ABSTRACT
The etiopathogenesis of multiple sclerosis (MS) is strongly affected by environmental factors such as diet and the gut microbiota. An isoflavone-rich (ISO) diet was previously shown to reduce the severity of MS in the animal model experimental autoimmune encephalomyelitis (EAE). Translation of this concept to clinical trial where dietary isoflavones may be recommended for MS patients will require preliminary evidence that providing the isoflavone-rich diet to people with MS (PwMS) who lack phytoestrogen-metabolizing bacteria has beneficial effects. We have previously shown that the gut microbiota of PwMS resembles the gut microbiota of mice raised under a phytoestrogen-free (phyto-free) diet in that it lacks phytoestrogen-metabolizing bacteria. To investigate the effects of phytoestrogens on the microbiota inflammatory response and EAE disease severity we switched the diet of mice raised under a phyto-free (PF) diet to an isoflavone-rich diet. Microbiota analysis showed that the change in diet from one that is ISO to one that is PF reduces beneficial bacteria such as Bifidobacterium species. In addition we observed functional differences in lipopolysaccharide (LPS) biosynthesis pathways. Moreover LPS extracted from feces of mice fed an ISO diet induced increased production of anti-inflammatory cytokines from bone marrow-derived macrophages relative to fecal-LPS isolated from mice fed a PF diet. Eventually mice whose diet was switched from a PF diet to an ISO diet trended toward reduced EAE severity and mortality. Overall we show that an isoflavone-rich diet specifically modulates LPS biosynthesis of the gut microbiota imparts an anti-inflammatory response and decreases disease severity.
KEYWORDS: Gut microbiota, isoflavones, lipopolysaccharides, diet, EAE
Introduction
Multiple sclerosis (MS) is an autoimmune neurodegenerative disease of the central nervous system affecting around 2.8 million people worldwide which is 30% higher compared to its prevalence in 2013.1 The etiopathogenesis of MS is complex and multifactorial involving both genetic and environmental factors.2–4 The interplay between several factors leads to chronic inflammation of the CNS which induces degradation of the myelin sheath a hallmark of MS. Genetic factors make up only ~30% of MS risk as shown by multiple previous studies5–7 which provides strong support that environmental factors play a crucial role in the etiopathogenesis of this disease. While there are numerous contributing environmental factors we and others have previously demonstrated that the gut microbiota is an important factor associated with MS.8–10 One of the key modulators of the gut microbiota is diet. Dietary habits are reported to shape the composition of the gut microbiota alter intestinal inflammation and effect the immune status of the host.11,12 Thus a shift in diet can modify the composition of gut bacteria and the metabolites produced by the gut microbiota which in turn can influence gut barrier integrity immune cell function host-microbe interactions and skew the gut environment toward a proinflammatory or anti-inflammatory response.13,14
Isoflavones represent a major class of phyto-estrogenic compounds that are found in legume-based diets and possess numerous anti-oxidative anti-bacterial anti-carcinogenic anti-inflammatory and cardioprotective effects.15–17 Some gut microbes such as Adlercreutzia equolifaciens,18 Eggerthella species19,20 Slackia isoflavoniconvertens,21 and Lactococcus garvieae22 specialize in metabolizing these phyto-estrogenic compounds. An isoflavone-rich (ISO) diet consists of the key compounds daidzein and genistein which are metabolized by the gut microbiota to produce biologically active S-equol to exert numerous health benefits.23 Using experimental autoimmune encephalomyelitis (EAE) a mouse model of MS we have previously shown that the gut microbiota is required for an ISO diet to mediate disease suppression.24 The same study demonstrated that isoflavone-metabolizing bacteria specifically Adlercreutzia equolifaciens and Parabacteroides distasonis and/or their metabolite S-equol are sufficient to ameliorate EAE. Moreover people with MS (PwMS) have a reduced abundance of isoflavone-metabolizing bacteria such as A. equolifaciens and P. distasonis in their fecal samples compared to healthy controls.8 These studies imply that the composition of the gut microbiota in mice kept on a phytoestrogen-free (PF) diet is more similar to the composition of the gut microbiota in PwMS specifically regarding bacteria that are capable of metabolizing isoflavones. As isoflavone metabolism is identified as one of the key microbial characteristics in a healthy microbiota it is important to understand whether changes to an ISO diet can modify the existing microbiota in PwMS to impart health benefits. To test the potential of an ISO diet to be translated into use in the clinic we modeled the microbiota of PwMS in mice. We reared mice that lacked phytoestrogen-metabolizing bacteria on a PF diet before switching them to an ISO diet and evaluated their ability to suppress inflammation and disease.
In the current study we observed that changing from a PF to an ISO diet modulated the taxonomic composition and function of the gut microbiota. Specifically we observed that mice that were switched to an ISO diet showed overall improved gut health in the context of microbiome composition and stability compared to mice whose diet was changed to one that was PF. Functional investigation revealed differences in lipopolysaccharide (LPS) biosynthesis after the change in diet. Also LPS extracted from feces of mice kept on ISO diet enhanced anti-inflammatory cytokine production from bone marrow-derived macrophages. Finally we observed that mice whose diet was switched from one that was PF to an ISO diet showed reduced disease severity and mortality even though statistically significant levels were not reached. Collectively our data show an alternative pathway (i.e. alteration of LPS biosynthesis) through which an ISO diet can induce an anti-inflammatory immune response and modulate CNS autoimmunity.
Results
An ISO diet enhances microbial diversity
To test if a change in diet can alter microbial evenness and diversity we analyzed the alpha diversity of the microbiome at baseline (D0) and on days 2 3 7 14 21 and 28 (D28) after the diet was switched (Figure 1a). On D0 fecal microbial communities of mice on a PF diet had a significantly higher Shannon diversity compared to mice on an ISO diet (p=.03; Figure 1b). The PF diet also supported higher phylogenetically diverse microbes but was statistically non-significant compared to the ISO diet (p=.12; Figure 1c). However Shannon diversity of mice kept on the ISO diet was significantly reduced over time after the diet was switched to one that was PF indicating that changing the diet from ISO to PF reduced the overall richness and evenness of microbial communities (Figure 1d). Such a significant change in the microbial community was not detected when the diet was changed from PF to ISO over time (p=.48) suggesting that the ISO diet supports the growth of the diverse microbial species present in the PF diet at D0 and over the course of 28days. Further there was a significant rise in phylogenetic diversity when the diet was switched from PF to ISO (p=.04) implying that the change in diet to one that was ISO incorporated phylogenetically diverse microbes in the community over time which was not evident when the diet was switched from an ISO one to a PF one (p=.74; Figure 1e). Furthermore the fecal microbial communities of the mice kept on ISO and PF diets were significantly different from one another at baseline (Figure 1f) and after D28 following a switch in diet (Figure 1g). Twenty-eight days after diets were switched (ISO to PF or PF to ISO) the microbial community was significantly altered relative to D0 (Figure 1(h,i)). There were no differences in the homogeneity of variances when the diet was switched from PF to ISO (p=.71) or ISO to PF (p=.68) indicating that samples had higher microbial evenness within groups. The diet-driven changes in the microbial community were evident for both alpha and beta diversity but the microbial communities on D28 did not radically change to be similar to the microbial communities at baseline for either the ISO or PF diet (Figure 1j) indicating that dietary change can only prime the microbiota originally present. Thus these data indicate that a change to an ISO diet enhances microbial diversity and evenness in the gut a sign of overall improved gut health.
Figure 1.
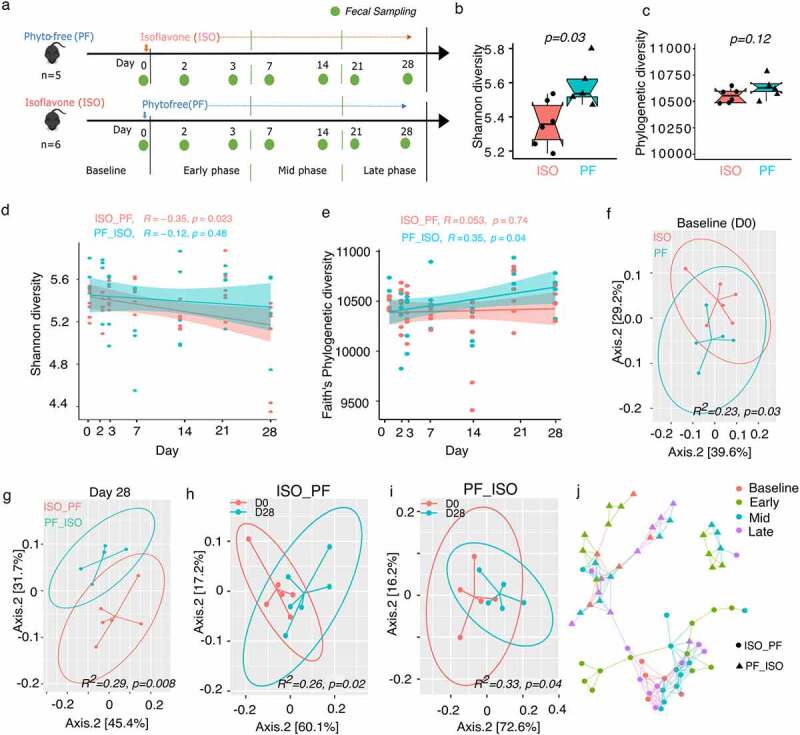
Diet change alters community composition and richness. a) Experimental schematic outline for diet change and fecal sample collection. Alpha diversity measures b) Shannon diversity c) Faith’s Phylogenetic diversity pre-diet change on day 0 (D0) from isoflavone-rich (ISO) to phyto-free (PF) or vice versa. d) Shannon diversity and e) Faith’s phylogenetic diversity changes after the diet was switched from PF to ISO or vice versa. Beta diversity at f) baseline (D0) and g) day 28 after diet change measured using weighted unifrac distance metrics. Beta diversity between D0 and day 28 (D28) for diet change from h) ISO to PF and i) PF to ISO. adonis2 test was performed to for statistical differentiation between the groups for beta diversity analysis. j) A network created using Jaccard dissimilarity matrix between the samples over time. The colors represent the different phases of microbial community change over time and the shape represents the change in diet.
