ABSTRACT
SARS-CoV-2 B.1.1.529.1 (Omicron BA.1) emerged in November 2021 and quickly became the predominant circulating SARS-CoV-2 variant globally. Omicron BA.1 contains more than 30 mutations in the spike protein, which contribute to its altered virological features when compared to the ancestral SARS-CoV-2 or previous SARS-CoV-2 variants. Recent studies by us and others demonstrated that Omicron BA.1 is less dependent on transmembrane serine protease 2 (TMPRSS2), less efficient in spike cleavage, less fusogenic, and adopts an altered propensity to utilize the plasma membrane and endosomal pathways for virus entry. Ongoing studies suggest that these virological features of Omicron BA.1 are in part retained by the subsequent Omicron sublineages. However, the exact spike determinants that contribute to these altered features of Omicron remain incompletely understood. In this study, we investigated the spike determinants for the observed virological characteristics of Omicron. By screening for the individual changes on Omicron BA.1 and BA.2 spike, we identify that 69–70 deletion, E484A, and H655Y contribute to the reduced TMPRSS2 usage while 25–27 deletion, S375F, and T376A result in less efficient spike cleavage. Among the shared spike mutations of BA.1 and BA.2, S375F and H655Y reduce spike-mediated fusogenicity. Interestingly, the H655Y change consistently reduces serine protease usage while increases the use of endosomal proteases. In keeping with these findings, the H655Y substitution alone reduces plasma membrane entry and facilitates endosomal entry when compared to SARS-CoV-2 WT. Overall, our study identifies key changes in Omicron spike that contributes to our understanding on the virological determinant and pathogenicity of Omicron.
KEYWORDS: SARS-CoV-2, Omicron BA.1 and BA.2, pathogenesis, entry, endosomal entry pathway, fusogenicity, spike protein cleavage
Introduction
Coronavirus Disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first reported in late 2019 [1–3]. The virus disseminated efficiently and has resulted in a pandemic at the global scale which is still ongoing as of today [4]. Importantly, SARS-CoV-2 continues to generate new variants by acquiring mutations that alter its transmissibility, infectivity, and resistance to neutralizing/therapeutic antibodies. SARS-CoV-2 Omicron BA.1 emerged from South Africa and Botswana in November 2021 and quickly became the most prevalent SARS-CoV-2 variant worldwide due to its high transmissibility and immune evasiveness [5–9]. More recently, continuous surveillance of SARS-CoV-2 evolution revealed additional Omicron lineages, including BA.2, BA.2.12.1, BA.4 and BA.5, which have become the dominant circulating SARS-CoV-2 variants [10,11].
Omicron BA.1 contains a large number of changes in comparison with the ancestral SARS-CoV-2, particularly at its spike protein, which carries 30 substitutions, 3 short deletions and 1 insertion. Among these changes, there were 8 and 15 mutations located at the N-terminal domain (NTD) and receptor-binding domain (RBD), respectively. In addition, several substitutions are located at or near the spike S1/S2 cleavage site. BA.2 spike shares 21 substitutions with that of BA.1 when compared to the ancestral SARS-CoV-2. Meanwhile, BA.2 spike carries 8 specific mutations, including 4 changes (T19I, L24S, 25–27deletion, V213G) in the NTD and 4 substitutions (S371F, T376A, D405N, R408S) in the RBD. BA.2.12.1 spike contains two substitutions, L452Q and S704L, when compared to BA.2. BA.4 and BA.5 share the same spike sequence, which contains three substitutions (L452R, F486V, R493Q) and the 69–70 deletion when compared with BA.2.
We and others recently demonstrated that Omicron BA.1 is less pathogenic compared to SARS-CoV-2 wildtype (WT) or previous variants of concerns (VOCs) [12–14], which is contributed by the unique virological features of Omicron BA.1 in comparison to other SARS-CoV-2 strains, including less efficient transmembrane serine protease 2 (TMPRSS2) usage, less spike cleavage, lower fusogenicity, and an altered entry mechanism [12,14–18]. More recent studies suggest that these virological features of Omicron BA.1 are in part retained by the subsequent Omicron sublineages [19–22]. However, the exact spike determinants that contribute to these observed virological features of Omicron and Omicron sublineages have not been extensively explored. Here, we evaluated the spike determinants for the observed virological characteristics of Omicron BA.1 and BA.2. Our study identified key changes in the spike that contributed to the lower TMPRSS2 usage, reduced spike cleavage, lower fusogenicity, and altered entry mechanism of Omicron BA.1 and BA.2. Together, these findings improved our current knowledge on the virological determinant and pathogenicity of Omicron and Omicron sublineages.
Materials and methods
Viruses and safety
SARS-CoV-2 HKU-001a (WT) (GenBank: MT230904), B.1.617.2/Delta (GenBank: OM212471) and B.1.1.529.1/Omicron BA.1 (GenBank: OM212472) were clinical isolate strains from laboratory-confirmed COVID-19 patients in Hong Kong [23,24]. SARS-CoV-2 HKU-001a S1/S2-10Del (GenBank: MT621560), which carries a 10 amino-acid deletion, was isolated by plaque purification from SARS-CoV-2 cultured in VeroE6 cells and subsequently sequenced [25]. SARS-CoV-2 HKU-001a, Delta and Omicron were cultured using VeroE6-TMPRSS2 cells and titrated by plaque assays [12,26]. All experiments with infectious SARS-CoV-2 were performed according to the approved standard operating procedures of the Biosafety Level 3 facility at Department of Microbiology, HKU [27].
Cell cultures
293T and VeroE6 were obtained from ATCC and maintained in Dulbecco’s modified Eagle’s medium (DMEM) (11965-092, Gibco, Amarillo, Texas, USA) according to supplier’s instructions. Calu3 was obtained from ATCC and maintained in DMEM/F12 (11320-033, Gibco). VeroE6-TMPRSS2 was obtained from the Japanese Collection of Research Bioresources (JCRB) Cell Bank and cultured in DMEM. All cell lines used are routinely tested for mycoplasma and are maintained mycoplasma-free.
Virus replication in cell lines
Calu3 and VeroE6 cells were challenged by WT, Delta, Omicron BA.1 and S1/S2-10Del at 0.5 MOI or 0.1 MOI. At 24 hpi, the cell lysates were harvested for qRT-PCR quantification of virus replication. Viral RNA from infected cells was extracted using QIAsymphony RNA Kit (931636, Qiagen, Germantown Road Germantown, MD, USA). Viral subgenomic RNA of E gene was quantified using the QuantiNova Probe RT–PCR Kit (208354, Qiagen) [28].
Production of SARS-CoV-2-spike pseudoviruses
All mutated SARS-CoV-2-spike pseudoviruses were packaged as described previously [25,29]. In brief, 293T cells were transfected with different spikes, or VSV-G plasmids with Lipofectamine 3000 (L3000015, Thermo Fisher Scientific, Waltham, MA, USA). At 24 h post-transfection, the cells were transduced with VSV-deltaG-firefly pseudotyped with VSV-G. At 2 h post-transduction, the cells were washed three times with PBS and cultured in fresh media with an anti-VSV-G (8G5F11) antibody (EB0010, kerafast, Boston, MA, USA). The pseudoviruses were then harvested 16 h post-transduction and titrated with TCID50. All single mutation plasmids of SARS-CoV-2 spike were constructed in Genscript (Nanjing, China). Q498R was not included in the study because of its low expression.
