ABSTRACT
Patients presenting with severe COVID-19 are predisposed to acquire secondary fungal infections such as COVID-19-associated candidemia (CAC), which are associated with poor clinical outcomes despite antifungal treatment. The extreme burden imposed on clinical facilities during the COVID-19 pandemic has provided a permissive environment for the emergence of clonal outbreaks of multiple Candida species, including C. auris and C. parapsilosis. Here we report the largest clonal CAC outbreak to date caused by fluconazole resistant (FLZR) and echinocandin tolerant (ECT) C. parapsilosis. Sixty C. parapsilosis strains were obtained from 57 patients at a tertiary care hospital in Brazil, 90% of them were FLZR and ECT. Although only 35.8% of FLZR isolates contained an ERG11 mutation, all of them contained the TAC1L518F mutation and significantly overexpressed CDR1. Introduction of TAC1L518F into a susceptible background increased the MIC of fluconazole and voriconazole 8-fold and resulted in significant basal overexpression of CDR1. Additionally, FLZR isolates exclusively harboured E1939G outside of Fks1 hotspot-2, which did not confer echinocandin resistance, but significantly increased ECT. Multilocus microsatellite typing showed that 51/60 (85%) of the FLZR isolates belonged to the same cluster, while the susceptible isolates each represented a distinct lineage. Finally, biofilm production in FLZR isolates was significantly lower than in susceptible counterparts Suggesting that it may not be an outbreak determinant. In summary, we show that TAC1L518F and FKS1E1393G confer FLZR and ECT, respectively, in CAC-associated C. parapsilosis. Our study underscores the importance of antifungal stewardship and effective infection control strategies to mitigate clonal C. parapsilosis outbreaks.
KEYWORDS: Candida parapsilosis, outbreak, candidemia, fluconazole resistance, echinocandin tolerance
Introduction
The global pandemic of COVID-19 during the last two years has had a profound impact on healthcare settings and predisposed a significant number of patients to develop secondary bacterial and fungal infections [1]. COVID-19-associated candidemia (CAC) is one of the most frequently observed fungal infections complicating COVID-19 [1]. Although the mortality rates among the COVID-19 patients admitted to ICUs are notably high, development of CAC further significantly increases these mortality rates [2]. More importantly, the limited availability of personal protective equipment and the crowdedness of hospital units have created a permissive environment for emergence of outbreaks due to Candida auris [3,4] and C. parapsilosis [5,6]. Indeed, a recent study from Brazil documented a large outbreak of fluconazole resistant C. parapsilosis (FLZR-CP) isolates involving 30 patients in a cardiology ward, which continued despite the application of ethanol-based disinfectant [6]. Similarly, persistence of such outbreaks even after extensive application of quaternary ammonium-based disinfectant has also been reported prior to COVID-19 pandemic [7]. Because FLZR-CP isolates are associated with significantly higher mortality, the emergence of such outbreaks could lead to poorer clinical outcomes [8,9]. In fact, the persistence of such infections has been a motive behind the change in clinical practice and replacement of fluconazole with echinocandins for patients infected with C. parapsilosis in centres dealing with clonal FLZR-CP outbreaks, which in turn is leading to the emergence of multidrug-resistant C. parapsilosis isolates [10]. The extensive use of antibiotics and antifungals during COVID-19 may also contribute to worsening the problem of the antimicrobial resistance in the aftermath of the pandemic [11,12]. In this scenario, identification of the source of infection in conjunction with effective infection control strategies and antifungal stewardship are instrumental in lowering the risk of antifungal resistance.
Although fluconazole resistance in C. parapsilosis is primarily mediated by ERG11 mutations affecting the binding of drug to its target, such as Y132F and K143R, other factors, including overexpression of efflux pumps (CDR1 and MDR1) and of ERG11 have been reported among FLZR-CP isolates [13–15]. Such overexpression is mainly driven by gain-of-function (GOF) mutations in transcription factors regulating CDR1, MDR1, and ERG11, namely TAC1, MRR1, and UPC2 [16]. Nonetheless, the potential direct contribution of such GOF mutations to the overexpression of their target genes is still poorly explored in C. parapsilosis [17]. Although echinocandin resistance (ECR) is less frequently encountered than FLZR in C. parapsilosis, a recent study identified R658G in the Fks1 hotspot 1 (HS1) of MDR in C. parapsilosis isolates [10], while other studies have identified either FKS mutation outside of the HS region [18] or no FKS mutations at all [19]. Although less studied in Candida species compared to pathogenic bacterial species, the concept of antifungal tolerance is increasingly being encountered in Medical Mycology, which has the potential to negatively impact therapeutic success, as well as pave the way for emergence of stable antifungal resistance [20,21]. Antifungal tolerance is defined as reduced in vitro susceptibility in the absence of known resistance mechanisms, and measurement strategies vary depending on the killing dynamic, where azole tolerance is defined as slow growth above the minimum inhibitory concentrations (MIC) after 48 h using E-test and broth microdilution assays [21]. Echinocandin tolerance, however, quantitatively measures the survival rate using colony forming unit (CFU) at any given time (arbitrary but typically up to 24 h) [20].
The advent of novel precise genetic tools, such as CRISPR-Cas9, has remarkably increased our understanding of fungal pathogenesis and antifungal resistance [22]. For instance, a recent study successfully employed this technique to confirm that Erg11-G458S, but not Erg11-L376I, confers azole resistance in C. orthopsilosis, a sibling species of C. parapsilosis [23]. However, such tools have not yet been broadly employed to dissect the role of specific mutations in antifungal drug resistance in C. parapsilosis.