Change to a PF diet reduces beneficial gut bacteria
As diet alone can shape the microbiota we investigated if the microbiome of mice originally kept on a PF diet and later switched to ISO diet can be enriched for isoflavone-metabolizing genes and the related bacteria. Isoflavone metabolism by the gut microbiota is known to produce the metabolite S-equol that exerts numerous health benefits.25 Thus we determined levels of S-equol in urine as well as the presence of isoflavone-metabolizing genes in the assembled metagenomes at baseline through the end of the experiment (Figure S1 Table S1). Phytoestrogen-metabolizing genes were completely absent in mice kept on a PF diet at D0 whereas all mice kept on ISO diet had at least one gene homolog linked to phytoestrogen metabolism except for one mouse. This finding resonated with S-equol levels where we observed no measurable quantity of S-equol in the urine of mice kept on a PF diet and significantly higher levels of S-equol in the urine of mice kept on an ISO diet before the diet switch (Figure S1). This demonstrates the presence of phytoestrogen-metabolizing bacteria in the metagenomes of mice kept on an ISO diet at baseline. However after the change from a PF to ISO diet the microbial community evolved to harbor phytoestrogen-metabolizing genes after 28days in two out of five mice and the S-equol levels were restored to similar levels as those in mice kept on an ISO diet before the diet change (Figure S1). On other hand phytoestrogen-metabolizing genes as well as levels of S-equol were absent after the diet was switched from ISO to PF indicating that the capability of phytoestrogen metabolism and the associated function is lost after the diet change (Table S1).
In addition to phytoestrogen-metabolizing genes we investigated the shift in the composition of the gut microbiota in mice at baseline and in mice on D28 after the diet switch. Taxonomically Bacteroidetes and Firmicutes were the two major phyla in fecal microbial communities in mice kept on an ISO or PF diet at D0 (Figure S2a). There was no significant difference between the Bacteroidetes-to-Firmicutes ratio (B:F ratio) for mice at D0 (p=.79). These two phyla also dominated the gut in both groups on D28 after the diet change. However change to either diet brought a reduction in the Bacteroidetes population (Figure S2a). This change was evident with a significant decrease in the B:F ratio over time (Figure S2b). The reduction in the B:F ratio was more pronounced when the diet was switched from PF to ISO. At the species level Faecalibacterium rodentium Phocaeicola vulgatus and Erysipelotrichaceae bacterium NYU-BL-E8 were dominant in mice subjected to both PF and ISO dietary conditions at D0. The top dominant species with > 5% abundance in the metagenomes on days 2 3 7 14 21 and 28 are shown in Figure S2c. Upon differential analysis at D0 bacteria enriched in the PF diet belonged exclusively to either the Bacteroidetes or Proteobacteria phylum (Figure 2a). However at least one member of bacteria belonging to Firmicutes Actinobacteria Bacteoridetes and Verrucomicrobia were found in mice fed an ISO diet (Figure 2a). At the species level Odoribacter splanchnicus Bacteroides sps Rhodospirillum sps and Rikenella microfusus were differentially abundant in mice fed a PF diet whereas Akkermansia muciniphila Bifidobacterium adolescentis Bacteorides thetaiotaomicron Candidatus Homeotherum arabinoxylanisolvens Kineothrix alysoides Firmicutes bacterium CAG:102 and B. longum were differentially abundant in mice fed an ISO diet at D0 (Figure 2a). On D28 after the diet was switched the majority of differentially expressed bacteria remained unaffected (Figure 2b). However the significant differences in the abundance of B. adolescentis B. thetaiotaomicron K. alysoides Firmicutes bacterium CAG:102 and B. longum were lost after the diet was changed from ISO to PF. Also Marinifilum fragile Desulfomicrobium escambiense Prevotella sp. KH2C16 Rikenellaceae bacterium DTU002 Bacteroidales bacterium Bact_10 Prevotella sp. CAG:755 Bacteroidetes bacterium GWD2_33_33 Desulfurispirillum indicum Prevotella sp. CAG:617 and Reichenbachiella agariperforans were lost 28days after the diet was changed to an ISO one. Instead B. pseudocatenulatum B. stellenboschense Parabacteroides gordonii Roseburia sp. CAG:309 Desulfovibrio cuneatus and D. gigas were differentially abundant on D28 after the diet was changed to ISO from PF (Figure 2b). Among these differentially abundant taxa eight species were significantly altered over time with diet change (Figure 3). Specifically B. adolescentis B. longum and K. alysoides were significantly reduced with a change in diet from ISO to PF whereas B. stellenboschense was significantly increased (Figure 3(a–d)). The change in diet from PF to ISO did not alter the abundance of these bacteria over time except for a significant increase in B. stellenboschense. However the change from PF to ISO saw a significant reduction in the abundance of Prevotella sp. KH2C16 Prevotella sp. CAG:755 Prevotella sp. CAG:617 and Parabacteroides gordonii (Figure 3(e–h)). As Bifidobacteria are designated as useful “probiotic” species beneficial for the human gut26–28 the loss of these species after the diet change to PF is indicative of deterioration in the overall homeostasis of the gut microbiome.
Figure 2.
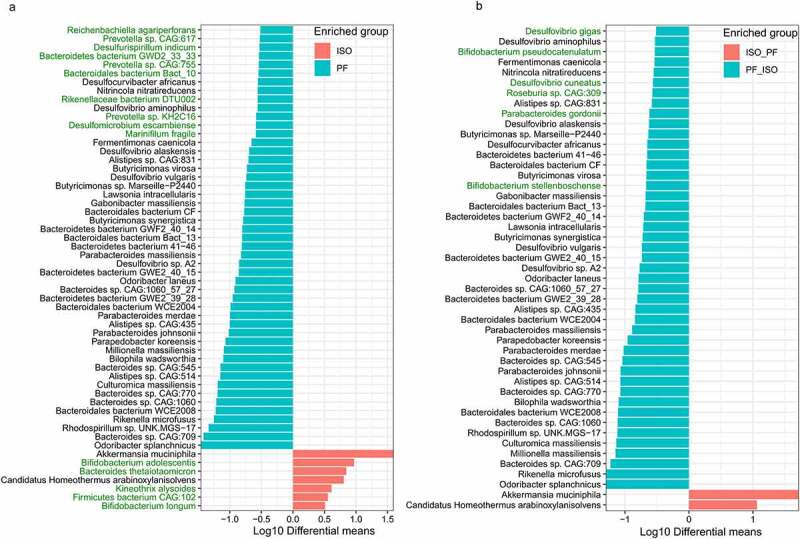
Differentially abundant microbial taxa after the diet switch from either phyto-free (PF) to isoflavones-rich (ISO) diet and vice-versa. Taxonomic differentiation of the fecal microbial taxa at a) baseline (D0) between the mice kept on ISO and PF diet. b) Day 28 (D28) after diet switch from ISO to PF (ISO_PF) or PF to ISO (PF_ISO). Differential analysis was performed using welch test from microbiomeMarker package in R with p-value_cutoff of 0.01 and p_adjust using “BH”.
Figure 3.
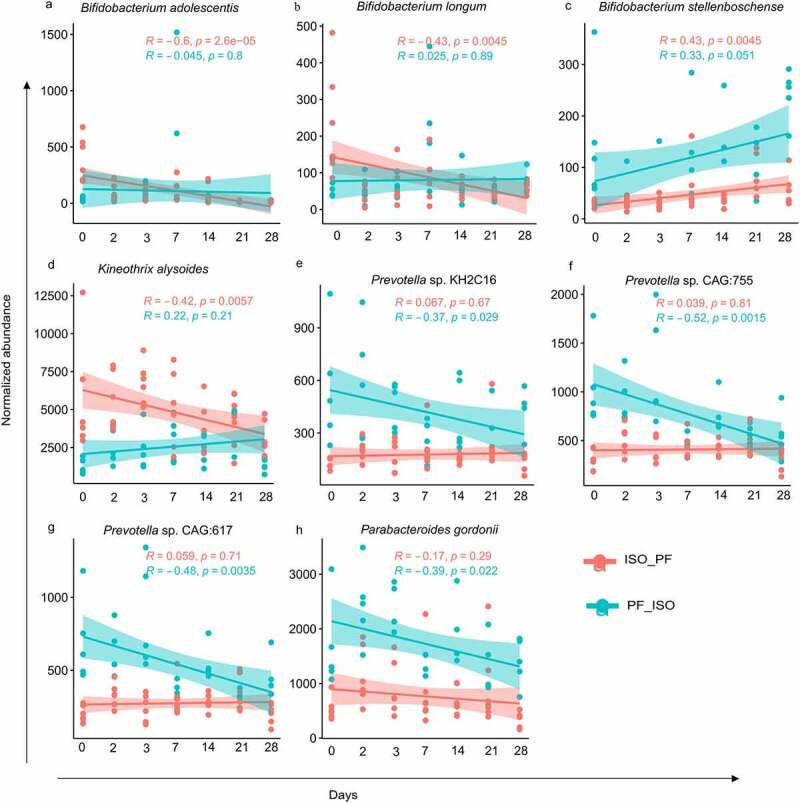
Scatterplots displaying correlation of normalized abundance of significantly altering taxa over time after the change from isoflavone-rich to phyto-free (ISO_PF) and phyto-free to isoflavone-rich (PF_ISO) diet. A positive or negative correlation is expressed by the values of correlation coefficient “R” calculated using the “Pearson” method with corresponding p values for both dietary changes.
