Abstract
Background
This study assessed the efficacy of using oral liquid‐based brush cytology (OLBC) coupled with immunostained cytology‐derived cell‐blocks, quantified using machine‐learning, in the diagnosis of oral lichen planus (OLP).
Methods
Eighty‐two patients diagnosed clinically with either OLP or oral lichenoid lesion (OLL) were included. OLBC samples were obtained from all patients before undergoing surgical biopsy. Liquid‐based cytology slides and cell‐blocks were prepared and assessed by cytomorphology and immunocytochemistry for four antibodies (Ki‐67, BAX, NF‐κB‐p65, and AMACR). For comparison purposes, a sub‐group of 31 matched surgical biopsy samples were selected randomly and assessed by immunohistochemistry. Patients were categorized according to their definitive diagnoses into OLP, OLL, and clinically lichenoid, but histopathologically dysplastic lesions (OEDL). Machine‐learning was utilized to provide automated quantification of positively stained protein expression.
Results
Cytomorphological assessment was associated with an accuracy of 77.27% in the distinction between OLP/OLL and OEDL. A strong concordance of 92.5% (κ = 0.84) of immunostaining patterns was evident between cell‐blocks and tissue sections using machine‐learning. A diagnostic index using a Ki‐67‐based model was 100% accurate in detecting lichenoid cases with epithelial dysplasia. A BAX‐based model demonstrated an accuracy of 92.16%. The accuracy of cytomorphological assessment was greatly improved when it was combined with BAX immunoreactivity (95%).
Conclusions
Cell‐blocks prepared from OLBC are reliable and minimally‐invasive alternatives to surgical biopsies to diagnose OLLs with epithelial dysplasia when combined with Ki‐67 immunostaining. Machine‐learning has a promising role in the automated quantification of immunostained protein expression.
Keywords: cell‐blocks, computer‐assisted diagnosis, epithelial dysplasia, liquid‐based brush cytology, oral lichen planus
1. INTRODUCTION
Oral lichen planus (OLP), is a relatively common chronic inflammatory condition of uncertain etiology, with an estimated global prevalence of 1.01%. 1 There have been a number of iterations of diagnostic criteria for OLP and this condition can be challenging to diagnose due to the lack of distinct clinical and histopathological features. 2 Oral lesions with microscopic lichenoid features are among the most common conditions seen in oral medicine clinics, 3 and as a consequence, other oral potentially malignant disorders (OPMDs) are readily misdiagnosed as OLP. 4 This relatively high rate of misdiagnosis is likely to contribute to the highly variable rate of malignant transformation among OLP cases in the literature (0%–14.3%). 4 , 5
Oral lichenoid lesions (OLLs), on the other hand, include a spectrum of lichenoid lesions with different aetiologies. 5 It is widely believed that OLLs are associated with a higher risk of malignancy than OLP, 6 although this has not been correlated with the risk of underlying predisposing factors in OLLs. 6 Oral lichenoid dysplasia was introduced to describe lesions displaying features of oral lichen planus and epithelial dysplasia, but is no longer recommended as a diagnostic entity. 7 , 8 Further, mucosal disorders with oral epithelial dysplasia (OED) carry a higher malignant transformation risk irrespective of the presence or absence of lichenoid features. 9
Accurate diagnosis of OLP relies on clinico‐pathological correlation, with histopathological evaluation of surgical biopsies representing the gold standard for diagnosis. 10 Nonetheless, significant numbers of OLP cases are diagnosed in the absence of a surgical biopsy, 4 particularly when clinicians rely on clinical judgment alone for features they deem as characteristic of the disease. 11
There is, therefore, a critical need to propose and validate a minimally invasive and user‐friendly adjunctive technique to stratify oral lesions with lichenoid features. Oral liquid‐based brush cytology (OLBC) has gained great attention as an adjunct to surgical biopsy. 12 The main advantage of OLBC is that it is a relatively straightforward, minimally‐invasive technique that does not need special training and is less sensitive in terms of site collection, is well tolerated by operators and patients alike, and adequately harvests representative cells from all epithelial layers. 12
Importantly, the efficacy of this technique in stratifying the potential risk of malignant transformation among OPMDs has shown promising results, especially when combined with molecular biomarkers. 13 , 14 , 15
OED and OLP are two distinct pathologies, but their microscopic features may overlap in some cases causing confusion and misdiagnosis. Apoptotic regulation of basal keratinocytes is a key feature in OLP, while OED in contrast, is characterized by increased mitotic activity. 8 Protein expression of Ki‐67 has been used as a marker of cellular proliferation and is strongly correlated with the severity of OED. 16 On the other hand, B‐cell lymphoma 2 (Bcl‐2) and apoptosis regulator BCL2 associated X (BAX) are notable players in the OLP apoptosis process.
Another major molecule involved in OLP pathogenesis is nuclear factor kappa‐B‐p65 subunit (NF‐κB‐p65). 17 , 18 Many studies have identified Alpha‐methylacyl‐CoA racemase (AMACR) as a valuable biomarker in the detection of epithelial dysplastic changes across a range of conditions such as chronic inflammatory bowel disease and colorectal polyps. 19 AMACR is a peroxisomal and mitochondrial enzyme with an important role in regulating the metabolism of lipids and drugs. Neither NF‐κB‐p65 nor AMACR have previously been assessed for their ability to distinguish oral lichenoid and dysplastic lesions.
The development of high‐quality brightfield and fluorescent slide scanners has enhanced the capabilities of digital pathology by improving diagnostic accuracy, enhancing patient care, and facilitating global collaboration. 20 Additionally, machine‐learning, has proven to be very promising in the field of oral diagnosis. 21 , 22 Collectively, these approaches have several advantages over conventional diagnostic methods, including the speed of analysis, minimal cost after initial installation, and the delivery of consistent outcomes due to standardization.
