Abstract
Biphenyl dioxygenase (Bph Dox) catalyzes the initial oxygenation of biphenyl and related compounds. Bph Dox is a multicomponent enzyme in which a large subunit (encoded by the bphA1 gene) is significantly responsible for substrate specificity. By using the process of DNA shuffling of bphA1 of Pseudomonas pseudoalcaligenes KF707 and Burkholderia cepacia LB400, a number of evolved Bph Dox enzymes were created. Among them, an Escherichia coli clone expressing chimeric Bph Dox exhibited extremely enhanced benzene-, toluene-, and alkylbenzene-degrading abilities. In this evolved BphA1, four amino acids (H255Q, V258I, G268A, and F277Y) were changed from the KF707 enzyme to those of the LB400 enzyme. Subsequent site-directed mutagenesis allowed us to determine the amino acids responsible for the degradation of monocyclic aromatic hydrocarbons.
Biphenyl-utilizing bacteria have been extensively studied in terms of the degradation of polychlorinated biphenyls (PCB), which have been recognized as some of the most significant environmental pollutants (7). These PCB-degrading bacteria exhibit substantial differences in the range of degradation ability and in congener selectivity for PCB. The biphenyl dioxygenases (Bph Dox) are involved in the initial oxygenation of biphenyl and thereby the cometabolic degradation of PCB (10). The Bph Dox of Pseudomonas pseudoalcaligenes KF707 and Burkholderia cepacia LB400 exhibit distinct differences in the substrate range for PCB (6, 9), although these two Bph Dox share over 95% identity in their amino acid sequences (5, 22). These Bph Dox are multicomponent enzymes encoded by four genes, bphA1A2A3A4, where bphA1 encodes a large subunit (BphA1) of the terminal dioxygenase (an iron-sulfur protein), bphA2 encodes a small subunit (BphA2) of the terminal dioxygenase, bphA3 encodes the ferredoxin (BphA3), and bphA4 encodes the ferredoxin reductase (BphA4) (5, 22). BphA1 contains a [2Fe-2S] Rieske center which is involved in electron transfer from the ferredoxin component to a mononuclear Fe2+, which is believed to activate molecular oxygen (1, 13, 17). Among these four subunits, BphA1 is crucially responsible for the recognition and binding of the substrates and thereby for substrate specificity (6, 10, 15). Previously, we constructed various bphA1 variants by using DNA shuffling between the KF707 and LB400 bphA1 genes (16). Some of the evolved Bph Dox thus obtained exhibited enhanced abilities to degrade PCB and some biphenyl-related compounds. Further screening of these clones allowed us to obtain evolved Bph Dox which exhibit extremely enhanced abilities to degrade benzene, toluene, and alkylbenzenes, such as ethylbenzene, isopropylbenzene, and butylbenzene.
A library of evolved bphA1 genes was created by DNA shuffling between the bphA1 genes of strains KF707 and LB400 as previously described (16). The shuffled bphA1 genes were digested with SacI and BglII, inserted just upstream of bphA2A3A4BC in pJHF18ΔMluI, and transformed into Escherichia coli XL1-Blue. The clones grown on Luria-Bertani agar plates containing ampicillin at 50 μg/ml and 0.1 mM isopropyl-β-d-thiogalactopyranoside were screened for the ability to produce yellow pigment from biphenyl (8). Eighty positive clones thus selected were then analyzed for the ability to degrade biphenyl-related compounds and monocyclic aromatic hydrocarbons. The production of yellow meta ring cleavage products from various aromatic compounds was monitored with the supernatants at the following absorbance wavelengths: biphenyl, 434 nm; diphenylmethane, 395 nm; dibenzofuran, 465 nm; benzene, 388 nm; toluene, 375 nm; ethylbenzene, 319 nm; isopropylbenzene, 321 nm; butylbenzene, 323 nm.