Protease usage assays
293T cells were transfected with hACE2 or co-transfected with hACE2 and different protease plasmids including TMPRSS2, TMPRSS11D, TMPRSS13, Cathepsin L or Cathepsin B plasmids. The transfected cells were inoculated with pseudoviruses for 24 h post-transfection and cultured in 1% FBS media for another 18 h, before washed and lysed for detection of luciferase signal with a luciferase assay system (E1501, Promega, Madison, WI, USA). The protease plasmids were obtained from OriGene (Rockville, MD, USA) or Sino Biological (Beijing, China).
Western blot analysis of spike cleavage
Spike plasmids of SARS-CoV-2 D614G, Omicron BA.1, BA.2, S1/S2-10Del and all single mutation plasmids were transfected with Lipofectamine 3000 (L3000015, Thermo Fisher Scientific) in 293T cells. Cell lysates were harvested 24 h post-transfection for Western blot analysis. Specific primary antibodies were incubated with the blocked membranes at 4°C overnight, followed by horseradish peroxidase (HRP) conjugated secondary antibodies (62-6520, Thermo Fisher Scientific) for 1 h at room temperature. The signal was developed by SuperSignal West Pico PLUS Chemiluminescent Substrate (34580, Thermo Scientific, USA) and detected using Alliance Imager apparatus (Uvitec, Cambridge, UK). The full-length spike and S2 were detected with a rabbit anti-SARS-CoV-2 spike S2 antibody (40590-T62, Sino Biological) (1:5000). β-actin was detected with a β-actin antibody (clone AC-74, A5316, Sigma, USA) (1:5000). The cleavage ratio of the spike was quantified by ImageJ.
Cell–cell fusion assay
293T cells were co-transfected with different SARS-CoV-2 spike plasmids with GFP1-10 plasmid (cat#68715, Addgene) as effector cells. Another population of 293T cells was co-transfected with human ACE2 (hACE2), TMPRSS2, and GFP11 (cat#68716, Addgene) as target cells. After 24 h post-transfection, the effector and target cells were digested by EDTA-Trypsin (25200072, Gibco) and mixed at a 1:1 ratio. The mixed cells were co-cultured at a 37°C incubator for another 24 h. The mixed cells were fixed in 10% formalin and then permeabilized with 0.1% Triton-X100 (11332481001, Sigma, USA) at room temperature. The antifade mounting medium with 4′,6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI, H-1200, Vector Laboratories) was used for mounting and DAPI staining. Images were taken with the Olympus BX73 fluorescence microscope (Olympus Life Science, Tokyo, Japan). The fusion area of images was quantified by ImageJ.
Protease inhibitor treatment assay
The serine protease inhibitor, camostat (HY-13512), and the cysteine protease inhibitor, E64D (HY-100229), were purchased from MedChemExpress (Monmouth Junction, NJ, USA). Calu3 or VeroE6-TMPRSS2 cells were treated with DMSO, Camostat, or E64D at concentrations of 1, 25, and 50 µM for 2 h before authentic virus infection. At 24 hpi, the cell lysates were harvested for qRT-PCR quantification of virus replication. For pseudovirus entry assays, VeroE6-TMPRSS2 cells were treated with DMSO, Camostat, or E64D at concentrations of 1, 25, and 50 µM for 2 h before pseudoviruses transduction. The cell lysates were lysed for detection of luciferase signal 18 h post-transduction.
Statistical analysis
Statistical comparison among three or more experiment groups was performed with one-way ANOVA with Tukey’s multiple comparison test. Differences were considered statistically significant when p < 0.05. * represented p < 0.05, ** represented p < 0.01, *** represented p < 0.001, and **** represented p < 0.0001. ns, not statistically significant. Data analysis was performed with Graphpad prism 8.0.
Results
Omicron BA.1 is substantially less dependent on TMPRSS2-mediated plasma membrane entry pathway and is highly dependent on the endosomal entry pathway for virus entry
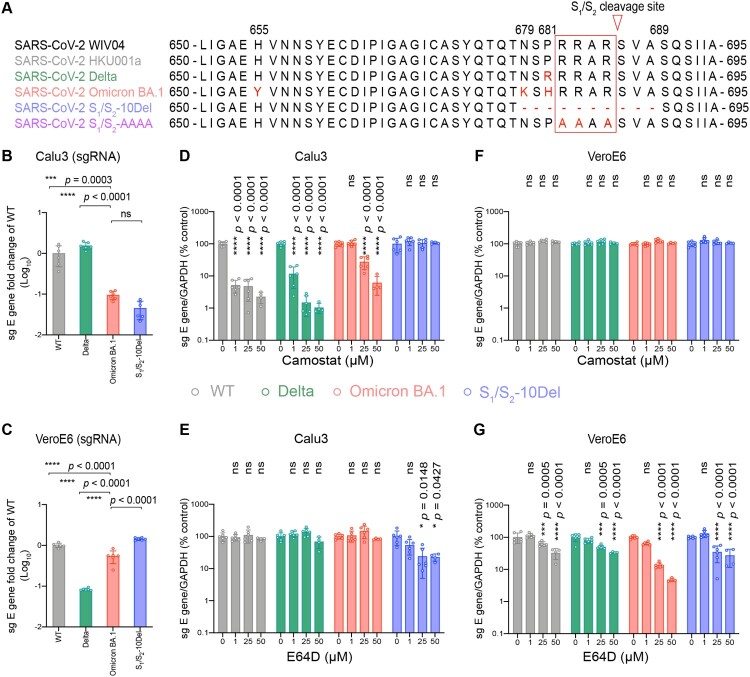
Ancestral SARS-CoV-2 infects lung cells predominantly through the TMPRSS2-mediated plasma membrane entry pathway [30–32]. In cells with low or no TMPRSS2 expression, ancestral SARS-CoV-2 can alternatively enter through endosomes mediated by endosomal proteases such as cathepsin L [33]. We and others recently demonstrated that SARS-CoV-2 Omicron BA.1 is less dependent on TMPRSS2 for virus entry [12,15,34]. In keeping with these findings, more recent reports have suggested an enhanced dependence of endosomal entry by Omicron BA.1 [18,35]. However, comprehensive analyses on Omicron BA.1 entry pathways in association with the contributing spike residues have not been carried out. To this end, we first evaluated the propensity of Omicron BA.1 in utilizing the plasma membrane entry pathway and the endosomal entry pathway for virus entry. For a more thorough investigation, we included SARS-CoV-2 WT, Delta, and S1/S2-10Del as comparison groups (Figure 1(A)). The Delta variant is highly efficient in using TMPRSS2-mediated plasma membrane entry pathway due to the P681R substitution in spike [36–39]. In contrast, the S1/S2-10Del isolate contained a 10 amino acid deletion flanking the S1/S2 cleavage site, resulting in substantially reduced efficiency in plasma membrane entry but gained efficiency in endosomal entry [25,30,40–42]. We first infected Calu3 and VeroE6 cells, which are model cell types for plasma membrane entry and endosomal entry, respectively, with SARS-CoV-2 WT, Delta, Omicron BA.1, and S1/S2-10Del, and quantified virus replication at 24 h post infection (hpi). In Calu3 cells, Omicron BA.1 replication was attenuated when compared to that of WT (P = 0.0003) and Delta (P < 0.0001), and was at similar level with that of S1/S2-10Del (P = ns) (Figure 1(B)). In VeroE6 cells, Omicron BA.1 replication was more efficient than that of Delta (P < 0.0001) but was lower when compared with that of WT (P < 0.0001) and S1/S2-10Del (P < 0.0001) (Figure 1(C)). These results indicate that Omicron BA.1 is less efficient in plasma membrane entry while is capable of efficient endosomal entry.