This study aimed to assess the efficacy of using OLBC coupled with immunostained cytology‐derived cell‐blocks, quantified using machine‐learning, in the diagnosis of OLP. We first demonstrated the feasibility of using OLBC as a reliable adjunct by displaying a strong concordance between tissue blocks and OLBC‐cell blocks obtained from the same patient. Second, we established the use of machine‐learning for reproducible automated quantification of immunostaining of protein biomarkers in cell blocks. Last, we show that a Ki‐67‐based diagnostic model is able to distinguish, with high accuracy, between oral lesions with clinical lichenoid features based on the presence or absence of microscopic epithelial dysplasia.
2. MATERIALS AND METHODS
2.1. Study design and data sets
This retrospective cross‐sectional study was approved by the Human Ethics Committee of the University of Western Australia (RA/4/20/4530 and RA/4/1/8562) and was conducted following the Standards for Reporting Diagnostic Accuracy Studies (STARD 2015).
Cases were selected from a cohort of patients diagnosed with OPMD and recruited by our research group between 2017 and 2021 to assess the utility of OLBC in the diagnosis of oral mucosal lesions based on defined inclusion and exclusion criteria (Table S1). 13 , 14 , 15
All cases were clinically and histopathologically re‐evaluated by two authors to confirm the diagnosis. Patients were classified according to their definitive diagnosis into three categories: (1) OLP, (2) OLL, and (3) clinically lichenoid, but histopathologically dysplastic lesions (OEDL). OLP cases were diagnosed based on clinical and microscopic features of OLP diagnostic criteria. 10 OLL was defined as any lesion with clinical and/or histopathological lichenoid features that are compatible (but not typical) with OLP. 5 The absence of epithelial dysplasia was a prerequisite to consider any case as OLL in this study. Cases that are clinically diagnosed as OLP or OLL but histopathologically exhibiting features of epithelial dysplasia, irrespective of the presence of microscopic lichenoid features, were categorized as “clinically lichenoid, but histopathologically dysplastic” lesions for the purpose of this study (designated as OEDL). Lesions with OED were graded using the binary grading system into low‐risk and high‐risk lesions. 23
Although all cases had surgical biopsies, 31 surgical formalin‐fixed paraffin‐embedded (FFPE) blocks related to the included patients were randomly selected for comparison purposes. Accordingly, patients were divided into two cohorts: (1) the cell‐block cohort, for patients with OLBC samples, and (2) the tissue‐block cohort, for patients with FFPE blocks of tissue sections. Figure 1 illustrates the basic design of the study.
FIGURE 1.
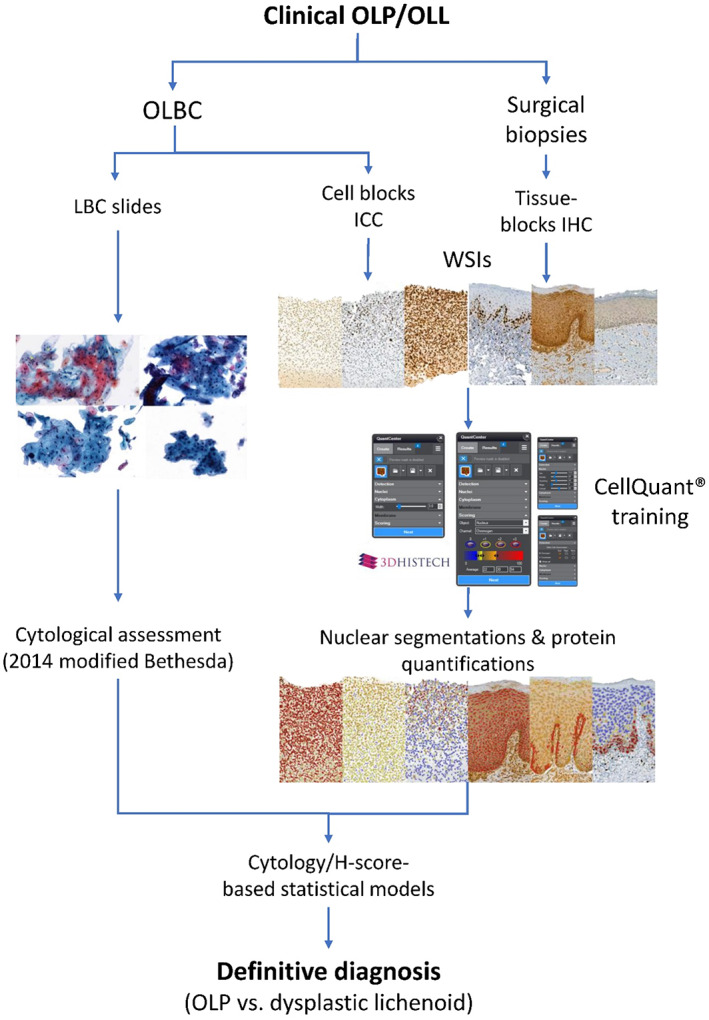
A representative flow diagram of the study's design
2.2. Sample collection and preparation
OLBC samples were obtained from all participants by oral medicine specialists using a defined protocol as outlined in our previous reports. 13 , 15 A liquid‐based cytology (LBC) slide was prepared for each OLBC sample using the ThinPrep® 2000 processor (Hologic Inc., MA) according to the manufacturer's protocol.
