ABSTRACT
Dysbiosis of gut microbiota is well-described in patients with coronavirus 2019 (COVID-19), but the dynamics of antimicrobial resistance genes (ARGs) reservoir, known as resistome, is less known. Here, we performed longitudinal fecal metagenomic profiling of 142 patients with COVID-19, characterized the dynamics of resistome from diagnosis to 6 months after viral clearance, and reported the impact of antibiotics or probiotics on the ARGs reservoir. Antibiotic-naive patients with COVID-19 showed increased abundance and types, and higher prevalence of ARGs compared with non-COVID-19 controls at baseline. Expansion in resistome was mainly driven by tetracycline, vancomycin, and multidrug-resistant genes and persisted for at least 6 months after clearance of SARS-CoV-2. Patients with expanded resistome exhibited increased prevalence of Klebsiella sp. and post-acute COVID-19 syndrome. Antibiotic treatment resulted in further increased abundance of ARGs whilst oral probiotics (synbiotic formula, SIM01) significantly reduced the ARGs reservoir in the gut microbiota of COVID-19 patients during the acute infection and recovery phase. Collectively, these findings shed new insights on the dynamic of ARGs reservoir in COVID-19 patients and the potential role of microbiota-directed therapies in reducing the burden of accumulated ARGs.
KEYWORDS: COVID-19, gut microbiome, antimicrobial resistance gene, synbiotic formula, SIM01
Introduction
Antimicrobial resistance (AMR) poses a major health threat, is responsible for more than 700,000 deaths per year, and is projected to increase to up to 10 million deaths globally by 2050.1 The coronavirus 2019 (COVID-19) pandemic has affected antimicrobial stewardship activities and driven AMR in various ways due to empirical use of antibiotics and increasing secondary bacterial infections among patients with COVID-19. Studies and meta-analysis have shown that approximately three-quarters of patients with COVID-19 received antibiotics but only less than 10% had bacterial or fungal co-infections.2,3
The gut microbiome serves as a reservoir of antimicrobial resistance genes (ARGs), known as the gut resistome,4–6 which can horizontally communicate between commensals and pathogens, contributing to the emergence of drug-resistant bacteria.7–10 The gut microbiome in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-infected subjects was characterized by enrichment of opportunistic pathogens and depletion of beneficial commensals.11–15 Gut dysbiosis reported in COVID-19 patients persisted at least 6 months after virus clearance and may be associated with post-acute COVID-19 syndrome (PACS).16–19 Use of antibiotics in COVID-19 patients contributes to resistome expansion with distinct ARGs profile compared to non-COVID-19 patients treated with antibiotics,20 suggesting specific effects of COVID-19 on the resistome.
Increasing interest has emerged to develop therapeutic strategies against COVID-19 by modulating gut microbiota via dietary intervention, probiotics, and prebiotics.21 Probiotics have been hailed as a mean to restore microbiome balance after perturbation by antibiotics and, consequently, prevent resistome expansion.22 In patients with COVID-19, probiotics have been shown to modulate host immune responses against SARS-CoV-2, restore microbiome balance and, consequently, improve outcome or reduce long-term complications in COVID-19 patients.23,24 A pilot study showed that 4 weeks of oral supplementation of a probiotic formula (S1M01) targeted to replenish bacteria species known to be depleted in COVID-19 subjects hastened recovery and antibody formation, reduced nasopharyngeal viral load, and suppressed serum proinflammatory cytokines in hospitalized COVID-19 patients.25 Others have shown that a probiotic mix can reduce the number of ARGs in colonization-permissive, antibiotic-naive individuals.5
We therefore hypothesize that patients with COVID-19 have a high risk of ARGs accumulation and that probiotic therapy may have a role in reducing the resistome in these COVID-19. In this study, we performed longitudinal metagenomic analysis of fecal samples of 142 patients with COVID-19 and characterized the dynamics of ARGs reservoir from admission to 6 months after viral clearance. We also demonstrated the potential of probiotics to reduce the number of ARGs accumulated during COVID-19 disease in antibiotic-naive individuals.
Results
Antibiotic-naive COVID-19 patients harbor more ARGs than non-COVID-19 controls
Between 1 March 2020 and 31 January 2021, we included 66 antibiotic-naive patients (no antibiotics treatment during SARS-CoV-2-positive period) and 24 antibiotic-experienced patients with confirmed SARS-CoV-2 infection and followed them up to 9 months after virus clearance. All these patients did not use antibiotics for at least 6 months before and after SARS-CoV-2 infection. We also included 66 non-COVID-19 controls matched for age, gender, and comorbidities with COVID-19 patients (Figure 1a). We performed metagenomic profiling of stool samples and characterized their resistome. We found that Bray–Curtis dissimilarities separated the resistome of baseline samples of COVID-19 patients (antibiotic-naive, n = 66) and non-COVID-19 controls (p < .0001, PERMANOVA test). The diversity (Kruskal–Wallis p = 3.6e-14 based on Shannon index, Figure 1c) and the number of observed ARGs (p = 7.3e-08 based on ARG types, Figure 1d; p = .0003 based on ARGs, Figure 1e) were significantly higher in samples from COVID-19 patients. After normalization to the total number of 16s reads, the overall abundance of ARGs remained significantly higher in samples from COVID-19 patients than non-COVID-19 controls (p = 5.2e-07, Figure 1f). The prevalence of all observed ARG types was also higher than that of non-COVID-19 controls (Figure 1g, Supplementary Table 1). The most prevalent ARG types in COVID-19 patients were that of tetracycline (77%), chloramphenicol (76%), and multi-drug resistance (74%). Resistome composition (ARG types or ARGs level) was, however, not associated with demographic factors (age, sex, comorbidities) and COVID-19 severity, suggesting that all COVID-19 patients who were antibiotic-naive were susceptible to ARGs (Supplementary Figure 1A). We analyzed the correlation between ARGs and the gut microbiota at the species level by Masslin2, and found that these ARGs were significantly positively correlated with a variety of Proteobacteria, Firmicutes, and Bacteroidetes (FDR < 0.05), indicating that these bacteria may be the main hosts of ARGs (Supplementary Figure 1B).
Figure 1.
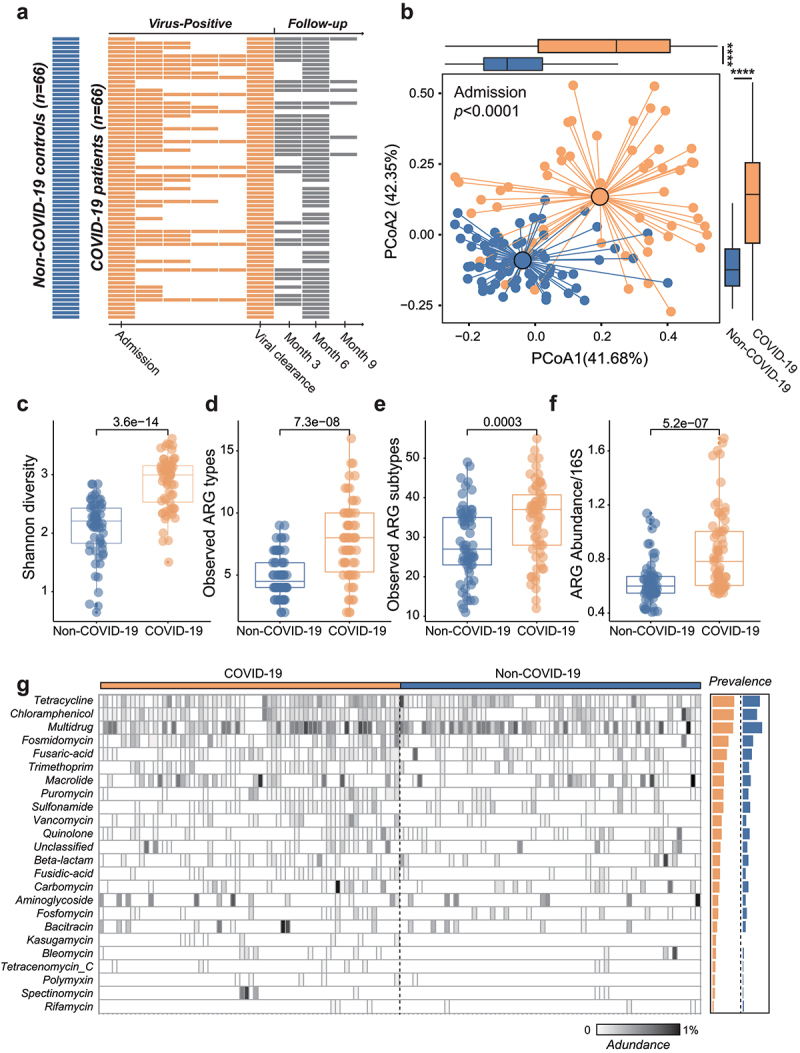
Antibiotic-naive COVID-19 patients harbor more ARGs than non-COVID-19 controls. (a) Schematic overview of the study design, depicting the total number of samples and participants from whom data were available. The horizontal bars represent the sample collected at specific time point from antibiotic-naive COVID-19 patients. (b) Bray–Curtis-based beta diversity of baseline stool samples from COVID-19 patients and non-COVID-19 subjects based on ARG subtypes. The alpha diversity (Shannon, C), observed ARG types (d), observed ARG subtypes (e), and the normalized abundance of ARGs (f) were all significantly higher in stool samples from COVID-19 patients than that of non-COVID-19 subjects. (g) The prevalence of ARG types in COVID-19 patients and non-COVID-19 subjects.
