Summary
Indole‐3‐acetic acid (IAA) is the main naturally occurring auxin and is produced by organisms of all kingdoms of life. In addition to the regulation of plant growth and development, IAA plays an important role in the interaction between plants and growth‐promoting and phytopathogenic bacteria by regulating bacterial gene expression and physiology. We show here that an IAA metabolizing plant‐associated Pseudomonas putida isolate exhibits chemotaxis to IAA that is independent of auxin metabolism. We found that IAA chemotaxis is based on the activity of the PcpI chemoreceptor and heterologous expression of pcpI conferred IAA taxis to different environmental and human pathogenic isolates of the Pseudomonas genus. Using ligand screening, microcalorimetry and quantitative chemotaxis assays, we found that PcpI failed to bind IAA directly, but recognized and mediated chemoattractions to various aromatic compounds, including the phytohormone salicylic acid. The expression of pcpI and its role in the interactions with plants was also investigated. PcpI extends the range of central signal molecules recognized by chemoreceptors. To our knowledge, this is the first report on a bacterial receptor that responds to two different phytohormones. Our study reinforces the multifunctional role of IAA and salicylic acid as intra‐ and inter‐kingdom signal molecules.
Introduction
The phytohormone indole‐3‐acetic acid (IAA) is the most common naturally occurring auxin and is key for plant growth, development and defence, playing essential roles in embryogenesis, de novo organogenesis, vascular formation as well as seed, root and flower development, among other processes (Zhao, 2018; Gallei et al., 2020). However, IAA is an ubiquitous signalling molecule, since bacteria (Kunkel and Harper, 2018; Duca and Glick, 2020), fungi (Fu et al., 2015), archaea (Aklujkar et al., 2014), algae (Bogaert et al., 2019; Laird et al., 2020) and animals (Oliveira et al., 2007) were found to produce IAA. This ubiquity, together with a growing body of experimental evidence, supports the role of IAA as an inter‐ and intra‐kingdom signal molecule. For example, IAA was found to regulate cell division and development in algae (Ohtaka et al., 2017; Bogaert et al., 2019) and IAA produced by various algae modulate different virulence traits in an aquatic bacterial pathogen (Yang et al., 2017). Alternatively, bacteria co‐occurring with marine diatoms were shown to promote diatom growth through the synthesis of IAA (Amin et al., 2015). In fungi, IAA affected growth, sporulation, spore germination as well as fungal competitiveness (Fu et al., 2015; Liu et al., 2016; Nicastro et al., 2021) and fungal IAA synthesis modulated growth, development and immune responses in plant hosts (Fu et al., 2015; Jahn et al., 2021).
Many plant‐associated bacteria synthesize IAA (Spaepen and Vanderleyden, 2011; Duca et al., 2014; Kunkel and Harper, 2018; Duca and Glick, 2020), which has been shown to play crucial roles during their interaction with their hosts. Indeed, IAA production was found to be involved in nodule formation and nitrogen fixation by rhizobia in legume plants as well as in the stimulation of plant growth by non‐symbiotic beneficial rhizobacteria (Spaepen and Vanderleyden, 2011; Duca and Glick, 2020). Furthermore, IAA plays an essential role in plant–phytobacteria interactions, typically promoting plant susceptibility and disease development by different mechanisms that include the alteration of the IAA balance in the plant, the suppression of host basal defence responses and the regulation of the synthesis of virulence factors in the bacterial pathogen (Kunkel and Johnson, 2021). Beyond the role of bacterial IAA in the interaction with plants, a number of studies have provided first insight into the molecular basis of IAA action in phytobacteria, as it was shown to modulate gene expression and numerous physiological processes such as stress tolerance, primary metabolism, production of virulence factors, antibiotic synthesis and biofilm formation (Duca et al., 2014; Kunkel and Harper, 2018; Matilla et al., 2018; Duca and Glick, 2020; Djami‐Tchatchou et al., 2021). In addition, there is also growing evidence for a role of IAA in the modulation of bacterial motility and chemotaxis in plant‐associated bacteria like Rhizobium etli (Spaepen et al., 2009), Bradyrhizobium japonicum (Donati et al., 2013) and Pseudomonas syringae (Soby et al., 1991; Djami‐Tchatchou et al., 2021). However, the molecular mechanisms behind most of these IAA‐mediated processes remain unknown.
Chemotaxis permits bacteria to adapt their swimming motility patterns in chemical gradients, thus favouring access to nutritional sources and preferred environments for growth (Matilla and Krell, 2018; Colin et al., 2021). Typically, chemotaxis signalling is initiated by the recognition of chemoeffectors by the ligand‐binding domain (LBD) of a chemoreceptor. Chemoeffector binding causes a molecular stimulus that modulates the autophosphorylation activity of the histidine kinase CheA, subsequently altering the transphosphorylation activity of the response regulator CheY. Phosphorylated CheY binds to the flagellar motor resulting in a change in the direction of flagellar rotation, ultimately causing a chemotactic response (Bi and Sourjik, 2018; Matilla et al., 2021a). To date, most chemoeffectors identified appear to be compounds of metabolic value such as sugars, amino acids and organic acids that can serve as nutrient and energy sources for bacteria (Sampedro et al., 2015; Matilla et al., 2021a; Matilla et al., 2022). However, other chemoeffectors like animal (Lopes and Sourjik, 2018) and plant (Kim et al., 2007; Antunez‐Lamas et al., 2009) hormones, quorum sensing molecules (Zhang et al., 2020), plant defence metabolites (Neal et al., 2012) and neurotransmitters (Pasupuleti et al., 2014; Corral‐Lugo et al., 2018) can alternatively provide information about favourable, and even highly specific, environmental niches.
The plant‐associated bacterium Pseudomonas putida 1290 was isolated from a pear plant due to its ability to efficiently use IAA as carbon, nitrogen and energy source (Leveau and Lindow, 2005). Indeed, P. putida 1290 was the first bacterium for which the gene cluster responsible for IAA degradation, named iacABCDEFGRHI, was described (Leveau and Gerards, 2008), and it is currently used as a model for the isolation and characterization of genes involved in IAA degradation (Laird et al., 2020). Significantly, the IAA catabolic properties of P. putida 1290 were shown to alleviate the detrimental effects that appear on plants caused by the exogenous addition of IAA (Leveau and Gerards, 2008) or due to the production of high levels of IAA by rhizosphere microbial competitors (Leveau and Lindow, 2005). Using swim plate motility assays, P. putida 1290 was shown to exhibit directed movement towards IAA (Scott et al., 2013). However, such plate‐based assays do not permit to distinguish between chemotaxis and energy taxis; a lower specific form of directed cell movement to environmental sites at which the cellular metabolism is optimal (Schweinitzer and Josenhans, 2010; Colin et al., 2021). Heterologous expression of the iac catabolic cluster in P. putida KT2440 provided this bacterium with the ability to use IAA as nutrient and energy source but did not confer the ability to migrate towards IAA, as determined by swim plate assays (Scott et al., 2013) – suggesting that a specific IAA chemoreceptor encoded in the genome of P. putida 1290 may be responsible for the observed behaviour. To our knowledge, no evidence of IAA chemotaxis has been reported in other bacterial strains.
We show here that IAA chemotaxis in P. putida 1290 is based on the action of the chemoreceptor PcpI that employs a mechanism that does not involve energy taxis. PcpI was also found to mediate taxis to additional chemoeffectors, including the phytohormone salicylic acid. The expression of pcpI and its role in plant root colonization was also studied. This work expands the range of chemoreceptors that are stimulated by important signal molecules of life.