The Countless II automated cell counter (ThermoFisher, MA) was then used to measure cell yields in each OLBC sample after preparing LBC slides. Any sample with at least 1 × 106 cells was considered adequate for further cell‐block preparation. The remaining cells in each ThinPrep® PreservCyt vial were processed to prepare an individual paraffin cell‐block for each case using the thromboplastin‐plasma method. 15 The quality of each cell‐block was evaluated in terms of cellular morphologic preservation by visually assessing 4 μm hematoxylin and eosin‐stained sections.
2.3. Cytomorphological assessment
LBC slides were Papanicolaou stained and assessed blindly by two authors who were unaware about the definitive diagnosis using the modified 2014 Bethesda cervical cytology grading system. 13 Cases were categorized into five groups: (1) atypical squamous cells of undetermined significance, (2) atypical squamous cells suggestive of OLP or lichenoid reaction, (3) low‐grade squamous intraepithelial lesion (low‐grade single group termed [SIL]), (4) high‐grade SIL, and (5) oral squamous cell carcinoma (OSCC). 13
2.4. Immunostaining of cell block and histology cohorts
Sections of 4 μm from paraffin blocks of both cell‐block and tissue‐block cohorts were cut. Standard horseradish peroxidase immunohistochemistry (IHC) was carried out to detect four antibodies: Ki‐67, BAX, NF‐κB‐p65, and AMACR. Appropriate positive and negative controls for cell‐blocks and tissue blocks were included. Full details about the IHC used materials are outlined in Table S1.
2.5. Digitization of slides and region of interest identification
All slides were scanned using the Pannoramic 250 Flash III® (3DHISTECH, Budapest, Hungary) at ×40 magnification and whole‐slide images (WSIs) saved in MRXS file format (CaseViewer®; version 2.5, 3DHISTECH).
Slides were automatically scanned with a seven‐layer extended focus algorithm to extract focused areas from each focal plane and assemble them into a single composite image. The average scanning time for each slide was 25 min. A pathologist detected a region of interest (ROI) or more for each WSI. For tissue sections, ROIs included the full epithelial thickness of the WSI excepting the keratin layer of not less than 250 000 μm2. For cell‐block images, the ROIs included all cellular communities of not less than 500 cells per WSI. Any case that did not meet these criteria was excluded.
2.6. Cellular identification and automated quantification of immunostaining using machine‐learning
QuantCenter® software version 2.3 (3DHistech, Budapest, Hungary) was used for cell recognition and quantification of stains. QuantCenter® uses integrated Quant algorithms called CellQuant based on colour deconvolution and cellular morphology for image analysis. These algorithms can be adapted by developing a “scenario” to act as a unique customized measurement profile to detect cells and provide quantification of the immunoreactions based on intensity. Table S1 shows the basic elements of each QuantCenter® algorithm for immunostaining quantifications.
The result of each case based on the appropriate scenario was reported as a H‐score. H‐score is a value between 0 and 300 to represent both the intensity and the proportion of the protein of interest and is calculated using the following formula:
However, as the CellQuant® training is only available on one slide, a representative slide (a slide containing a variety of IHC staining intensities) from each cohort and for each antibody was selected for this purpose. The values that were used for each scenario are shown in Table S2.
2.7. Statistical analysis
Full details of the performed statistical analyses are listed in Table S1.
3. RESULTS
3.1. General characteristics of the included cohorts
A total of 106 cell‐block cases were initially H&E stained and checked for quality assurance. Of these, 24 cases (22.6%) were excluded due to inadequate cellularity. A total of 82 patients included in this study; 43 (52.4%) females and 39 males (47.6%). An individual cell‐block was prepared for each patient and checked for quality assurance by examining its related H&E stained slide. The mean age of the participants in both cohorts was 65.78 (±11.1) years, with no significant difference between genders, p = 0.542. Of these cases, 31 FFPE tissue blocks matched with their cell‐block counterparts were included in the tissue‐block cohort, 16 females (51.6%) and 15 males (48.4%).
The majority (41.6%) of cases were diagnosed as OLP, followed by 29.2% for OLL and the remaining 29.2% as OEDL. As the majority of OEDL cases were graded as low‐risk epithelial dysplasia (84.8% low‐risk OED vs. 15.2% high‐risk OED), they were grouped together. Neither the gender nor the age of the study subjects was associated with the definitive diagnosis (p = 0.215 and p = 0.536, respectively). Table S3 illustrates the general characteristics of the study subjects in both cohorts.
A Pearson chi‐square test of independence showed significant associations between the definitive diagnosis and the lesion site in both the cell‐block cohort (χ 2(9) = 38.75, p <0.0005), and the tissue‐block cohort (χ 2(9) = 13.1, p = 0.041; Table S3).
3.2. Accuracy of the cytomorphological assessment in predicting the diagnosis
Of 82 cases, a single case was excluded due to a human error in Papanicolaou staining. Of the remaining 81 cases, 33 (40.7%) cases were cytomorphologically assessed as atypical squamous cells suggestive of OLP or lichenoid reaction, followed by 22 (27.2%) cases of low‐grade SIL and 7 (8.6%) cases of high‐grade SIL. However, for the purposes of this study, and for agreement with the histopathological assessment, all SIL cases were combined in a SIL (Table 1).
TABLE 1.
Associations between cytomorphological assessment and definitive diagnosis
| Cytomorphological assessment | Definitive diagnosis | Total | p‐value | ||
|---|---|---|---|---|---|
| OLP | OLL | OEDL | |||
| Undetermined significance | 8 | 9 | 2 | 19 | <0.0005 |
| OLP/lichenoid reactions | 24 a | 8 | 1 a | 33 | |
| Squamous intraepithelial lesion | 2 a | 6 | 21 a | 29 | |
| Total | 34 | 23 | 24 | 81 | |
Significant pairwise comparison (post hoc adjusted p‐value <0.0056).
