Abstract
Primary cilium (PC) is a microtubule‐based organelle found on the apical surface of most mammalian cell types, playing a role in development and tissue homeostasis. Ciliopathies are a rapidly growing group of human diseases characterized by disordered cilium. PC plays an important role in pathogenesis of basal cell cancer, the most common human malignancy. A significant increase in ciliation has been observed in the epidermis of atopic dermatitis and psoriasis patients. Spontaneously immortalized human keratinocytes, HaCaT are a model to study the epidermal homeostasis and pathophysiology. In contrast to what has been previously described, here, we show that HaCaT can be efficiently ciliated. In HaCaT cells, differentiation significantly increased the number of ciliated cells and we were able to analyse in detail the ciliary length progression with duration of differentiation. As the number of recognized ciliopathies continues to increase, the importance of ciliary models also rises. Even though keratinocytes do not become as highly and rapidly ciliated as cell lines frequently used in ciliary studies, they are a better model for the study of skin ciliopathies. Detailed progression of ciliation in HaCaT could serve as the basis for ciliary studies in this cell line.
Keywords: cancer, differentiation, keratinocyte biology, keratinocytes, primary cilium
Abbreviations
- Ac‐tubul
acetylated tubulin
- BCC
basal cell carcinoma
- D
day of differentiation
- GFP
green fluorescent protein
- HH
hedgehog
- HHSP
hedgehog signalling pathway
- HKGS
human keratinocyte growth supplement
- IB
immunoblot
- IF
immunofluorescence
- K
keratin
- LV
lentiviral particle
- ND
non‐differentiated
- O/N
over night
- PBS
phosphate‐buffered saline
- PC
primary cilium
- RPE
retinal pigment epithelium
- RT
room temperature
- SMO
Smoothened
- T
Tween
- TBS
Tris‐buffered saline
- WB
Western blot
1. INTRODUCTION
Primary cilium (PC) is a nonmotile microtubule‐based organelle found on the apical surface of most mammalian cell types, where it serves as cellular antenna, sensing diverse extracellular signals. PC is implicated in development and tissue homeostasis and possesses mechanosensory, osmosensory, chemosensory, as well as photosensory functions. 1 , 2 Core of the PC is called ciliary axoneme, composed of nine sets of parallel doublets of microtubules. In nonmitotic cells, the centrosome, composed of mother and daughter centriole, moves to the apical surface of the cell. 3 , 4 Mother centriole docks into the plasmatic membrane, which allows its transformation into the basal body. 5
Ciliopathies are a rapidly growing group of genetic human diseases characterized by disordered cilium. As cilia can be found on most cell types of the human body, ciliary malfunction has widespread consequences. 6 We recently published the role of ARP‐T1 in Bazex‐Dupré‐Christol syndrome, which could be considered a skin ciliopathy. 7
Multiple signalling pathways have been linked to the PC, either as authentic ciliary pathways, such as canonical hedgehog signalling pathway (HHSP), or as PC‐associated ones. 8 Epidermal PC plays a role in epidermal homeostasis, with possible implication in wound healing and scarring. 9 , 10 With growing evidence on widespread roles of PC, its involvement in skin cancer is consistently becoming of high interest. PC is considered a prerequisite for functioning canonical HHSP, whose aberrant activation is found in many cancers, most notably in basal cell cancer (BCC) and medulloblastoma. 11 Smoothened (SMO) inhibitors target HH signalling through the PC. PC has been recently shown to be at origin of one of the resistance mechanisms in advanced BCC treated with SMO inhibitors. 12
HaCaT are a spontaneously immortalized keratinocyte cell line, induced by bi‐allelic UV‐specific mutations of the tumour‐suppressor gene p53 followed by additional loss of senescence genes. 13 They are used as a model to study the epidermal homeostasis and its pathophysiology. 14 Although the growth and differentiation of HaCaT keratinocytes have been described previously, 15 , 16 very little information is available concerning their ciliation. In 2019, Choi et al. 17 claim that HaCaT do not form PC, arguing that differentiated skin keratinocytes are rarely ciliated after serum starvation. We reexamined their possible ciliation under different conditions.