Results
IAA chemotaxis of Pseudomonas putida 1290 does not dependent on auxin metabolism
To investigate the chemotactic behaviour of P. putida 1290 towards IAA, we conducted quantitative capillary chemotaxis assays – an experimental approach that primarily monitors chemotaxis and to a much lower degree energy taxis. IAA was tested at concentrations ranging from 0.01 to 10 mM, with optimal chemotactic responses at 10 mM and an onset at 100 μM of IAA (Fig. 1A). These concentrations do not necessarily account for the minimum threshold for chemotaxis since the chemoeffector concentration decreases sharply from the capillary source (Raina et al., 2019; Tunchai et al., 2021). In analogy to P. putida KT2440 (Lopez‐Farfan et al., 2019), P. putida 1290 has three chemosensory pathways and a mutant defective in cheA, present within the chemotaxis signalling gene cluster, failed to respond to IAA (Fig. 1B).
Fig. 1.
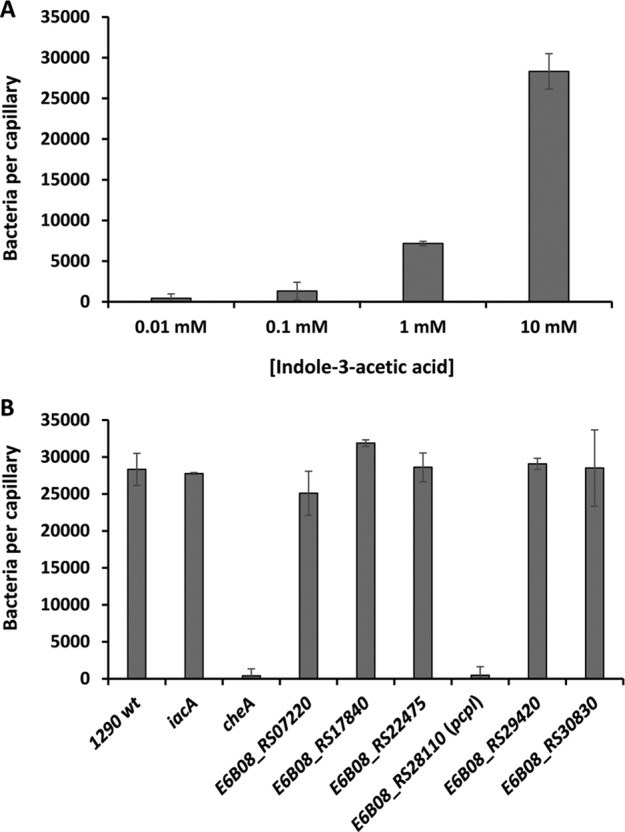
Chemotaxis of Pseudomonas putida 1290 wild type and mutant strains towards indole‐3‐acetic acid (IAA).
A. Quantitative capillary chemotaxis assays of the wild type strain to different concentrations of IAA.
B. Chemotaxis to 10 mM IAA of different strains of P. putida 1290. In all cases, data were corrected with the number of cells that swam into buffer containing capillaries. Shown data are means and standard deviations from three independent experiments conducted in triplicate.
In order to rule out the involvement of IAA metabolism in the observed chemotactic response, we generated a polar mutant in the first gene of the IAA catabolic operon, iacA (Leveau and Gerards, 2008). Mutation of iacA resulted in the inability to grow on IAA as sole carbon source (Supp. Figs. S1A and S2) and this mutant strain showed wild type like chemotaxis towards IAA using quantitative capillary chemotaxis assays (Fig. 1B) – confirming that the observed response is not based on energy taxis.
The chemoreceptor repertoire of P. putida 1290
The genome of P. putida 1290 (Laird and Leveau, 2019) encodes 27 chemoreceptors (Fig. 2), which corresponds or is similar to the number of chemoreceptors encoded in two Pseudomonas chemotaxis model strains, namely P. putida KT2440 (Lopez‐Farfan et al., 2019) and P. aeruginosa PAO1 (26 chemoreceptors) (Matilla et al., 2021a), respectively. At least 10 different types of LBDs were identified in P. putida 1290 chemoreceptors, including LBDs consisting of parallel helices (e.g. 4HB_MCP‐1, HBM, PilJ) and α/β folds (e.g. sCache_2, dCache_1, Cache_3‐Cache_2, PAS_3, PAS_9) (Fig. 2). The most abundant LBD types were dCache_1 and 4HB_MCP_1, which are also the most abundant LBDs in bacterial chemoreceptors (Upadhyay et al., 2016; Ortega et al., 2017). Twenty‐one (i.e. 78%) of the P. putida 1290 chemoreceptors showed the canonical topology and are transmembrane proteins with their LBD located in the periplasm. However, the structural and topological diversity of P. putida 1290 chemoreceptors was reflected by the presence of a transmembrane chemoreceptor that lacks an LBD, three membrane‐associated receptors with cytosolic PAS_3 and dCache_1 LBDs as well as two entirely cytosolic receptors composed of two tandem PAS domains (Fig. 2). The latter two receptors are likely to be involved in the sensing of cytosolic signals like redox‐active cofactors or oxygen (Collins et al., 2014).
Fig. 2.
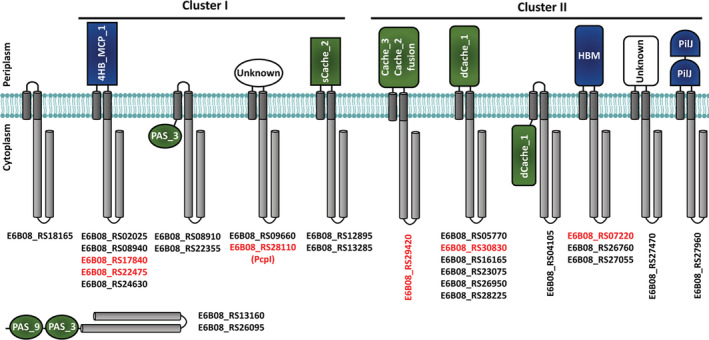
The chemoreceptor repertoire of Pseudomonas putida 1290. Predicted receptor topology and locus tags are shown. Annotation was based on the Pfam database and, in case of un‐annotated LBDs, domain type was defined by visual inspection of homology models generated by the Phyre2 algorithm (Kelley et al., 2015). Topologies are based on the prediction of transmembrane regions using the DAS algorithm (Cserzo et al., 1997). Chemoreceptors were organized into cluster I and cluster II based on the length of their LBDs, as described previously (Lacal et al., 2010b). Ligand binding domains with α/β folds or parallel helices are shown in green and blue, respectively. Chemoreceptor names in red indicate receptors that do not have homologues in P. putida KT2440 and P. aeruginosa PAO1 (i.e. LBDs with <41% sequence identity). 4‐HB, 4‐helix bundle domain; HBM, helical bimodular domain; PAS, Per‐Arnt‐Sim domain; PilJ, Type IV pili domain; Unknown, LBDs of unknown type.
Identification of PcpI as the chemoreceptor responsible for IAA chemotaxis
Around half of the chemoreceptors of KT2440 and PAO1 have been characterized and some of their ligands include amino acids, organic acids, phytohormones, polyamines and inorganic nutrients, among others (Ortega et al., 2017; Matilla et al., 2021a). Quantitative capillary chemotaxis assays of KT2440 and PAO1 showed that both strains failed to respond to different concentrations of IAA (Supp. Fig. S3). Based on these results, we hypothesized that a receptor that was absent in KT2440 and PAO1 would be responsible for IAA taxis in P. putida 1290.