The association between the cytomorphological assessment and the definitive diagnosis was significant χ 2(4) = 47.288 (p <0.0005). The level of association was relatively strong (Cramer's V = 0.532). The post hoc analysis of pairwise comparisons showed statistically significant associations among four groups of comparison (Table 1). The accuracy of this approach was 77.27% while the associated sensitivity and specificity were 66.23% and 88.31% respectively, and the area under the curve (AUC) was 84.4% as shown in Table 3.
TABLE 3.
Proposed diagnostic indices and their associated statistics
| Cohort/cytological assessment | Diagnostic model | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) | AUC (%) |
|---|---|---|---|---|---|---|---|
| Cytological assessment | — | 66.23 | 88.31 | 85 | 72.34 | 77.27 | 84.4 |
| Cell‐block |
Ki‐67 OLP/OLL <20.09 |
100 | 100 | 100 | 100 | 100 | 100 |
|
BAX OLP/OLL >101.22 |
93.55 | 90 | 93.55 | 90 | 92.16 | 91.8 | |
| Tissue‐block |
Ki‐67 OLP/OLL <86.16 |
100 | 100 | 100 | 100 | 100 | 100 |
|
BAX OLP/OLL >188.9 |
88.24 | 100 | 100 | 66.67 | 90.48 | 94.1 | |
| Regression models | Ki‐67/BAX OLP/OLL <1.59 | 100 | 100 | 100 | 100 | 100 | 100 |
| Cytology/BAX OLP/OLL <1.51 | 90 | 100 | 100 | 93.1 | 95.74 | 95 |
Abbreviations: AUC, area under the curve; NPV, negative predictive value; PPV, positive predictive value.
3.3. Automated quantification of protein immuno‐expression
Ki‐67, BAX, and NF‐κB‐p65 proteins were all expressed in both cell‐blocks and tissue‐blocks displaying specific patterns. AMACR showed very weak staining in the tissue‐block cohort with no potential diagnostic utility, although this protein was associated with strong expression in both the cell‐block and positive tissue controls. Therefore, AMACR was excluded from any further analysis for the cell‐block cohort. Figure S1 shows the expression of each protein on its cell‐block positive control and tissue positive control.
3.3.1. Immunocytochemical expression of the cell‐block cohort
Ki‐67 was negative in 26 cell blocks (14 OLP and 12 OLL) but all OEDL cases (24) were associated with positive expression. NF‐κB‐p65 was expressed in all cell‐block sections, while only one OEDL case was negative for BAX. A H‐score was provided for each case based on the proposed customized scenario for each antibody. There were no outliers in the values of automated H‐scores for the included antibodies as assessed by inspection of their boxplots (Figure S2).
The H‐scores for Ki‐67 and BAX were statistically significantly associated with the definitive diagnosis (Welch's F (2,17.905) = 45.254, p <0.0005) and (F (2,48) = 38.855, p <0.005), respectively. Post hoc analysis showed significant associations between the H‐scores of OLP and OEDL groups and between the H‐scores of OLL and OEDL groups for both Ki‐67 and BAX (Table 2). There was no significant difference in the H‐score between the OLP and OLL groups. In contrast, the association between NF‐κB‐p65 and the definitive diagnosis was not significant, (F (2,42) = 1.603, p = 0.213). Figure 2 shows the immunocytochemical expression of the investigated proteins in representative cell‐block samples and the performance of the proposed immunostain quantification scenarios.
TABLE 2.
Mean difference in the H‐score of diagnostic groups for each antibody
| Cohort | Group A | Group B | ||
|---|---|---|---|---|
| Mean difference in the H‐score (Group A – Group B) (Standard error) | ||||
| OLL | OEDL | |||
| Cell‐block | Ki‐67 | OLP | −0.04 (0.6) p = 0.998 | −49.72 (5.1) p <0.0005 a |
| OLL | — | −49.68 (5.1) p <0.0005 a | ||
| BAX | OLP | 4.86 (13.3) p = 0.929 | 92.74 (11.5) p <0.0005 b | |
| OLL | ‐ | 87.88 (13.1) p <0.0005 b | ||
| NF‐κB‐p65 | OLP | −14.66 (16.4) p = 0.647 | −32.61 (18.3) p = 0.187 | |
| OLL | — | −17.94 (18.7) p = 0.607 | ||
| Tissue‐block | Ki‐67 | OLP | 2.86 (5.6) p = 0.882 | −64.4 (5.9) p <0.0005 b |
| OLL | — | −67.1 (6.5) p <0.0005 b | ||
| BAX | OLP | 12.26 (16.7) p = 0.748 | 83.73 (20.7) p = 0.002 b | |
| OLL | — | 71.47 (21.1) p = 0.009 b | ||
| NF‐κB‐p65 | OLP | 3.6 (13.2) p = 0.96 | −3.89 (15.3) p = 15.3 | |
| OLL | — | −7.49 (16.2) p = 0.889 | ||
| AMACR | OLP | 0.812 (12.3) p = 0.998 | −27.27 (14.6) p = 0.179 | |
| OLL | — | −28.08 (14.9) p = 0.176 | ||
Games–Howell post hoc significant association.
Tukey post hoc significant association.
FIGURE 2.