Resistome expansion persists for 6 months in antibiotic-naive COVID-19 patients
To characterize the dynamics of resistome in COVID-19 patients, we performed longitudinal analysis of stool samples collected during the SARS-CoV-2-positive period and after clearance of SARS-CoV-2. The number of observed ARG showed a significant rise from baseline to viral clearance (Kruskal–Wallis p < .001 based on ARG types, Figure 2a; p < .001 based on ARGs, Figure 2b), demonstrating that ARGs further expanded during the virus-positive period. A similar trend was also seen for the overall abundance of ARGs (p < .001, Figure 1c). The increment of ARGs (samples collected at viral clearance compared to baseline samples) during the virus-positive period was not significantly associated with other potential confounders (Supplementary Figure 1C). Besides, in 42 COVID-19 patients who provided serial stool samples during hospitalization enabled us to further depict the longitudinal dynamics of resistome during the virus-positive period (Figure 1a). We found that the resistome (including type, number, and abundance of ARGs) gradually expanded in the gut microbiome over time (Supplementary Figure 2A–C). The increment of resistome also exhibited significant associations with the length of virus-positive period (Supplementary Figure 2E and F).
Figure 2.
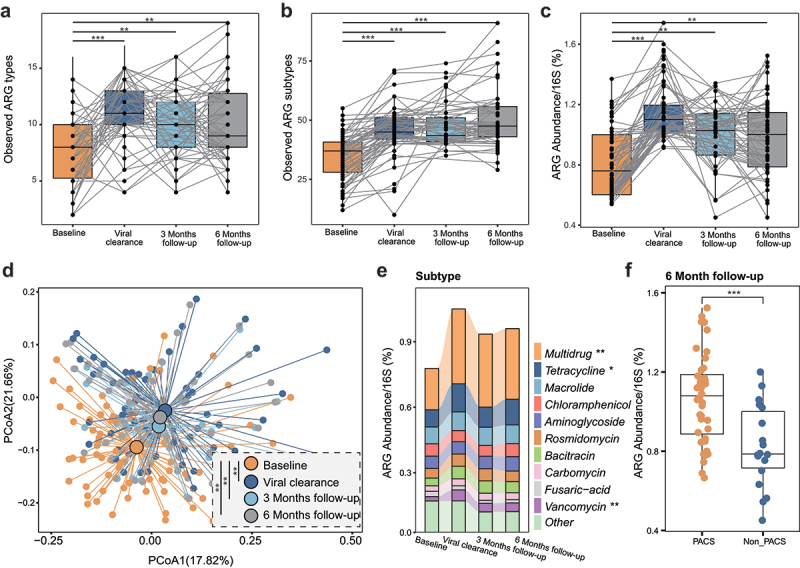
Resistome expansion persists for 6 months in antibiotic-naive COVID-19 patients. Dynamics of the observed ARG types (a), subtypes (b) and normalized abundance (c) in antibiotic-naive COVID-19 patients from baseline to 6-month follow-up. (d) Bray–Curtis dissimilarity of ARGs (‘types’) in stool samples from baseline to 6-month follow-up. (e) Abundance of antibiotic resistance ‘types’ in stool samples from baseline to 6-month follow-up. (f) The normalized abundance of ARGs in subjects with PACS is significantly higher than that of subjects without PACS at 6-month follow-up.
Unlike previous studies showing partial or complete recovery of the gut microbiome in COVID-19 patients after SARS-CoV-2 viral clearance,17,18 we did not observe significant reduction in resistome during 6-month follow-up in COVID-19 patients (Figure 2a-c). In particular, the observed ARG types and/or the total abundance of ARGs were significantly higher at 3- and 6-month follow-up compared with baseline samples (all p < .01, Figure 2a-c). In six patients followed up to 9 months, the resistome in their stool samples remained significantly higher compared with their baseline samples (Supplementary Figure 3). Beyond this, Bray–Curtis dissimilarities clearly separated the resistome of baseline samples from that of all follow-up samples (p < .01, Figure 2d). Furthermore, the expansion of resistome of COVID-19 patients during the virus-positive period was primarily driven by ARGs against multidrug resistance (p < .01), tetracycline (p < .05), and vancomycin (p < .01, Figure 2e). There was no reduction in these ARG types during follow-up (Figure 2e). These data illustrated that the expanded resistome in COVID-19 patients persisted for at least 6 months despite recovery and clearance of SARS-CoV-2.
Expanded resistome predicts risk of post-acute COVID-19 syndrome
It has been reported that a substantial number of patients developed PACS after the acute infection consisting of symptoms affecting multiple systems that can last for months, although the exact cause for this is not clear.26 We found that the overall ARGs abundance in patients with PACS (n = 48) was significantly higher than those without PACS at 6-month follow-up (n = 18, p < .0001, Figure 2f). Then, we retrospectively analyzed AMR profiles of patients when their SARS-CoV-2 turned negative. At the ARG level, we used eigengap method to analyze the similarity of resistome of the 66 patients and divided them into two groups based on the overall composition of resistome and the two clusters were verified by Bray–Curtis dissimilarities (p < .001, PERMANOVA test, Figure 3a). The observed numbers (Kruskal–Wallis p = .00093 based on ARG types, Figure 3b; p = .0079 based on ARGs, Figure 3c) and total abundance of ARGs (p = 2.4e-05) were significantly higher in subjects in cluster 1 (n = 34) than cluster 2 (n = 32), suggesting that subjects in cluster 1 exhibited a more expanded resistome than those in cluster 2 at viral clearance. Importantly, such differences still existed in samples analyzed at 3-month and 6-month follow-up (Supplementary Figure 4). A previous work showed the associations between Klebsiella and several symptoms of PACS.18 In this study, the gut metagenome of subjects in cluster 1 showed higher prevalence of pathogenic Klebsiella than cluster 2 after virus clearance, and the overall prevalence of Klebsiella in cluster 1 rose from 35% to 70% (3-month follow-up, Figure 3e), while such rise was much smaller in cluster 2 (28% to 31%), suggesting that these individuals with higher resistome may be more susceptible to opportunistic or secondary Klebsiella infections.27,28 Besides, we observed that the overall incidence of PACS at 6 months in cluster 1 (85%) was 1.5 times higher than that of cluster 2 (56%, FDR<0.1, figure 3f). Specifically, patients in cluster 1 were 1.5, 2.2, 1.9, 2, and 1.5 times more likely to develop symptoms of anxiety, diarrhea, memory problems, insomnia, and fatigue, respectively, than patients in cluster 2 (all FDR<0.1, Figure 3f). Overall, these findings suggest that expanded resistome may predict a higher risk of PACS, though the exact mechanisms for this are unclear.