Dietary change modifies LPS biosynthesis by the gut microbiota
We also investigated the presence of active enzymes in the metagenomes of the microbiota before diet change (D0) and on D28 after the diet change in both groups (Figure 4). The relative abundances of auxiliary activities (AAs p =.68) carbohydrate-binding modules (CBMs p =.17) carbohydrate esterases (CEs p =.33) glycoside hydrolases (GHs p =.16) and glycosyltransferases (GTs) (p=.096 Figure 4a) were similar at D0 between the mice fed an ISO or PF diet. However mice fed an ISO diet had significantly higher polysaccharide lyase (PL) activity along with a reduction in the activity of S-layer homology domains (SLHs; Figure 4b 4c). No significant changes in the relative abundances of AAs CBMs CEs and GHs were observed on D28 after diet change between the groups. Yet there was a significant reduction in GT activity when the diet was changed from ISO to PF (Figure 4d). Within GTs only GT2_glyco_tranf_2 activity was significantly higher on D28 in mice in which the diet changed from PF to ISO compared to mice in which the diet changed from ISO to PF (p =.00045). The GT2 family of enzymes are diverse and are involved in transferring the sugar from UDP-glucose UDP-N-acetyl- galactosamine GDP-mannose or CDP-abequose to a range of substrates including cellulose dolichol phosphate and teichoic acids (https://pfam.xfam.org/family/PF00535) which form the building blocks of LPSs implying that ISO dietary conditions may contribute to formation of structurally different LPS compared to a PF diet. Similarly the relative abundance of PL activity was significant on D28 when the diet was changed to PF from ISO but the change was also evident on D0 indicating that PLs were not affected overall by the change in diet (Figure 4e). The significantly high SLH activity at D0 in mice fed a PF diet was lost by D28 when the diet was changed to ISO (Figure 4f) suggesting that the ISO diet reduces SLH activity. SLHs are shown to mediate the binding of exocellular proteins to the cell surface in vivo and in vitro29–31 and thus can affect cellular signaling. Taken together differences in active carbohydrate enzymatic activity after dietary change suggest differences in LPS and exocellular protein composition and structure of the microbiota.
Figure 4.
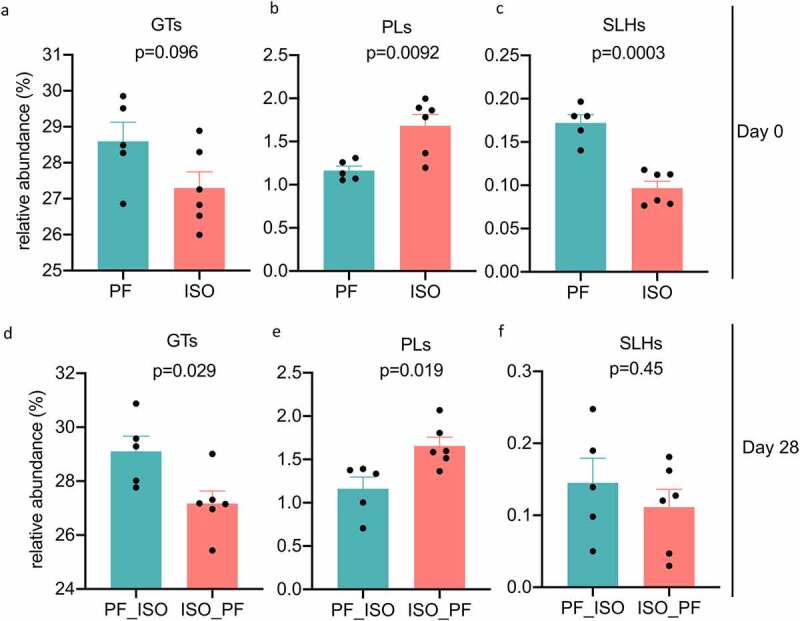
Relative abundances of significantly altered active carbohydrate enzyme families identified using dbCAN2 and CaZy database in the metagenomes of mice before diet switch at day 0 and after diet switch at day 28. Diet change from isoflavone-rich (ISO) to phyto-free (PF) diet is represented as ISO_PF and PF to ISO diet as PF_ISO. GTs represent glycosyltransferases PLs represent polysaccharide lyases and SLHs represent S-Layer homology domain active carbohydrate enzymatic families.
While we found no significant functional differences in the microbial communities after the diet switch from ISO to PF and PF to ISO when comparing D0 and D28 separately (Figure S3a S3b) there were significant differences between D0 and D28 over time for both diet changes (Figure S3c S3d). Functionally on D0 the ISO diet had significant enrichment of mixed acid fermentation and TCA cycle VI pathways suggesting a higher abundance of short-chain fatty acid pathways (Figure 5a). The folate transformation pathway was only significantly enriched in mice on a PF diet. However on D28 the diet change from PF to ISO had significant enrichment of chorismate biosynthesis pathways and L-lysine biosynthesis VI pathway (Figure 5b). The lysine biosynthesis pathway VI is crucial as it produces an important metabolite meso-diaminopimelate which is a constituent of the bacterial cell wall peptidoglycan. The chorismite biosynthesis pathways produce chorismite which is an important intermediate leading to the synthesis of essential metabolites such as L-phenylalanine L-tyrosine L-tryptophan vitamins E and K ubiquinone and certain siderophores. Especially these amino acids act as substrates for the production of secondary metabolites from alkaloids flavonoids lignin coumarin and other phenolic compounds implying that a diet switch to PF from ISO but not ISO from PF enhances secondary metabolite production from dietary flavonoids.
Figure 5.
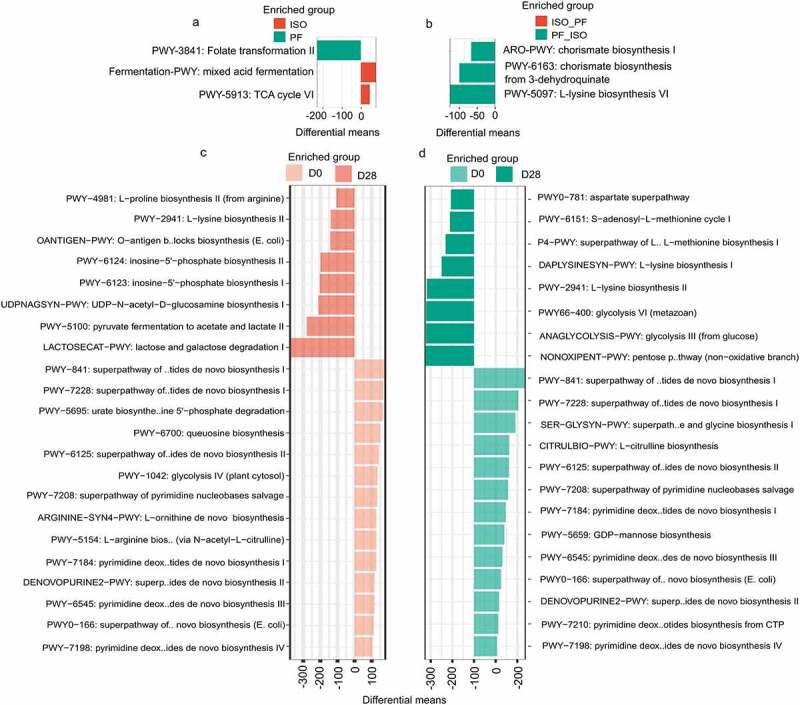
Functional alteration of the gut microbiota a) at baseline (D0) between mice kept on either isoflavone-rich (ISO) or phyto-free (PF) diet and b)at day 28 (D28) after diet switch from ISO to PF (ISO_PF) and PF to ISO (PF_ISO). The differentially abundant pathways were identified using the Welch test with pvalue_cutoff of 0.1 p_adjust=”BH” and nperm=1000 between the two groups. For the change in functional profile over time D0 and D28 were compared using the welch test with pvalue_cutoff of 0.2 p_adjust=”BH” and nperm=1000 between the groups of c) diet change from ISO to PF and d) diet change from PF to ISO. The functional profile was obtained using humann3 from the MetaCyc database.