Figure 1.
Omicron BA.1 is less dependent on TMPRSS2-mediated plasma membrane entry pathway and is highly dependent on the endosomal entry pathway for virus entry. (A) Amino-acid sequence alignment of residues around the S1/S2 cleavage site of SARS-CoV-2 reference strain WIV04, HKU001a, Delta, Omicron BA.1, S1/S2-10Del, and S1/S2-AAAA. Amino acid positions are designated based on SARS-CoV-2 reference strain. (B–C) Calu3 and VeroE6 cells were challenged with SARS-CoV-2 WT, Delta, Omicron BA.1, or S1/S2-10Del. Cell lysates were harvested at 24 hpi for quantification of the subgenomic RNA of the envelope (sgE) gene (n = 6). (D–G) Calu3 and VeroE6 cells were pre-treated with indicated concentrations of camostat or E64D for 2 h followed by the authentic SARS-CoV-2 variants infection. The amount of viral subgenomic envelope RNA in harvested cell lysates at 24 hpi was determined by qRT-PCR (n = 4 for 50uM of camostat and E64D groups, n = 6 for the other groups). Error bars were calculated by using log-transformed data and represented mean ± SD from the indicated number of biological repeats. Statistical significances were determined with one way-ANOVA. Data were obtained from three independent experiments. Each data point represents one biological repeat. * represented p < 0.05, ** represented p < 0.01, *** represented p < 0.001, and **** represented p < 0.0001. ns, not statistically significant; WT, wildtype SARS-CoV-2.
Next, we analyzed the dependency of SARS-CoV-2 WT, Delta, Omicron BA.1, and S1/S2-10Del on the plasma membrane entry pathway and the endosomal entry pathway by treating the infected cells with camostat (a serine protease inhibitor that inhibits plasma membrane entry) or E64D (a cysteine protease inhibitor that inhibits endosomal entry). In Calu3 cells, camostat treatment inhibited the replication of WT, Delta, and Omicron BA.1 in a dose-dependent manner, but not S1/S2-10Del. In keeping with previous findings [12,15], Omicron BA.1 was less sensitive to camostat inhibition as 1μM camostat reduced WT and Delta replication by 94.9% (P < 0.0001) and 88.3% (P < 0.0001), respectively, but did not reduce Omicron BA.1 replication (P = ns) (Figure 1(D)). At high camostat concentration, 50μM camostat reduced WT, Delta, and Omicron replication to 2.25% (P < 0.0001), 1.04% (P < 0.0001), and 6.11% (P < 0.0001) when compared to that of their controls, suggesting that Omicron BA.1 is 2.7-folds and 5.9-folds less sensitive than WT and Delta, respectively, at this high camostat concentration (Figure 1(D)). E64D treatment in Calu3 reduced the replication of S1/S2-10Del, but not that of WT, Delta, and Omicron BA.1 (Figure 1(E)). In VeroE6 cells, camostat treatment did not reduce the replication of all evaluated viruses since this cell type is deficient in TMPRSS2 expression (Figure 1(F)). Meanwhile, E64D treatment in VeroE6 cells most substantially inhibited the replication of Omicron BA.1 among all evaluated viruses (Figure 1(G)). 50μM E64D reduced the replication of WT, Delta, Omicron BA.1, and S1/S2-10Del by 57.7% (P < 0.0001), 65.1% (P < 0.0001), 95.2% (P < 0.0001), and 72.9% (P < 0.0001), respectively (Figure 1(G)). Collectively, these results indicate that Omicron BA.1 is substantially less dependent on TMPRSS2-mediated plasma membrane entry pathway and is highly dependent on the endosomal entry pathway for virus entry.
Spike determinants for the reduced TMPRSS2 usage of Omicron BA.1 and BA.2
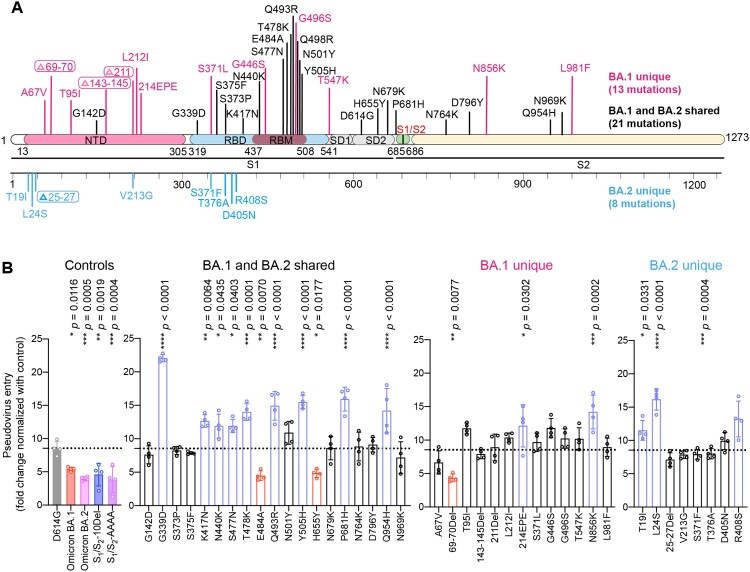
Recent studies identified altered virological features of Omicron BA.1 in comparison to SARS-CoV-2 WT and previous variants, including less efficient TMPRSS2 usage, less spike cleavage, and lower fusogenicity, which may explain its altered entry mechanism and change in pathogenicity [12–15,18,34]. Additional studies suggest that these virological features of Omicron BA.1 are in part retained by the subsequent Omicron sublineages, including BA.2 [19–22]. Omicron BA.1 spike carries 34 changes when compared to the ancestral SARS-CoV-2 spike. Omicron BA.2 spike shares 21 substitutions with that of Omicron BA.1 when compared to the ancestral SARS-CoV-2. Meanwhile, Omicron BA.2 spike carries 8 specific mutations in its NTD and RBD (Figure 2(A)). To delineate the specific spike changes that contribute to the observed virological features in Omicron BA.1 and BA.2, we constructed vesicular stomatitis virus (VSV)-based SARS-CoV-2-spike pseudoviruses carrying individual mutations present on Omicron BA.1 and BA.2 spike with the D614G background. First, we compared the efficiency of TMPRSS2 usage of this panel of pseudoviruses with Omicron BA.1-, BA.2-, S1/S2-10Del-, and S1/S2-AAAA-pseudoviruses included as controls. S1/S2-AAAA was an additional control that contained three amino acid changes (R682A, R683A and R685A) that changed the S1/S2 multibasic cleavage site from RRAR [43] to AAAA (Figure 1(A)). We transfected 293T cells with hACE2 or hACE2 and TMPRSS2, followed by transducing the cells with the panel of pseudoviruses and quantified virus entry at 18 h post transduction. Our results showed that TMPRSS2 overexpression increased D614G-pseudovirus entry by 8.54-fold. In keeping with previous literature [12,15,30,41], Omicron BA.1-, Omicron BA.2-, S1/S2-10Del-, and S1/S2-AAAA-pseudoviruses were attenuated in TMPRSS2 usage in comparison to that of D614G-pseudoviruses. Importantly, our results revealed three spike changes that significantly attenuated TMPRSS2 usage when compared to that of D614G, including 69-70Del (4.35-fold; P = 0.0077), E484A (4.47-fold; P = 0.0070), and H655Y (4.82-fold; P = 0.0177) (Figure 2(B)). Interestingly, these three changes facilitated ACE2-mediated entry into 293T cells (Supplementary Figure 1). Since 69-70Del is only present in Omicron BA.1 but not BA.2 spike, our results indicate that changes at E484A and H655Y contribute to the reduced TMPRSS2 usage of Omicron BA.1 and BA.2.