Figure 3.
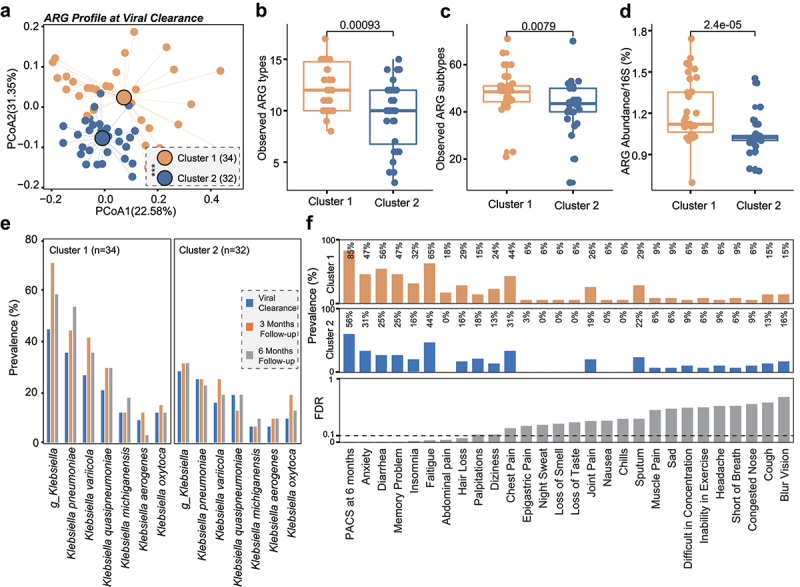
Expanded resistome predicts risk of post-acute COVID-19 syndrome. (a) Bray–Curtis dissimilarity of ARGs (‘subtypes’) in stool samples from cluster 1 and cluster 2 at viral clearance. The observed ARG types (b), subtypes (c), and the normalized abundance of ARGs (d) were all significantly higher in stool samples from COVID-19 patients in cluster 1 than that of cluster 2 at viral clearance. (e) The prevalence of Klebsiella in COVID-19 patients of cluster 1 and cluster 2 at viral clearance, 3-month and 6-month follow-up. (f) The prevalence of PACS in COVID-19 patients of cluster 1 and cluster 2 at 6-month follow-up. Eigengap method was used to analyze the similarity of resistome of the 66 antibiotic-naive patients and divided them into two clusters based on the overall composition of resistome.
Antibiotics further expand the resistome in COVID-19 patients
According to various statistics, about 30–70% of COVID-19 patients were prescribed antibiotics.2,3,29 It is of paramount importance to understand the effect of antibiotics on resistome in COVID-19 patients. In 24 COVID-19 patients who were prescribed antibiotics during hospitalization, we calculated their increment of ARGs at type, subtype, and abundance levels from admission to virus clearance and compared them with the corresponding increments in 66 antibiotic-naive patients. Increase in the observed number of ARGs in antibiotic-treated patients was significantly higher than that of antibiotic-naive patients (Kruskal–Wallis p = .027 based on ARG type, Figure 4a; p = .026 based on ARGs, Figure 4b). The use of antibiotics also drove the increment of ARGs abundance by approximately threefold in antibiotic-treated compared with antibiotic-naive patients (p = 2.1e-11; Figure 4c). The increase in abundance of ARGs was not associated with the duration of antibiotics treatment (p > .05, Figure 4d). However, patients who received two types of antibiotics (ceftriaxone plus doxycycline) had a larger ARG increments than those receiving only one type of antibiotic (amoxicillin clavulanate, p = .0014, Figure 4e), indicating that multiple antibiotics further promoted the expansion of the resistome. Expanded resistome at the type and subtype levels induced by antibiotics also persisted for at least 6 months in these patients (Figure 4d; Supplementary Figure 5). Overall, these findings supported that antibiotic use further expands fecal resistome in COVID-19 patients.
Figure 4.
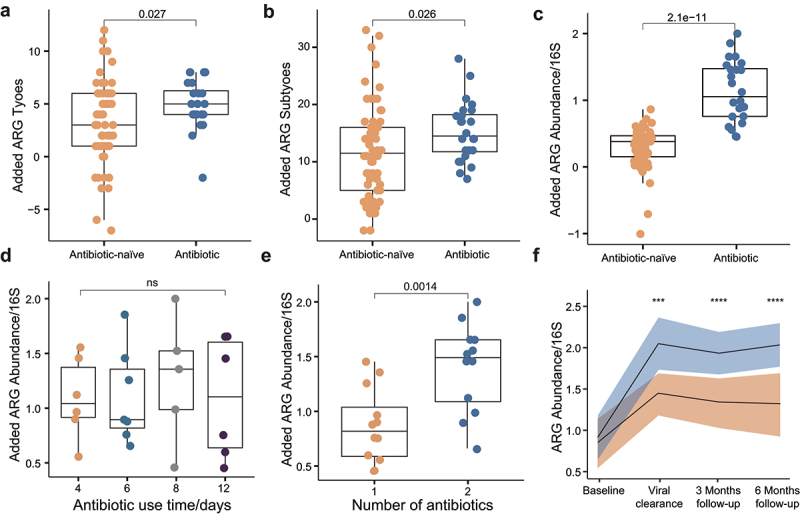
Antibiotics further expand the resistome in COVID-19 patients. The increment of the observed ARG types (a), subtypes (b), and abundance (c) in antibiotic-naive COVID-19 patients is significantly higher than that of antibiotic-experienced COVID-19 patients. Increment refers to the expansion of resistome from baseline to viral clearance. (d) The increment of ARGs abundance in antibiotic-experienced COVID-19 patients is not associated with the length of antibiotic use time. (e) The increment of ARGs abundance in subjects who received two antibiotics is significantly higher than that of subjects who received one antibiotic. (f) Dynamics of ARGs abundance in antibiotic-naive COVID-19 patients and antibiotic-experienced COVID-19 patients. *P < .05, **P < .01, ***P < .001, ****P < .0001, Kruskal–Wallis and Dunn’s tests (all panels).
Probiotics reduce ARGs in COVID-19 patients during virus-positive period
To determine the effects of probiotics on the gut resistome of COVID-19 patients, we analyzed metagenomic data from our previous open-label study of probiotic treatment in consecutive hospitalized patients with a confirmed diagnosis of SARS-CoV-2 (registration number: NCT04581018).25 We analyzed stool samples of 22 COVID-19 patients treated with the oral probiotics (synbiotic formula SIM01, containing three probiotics of Bifidobacterium strains, galactooligosaccharides, xylooligosaccharide, and resistant dextrin) for 4 weeks and 10 subjects without SIM01 as controls (Figure 5a). All patients received standard treatments and diets for COVID-19 according to the hospital protocol, and provided longitudinal stool samples from baseline to 12 weeks. Probiotics was associated with an increased dissimilarity of resistome configuration compared with pre-treatment ARGs (PERMANOVA test, p = 2.24e-8, Figure 5b). There was no difference in resistome dissimilarity in the control group (p = .0892). Furthermore, we observed significant decreases in the observed number (p = 2.85e-05 based on ARG types, Figure 5c; p = 4.09e-10 based on ARGs) and relative abundance of ARGs (p = 1.81e-10, Figure 5e) after probiotic supplementation. However, there was no change in ARGs number and abundance in the control group (all p > .05, Figure 5c-e). Specifically, in the SIM01 group, taking probiotics for 2 weeks was associated with a significantly reduced resistome (p < .0001 comparing baseline and week 2 samples, Supplementary Figure 6A-C), and there was no rebound after stopping SIM01 until 12-week follow-up (p > .05 comparing week 4 and follow-up samples, Supplementary Figure 6A–C). These results suggest that SIM01 not only inhibits the expansion of resistome during the virus positive period but also effectively removes the accumulated ARGs.
Figure 5.
