. (2018) also examined promoter CpG methylation among female workers in healthcare (n = 347 rotating shift workers, n = 363 day workers) and observed that women with a longer lifetime duration of shift work (>10 years) had lower methylation in ARNTL, compared to those with 10 or less years of shift work duration [14]. Other studies have found that night shift work is associated with differential in methylation in other core circadian genes, including CLOCK, PER1, PER2, CRY1, and CRY2− [14,16].
Differences across studies may be attributable to multiple factors, including lack of adjustment for white blood cell composition, differences in confounder adjustment, and timing of blood collection. In addition, the majority of studies have compared associations among long-term night shift workers and day workers. It remains unclear how various parameters of night shift work such as cumulative night shift work (combining intensity and duration of shift work exposure) may influence circadian gene methylation. Epidemiologic studies examining night shift work and breast cancer risk suggest that female workers with a long duration of shift work (over 20 years) and a higher intensity of night shifts have the highest risk of breast cancer [4,5]. In our sample, 9 of 11 current night shift workers who reported working ≥3 night shifts a week also reported ≥10 years of night shift work duration. However, night shift workers in our sample were typically scheduled to work a rotating schedule that included two night shifts per week, meaning Individuals who opted-in to working ≥3 consecutive nights could be systematically different than those who do not. Well-powered studies with detailed assessment of night shift work and longitudinal follow-up are therefore needed to fully assess the relationship between night shift work and circadian gene methylation.
The evaluation of methylation for 22 circadian genes using microarrays is a major strength of this study. We were able to assess methylation at 1150 CpG sites within 22 circadian genes at single-nucleotide resolution. This includes extensive coverage of CpG Islands, genes, and enhancers. The use of several night shift work parameters is also a strength allowing for an assessment of how different aspects of night shift work, including years of night shift work and night shift intensity, may relate to methylation in circadian genes [1]. In general, self-reported exposure to night shift work is shown to be highly valid, limiting the potential for exposure misclassification and bias [46]. Furthermore, we carefully selected confounders using a causal model that was constructed based on hypothesized and tested relationships in the literature.
Limitations include a modest sample size, however, this is an exploratory analysis designed to explore the relationship between shift work and DNA methylation and inform future larger-scale studies. DNA methylation was only measured at one point in time, which may not reflect long-term impacts [47]. Considering DNA methylation is known to change over a person’s life course and is susceptible to environmental influence, future studies should examine the relationship longitudinally at multiple time points [48]. Although differential gene expression in the identified genes has been linked to cancer, it remains unclear if the magnitude of methylation changes observed at each loci would impact gene expression and/or have downstream carcinogenic effects. Due to the cross-sectional design, selection bias (i.e., the healthy shift worker effect) is also possible [49]. Older workers who have remained in night shift work for many years may represent a cohort of healthier individuals that are tolerant to shift work, and workers more susceptible to circadian misalignment (and potentially more susceptible to disease risk) may select out of working night shifts [49]. If present, this may have attenuated our effect estimates. This also means we cannot rule out that our observed associations could be explained by reverse causation; it is possible that methylation patterns influencing adaptability to shift work (e.g., chronotype) may have affected a participant’s willingness to work night shifts long-term.
In conclusion, this exploratory study suggests that various night shift work parameters are associated with differential methylation in core circadian genes, including CSNK1E, ARNTL and NR1D1. In order to better understand how night shift work may impact circadian gene methylation, large, well-powered studies with detailed assessments of night shift work and long-term follow-up are needed.
Supplementary Material
Acknowledgments
We thank study participants, staff, and our funding sources (Cancer Research Society, Garfield-Kelly from Queen’s University). Epigenetics data analyses were performed on resources and with support provided by the Centre for Advanced Computing (CAC) at Queen’s University in Kingston, Ontario. The CAC is funded by: the Canada Foundation for Innovation, the Government of Ontario, and Queen’s University. This study was completed in partial fulfillment of degree requirements for Jennifer A. Ritonja’s PhD at Queen’s University, Kingston, ON, Canada. JA Ritonja would like to thank her personal funding sources: S. Leonard Syme Training Fellowship in Work and Health (Institute for Work & Health), Ontario Graduate Scholarship, Queen Elizabeth II Graduate Scholarship in Science and Technology.