Here, we show that HaCaT can be efficiently ciliated. Induction of ciliogenesis has been correlated to the arrest of cell proliferation, 18 we therefore hypothesized that keratinocyte differentiation will result in increased ciliation. HaCaT differentiation was reached through their cultivation up to confluency and subsequent high‐calcium switch lasting from 5 to 7 days. As keratinocytes, which engage to terminal differentiation switch from keratin (K)‐5 and K14 to expression of K1 and K10, we used K10 as a marker of cell differentiation (Figure 1A). Confocal microscopy analysis was performed using acetylated tubulin as a marker of the ciliary axoneme and rootletin as a marker of the basal body (Figure 1B).
FIGURE 1.
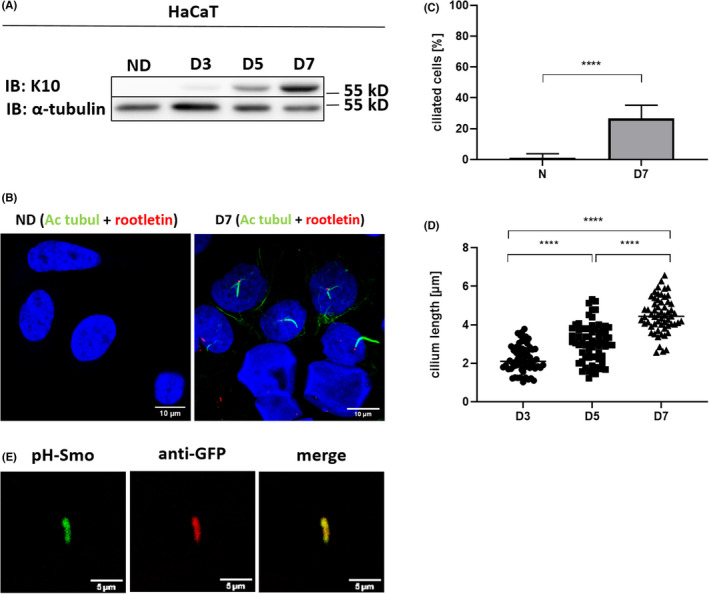
Differentiation of HaCaT keratinocytes increases their ciliation. (A) Western blot showing a representative image of K10 protein expression during differentiation of HaCaT. α‐tubulin used as a loading control. (B) Representative immunofluorescence images of acetylated tubulin (green) and rootletin (red) co‐staining in non‐differentiated and 7 days‐differentiated HaCaT. Scale bar, 10 μm. (C) Quantification of ciliated HaCaT upon non‐differentiated and 7 days differentiated conditions. Unpaired t test with Welch's correction (N = 10, 16). (D) Cilium length measured in HaCaT at D3, D5 and D7 of differentiation. Ordinary one‐way ANOVA test with Holm‐Šidák's correction (N = 64, 68, 59). (E) Representative images of SMO IN/OUT assay in 7 days‐differentiated HaCaT. Scale bar, 5 μm. Ac‐tubul, acectylated tubulin; IB, immunoblot; ND, non‐differentiated, D3, D5, D7: day 3, 5 and 7 of differentiation, respectively
Indeed, in HaCaT cells, differentiation significantly increased the number of ciliated cells, with median of 26% of ciliated cells, compared to very rare (<1%) ciliated cells upon non‐differentiated conditions (Figure 1C). Comparable results were obtained in primary human keratinocytes NHEK‐1, established from human foreskins in our laboratory (data not shown). Discrepancy between our findings and results published by Choi et al. could be explained by different methods used to induce differentiation. They tried to induce ciliation in HaCaT cells via serum‐starvation as it is necessary for retinal pigment epithelium (RPE) cells. RPE cells are often used as a ciliary model, and serum‐starvation was described to modulate the cell metabolism. 19 According to Choi et al., HaCaT cells are not responsive to serum‐starvation, at least, not for ciliation. We searched for a protocol, which could modify HaCaT metabolism. As HaCaT cells are known to differentiate at confluency under a calcium shift, we studied ciliation in these conditions. We now provide a detailed protocol allowing PC induction in HaCaT. To our knowledge for the first time in keratinocyte culture, we were able to analyse in detail the ciliary length progression with duration of differentiation (Figure 1D). An average ciliary length varied from 2.2 μm on day (D) 3, to 3.1 μm on D5 and finally 4.5 μm on D7.