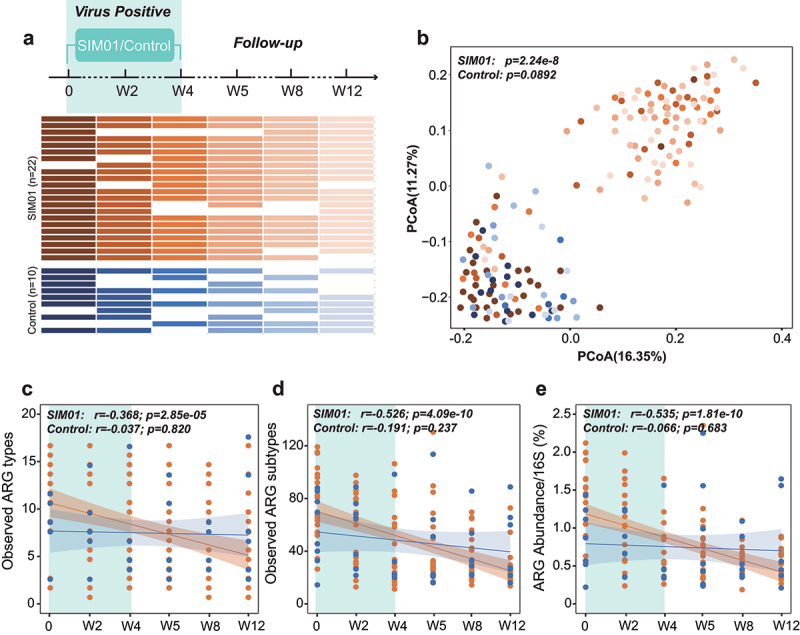
Probiotics reduce ARGs in COVID-19 patients during virus-positive period. (a) Schematic overview of the study design, depicting the total number of samples and participants from whom data were available. The horizontal bars represent the sample collected at specific time point. (b) Probiotics were associated with an increased dissimilarity of resistome configuration compared with pre-treatment ARGs. The observed ARG types (c), subtypes (d), and abundance (e) in COVID-19 patients exhibited significant decrease after taking probiotics, but no significant trend was found in control group (Pearson Correlation).
Probiotics reduce ARGs in COVID-19 after virus clearance
Our data showed that the expanded resistome persisted in COVID-19 patients for over 6 months after virus clearance (Figure 2). We next determined whether taking probiotics after virus clearance can also help reduce ARGs. To this purpose, we performed a separate open-label pilot study of 20 COVID-19 patients who received SIM01 for 4 weeks after viral clearance (defined as negative SARS-CoV-2 qPCR on nasopharyngeal samples; registration number: NCT04730284) and analyzed their stool samples for up to 12 weeks (Figure 6a). Interestingly, Bray–Curtis dissimilarities clearly separated the resistome at baseline and that after probiotics supplementation in subjects taking SIM01 (PERMANOVA test, p = 1.57e-7, Figure 6b). The observed number (p < .01 based on ARG types, Figure 6c; p < .01 based on ARGs, Figure 6d) and relative abundance (p < .001, Figure 6e) of ARGs were significantly lower in week 2 to week 12 samples compared to that of pre-supplementation baseline. The resistome reduced with time during probiotic use (p < .0001 from baseline to week 4, Supplementary Figure 7A–C). When patients were followed up to 12 weeks, we found that their resistome remained stable and did not expand spontaneously despite stopping probiotics (all p > .05 comparing week 4 and follow-up samples, Supplementary Figure 7A-C). These data highlight that SIM01 was associated with reduction of ARGs in samples of patients with COVID-19 who had recovered.
Figure 6.
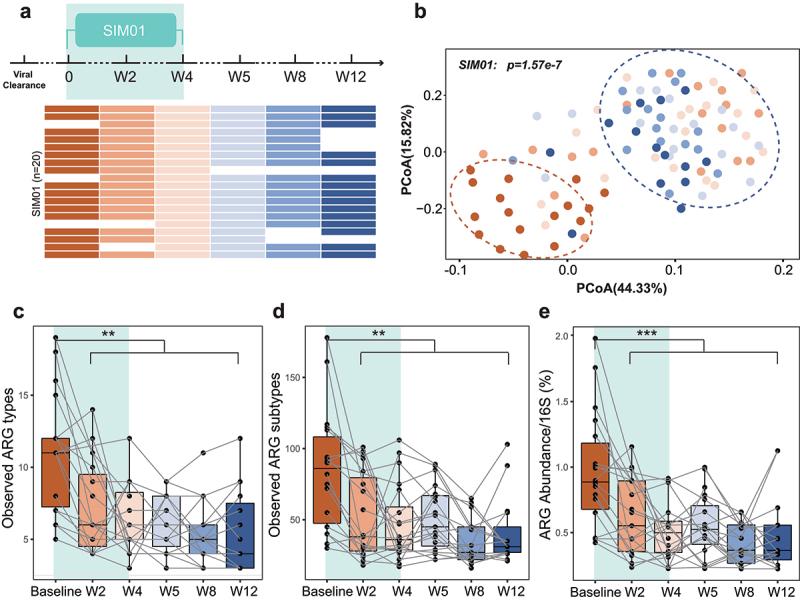
Probiotics reduce ARGs in COVID-19 patients after viral clearance. (a) Schematic overview of the study design, depicting the total number of samples and participants from whom data were available. The horizontal bars represent the sample collected at specific time point. (b) Bray–Curtis dissimilarities clearly separated the resistome at baseline and that after probiotics supplementation in subjects taking SIM01. The dynamics of the observed ARG types (c), subtypes (d), and abundance (e) in subjects taking SIM01 from baseline to week 12. *P < .05, **P < .01, ***P < .001, ****P < .0001, Kruskal–Wallis and Dunn’s tests (all panels).
Discussion
Understanding factors that shape human gut resistome and devising means to circumvent resistome expansion can facilitate the battle against AMR. In this study, we report longitudinal dynamics of the intestinal reservoir of ARGs in COVID-19 patients from admission to 6 months after viral clearance, and characterized the effects of antibiotics and probiotics on the resistome by analyzing shotgun metagenomic sequencing data from several human cohorts. We showed a significant expansion of resistome in COVID-19 at admission, which further increased during the virus-positive period and persisted for 6 months after viral clearance. Antibiotics use during the virus-positive period further led to expansion of resistome, while supplementation with a commercial probiotic (synbiotic formula, SIM01) before or after viral clearance was associated with reduction of the number and abundance of ARGs accumulated in COVID-19 patients, and there was no rebound during the 12-week follow-up.
These novel findings highlight that COVID-19 patients are susceptible to ARG accumulation, which may be due to gut dysbiosis, enrichment of opportunistic bacteria and superbugs from the hospital environment.30,31 Gut dysbiosis in COVID-19 patients characterized by enrichment of pathogenic bacteria like Klebsiella sp. will not only potentially attack the host and aggravate the disease11–13,18,27,28 but are also the most common carriers of ARGs.10,32,33 Some Klebsiella can drive horizontal gene transfer via outer membrane vesicles,34 thus accelerating the transfer of ARGs from pathogenic bacteria to commensal, posing a long-lasting threat to human health. Unlike previous studies that reported partial or complete recovery of the gut microbiome after SARS-CoV-2 clearance,17,18,35,36 we found that the expanded resistome in post-acute COVID-19 subjects did not show any downward trend until 6-month follow-up, reflecting the stability of ARGs in the gut. Furthermore, the effect of antibiotics on gut microbiome is generally considered to subside within 3 to 6 months, based on compositional analysis.18,37 However, when we probed deeper into the ARGs level, resistome expansion caused by antibiotics maintained for a longer time in the patients which led us to reconsider the interaction between antibiotics and gut microbiome at the functional level.
The genes that expanded the most in COVID-19 patients confer resistance against tetracycline, vancomycin, and multidrug resistance. Previous studies have demonstrated horizontal transfer of these genes among microbes,8,38,39 and pointed out their harm to human health.40,41 Similarly, our analysis showed that the expanded resistome may predict a higher risk of PACS after SARS-CoV-2 clearance but the causal relationship between them should be further explored. All these findings suggest an urgent need for reducing ARGs in COVID-19 patients as our analysis showed that all patients were susceptible to ARGs during SARS-CoV-2-positive period despite differences in age, gender, COVID-19 severity, and so on.