Funding Statement
This study was funded by the Cancer Research Society (Montreal), under Cancer Research Society/Environment Cancer Fund – Read for the Cure Operating Grant (number 23218), and the Queen’s University, under the Queen’s University Faculty of Health Sciences Garfield Kelly Cardiovascular Research & Development Fund (number 6026402);Faculty of Health Sciences, Queen’s University [Garfield Kelly Cardiovascular Research & Development Fund (6026402)].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are available from the corresponding author, JA Ritonja, upon reasonable request.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Stevens RG, Hansen J, Costa G, et al. Considerations of circadian impact for defining ‘shift work’ in cancer studies: IARC working group report. Occup Environ Med. 2011;68(2):154–162. [DOI] [PubMed] [Google Scholar]
- [2].Rydz E, Hall AL, Peters CE.. Prevalence and recent trends in exposure to night shiftwork in Canada. Ann Work Expo Health. 2020;64(3):270–281. [DOI] [PubMed] [Google Scholar]
- [3].IARC . Night shift work. IARC Monogr Eval Carcinog Risks Hum. 2020;124:1–371. [Google Scholar]
- [4].Cordina-Duverger E, Menegaux F, Popa A, et al. Night shift work and breast cancer: a pooled analysis of population-based case–control studies with complete work history. Eur J Epidemiol. 2018;33(4):369–379. [DOI] [PubMed] [Google Scholar]
- [5].Wegrzyn LR, Tamimi RM, Rosner BA, et al. Rotating night-shift work and the risk of breast cancer in the nurses’ health studies. Am J Epidemiol. 2017;186(5):532–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Fritschi L, Glass DC, Heyworth JS, et al. Hypotheses for mechanisms linking shiftwork and cancer. Med Hypotheses. 2011;77(3):430–436. [DOI] [PubMed] [Google Scholar]
- [7].Haus EL, Smolensky MH. Shift work and cancer risk: potential mechanistic roles of circadian disruption, light at night, and sleep deprivation. Sleep Med Rev. 2013;17(4):273–284. [DOI] [PubMed] [Google Scholar]
- [8].Hoffman AE, Yi CH, Zheng T, et al. CLOCK in breast tumorigenesis: genetic, epigenetic, and transcriptional profiling analyses. Cancer Res. 2010;70(4):1459–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hoffman AE, Zheng T, Ba Y, et al. The circadian gene NPAS2, a putative tumor suppressor, is involved in DNA damage response. Mol Cancer Res. 2008;6(9):1461–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lesicka M, Jabłońska E, Wieczorek E, et al. A different methylation profile of circadian genes promoter in breast cancer patients according to clinicopathological features. Chronobiol Int. 2019;36(8):1103–1114. [DOI] [PubMed] [Google Scholar]
- [11].Kuo SJ, Chen ST, Yeh KT, et al. Disturbance of circadian gene expression in breast cancer. Virchows Arch. 2009;454(4):467–474. [DOI] [PubMed] [Google Scholar]
- [12].Samulin Erdem J, Skare Ø, Petersen-Øverleir M, et al. Mechanisms of breast cancer in shift workers: DNA methylation in five core circadian genes in nurses working night shifts. J Cancer. 2017;8(15):2876–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhu Y, Stevens RG, Hoffman AE, et al. Epigenetic impact of long-term shiftwork: pilot evidence from circadian genes and whole-genome methylation analysis. Chronobiol Int. 2011;28(10):852–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Reszka E, Wieczorek E, Przybek M, et al. Circadian gene methylation in rotating-shift nurses: a cross-sectional study. Chronobiol Int. 2018;35(1):111–121. [DOI] [PubMed] [Google Scholar]
- [15].Bhatti P, Zhang Y, Song X, et al. Nightshift work and genome-wide DNA methylation. Chronobiol Int. 2015;32(1):103–112. [DOI] [PubMed] [Google Scholar]
- [16].Adams CD, Jordahl KM, Copeland W, et al. Nightshift work, chronotype, and genome-wide DNA methylation in blood. Epigenetics. 2017;12(10):833–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ritonja J, Tranmer J, Aronson KJ. The relationship between night work, chronotype, and cardiometabolic risk factors in female hospital employees. Chronobiol Int. 2019;36(5):616–628. [DOI] [PubMed] [Google Scholar]
- [18].Maksimovic J, Phipson B, Oshlack A. A cross-package Bioconductor workflow for analysing methylation array data. F1000Res. 2016;5:1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Pidsley R, Zotenko E, Peters TJ, et al. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016;17(1):208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fujita PA, Rhead B, Zweig AS, et al. The UCSC genome browser database: update 2011. Nucleic Acids Res. 2010;39(Database):D876–D882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2017;18(3):164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Salas LA, Koestler DC, Butler RA, et al. An optimized library for reference-based deconvolution of whole-blood biospecimens assayed using the Illumina human methylationepic BEADARRAY. Genome Biol. 2018;19(1):64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- [24].McAlpine CS, Swirski FK. Circadian influence on metabolism and inflammation in atherosclerosis. Circ Res. 2016;119(1):131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Houseman EA, Kim S, Kelsey KT, et al. DNA methylation in whole blood: uses and challenges. Curr Environ Health Rep. 2015;2(2):145–154. [DOI] [PubMed] [Google Scholar]