The ligand specificity of most chemoreceptors is determined by their rapidly evolving LBDs (Ortega et al., 2017; Gavira et al., 2020; Matilla et al., 2021a). None of the 27 chemoreceptors of P. putida 1290 has been characterized and to identify the IAA chemoreceptor, we performed homology comparisons between LBD sequences of P. putida 1290 chemoreceptors with those of KT2440 and PAO1. These analyses revealed that P. putida 1290 has 19 and 15 chemoreceptors that are homologous (i.e. LBDs with more than 41% sequence identity) to the receptors present in KT2440 and PAO1 respectively (Table 1). Homologous chemoreceptors were found to mediate taxis towards amino acids (e.g. PctA, PctC, McpA), organic acids (e.g. McpR, McpP, McpS, PA2652), polyamines (e.g. TlpQ, McpU) and inorganic phosphate (Pi) (e.g. CtpH, CtpL). Furthermore, a receptor homologous to the energy taxis chemoreceptor Aer or to proteins that mediate alternative cellular functions such as the modulation of intracellular levels of second messengers (e.g. WspA, PilJ, BdlA) were also found (Table 1). Notably, we identified six P. putida 1290 chemoreceptors that were either not present in KT2440 or PAO1 (e.g. E6B08_RS07220, E6B08_RS17840, E6B08_RS22475, E6B08_RS28110, E6B08_RS29420) or which LBD had low level of sequence identity (e.g. E6B08_RS30830). These chemoreceptors have different types of LBDs, including 4HB_MCP_1, sCache_3‐sCache_2, dCache_1 and HBM (Fig. 2; Table 1).
Table 1.
Pseudomonas putida 1290 chemoreceptors and their characterized homologues of P. putida KT2440 and P. aeruginosa PAO1.
| Chemoreceptor | LBD name (Pfam) | Closest homologue in KT2440 (% identity) | Closest homologue in PAO1 (% identity) | Chemoeffector(s)/comment(s) | Reference(s) |
|---|---|---|---|---|---|
| E6B08_RS02025 | 4HB_MCP_1 (PF12729) | PP_0317/McpR (67.3%) | – | Succinate, malate, fumarate | Parales et al. (2013) |
| E6B08_RS04105 | Not annotated (dCache_1‐like) a | PP_3950 (76.4%) | – | Unknown | – |
| E6B08_RS05770 | dCache_1 (PF02743) | PP_2249/McpA (45.1%) | PA4309/PctA (56.0%) | Amino acids | Rico‐Jimenez et al. (2013), Corral‐Lugo et al. (2016), Gavira et al. (2020) |
| E6B08_RS07220 | HBM (PF16591) | – | – | Unknown | – |
| E6B08_RS08910 | PAS_3 (PF08447) | PP_2111/Aer2 (89.5%) | PA1561/Aer/TlpC (76.5%) | Energy taxis | Hong et al. (2004a), Hong et al. (2004b), Sarand et al. (2008) |
| E6B08_RS08940 | Not annotated (4HB_MCP_1‐like) a | PP_2120/CtpH_PP (82.4%) | PA2561/CtpH (50.6%) | Inorganic phosphate | Wu et al. (2000), Rico‐Jimenez et al. (2016) |
| E6B08_RS09660 | Small unknown | PP_2310 (68.2%) | PA2867 (40.5%) | Mutation in PP2310 increases biofilm formation | Corral‐Lugo et al. (2016) |
| E6B08_RS12895 | sCache_2 (PF17200) | – | PA2652 (45.2%) | l‐malate, bromosuccinate, citramalate | Martin‐Mora et al. (2018b) |
| E6B08_RS13160 | PAS_9‐PAS_3 (PF13426–PF08447) | PP_3414/Aer (71.4%) | BldA (51.3%) | BdlA is involved in biofilm dispersion | Morgan et al. (2006), Petrova and Sauer (2012b), Petrova and Sauer (2012a) |
| E6B08_RS13285 | sCache_2 (PF17200) | PP_2861/McpP (88.1%) | – | Pyruvate, l‐lactate, propionate, acetate | Garcia et al. (2015) |
| E6B08_RS16165 | dCache_1 (PF02743) | PP_3557 (80.2%) | PA2654/TlpQ (53.4%) | Polyamines | Corral‐Lugo et al. (2018) |
| E6B08_RS17840 | 4HB_MCP_1 (PF12729) | – | – | Unknown | – |
| E6B08_RS18165 | No LBD | – | – | Unknown | – |
| E6B08_RS22355 | PAS_3 (PF08447) | PP_4521/Aer3 (81.4%) | PA1561/Aer (60.8%) | Energy taxis? | Sarand et al. (2008) |
| E6B08_RS22475 | 4HB_MCP_1 (PF12729) | – | – | Unknown | – |
| E6B08_RS23075 | dCache_1 (PF02743) | PP_1228/McpU (76.8%) | PA2654/TlpQ (49.0%) | Polyamines | Corral‐Lugo et al. (2016), Corral‐Lugo et al. (2018) |
| E6B08_RS24630 | 4HB_MCP_1 (PF12729) | PP_1488/WspA_PP (68.1%) | PA3708/WspA (36.7%) | Surface sensing, modulation of c‐di‐GMP levels | O'Connor et al. (2012), Chen et al. (2014), Corral‐Lugo et al. (2016) |
| E6B08_RS26095 | PAS_9‐PAS_3 (PF13426‐PF08447) | PP_0779 (72.3%) | BldA (38.3%) | BdlA is involved in biofilm dispersion | Morgan et al. (2006), Petrova and Sauer (2012b), Petrova and Sauer (2012a) |
| E6B08_RS26760 | HBM (PF16591) | PP_4658/McpS (73.6%) | – |
Malate, fumarate, oxaloacetate, succinate, citrate, isocitrate , butyrate |
Lacal et al. (2010a), ), Pineda‐Molina et al. (2012) |
|
E6B08_RS26950 |
dCache_1 (PF02743) |
PP_0584 /McpC (82.9%) |
– | Cytosine?, nicotinic acid? | Liu et al. (2009), Parales et al. (2014) |
| E6B08_RS27055 | Not annotated (HBM‐like) a | PP_0562/CtpL_PP (82.5%) | PA4844/CtpL (55.6%) | Inorganic phosphate | Wu et al. (2000), Rico‐Jimenez et al. (2016) |
| E6B08_RS27470 | Large unknown | PP_4888 (84.9%) | – | Expression regulated by benzoxazinoids | Neal et al. (2012) |
| E6B08_RS27960 | PilJ‐PilJ (PF13675) | PP_4989/PilJ (93.4%) | PA0411/PilJ (73.5%) | Surface sensing, modulation of c‐di‐GMP and cAMP levels | Fulcher et al. (2010), Luo et al. (2015), Jansari et al. (2016) |
| E6B08_RS28110 (PcpI) | Small unknown | – | – | IAA, salicylate, benzoate, 3‐methylbenzoate | This study |
| E6B08_RS28225 | dCache_1 (PF02743) | PP_2249/McpA (40.7%) | PA4307/PctC (43.5%) | Amino acids | Rico‐Jimenez et al. (2013), Corral‐Lugo et al. (2016), Gavira et al. (2020) |
| E6B08_RS29420 | Cache_3‐Cache_2 (PF17201) | – | – | Unknown | – |
| E6B08_RS30830 | dCache_1 (PF02743) | PP_1228/McpU (38.3%) | PA2654/TlpQ (40.2%) | Polyamines | Corral‐Lugo et al. (2016), Corral‐Lugo et al. (2018) |
Domain type un‐annotated in Pfam and defined by visual inspection of a homology model generated using the Phyre2 algorithm (Kelley et al., 2015).