Herein, we describe the largest to date clonal fungal outbreak in COVID-19 patients due to FLZR and echinocandin tolerant (ECT) C. parapsilosis in a single referral hospital in Salvador, Brazil. We also use the CRISPR-Cas9 technology in C. parapsilosis to further our understanding of fluconazole resistance and echinocandin tolerance in this fungal pathogen. Collectively, our study cautions against the extensive use of antifungal drugs in severely ill COVID-19 patients and suggests that the implementation of strict antifungal stewardship and effective infection control strategies are required to prevent the occurrence of antifungal drug-resistant fungal outbreaks. Importantly, our study also advocates for FKS sequencing even among susceptible Candida isolates, showing that mutations outside the canonical HS regions should not be overlooked.
Methods
Patients, isolate collection, and identification
Severely ill COVID-19 patients referred to São Rafael hospital located in Salvador, Brazil, who presented with candidemia due to C. parapsislosis were recruited to the current study. Candidemia was defined when C. parapsilosis was recovered from blood samples. Our hospital has 329 beds and gives care to adult and paediatric patients, and it was also one of the major referral centres during the COVID-19 pandemic, with an admission rate of 1315 and 1644 patients during 2020 and 2021, respectively. C. parapsilosis isolates recovered from the blood samples of patients placed in a COVID-19 ICU were identified by ITS1 and ITS4 primers as described previously [24]. Any C. parapsilosis isolates recovered from the blood samples of COVID-19 patients, including sequential isolates, were included and investigated in the current study. This study was approved by local ethical committee of our centre (5.412.257).
Antifungal susceptibility testing (AFST)
AFST used the broth microdilution of CLSI M27/A3 protocol [25]. Fluconazole, voriconazole, Amphotericin B (AMB) (all from Sigma-Aldrich, St. Louis, MO, United States), micafungin, anidulafungin (both from Pfizer, New York, NY, United States) were included. MICs were assessed visually after 24 h incubation at 37°C. Isolates with MIC≥ 8 µg/ml were considered as fluconazole, micafungin, and anidulafungin resistant, while those with MIC≥ 1 µg/ml were defined as voriconazole resistant [26].
Multi-locus microsatellite typing (MLMT)
C. parapsilosis sensu stricto isolates and the reference strain, ATCC 22019, were subjected to a previously described MLMT approach [27], which PCR amplified eight different loci. After separation on 3% agarose gel, PCR products were stained with GelRed™ (Biotium, Fremont, CA, USA), and visualized with the UVITEC gel documentation system (Cleaver Scientific, Rugby, Warks, UK). Dice coefficient was used to examine the allelic profiles and Bionumerics software v. 7.6 (Applied Maths, Sint-Martens-Latem, Belgium) was used for clustering using unweighted pair group method with arithmetic mean (UPGMA) employing the. Cluster was defined, when ≥2 isolates showed an identical allelic profile [28,29].
Analysis of biofilm production
To assess biofilm formation, we used a previously described protocol [30] with a few modifications. Briefly, C. parapsilosis isolates were grown on YPD-agar overnight at 37°C and a single colony was inoculated into 5 ml YPD broth and incubated at 37°C for overnight (150 rpm). The next day, the OD600 nm of the isolates was adjusted to 1 in YPD broth, and 200 µl from each culture (in triplicate) were transferred to a 96-well microtiter plate and incubated at 37°C for 24 without shaking. After 24 h, the nonadherent cells were removed by washing with distilled water three times, and after air-drying the biofilms, 100 µl of 0.1% (w/v) crystal violet were added to each well and incubated at 37°C for 30 min. Subsequently, the plates were washed three times with distilled water, air-dried, and 200 µl of a solution containing 1% (w/v) SDS and 50% ethanol was added to release the biofilms. Finally, using a plate reader (Infinite®PRO, TECAN) the crystal violet absorbance was measured at OD490nm. The biofilm formation for each isolate was measured using two biological replicates in triplicate.
Sequencing
PCR amplification and sequencing of ERG11, HS1 and HS2 of FKS1, TAC1, UPC2, and MRR1 were performed as described previously [31]. After assembly and curation of the sequence data, they were aligned against their WT sequences (ERG11 = GQ302972, TAC1 = HE605204, MRR1 = HE605205, UPC2 = HE605206, and FKS1 = EU221325.1).
RNA extraction and gene expression analysis
Overnight C. parapsilosis cultures (150 rpm and 37°C) were washed with PBS once, and the OD600nm of the cultures was adjusted at 0.5 using fresh YPD, followed by incubation at 37°C and 250 rpm for another 6 h. Upon washing with PBS, C. parapsilosis isolates (105 cells/ml), were incubated in RPMI 1640 containing fluconazole one dilution below the minimum inhibitory concentration (MIC) at 37°C and 250 rpm for 90 min. The pellets were then collected by centrifugation (13,000 rpm for 5 min) and stored at −80°C. RNA samples were extracted using a previously described approach [32], subjected to DNase treatment (QIAGEN), and finally repurified using an RNeasy mini-Kit (QIAGEN) as per the manufacturer's suggestion.
qPCR was performed using the primers described previously [33], which included One-Step TB Green PrimeScript RT–PCR Kit II (Perfect Real Time, TaKaRa, Shiga, Japan). qPCRs containing 40 ng of RNA samples, 0.4 µM of primers, 0.8 µl of enzyme and 10 µl of buffer in a final volume of 20 µl were subjected to an Mx3005P qPCR System (Agilent Technologies, Santa Clara, USA).
Experiments were carried out in two biological and at least two technical replicates, and gene expression data were normalized against ACT1 gene [33]. Fold changes were determined using normalized data of C. parapsilosis cells treated with fluconazole relative to untreated initial inoculums of each sample using 2−ΔΔCT as described previously [34]. Overexpression was defined as a fold change ≥2 relative to the untreated cells. Basal expression values for each untreated samples were calculated using the following formula: 2−ΔCt, where the ΔCt refers to Ct gene of target minus the Ct ACT1.