On D28 the diet change from ISO to PF enriched the following biosynthesis pathways compared to D0: lactose and galactose degradation pyruvate fermentation inosine-5’ phosphate biosynthesis UDP-N-acetyl-D-glucosamine biosynthesis I and O-antigen building block biosynthesis (Figure 5c). UDP-N-acetyl-D-glucosamine is the precursor of cell wall peptidoglycan LPS and enterobacterial common antigen. Similarly the O-antigen specific chains are a part of LPSs that are known to evoke a specific immune response. Thus it appears that the diet switch from ISO to PF enhances LPS biosynthesis in the gut to induce an immune response. However enrichment of no such LPS biosynthesis pathway was observed when the diet was changed from ISO to PF on D28 when compared to D0. The differences in the carbohydrate enzymatic activity that we observed after dietary change could change the LPS and exocellular protein structure produced by the microbiota. Thus LPS synthesis may be altered because of dietary ISO or PF conditions after changes in the diet. In fact the differences in the gut microbiota have been previously described to affect immune signaling and immunogenicity in humans because of differences in LPS.32 Also we observed enrichment of methionine aspartate and lysine pathways along with glycolysis and non-oxidant pentose phosphate pathways on D28 compared to D0 when the diet was switched from PF to ISO (Figure 5d). This indicates that changing the diet to one that is PF alters the functions of the gut microbiota and may promote the synthesis of LPS-based antigenic molecules to evoke a different immune response.
An ISO diet promotes anti-inflammatory cytokines
From our functional analysis of the gut microbiota we found that LPS biosynthesis was altered and that O-antigen biosynthesis (E. coli) and UDP-N-acetyl-D-glucosamine biosynthesis pathways were increased after four weeks when the diet was switched from ISO to PF. First we quantified the fecal endotoxin levels in the fecal lysate obtained from mice from both groups. We observed that the fecal endotoxin levels were significantly higher in mice kept on a PF diet (Figure 6a). To test the differences on inflammation induced by LPS that is synthesized after the diets are switched we treated bone marrow-derived macrophages with LPS isolated from the fecal lysate of mice kept on either a PF or ISO diet for four weeks and quantified expression of the cytokines TNF-α IL-6 IL-1β CXCL1 IL-10 and IL-12/23 in the supernatant. The LPS derived from the feces of mice kept on the PF diet produced significantly higher levels of TNF-α IL-6 IL-12/23 and IL-1β compared to the LPS derived from feces of mice kept on the ISO diet (Figure 6(b–e)). There was no difference in the CXCL1 levels between mice fed a PF or ISO diet (Figure 6f). On the other hand IL-10 production was significantly higher in the mice kept on an ISO diet (Figure 6g). Higher levels of TNF-α IL-6 IL-1β and IL-12/23 and lower levels of IL-10 indicate that the PF diet may contribute to a pro-inflammatory effect in the gut. Additionally the ratio of IL-10 to IL-12/23 in BMDM induced with LPS isolated from the feces of mice kept on an ISO diet was significantly higher than those on a PF diet (Figure 6h) highlighting the potent anti-inflammatory effect of ISO diet-altered LPS. These results suggest there are pro-inflammatory consequences in the mouse gut for those fed a PF diet and anti-inflammatory outcomes in the gut for mice fed an ISO diet.
Figure 6.
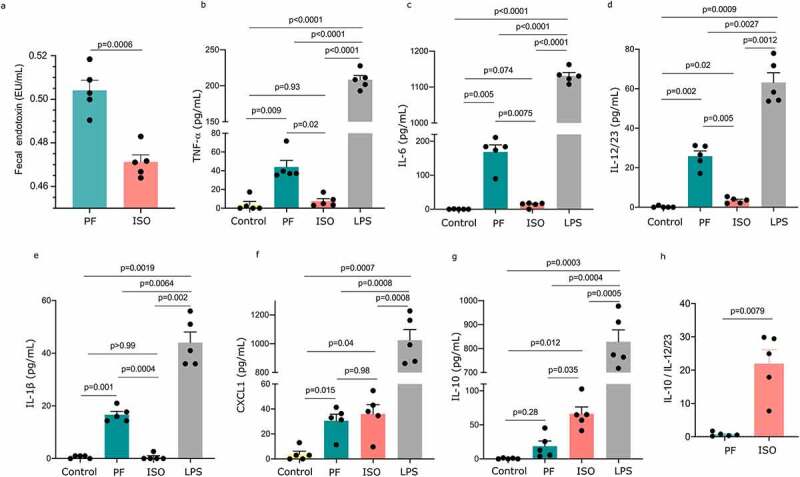
Anti-inflammatory effect of LPS isolated from feces of mice kept on isoflavone-rich (ISO) diet. LPS was extracted from the feces of mice kept on phyto-free (PF) or ISO diet for four weeks and was used to treat bone-marrow-derived macrophages. a) Fecal endotoxin levels were determined using LAL endotoxin assay. The concentration of b) TNF-α c) IL-6 d) IL-12/23 e) IL-1β f) CXCL1 and g) IL-10 was determined using ELISA. h) Ratio of IL-10 to IL-12/23. Mann-Whitney test was performed for (a) and (h) while Brown-Forsythe and Welch ANOVA tests followed by Dunnett T3 for multiple comparisons were performed for bcdef and g to determine the statistical significance.
Changing from a PF to ISO diet reduces EAE severity
We have previously shown that the ISO diet significantly reduces EAE severity.24 Here we observed that the ISO diet exerts an anti-inflammatory effect. Thus we investigated if the diet change from PF to ISO or ISO to PF can affect EAE outcomes. We induced EAE in mice four weeks after changing their diet (Figure 7a). After EAE induction mice were monitored for disease for 41days. We observed that the onset of disease for mice originally on a PF diet but later switched to an ISO diet showed earlier disease onset compared to mice that were originally on the ISO diet and later switched to the PF diet (Figure 7b). However mice switched to the ISO diet from the PF diet consistently trended toward lower average daily clinical scores over the course of 41days after immunization compared to mice in which the diet was switched to PF from the ISO diet. This indicates that switching a PF diet to an ISO one helps to reduce EAE severity (Figure 7c). Interestingly the probability of survival was lower (83.1%) in mice whose diet was switched to PF from ISO compared to mice whose diet was switched to ISO from PF indicating that a switch to an ISO diet confers protection from both EAE severity and fatality (Figure 7c). Thus our data suggest that an ISO diet suppresses EAE disease severity and overall disease mortality compared to mice switched to a PF diet.
Figure 7.
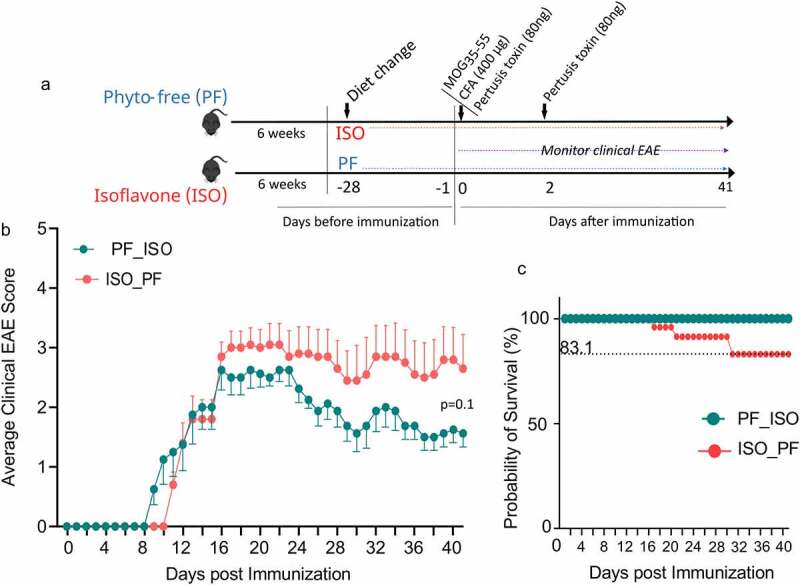
EAE severity in mice after diet switch from phyto-free (PF) to isoflavone-rich (ISO) and ISO to PF diet. Mice were kept for six weeks on PF/ISO diet until day −28 where diet was switched for four weeks. After the diet switch EAE was induced using MOG35-55.a) Experimental outline of dietary regime and EAE induction in mice and clinical monitoring of EAE scores. b) Average clinical EAE scores of mice in which diet was changed from PF to ISO (PF_ISO n =8) and ISO to PF (ISO_PF n =10) for 41days post EAE induction. c) Kaplan-Meier survival analysis of mice after EAE induction for 41days.
Discussion
As PwMS have gut dysbiosis in which beneficial bacteria are depleted restoring their microbiota to one with a healthy balance of microorganisms is as a potential treatment option that is currently being explored. Diet has been shown to have the strongest effect on the composition of gut bacteria33,34 and thus can provide an exciting option to correct gut dysbiosis in PwMS as well as other diseases. We have previously shown that PwMS lack bacteria with the ability to metabolize dietary phytoestrogen and most importantly that an ISO diet can suppress disease in an animal model of MS.24 The disease protection was dependent on the presence of isoflavone-metabolizing bacteria and their metabolite. Translating this concept to a treatment for MS will require clinical trials that assess the effect of providing an ISO diet to PwMS who lack phytoestrogen-metabolizing bacteria and most probably dietary phytoestrogen. Therefore the first step toward reaching this goal is to determine the extent to which the dietary regimen change to an ISO or PF diet affects the composition of the microbiota and its functions and the effect of this on modulating disease. Here we demonstrate that changing the diet from a PF one to an ISO one enhances evenness and phylogenetic diversity in the gut microbiota. Our data reveal a shift in microbial communities and functional capabilities after a short-term diet switch which affected LPS biosynthesis. Furthermore the impact of differences in LPS produced after the diet switch was evident by enhanced anti-inflammatory cytokine production when the diet was changed from PF to ISO compared to when the diet was changed from ISO to PF. Consequently the changes in the microbial structure and function ultimately affected the phenotypic outcome by reducing the production of pro-inflammatory cytokines and the severity of EAE in mice in which the diet was changed from a PF one to an ISO one. Our data showing the ability of an ISO diet to regulate pro-inflammatory responses and protect against CNS inflammatory disease by modulating LPS biosynthesis pathways points toward a previously unknown mechanism through which isoflavones can modulate host physiology. Taken together these results are proof of principle that switching from a PF diet to an ISO one can reduce pro-inflammatory responses and disease severity through the previously unknown effects of phytoestrogen metabolism on LPS biosynthesis.