To confirm these findings, we decided to test SMO IN/OUT assay developed by Kukic et al. 20 as an additional tool for PC visualization. This assay, said to be able to easily and accurately identify a PC, was developed in hTERT‐RPE1 cells and was not yet tested in keratinocytes. Differentiated keratinocytes were transduced with SMO IN/OUT lentivirus pLVX‐ss‐pH‐Smo, allowing cells to stably express the N‐terminally pHluorin tagged Smo construct. PC can then be observed directly after fixing cells in paraformaldehyde using confocal microscopy (in green, Figure 1E). Measured ciliary lengths were consistent with findings using double ciliary staining with acetylated tubulin and rootletin. As no antibody use is necessary, this assay is not only a rapid but also a cost‐convenient alternative to assess ciliary length.
Additionally, SMO IN/OUT tool can differentiate between inside and outside ciliary portions. Using anti‐GFP antibody in non‐permeabilized cells, outer part of the PC will be visualized in red (Figure 1E). Two pathways of ciliogenesis have been described, intracellular and extracellular. 21 In intracellular route, mother centriole and associated ciliary vesicle initiate the elongation of the nascent axoneme while still in the cytosol. In extracellular pathway, the basal body, transformed from the mother centriole, docks at the apical surface of the cell prior to axoneme extension. 22 An absolute majority of PC in keratinocytes were in the ‘OUT’ stage, which is consistent with extracellular pathway being typically employed in the cilia formation of polarized epithelial cells. 23
As the number of recognized ciliopathies continues to increase, the importance of ciliary models also rises. Even though keratinocytes do not become as highly and rapidly ciliated as cell lines frequently used in ciliary studies, such as hTERT‐RPE1 cells, they are a better model for the study of skin ciliopathies. PC has been implicated not only in BCC onset, but recently a significant increase in ciliation has been observed in the epidermis of atopic dermatitis and psoriasis patients. 24 Detailed progression of ciliation in HaCaT could serve as the basis for ciliary studies in this cell line.
2. MATERIALS AND METHODS
2.1. Cell culture
Immortalized HaCaT keratinocytes (kindly given by Pr. Boukamp, German Cancer Research Center, Heidelberg, Germany) were cultured in EpiLife medium (11684842; Gibco, Invitrogen), supplemented with Human Keratinocyte Growth Supplement (HKGS) (10761364; Gibco, Invitrogen), 100 U/ml penicillin and 100 μg/ml streptomycin (BioConcept AG, Allschwil, Switzerland). Cells were cultured in 5% CO2 at 37°C.
To induce ciliogenesis, HaCaT were cultured up to 7 days in EpiLife medium supplemented with HKGS, 100 U/ml penicillin, 100 μg/ml streptomycin and 2 mM CaCl2 (208 291, Milipore).
2.2. Lentivirus production and transduction
Lentivirus particles (LVs) pLVX‐ss‐pH‐Smo for SMO IN/OUT assay were a kind gift from laboratory of Pr. Toomre lab and were produced using calcium phosphate transfection method. HEK293T cells were transiently co‐transfected with psPAX2 (Addgene, Cambridge, MA), pMD2.G (Addgene) and specific constructs. LVs were harvested 48 h later. Titre was determined in HeLa cells or using Lenti‐X GoStix Plus (631280; Takara). HeLa and HaCaT cells were transduced using polybrene (8 μg/ml, TR1003; Millipore), with LVs O/N at 37°C in 5% CO2. The next day, LVs were removed and cells grew in their medium. Transduced HaCaT were selected with puromycin (2.5 μg/ml, 540 411; Calbiochem).