Micafungin tolerance
Overnight cultures of C. parapsilosis isolates (37°C and 150 rpm) were washed twice with PBS and 50 µl of 2 × 108 cells were inoculated in 1 ml of RPMI1640 containing 4 µg/ml of micafungin. We used this concentration since it differentiates the susceptible from non-susceptible C. parapsilosis isolates. Cultures were incubated at 37°C and 150 rpm and plating was performed at each time-points (3, 6, and 24 h). Colony forming units (CFUs) of treated isolates were normalized against untreated positive controls. This experiment involved three biological replicates of two independent FKS1 mutants carrying G1393E and the wild-type (WT) parental strains.
Introduction of single nucleotide polymorphisms (SNPs) in the ATCC22019 background using CRISPR-Cas9
We used the pCP-tRNA CRISPR-Cas9 plasmid-based system [35] to introduce mutations in the sequences of TAC1 and FKS1 into the C. parapsilosis ATCC 22019 background. Suitable protospacer adjacent motif (PAM) sequences targeting TAC1 and FKS1 were selected using EuPaGDT [36], based on their specificity and proximity to the desired cut site (TAC1-g1 and FKS1-g1, see Table 1). Each guide RNA was then generated by annealing of two 23-bp oligonucleotides carrying appropriate overhanging ends and was cloned into the SapI-digested pCP-tRNA plasmid (see Table 1). Each Repair Template (RT; TAC1-RT1 and FKS-RT1, Table 1) was designed to: (i) introduce the desired amino acid change (L518F into Tac1; E1393G into Fks1), and (ii) introduce synonymous SNPs to prevent Cas9 from cutting the edited site by changing the seed sequence [37]. The 100-nt RTs were generated with ExTaq DNA polymerase (TaKaRa Bio, USA) by primer extension from two oligonucleotide primers with 20 bp overlaps at the 3′-ends.
Table 1.
Oligonucleotides used to generate mutant C. parapsilosis isolates carrying TAC1L518F and FKS1E1393G.
|
Production of the guide RNA The protospacer sequence/PAM site is indicated, followed by the 23-mers (TOP and BOT oligos) for generating the guide RNA. Overhangs are highlighted in bold | ||
| TAC1-g1 (sequence) | 5′-GTGGCTGATGAGGCATTACT/TGG-3′ | |
| TAC1-g1-TOP | 5′-CCAGTGGCTGATGAGGCATTACT-3′ | Annealing of the 23-mer to produce the guide RNA that was cloned into SapI-digested pCP-tRNA |
| TAC1-g1-BOT | 5′-AACAGTAATGCCTCATCAGCCAC-3′ | |
| FKS1-g1 (sequence) | 5′-AGTTGATTGAAAGAGGTGTG/TGG-3′ | |
| FKS1-g1-TOP | 5′-CCAAGTTGATTGAAAGAGGTGTG-3′ | Annealing of the 23-mer to produce the guide RNA that was cloned into SapI-digested pCP-tRNA |
| FKS1-g1-BOT | 5′-AACCACACCTCTTTCAATCAACT-3′ | |
|
Synthesis of RTs The sequence of each RT is indicated, followed by the long oligos (TOP and BOT) used for primer extension. The SNPs are highlighted in bold, and described on the right (the numbering refers to the ORF); syn = synonymous | ||
| TAC1-RT-1 (sequence) | 5′-GGCGACGAATTGGATCGTCAAATGTCGATTGCAGTGGCTGATGAGGCATTGTTTGGAGATCCAGCACTACCACTCAGTTTTCGATTGTTGAAAAAGTTGA-3′ | SNPs introduced: A1551G [syn SNP]; C1552T [non syn SNP] |
| TAC1-RT-1-TOP | 5′-GGCGACGAATTGGATCGTCAAATGTCGATTGCAGTGGCTGATGAGGCATTGTTTGGAGAT-3′ | |
| TAC1-RT-1-BOT | 5′-TCAACTTTTTCAACAATCGAAAACTGAGTGGTAGTGCTGGATCTCCAAACAATGCCTCAT-3′ | |
| FKS1-RT-1 (sequence) | 5′-TCTTCATTTCGTTCATTCCATTGGTTGTTCAAGGGTTGATTGAAAGAGGAGTCTGGAAAGCTTGTCAAAGATTTGTTAGACATTTCATTTCGTTGTCACC-3′ | A4178G [non syn SNP]; G4197C [syn SNP]; T4194A [syn SNP] |
| FKS1-RT-1-TOP | 5′-TCTTCATTTCGTTCATTCCATTGGTTGTTCAAGGGTTGATTGAAAGAGGAGTCTGGAAAG-3′ | |
| FKS1-RT-1-BOT | 5′-GGTGACAACGAAATGAAATGTCTAACAAATCTTTGACAAGCTTTCCAGACTCCTCTTTCA-3′ | |
| Sequencing of mutated loci | ||
| sTAC1-F | 5′-GGTATGCTCAGGAGATTGGA-3′ | Sequencing of TAC1 edited site |
| sTAC1-R | 5′-ATAGTTCCACGTTCAGGCTC-3′ | |
| sFKS1-F | 5′-CGGACATCCTGGTTTCCATA-3′ | Sequencing of FKS1 edited site |
| sFKS1-R | 5′-CAATGAAGACAACGAAGCCC-3′ | |
Yeast cells were transformed with 5 μg of the relevant plasmid and 25 μl of unpurified RT using the lithium acetate method described in ref. [38], with minor modifications (starting OD600nm of YPD culture 0.1 instead of 0.05). Transformants were plated onto parallel YPD agar plates containing 200 μg/ml nourseothricin (Jena Bioscience GmbH, Germany) and incubated at 30°C for 48 h. Representative mutants were sequenced by Sanger sequencing (MWG/Eurofins). For each mutant strain (LL-TAC1 and LL-FKS1), two independent lineages (A and B) carrying the desired mutations were patched onto YPD agar without selection twice to induce the loss of the pCP-tRNA plasmid. The resulting strains carried the desired mutations and did not contain the Cas9-expressing plasmid anymore.
Statistical analysis
Excel (Microsoft, Redmond, WA, USA) was used to carry out the statistical analysis. T-test (tow-tailed) was used to define the statistical significance for biofilm formation and echinocandin tolerance. Values < 0.05 were considered as statistically significant.