Multiple studies have shown that several gut bacteria are differentially abundant in PwMS compared to healthy controls.8,10 The abundance of gut bacteria that can metabolize isoflavones such as Adlercruetzia equolifaciens and Parabacteroides distasonis are reduced in MS patients indicating that isoflavone metabolism may be compromised.8,35 Moreover we have previously reported that an ISO diet can ameliorate EAE in several mouse models.24 Similarly this current study shows that the change in diet from PF to ISO reduces pro-inflammatory responses EAE severity and mortality in mice compared to a change in diet from ISO to PF confirming that isoflavones are protective against EAE. Moreover changing the diet from a PF one to an ISO one altered the gut microbiota and enhanced phylogenetic richness without affecting microbial evenness. However a change in diet from one that is ISO to one that is PF significantly reduced the evenness of the microbiota and did not alter phylogenetic diversity. These findings validate our previous results that demonstrate the ISO diet enhances species richness.24 Indeed healthy individuals harbor diverse gut bacteria and an increase in microbial diversity has been linked to gut homeostasis and better overall health.36 Similarly the composition of the microbiome showed a longitudinal shift after the diet changed for both conditions over time. The change in microbiota was evident after switching the diet from an ISO one to a PF one as it resulted in the loss of isoflavone-metabolizing genes as early as day seven. These isoflavone-metabolizing genes convert isoflavones to S-equol and were absent at D0 in mice fed a PF diet but were later found in some mice after D28 following a change to an ISO diet. This finding suggests that microbial species harboring these genes are affected by changes in the diet and thus might influence potential S-equol production.
S-equol is produced as a result of the breakdown of dietary isoflavones such as daidzein and genistein by gut bacteria.37 S-equol production requires a complete set of enzymes daidzein reductase dihydrodaidzein reductase and tetrahydrodaidzein reductase to metabolize daidzein to S-equol.38 Genistein was found to be catalyzed at an even higher rate by daidzein reductase compared to daidzein.39 Our previous study showed that both isoflavone-metabolizing bacteria and their product is required for EAE suppression.24 Even though rodents can efficiently produce S-equol from phytoestrogen metabolism40,41 we did not find well-known bacterial “equol producers” such as Adlercreutzia equolifaciens,18 Eggerthella species19,20 Slackia isoflavoniconvertens21 and Lactococcus garvieae22 that were differentially enriched at baseline between the ISO and PF fed mice or at endpoint after the diet change. However we found significant loss of B. adolescentis and B. longum after the diet change to PF and gain of B. stellenboschense after the diet change to ISO. Bifidobacterium species were previously described to efficiently produce S-equol from isoflavones42 suggesting a loss of S-equol production after the diet change from ISO to PF. Importantly the loss of S-equol levels after a diet switch from ISO to PF validated that dietary change was critical for restoring levels of S-equol. Albeit we did not find the presence of homologs of isoflavone-metabolizing enzymes in reference genomes of B. adolescentis B. longum and B. stellenboschense. This indicates that the genomes of these bacteria need further investigation for the presence of non-canonical genes responsible for metabolizing isoflavone. Also on D28 after a diet switch from PF to ISO we did not observe restoration of the complete set of genes required for isoflavone degradation to S-equol as shown in Table S1. However S-equol levels were restored indicating that S-equol production is revived after changing to an ISO diet. Also the production rate of S-equol is very low in human intestines43 and only 30–50% of humans can convert isoflavones to S-equol.44,45 This implies that either there are undefined pathways for the production of S-equol or there are other possible methods by which dietary isoflavones are acted upon by gut microbes to benefit overall health.
Bifidobacteria are designated as useful “probiotic” species for the human gut and harbor β-glucosidase enzyme activity required for initial hydrolysis of isoflavone glucosides26–28 suggesting that the ISO diet likely selected a metabolic niche of β-glucosidase activity. In addition Bifidobacterium species are better known for their diverse carbohydrate metabolism SCFAs production interaction with the host secondary metabolite production and probiotic effect that influence inflammation and homeostasis.46 B. adolescentis and B. longum species can produce exopolysaccharides which increase the production of anti-inflammatory cytokines such as IL-10 in the gut47,48 suggesting that a change in diet might have altered polysaccharide assimilation by the gut microbiota. Indeed we observed a significant reduction in Bifidobacterium species along with changes in CHO-active enzymes: glycosyltransferase polysaccharide lyase and S-Layer-homology activity 28days after diet change from PF to ISO. Also a change in diet to one that was PF permitted specific enrichment of certain LPS biosynthesis pathways: UDP-N-acetyl-D-glucosamine biosynthesis I and O-antigen building blocks biosynthesis. Differences in the microbiota have been previously described to affect immune signaling due to their differences in LPS.32 These findings prompted us to investigate the properties of LPS present in the gut microbiota for both diets. Interestingly treatment of bone marrow-derived macrophages with LPS derived from mice fed with the ISO diet showed significantly higher anti-inflammatory IL-10 and lower proinflammatory TNFα IL-6 IL-1β and IL-12/23 cytokine production. In fact mice fed with phytoestrogens has been shown to increase the anti-inflammatory effect in the CNS24 and LPS-inducedmacrophages.49,50 Similarly when the diet was changed to PF from ISO we found a significant loss of K. alysoides a saccharolytic butyrate producer51 that may promote a proinflammatory environment in the gut. Butyrate has been shown to increase the effectiveness and production of circulating Treg cells in mice which is important for peripheral tolerance to prevent autoimmune diseases.52 On other hand Prevotella species were lost after the diet was switched to one that was ISO. The loss of Prevotella species could be due to alterations in the nutritional niche or nutritional competition. Prevotella species presence is highly debated in terms of the role they play in health and disease. The abundance of Prevotella species were found to decrease in MS patients8 and importantly P. histicola was shown to suppress EAE.53 However Prevotella species have been associated with several other inflammatory diseases and are proposed as a marker of that defines an inflammatory state of microbiome promoting inflammation rather than being the cause of inflammation.54,55 Further detailed studies are necessary to ascertain the role of Prevotella species in ISO dietary conditions. Taken together these findings suggest an alternative mechanism by which the ISO diet supports the enrichment of beneficial bacteria and alters LPS assimilation to modulate the severity of EAE in mice. Although switching the diet from one that is PF to one that is ISO resulted in lower average clinical scores of EAE it did not reach statistical significance. Thus it is possible that a longer timeframe after switching diets is required for significant disease suppression. Further analysis of T cell subsets such as Th1 and Th17 which are critical in driving EAE disease56 will be necessary to determine the role of Th1 and Th17 in disease modulation after switching the diet. Finally future studies investigating the disease suppressive effect of switching the diet in already established EAE disease will strengthen the therapeutic potential of isoflavone-rich diet.
In conclusion we show that changing to an ISO diet induces a change in the composition of the gut microbiota and LPS production pathways. Such changes were evident with ISO-induced LPS which enhanced the expression of anti-inflammatory cytokines regardless of the presence of isoflavone-metabolizing bacteria. This provides justification for the beneficial effects of isoflavones through their ability to metabolize phytoestrogens and alter LPS biosynthesis. However further studies are necessary to determine the metabolism of isoflavones along the non-equol pathway by gut bacteria to gain a comprehensive understanding of the role of dietary isoflavones in MS and other inflammatory disorders.
Materials and methods
Mice procurement and dietary regiment
C57BL/6J female mice (4 to 6weeks old) were purchased from Jackson Laboratories (Bar Harbor ME) and maintained at the University of Iowa mouse facility. At the age of 4–6weeks pups were placed on either an ISO diet [genistein (0.24g/kg) and daidzein (0.22g/kg of diet)] (n=10) or a PF diet (Envigo Indianapolis IN) (n=8) ad libitum for 6weeks before the diet switch was made.