To assess the potential involvement of these receptors in IAA chemotaxis of P. putida 1290, we constructed mutants in the corresponding genes which were subsequently phenotypically characterized using quantitative capillary chemotaxis assays. We found that the mutant defective in the E6B08_RS28110 chemoreceptor was the only strain that showed no chemotaxis to IAA (Fig. 1B) – a tactic phenotype that was indistinguishable to that of a mutant defective in cheA (Fig. 1B). Control experiments showed that the E6B08_RS28110 mutant showed wild type like chemotaxis to casamino acids (Supp. Fig. S4), indicating that the E6B08_RS28110 mutation does not cause a general chemotactic defect. Swim plate chemotaxis assays containing IAA as sole carbon source revealed only a slight decrease in the motility of the E6B08_RS28110 mutant compared to the parental strain (Supp. Fig. S2) – supporting that IAA energy taxis masks to a large degree IAA chemotaxis and that the initial tactic phenotype observed in swim plate assays (Scott et al., 2013) was primarily driven by energy taxis. As observed here, energy taxis was previously shown to mask chemotaxis using swim plate assays (Alvarez‐Ortega and Harwood, 2007; Parales et al., 2013).
To confirm the association between the E6B08_RS28110 mutation and the loss of IAA chemotaxis, we cloned the E6B08_RS28110 gene into a pBBR1MCS‐based medium copy number plasmid. In trans expression of E6B08_RS28110 not only restored chemotaxis to IAA in the mutant strain but also increased the magnitude of chemotaxis more than 10‐fold compared to the wild type strain (Fig. 3A). These results imply that enhanced cellular chemoreceptor levels were responsible for an increased chemotactic behaviour towards IAA, as described previously for other chemoreceptors (Fernández et al., 2016; Hida et al., 2020). To determine whether E6B08_RS28110 can confer the IAA chemotaxis phenotype to KT2440 and PAO1, we expressed heterologously the E6B08_RS28110 gene in these bacterial strains. We found that E6B08_RS28110 conferred IAA chemotaxis to both strains (Fig. 3B and C), inducing a particularly strong response in PAO1 (Fig. 3B). Based on these results, the chemoreceptor E6B08_RS28110 was named PcpI ( Pseudomonas chemoreceptor protein IAA).
Fig. 3.
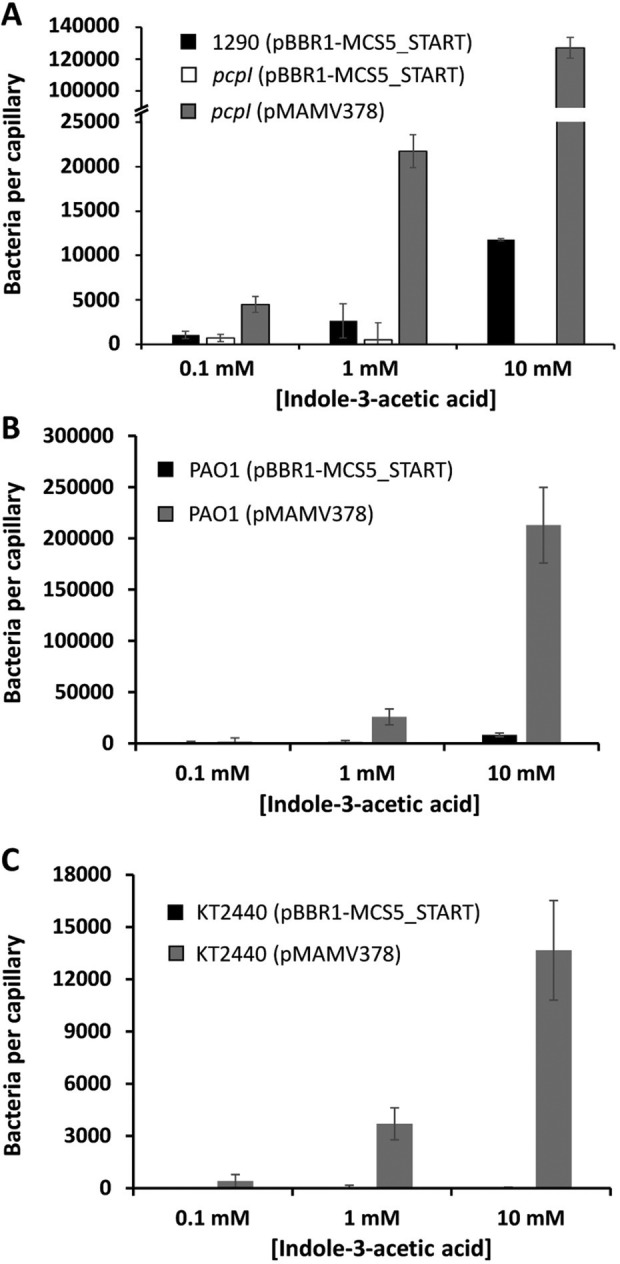
In trans expression of pcpI in different Pseudomonas strains. Multicopy expression of pcpI from the pBBR1‐MCS5_START derivative plasmid pMAMV378 increases the magnitude of IAA chemotaxis in P. putida 1290 (A) and confers IAA taxis to P. aeruginosa PAO1 (B) and P. putida KT2440 (C). Data are means and standard deviations from three independent experiments conducted in triplicate.
Expression of pcpI correlates with the magnitude of IAA chemotaxis
The observation that multicopy expression of the pcpI gene dramatically increased chemotactic responses towards IAA encouraged us to investigate the expression of pcpI in comparison with other chemoreceptor genes present in the genome of 1290. Since we found in P. putida 1290 homologous chemoreceptors that respond to amino acids, polyamines, organic acids and Pi (Table 1), we first conducted chemotaxis assays to 1 mM concentrations of arginine, putrescine, propionate, oxaloacetate and Pi. Quantitative chemotaxis assays revealed that P. putida 1290 showed strong chemotactic responses to polyamines, amino and organic acids (Supp. Fig. S5), whereas only minor responses to Pi were observed (Supp. Fig. S5), which may be due to the low expression of the corresponding chemoreceptor genes under conditions of Pi excess (Wu et al., 2000; Bains et al., 2012). We subsequently analyzed the transcript levels of pcpI under the same growth conditions used to conduct chemotaxis assays, namely mid‐logarithmic growth phase in M9 minimal medium supplemented glucose as carbon source, and compared these to the transcript levels of the chemoreceptor genes homologous to receptors involved in amino acid, organic acid, polyamine and Pi chemotaxis in another model Pseudomonas. The results showed that pcpI transcript levels were between 2.1‐ and 143.2‐fold lower than those of E6B08_RS05770, E6B08_RS13285, E6B08_RS23075 and E6B08_RS26760 (Fig. 4) – chemoreceptor genes homologous to pctA, mcpP, mcpU and mcpS, respectively (Table 1). In contrast, the expression of pcpI was 5.0 times higher than E6B08_RS27055, a ctpL homologue, which is in accordance with the very low chemotactic responses to Pi (Supp. Fig. S5). Taken together, these results correlate pcpI expression with the chemotactic responses observed towards IAA.
Fig. 4.
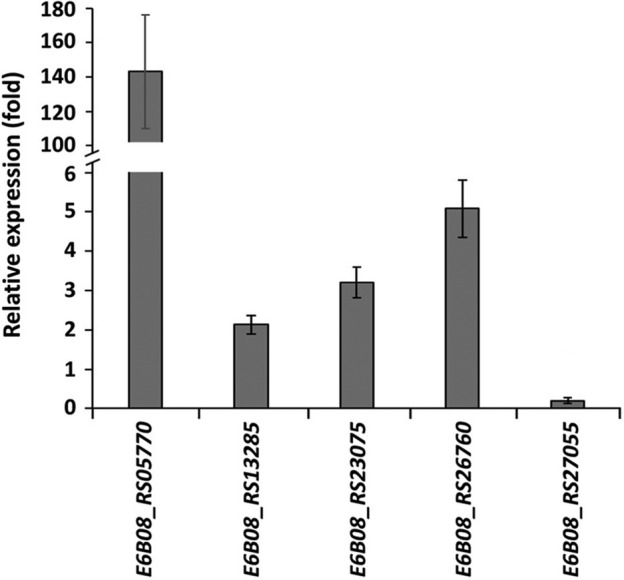
Transcript levels of P. putida 1290 chemoreceptor genes in comparison to transcript levels of pcpI measured by quantitative real‐time PCR. The values showed the expression of five chemoreceptor genes relative to pcpI expression. Data are the means and standard deviations from three biological replicates conducted in triplicate.