Results and discussion
We evaluated a total of 60 isolates cultured from 57 patients who had developed C. parapsilosis candidemia in the COVID-19 ICU with three of them had two sequential positive blood bottles. Four (7%) of the patients had previous exposure to fluconazole, while 42 (73.7%) had received an echinocandin before fungemia (median 14 days, interquartile range 9–14 days) (Supplementary Table 1). Most of the patients (n = 54, 93%) had a central venous catheter when fungemia was diagnosed, and the 30-day overall mortality rate was 59.6%.
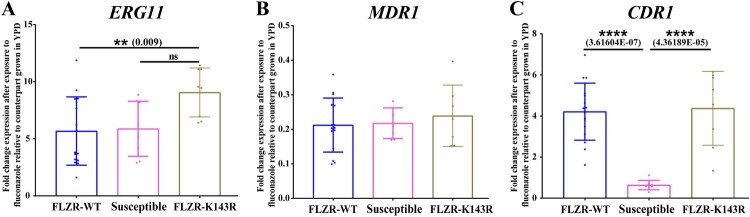
Our AFST data revealed that 53 of 60 (88.3%) isolates tested were FLZR (≥ 8 µg/ml), which all had intermediate phenotype against voriconazole (Table 2 and supplementary Table 1); while none of the isolates showed resistance to echinocandins and AMB. Although susceptible to both micafungin and anidulafungin, all FLZR isolates had one or two dilution higher MICs compared to FLZS counterparts (Supplementary Table 1). Interestingly, the four patients with prior exposure to fluconazole were infected with FLZR isolates. To delineate the mechanism of FLZR, we first sequenced ERG11 and to our surprise, only 35.1% of these isolates carried a ERG11 mutation, K143R, which is a well-known mutation conferring fluconazole resistance in numerous Candida species [16]. Of note, the fluconazole MIC of FLZR isolates carrying K143R was >8 µg/ml. Because the vast majority of the FLZR isolates were WT for ERG11 (FLZR-WT hereafter), we suspected the involvement of GOF mutations in major transcription factors (TFs), i.e. TAC1, UPC2, and MRR1. Therefore, we selected 16 C. parapsilosis isolates, including 10 FLZR without ERG11 mutation, 3 FLZR carrying K143R and 3 FLZS, and sequenced the aforementioned TFs. Interestingly, all the FLZR isolates sequenced contained a mutation in TAC1, L518F, which was missing in reference and susceptible strains, while all the isolates were WT for UPC2 and MRR1. This mutation is located in the middle homology region, which is thought to negatively control the activating domain in the C terminus [39]. Therefore, such mutations could potentially render Tac1 active, which subsequently results in overexpression of genes under the control of this transcription factor, including CDR1 [39]. Consequently, we suspected that FLZR isolates, but not FLZS counterparts, should overexpress CDR1 after fluconazole exposure. To test this hypothesis, we selected eight of the sequenced strains, including four FLZR without ERG11 mutations and two FLZS isolates, exposed them to fluconazole at half the MIC for 90 min and measured the level of expression of CDR1, ERG11, and MDR1 (Table 3). While ERG11 was highly expressed among both FLZR and FLZS isolates (Figure 1A) and MDR1 was downregulated in all isolates tested (Figure 1B), only FLZR isolates overexpressed CDR1 (Figure 1C). On the other hand, the basal expression of CDR1 was significantly lower in FLZR isolates than in FLZS isolates (Supplementary Figure 1A–C). This observation further reinforced our hypothesis that TAC1L518F could confer the overexpression of CDR1 upon exposure to fluconazole and therefore confers fluconazole resistance.
Table 2.
Minimum inhibitory concentration of antifungal agents used against Candida parapsilosis isolates (n = 60). The number of isolates for each concentration of a given drug is indicated.
| Antifungal agent | Minimum inhibitory concentration (µg/ml) | MIC50 | MIC90 | GM | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.016 | 0.032 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | ||||
| Fluconazole | 4 | 3 | 53a | 8 | 16–32 | 6.64 | ||||||||||
| Voriconazole | 4 | 1 | 2 | 53 | 0.032 | 0.5 | 0.192 | |||||||||
| Micafungin | 3 | 4 | 53 | 4 | 8 | 3.52 | ||||||||||
| Anidulafungin | 1 | 6 | 53 | 4 | 8 | 3.56 | ||||||||||
| Amphotericin B | 1 | 47 | 12 | 0.5 | 1 | 0.567 | ||||||||||
Note that 19 strains carrying K143R in Erg11 and L518F in Tac1 had fluconazole MICs >8 µg/ml.
Table 3.
The characteristics of Candida parapsilosis isolates selected for sequencing and gene expression analysis. The expression profile values of the genes studied are based on the average ± standard deviation.
| Strain # | FLZ (µg/ml) | VOR (µg/ml) | MICA (µg/ml) | ANI (µg/ml) | HS2-Fks1 | Erg11 | CDR1 expression | ERG11 expression | MDR1 expression | Tac1 | Upc2 | Mrr1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 8 | 0.25 | 2 | 2 | E1393G | WT | 2.64 ± 0.94 | 0.19 ± 0.004 | 9.32 ± 1.86 | L518F | WT | WT |
| 33 | 16 | 0.25 | 2 | 2 | E1393G | WT | 5.91 ± 0.80 | 0.11 ± 0.02 | 7.16 ± 1.37 | L518F | WT | WT |
| 40 | 8 | 0.25 | 2 | 2 | E1393G | WT | 4.57 ± 0.39 | 0.31 ± 0.03 | 3.25 ± 0.38 | L518F | WT | WT |
| 51 | 8 | 0.25 | 2 | 2 | E1393G | WT | 3.69 ± 0.53 | 0.24 ± 0.03 | 2.95 ± 0.98 | L518F | WT | WT |
| 7 | 8 | 0.25 | 2 | 2 | E1393G | K143R | 2.93 ± 1.36 | 0.16 ± 0.01 | 8.6 ± 2.3 | L518F | WT | WT |
| 27 | 16 | 0.25 | 2 | 2 | E1393G | K143R | 5.81 ± 0.37 | 0.29 ± 0.07 | 9.50 ± 2.2 | L518F | WT | WT |
| 10 | 1 | 0.03 | 1 | 1 | WT | WT | 0.52 ± 0.16 | 0.24 ± 0.02 | 7.91 ± 1.11 | WT | WT | WT |
| 20 | 0.5 | 0.015 | 0.5 | 0.5 | WT | WT | 0.74 ± 0.25 | 0.20 ± 0.205 | 3.83 ± 1.08 | WT | WT | WT |
FLZ: Fluconazole; VOR: Voriconazole; MICA: Micafungin; ANI: Anidulafungin.