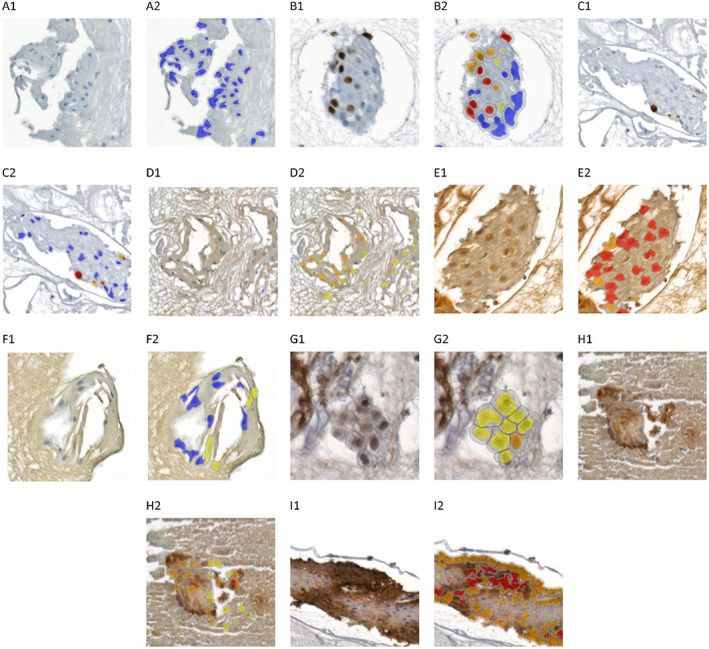
Immunocytochemical expression of investigated proteins and the performance of proposed immunostaining quantification scenarios in representative cell‐block samples. (A1,A2) Ki‐67 in OLP, (B1,B2) Ki‐67 in OEDL, (C1,C2) Ki‐67 in OLL, (D1,D2) BAX in OLL, (E1,E2 BAX in OLP, (F1,F2) BAX in OEDL, (G1,G2) NF‐kP‐p65 in OLL, (H1,H2) NF‐kP‐p65 in OEDL, (I1,I2) NF‐kP‐p65 in OLP
3.3.2. Immunohistochemical expression of the tissue‐block cohort
Ki‐67, BAX, NF‐κB‐p65, and AMACR were expressed in all cases of the histology cohort. Ki‐67 was predominantly nuclear and expressed exclusively in the basal layers. Other proteins were expressed in all epithelial layers and their expression was both nuclear and cytoplasmic. The associated boxplots showed no outliers in H‐score values (Figure S2).
The H‐scores for Ki‐67 and BAX were statistically significantly associated with the definitive diagnosis, (F (2,21) = 71.812, p <0.0005), and (F (2,18) = 8.577, p = 0.002), respectively. Tukey post hoc analysis revealed significant differences between OLP and OEDL and between OLL and OEDL for both Ki‐67 and BAX as shown in Table 2. On the other hand, neither NF‐κB‐p65 nor AMACR were statistically significantly associated with the definitive diagnosis, (F (2,21) = 0.109, p = 0.897), and (F (2,16) = 2.136, p = 0.151), respectively. The performance of the proposed immunostain quantification scenarios is shown for representative images in the tissue‐block cohort in Figure 3 and Figure S3.
FIGURE 3.
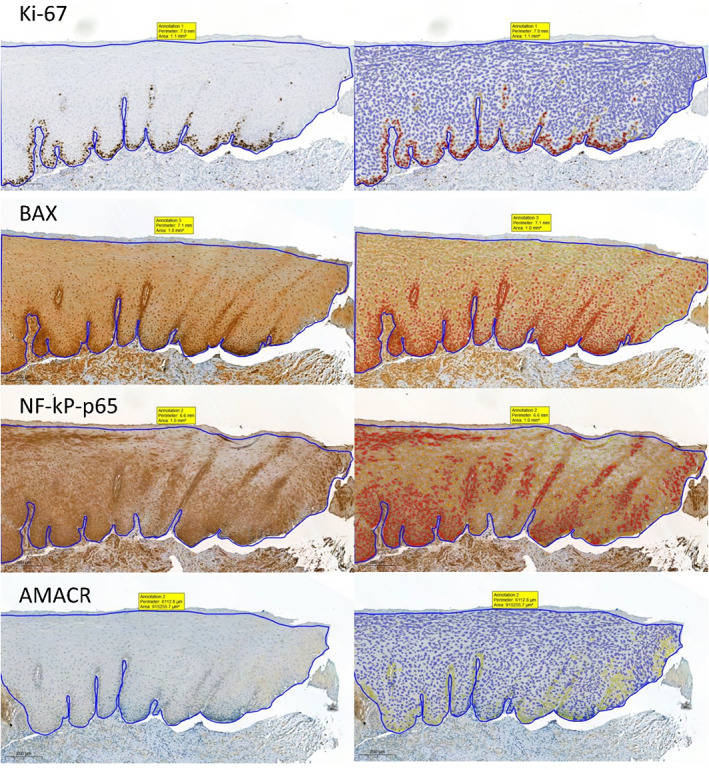
Representative images showing immunohistochemical expression of investigated proteins and the performance of the proposed immunostaining quantification scenarios in a case of OLP. Magnification ×200
3.3.3. Concordance between cell‐blocks and tissue‐blocks of paired cases
The overall concordance rate between cell‐blocks and tissue‐blocks for Ki‐67, BAX, and NF‐κB‐p65 was 92.5% (86 of 93), the level of agreement was very good (Cohen's kappa κ = 0.84, p <0.0005). No statistically significant difference was observed between the two cohorts (McNemar test p = 0.125; Table S4).
When proteins were considered individually, the concordance rate was 100% for NF‐κB‐p65, 93.5% for BAX, and 83.9% for Ki‐67. The difference between cell‐blocks and the tissue‐blocks was not significant for BAX and Ki‐67 (McNemar test, p = 0.5 and p = 0.063, respectively). Table S4 shows all pair comparisons for concordance.