Whether probiotics can effectively eliminate ARGs of gut microbiome is still controversial.42 In particular, several commercial probiotics have been proven to carry multiple ARGs,43,44 and a previous study reported that a commercially available probiotics containing 11 different strains further exacerbated resistome expansion after antibiotics using,5 highlighting the importance of developing personalized probiotics for specific disease or condition.23 SIM01 is a synbiotic product customized for COVID-19 patients aiming to replenish the missing bacteria associated with immune defense.11,45 Our previous work confirmed its efficacy as an adjuvant therapy on immunologic response and gut microbiota among patients hospitalized for COVID-19,25 while this study further proved that it can significantly clear the ARGs no matter used during the virus-positive period or after virus clearance. Efficient ARGs clearance that was sustained after discontinuation of probiotics during 12-week follow-up may be related to the higher colonization ability of SIM01,25 considering that colonization is important to support a beneficial and clinically relevant effect on resistome reduction.5
This work also has some limitations. First, the sample size is small, especially for patients with antibiotic or probiotic usage, and the sample collection from control cases varies from 1 to 3 months prior to the COVID-19 pandemic in Hong Kong. Therefore, there may be unknown confounding factors that could potentially impact the results, and our findings should be confirmed in larger cohorts across different populations. Hospitalization is an important factor in the ARGs accumulation,46 thus using non-COVID-19 subjects hospitalized in the same period as controls with longitudinal data can further enhance the robustness of the results. PACS or long COVID-19 is a top concern in the current pandemic and will inevitably lead to long-term disability and increase healthcare burden;26 thus, the potential causality between expanded resistome and PACS requires further verification. Next, whether PACS can be alleviated after removal of accumulated ARGs also requires further clinical investigations.
COVID-19 with emerging variants continues to cause a global toll. It is imperative to gain better understanding of the role of gut microbiome in COVID-19 and PACS, and to develop efficient treatments. Our data highlighted the susceptibility of the gut microbiome to ARGs in hospitalized COVID-19 patients and demonstrated the efficiency of probiotics in reducing resistome. Our findings provide new insights into future gut microbiome-based therapeutics to reduce health risks posed by expansion of resistome.
Patients and methods
Study subjects
Consecutive participants were recruited under the Clinical Research Ethics Committee (REC) no. 2020.076, and all subjects provided informed consent. Our first cohort was a longitudinal cohort of 90 subjects (66 antibiotic-naive patients, no antibiotics treatment during SARS-CoV-2-positive period; 24 antibiotic-experienced patients, 12 patients were on amoxicillin clavulanate and 12 patients were taking ceftriaxone plus doxycycline) with a confirmed diagnosis of COVID-19 (defined as positive RT-PCR test for SARS-CoV-2 in nasopharyngeal swab, deep throat saliva, sputum, or tracheal aspirate) who were hospitalized at three regional hospitals (Prince of Wales Hospital, United Christian Hospital, and Yan Chai Hospital) in Hong Kong, China, between 1 March 2020 and 31 January 2021. We collected stool samples at admission (n = 90), at viral clearance (n = 90), and at 3-month (n = 58) and 6-month (n = 82) follow-up. A total of 42 patients provided additional samples (n = 100) during the virus-positive period and six patients provided stool samples at 9-month follow-up (n = 6). The second cohort consisted of patients with COVID-19 who were recruited into two open-label pilot studies of probiotics (SIM01). A total of 22 subjects were treated with 4 weeks of probiotics in the acute infection, and 10 subjects were involved as controls; 20 subjects received 4 weeks of probiotics after viral clearance. A total of 52 participants in this study provided longitudinal stool samples at baseline, week 2, week 4, week 5, week 8, and week 12.
Disease severity at admission was defined based on a clinical score of 1−5: (1) asymptomatic, individuals who tested positive for SARS-CoV-2 but who had no symptoms consistent with COVID-19; (2) mild, individuals who had any signs of COVID-19 (e.g. fever, cough, sore throat, malaise, headache, muscle pain) but no radiographic evidence of pneumonia; (3) moderate, if pneumonia was present along with fever and respiratory tract symptoms; (4) severe, if respiratory rate ≥30/min, oxygen saturation ≤93% when breathing ambient air, or PaO2/FiO2 ≤300 mm Hg (1 mm Hg = 0.133 kPa); or (5) critical, if there was respiratory failure requiring mechanical ventilation, shock, or organ failure requiring intensive care. We defined post-acute COVID-19 syndrome (PACS) as at least one persistent symptom or long-term complications of SARS-CoV-2 infection beyond 4 weeks from the onset of symptoms that could not be explained by an alternative diagnosis. We assessed the presence of the 30 most-commonly reported symptoms post-COVID at 6 months after illness onset18 (Supplementary Table 2).
Patients who fulfilled the following criteria were eligible for analyses: (i) 18−70 years of age, (ii) no gastrointestinal symptoms during acute infection, and (iii) no antibiotic was used before and 6 months after the SARS-CoV-2-positive period. Dietary data were documented for all COVID-19 patients during the time of hospitalization (whereby standardized meals were provided by the hospital catering service of each hospital) and individuals with special eating habits such as vegetarians were excluded. After discharge, patients with COVID-19 were advised to continue a diverse and standard Chinese diet that was consistent with habitual daily diets consumed by Hong Kong Chinese.
Controls were recruited before the COVID-19 pandemic (between Sep 2019 and Nov 2019) from the community through advertisement and from the endoscopy center at the Prince of Wales Hospital in subjects who had a normal colonoscopy (stools collected before bowel preparation) with the same collection protocol. We selected age-, gender- and comorbidity-matched controls with standard dietary patterns for cross-sectional comparison of gut resistome composition between subjects with and without COVID-19 infection. The exclusion criteria for non-COVID-19 controls were (1) the use of antibiotics in the last 6 months; (2) the use of laxatives or anti-diarrheal drugs in the last 3 months; (3) recent dietary changes (e.g. becoming vegetarian/vegan); (4) known complex infections or sepsis; (5) known history of severe organ failure (including decompensated cirrhosis, malignant disease, kidney failure, epilepsy, active serious infection, acquired immunodeficiency syndrome); (6) bowel surgery in the last 6 months (excluding colonoscopy/procedure related to perianal disease); (7) presence of an ileostomy/stoma; and (8) current pregnancy. Finally, demographics and comorbidities were comparable between subjects with COVID-19 and controls (Supplementary Tables 3 and 4).
Synbiotic formula (SIM01)
SIM01 is an oral encapsulated formulation of three lyophilized Bifidobacteria found to be depleted in COVID-19 patients from a previous study11,45 and three prebiotics (galactooligosaccharides, xylooligosaccharide, and resistant dextrin) that are beneficial for growth of these bacteria. The ratio of three bacteria was derived from the average natural accruing ratio in over 3000 healthy population, and this formula aims to replenish the missing bacteria associated with immune defense in COVID-19 patients.25 The three Bifidobacterium strains are commercially available from Chambio Co., Ltd. and WECAREPROBIOTICS. A blend of Bifidobacterium strains (25 billion CFU per capsule) and prebiotics (galactooligosaccharides, xylooligosaccharide, resistant dextrin) was prepared by the Center for Gut Microbiota Research, The Chinese University of Hong Kong according to the hospital policy. Sealed bottles of the study capsules were stored in a double-locked cabinet with temperature and humidity control in the Clinical Research Pharmacy. Trained pharmacist was responsible for dispensing the study capsules.
Pilot study of probiotics for acute COVID-19 infection
Details of this study can be found in Zhang et al.25 Briefly, 22 COVID-19 patients received two doses of SIM01 (2 × 1011 billion CFU per day) for 28 consecutive days together with standard meals provided by the hospital catering service. The control group consisted of 10 COVID-19 patients who were admitted under an independent infectious disease team during the same period without receiving SIM01. Both study group and control group received the same treatment protocol for COVID-19 endorsed by the local health authority. This study was approved by the Clinical Research Ethics Committees (2020.407) and was registered in the Clinical Trials Registry (NCT04581018). Written informed consents were obtained from all patients. Demographics of these participants can be found in Supplementary Table 5.