Figure 1.
Expression profile of ERG11 (A), MDR1 (B), and CDR1 (C) from a selected number of C. parapsilosis isolates (n = 8) after exposure to fluconazole, which showed that fluconazole-resistant isolates (FLZR) significantly overexpressed CDR1 relative to susceptible ones. C. parapsilosis isolates grown at logarithmic phase were subjected to one dilution below MIC of fluconazole for 90 min, and after RNA extraction, relative gene expression was assessed as described in methods section.
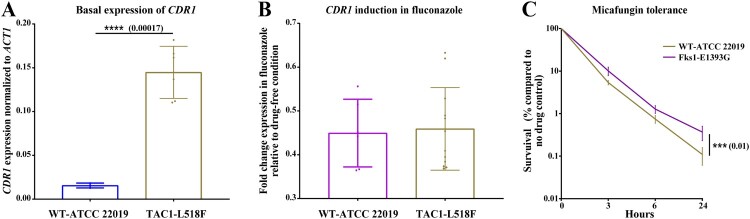
To ascertain if TAC1L518F confers fluconazole resistance by inducing CDR1 overexpression, we used a plasmid-based CRISPR-Cas9 system to introduce this mutation into a susceptible tester strain background, ATCC 22019 [23]. Indeed, the fluconazole and voriconazole MIC of mutants carrying this mutation was increased 8- and 4-fold, 0.5 µg/ml vs 4 µg/ml and 0.03 µg/ml vs 0.125 µg/ml, respectively (Table 4). Unexpectedly, two independent mutants carrying TAC1L518F showed only basal CDR1 overexpression (Figure 2A), while fluconazole exposure at three different concentrations (2, 4, and 8 µg/ml) did not cause CDR1 overexpression (Figure 2B). Therefore, we reasoned that the increase in the basal expression of CDR1 was sufficient to confer fluconazole resistance. We speculate that basal overexpression of CDR1 may carry a fitness-cost given that such isolates would constitutively overexpress CDR1, which carries a high energy demand. However, given the nutritional immunity imposed by host restrict ATP production, infecting several patients may allow the clinical isolates to overcome this fitness cost by controlling the high basal expression of CDR1 and only induce overexpression in the presence of azole. This observation deserves deeper investigation, including application of whole-genome sequencing and transcriptomic analysis to unravel the mechanisms underpinning differential expression of CDR1 in FLZR C. parapsilosis.
Table 4.
The minimum inhibitory concentration of antifungal drugs against the mutants carrying L518F in Tac1 and E1393G in Fks1 and their parental wild-type (WT) strain.
| Strain | Phenotype | Fluconazole (µg/ml) | Voriconazole (µg/ml) | Micafungin (µg/ml) |
|---|---|---|---|---|
| WT | Susceptible | 0.5 | 0.03 | 2 |
| L518F | Fluconazole non-susceptible | 4 | 0.125 | 2 |
| E1393G | Micafungin tolerant | 0.5 | 0.03 | 2 |
Figure 2.
The expression profile of CDR1 for parental strain ATCC 22019 and its mutants carrying L518F in Tac1 and the micafungin tolerance of ATCC 22019 and it mutant carrying E1393G in Fks1. Mutants carrying TAC1L518F (from two independent mutants) had a significantly higher basal expression of CDR1, which was not induced upon fluconazole exposure (A and B). Mutants carrying FKS1E1393G had a significantly higher tolerance to micafungin (4 µg/ml), which shows the average of two independent mutants.
Because our FLZR isolates also had higher echinocandin MICs compared to susceptible isolates, we sought to sequence HS1 and HS2 of FKS1. Surprisingly, all the FLZR isolates carried a nonsynonymous mutation outside of the HS2, E1393G (Supplementary Table 1). Interestingly, E1393 is a highly conserved amino acid across a wide range of fungal species ranging from Saccharomyces cerevisiae and Aspergillus fumigatus to C. albicans; therefore, we wondered if this mutation could confer a higher echinocandin MIC once introduced into a susceptible background. Using CRISPR-Cas9, we introduced this mutation into C. parapsilosis ATCC 22019, selected two independent mutants carrying this mutation and subjected them to AFST using micafungin and anidulafungin. We found that the mutants and susceptible parental strain showed the same MIC for both echinocandins (Table 4). Bearing in mind that AFST is a qualitative growth/no growth test and that echinocandins are fungicidal in Candida, we sought to gain a deeper insight into the impact of E1393G on killing by echinocandins. To this end, we exposed the mutants and the parental strains to 4 µg/ml micafungin, which is an intermediate concentration (i.e. below the MIC of resistant strains but above the sensitivity of the wild type strain), and measured survival using CFU counts at different time-points (3, 6, and 24 h) after drug exposure. Interestingly, we found that the FKS1E1393G mutants had a significantly increased survival at all time-points, especially at 24 h (Figure 2C). Therefore, although this FKS1E1393G does not confer echinocandin resistance as clinically defined, it renders isolates carrying this mutation significantly more tolerant to echinocandins, if we define tolerance as the ability to survive better in a given echinocandin concentration. Although such mutations could potentially modulate the echinocandins binding to β-glucan synthase, gaining a deeper understanding on this matter requires RNAseq studies involving both WT and mutants.