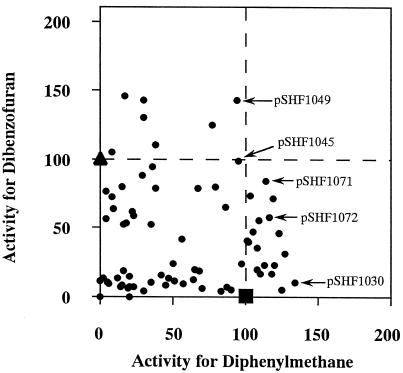
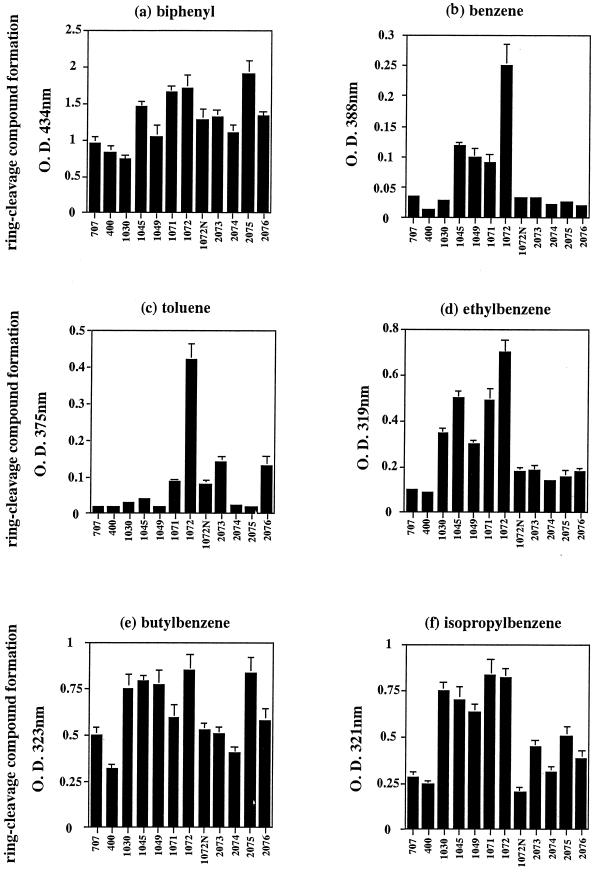
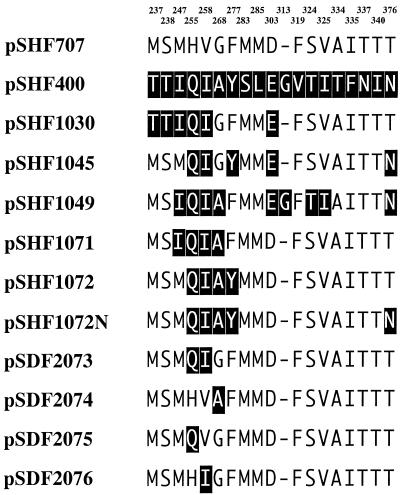
The degradative activities of the 80 positive clones toward diphenylmethane and dibenzofuran are presented in Fig. 1. First, it is noted that Escherichia coli(pSHF707) expressing the original BphA1 (KF707 enzyme) and E. coli(pSHF400) expressing the original LB400 BphA1 (LB400 enzyme) exhibited major differences in the formation of the meta ring cleavage yellow products for these two compounds. The yellow compound 2-hydroxy-6-oxo-6-benzylhexa-2,4-dienoic acid was produced from diphenylmethane by the KF707 enzyme but not by the LB400 enzyme. In contrast, the yellow compound 2-hydroxy-6-oxo-6-(2-hydroxyphenyl)-hexa-2,4-dienoic acid was produced from dibenzofuran by the LB400 enzyme but not by the KF707 enzyme. The formation of yellow meta ring-cleavage products from diphenylmethane and dibenzofuran were monitored with the supernatants at the corresponding absorption maxima. The amount of yellow meta ring cleavage product from diphenylmethane was 4.64 nmol/mg of protein/min by E. coli expressing the original KF707 Bph Dox, and that from dibenzofuran by the original LB400 Bph Dox was 3.16 nmol/mg of protein/min. The 80 positive clones which exhibited both diphenylmethane and dibenzofuran degradation activities were screened, and the relative abilities to degrade these two compounds were determined, where the activities of E. coli expressing the above original Bph Dox were set to 100 as the basal activities. Among these, we selected five clones, E. coli carrying pSHF1030, pSHF1045, pSHF1049, pSHF1071, and pSHF1072, which produced indigo on Luria-Bertani agar plates, because it is known that oxygenases capable of metabolizing monocyclic aromatic compounds such as toluene, phenol, and styrene transform indole to indigo (3, 4, 11, 18). As expected, these clones exhibited enhanced abilities to degrade ethylbenzene, isopropylbenzene, and butylbenzene (Fig. 2). These clones produced almost the same amount of Bph Dox, as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). In particular, E. coli(pSHF1072) was interesting because this clone exhibited wide and enhanced capabilities to degrade not only the alkylbenzenes but also benzene and toluene, compounds scarcely attacked by the original Bph Dox. The amounts of yellow compounds produced from ethylbenzene, isopropylbenzene, and butylbenzene by this clone were two to seven times those produced by E. coli(pSHF707) and E. coli(pSHF400) expressing the respective original Bph Dox. Sequence analyses of the evolved BphA1 that acquired the higher activities for the alkylbenzenes gained the same two amino acids from the LB400 enzyme, i.e., His255Gln and Val258Ile (KF707 numbering) (Fig. 3). These results suggest that alterations of His-255 and Val-258 in the KF707 enzyme are important for the enhancement of substrate specificity toward monocyclic aromatic hydrocarbons. In order to investigate this point, site-directed mutagenesis was applied to pSHF1072, in which two or three amino acids of the four substituted amino acids from the LB400 enzyme were changed to those of the KF707 enzyme.