Figure 2.
Spike determinants for the reduced TMPRSS2 usage of Omicron BA.1 and BA.2. (A) Schematic of all amino acid mutation sites on Omicron BA.1 and BA.2 spike when compared to ancestral SARS-CoV-2 spike. (B) 293T cells were transfected with hACE2 or co-transfected with hACE2 and TMPRSS2, followed by transduction with pseudoviruses bearing the spike of SARS-CoV-2 D614G, Omicron BA.1, Omicron BA.2, S1/S2-10Del, S1/S2-AAAA and individual mutation at 24 h post-transfection. Pseudovirus entry was quantified by measuring the luciferase signal (n = 4). Fold changes in the luciferase signal were normalized to the mean luciferase readouts of cells with only hACE2 overexpression. Data represent mean ± SD from the indicated number of biological repeats. Statistical significance was determined with one way-ANOVA. Data were obtained from three independent experiments. Each data point represents one biological repeat. * represented p < 0.05, ** represented p < 0.01, *** represented p < 0.001, and **** represented p < 0.0001. ns, not statistically significant.
Spike determinants for the reduced spike cleavage of Omicron BA.1 and BA.2
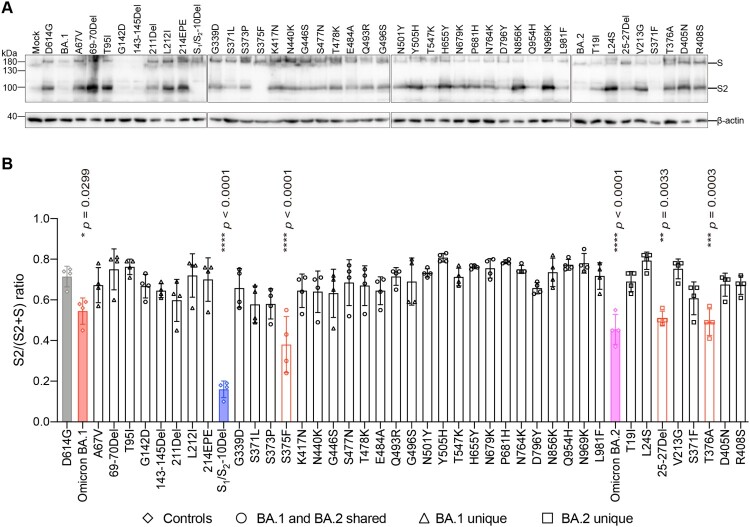
Omicron BA.1 and BA.2 spike carry two substitutions at the S1/S2 cleavage site (N679K, P681H) and three substitutions near the S1/S2 cleavage site (H655Y, N764K, D796Y) (Figure 2(A)). Previous reports suggested that mutations in the N-terminal domain (NTD) and receptor-binding domain (RBD) of spike can also alter spike cleavage [44–46]. To investigate the spike mutation that may contribute to the reduced spike cleavage of Omicron BA.1 and BA.2, we transfected 293T cells with the panel of spike constructs and harvested cell lysates at 24 h post transfection for Western blot analysis. Our results revealed that the cleavage of Omicron BA.1, BA.2, and S1/S2-10Del spike was significantly reduced in comparison that of D614G spike (Figure 3(A,B)), in keeping with previous reports [12,14,15,34,47]. Intriguingly, among the panel of spike constructs, the 25-27Del, S375F, and T376A changes significantly reduced spike cleavage when compared to that of D614G spike (Figure 3(A,B)). Since 25-27Del and T376A are present only in Omicron BA.2 but not in Omicron BA.1 spike, our results indicate that the reduced spike cleavage detected in Omicron BA.1 and BA.2 is associated with the S375F substitution.
Figure 3.
Spike determinants for the reduced spike cleavage of Omicron BA.1 and BA.2. (A) 293T cells were transfected with the indicated spike plasmids. Cell lysates were harvested at 24 h post-transfection for detection of SARS-CoV-2 spike cleavage using an anti-spike S2 antibody. Representative images of spike were shown with β-actin added as a sample processing control. Spike and β-actin were run on different gels and detected on different membranes. The experiment was repeated four times independently with similar results. (B) The cleavage ratio of different spikes from four times independent experiments was quantified by ImageJ. Data represent mean ± SD from the indicated number of biological repeats. Statistical significance was determined with one way-ANOVA. Data were obtained from four independent experiments. Each data point represents one biological repeat. * represented p < 0.05, ** represented p < 0.01, *** represented p < 0.001, and **** represented p < 0.0001. ns, not statistically significant.
Determinants for the reduced spike-mediated cell–cell fusion of Omicron BA.1 and BA.2
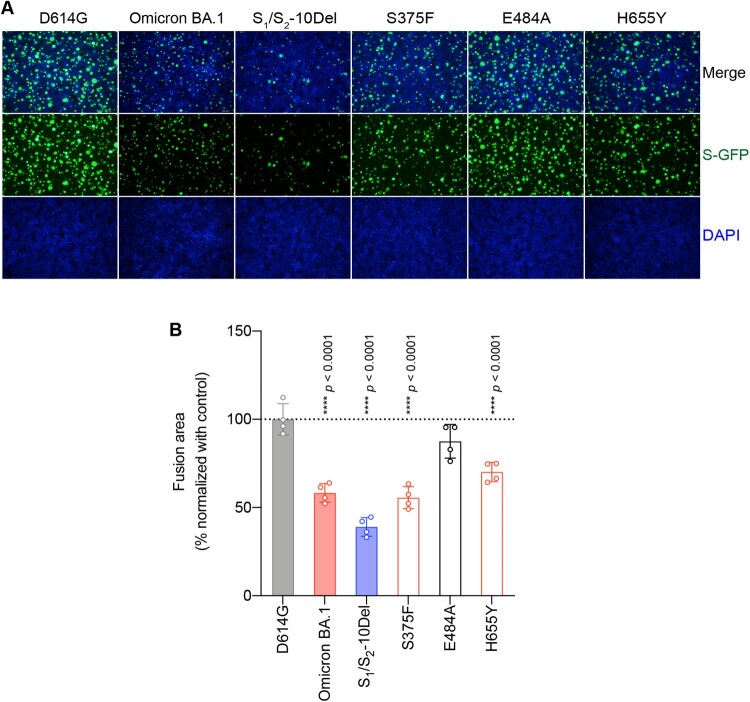
Next, we evaluated the spike mutations that may contribute to the reduced spike-mediated cell–cell fusion of Omicron BA.1 and BA.2. To this end, we analyzed spike-mediated cell–cell fusion assays in 293T cells using the split GFP system [48–50]. We focused our analysis on S375F, E484A, and H655Y since these changes are shared by both Omicron BA.1 and BA.2, and contributed to either reduced TMPRSS2 usage or reduced spike cleavage. Our results suggested that cell–cell fusion induced by Omicron BA.1 and S1/S2-10Del spike was significantly reduced in comparison to that of D614G spike (Figure 4(A,B)), in keeping with previous reports [14,15,30]. In addition, we found that spike-mediated cell–cell fusion was significantly attenuated for S375F (P < 0.0001) and H655Y (P < 0.0001), suggesting that these mutations are associated with the decreased fusogenicity of Omicron BA.1 and BA.2 spike.