PcpI does not recognize IAA directly but the phytohormone salicylic acid
To delve into the molecular mechanisms of IAA chemotaxis in P. putida 1290, we cloned the DNA fragment encoding the LBD of PcpI into an expression vector and purified the protein by affinity chromatography. Subsequently, recombinant PcpI‐LBD was submitted to microcalorimetric titrations with IAA. We did not observe binding heats in microcalorimetric titrations conducted at two different temperatures, 25°C and 10°C, indicative of an absence of binding (Fig. 5). To assess the possibility that PcpI may be stimulated by the binding of an IAA‐loaded solute binding protein (SBP), we conducted pulldown assays with immobilized PcpI‐LBD and P. putida 1290 protein extracts but found no evidence for an SBP involved (Supp. Fig. S6).
Fig. 5.
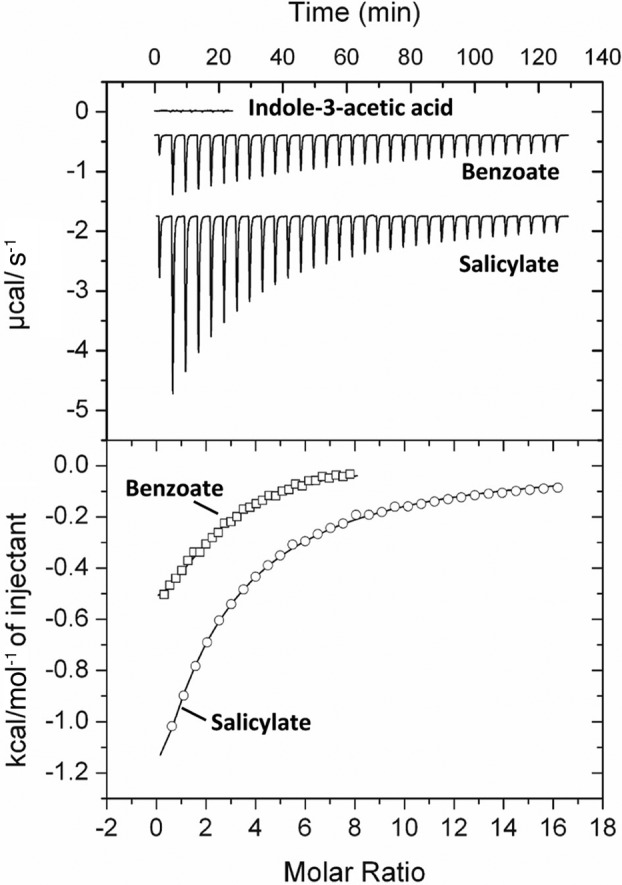
Isothermal titration calorimetry analysis of ligand binding to PcpI‐LBD. Upper panel: Raw data for the titration of PcpI‐LBD with 9.6 μl aliquots of indole‐3‐acetic acid (3 mM), salicylate (2 mM) and benzoate (5 mM). Lower panel: Integrated, dilution heat‐corrected and concentration‐normalized peak areas of the titration data for PcpI‐LBD. Data were fitted using the ‘one binding site’ model of the MicroCal version of ORIGIN. The derived thermodynamic parameters are provided in Suppl. Table S1.
Typically, SBPs that interact with chemoreceptors are encoded in transporter gene clusters (Matilla et al., 2021b). Genome analysis of P. putida 1290 revealed the presence of an ABC type transporter gene cluster, E6B08_RS28115‐E6B08_RS28125, immediately downstream of pcpI. The TransportDB database (Elbourne et al., 2017) predicted this ABC transporter to be involved in the uptake of amino acids. Given that there are transcriptional regulators (Marmorstein and Sigler, 1989; Herud‐Sikimić et al., 2021) as well as SBPs (Vetting et al., 2015) that bind both, amino acids and IAA, we purified the SBP of this transporter, E6B08_RS28125, and isothermal titration calorimetry (ITC) assays with IAA revealed no binding (Supp. Fig. S7). Subsequently, we used differential scanning fluorimetry (DSF) (Martin‐Mora et al., 2018a) and microcalorimetric titrations to analyze the ligand of profile of E6B08_RS28125 and found that E6B08_RS28125 binds l‐ornithine, l‐His and l‐Arg with dissociation constants (K D) of 0.9 ± 0.1, 3.3 ± 0.3 and 29.5 ± 3 μM, respectively (Supp. Figs S7 and S8; Supp. Table S1). Further protein–protein interaction assays using ITC revealed no evidence of protein complex formation between PcpI‐LBD and E6B08_RS28125 (Supp. Fig. S9).
To identify ligands that are directly recognized by PcpI, the LBD of PcpI was submitted to high‐throughput ligand screening using DSF. We screened ~480 compounds from the Biolog Compound arrays PM1, PM2A, PM3B, PM4A and PM5 that contain multiple carbon, nitrogen, sulfur and phosphorus sources. We found that ligand‐free PcpI‐LBD has a midpoint of protein unfolding transition (Tm) of 39.6°C and that salicylate caused an increase in the Tm of PcpI‐LBD of 2.6°C (Supp. Fig. S10). No additional compounds causing Tm shifts were identified. To confirm binding, PcpI‐LBD was titrated with salicylate. Exothermic heats were observed that decreased as protein saturation progressed and a K D of 826 ± 34 μM was derived (Fig. 5; Supp. Table S1). We subsequently analyzed 14 additional aromatic and non‐aromatic C6‐ring containing molecules (listed in the legend to Supp. Table S1) and found binding for benzoate and 3‐methylbenzoate (3‐MBA) with affinities of 171 ± 14 and 91 ± 8 μM, respectively (Fig. 5; Supp. Table S1). We therefore conclude that PcpI directly binds the carboxylic acid aromatic compounds salicylate, benzoate and 3‐MBA.
PcpI mediates chemotaxis to benzoate, 3‐MBA and salicylate
To assess the relevance of benzoate, 3‐MBA and salicylate on the physiology of P. putida 1290, we first conducted quantitative capillary assays. The strain 1290 exhibited chemotaxis towards the three ligands with an onset of chemotaxis at 10 μM and a maximal response at 1 mM for all three compounds (Fig. 6). The magnitude of the response was similar for the three PcpI ligands, although a slightly greater tactic response was observed for benzoate at concentrations above 1 mM (Fig. 6). Contrary to what was previously observed for other chemoreceptors (Reyes‐Darias et al., 2015; Fernandez et al., 2017), no correlation was observed between the affinity of the chemoreceptor LBD for the ligands and the magnitude of the chemotactic response. The in vivo response occurred at concentrations well below the K D for ligand recognition (Figs. 5 and 6).
Fig. 6.