FIG. 1.
Formation of yellow meta ring cleavage products from diphenylmethane and dibenzofuran by E. coli expressing chimeric biphenyl dioxygenases. The cells were incubated with 0.1 mM substrate at 30°C for 1 h. The formation of yellow compounds was measured at the corresponding absorption maximum. The degradation activities of KF707 Bph Dox and LB400 Bph Dox were used as the basal activities (set to 100) toward diphenylmethane and dibenzofuran, respectively. The activity of KF707 Bph Dox is shown by the larger square on the x axis, and that of LB400 Bph Dox is shown by the larger triangle on the y axis. The closed circles indicate the activities in E. coli expressing the evolved Bph Dox. The relative degradation activities of 80 clones were plotted for the basal activities.
FIG. 2.
Formation of meta-cleaved yellow compounds from a variety of aromatic hydrocarbons by E. coli expressing chimeric Bph Dox. Equal amounts of E. coli cells expressing evolved Bph Dox were incubated with the substrate at 30°C for 1 h (biphenyl, isopropylbenzene, butylbenzene, and ethylbenzene) or 8 h (benzene and toluene). The formation of the yellow compounds was measured at the corresponding absorption maximum. The results are shown as average values ± the standard deviation of three independent experiments. O.D., optical density.
FIG. 3.
Sequence analyses of the resultant shuffled bphA1 genes. Twenty amino acids that differ between KF707 BphA1 and LB400 BphA1 are shown with KF707 numbering at the top. The dash indicates an amino acid lacking in KF707 relative to the LB400 sequence. The pSDF series plasmids were constructed by site-directed mutagenesis.
Bph Dox from pSDF2073 and pSDF2076 possess the amino acids Q255 and I258 and the amino acid I258, respectively, from the LB400 enzyme. E. coli expressing these enzymes exhibited slightly greater activities against all of the substrates tested than did the KF707 enzyme but much less activity than the pSHF1072 enzyme. However, these enzymes retained relatively greater activity against toluene. Bph Dox from pSDF2075, in which only Gln-255 was derived from the LB400 enzyme, exhibited almost the same activity for biphenyl and butylbenzene as did the pSHF1072 enzyme but much lower activities against benzene and toluene. Bph Dox from pSDF2074 exhibited activities similar to those of the KF707 enzyme, in which three amino acids at positions 255, 258, and 277 were the reverse of those of the KF707 enzyme.
It was previously shown that the replacement of Thr-376 with Asn led to the acquisition of 3,4-dioxygenase activity for 2,5,4′-trichlorobiphenyl and 2,5,2′,5′-tetrachlorobiphenyl (15, 20). Furthermore, a change to Val at the same position produced the novel abilities to degrade dibenzofuran, dibenzo-p-dioxin, dibenzothiophene, and fluorene (21). These results indicated that the amino acid at position 376 in BphA1 plays a very important role in determining substrate selectivity. Therefore, a variant, pSHF1072N, in which Thr-376 in the pSHF1072 enzyme was changed to Asn, was created. The Bph Dox from pSHF1072N exhibited almost the same degradation ability as the original Bph Dox from pSHF707 and pSHF400 for the substrates tested (Fig. 2). The Bph Dox from pSHF34 (20) and pSHF1046, in which Thr-376 in the KF707 enzyme was replaced with Asn and Asn-376 in the LB400 enzyme was replaced with Thr, respectively, hardly attacked benzene, toluene, and ethylbenzene (data not shown). These results suggest that even one amino acid change at position 376 in BphA1 significantly affected the ability to degrade monocyclic aromatic hydrocarbons and that the combination of Gln-255, Ile-258, Ala-268, Tyr-277, and Thr-376 is important for the enhanced degradation of benzene and toluene, as seen in pSHF1072.