Figure 4.
Determinants for the reduced spike-mediated cell–cell fusion of Omicron BA.1 and BA.2. (A) Representative images of spike-mediated cell–cell fusion. 293T cells (effectors cells) were co-transfected with the indicated spike with GFP1-10, and were co-cultured with 293T cells co-transfected with human ACE2 (hACE2), TMPRSS2, and GFP11 (target cells). The co-cultured cells were fixed with 10% formalin and stained with DAPI. Representative images were from four independent experiments with similar results. (B) The fusion area was normalized with the SARS-CoV-2-D614G spike-mediated cell–cell fusion group by ImageJ. Data represent mean ± SD from the indicated number of biological repeats. Statistical significance was determined with one way-ANOVA. Data were obtained from four independent experiments. Each data point represents one biological repeat. * represented p < 0.05, ** represented p < 0.01, *** represented p < 0.001, and **** represented p < 0.0001. ns, not statistically significant.
Spike determinants for the altered entry mechanism of Omicron BA.1 and BA.2
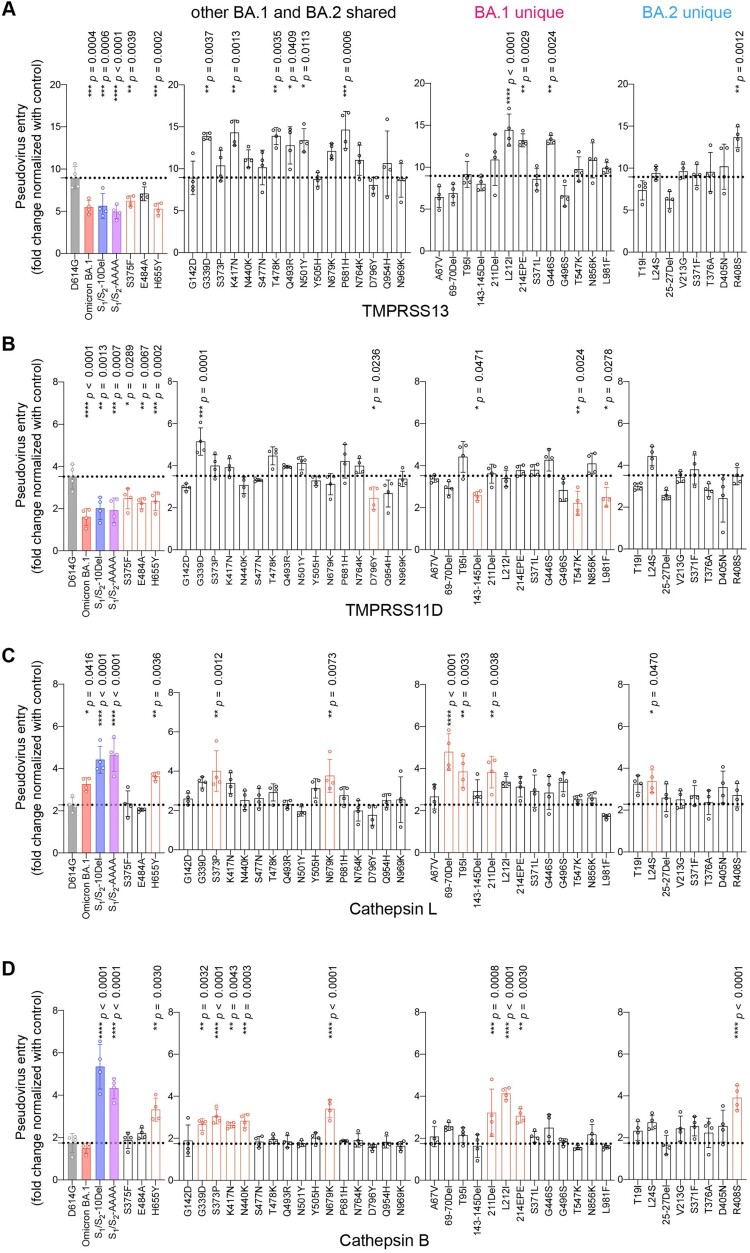
Recent evidence revealed that additional transmembrane serine proteases TMPRSS11D and TMPRSS13 can similarly activate SARS-CoV-2 spike and facilitate SARS-CoV-2 spike pseudovirus entry at the plasma membrane [51–53]. Besides the plasma membrane pathway, SARS-CoV-2 alternatively enters through the endosomal pathway where the spike proteins on virus particles are activated by cathepsin L or cathepsin B in the endosomes [30,31,33]. However, whether or not TMPRSS11D, TMPRSS13, cathepsin L, and cathepsin B are differentially involved in Omicron BA.1 entry is currently incompletely understood. To this end, we first evaluated TMPRSS11D and TMPRSS13 on their capacities to mediate Omicron BA.1 pseudovirus entry. We found that Omicron BA.1 pseudovirus utilized TMPRSS11D and TMPRSS13 at a decreased efficiency when compared to SARS-CoV-2 D614G pseudovirus, and was at comparable level with that of S1/S2-10Del and S1/S2-AAAA pseudoviruses (Figure 5(A,B)). In keeping with our findings described earlier, changes at S375F, E484A, and H655Y reduced TMPRSS11D and/or TMPRSS13 usage, with S375F and H655Y consistently reduced both TMPRSS11D and TMPRSS13 usage (Figure 5(A,B)). Next, we evaluated the role of cathepsin L and cathepsin B on their capacities to facilitate Omicron BA.1 pseudovirus entry and we found that cathepsin L but not cathepsin B promoted Omicron BA.1 pseudovirus entry at an increased efficiency over that of SARS-CoV-2 D614G pseudovirus (Figure 5(C,B)). Interestingly, both cathepsin L and cathepsin B promoted the entry of S1/S2-10Del and S1/S2-AAAA pseudoviruses at significantly increased efficiency when compared with SARS-CoV-2 D614G pseudoviruses (Figure 5(C,D)). Under this setting, the H655Y substitution consistently increased both cathepsin L and cathepsin B usage (Figure 5(C,D)). In parallel, we extended our analysis to the entire panel of Omicron BA.1 and BA.2 individual mutations. We found that although a number of the spike mutations reduced TMPRSS13/11D usage while others increased cathepsin L/B usage, the H655Y substitution was the one that consistently altered serine protease (TMPRSS2, TMPRSS11D, TMPRSS13) and cysteine protease (cathepsin L and cathepsin B) usage (Figures 2(B) and 5(A–D)).