As we and others have previously hypothesized [20,21], the higher level of echinocandin tolerance could translate into a higher level of persistent colonization and a greater likelihood of emergence of stable ECR isolates carrying FKS1 HS mutations during treatment. We propose that due to the prevalence of ECT FKS1E1393G mutation, the emergence of such ECR isolates in our centre may only be a matter of time, underscoring the importance of using antifungals only when they are necessary. Indeed, the current guidelines advocate using echinocandins as the first line therapy to treat patients with candidemia because they exert fungicidal activity against several species of Candida, have a favourable safety profile, and are associated with better survival in a large patient-level quantitative review of randomized clinical trials [40,41]. Considering the inherent reduced susceptibility and the high rate of tolerance to echinocandins observed in our cohort of patients infected by FLZR C. parapsilosis, as well as the emerging reports of MDR C. parapsilosis isolates [10], lipid formulations of amphotericin B potentially may be a better choice for therapy [42,43]. The establishment of strict antifungal stewardship and appropriate use of antifungals for Candida species, especially those causing outbreaks, is of paramount importance to minimize the risk of emergence of antifungal resistance. Importantly, in vitro studies have found that caspofungin treatment is associated with the highest mutation frequency in FKS compared to micafungin and anidulafungin and the emergence of ECR in C. glabrata [44], which may also warn against the high use of this drug in our centre, as its use may further consolidate the prevalence of such tolerant C. parapsilosis isolates.
Consistent with the present study, a recent report identified a few mutations outside of FKS1 HS1 and HS2 in C. parapsilosis but the authors only performed AFST and did not introduce this mutation into a sensitive strain to explore its effect on echinocandin-mediated killing quantitatively [18]. Moreover, ECR isolates lacking FKS1 mutations have also been reported [19], which implicate the involvement of as yet unknown mechanisms underlying echinocandin resistance. Mutations outside of the FKS HS regions in C. glabrata have been reported to confer ECR and therapeutic failure in vivo [45]. The phenomenon of ECR C. parapsilosis is becoming more predominant due to the heavy use of echinocandins in routine clinical practice. Collectively, these observations reinforce the importance of FKS sequencing even in echinocandin susceptible isolates. Furthermore, the importance of mutations outside of the FKS HS may be currently underestimated, and they may have a profound impact on in-host survival of such isolates during echinocandin exposure, facilitating and accelerating the emergence of echinocandin resistance.
Given the large number of patients infected over a short period of time and that approximately 90% of the isolates were both FLZR and ECT and harboured the same mutations, we suspected a large clonal outbreak. MLMT was performed, which identified nine minor clusters each represented by a single isolate (Figure 3) and two major clusters close to each other, one containing 80% of the isolates (48/60), and others comprising 2% of the isolates (3/60). Interestingly, each of the fluconazole susceptible isolates had a unique genotype. Therefore, our MLMT findings not only point to a severe clonal outbreak due to FLZR and ECT C. parapsilosis isolates, but they also document the largest clonal outbreak due to human fungal pathogens in the context of severely ill COVID-19 patients.
Figure 3.
Minimum spanning tree of C. parapsilosis isolates.
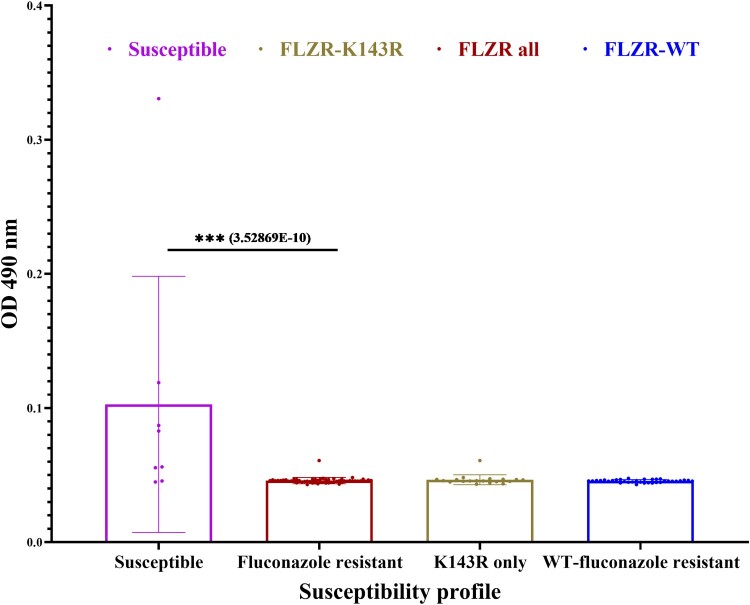
Because clonal outbreaks have been associated with increased biofilm production [46], we tested biofilm production of all isolates. To our surprise, none of the FLZR isolates produced biofilms, while the FLZS counterparts produced a higher biofilm level (Figure 4). It should be noted mature biofilm structure is intrinsically resistant to all classes of antifungal drugs, and even immune system, in the absence of genetic changes, whereas the mutation found in our FLZR isolates have been selected for in the presence of fluconazole and only confers protection against azoles in the planktonic conditions. This observation not only challenges the notion that biofilm production is a primary determinant of clonal outbreaks [46] but also questions the findings that biofilms could predict mortality [47]. Indeed, this observation is in line with recent findings obtained with Turkish FLZR C. parapsilosis isolates carrying ERG11Y132F, which did not produce biofilm but were associated with a significantly increased mortality rates in infected patients [8,9]. Since biofilm production is required for survival in hospital environments, such as on abiotic surfaces, we speculate that our outbreak was primarily transferred through skin. Of note, the skin samples for patients recruited in the current study were not available to ascertain this hypothesis. We acknowledge that the in-vitro biofilm formation tested in this study may not fully recapitulate the real-life conditions and therefore the application of the in-vivo catheter model is required to prove this observation.