3.4. Proposed index scores to differentiate between oral lesions with lichenoid features based on the presence or absence of epithelial dysplasia and their associated accuracy
As both Ki‐67 and BAX were significantly associated with the definitive diagnosis, indices were proposed and assessed based on H‐scores of the cell‐block and tissue‐block cohorts. The accuracy of the Ki‐67‐based indices was 100% for both the cell‐block and the tissue‐block cohorts. The BAX‐based index was associated with an accuracy of 92.16% for the cell‐block cohort and 90.48% for the tissue‐block cohort (Table 3).
To further validate the utility of OLBC in predicting the definitive diagnosis, several regression models were built including two variables. A regression model based on Ki‐67 and BAX H‐scores in the cell‐blocks was statistically significant for predicting the definitive diagnosis (F (2,33) = 250.24, p <0.0005). Both H‐scores of Ki‐67 and BAX added significant value to this prediction, p <0.0005. The following formula was created based on this model:
Another two regression models were assessed by combining the cytomorphological findings with the immunoexpression results. This first model was statistically significant in terms of the ability to predict the definitive diagnosis based on the cytomorphological findings and the H‐score of BAX (F (2,44) = 53.5, p <0.0005), both inputs were associated with statistical values, p <0.0005. The following formula was proposed for this prediction:
A second regression model combining cytomorphological findings with the H‐score of Ki‐67 to predict the definitive diagnosis was not possible as there was no significant value for the cytomorphological assessment in this equation, p = 0.891.
The accuracy of these models was analyzed by calculating cut‐off values to discriminate between oral lesions with lichenoid features with and without dysplasia. Accordingly, the combined Ki‐67‐BAX model was associated with an accuracy of 100%. Moreover, combining the cytomorphological assessment and the H‐score of BAX enhanced the accuracy and the AUC of these variables to 95.75% and 95%, respectively as shown in Table 3.
4. DISCUSSION
This project introduces a novel approach to distinguish between oral lesions with clinical lichenoid features with and without microscopic dysplastic changes utilizing highly objective and minimally invasive techniques. By using OLBC and immunocytology from cell blocks, we have produced a valuable clinical tool that can be applied by clinicians to further assess any clinically suspicious lesion. To the best of our knowledge, this is the first application using paraffinized cell blocks to detect immunoreactive protein expression in oral lesions with clinical lichenoid features. This work also represents the first implementation of machine learning to analyze immunostaining of cell‐blocks from cytological samples.
Whilst there have been several attempts to assess the utility of oral cytology in the diagnosis of OLP, the results have not been consistent due to the lack of standardized criteria to report cytological findings of OPMDs in general, and OLP in particular. 13 Therefore, in 2018 our group proposed modifications to the 2014 Bethesda System for Reporting Cervical Cytology to report oral mucosal lesions. These modifications showed promising results in the diagnosis of OPMDs with an accuracy of 75%, 13 which is comparable with the findings of this study. The level of association between the cytomorphological assessment and the definitive diagnosis was relatively strong. However, there is a possibility that the relatively small patient cohort and the absence of various diagnostic categories of OPMDs underestimate the actual utility of this approach, therefore, we recommend further validation in a large multicenter study.
OLBC as a cytological technique shares the major disadvantage of conventional cytology in that it provides a limited amount of cellular material for ancillary analysis. 15 Preparing cell‐blocks overcomes this limitation and provides archival material that can be treated in a similar manner to FFPE tissue‐blocks. 15 Several reports have found cell‐blocks to provide superior results for immunostaining in comparison to other cytological preparations in terms of diagnostic accuracy and morphological proximity to surgical pathology. 24 The results of this study add additional evidence supporting the efficacy of cell‐block immunocytochemistry in the diagnosis of oral mucosal lesions, which is in agreement with previous findings in this field. 15
It is worth noting that our results demonstrate an overall strong consistency in the expression of examined proteins in both cell‐blocks and tissue‐blocks, although immunostaining intensities were reduced in cell‐blocks compared to tissue‐blocks. The concordance for individual proteins in this study is consistent with the literature where concordance rates have ranged between 84% and 100%. 25 This highlights the significance of this method as a reliable, minimally‐invasive tool for molecular analysis.
Our study revealed a significant increase in the immunoreactivity of Ki‐67 in dysplastic cases compared to OLP and OLL. Interestingly, OEDL cases were associated with a significant reduction in the expression of BAX (p <0.05). This finding was not surprising as it is consistent with the molecular pathogenesis of these conditions. 26 , 27 Most importantly, it provides molecular evidence that cell‐blocks are reliable representatives of transepithelial tissue sections.
Our results did not find diagnostic utility for NF‐κB‐p65 and AMACR in the detection of microscopic dysplastic changes among oral lesions with lichenoid features.
Interestingly, OLP and OLL were associated with very similar protein expression for all investigated biomarkers. A similar pattern of expression among OLP and OLL was previously reported for Ki‐67. 28 Nonetheless, our previous study found a significant increase in the number of inflammatory cells in OLP in comparison to OLL. 21 Accordingly, some authors have argued that idiopathic OLL should be considered as “atypical OLP lesions” rather than OLL, while the term OLL should only be used to describe lichenoid reactions induced by drugs or contact sensitivity. 29 Further investigations with longer term follow‐up of OLLs are required to assess potential microscopic changes over time.
The current method of IHC scoring relies on the visual examination of stain intensity and percentage. However, these criteria are judged subjectively as there is no clear‐cut method to differentiate between different stain intensities, and it is almost impossible to identify the exact percentage of each staining intensity in the whole section. Therefore, in this project we used machine learning algorithms for immunostain quantification to minimize potential human error. This approach is not novel in itself in tissue sections; however, this is the first application of this approach using cell‐block sections. The outcomes of this study support the reliability of this technique as an adjunctive tool to surgical biopsy for lesion surveillance.