Figure 5.
Spike determinants for the altered entry mechanism of Omicron BA.1 and BA.2. A-D. 293T cells were transfected with hACE2 or co-transfected with hACE2 and TMPRSS13 (A), TMPRSS11D (B), Cathepsin L (C), or Cathepsin B (D), followed by transduction with pseudoviruses expressing the indicated spike at 24 h post-transfection. Pseudovirus entry was quantified by measuring the luciferase signal (n = 4). Fold changes in the luciferase signal were normalized to the mean luciferase readouts of cells with only hACE2 overexpression. Data represent mean ± SD from the indicated number of biological repeats. Statistical significance was determined with one way-ANOVA. Data were obtained from three independent experiments. Each data point represents one biological repeat. * represented p < 0.05, ** represented p < 0.01, *** represented p < 0.001, and **** represented p < 0.0001. ns, not statistically significant.
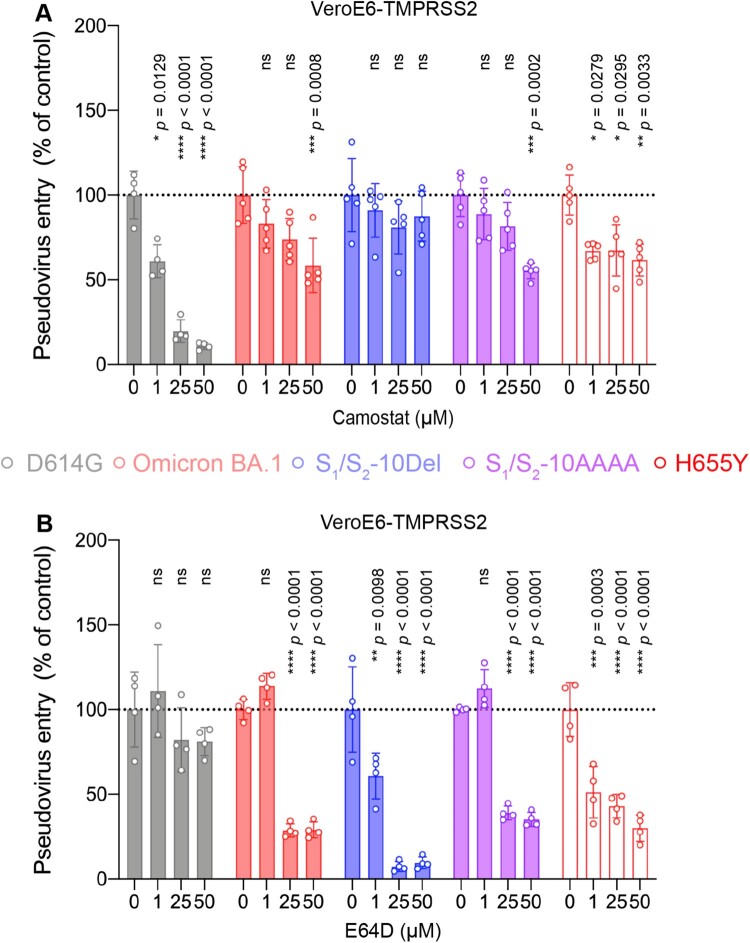
We next asked if H655Y plays a role in modulating the entry pathways utilized by Omicron BA.1. To this end, we compared SARS-CoV-2 D614G, Omicron BA.1, S1/S2-10Del, S1/S2-AAAA, and H655Y pseudovirus entry in VeroE6-TMPRSS2 cells, which supports both plasma membrane entry and endosomal entry, in the presence of camostat or E64D. Our results demonstrated that 50μM camostat reduced SARS-CoV-2 D614G pseudovirus entry by 89.11% (P < 0.0001) while 50μM E64D did not significantly reduce SARS-CoV-2 D614G pseudovirus entry (P = ns) (Figure 6(A,B)). In sharp contrast, 50μM camostat only reduced H655Y pseudovirus entry by 34.81% (P = 0.0033) while 50μM E64D significantly reduced H655Y pseudovirus entry by 69.99% (P < 0.0001) (Figure 6(A,B)). These results indicated that the H655Y pseudoviruses are more dependent on the endosomal entry pathway for virus entry, which are similar to Omicron BA.1, S1/S2-10Del, and S1/S2-AAAA pseudoviruses, and are different from the SARS-CoV-2 D614G pseudovirus (Figure 6(A,B)).
Figure 6.
The H655Y substitution in Omicron BA.1 and BA.2 spike promotes endosomal entry. (A–B) VeroE6-TMPRSS2 cells were pre-treated with 1, 25, or 50μM camostat (A) or E64D (B) or DMSO (A–B) for 2 h followed by transduction with pseudoviruses expressing the indicated spike at 24 h post-transfection. Pseudovirus entry was quantified by measuring the luciferase signal (n = 5 for camostat treatment of Omicron BA.1, S1/S2-10Del, S1/S2-AAAA, and H655Y; n = 4 for the other groups). Data represent mean ± SD from the indicated number of biological repeats. Statistical significances were determined with one way-ANOVA. Data were obtained from three independent experiments. Each data point represents one biological repeat. * represented p < 0.05, ** represented p < 0.01, *** represented p < 0.001, and **** represented p < 0.0001. ns, not statistically significant.
Discussion
Omicron BA.1 and BA.2 carry several unique features that distinguish it from the ancestral SARS-CoV-2 or other previous VOCs [12–17]. These features, including the less efficient TMPRSS2 usage, less spike cleavage, lower fusogenicity, and altered entry mechanism together contributed to its lower pathogenicity in animal models and humans. As of today, the specific amino acid changes contributing to each of these observed phenotypes remain incompletely understood. In this study, we investigated on the spike determinants that contributed to these important virological features of Omicron BA.1 and BA.2. By screening each individual change on Omicron BA.1 and BA.2 spike, we identified that 69–70 deletion, E484A, and H655Y contributed to reduced TMPRSS2 usage while 25–27 deletion, S375F, and T376A resulted in less efficient spike cleavage. Among the shared mutations of BA.1 and BA.2 spike, S375F and H655Y reduced fusogenicity. Interestingly, the H655Y change consistently reduced serine protease (TMPRSS2, TMPRSS13, TMPRSS11D) usage while increased the use of endosomal proteases (cathepsin L and cathepsin B). In keeping with these findings, the H655Y substitution alone reduced plasma membrane entry and facilitated endosomal entry when compared to SARS-CoV-2 WT. Overall, our study identified key changes in Omicron BA.1 and BA.2 spike that contribute to our current knowledge on the virological determinant and pathogenicity of Omicron and Omicron sublineages.