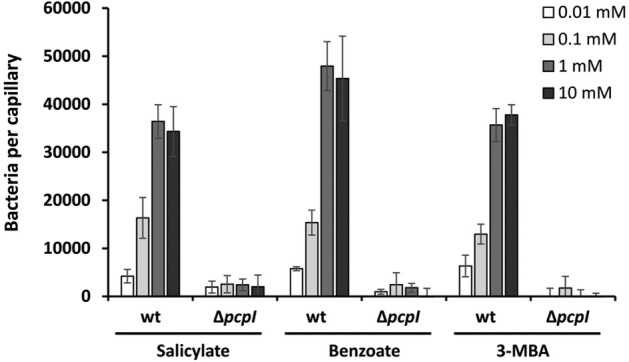
Quantitative capillary chemotaxis assays of Pseudomonas putida 1290 wild type and a pcpI mutant to different carboxylic acid aromatic ligands of PcpI. In all cases, data were corrected with the number of cells that swam into buffer containing capillaries. Shown data are means and standard deviations from three independent experiments conducted in triplicate. 3‐MBA, 3‐methylbenzoate.
To determine the role of PcpI in the observed tactic responses to aromatic compounds, quantitative capillary assays with a mutant defective in pcpI were carried out. The results showed that the deletion of pcpI caused the complete disappearance of chemotaxis to all three ligands over the entire concentration range (Fig. 6), indicating that PcpI is the sole P. putida 1290 chemoreceptor for benzoate, 3‐MBA and salicylate under the conditions tested.
We subsequently analyzed the metabolic relevance of the three PcpI ligands by conducting growth experiments in minimal medium containing each of the chemoattractants as sole carbon source. We found that benzoate and salicylate served as growth substrates for P. putida 1290 (Supp. Fig. S1B,C), whereas 3‐MBA did not support the growth of strain 1290 (Supp. Fig. S1D).
Role of PcpI in the chemotaxis towards root exudates and plant colonization
To evaluate the relevance of PcpI for establishing interactions with plants, we conducted competitive root colonization assays. In these assays, P. putida 1290 wild type and a pcpI mutant were inoculated at a certain distance from the maize seedlings and the number of wild type and mutant bacteria that colonized the roots 10 days post‐inoculation were quantified. We determined that P. putida 1290 colonizes maize roots at a density of around 7 × 107 bacteria per gram of root and that a mutant defective in pcpI was equally competitive than the wild strain in the colonization of the total root and root tips (Supp. Fig. S11). Subsequently, we evaluated in vitro whether maize root exudates serve as attractants for P. putida 1290. Quantitative capillary assays revealed that root exudates strongly attracted P. putida 1290 and that the magnitude of this attraction increased with the concentration of root exudates (Supp. Fig. S12). However, the pcpI mutant and the wild type strain exhibited similar chemotaxis to maize root exudates (Supp. Fig. S12).
Discussion
IAA is one of the central signal molecules of life. This auxin is synthesized in all kingdoms of life (Oliveira et al., 2007; Aklujkar et al., 2014; Bogaert et al., 2019; Duca and Glick, 2020; Gallei et al., 2020) and exerts a variety of different biological functions, including the regulation of (i) inflammatory responses in humans (Addi et al., 2019): (ii) growth and development in plants (Zhao, 2018; Gallei et al., 2020) and algae (Ohtaka et al., 2017; Bogaert et al., 2019); (iii) hyphal growth and sporulation in fungi (Fu et al., 2015; Nicastro et al., 2021); and (iv) bacterial physiology and metabolism (Duca and Glick, 2020). Notably, the role of IAA as an intra‐ and inter‐kingdom signal molecule has been investigated primarily in model systems based on bacteria–plant interactions, where it has been shown to act as a key signal in the modulation of various phytostimulatory and phytopathogenic processes through various mechanisms that include the alteration of auxin homeostasis and disturbances of auxin signalling in their plant hosts (Spaepen and Vanderleyden, 2011; Duca et al., 2014; Kunkel and Harper, 2018; Duca and Glick, 2020).
We identify here the first bacterial IAA chemoreceptor; a finding that expands the range of chemoreceptors that recognize central signal molecules of life, such as receptors for histamine (Corral‐Lugo et al., 2018), putrescine (Corral‐Lugo et al., 2016) or γ‐aminobutyrate (Rico‐Jimenez et al., 2013). Importantly, PcpI recognized and mediated chemoattraction to another important signal molecule, salicylate. Salicylate is an essential phytohormone that promotes plant immune responses against pathogens, as well as regulates plant growth, flowering and senescence (Bakker et al., 2014; Peng et al., 2021). Salicylate production has been described in bacteria and fungi (Bakker et al., 2014; Mishra and Baek, 2021) and its biosynthesis in bacteria is mainly associated with the production of salicylate‐based siderophores (Miethke and Marahiel, 2007; Bakker et al., 2014). However, current data support the role of salicylate as a central bacterial signal molecule, since it was shown to regulate antibiotic resistance, secondary metabolism, biofilm formation and virulence, among other processes (Price et al., 2000; Bakker et al., 2014; Lowe‐Power et al., 2016; Matilla et al., 2022). Notably, we have published recently a catalogue of signal molecules that are recognized by bacterial chemoreceptors, sensor kinases and transcriptional regulators, and salicylate was among the signal molecules for which the highest number of different sensor domains has been identified, namely, domains that belong to seven different Pfam families (Matilla et al., 2022). The PcpI LBD is un‐annotated in Pfam, suggesting that the diversity of salicylate binding domains can be even larger. Although PcpI‐LBD recognized salicylate with a modest affinity (K D = 826 ± 34 μM), the onset of chemotactic responses occurred at much lower concentrations, namely, 10 μM (Fig. 6). These discrepancies may be due to signal amplification in chemosensory arrays observed previously in Escherichia coli (Sourjik and Berg, 2002), the model bacterium for studying chemotaxis signal transduction (Parkinson et al., 2015). Salicylate can be detected in plant fluids and tissues at concentrations of up to 600 μM (Smith‐Becker et al., 1998; Huang et al., 2006; Ratzinger et al., 2009), indicating that PcpI mediates chemotaxis to physiological concentrations of this plant hormone.
Chemotaxis towards different phytohormones, including salicylate (Fernandez et al., 2017), ethylene (Kim et al., 2007) and jasmonic acid (Antunez‐Lamas et al., 2009) has been described in several plant‐associated bacteria, and the corresponding chemoreceptors involved identified (Kim et al., 2007; Rio‐Alvarez et al., 2015; Fernandez et al., 2017). However, to the best of our knowledge, PcpI is the first chemoreceptor that mediates chemotaxis towards two different phytohormones. The mechanisms by which IAA is sensed by bacteria remain mostly unknown. In E. coli, the tryptophan repressor TrpR recognizes IAA with low affinity (Marmorstein et al., 1987) and antibiotic synthesis in Serratia plymuthica is controlled by the transcriptional regulator AdmX, which binds IAA with significant affinity (K D = 15.2 μM) (Matilla et al., 2018). Our data strongly indicate that IAA and salicylic acid employ two different mechanisms to activate PcpI. Whereas salicylate activates PcpI by binding to the LBD, the mode of receptor stimulation by IAA is different since it does not involve direct recognition by the LBD (Fig. 5). Chemotaxis towards the hormone norepinephrine in E. coli was found to require its metabolization to 3,4‐dihydroxymandelic acid – a metabolite that was proposed to be the chemoeffector recognized by the Tsr chemoreceptor (Pasupuleti et al., 2014). However, the fact that mutation of the iac gene cluster does not affect the chemotactic properties of 1290 towards IAA, as well as the finding that in trans expression of pcpI in KT2440 and PAO1 conferred IAA chemotaxis to both strains strongly indicates that this tactic behaviour is not dependent on the sensing of an IAA catabolic intermediate.