Pilot study of probiotics after viral clearance
This is an open-label, proof-of-concept clinical trial of COVID-19 patients after viral clearance. Patients exclusion criteria were as follows: below the age of 18; known allergy or intolerance to the intervention product or its components; any known medical condition that would prevent taking oral probiotics or increase risks associated with probiotics including but not limited to inability to swallow/aspiration risk and no other methods of delivery (e.g., no G/J tube); known increased infection risk due to immunosuppression such as prior organ or hematopoietic stem cell transplant, neutropenia (ANC <500 cells/µl), HIV, and CD4 <200 cells/µl; known increased infection risk due to endovascular or rheumatic heart disease, congenital heart defect, mechanical heart valves, endocarditis, endovascular grafts, permanent endovascular devices such as permanent (not short-term) hemodialysis catheters, pacemakers, or defibrillators; and documented pregnancy. Finally, 20 subjects who had been infected with SARS-CoV-2 but have turned negative participated in the study. All patients received two doses of the commercially available SIM01 (2 × 1011 billion CFU per day) for 28 consecutive days together with standard Hong Kong Chinese meals. This study was approved by the Clinical Research Ethics Committees (2020.400) and was registered in the Clinical Trials Registry (NCT04730284). Written informed consents were obtained from all patients. Demographics of these participants can be found in Supplementary Table 6.
Stool samples
Stool samples from in-hospital patients were collected by hospital staff while discharged patients provided stools on the day of follow-up or self-sampled at home and had samples couriered to the hospital within 24 hours of collection. Baseline (stools collected at admission) samples were collected before any treatment. All samples were collected in tubes containing preservative media (cat. 63700, Norgen Biotek Corp, Ontario Canada) and stored immediately at −80°C until processing. We have previously shown that data of gut microbiota composition generated from stools collected using this preservative medium are comparable to data obtained from samples that are immediately stored at −80°C.47
Stool DNA extraction and sequencing
Detailed methods for extracting microbial DNA are described in Zuo et al.11 Briefly, the fecal pellet (100 mg) was added to 1 mL of CTAB buffer and vortexed for 30 seconds, and then the sample was heated at 95°C for 5 minutes. After that, the samples were vortexed thoroughly with beads at maximum speed for 15 minutes. Then, 40 μL of proteinase K and 20 μL of RNase A was added to the sample and the mixture was incubated at 70°C for 10 minutes. The supernatant was then obtained by centrifuging at 13,000 g for 5 minutes and was added to the Maxwell RSC machine for DNA extraction. After the quality control procedures by Qubit 2.0, agarose gel electrophoresis, and Agilent 2100, extracted DNA was subject to DNA libraries construction, completed through the processes of end repairing, adding A to tails, purification and PCR amplification, using Nextera DNA Flex Library Preparation kit (Illumina, San Diego, CA). Libraries were subsequently sequenced on our in-house sequencer Illumina NextSeq 550 (150 base pairs paired-end) at the Center for Microbiota Research, The Chinese University of Hong Kong. Raw sequence data generated for this study are available in the Sequence Read Archive under BioProject accession: PRJNA841789.
Bioinformatics
Raw sequence data were quality filtered using Trimmomatic V.39 to remove the adapter, low-quality sequences (quality score <21), reads shorter than 100 base pairs. Contaminating human reads were filtered using Kneaddata (V.0.7.2 https://bitbucket.org/biobakery/kneaddata/wiki/Home, Reference database: GRCh38 p12) with default parameters. The mean sequencing depth was 41918852.26. The cleared FASTQ files were randomly subsampled into 30 M using Seqtk v.1.3-r114 (https://github.com/lh3/seqtk). Following this, subsampled quality-controlled reads were processed using ARG-OAP v.2.5 (singularity version, https://github.com/biofuture/Ublastx_stageone) to obtain the annotation of ARG profiles. ARG-OAP v.2.5 provides model-based identification of assembled sequences using SARGfam, a high-quality profile Hidden Markov Model containing profiles of ARG subtypes and including 16S reads quantification.48 We used ARG-OAP with default settings. ARG abundances were normalized by the number of 16S reads. Similarly, each reference sequence was tagged with its functional gene annotation (ARG subtype) and membership within a class of antibiotics targeted by the gene (ARG type). Subsampled quality-controlled reads were de novo assembled to contigs using Megahit (1.2.949) and then processed with ABRicate (https://github.com/tseemann/abricate) using CARD v.1.0550 and NCBI51 as a reference database for cross-validation with default parameters. ARGs under 80% coverage and 95% identity will be removed from the final abundance table to reduce some ARGs with low coverage and identity mistakenly recognized by ARGs-OAP since it is designed for short reads. Then, microbiota composition profiles were inferred from quality-filtered forward reads using MetaPhlAn3 version 3.0.5.
Statistical analysis
Continuous variables were expressed in median (interquartile range), whereas categorical variables were presented as numbers (percentage). Qualitative and quantitative differences between subgroups were analyzed using chi‐squared or Fisher’s exact tests for categorical parameters and Kruskal–Wallis test for continuous parameters, as appropriate. The relative abundance tables were input into R V.3.5.1 for statistical analysis. Principal Coordinates Analysis (PCoA) was used to visualize the clustering of samples based on their ARGs-level compositional profiles. Associations between gut resistome composition and patients’ parameters were assessed using permutational multivariate analysis of variance (PERMANOVA). Benjamini–Hochberg correction (FDR <0.1) was used for multiple hypothesis testing. Analyses of alpha and beta diversity, PCoA, PERMANOVA are implemented in the vegan R package V.2.5-7.
Supplementary Material
Acknowledgments
This work was supported by the Health and Medical Research Fund, the Food and Health Bureau, and InnoHK, The Government of Hong Kong, Special Administrative Region of the People’s Republic of China. SCN is also supported by the Croucher Senior Medical Research Fellowship. We would like to thank all healthcare workers working in isolation wards of the Prince of Wales Hospital, United Christian Hospital and Yan Chai Hospital, Hong Kong SAR, China. We thank Kitty Cheung, Kristy Ho, Chun Pan Cheung, Joey Chan, Dai Min, Lok Cheung Chu, and other staff/students for their technical contribution to this study, including sample collection, inventory, and processing; and Hui Zhan for assistance with DNA extraction and sequencing.
Funding Statement
This work was supported by the Health and Medical Research Fund, the Food and Health Bureau, and InnoHK, The Government of Hong Kong, Special Administrative Region of the People’s Republic of China.
Data availability
All shotgun metagenomic sequencing data analysed in this work will be available in the Sequence Read Archive (SRA) under BioProject accession PRJNA841789. Access will be granted after appropriate request to corresponding author.
Disclosure statement
FKLC and SCN are the scientific co-founders and sit on the board of Directors of GenieBiome Ltd. SCN, FKLC and ZX are inventors of patent applications related to SIM01. SCN has served as an advisory board member for Pfizer, Ferring, Janssen, and Abbvie and a speaker for Ferring, Tillotts, Menarini, Janssen, Abbvie, and Takeda. She has received research grants from Olympus, Ferring, and Abbvie. FKLC has served as an advisor and lecture speaker for Eisai Co. Ltd., AstraZeneca, Pfizer Inc., Takeda Pharmaceutical Co., and Takeda (China) Holdings Co. Ltd. All other co-authors have no conflict of interest.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19490976.2022.2128603
References
- 1.Resistance, I.C.G.o.A . 2019. No time to Wait. Securing the future from drug-resistant infections.