Fecal sample collection and sequencing
Fecal pellets from six individual mice were collected at different time points (Figure 1a). After switching diets mice were reared for four weeks before being euthanized. A total of 77 fecal pellet samples were collected on D0 (baseline) and on the following days after the diets were switched: day 2 day 3 day 7 day 14 day 21 and day 28. Samples were stored at −80°C until further analysis. DNA was isolated using DNesay PowerLyzer PowerSoil Kit (Qiagen) following the manufacturer’s instructions. DNA samples were quantified using Qubit (Qiagen) and sequenced using a MiSeq Illumina platform at CosmosID Inc (Rockville MD). The raw sequences were quality controlled using metawrap v =1.3.2 read_qc pipeline.57 Briefly adaptor sequences were trimmed low-quality bases (Phred score <20) were removed and host reads were removed by mapping reads to the reference mouse genome (mm39) using default parameters. After quality control and host reads were removed the clean reads were mapped to proGenomes reference database58 for taxonomic assignment of the sequences using Kaiju greedy mode.59
Microbiota analysis
The downstream analysis of the taxonomic table was performed in R v4.1.060 using phyloseq microbiome and vegan packages after removing the reads assigned to either “unclassified” “viruses” or “cannot be assigned to non-viral species.” One of the samples from the PF group was removed because of a low number of sequences. Thus the microbiota analysis includes five mice from the PF group and six mice from the ISO diet group respectively for the downstream analysis. Shannon diversity and Faith’s phylogenetic diversity were calculated as alpha diversity measures. The taxa were then filtered by removing those that are not observed at least 100 times in at least 20% of the samples and normalized to median sequencing depth. Weighted unifrac distances and Bray-Curtis dissimilarity metrics were used for beta diversity analysis. For time points D0 (baseline) day 2 day 3 day 7 day 14 day 21 and day 28 were considered based on fecal collection. Differential microbiota analysis was performed using the Welch test from the microbiomemarkerpackage.61 HUMANn362 was used with default parameters to profile functional pathway abundances in the clean reads. Pathways that were not observed at least 100 times in at least 10% of the samples were removed normalized to median depth and analyzed downstream in R.
Gene homology
Multiple microbial species are known to possess isoflavone-metabolizing daidzein reductase (DR) dihydrodaidzein reductase (DHDR) and tetrahydrodaidzein reductase (THDR) genes. The nucleotide sequences of these genes from Adlercreutzia equolifaciens (DR: RFT81436.1 DHDR: RFT81438.1 THDR: RFT81439.1) Eggerthella sp. YY7918 (DR: BAK44713.1 DHDR: BAK44715.1 THDR: BAK44716.1) Lactococcus garvieae (DR: BAJ72750.1 DHDR: BAJ72748.1 THDR: BAJ72744.1) Senegalimassilia sp. KGMB04484 (DR: RXZ54824.1 DHDR: RXZ54822.1 THDR: RXZ54821.1) Slackia equolifaciens (DR: RNL39925.1 DHDR: RNL39927.1 THDR: RNL39927.1) Slackia sp. NATTS (DR: BAL46930.1 DHDR: BAL46929.1 THDR: BAL46928.1) and Slackia isoflavoniconvertens (DR: AFV15453.1 DHDR: AFV15451.1 THDR: AFV15450.1) were obtained from NCBI. We also obtained the daidzein and genistein reductase (dgr) gene (KJ452760.1) from Slackia sp. AUH-JLC159 and combined with DR DHDR and THDR genes from the aforementioned bacterial species to create a custom database. The assembled fecal metagenomes from each mouse were searched for the presence of isoflavone-metabolizing genes in a custom database using nucmer with default parameters.63 Similarly bacterial reference genomes of B. adolescentis (NZ_CP028341.1) B. longum (NZ_AKCA01000001.1) and B. stellenboschense (NZ_JGZP01000001.1) were obtained from NCBI and searched for the presence of isoflavone-metabolizing genes.
Carbohydrate metabolizing active enzymes annotation and quantification
Carbohydrate metabolizing active enzymes were predicted from the shotgun metagenomic sequences. After quality control and host reads removal clean paired reads were assembled using Megahit using default parameters.64 The obtained assembly of each sample was annotated using Prokka65 and .fna was used for identifying the active carbohydrate metabolizing enzymes in dbCAN2 with default parameters.66 dbCAN2 aligns the sequences to CAZy database67 to identify the enzymatic families. All enzymes identified by at least one of the HMMR Hotpep or DIAMOND algorithms within dbCAN2 were used for analysis. The relative abundance of the obtained modules was further analyzed using an unpaired t-test with Welch correction in GrapPad Prism 10.0.
Extraction of LPS from fecal lysate
Fecal samples were collected from mice (n=5) kept on a PF or ISO diet for four weeks. LPS was isolated from the fecal samples following methods described previously with slight modification.68 Briefly 100 mg of feces was mixed with 25ml of PBS in a pyrogen-free tube and sonicated for one hour. The mixture was centrifuged at 400g for 15minutes and 15ml of supernatant was collected. The supernatant was sterilized by filtration through a 0.22μm non-positively charged filter and inactivated for 10minutes at 70°C. The filtrate was then kept at −80°C until further use. The fecal lysate was thawed and endotoxin levels were quantified in 10μl of filtrates using ToxinSensor Chromogenic LAL Endotoxin Assay Kit (Cat. No. L00350 GenScript NJ) following the manufacturer’s protocol.
Bone marrow-derived macrophages culture and treatment with fecal LPS
Bone marrow extraction and subsequent culture to yield macrophages were performed following methods previously described.69 Briefly mice were euthanized by exposure to carbon dioxide and bone marrow was extracted from femur and tibia bones aseptically. Clumps of marrow were gently disintegrated using a needleless syringe and passed through a 70μm cell strainer. The cell suspension was then centrifuged at 250g for 5minutes at room temperature the supernatant was discarded and the cells were suspended in DMEM F12 medium containing L-glutamine (2mM) penicillin (100 units/ml) streptomycin (0.1 mg/ml) 5% FBS and recombinant MCSF (25ng/ml) growth factor (DMEM F12 MCSF growth media). 200000 cells/well were seeded into a non-tissue culture treated 12 well plate in 1ml DMEM F12 MCSF growth media on D0. The cells were cultured for 7days and fed with an addition of 500μl of DMEM F12 MCSF growth media containing MCSF (50ng/ml) on day 5 of culture. On day 7 the cells were washed with PBS to remove unadhered cells and 1ml of media containing DMEM F12 containing l-glutamine (2mM) penicillin (100 units/ml) streptomycin (0.1 mg/ml) was added. Cells were then either left unstimulated (control) or were stimulated with 10μl of LPS isolated from the fecal lysate of mice kept on a PF or ISO diet or 10μl of LPS standard control solution (0.6μg/μl) (LPS Escherichia coli 055.B5 EMD Chemicals CA) for 3hours at 37°C. Subsequently the cells were washed with PBS and the media was replaced with DMEM F12 containing l-glutamine (2mM) penicillin (100 units/ml) streptomycin (0.1 mg/ml) and incubated at 37°C for 16hours before collecting supernatants. The supernatants collected from all four groups control PF ISO and LPS standard treated group and were kept at −80°C until further use. The levels of cytokines TNF-α IL-6 IL-12/23 (Biolegend CA) and IL-10 (BD Biosciences Pharmigen CA) were quantified in the supernatants using ELISA as per manufacturer’s instructions. GraphPad Prism 10.0 was used for visualization and statistical tests.
S-equol quantification
S-equol was quantified from the urine of mice before and 28days after diets were switched. Urine samples were collected and sent to the Metabolomics Core Henry Ford Health System (Detroit MI) for quantification using Liquid Chromatography Mass Spectrometry (LC-MS/MS) where S-equol was quantified following methods described previously with modification.70 Briefly Mobile phase A (2 mm Amonium acetate +0.2% acetic acid) and B (Acetonitrile+0.2% acetic acid) at a flow rate of 0.30ml/min were used to separate the metabolites in gradient mode. Best separation of S-Equol was achieved using Waters Acquity Column CSH 1.7µm 2.1 mm × 150 mm kept at 65°C using 5minutes linear gradient. The column effluent was monitored by Negative Electrospray Ionization (ESI) using multiple reaction monitoring (MRM) workflow. S-Equol peak area under the curve (AUC) of the analyte was then used for quantification.
EAE induction and scoring
For EAE studies EAE was induced and evaluated as described previously.71 Briefly mice were immunized subcutaneously on D0 on the left and right flank with 100μg of MOG35-55 emulsified in 200μg of CFA followed by 80 ng of pertussis toxin (PTX) intraperitoneally on D0 and D2. Disease severity was scored as follows: 0 no clinical symptoms; 1 loss of tail tonicity; 2 hindlimb weakness; 3 hindlimb paralysis; 4 forelimb weakness; and 5 moribund or death. All procedures were done according to the Institutional Animal Care and Use Committee guidelines at the University of Iowa.
Supplementary Material
Acknowledgments
We thank members of the Karandikar laboratory for helpful discussions.
Funding Statement
This work was supported by the National Institutes of Health/NIAID 1R01AI137075 (AKM) Veteran Affairs Merit Award 1I01CX002212 (AKM) University of Iowa Environmental Health Sciences Research Center NIEHS/NIH P30 ES005605 (AKM) Gift from P. Heppelmann and M. Wacek to (AKM) and Carver Trust Pilot Grant (AKM). SRP was supported by a T32 predoctoral training grant T32AI007485 (PI Gail Bishop).
Data availability statement
The shotgun metagenomic sequences are deposited in NCBI under BioProject PRJNA834824. All other data needed to evaluate the conclusions in the manuscript are present in the manuscript and/or the Supplementary Materials.