- [26].Rosen AD, Robertson KD, Hlady RA, et al. DNA methylation age is accelerated in alcohol dependence. Transl Psychiatry. 2018;8(1):182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gao X, Zhang Y, Breitling LP, et al. Relationship of tobacco smoking and smoking-related DNA methylation with epigenetic age acceleration. Oncotarget. 2016;7(30):46878–46889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Narasimamurthy R, Hunt SR, Lu Y, et al. CK1δ/ε protein kinase primes the PER2 circadian phosphoswitch. Proc Natl Acad Sci USA. 2018;115(23):5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhou L, Bryant CD, Loudon A, et al. The circadian clock gene Csnk1e regulates rapid eye movement sleep amount, and nonrapid eye movement sleep architecture in mice. Sleep. 2014;37(4):785–793C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Möller-Levet CS, Archer SN, Bucca G, et al. Effects of insufficient sleep on circadian rhythmicity and expression amplitude of the human blood transcriptome. Proc Natl Acad Sci USA. 2013;110(12):E1132–E1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Martino D, Saffery R. Characteristics of DNA methylation and gene expression in regulatory features on the Infinium 450k Beadchip. bioRxiv. 2015;032862. [Google Scholar]
- [32].Yang WS, Stockwell BR. Inhibition of casein kinase 1-epsilon induces cancer-cell-selective, PERIOD2-dependent growth arrest. Genome Biol. 2008;9(6):R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chen S-T, Choo K-B, Hou M-F, et al. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis. 2005;26(7):1241–1246. [DOI] [PubMed] [Google Scholar]
- [34].Huisman SA, Ahmadi AR, Ijzermans JNM, et al. Disruption of clock gene expression in human colorectal liver metastases. Tumour Biol. 2016;37(10):13973–13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Winter SL, Bosnoyan-Collins L, Pinnaduwage D, et al. Expression of the circadian clock genes Per1 and Per2 in sporadic and familial breast tumors. Neoplasia. 2007;9(10):797–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ikeda R, Tsuchiya Y, Koike N, et al. REV-ERBα and REV-ERBβ function as key factors regulating mammalian circadian output. Sci Rep. 2019;9(1):10171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mendizabal I, Zeng J, Keller TE, et al. Body-hypomethylated human genes harbor extensive intragenic transcriptional activity and are prone to cancer-associated dysregulation. Nucleic Acids Res. 2017;45:4390–4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1(2):239–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang X, Wang N, Wei X, et al. REV-ERBα reduction is associated with clinicopathological features and prognosis in human gastric cancer. Oncol Lett. 2018;16(2):1499–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang Y, Kojetin D, Burris TP. Anti-proliferative actions of a synthetic REV-ERBα/β agonist in breast cancer cells. Biochem Pharmacol. 2015;96(4):315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hernández-Rosas F, López-Rosas CA, Saavedra-Vélez MV. Disruption of the molecular circadian clock and cancer: an epigenetic link. Biochem Genet. 2020;58(1):189–209. [DOI] [PubMed] [Google Scholar]
- [42].Korkmaz T, Aygenli F, Emisoglu H, et al. Opposite carcinogenic effects of circadian clock gene BMAL1. Sci Rep. 2018;8(1):16023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wang J, Li S, Li X, et al. Circadian protein BMAL1 promotes breast cancer cell invasion and metastasis by up-regulating matrix metalloproteinase9 expression. Cancer Cell Int. 2019;19(1):182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Yeh CM, Shay J, Zeng TC, et al. Epigenetic silencing of ARNTL, a circadian gene and potential tumor suppressor in ovarian cancer. Int J Oncol. 2014;45(5):2101–2107. [DOI] [PubMed] [Google Scholar]
- [45].Taniguchi H, Fernández AF, Setién F, et al. Epigenetic inactivation of the circadian clock gene BMAL1 in hematologic malignancies. Cancer Res. 2009;69(21):8447–8454. [DOI] [PubMed] [Google Scholar]
- [46].Härmä M, Koskinen A, Ropponen A, et al. Validity of self-reported exposure to shift work. Occup Environ Med. 2017;74(3):228–230. [DOI] [PubMed] [Google Scholar]
- [47].Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13(1):86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Field AE, Robertson NA, Wang T, et al. DNA methylation clocks in aging: categories, causes, and consequences. Mol Cell. 2018;71(6):882–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Knutsson A, Akerstedt T. The healthy-worker effect: self-selection among Swedish shift workers. Work Stress. 1992;6(2):163–167. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, JA Ritonja, upon reasonable request.