- 2.Langford BJ, So, M, Raybardhan, S, Leung, V, Soucy, JR, Westwood, D, Daneman, N, MacFadden, DR.. Antibiotic prescribing in patients with COVID-19: rapid review and meta-analysis. Clin Microbiol Infect. 2021;27:520–531. doi: 10.1016/j.cmi.2020.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rawson TM, Moore LS, Zhu N, Ranganathan N, Skolimowska K, Gilchrist M, Satta G, Cooke G, Holmes A, et al. Bacterial and fungal coinfection in individuals with coronavirus: a rapid review to support COVID-19 antimicrobial prescribing. Clin Infect Dis. 2020;71:2459–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sommer MOA, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–1131. doi: 10.1126/science.1176950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montassier E, Valdés-Mas R, Batard E, Zmora N, Dori-Bachash M, Suez J, Elinav E, et al. Probiotics impact the antibiotic resistance gene reservoir along the human GI tract in a person-specific and antibiotic-dependent manner. Nat Microbiol. 2021;6:1043–1054. doi: 10.1038/s41564-021-00920-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell TP, Sun X, Patel VH, Sanz C, Morgan D, Dantas G. The microbiome and resistome of chimpanzees, gorillas, and humans across host lifestyle and geography. ISME J. 2020;14:1584–1599. doi: 10.1038/s41396-020-0634-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stecher B, Denzler R, Maier L, Bernet F, Sanders MJ, Pickard DJ, Barthel M, Westendorf AM, Krogfelt KA, Walker AW, Ackermann M, Dobrindt U, Thomson NR, Hardt WD, et al. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc Natl Acad Sci U S A. 2012;109:1269–1274. doi: 10.1073/pnas.1113246109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Launay A, Ballard SA, Johnson PD, Grayson ML, Lambert T. Transfer of vancomycin resistance transposon Tn1549 from Clostridium symbiosum to Enterococcus spp. in the gut of gnotobiotic mice. Antimicrob Agents Chemother. 2006;50:1054–1062. doi: 10.1128/AAC.50.3.1054-1062.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottig S, Gruber TM, Stecher B, Wichelhaus TA, Kempf VA. In vivo horizontal gene transfer of the carbapenemase OXA-48 during a nosocomial outbreak. Clin Infect Dis. 2015;60:1808–1815. doi: 10.1093/cid/civ191 [DOI] [PubMed] [Google Scholar]
- 10.Goren MG, Carmeli Y, Schwaber MJ, Chmelnitsky I, Schechner V, Navon-Venezia S. Transfer of carbapenem-resistant plasmid from Klebsiella pneumoniae ST258 to Escherichia coli in patient. Emerg Infect Dis. 2010;16:1014–1017. doi: 10.3201/eid1606.091671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tao Zuo FZ, Lui GCY, Yeoh YK, Li AYL, Zhan H, Wan Y, Chung ACK, Cheung CP, Chen N, Lai CKC, et al. Alterations in Gut Microbiota of Patients With COVID-19 During Time of Hospitalization. Gastroenterology. 2020;159:944–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren Z, Wang H, Cui G, Lu H, Wang L, Luo H, Chen X, Ren H, Sun R, Liu W, Liu X, Liu C, Li A, Wang X, Rao B, Yuan C, Zhang H, Sun J, Chen X, Li B, Hu C, et al. Alterations in the human oral and gut microbiomes and lipidomics in COVID-19. Gut. 2021;70:1253–1265. doi: 10.1136/gutjnl-2020-323826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu S, Chen Y, Wu Z, Chen Y, Gao H, Lv L, Guo F, Zhang X, Luo R, Huang C, Lu H, Zheng B, Zhang J, Yan R, Zhang H, Jiang H, Xu Q, Guo J, Gong Y, Tang L, Li L, et al. Alterations of the Gut Microbiota in Patients With Coronavirus Disease 2019 or H1N1 Influenza. Clin Infect Dis. 2020;71:2669–2678. doi: 10.1093/cid/ciaa709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, Ai JW, Yang W, Zhou X, He F, Xie S, Zeng W, Li Y, Yu Y, Gou X, Li Y, Wang X, Su H, Zhu Z, Xu T, Zhang W, et al. Metatranscriptomic Characterization of Coronavirus Disease 2019 Identified a Host Transcriptional Classifier Associated With Immune Signaling. Clin Infect Dis. 2021;73:376–385. doi: 10.1093/cid/ciaa663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao J, Wang C, Zhang Y, Lei G, Xu K, Zhao N, Lu J, Meng F, Yu L, Yan J, Bai C, Zhang S, Zhang N, Gong Y, Bi Y, Shi Y, Chen Z, Dai L, Wang J, Yang P, et al. Integrated gut virome and bacteriome dynamics in COVID-19 patients. Gut Microbes. 2021;13:1–21. doi: 10.1080/19490976.2021.1887722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang F, Wan Y, Zuo T, Yeoh YK, Liu Q, Zhang L, Zhan H, Lu W, Xu W, Lui GCY, Li AYL, Cheung CP, Wong CK, Chan PKS, Chan FKL, Ng SC, et al. Prolonged Impairment of Short-Chain Fatty Acid and L-Isoleucine Biosynthesis in Gut Microbiome in Patients With COVID-19. Gastroenterology. 2022;162548–561:e544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yanfei Chen SG, Chen Y, Haifeng L, Shi D, Guo J, Wu WR, Yang Y, Li Y, Xu KJ, Ding C, Luo R, Huang C, Yu L, Xu M, Yi P, Liu J, Tao JJ, Zhang Hua, Lv L, Wang B, Sheng J, et al. Six-month follow-up of gut microbiota richness in patients with COVID-19. Gut. 2022;71:222–225. doi: 10.1136/gutjnl-2021-324090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Q, Mak JWY, Su Q, Yeoh YK, Lui GC, Ng SSS, Zhang F, Li AYL, Lu W, Hui DS, Chan PK, Chan FKL, Ng SC, et al. Gut microbiota dynamics in a prospective cohort of patients with post-acute COVID-19 syndrome. Gut. 2022;71:544–552. doi: 10.1136/gutjnl-2021-325989 [DOI] [PubMed] [Google Scholar]
- 19.Su Q, Lau RI, Liu Q, Chan FKL, Ng SC, Post-acute COVID-19. syndrome and gut dysbiosis linger beyond 1 year after SARS-CoV-2 clearance. Gut. 2022;gutjnl-2022–328319. doi: 10.1136/gutjnl-2022-328319 [DOI] [PubMed] [Google Scholar]
- 20.Kang Y, Chen S, Chen Y, Tian L, Wu Q, Zheng, M. Alterations of fecal antibiotic resistome in COVID-19 patients after empirical antibiotic exposure. Int J Hyg Environ Health. 2022;240:113882. doi: 10.1016/j.ijheh.2021.113882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lau HC, Ng SC, Yu J. Targeting the Gut Microbiota in Coronavirus Disease 2019: hype or Hope? Gastroenterology. 2022;162:9–16. doi: 10.1053/j.gastro.2021.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsigalou C, Konstantinidis T, Stavropoulou E, Bezirtzoglou EE, Tsakris A. Potential Elimination of Human Gut Resistome by Exploiting the Benefits of Functional Foods. Front Microbiol. 2020;11: doi: 10.3389/fmicb.2020.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mak JWY, Chan FKL, Ng SC. Probiotics and COVID-19: one size does not fit all. Lancet Gastroenterol Hepatol. 2020;5:644–645. doi: 10.1016/S2468-1253(20)30122-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu C, Xu Q, Cao Z, Pan D, Zhu Y, Wang S, Liu D, Song Z, Jiang W, Ruan Y, Huang Y, Qin N, Lu H. Qin H . The volatile and heterogeneous gut microbiota shifts of COVID-19 patients over the course of a probiotics-assisted therapy. Clin Transl Med. 2021;11:e643. doi: 10.1002/ctm2.643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, Xu Z, Mak JWY, Chow KM, Lui G, Li TCM, Wong CK, Chan PKS, Ching JYL, Fujiwara Y, Chan FKL, Ng SC, et al. Gut microbiota-derived synbiotic formula (SIM01) as a novel adjuvant therapy for COVID-19: an open-label pilot study. J Gastroenterol Hepatol. 