Disclosure statement
AKM is inventor of a technology claiming the use of Prevotella histicola for the treatment of autoimmune diseases. The patent for the technology is owned by Mayo Clinic, who has given exclusive license to Evelo Biosciences. AKM received royalties from Mayo Clinic (paid by Evelo Biosciences). However, no fund or product from the patent were used in the present study. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19490976.2022.2127446
References
- 1.Walton C, King R, Rechtman L, Kaye W, Leray E, Marrie RA, Robertson N, La Rocca N, Uitdehaag B, van der Mei I, et al. Rising prevalence of multiple sclerosis worldwide: insights from the atlas of MS. Mult Scler. 2020;26:1816–19. third edition. doi: 10.1177/1352458520970841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mangalam A, Rodriguez M, David C.. A new humanized HLA transgenic mouse model of multiple sclerosis expressing class II on mouse CD4 T cells. Ann N Y Acad Sci. 2007;1103:112–117. doi: 10.1196/annals.1394.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koch MW, Metz LM, Agrawal SM, Yong VW. Environmental factors and their regulation of immunity in multiple sclerosis. J Neurol Sci. 2013;324:10–16. doi: 10.1016/j.jns.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giordano M, D’Alfonso S, Momigliano-Richiardi P. Genetics of multiple sclerosis: linkage and association studies. Am J Pharmacogenomics. 2002;2:37–58. doi: 10.2165/00129785-200202010-00004. [DOI] [PubMed] [Google Scholar]
- 5.Willer CJ, Dyment DA, Risch NJ, Sadovnick AD, Ebers GC. Canadian Collaborative Study G. Twin concordance and sibling recurrence rates in multiple sclerosis. Proc Natl Acad Sci U S A. 2003;100:12877–12882. doi: 10.1073/pnas.1932604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen T, Skytthe A, Stenager E, Petersen HC, Bronnum-Hansen H, Kyvik KO. Concordance for multiple sclerosis in Danish twins: an update of a nationwide study. Mult Scler. 2005;11:504–510. doi: 10.1191/1352458505ms1220oa. [DOI] [PubMed] [Google Scholar]
- 7.Kuusisto H, Kaprio J, Kinnunen E, Luukkaala T, Koskenvuo M, Elovaara I. Concordance and heritability of multiple sclerosis in Finland: study on a nationwide series of twins. Eur J Neurol. 2008;15:1106–1110. doi: 10.1111/j.1468-1331.2008.02262.x. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Chia N, Kalari KR, Yao JZ, Novotna M, Paz Soldan MM, et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci Rep. 2016;6:28484. doi: 10.1038/srep28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colpitts SL, Kasper EJ, Keever A, Liljenberg C, Kirby T, Magori K, Kasper LH, Ochoa-Repáraz J. A bidirectional association between the gut microbiota and CNS disease in a biphasic murine model of multiple sclerosis. Gut Microbes. 2017;8:561–573. doi: 10.1080/19490976.2017.1353843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jangi S, Gandhi R, Cox LM, Li N, von Glehn F, Yan R, Patel B, Mazzola MA, Liu S, Glanz BL, et al. Alterations of the human gut microbiome in multiple sclerosis. Nat Commun. 2016;7:12015. doi: 10.1038/ncomms12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nat Immunol. 2011;12:5–9. doi: 10.1038/ni0111-5. [DOI] [PubMed] [Google Scholar]
- 12.Bibbo S, Ianiro G, Giorgio V, Scaldaferri F, Masucci L, Gasbarrini A, Cammarota G. The role of diet on gut microbiota composition. Eur Rev Med Pharmacol Sci. 2016;20:4742–4749. [PubMed] [Google Scholar]
- 13.Rinninella E, Cintoni M, Raoul P, Lopetuso LR, Scaldaferri F, Pulcini G, et al. Food components and dietary habits: keys for a healthy gut microbiota composition. Nutrients. 2019;11:2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forgie AJ, Fouhse JM, Willing BP, Forgie AJ, Fouhse JM, Willing BP . Diet-microbe-host interactions that affect gut mucosal integrity and infection resistance. Front Immunol. 2019;10:1802. doi: 10.3389/fimmu.2019.01802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rimbach G, Boesch-Saadatmandi C, Frank J, Fuchs D, Wenzel U, Daniel H, Hall WL, Weinberg PD. Dietary isoflavones in the prevention of cardiovascular disease–a molecular perspective. Food Chem Toxicol. 2008;46:1308–1319. doi: 10.1016/j.fct.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 16.Hertog MG, Feskens EJ, Hollman PC, Katan MB, Kromhout D, Hertog MGL, Katan MB. Dietary antioxidant flavonoids and risk of coronary heart disease: the Zutphen elderly study. Lancet. 1993;342:1007–1011. doi: 10.1016/0140-6736(93)92876-U. [DOI] [PubMed] [Google Scholar]
- 17.Larrosa M, Luceri C, Vivoli E, Pagliuca C, Lodovici M, Moneti G, Dolara P. Polyphenol metabolites from colonic microbiota exert anti-inflammatory activity on different inflammation models. Mol Nutr Food Res. 2009;53:1044–1054. doi: 10.1002/mnfr.200800446. [DOI] [PubMed] [Google Scholar]
- 18.Maruo T, Sakamoto M, Ito C, Toda T, Benno Y. Adlercreutzia equolifaciens gen. nov., sp. nov., an equol-producing bacterium isolated from human faeces, and emended description of the genus Eggerthella. Int J Syst Evol Microbiol. 2008;58:1221–1227. doi: 10.1099/ijs.0.65404-0. [DOI] [PubMed] [Google Scholar]
- 19.Kim M, Kim SI, Han J, Wang XL, Song DG, Kim SU. Stereospecific biotransformation of dihydrodaidzein into (3S)-equol by the human intestinal bacterium Eggerthella strain Julong 732. Appl Environ Microbiol. 2009;75:3062–3068. doi: 10.1128/AEM.02058-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yokoyama S, Suzuki T. Isolation and characterization of a novel equol-producing bacterium from human feces. Biosci Biotechnol Biochem. 2008;72:2660–2666. doi: 10.1271/bbb.80329. [DOI] [PubMed] [Google Scholar]
- 21.Schroder C, Matthies A, Engst W, Blaut M, Braune A. Identification and expression of genes involved in the conversion of daidzein and genistein by the equol-forming bacterium Slackia isoflavoniconvertens. Appl Environ Microbiol. 2013;79:3494–3502. doi: 10.1128/AEM.03693-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uchiyama S, Ueno T, Suzuki T. Identification of a newly isolated equal-producing lactic acid bacterium from the human feces. J Intestinal Microbiol. 2007;21:217. [Google Scholar]
- 23.Bowey E, Adlercreutz H, Rowland I. Metabolism of isoflavones and lignans by the gut microflora: a study in germ-free and human flora associated rats. Food Chem Toxicol. 2003;41:631–636. doi: 10.1016/S0278-6915(02)00324-1. [DOI] [PubMed] [Google Scholar]
- 24.Jensen SN, Cady NM, Shahi SK, Peterson SR, Gupta A, Gibson-Corley KN, et al. Isoflavone diet ameliorates experimental autoimmune encephalomyelitis through modulation of gut bacteria depleted in patients with multiple sclerosis. Sci Adv. 2021;7:eabd4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayo B, Vazquez L, Florez AB. Equol: a bacterial metabolite from the daidzein isoflavone and its presumed beneficial health effects. Nutrients. 2019;11:2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sullivan A, Nord CE. Probiotics and gastrointestinal diseases. J Intern Med. 2005;257:78–92. doi: 10.1111/j.1365-2796.2004.01410.x. [DOI] [PubMed] [Google Scholar]
- 27.Gibson GR. Dietary modulation of the human gut microflora using the prebiotics oligofructose and inulin. J Nutr. 1999;129:1438S–41S. doi: 10.1093/jn/129.7.1438S. [DOI] [PubMed] [Google Scholar]
- 28.Tsangalis D, Ashton JF, Mcgill AEJ, Shah NP. Enzymic transformation of isoflavone phytoestrogens in soymilk by b-glucosidase-producing bifidobacteria. J Food Sci. 2002;67:3104–3113. doi: 10.1111/j.1365-2621.2002.tb08866.x. [DOI] [Google Scholar]
- 29.Lemaire M, Miras I, Gounon P, Beguin P. Identification of a region responsible for binding to the cell wall within the S-layer protein of Clostridium thermocellum. Microbiology (Reading). 1998;144(Pt 1):211–217. doi: 10.1099/00221287-144-1-211. [DOI] [PubMed] [Google Scholar]
- 30.Olabarria G, Carrascosa JL, de Pedro MA, Berenguer J. A conserved motif in S-layer proteins is involved in peptidoglycan binding in thermus thermophilus. J Bacteriol. 1996;178:4765–4772. doi: 10.1128/jb.178.16.4765-4772.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemaire M, Ohayon H, Gounon P, Fujino T, Beguin P. OlpB, a new outer layer protein of clostridium thermocellum, and binding of its S-layer-like domains to components of the cell envelope. J Bacteriol. 1995;177:2451–2459. doi: 10.1128/jb.177.9.2451-2459.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, Yassour M, Kolde R, Vlamakis H, Arthur T, Hämäläinen A-M, et al. Variation in microbiome lps immunogenicity contributes to autoimmunity in humans. Cell. 2016;165:842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 34.Zhang C, Zhang M, Wang S, Han R, Cao Y, Hua W, Mao Y, Zhang X, Pang X, Wei C, et al. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010;4:232–241. doi: 10.1038/ismej.2009.112. [DOI] [PubMed] [Google Scholar]