There were several potential limitations associated with this study. First, the number of included cases in both cohorts was relatively low, which limits the ability to further validate the proposed techniques. Second, the cohort did not include patients with oral lichenoid reactions, thus, it was not possible to assess the performance of this technique in a broad clinical setting. Finally, the thresholds used for staining intensities were optimized according to our staining protocols and reagents and there is a possibility that using antibodies from different vendors or different antibody dilutions could impact the accuracy of these thresholds. Further investigations and optimizations are recommended when considering future studies.
In conclusion, OLBC is a reliable and efficient tool for OLP diagnosis. Combining cytomorphological findings with cell‐block‐based OLBC shows diagnostic utility comparable to surgical biopsies and can be adopted as a minimally‐invasive approach to stratify patients with clinical lichenoid lesions. The use of machine learning for immunostain quantification is effective in ensuring objective assessment.
CONFLICT OF INTEREST
All authors declare that they do not have any conflict of interest.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1111/jop.13301.
Supporting information
Data S1 Supporting Information
ACKNOWLEDGMENTS
We acknowledge funding support from the University of Western Australia. We also wish to acknowledge the Australian Dental Research Foundation Inc. and the Australian Dental Association (WA Branch) for the clinical dentistry research grant awarded to support this study. Open access publishing facilitated by The University of Western Australia, as part of the Wiley ‐ The University of Western Australia agreement via the Council of Australian University Librarians.
Idrees M, Shearston K, Farah CS, Kujan O. Immunoexpression of oral brush biopsy enhances the accuracy of diagnosis for oral lichen planus and lichenoid lesions. J Oral Pathol Med. 2022;51(6):563‐572. doi: 10.1111/jop.13301
Funding information Australian Dental Research Foundation; University of Western Australia
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author.
REFERENCES
- 1. Gonzalez‐Moles M, Warnakulasuriya S, Gonzalez‐Ruiz I, et al. Worldwide prevalence of oral lichen planus: a systematic review and meta‐analysis. Oral Dis. 2021;27(4):813‐828. doi: 10.1111/odi.13323 [DOI] [PubMed] [Google Scholar]
- 2. Idrees M, Farah CS, Khurram SA, Firth N, Soluk‐Tekkesin M, Kujan O. Observer agreement in the diagnosis of oral lichen planus using the proposed criteria of the American Academy of Oral and Maxillofacial Pathology. J Oral Pathol Med. 2021;50(5):520‐527. doi: 10.1111/jop.13170 [DOI] [PubMed] [Google Scholar]
- 3. Czerninski R, Zeituni S, Maly A, Basile J. Clinical characteristics of lichen and dysplasia vs lichen planus cases and dysplasia cases. Oral Dis. 2015;21(4):478‐482. doi: 10.1111/odi.12306 [DOI] [PubMed] [Google Scholar]
- 4. Idrees M, Kujan O, Shearston K, Farah CS. Oral lichen planus has a very low malignant transformation rate: a systematic review and meta‐analysis using strict diagnostic and inclusion criteria. J Oral Pathol Med. 2020;50(3):287‐298. doi: 10.1111/jop.12996 [DOI] [PubMed] [Google Scholar]
- 5. Warnakulasuriya S, Kujan O, Aguirre‐Urizar JM, et al. Oral potentially malignant disorders: a consensus report from an international seminar on nomenclature and classification, convened by the WHO Collaborating Centre for Oral Cancer. Oral Dis. 2020;27(8):1862‐1880. doi: 10.1111/odi.13704 [DOI] [PubMed] [Google Scholar]
- 6. Iocca O, Sollecito TP, Alawi F, et al. Potentially malignant disorders of the oral cavity and oral dysplasia: a systematic review and meta‐analysis of malignant transformation rate by subtype. Head Neck. 2019;42(3):539‐555. doi: 10.1002/hed.26006 [DOI] [PubMed] [Google Scholar]
- 7. Farah CS, Fox S, Shearston K, Newman L, Babic S, Vacher M. Lichenoid dysplasia is not a distinct pathological entity. Oral Oncol. 2021;119:105362. doi: 10.1016/j.oraloncology.2021.105362 [DOI] [PubMed] [Google Scholar]
- 8. Odell E, Kujan O, Warnakulasuriya S, Sloan P. Oral epithelial dysplasia: recognition, grading and clinical significance. Oral Dis. 2021;27(8):1947‐1976. doi: 10.1111/odi.13993 [DOI] [PubMed] [Google Scholar]
- 9. Shearston K, Fateh B, Tai S, Hove D, Farah CS. Oral lichenoid dysplasia and not oral lichen planus undergoes malignant transformation at high rates. J Oral Pathol Med. 2019;48(7):538‐545. doi: 10.1111/jop.12904 [DOI] [PubMed] [Google Scholar]
- 10. Cheng Y, Gould A, Kurago Z, Fantasia J, Muller S. Diagnosis of oral lichen planus: a position paper of the American Academy of Oral and Maxillofacial Pathology. Oral Surg Oral Med Oral Pathol Oral Radiol. 2016;122(3):332‐354. doi: 10.1016/j.oooo.2016.05.004 [DOI] [PubMed] [Google Scholar]
- 11. Lee J, Hung H, Cheng S, et al. Factors associated with underdiagnosis from incisional biopsy of oral leukoplakic lesions. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2007;104(2):217‐225. doi: 10.1016/j.tripleo.2007.02.012 [DOI] [PubMed] [Google Scholar]