SARS-CoV-2 spike is first processed during virus egress by furin at the S1/S2 site [41,54–56]. Upon binding host angiotensin-converting enzyme 2 (ACE2) on target cells, the pre-processed spike will be further cleaved by TMPRSS2 or other transmembrane serine proteases to facilitate virus fusion with the plasma membrane [51–53,57,58]. Interestingly, in the current study, the identified amino acid changes (25–27 deletion, 69–70 deletion, S375F, T376A, E484A, H655Y) that modulated Omicron BA.1/BA.2 entry-related events, including spike cleavage and TMPRSS2 usage, were not located at the S1/S2 site. In this regard, the exact mechanisms of how these amino acid changes dictate Omicron BA.1 and BA.2 entry warrant further investigation. The findings of our study are in keeping with that from a recent preprint, which suggested the spike S375F substitution could attenuate spike cleavage and fusogenicity due to an interprotomer pi-pi interaction with the H505 residue in another protomer in the spike trimer [45]. A separate preprint similarly suggested the contribution of the spike H655Y substitution in the altered capacity of endosomal entry of Omicron BA.1 without modulating BA.1 spike cleavage [16]. In contrast, a previous report suggested that the H655Y substitution in a clinical isolate could increase spike cleavage and fusion, leading to more efficient virus replication and transmission [59]. The reason behind the discrepancy on the role of H655Y between these studies is currently unknown but may in part attribute to the additional mutations present in the clinical isolate. Alternatively, spike cleavage may be differentially presented in virus-infected and spike-overexpressed cells. The pattern of spike cleavage may also differ based on the models of investigation since the endogenous expression of spike-processing proteases may be different. Importantly, H655Y is also carried by the Gamma variant (P.1), yet Gamma is not known to demonstrate the entry preference of Omicron BA.1 and BA.2. In this regard, the combined effect of the changes on Omicron BA.1 and BA.2 spike should be further evaluated.
Our study has a number of limitations. First, the current study focused on single spike mutations. It is possible that combined analysis of two or more spike mutations is important to better understand the TMPRSS2 usage, spike cleavage, and cell–cell fusion of Omicron. Second, the findings of the current study should be further verified in the context of infectious recombinant viruses.
Overall, by systemically screening the individual mutations on Omicron BA.1 and BA.2 spike, our study reveals key determinants in Omicron BA.1 and BA.2 spike that contribute to our understanding on the virological determinant and pathogenicity of Omicron and Omicron sublineages.
Supplementary Material
Funding Statement
This work was partly supported by funding from the Health and Medical Research Fund [CID-HKU1-5, COVID1903010-Projects 7 and 14, 20190652, and COVID190214], the Food and Health Bureau, the Government of the Hong Kong Special Administrative Region; the General Research Fund [17118621 and 17123920], Collaborative Research Fund [C7060-21G], and Theme-Based Research Scheme [T11-709/21-N and T11-706/18-N], the Research Grants Council of the Hong Kong Special Administrative Region; Health@InnoHK, Innovation and Technology Commission, the Government of the Hong Kong Special Administrative Region; National Natural Science Foundation of China Excellent Young Scientists Fund (Hong Kong and Macau) [32122001]; National Program on Key Research Project of China [grant number 2020YFA0707500 and 2020YFA0707504]; the Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Diseases and Research Capability on Antimicrobial Resistance for Department of Health of the Hong Kong Special Administrative Region Government, Sanming Project of Medicine in Shenzhen, China [No. SZSM201911014]; the High Level-Hospital Program, Health Commission of Guangdong Province, China; the University of Hong Kong Li Ka Shing Faculty of Medicine Enhanced New Staff Start-up Fund; the University of Hong Kong Outstanding Young Researcher Award; the University of Hong Kong Research Output Prize (Li Ka Shing Faculty of Medicine); the Major Science and Technology Program of Hainan Province [ZDKJ202003]; the research project of Hainan Academician Innovation Platform [YSPTZX202004]; the Hainan Talent Development Project [SRC200003]; Emergency Collaborative Project [EKPG22-01] of Guangzhou Laboratory; and Emergency COVID-19 Project [2021YFC0866100], Major Projects on Public Security, National Key Research and Development Program; and the donations of the Shaw Foundation Hong Kong, Richard Yu and Carol Yu, May Tam Mak Mei Yin, Michael Seak-Kan Tong, the Providence Foundation Limited (in memory of the late Lui Hac Minh), Lee Wan Keung Charity Foundation Limited, Hui Ming, Hui Hoy and Chow Sin Lan Charity Fund Limited, The Chen Wai Wai Vivien Foundation Limited, Hong Kong Sanatorium & Hospital, Chan Yin Chuen Memorial Charitable Foundation, Marina Man-Wai Lee, the Hong Kong Hainan Commercial Association South China Microbiology Research Fund, the Jessie & George Ho Charitable Foundation, Perfect Shape Medical Limited, Kai Chong Tong, Foo Oi Foundation Limited, Tse Kam Ming Laurence, Betty Hing-Chu Lee, Ping Cham So, and Lo Ying Shek Chi Wai Foundation. The funding sources had no role in the study design, data collection, analysis, interpretation, or writing of the report.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Chan JF, Yuan S, Kok KH, et al. . A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020 Feb 15;395(10223):514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P, Yang XL, Wang XG, et al. . A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 Mar;579(7798):270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan JF, Kok KH, Zhu Z, et al. . Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9(1):221–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.To KK, Sridhar S, Chiu KH, et al. . Lessons learned 1 year after SARS-CoV-2 emergence leading to COVID-19 pandemic. Emerg Microbes Infect. 2021 Dec;10(1):507–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Viana R, Moyo S, Amoako DG, et al. . Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022 Mar;603(7902):679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao Y, Wang J, Jian F, et al. . Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022 Feb;602(7898):657–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carreño JM, Alshammary H, Tcheou J, et al. . Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022 Feb;602(7898):682–688. [DOI] [PubMed] [Google Scholar]
- 8.Cele S, Jackson L, Khoury DS, et al. . Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022 Feb;602(7898):654–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu L, Iketani S, Guo Y, et al. . Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022 Feb;602(7898):676–681. [DOI] [PubMed] [Google Scholar]
- 10.Hachmann NP, Miller J, Collier AY, et al. . Neutralization escape by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4, and BA.5. N Engl J Med. 2022 Jul 7;387(1):86–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arora P, Kempf A, Nehlmeier I, et al. . Augmented neutralisation resistance of emerging omicron subvariants BA.2.12.1, BA.4, and BA.5. Lancet Infect Dis. 2022 Aug;22(8):1117–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shuai H, Chan JF, Hu B, et al. . Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature. 2022 Mar;603(7902):693–699. [DOI] [PubMed] [Google Scholar]
- 13.Halfmann PJ, Iida S, Iwatsuki-Horimoto K, et al. . SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022 Mar;603(7902):687–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki R, Yamasoba D, Kimura I, et al. . Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature. 2022 Mar;603(7902):700–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meng B, Abdullahi A, Ferreira I, et al. . Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature. 2022 Mar;603(7902):706–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto M, Tomita K, Hirayama Y, et al. SARS-CoV-2 Omicron spike H655Y mutation is responsible for enhancement of the endosomal entry pathway and reduction of cell surface entry pathways. bioRxiv. 2022:2022.03.21.485084.
- 17.Peacock TP, Brown JC, Zhou J, et al. The altered entry pathway and antigenic distance of the SARS-CoV-2 Omicron variant map to separate domains of spike protein. bioRxiv. 2022:2021.12.31.474653.