For the large majority of the characterized chemoreceptors a single mode of activation, namely, by signal binding to the receptor LBD, has been reported (Ortega et al., 2017; Matilla et al., 2021a). However, studies of the two primary chemoreceptor models, E. coli Tar and Tsr, has revealed that both receptors can be activated by the direct binding of l‐Asp and l‐Ser, as well as by the recognition of the SBPs MBP and LsrB in complex with maltose and autoinducer‐2, respectively (Zhang et al., 1999; Hegde et al., 2011; Laganenka et al., 2016). Further research is necessary to identify the mode of PcpI activation by IAA, but current data indicate a convergent evolution of two different mechanisms that permit the sensing of two phytohormones. In accordance, an IAA binding SBP, Dde_0634, has been identified in an environmental isolate of Desulfovibrio desulfuricans (Vetting et al., 2015) and the SBP IaaM from the IAA‐degrading bacterium Azoarcus evansii was predicted to be involved in the uptake of IAA (Ebenau‐Jehle et al., 2012). However, the analysis of the genome of P. putida 1290 did not reveal the presence of any SBP homologous to Dde_0634 or IaaM, making targeted analysis of any candidate IAA binding SBPs unfeasible. SBP expression is tightly regulated (Matilla et al., 2021b) and the failure of our pull‐down experiments to detect an SBP that interacts with PcpI may be due to a very low cellular abundance. SBP‐mediated receptor stimulation has been proposed to expand the diversity of chemoeffectors recognized by chemoreceptors as well as their ligand concentration range (Matilla et al., 2021b). IAA can be found in plant cells, organic soils and in the rhizosphere at concentrations in the micromolar range (Brandl and Lindow, 1998; Petersson et al., 2009; Greenhut et al., 2018); values that are in the same range as the IAA concentrations for which taxis was observed (Figs. 1 and 3).
Current data support that chemotaxis represents an evolutionary advantage for bacteria that establish interactions with plants, being essential for plant colonization and infection in several bacterial species (Corral‐Lugo et al., 2016; Matilla and Krell, 2018; Compton and Scharf, 2021; Sanchis‐López et al., 2021). Indeed, 81% of the plant‐associated bacteria have chemoreceptor genes, which are superior to the bacterial average of 47% (Sanchis‐López et al., 2021). Furthermore, phytobacteria possess twice as many chemoreceptors than bacteria classified as non‐plant‐associated (Sanchis‐López et al., 2021). This prevalence of chemoreceptor genes in phytobacteria may be linked to the physical and chemical complexity of the plant environment as well as to the high competitiveness that exists in plant‐associated niches such as the rhizosphere (Raina et al., 2019; Fitzpatrick et al., 2020; Sanchis‐López et al., 2021). In this regard, a growing body of data reveals the importance of chemotaxis towards specific nutrients for an efficient plant colonization by beneficial and pathogenic phytobacteria. In this chemotaxis‐mediated host colonization, amino acids, organic acids and sugars were found to play major roles (Oku et al., 2012, 2014; Hida et al., ; Cerna‐Vargas et al., 2019; Feng et al., 2019; O'Neal et al., 2020; Compton and Scharf, 2021). However, determining the role of chemotaxis towards alternative plant molecules (e.g. fatty acids, nucleotides, host hormones, inorganic nutrients) and the biological function of specific chemoreceptors remains challenging. For example, chemotaxis to root exudates required multiple chemoreceptors in Bacillus subtilis, namely McpB, McpC and TlpC. In contrast, a triple deletion mutant defective in these chemoreceptors colonized plant roots at the wild type levels (Allard‐Massicotte et al., 2016). Root colonization is a multifactorial process (Jones et al., 2019; Knights et al., 2021) and current research supports that the combined action of chemoreceptors with complementary functions is responsible for chemotaxis towards roots as a prior step for plant colonization (Allard‐Massicotte et al., 2016; Feng et al., 2019). In this context, under the experimental conditions tested, PcpI did not play a relevant role in plant root colonization (Suppl. Fig. S11). This aspect may be associated with the remarkable number and diversity of chemoreceptors encoded in the genome of P. putida 1290 and the chemical composition of maize root exudates; which major constituents are sugars, amino and organic acids (Fan et al., 2012; da Silva Lima et al., 2014; Lopez‐Farfan et al., 2019). However, the composition of plant exudates varies qualitatively and quantitatively according to physical, chemical and biological factors (Sasse et al., 2018; Vives‐Peris et al., 2020; Compton and Scharf, 2021). Alterations in metabolite exudation influences plant microbiome composition (Sasse et al., 2018; Pascale et al., 2020) and chemotactic recruitment of bacteria is dependent on variations in the composition of plant exudates (Feng et al., 2019; Compton and Scharf, 2021). It can therefore be hypothesized that PcpI may play a role under plant‐specific physiological conditions, for example, during the induction of systemic acquired resistance when strong increases in salicylic acid levels have been measured in plant fluids (Smith‐Becker et al., 1998).
Salicylate and IAA served as nutrient source for P. putida 1290 (Supp. Fig. S1) and migration mediated by chemotaxis or energy taxis towards these compounds may confer a selective advantage over microbial competitors in specific niches with significant concentrations of these PcpI ligands. In accordance, bacterial IAA metabolism was demonstrated to act as a metabolic signal interference altering the communication networks between competitor bacteria and their plant hosts (Finkel et al., 2020). The wide distribution of IAA catabolic genes in bacteria (Li et al., 2016; Laird et al., 2020) has raised questions about their ecological role and further research will establish whether chemotaxis to IAA is a general feature of IAA degrading bacteria.
Experimental procedures
Bacterial strains, plasmids and culture conditions
Bacterial strains and plasmids are listed in Supp. Table S2. Pseudomonas putida and P. aeruginosa strains were grown routinely at 30°C and 37°C, respectively, in LB or M9 minimal medium supplemented with 1 mM MgSO4, 6 mg L−1 Fe‐citrate, 15 mM glucose as carbon source and trace elements as described previously (Abril et al., 1989). Escherichia coli strains were grown at 37°C. Escherichia coli DH5α was used as a host for gene cloning. Media for propagation of E. coli β2163 were supplemented with 300 μM 2,6‐diaminopimelic acid. When necessary, antibiotics were used at the following final concentrations: kanamycin, 50 μg ml−1, ampicillin, 100 μg ml−1, gentamycin 10 μg ml−1 (E. coli) or 100 μg ml−1 (P. putida and P. aeruginosa), streptomycin, 50 μg ml−1. Sucrose was added to a final concentration of 10% (wt/vol) when required to select derivatives that had undergone a second crossover event during marker‐exchange mutagenesis.
Construction of bacterial strains and complementation plasmid
Mutants defective in iacA, E6B08_RS07220, E6B08_RS17840, E6B08_RS22475, E6B08_RS29420 and E6B08_RS30830 were constructed using derivate plasmids of pCHESIΩKmGm. These plasmids are listed in Supp. Table S2 and were generated by amplifying a 0.6–0.9 kb region of the gene to be mutated using primers listed in Supp. Table S3. The PCR products were then cloned into pCHESIΩKmGm in the same transcriptional direction as the P lac promoter using the enzymes specified in Supp. Table S2. A plasmid‐free mutant defective in pcpI was constructed by homologous recombination using a derivative plasmid of the suicide vector pKNG101. The plasmid for the construction of this pcpI deletion mutant was generated by amplifying the up‐ and downstream flanking regions of the pcpI gene using the primers listed in Supp. Table S3. The resulting PCR products were digested with the enzymes specified in Supp. Table S2 and ligated in a three‐way ligation into pUC18Not, previously cloned into the marker exchange vector pKNG101. In all cases, plasmids for mutagenesis were transferred to P. putida strains by biparental conjugation using E. coli β2163. For the construction of the plasmid for complementation assays, the pcpI gene was amplified using primers listed in Supp. Table S3 and cloned into pBBR1MCS‐5_START to generate the plasmid pMAMV378. The resulting plasmid was transformed into P. aeruginosa and P. putida strains by electroporation. All plasmids and mutations were confirmed by PCR and sequencing.