Figure 4.
Biofilm formation of fluconazole resistant (FLZR-WT and FLZR-K143R) and fluconazole susceptible isolates. The Y-axis represents biofilm production as a function of absorption at OD490.
In conclusion, our study documented the largest clonal outbreak of candidemia due to fluconazole resistant and echinocandin tolerant C. parapsilosis isolates among COVID-19 patients, underscoring the importance of rigorous antifungal stewardship to minimize the risk of dangerous outbreaks due to MDR C. parapsilosis. Furthermore, our study determined the role of TAC1L518Fand FKS1E1393G in fluconazole resistance and echinocandin tolerance, respectively, through the application of CRISPR-Cas9 precise genome editing.
Supplementary Material
Acknowledgements
The authors are thankful to Dr. Carlos Henrique Camargo for helping us with the preparation of the MLMT figures. ALC received educational grants from Amgen, Eurofarma, Knight-United Medical, and Pfizer.
Funding Statement
This investigation was partially supported by Fundação de Amparo a Pesquisa do Estado de São Paulo (Grant FAPESP 2017/022037), Science Foundation Ireland (grant number 19/FFP/6668), and NIH grant R01AI109025 to DSP. This work was supported by internal support by the Center for Discovery and Innovation to D.S.P.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Arastehfar A, Carvalho A, Nguyen MH, et al. . COVID-19-associated candidiasis (CAC): an underestimated complication in the absence of immunological predispositions? . J Fungi (Basel, Switzerland). 2020;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arastehfar A, Shaban T, Zarrinfar H, et al. . Candidemia among Iranian patients with severe COVID-19 admitted to ICUs. J Fungi (Basel, Switzerland). 2021;7:56–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prestel C, Anderson E, Forsberg K, et al. . Candida auris outbreak in a COVID-19 Specialty Care Unit - Florida, July-August 2020. MMWR Morb Mortal Wkly Rep. 2021;70:56–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajni E, Singh A, Tarai B, et al. . A high frequency of Candida auris blood stream infections in Coronavirus disease 2019 patients admitted to intensive care units, northwestern India: a case control study. Open Forum Infect Dis. 2021;8:ofab452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arastehfar A, Ünal N, Hoşbul T, et al. . Candidemia among Coronavirus disease 2019 patients in Turkey admitted to intensive care units: a retrospective multicenter study. Open Forum Infect Dis. 2022;9:ofac078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomaz DY, Del Negro GMB, Ribeiro LB, et al. . A Brazilian inter-hospital candidemia outbreak caused by fluconazole-resistant Candida parapsilosis in the COVID-19 Era. J Fungi (Basel, Switzerland). 2022;8:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomaz DY, de Almeida JN Jr, Sejas ONE, et al. . Environmental clonal spread of azole-resistant Candida parapsilosis with Erg11-Y132F mutation causing a large candidemia outbreak in a Brazilian Cancer Referral Center. J Fungi. 2021;64(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arastehfar A, Daneshnia F, Hilmioğlu-Polat S, et al. . First report of candidemia clonal outbreak caused by emerging fluconazole-resistant Candida parapsilosis isolates harboring Y132F and/or Y132F+K143R in Turkey. Antimicrob Agents Chemother. 2020. doi: 10.1128/AAC.01001-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arastehfar A, Hilmioğlu-Polat S, Daneshnia F, et al. . Clonal candidemia outbreak by Candida parapsilosis carrying Y132F in Turkey: evolution of a persisting challenge. Front Cell Infect Microbiol. 2021;11:676177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arastehfar A, Daneshnia F, Hilmioglu-Polat S, et al. . Genetically related micafungin-resistant Candida parapsilosis blood isolates harbouring novel mutation R658G in hotspot 1 of Fks1p: a new challenge? J Antimicrob Chemother. 2021;76:418–422. [DOI] [PubMed] [Google Scholar]
- 11.Clancy CJ, Buehrle DJ, Nguyen MH.. PRO: the COVID-19 pandemic will result in increased antimicrobial resistance rates. JAC-antimicrobial Resist. 2020;2:dlaa049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai C-C, Chen S-Y, Ko W-C, et al. . Increased antimicrobial resistance during the COVID-19 pandemic. Int J Antimicrob Agents. 2021;57:106324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Souza ACR, Fuchs BB, Pinhati HMS, et al. . Candida parapsilosis resistance to fluconazole: molecular mechanisms and in vivo impact in infected Galleria mellonella Larvae. Antimicrob Agents Chemother. 2015;59:6581–6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Xiao M, Watts MR, et al. . Development of fluconazole resistance in a series of Candida parapsilosis isolates from a persistent candidemia patient with prolonged antifungal therapy. BMC Infect Dis. 2015;15:340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grossman NT, Pham CD, Cleveland AA, et al. . Molecular mechanisms of fluconazole resistance in Candida parapsilosis isolates from a U.S. surveillance system. Antimicrob Agents Chemother. 2015;59:1030–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arastehfar A, Lass-flörl C, Garcia-rubio R, et al. . The quiet and underappreciated rise of drug-resistant invasive fungal pathogens. J Fungi. 2020;59(10):6629–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Branco J, Silva AP, Silva RM, et al. . Fluconazole and voriconazole resistance in Candida parapsilosis is conferred by gain-of-function mutations in MRR1 transcription factor gene. Antimicrob Agents Chemother. 2015;59:6629–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martí-Carrizosa M, Sánchez-Reus F, March F, et al. . Implication of Candida parapsilosis FKS1 and FKS2 mutations in reduced echinocandin susceptibility. Antimicrob Agents Chemother. 2015;59:3570–3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang J, Macesic N, Blakeway L.. Comment on: genetically related micafungin-resistant Candida parapsilosis blood isolates harbouring novel mutation R658G in hotspot 1 of Fks1p: a new challenge? J Antimicrob Chemother. 2022. doi: 10.1093/jac/dkac091. [DOI] [PubMed] [Google Scholar]