2.3. SDS‐PAGE and Western blotting
HaCaT were washed in Phosphate‐buffered saline (PBS) and harvested in ice‐cold lysis buffer (50 mM Tris/HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X‐100) with protease inhibitor cocktail [cOmplete™ MiniTM tablette, 11836153 001; Roche Diagnostics] and phosphatase inhibitor cocktail [PhosSTOP, PHOSS‐RO, 4906837001; Roche]. Cells were incubated in lysis buffer for 30 min on an agitator at 4°C, scraped and sonicated on ice, twice for 10 s, followed by 20 min centrifugation at 12 000 g at 4°C. Supernatants were collected and total protein concentration determined by BCA assay (PierceTM BCA Protein Assay Kit, 23225; Thermo Fisher Scientific, Waltham, MA, USA). Same amount of cell lysate proteins were loaded onto a 10% SDS‐PAGE gel. Proteins were electroblotted onto an Immobilon® PVDF Membrane (IPVH00010; Merck, Millipore). Membrane was saturated for 1 h in 5% BSA or 10% powdered skimmed milk in 0.05% Tris‐buffered saline (TBS)‐Tween 20 (T). Membrane was incubated with the primary antibody of interest on an agitator, 2 h at room temperature (RT) or O/N at 4°C. Three consecutive washes with TBS‐T were followed by incubation with the secondary antibody ‐ 1 h at RT. List of primary and secondary antibodies used can be found in Table 1. After three TBS‐T washes, membrane was revealed using a WB detection kit WesternBright™ Quantum (K‐12042‐D10; Advansta) according to the manufacturer's instructions. Images were acquired using the Luminescent Image Analyser LAS‐4000 mini (Fujifilm, Tokyo, Japan). Images were cropped and quantified using Image J (https://imagej.nih.gov/ij/).
TABLE 1.
List of primary and secondary antibodies used
| Target | Type | Host | Concentration | Supplier | Accession number |
|---|---|---|---|---|---|
| Acetylated α‐tubulin (clone 6‐11B‐1) | Primary, monoclonal | Mouse | IF: 1/1000 | Sigma‐Aldrich | 6793 |
| Keratin 10 (clone DE‐K10) | Primary, monoclonal | Mouse | WB: 1/500 | NeoMarkers, Thermo Scientific | MS‐611‐P0 |
| Rootletin (CROCC) | Primary, polyclonal | Rabbit | IF: 1/200 | NovusBio | NBP1‐80820 |
| GFP (clone 3E6) | Primary, monoclonal | Mouse | IF: 1/250 | Invitrogen | A‐11120 |
| α‐tubulin (clone DM1A) | Primary, monoclonal | Mouse | WB: 1/5000 | Sigma‐Aldrich | T9026 |
| ECL™ Anti‐mouse IgG, HRP‐conjugated | Secondary, polyclonal | Sheep | WB: 1/5000 | GE Healthcare UK Limited | NA931V |
| AlexaFluor‐488 conjugated anti‐mouse | Secondary, polyclonal | Chicken | IF: 1/500 | Invitrogen | A21200 |
| AlexaFluor‐488 conjugated anti‐rabbit | Secondary, polyclonal | Donkey | IF: 1/500 | Invitrogen | A10040 |
| AlexaFluor‐546 conjugated anti‐mouse secondary | Secondary, polyclonal | Goat | IF: 1/500 | Invitrogen | A11003 |
Abbreviations: IF, immunofluorescence; WB, Western blot.
2.4. Immunofluorescence
Cells were grown on Lab‐Tek II Chamber Slides (154526; Thermo Fisher Scientific). Cells were washed three times with PBS and fixed in 4% paraformaldehyde in PBS for 20 min at RT. After one PBS wash, cells were permeabilized with 0.05% Triton‐X‐100 in PBS for 10 min at RT. Cells were washed in PBS and blocked using DAKO Real™ Antibody Diluent (S2022; Agilent Technologies) for 1 h at RT. In case of the use of SMO IN/OUT assay visualizing outer ciliary portion, no permeabilization was applied. Primary antibodies (Table 1) were diluted in DAKO Real™ Antibody Diluent. Cells were incubated with primary antibody for 2 h at RT or O/N at 4°C. Three repeated washes with PBS, each lasting 5 min, were applied between the use of primary and secondary antibody. Cells were incubated with secondary antibodies (Table 1) for 1 h at RT. After three repeated washes, nuclei were stained using DAPI (0.5 μg/ml PBS) for 5 min at RT. Slides were then mounted with fluorescent Dako mounting medium (S3023; Dako Schweitz AG, Baar, Switzerland) and assessed at RT using an inverted Zeiss LSM 700 laser scanning confocal microscope equipped with laser diode 405/488/555, SP490/SP555/LP560 emission filters, 2 PMT detectors and Zen2010 software (Zeiss, Feldbach, Switzerland).