It has been reported that the toluene dioxygenase of Pseudomonas putida F1 shows a wide range of substrate specificities for various aromatic compounds (2, 12, 23). In this study, we found that the toluene dioxygenase from F1 exhibited high oxygenation activities toward benzene (316% of pSHF1072 Bph Dox) and toluene (142%) but relatively low activities for ethylbenzene (29%), butylbenzene (25%), and isopropylbenzene (32%), of which the latter three compounds have bulky side groups. In naphthalene dioxygenase, replacement of Thr-351, corresponding to Thr-376 in KF707 BphA1, with Arg had a large effect on product formation from phenanthrene (19). Although the three-dimensional structure of Bph Dox is not available yet, that of naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4, whose structure is similar to that of Bph Dox, was solved by X-ray analysis (14). Based on the structural information from the naphthalene dioxygenase, we tried to analyze the possible structure of KF707 BphA1 (data not shown). The results indicate that the four amino acids in pSHF1072 BphA1 are situated surrounding a mononuclear iron center which is supposed to be an active site. The flexibility of amino acids near the active site may lead to the relaxation of substrate binding and allow expansion of the abilities of the pSHF1072 enzyme to degrade a variety of aromatic hydrocarbons.
Thus, more appropriate combinations of amino acids involved in substrate recognition will allow us to evolve new and novel enzymes with much wider and enhanced oxygenation capacities. DNA shuffling is an effective approach by which to get such evolved enzymes.
REFERENCES
- 1.Batie C J, Ballou D P, Correll C J. Phthalate dioxygenase reductase and related flavin-iron-sulfur containing electron transferases. In: Muller F, editor. Chemistry and biochemistry of flavoenzymes. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 544–554. [Google Scholar]
- 2.Cho M C, Kang D-O, Yoon B D, Lee K. Toluene degradation pathway from Pseudomonas putida F1: substrate specificity and gene induction by 1-substituted benzenes. J Ind Microbiol Biotechnol. 2000;25:163–170. [Google Scholar]
- 3.Eaton R W, Chapman P J. Formation of indigo and related compounds from indolecarboxylic acids by aromatic acid-degrading bacteria: chromogenic reactions for cloning genes encoding dioxygenases that act on aromatic acids. J Bacteriol. 1995;177:6983–6988. doi: 10.1128/jb.177.23.6983-6988.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ensley B D, Ratzkin B J, Ossulund T D, Simon M J, Wackett L P, Gibson D T. Expression of naphthalene oxidation genes in Escherichia coli results in the biosynthesis of indigo. Science. 1983;22:167–169. doi: 10.1126/science.6353574. [DOI] [PubMed] [Google Scholar]
- 5.Erickson B D, Mondello F J. Nucleotide sequencing and transcriptional mapping of the genes encoding biphenyl dioxygenase, a multicomponent polychlorinated-biphenyl-degrading enzyme in Pseudomonas strain LB400. J Bacteriol. 1992;174:2903–2912. doi: 10.1128/jb.174.9.2903-2912.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erickson B D, Mondello F J. Enhanced biodegradation of polychlorinated biphenyls after site-directed mutagenesis of a biphenyl dioxygenase gene. Appl Environ Microbiol. 1993;59:3858–3862. doi: 10.1128/aem.59.11.3858-3862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furukawa K. Microbial degradation of polychlorinated biphenyls. In: Chakrabarty A M, editor. Biodegradation and detoxification of environmental pollutants. Boca Raton, Fla: CRC Press, Inc.; 1982. pp. 33–57. [Google Scholar]