- 18.Zhao H, Lu L, Peng Z, et al. . SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg Microbes Infect. 2022 Dec;11(1):277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamasoba D, Kimura I, Nasser H, et al. . Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike. Cell. 2022 Jun 9;185(12):2103–2115.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uraki R, Kiso M, Iida S, et al. . Characterization and antiviral susceptibility of SARS-CoV-2 Omicron BA.2. Nature. 2022 Jul;607(7917):119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura I, Yamasoba D, Tamura T, et al. Virological characteristics of the novel SARS-CoV-2 Omicron variants including BA.2.12.1, BA.4 and BA.5. bioRxiv. 2022:2022.05.26.493539.
- 22.Qu P, Faraone JN, Evans JP, et al. Differential evasion of delta and omicron immunity and enhanced fusogenicity of SARS-CoV-2 Omicron BA.4/5 and BA.2.12.1 Subvariants. bioRxiv. 2022:2022.05.16.492158.
- 23.Lu L, Mok BW, Chen LL, et al. . Neutralization of SARS-CoV-2 Omicron variant by sera from BNT162b2 or Coronavac vaccine recipients. Clin Infect Dis. 2021 Dec 16;75(1):e822–e826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu H, Chan JF, Yuen TT, et al. . Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020 May;1(1):e14–e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu H, Hu B, Huang X, et al. . Host and viral determinants for efficient SARS-CoV-2 infection of the human lung. Nat Commun. 2021 Jan 8;12(1):134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang BZ, Shuai H, Gong HR, et al. . Bacillus Calmette-Guérin-induced trained immunity protects against SARS-CoV-2 challenge in K18-hACE2 mice. JCI Insight. 2022 Jun 8;7(11):e157393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu H, Hou Y, Yang D, et al. . Coronaviruses exploit a host cysteine-aspartic protease for replication. Nature. 2022 Aug 3. doi: 10.1038/s41586-022-05148-4. [DOI] [PubMed] [Google Scholar]
- 28.Yuen TT, Chan JF, Yan B, et al. . Targeting ACLY efficiently inhibits SARS-CoV-2 replication. Int J Biol Sci. 2022;18(12):4714–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shuai H, Chan JF, Yuen TT, et al. . Emerging SARS-CoV-2 variants expand species tropism to murines. EBioMedicine. 2021 Nov;73:103643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffmann M, Kleine-Weber H, Pöhlmann S.. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell. 2020 May 21;78(4):779–784.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beumer J, Geurts MH, Lamers MM, et al. . A CRISPR/Cas9 genetically engineered organoid biobank reveals essential host factors for coronaviruses. Nat Commun. 2021 Sep 17;12(1):5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ou X, Liu Y, Lei X, et al. . Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020 Mar 27;11(1):1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koch J, Uckeley ZM, Doldan P, et al. . TMPRSS2 expression dictates the entry route used by SARS-CoV-2 to infect host cells. EMBO J. 2021 Aug 16;40(16):e107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du X, Tang H, Gao L, et al. . Omicron adopts a different strategy from Delta and other variants to adapt to host. Signal Transduct Target Ther. 2022 Feb 10;7(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poston D, Weisblum Y, Hobbs A, et al. . VPS29 exerts opposing effects on endocytic viral entry. mBio. 2022 Apr 26;13(2):e0300221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mlcochova P, Kemp SA, Dhar MS, et al. . SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021 Nov;599(7883):114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, Liu J, Johnson BA, et al. . Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. Cell Rep. 2022 May 17;39(7):110829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saito A, Irie T, Suzuki R, et al. . Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature. 2022 Feb;602(7896):300–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neerukonda SN, Vassell R, Lusvarghi S, et al. . SARS-CoV-2 delta variant displays moderate resistance to neutralizing antibodies and spike protein properties of higher soluble ACE2 sensitivity, enhanced cleavage and fusogenic activity. Viruses. 2021 Dec 11;13(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson BA, Xie X, Bailey AL, et al. . Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature. 2021 Mar;591(7849):293–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peacock TP, Goldhill DH, Zhou J, et al. . The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol. 2021 Jul;6(7):899–909. [DOI] [PubMed] [Google Scholar]
- 42.Walls AC, Park YJ, Tortorici MA, et al. . Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020 Apr 16;181(2):281–292.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coutard B, Valle C, de Lamballerie X, et al. . The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020 Apr;176:104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meng B, Kemp SA, Papa G, et al. . Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep. 2021 Jun 29;35(13):109292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kimura I, Yamasoba D, Nasser H, et al. SARS-CoV-2 spike S375F mutation characterizes the Omicron BA.1 variant. bioRxiv. 2022:2022.04.03.486864. [DOI] [PMC free article] [PubMed]
- 46.Meng B, Datir R, Choi J, et al. SARS-CoV-2 spike n-terminal domain modulates TMPRSS2-dependent viral entry and fusogenicity. bioRxiv. 2022:2022.05.07.491004. [DOI] [PMC free article] [PubMed]
- 47.Pastorio C, Zech F, Noettger S, et al. . Determinants of spike infectivity, processing, and neutralization in SARS-CoV-2 Omicron subvariants BA.1 and BA.2. Cell Host Microbe. 2022 Jul 18;S1931-3128(22)00352-3. doi: 10.1016/j.chom.2022.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ren W, Ju X, Gong M, et al. . Characterization of SARS-CoV-2 variants B.1.617.1 (Kappa), B.1.617.2 (Delta), and B.1.618 by cell entry and immune evasion. mBio. 2022 Apr 26;13(2):e0009922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kamiyama D, Sekine S, Barsi-Rhyne B, et al. . Versatile protein tagging in cells with split fluorescent protein. Nat Commun. 2016 Mar 18;7:11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buchrieser J, Dufloo J, Hubert M, et al. . Syncytia formation by SARS-CoV-2-infected cells. Embo J. 2021 Feb 1;40(3):e107405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoffmann M, Hofmann-Winkler H, Smith JC, et al. . Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. EBioMedicine. 2021 Mar;65:103255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laporte M, Raeymaekers V, Van Berwaer R, et al. . The SARS-CoV-2 and other human coronavirus spike proteins are fine-tuned towards temperature and proteases of the human airways. PLoS Pathog. 2021 Apr;17(4):e1009500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kishimoto M, Uemura K, Sanaki T, et al. . TMPRSS11D and TMPRSS13 activate the SARS-CoV-2 spike protein. Viruses. 2021 Feb 28;13(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bestle D, Heindl MR, Limburg H, et al. . TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci Alliance. 2020 Jul 23;3(9):e202000786. doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Millet JK, Whittaker GR.. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci USA. 2014 Oct 21;111(42):15214–15219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park JE, Li K, Barlan A, et al. . Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc Natl Acad Sci USA. 2016 Oct 25;113(43):12262–12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoffmann M, Kleine-Weber H, Schroeder S, et al. . SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 Apr 16;181(2):271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zang R, Gomez Castro MF, McCune BT, et al. . TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol. 2020 May 13;5(47):eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Escalera A, Gonzalez-Reiche AS, Aslam S, et al. . Mutations in SARS-CoV-2 variants of concern link to increased spike cleavage and virus transmission. Cell Host Microbe. 2022 Mar 9;30(3):373–387.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.