To assess PC, three‐dimensional z‐stack imaging technique was used. Confocal images were taken using 63×/1.40 oil objectives, with intervals ranging from 0.33 to 1 μm. For all images within one individual experiment, the microscope settings (laser intensities, exposure time) remained identical. Using Image J, sum slices projection method was applied (Image J: Stacks: Z project: Projection type: Sum slices). Ciliary length was measured manually, using the straight, segmented or freehand line tool corresponding to fluorescent signal of acetylated tubulin, marker of the ciliary axoneme. Twenty to 50 cilia were measured for each condition. The mean cilia prevalence was expressed as the percentage of ciliated cells and the average PC length was expressed in μm.
2.5. Data analysis and reproducibility
Data analyses were performed using Prism GraphPad V 9.1.0 (GraphPad Software, Inc., La Jolla, CA, USA). All experiments were performed at least three times. Statistical significance was depicted as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. To compare an independent variable in more than two groups, we used one‐way ANOVA test. The equality of variances was assessed by Brown‐Forsynthe and Bartlett's test. If variances differed significantly, we used Holm‐Šidák's correction. If two groups were compared, unpaired t‐test was used. If F test to compare variances showed significant difference, Welch's correction was applied.
AUTHOR CONTRIBUTIONS
G.B. designed the experiments, analysed the results and wrote the manuscript. C.P. and D.H. supervised the project.
CONFLICT OF INTEREST
The authors declare no competing interests.
ACKNOWLEDGEMENTS
This study was funded by Swiss National Science Foundation (grant 310030‐173102) and “Fondation Dind Cottier pour la recherche sur la peau”. Open Access funding enabled and organized by Projekt DEAL.
Blanchard G, Pich C, Hohl D. HaCaT cells as a model system to study primary cilia in keratinocytes. Exp Dermatol. 2022;31:1276‐1280. doi: 10.1111/exd.14626
DATA AVAILABILITY STATEMENT
The authors confirm that the data supporting the findings of this study are available within the article.
REFERENCES
- 1. Goetz SC, Anderson KV. The primary cilium: a signalling Centre during vertebrate development. Nat Rev Genet. 2010;11(5):331‐344. doi: 10.1038/nrg2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Singla V, Reiter JF. The primary cilium as the cell's antenna: signaling at a sensory organelle. Science. 2006;313(5787):629‐633. doi: 10.1126/science.1124534 [DOI] [PubMed] [Google Scholar]
- 3. Verhey KJ, Gaertig J. The tubulin code. Cell Cycle. 2007;6(17):2152‐2160. doi: 10.4161/cc.6.17.4633 [DOI] [PubMed] [Google Scholar]
- 4. Yu I, Garnham CP, Roll‐Mecak A. Writing and reading the tubulin code. J Biol Chem. 2015;290(28):17163‐17172. doi: 10.1074/jbc.R115.637447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Malicki JJ, Johnson CA. The cilium: cellular antenna and central processing unit. Trends Cell Biol. 2017;27(2):126‐140. doi: 10.1016/j.tcb.2016.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reiter JF, Leroux MR. Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol. 2017;18(9):533‐547. doi: 10.1038/nrm.2017.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Park HS, Papanastasi E, Blanchard G, et al. ARP‐T1‐associated Bazex‐Dupré‐Christol syndrome is an inherited basal cell cancer with ciliary defects characteristic of ciliopathies. Commun Biol. 2021;4(1):544. doi: 10.1038/s42003-021-02054-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Anvarian Z, Mykytyn K, Mukhopadhyay S, Pedersen LB, Christensen ST. Cellular signalling by primary cilia in development, organ function and disease. Nat Rev Nephrol. 