2022. doi: 10.1111/jgh.15796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nalbandian A, Sehgal K, Gupta A, Madhavan MV, McGroder C, Stevens JS, Cook JR, Nordvig AS, Shalev D, Sehrawat TS, Ahluwalia N, Bikdeli B, Dietz D, Der-Nigoghossian C, Nordvig S, Shalev D, Sehrawat TS, Ahluwalia N, Bikdeli B, Dietz D, Der-Nigoghossian C, et al. Post-acute COVID-19 syndrome. Nat Med. 2021;27:601–615. doi: 10.1038/s41591-021-01283-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Y, Patel A, SantaLucia E, Roberts E, Zhao L, Kaye K, Rao K, Bachman MA, et al. Measurement of Klebsiella Intestinal Colonization Density To Assess Infection Risk. mSphere. 2021;6:e0050021. doi: 10.1128/mSphere.00500-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorrie CL, Mirceta M, Wick RR, Edwards DJ, Thomson NR, Strugnell RA, Pratt NF, Garlick JS, Watson KM, Pilcher DV, McGloughlin SA, Spelman DW, Jenney AWJ, Holt KE, et al. Gastrointestinal Carriage Is a Major Reservoir of Klebsiella pneumoniae Infection in Intensive Care Patients. Clin Infect Dis. 2017;65:208–215. doi: 10.1093/cid/cix270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwok KO, Wei WI, Ma BHM, Ip M, Cheung H, Hui E, Tang A, McNeil E, Wong SYS, Yeoh EK, et al. Antibiotic use among COVID-19 patients in Hong Kong, January 2018 to March 2021. J Infect. 2022. doi: 10.1016/j.jinf.2022.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dhar D, Mohanty A. Gut microbiota and Covid-19- possible link and implications. Virus Res. 2020;285:198018. doi: 10.1016/j.virusres.2020.198018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Domingues CPF, Rebelo JS, Dionisio F, Botelho A, Nogueira T. The Social Distancing Imposed To Contain COVID-19 Can Affect Our Microbiome: a Double-Edged Sword in Human Health. mSphere. 2020;5. doi: 10.1128/mSphere.00716-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Partridge SR, Neville Firth SMK, Jensen SO. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin Microbiol Rev. 2018;31(4):e00088–17. doi: 10.1128/CMR.00088-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schjorring S, Struve C, Krogfelt KA. Transfer of antimicrobial resistance plasmids from Klebsiella pneumoniae to Escherichia coli in the mouse intestine. J Antimicrob Chemother. 2008;62:1086–1093. doi: 10.1093/jac/dkn323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dell’Annunziata F, Doti N, Donadio G, Dal Piaz F, Izzo V, De Filippis A, Galdiero M, Altucci L, Boccia G, Galdiero M, et al. Outer Membrane Vesicles Derived from Klebsiella pneumoniae Are a Driving Force for Horizontal Gene Transfer. Int J Mol Sci. 2021;22. doi: 10.3390/ijms22168732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim HN, Joo EJ, Lee CW, Ahn KS, Kim HL, Park DI, Park SK. Reversion of Gut Microbiota during the Recovery Phase in Patients with Asymptomatic or Mild COVID-19: longitudinal Study. Microorganisms. 2021;9. doi: 10.3390/microorganisms9061237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newsome RC, Gauthier J, Hernandez MC, Abraham, GE, Robinson, TO, Williams, HB, Sloan, M, Owings, A, Laird, H, Christian, T, Pride, Y, Wilson, KJ, Parker, A, Senitko, M, Owings, A, Laird, H, Christian, T, Pride, Y, Wilson, KJ, Hasan, M, Parker, M, et al. The gut microbiome of COVID-19 recovered patients returns to uninfected status in a minority-dominated United States cohort. Gut Microbes. 2021;13:1–15. doi: 10.1080/19490976.2021.1926840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacPherson CW, Mathieu O, Tremblay J, Champagne J, Nantel, A, Girard SA, Tompkins TA. Gut Bacterial Microbiota and its Resistome Rapidly Recover to Basal State Levels after Short-term Amoxicillin-Clavulanic Acid Treatment in Healthy Adults. Sci Rep. 2018;8:11192. doi: 10.1038/s41598-018-29229-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hegstad K, Mylvaganam H, Janice J, Josefsen E, Sivertsen, A, Skaare D. Role of Horizontal Gene Transfer in the Development of Multidrug Resistance in Haemophilus influenzae. mSphere. 2020;5(1):e00969-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conwell M, Daniels V, Naughton PJ, Dooley JS. Interspecies transfer of vancomycin, erythromycin and tetracycline resistance among Enterococcus species recovered from agrarian sources. BMC Microbiol. 2017;17:19. doi: 10.1186/s12866-017-0928-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perl TM. The threat of vancomycin resistance. Am J Med. 1999;106(5A):26S–37S. doi: 10.1016/S0002-9343(98)00354-4 [DOI] [PubMed] [Google Scholar]
- 41.Huemer M, Mairpady Shambat S, Brugger SD, Zinkernagel AS. Antibiotic resistance and persistence-Implications for human health and treatment perspectives. EMBO Rep. 2020;21:e51034. doi: 10.15252/embr.202051034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suez J, Zmora N, Segal E, Elinav E. The pros, cons, and many unknowns of probiotics. Nat Med. 2019;25:716–729. doi: 10.1038/s41591-019-0439-x [DOI] [PubMed] [Google Scholar]
- 43.Selvin J, Maity D, Sajayan A, Kiran GS. Revealing antibiotic resistance in therapeutic and dietary probiotic supplements. J Glob Antimicrob Resist. 2020;22:202–205. doi: 10.1016/j.jgar.2020.02.007 [DOI] [PubMed] [Google Scholar]
- 44.Sharma P, Tomar SK, Goswami P, Sangwan V, Singh R. Antibiotic resistance among commercially available probiotics. Food Res J. 2014;57:176–195. doi: 10.1016/j.foodres.2014.01.025 [DOI] [Google Scholar]
- 45.Yeoh YK, Zuo T, Lui GC, Zhang F, Liu Q, Li AY, Chung AC, Cheung CP, Tso EY, Fung KS, Chan V, Ling L, Joynt G, Hui DS, Chow KM, Ng SSS, Li TC, Ng RW, Yip TC, Wong GL, Chan FK, et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698–706. doi: 10.1136/gutjnl-2020-323020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mindy Flanagan RR, Sutherland J, Vaughn T, Diekema D, Doebbeling BN. Development and validation of measures to assess prevention and control of AMR in hospitals. Med Care. 2007;45(6):537–544. doi: 10.1097/MLR.0b013e31803bb48b [DOI] [PubMed] [Google Scholar]
- 47.Zigui Chen PCH, Hui M, Yeoh YK, Wong PY, Chan MCW, Wong MCS, Ng SC, Chan FKL, Chan PKS. Impact of Preservation Method and 16S rRNA Hypervariable Region on Gut Microbiota Profiling. mSystems. 4(1):e00271–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y, Jiang X, Chai B, Ma L, Li B, Zhang A, Cole JR, Tiedje JM, Zhang T. ARGs-OAP: online analysis pipeline for antibiotic resistance genes detection from metagenomic data using an integrated structured ARG-database. Bioinformatics. 2016;32:2346–2351. doi: 10.1093/bioinformatics/btw136 [DOI] [PubMed] [Google Scholar]
- 49.Li D, Liu CM, Luo R, Sadakane K, Lam TW. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033 [DOI] [PubMed] [Google Scholar]
- 50.Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen AV, Cheng AA, Liu S, Min SY, Miroshnichenko A, Tran HK, Werfalli RE, Nasir JA, Oloni M, Speicher DJ, Florescu A, Singh B, Faltyn M, Hernandez-Koutoucheva A, et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48:D517–D525. doi: 10.1093/nar/gkz935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feldgarden M, Brover, V, Haft, DH, Prasad, AB, Slotta, DJ, Tolstoy, I, Tyson, GH, Zhao, S, Hsu, CH, McDermott, PF, Tadesse, DA, Morales, C, Simmons, M, Tillman, G, Wasilenko, J, Folster, JP, Klimke, W, et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob Agents Chemother. 2019;63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All shotgun metagenomic sequencing data analysed in this work will be available in the Sequence Read Archive (SRA) under BioProject accession PRJNA841789. Access will be granted after appropriate request to corresponding author.