- 12. Alsarraf A, Kujan O, Farah CS. The utility of oral brush cytology in the early detection of oral cancer and oral potentially malignant disorders: a systematic review. J Oral Pathol Med. 2018;47(2):104‐116. doi: 10.1111/jop.12660 [DOI] [PubMed] [Google Scholar]
- 13. Alsarraf A, Kujan O, Farah CS. Liquid‐based oral brush cytology in the diagnosis of oral leukoplakia using a modified Bethesda Cytology system. J Oral Pathol Med. 2018;47(9):887‐894. doi: 10.1111/jop.12759 [DOI] [PubMed] [Google Scholar]
- 14. Kujan O, Huang G, Ravindran A, Vijayan M, Farah CS. CDK4, CDK6, cyclin D1 and Notch1 immunocytochemical expression of oral brush liquid‐based cytology for the diagnosis of oral leukoplakia and oral cancer. J Oral Pathol Med. 2019;48(7):566‐573. doi: 10.1111/jop.12902 [DOI] [PubMed] [Google Scholar]
- 15. Kujan O, Idrees M, Anand N, Soh B, Wong E, Farah CS. Efficacy of oral brush cytology cell block immunocytochemistry in the diagnosis of oral leukoplakia and oral squamous cell carcinoma. J Oral Pathol Med. 2020;50(5):451‐458. doi: 10.1111/jop.13153 [DOI] [PubMed] [Google Scholar]
- 16. Takkem A, Barakat C, Zakaraia S, et al. Ki‐67 prognostic value in different histological grades of oral epithelial dysplasia and oral squamous cell carcinoma. Asian Pac J Cancer Prev. 2018;19(11):3279‐3286. doi: 10.31557/APJCP.2018.19.11.3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gerondakis S, Fulford TS, Messina NL, Grumont RJ. NF‐kappaB control of T cell development. Nat Immunol. 2014;15(1):15‐25. doi: 10.1038/ni.2785 [DOI] [PubMed] [Google Scholar]
- 18. Zhang D, Wang J, Li Z, et al. The activation of NF‐kappaB in infiltrated mononuclear cells negatively correlates with Treg cell frequency in oral lichen planus. Inflammation. 2015;38(4):1683‐1689. doi: 10.1007/s10753-015-0145-x [DOI] [PubMed] [Google Scholar]
- 19. Brahim E, Mrabet A, Jouini R, et al. Immunohistochemistry in the diagnosis of dysplasia in chronic inflammatory bowel disease colorectal polyps. Arab J Gastroenterol. 2016;17(3):121‐126. doi: 10.1016/j.ajg.2016.06.003 [DOI] [PubMed] [Google Scholar]
- 20. Cui M, Zhang DY. Artificial intelligence and computational pathology. Lab Invest. 2021;101(4):412‐422. doi: 10.1038/s41374-020-00514-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Idrees M, Farah CS, Shearston K, Kujan O. A machine‐learning algorithm for the reliable identification of oral lichen planus. J Oral Pathol Med. 2021;50(9):946‐953. doi: 10.1111/jop.13226 [DOI] [PubMed] [Google Scholar]
- 22. Shaban M, Khurram SA, Fraz MM, et al. A novel digital score for abundance of tumour infiltrating lymphocytes predicts disease free survival in oral squamous cell carcinoma. Sci Rep. 2019;9(1):13341. doi: 10.1038/s41598-019-49710-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kujan O, Oliver RJ, Khattab A, Roberts SA, Thakker N, Sloan P. Evaluation of a new binary system of grading oral epithelial dysplasia for prediction of malignant transformation. Oral Oncol. 2006;42(10):987‐993. doi: 10.1016/j.oraloncology.2005.12.014 [DOI] [PubMed] [Google Scholar]
- 24. Qin SY, Zhou Y, Li P, Jiang HX. Diagnostic efficacy of cell block immunohistochemistry, smear cytology, and liquid‐based cytology in endoscopic ultrasound‐guided fine‐needle aspiration of pancreatic lesions: a single‐institution experience. PLoS One. 2014;9(9):e108762. doi: 10.1371/journal.pone.0108762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Song SG, Lee J, Koh J, Kim S, Chung DH, Jeon YK. Utility of PD‐L1 immunocytochemistry using body‐fluid cell blocks in patients with non‐small‐cell lung cancer. Diagn Cytopathol. 2020;48(4):291‐299. doi: 10.1002/dc.24379 [DOI] [PubMed] [Google Scholar]
- 26. Rosa EA, Hurtado‐Puerto AM, Falcao DP, et al. Oral lichen planus and malignant transformation: the role of p16, Ki‐67, Bub‐3 and SOX4 in assessing precancerous potential. Exp Ther Med. 2018;15(5):4157‐4166. doi: 10.3892/etm.2018.5971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. de Sousa FA, Paradella TC, Carvalho YR, Rosa LE. Comparative analysis of cell proliferation ratio in oral lichen planus, epithelial dysplasia and oral squamous cell carcinoma. Med Oral Patol Oral Cir Bucal. 2009;14(11):e563‐e567. doi: 10.4317/medoral.14.e563 [DOI] [PubMed] [Google Scholar]
- 28. Acay R, Felizzola C, de Araujo N, de Sousa S. Evaluation of proliferative potential in oral lichen planus and oral lichenoid lesions using immunohistochemical expression of p53 and Ki67. Oral Oncol. 2006;42(5):475‐480. doi: 10.1016/j.oraloncology.2005.09.012 [DOI] [PubMed] [Google Scholar]
- 29. Gonzalez‐Moles M, Ramos‐Garcia P, Warnakulasuriya S. The importance of understanding the terminology on oral lichenoid lesions for future research: in reply. Oral Oncol. 2021;117:105282. doi: 10.1016/j.oraloncology.2021.105282 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1 Supporting Information
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author.