2019;15(4):199‐219. doi: 10.1038/s41581-019-0116-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Croyle MJ, Lehman JM, O'Connor AK, et al. Role of epidermal primary cilia in the homeostasis of skin and hair follicles. Development. 2011;138(9):1675‐1685. doi: 10.1242/dev.060210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hosio M, Jaks V, Lagus H, Vuola J, Ogawa R, Kankuri E. Primary ciliary signaling in the skin‐contribution to wound healing and scarring. Front Cell Dev Biol. 2020;8:578384. doi: 10.3389/fcell.2020.578384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hassounah NB, Bunch TA, McDermott KM. Molecular pathways: the role of primary cilia in cancer progression and therapeutics with a focus on hedgehog signaling. Clin Cancer Res. 2012;18(9):2429‐2435. doi: 10.1158/1078-0432.CCR-11-0755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kuonen F, Huskey NE, Shankar G, et al. Loss of primary cilia drives switching from hedgehog to Ras/MAPK pathway in resistant basal cell carcinoma. J Invest Dermatol. 2019;139(7):1439‐1448. doi: 10.1016/j.jid.2018.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fusenig NE, Boukamp P. Multiple stages and genetic alterations in immortalization, malignant transformation, and tumor progression of human skin keratinocytes. Mol Carcinog. 1998;23(3):144‐158. doi: [DOI] [PubMed] [Google Scholar]
- 14. Colombo I, Sangiovanni E, Maggio R, et al. HaCaT cells as a reliable in vitro differentiation model to dissect the inflammatory/repair response of human keratinocytes. Mediators Inflamm. 2017;2017:7435621. doi: 10.1155/2017/7435621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106(3):761‐771. doi: 10.1083/jcb.106.3.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wilson VG. Growth and differentiation of HaCaT keratinocytes. Methods Mol Biol. 2014;1195:33‐41. doi: 10.1007/7651_2013_42 [DOI] [PubMed] [Google Scholar]
- 17. Choi YJ, Laclef C, Yang N, et al. RPGRIP1L is required for stabilizing epidermal keratinocyte adhesion through regulating desmoglein endocytosis. PLoS Genet. 2019;15(1):e1007914. doi: 10.1371/journal.pgen.1007914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Takahashi K, Nagai T, Chiba S, Nakayama K, Mizuno K. Glucose deprivation induces primary cilium formation through mTORC1 inactivation. J Cell Sci. 2018;131(1):jcs208769. doi: 10.1242/jcs.208769 [DOI] [PubMed] [Google Scholar]
- 19. Rajapakse D, Peterson K, Mishra S, Wistow G. Serum starvation of ARPE‐19 changes the cellular distribution of cholesterol and Fibulin3 in patterns reminiscent of age‐related macular degeneration. Exp Cell Res. 2017;361(2):333‐341. doi: 10.1016/j.yexcr.2017.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kukic I, Rivera‐Molina F, Toomre D. The IN/OUT assay: a new tool to study ciliogenesis. Cilia. 2016;5:23. doi: 10.1186/s13630-016-0044-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bernabé‐Rubio M, Alonso MA. Routes and machinery of primary cilium biogenesis. Cell Mol Life Sci. 2017;74(22):4077‐4095. doi: 10.1007/s00018-017-2570-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mirvis M, Stearns T, James NW. Cilium structure, assembly, and disassembly regulated by the cytoskeleton. Biochem J. 2018;475(14):2329‐2353. doi: 10.1042/BCJ20170453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Labat‐de‐Hoz L, Rubio‐Ramos A, Casares‐Arias J, Bernabé‐Rubio M, Correas I, Alonso MA. A model for primary cilium biogenesis by polarized epithelial cells: role of the midbody remnant and associated specialized membranes. Front Cell Dev Biol. 2021;8:622918. doi: 10.3389/fcell.2020.622918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rizaldy D, Toriyama M, Kato H, et al. Increase in primary cilia in the epidermis of patients with atopic dermatitis and psoriasis. Exp Dermatol. 2021;30(6):792‐803. doi: 10.1111/exd.14285 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.