- 20.Healey KR, Perlin DS.. Fungal resistance to echinocandins and the MDR phenomenon in Candida glabrata. J Fungi (Basel, Switzerland). 2018;4:319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berman J, Krysan DJ.. Drug resistance and tolerance in fungi. Nat Rev Microbiol. 2020;18:319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morio F, Lombardi L, Butler G.. The CRISPR toolbox in medical mycology: state of the art and perspectives. PLoS Pathog. 2020;16:e1008201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morio F, Lombardi L, Binder U, et al. . Precise genome editing using a CRISPR-Cas9 method highlights the role of CoERG11 amino acid substitutions in azole resistance in Candida orthopsilosis. J Antimicrob Chemother. 2019;74:2230–2238. [DOI] [PubMed] [Google Scholar]
- 24.Stielow JB, Lévesque CA, Seifert KA, et al. . One fungus, which genes? development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia. 2015;35:242–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clinical and Laboratory Standards Institute . Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard M27-A3. 3rd ed. Wayne (PA: ): CLSI; 2008. [Google Scholar]
- 26.Pfaller MA, Diekema DJ.. Progress in antifungal susceptibility testing of candida spp. by use of Clinical and Laboratory Standards Institute broth microdilution methods, 2010 to 2012. J Clin Microbiol. 2012;50:2846–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pulcrano G, Roscetto E, Iula VD, et al. . MALDI-TOF mass spectrometry and microsatellite markers to evaluate Candida parapsilosis transmission in neonatal intensive care units. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol. 2012;31:2919–2928. [DOI] [PubMed] [Google Scholar]
- 28.Choi YJ, Kim Y-J, Yong D, et al. . Fluconazole-resistant Candida parapsilosis bloodstream isolates with Y132F mutation in ERG11 gene, South Korea. Emerg Infect Dis. 2018;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Yu S-Y, Chen SC-A, et al. . Molecular characterization of Candida parapsilosis by microsatellite typing and emergence of clonal antifungal drug resistant strains in a multicenter surveillance in China. Front Microbiol. 2020;11:1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carreté L, Ksiezopolska E, Pegueroles C, et al. . Patterns of genomic variation in the opportunistic pathogen Candida glabrata suggest the existence of mating and a secondary association with humans. Curr Biol. 2018;28:15–27.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arastehfar A, Daneshnia F, Najafzadeh MJ, et al. . Evaluation of molecular epidemiology, clinical characteristics, antifungal susceptibility profiles, and molecular mechanisms of antifungal resistance of Iranian Candida parapsilosis species complex blood isolates. Front Cell Infect Microbiol. 2020;10:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pekmezovic M, Hovhannisyan H, Gresnigt MS, et al. . Candida pathogens induce protective mitochondria-associated type I interferon signalling and a damage-driven response in vaginal epithelial cells. Nat Microbiol. 2021;6:643–657. [DOI] [PubMed] [Google Scholar]
- 33.Daneshnia F, Hilmioğlu-Polat S, Ilkit M, et al. . Determinants of fluconazole resistance and the efficacy of fluconazole and milbemycin oxim combination in sensitizing Candida parapsilosis isolates from Brazil and Turkey. Front Fungal Biol. 2022;25(4):402–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- 35.Lombardi L, Oliveira-Pacheco J, Butler G.. Plasmid-based CRISPR-Cas9 gene editing in multiple Candida species. mSphere. 2019;4:e000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng D, Tarleton R.. EuPaGDT: a web tool tailored to design CRISPR guide RNAs for eukaryotic pathogens. Microb Genomics. 2015;1:e000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu X, Kriz AJ, Sharp PA.. Target specificity of the CRISPR-Cas9 system. Quant Biol (Beijing, China). 2014;2:59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Németh T, Nosanchuk J D, Vagvolgyi C, et al. . Enhancing the chemical transformation of Candida parapsilosis. Virulence. 2021;12:937–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishimoto AT, Sharma C, Rogers PD.. Molecular and genetic basis of azole antifungal resistance in the opportunistic pathogenic fungus Candida albicans. J Antimicrob Chemother. 2020;75:257–270. doi: 10.1093/jac/dkz400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andes DR, Safdar N, Baddley JW, et al. . Impact of treatment strategy on outcomes in patients with candidemia and other forms of invasive candidiasis: a patient-level quantitative review of randomized trials. Clin Infect Dis an Off Publ Infect Dis Soc Am. 2012;54:1110–1122. [DOI] [PubMed] [Google Scholar]
- 41.Pappas PG, Kauffman CA, Andes DR, et al. . Clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis. 2016;62:1–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pinhati HMS, Casulari LA, Souza ACR, et al. . Outbreak of candidemia caused by fluconazole resistant Candida parapsilosis strains in an intensive care unit. BMC Infect Dis. 2016;16:433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Binder U, Arastehfar A, Schnegg L, et al. . Efficacy of LAMB against emerging azole- and multidrug-resistant Candida parapsilosis isolates in the Galleria mellonella model. J Fungi (Basel, Switzerland). 2020;6:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shields RK, Kline EG, Healey KR, et al. . Spontaneous mutational frequency and FKS mutation rates vary by echinocandin agent against Candida glabrata. Antimicrob Agents Chemother. 2019;63:e01692-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hou X, Healey KR, Shor E, et al. . Novel FKS1 and FKS2 modifications in a high-level echinocandin resistant clinical isolate of Candida glabrata. Emerg Microbes Infect. 2019;8:1619–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tóth R, Nosek J, Mora-Montes HM, et al. . Candida parapsilosis: from genes to the bedside. Clin Microbiol Rev. 2019;32:87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajendran R, Sherry L, Nile CJ, et al. . Biofilm formation is a risk factor for mortality in patients with candida albicans bloodstream infection-Scotland, 2012-2013. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2016;22:87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.