- 35.Mangalam AK, Yadav M, Yadav R. The emerging world of microbiome in autoimmune disorders: opportunities and challenges. Indian J Rheumatol. 2021;16:57–72. doi: 10.4103/injr.injr_210_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shreiner AB, Kao JY, Young VB. The gut microbiome in health and in disease. Curr Opin Gastroenterol. 2015;31:69–75. doi: 10.1097/MOG.0000000000000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matthies A, Blaut M, Braune A. Isolation of a human intestinal bacterium capable of daidzein and genistein conversion. Appl Environ Microbiol. 2009;75:1740–1744. doi: 10.1128/AEM.01795-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawada Y, Yokoyama S, Yanase E, Niwa T, Suzuki T. The production of S-equol from daidzein is associated with a cluster of three genes in Eggerthella sp. YY7918. Biosci Microbiota Food Health. 2016;35:113–121. doi: 10.12938/bmfh.2015-023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawada Y, Goshima T, Sawamura R, Yokoyama SI, Yanase E, Niwa T, Ebihara A, Inagaki M, Yamaguchi K, Kuwata K, et al. Daidzein reductase of Eggerthella sp. YY7918, its octameric subunit structure containing FMN/FAD/4Fe-4S, and its enantioselective production of R-dihydroisoflavones. J Biosci Bioeng. 2018;126:301–309. doi: 10.1016/j.jbiosc.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 40.Brown NM, Setchell KD. Animal models impacted by phytoestrogens in commercial chow: implications for pathways influenced by hormones. Lab Invest. 2001;81:735–747. doi: 10.1038/labinvest.3780282. [DOI] [PubMed] [Google Scholar]
- 41.Thigpen JE, Setchell KD, Ahlmark KB, Locklear J, Spahr T, Caviness GF, Goelz MF, Haseman JK, Newbold RR, Forsythe DB, et al. Phytoestrogen content of purified, open- and closed-formula laboratory animal diets. Lab Anim Sci. 1999;49:530–536. [PubMed] [Google Scholar]
- 42.Elghali S, Mustafa S, Amid M, Manap MYABD, Ismail A, Abas F. Bioconversion of daidzein to equol by Bifidobacterium breve 15700 and Bifidobacterium longum BB536. J Funct Foods. 2012;4:736–745. doi: 10.1016/j.jff.2012.04.013. [DOI] [Google Scholar]
- 43.Heng Y, Kim MJ, Yang HJ, Kang S, Park S. Lactobacillus intestinalis efficiently produces equol from daidzein and chungkookjang, short-term fermented soybeans. Arch Microbiol. 2019;201:1009–1017. doi: 10.1007/s00203-019-01665-5. [DOI] [PubMed] [Google Scholar]
- 44.Liu B, Qin L, Liu A, Uchiyama S, Ueno T, Li X, et al. Prevalence of the equol-producer phenotype and its relationship with dietary isoflavone and serum lipids in healthy Chinese adults. J Epidemiol. 2010;20:377–384. doi: 10.2188/jea.JE20090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rowland IR, Wiseman H, Sanders TA, Adlercreutz H, Bowey EA. Interindividual variation in metabolism of soy isoflavones and lignans: influence of habitual diet on equol production by the gut microflora. Nutr Cancer. 2000;36:27–32. doi: 10.1207/S15327914NC3601_5. [DOI] [PubMed] [Google Scholar]
- 46.Bottacini F, Ventura M, van Sinderen D, O’Connell Motherway M. Diversity, ecology and intestinal function of bifidobacteria. Microb Cell Fact. 2014;13:S4. doi: 10.1186/1475-2859-13-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu R, Zuo F, Ma H, Chen S. Exopolysaccharide-producing Bifidobacterium adolescentis strains with similar adhesion property induce differential regulation of inflammatory immune response in Treg/Th17 axis of DSS-colitis mice. Nutrients. 2019;11:782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan S, Yang B, Zhao J, Zhao J, Stanton C, Ross RP, Zhang H, Chen W. A ropy exopolysaccharide producing strain Bifidobacterium longum subsp. longum YS108R alleviates DSS-induced colitis by maintenance of the mucosal barrier and gut microbiota modulation. Food Funct. 2019;10:1595–1608. doi: 10.1039/C9FO00014C. [DOI] [PubMed] [Google Scholar]
- 49.Dia VP, Berhow MA, Gonzalez De Mejia E. Bowman−birk inhibitor and genistein among soy compounds that synergistically inhibit nitric oxide and prostaglandin E 2 pathways in lipopolysaccharide-induced macrophages. J Agric Food Chem. 2008;56:11707–11717. doi: 10.1021/jf802475z. [DOI] [PubMed] [Google Scholar]
- 50.Abron JD, Singh NP, Price RL, Nagarkatti M, Nagarkatti PS, Singh UP, Nakano H. Genistein induces macrophage polarization and systemic cytokine to ameliorate experimental colitis. PLoS One. 2018;13:e0199631. doi: 10.1371/journal.pone.0199631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haas KN, Blanchard JL. Kineothrix alysoides, gen. nov., sp. nov., a saccharolytic butyrate-producer within the family Lachnospiraceae. Int J Syst Evol Microbiol. 2017;67:402–410. doi: 10.1099/ijsem.0.001643. [DOI] [PubMed] [Google Scholar]
- 52.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic T reg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shahi SK, Freedman SN, Murra AC, Zarei K, Sompallae R, Gibson-Corley KN, Karandikar NJ, Murray JA, Mangalam AK. Prevotella histicola, A human gut commensal, is as potent as COPAXONE(R) in an animal model of multiple sclerosis. Front Immunol. 2019;10:462. doi: 10.3389/fimmu.2019.00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iljazovic A, Amend L, Galvez EJC, de Oliveira R, Strowig T. Modulation of inflammatory responses by gastrointestinal prevotella spp. - from associations to functional studies. Int J Med Microbiol. 2021;311:151472. doi: 10.1016/j.ijmm.2021.151472. [DOI] [PubMed] [Google Scholar]
- 55.Lucke K, Miehlke S, Jacobs E, Schuppler M. Prevalence of bacteroides and prevotella spp. in ulcerative colitis. J Med Microbiol. 2006;55:617–624. doi: 10.1099/jmm.0.46198-0. [DOI] [PubMed] [Google Scholar]
- 56.Hart BA, Gran B, Weissert R. EAE: imperfect but useful models of multiple sclerosis. Trends Mol Med. 2011;17:119–125. doi: 10.1016/j.molmed.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 57.Uritskiy GV, DiRuggiero J, Taylor J. MetaWRAP-a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome. 2018;6:158. doi: 10.1186/s40168-018-0541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mende DR, Letunic I, Huerta-Cepas J, Li SS, Forslund K, Sunagawa S, Bork P. proGenomes: a resource for consistent functional and taxonomic annotations of prokaryotic genomes. Nucleic Acids Res. 2017;45:D529–D34. doi: 10.1093/nar/gkw989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Menzel P, Ng KL, Krogh A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun. 2016;7:11257. doi: 10.1038/ncomms11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.R Core Team . R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2021. [Google Scholar]
- 61.Yang C. microbiomeMarker: microbiome biomarker analysis toolkit. 2020. [Google Scholar]
- 62.Beghini F, McIver LJ, Blanco-Miguez A, Dubois L, Asnicar F, Maharjan S, et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife. 2021;10:e65088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marcais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A, Darling AE. MUMmer4: a fast and versatile genome alignment system. PLoS Comput Biol. 2018;14:e1005944. doi: 10.1371/journal.pcbi.1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li D, Liu CM, Luo R, Sadakane K, Lam TW. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 65.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 66.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, Busk PK, Xu Y, Yin Y. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018;46:W95–W101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Drula E, Garron ML, Dogan S, Lombard V, Henrissat B, Terrapon N. The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 2022;50:D571–D7. doi: 10.1093/nar/gkab1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim KA, Jeong JJ, Yoo SY, Kim DH. Gut microbiota lipopolysaccharide accelerates inflamm-aging in mice. BMC Microbiol. 2016;16:9. doi: 10.1186/s12866-016-0625-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bailey JD, Shaw A, McNeill E, Nicol T, Diotallevi M, Chuaiphichai S, Patel J, Hale A, Channon KM, Crabtree MJ, et al. Isolation and culture of murine bone marrow-derived macrophages for nitric oxide and redox biology. Nitric Oxide. 2020;100-101:17–29. doi: 10.1016/j.niox.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raju KSR, Rashid M, Gundeti M, Taneja I, Malik MY, Singh SK, Chaturvedi S, Challagundla M, Singh SP, Gayen JR, et al. LC-ESI-MS/MS method for the simultaneous determination of isoformononetin, daidzein, and equol in rat plasma: application to a preclinical pharmacokinetic study. J Chromatogr B Analyt Technol Biomed Life Sci. 2019;1129:121776. doi: 10.1016/j.jchromb.2019.121776. [DOI] [PubMed] [Google Scholar]
- 71.Tyler AF, Mendoza JP, Firan M, Karandikar NJ, Platten M. CD8(+) T cells are required for glatiramer acetate therapy in autoimmune demyelinating disease. PLoS One. 2013;8:e66772. doi: 10.1371/journal.pone.0066772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The shotgun metagenomic sequences are deposited in NCBI under BioProject PRJNA834824. All other data needed to evaluate the conclusions in the manuscript are present in the manuscript and/or the Supplementary Materials.