- 8.Furukawa K, Miyazaki T. Cloning of gene cluster encoding biphenyl degradation in Pseudomonas pseudoalcaligenes. J Bacteriol. 1986;166:392–398. doi: 10.1128/jb.166.2.392-398.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibson D T, Cruden D L, Haddock J D, Zylstra G J, Brand J M. Oxidation of polychlorinated biphenyls by Pseudomonas sp. strain LB400 and Pseudomonas pseudoalcaligenes KF707. J Bacteriol. 1993;175:4561–4564. doi: 10.1128/jb.175.14.4561-4564.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haddok J D, Horton J R, Gibson D T. Dihydroxylation and dechlorination of chlorinated biphenyls by purified biphenyl 2,3-dioxygenase from Pseudomonas sp. strain LB400. J Bacteriol. 1995;177:20–26. doi: 10.1128/jb.177.1.20-26.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hart S, Koch K R, Woods D R. Identification of indigo related pigments produced by Escherichia coli containing a clone Rhodococcus gene. J Gen Microbiol. 1992;138:211–216. doi: 10.1099/00221287-138-1-211. [DOI] [PubMed] [Google Scholar]
- 12.Hirose J, Suyama A, Hayashida S, Furukawa K. Construction of hybrid biphenyl (bph) and toluene (tod) genes for functional analysis of aromatic ring dioxygenases. Gene. 1994;138:27–33. doi: 10.1016/0378-1119(94)90779-x. [DOI] [PubMed] [Google Scholar]
- 13.Jiang H, Parales R E, Lynch N A, Gibson D T. Site-directed mutagenesis of conserved amino acids in the α subunit of toluene dioxygenase: potential mononuclear non-heme iron coordination sites. J Bacteriol. 1996;178:3133–3139. doi: 10.1128/jb.178.11.3133-3139.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kauppi B, Lee K, Carredano E, Parales R E, Gibson D T, Eklund H, Ramaswamy S. Structure of an aromatic ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure. 1998;6:571–586. doi: 10.1016/s0969-2126(98)00059-8. [DOI] [PubMed] [Google Scholar]
- 15.Kimura N, Nishi A, Goto M, Furukawa K. Functional analyses of a variety of chimeric dioxygenases constructed from two biphenyl dioxygenases that are similar structurally but different functionally. J Bacteriol. 1997;179:3936–3943. doi: 10.1128/jb.179.12.3936-3943.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumamaru T, Suenaga H, Mitsuoka M, Furukawa K. Enhanced degradation of polychlorinated biphenyls by directed evolution of biphenyl dioxygenase. Nat Biotechnol. 1998;16:663–666. doi: 10.1038/nbt0798-663. [DOI] [PubMed] [Google Scholar]
- 17.Mason J R, Cammack R. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu Rev Microbiol. 1992;46:277–305. doi: 10.1146/annurev.mi.46.100192.001425. [DOI] [PubMed] [Google Scholar]
- 18.Murdock D, Ensley B D, Serdar C, Thalen M. Construction of metabolic operons catalyzing the de novo synthesis of indigo in Escherichia coli. Bio/Technology. 1993;11:381–386. doi: 10.1038/nbt0393-381. [DOI] [PubMed] [Google Scholar]
- 19.Parales R E, Lee K, Resnick S M, Jiang H, Lessner D J, Gibson D T. Substrate specificity of naphthalene dioxygenase: effect of specific amino acids at the active site of the enzyme. J Bacteriol. 2000;182:1641–1649. doi: 10.1128/jb.182.6.1641-1649.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suenaga H, Nishi A, Watanabe T, Sakai M, Furukawa K. Engineering a hybrid pseudomonad to acquire 3,4-dioxygenase activity for polychlorinated biphenyls. J Biosci Bioeng. 1999;87:430–435. doi: 10.1016/s1389-1723(99)80090-5. [DOI] [PubMed] [Google Scholar]
- 21.Suenaga H, Goto M, Furukawa K. Emergence of multifunctional oxygenase activities by random priming recombination. J Biol Chem. 2001;276:22500–22506. doi: 10.1074/jbc.M101323200. [DOI] [PubMed] [Google Scholar]
- 22.Taira K, Hirose J, Hayashida S, Furukawa K. Analysis of bph operon from the polychlorinated biphenyl-degrading strain of Pseudomonas pseudoalcaligenes KF707. J Biol Chem. 1992;267:4844–4853. [PubMed] [Google Scholar]
- 23.Zylstra G J, Gibson D T. Toluene degradation by Pseudomonas putida F1: nucleotide sequence of the todC1C2BADE genes and their expression in Escherichia coli. J Biol Chem. 1989;264:14940–14946. [PubMed] [Google Scholar]