Swimming motility assays
Pseudomonas putida 1290 strains were grown overnight in M9 minimal medium containing 5 mM IAA as carbon source and adjusted to an OD660 of 1. Two microliters of these cultures were spotted onto minimal medium‐Difco agar [0.3% (wt/vol)] plates containing 5 mM IAA acid as sole carbon source and incubated at 30°C.
Chemotaxis assays
Overnight cultures in M9 minimal medium were used to inoculate fresh medium to reach an OD660 of 0.075. Cells were cultured at 30°C (P. putida) or 37°C (P. aeruginosa) until an OD660 of 0.4–0.5 was reached. Subsequently, cells were washed twice by centrifugation (1667 xg for 5 min at room temperature) and re‐suspension in chemotaxis buffer [50 mM KH2PO4 /K2HPO4, 20 mM EDTA, 0.05% (vol/vol) glycerol, pH 7.0], and then re‐suspended in the same buffer to reach an OD660 of 0.1. Aliquots (230 μl) of the resulting cell suspension were placed into the wells of a 96‐well microtiter plate. One microliter capillaries (Microcaps, Drummond Scientific, Ref. P1424) were heat‐sealed at one end and filled with buffer (control) or chemoeffector solutions prepared in chemotaxis buffer. The capillaries were rinsed with sterile water and immersed into the bacterial suspensions at its open end. After 30 min, capillaries were removed from the wells, rinsed with sterile water, and emptied into 1 ml of chemotaxis buffer. Serial dilutions were plated onto M9 minimal medium plates supplemented with 15 mM glucose and incubated at 30°C or 37°C. Colony forming units (CFU) counts were determined and corrected with the number of cells that swam into buffer containing capillaries. Data are means and standard deviations of three biological replicates conducted in triplicate.
RNA extraction, cDNA synthesis and quantitative real‐time PCR analyses
RNA was extracted from mid‐logarithmic growth phase cultures grown in minimal medium by the hot phenol method using the TRI® Reagent protocol (Ambion) according to the manufacturer's instructions. RNA concentration was determined spectrophotometrically using a NanoDrop spectrophotometer (Thermo Scientific) and RNA integrity was assessed by agarose gel electrophoresis. Genomic DNA contamination was eliminated by treating total RNA with Turbo DNA‐free (Ambion), followed by a purification with RNeasy mini kit (Qiagen). The synthesis of cDNA was performed using 200 ng of random hexamer primers (Roche) and SuperScript II reverse transcriptase (Invitrogen) in a 20 μl reaction with 1 μg of total RNA and incubation at 42°C for 1.5 h. Quantitative real‐time PCR amplifications were performed using the iQ™ SYBR® Green supermix (Bio‐Rad) in a MyiQ2 system (Bio‐Rad) associated with iQ5 optical system software (version 2.1.97.1001). PCR reactions contained 6.25 μl of 2× SYBR Green supermix, 400 nM of each primer and 0.5 μl of cDNA in a final volume of 12.5 μl. The PCR protocol used was as follows: one cycle at 95°C for 5 min followed by 40 cycles at 95°C for 15 s, 63°C for 30 s, and 72°C for 20 s and melting curve analysis from 55°C to 95°C, with an increment of 0.5°C/10 s for 80 cycles. The primers used in this study were designed using the Clone Manager software 6.0 (Sci‐Ed Software) and are listed in Supp. Table S3. Standard curves for each primer pair were generated with serial dilutions of genomic DNA to determine PCR efficiency and melting curve analyses were conducted to ensure amplification of a single product. The relative gene expression was calculated using the critical threshold (∆Ct) method (Silver et al., 2006) using gyrB as the internal control to normalize the data. Data are the means and standard deviations of three biological replicates conducted in triplicate.
Construction of overexpression plasmids, protein expression and purification
The DNA fragments encoding the LBD of the chemoreceptor PcpI (amino acids 38–174) and the SBP E6B08_RS28125 were amplified by PCR from genomic DNA and primers listed in Supp. Table S3. The PCR products were then cloned into the NdeI and BamHI sites of pET28b(+) to generate plasmids pMAMV365 and pMAMV385, respectively. The sequence predicted to be signal peptide was not included in pMAMV385. Escherichia coli BL21 (DE3) harbouring plasmids pMAMV365 and pMAMV385 were grown under continuous shaking (200 rpm) at 30°C in 2 L Erlenmeyer flasks containing 500 ml LB medium supplemented with kanamycin. At an OD660 of 0.6, PcpI‐LBD and E6B08_RS28125 expression was induced by the addition of 0.25 mM isopropyl ß‐D‐1‐thiogalactopyranoside (IPTG). Growth was continued at 18°C overnight and cells were harvested by centrifugation at 10 000 xg for 20 min at 4°C. Proteins were purified by metal affinity chromatography using standard procedures. Briefly, cell pellets for the purification of PcpI‐LBD and E6B08_RS28125 were re‐suspended in buffer A [20 mM Tris, 500 mM NaCl, 10 mM imidazole, 1 mM EDTA, 5% (vol/vol) glycerol, pH 8.0] and buffer B [50 mM Tris, 150 mM NaCl, 10 mM imidazole, 10% (vol/vol) glycerol, pH 8.0] respectively, containing cOmplete™ protease inhibitor cocktail (Roche) and benzonase (Sigma‐Aldrich). Cells were broken by French press treatment at a gauge pressure of 62.5 lb in−2. After centrifugation at 10 000 xg for 1 h, the supernatants were loaded onto a 5‐ml HisTrap column (Amersham Bioscience) equilibrated with the corresponding buffers A and B, and proteins were eluted by a linear gradient of 40–500 mM imidazole in the same buffers.
Differential scanning fluorimetry‐based thermal shift assays
Using DSF, changes in the midpoint of protein unfolding transition (Tm) of a protein can be recorded. Typically, ligand binding stabilizes the protein and the identification of compounds that cause an increase in the Tm value is an evidence for ligand binding (Martin‐Mora et al., 2018a). DSF assays were performed using a Bio‐Rad MyiQ2 Real‐Time PCR instrument. Ligands from different compound arrays (Biolog, Hayward, CA, USA; for further information, refer to http://www.biolog.com/) were dissolved in 50 μl of Milli‐Q water, which, according to the manufacturer, corresponds to a concentration of 10–20 mM. Assay mixtures (25 μl) contained 20–50 μM protein dialyzed in buffer C [50 mM Tris, 150 mM NaCl, 5% (vol/vol) glycerol, pH 8.0; PcpI‐LBD] or buffer D [5 mM Tris, 5 mM Pipes, 5 mM Mes, 10% glycerol (vol/vol), 150 mM NaCl, pH 8; E6B08_RS28125], SYPRO® Orange (Life Technologies) at 5× concentration and ligands at final concentrations of 1–2 mM. Samples were heated from 23°C to 85°C at a rate of 1°C min−1. The protein unfolding curves were obtained by monitoring the changes in SYPRO Orange fluorescence. Tm values correspond to the minima of the first derivatives of the raw fluorescence data.
Isothermal titration calorimetry
Measurements were made using a VP‐ITC titration calorimeter (Microcal, Northampton, MA, USA) at a temperature of 25°C. PcpI‐LBD and E6B08_RS28125 were dialyzed into buffer C and buffer D, respectively, and proteins at 40–226 μM were placed into the sample cell and titrated with 3.2–9.6 μl aliquots of 0.5–5 mM ligand solutions freshly made up in dialysis buffer. In the absence of binding, the experiment was repeated at an analysis temperature of 10°C. The mean enthalpies measured from the injection of effectors into the buffer were subtracted from raw titration data prior to data analysis with the MicroCal version of ORIGIN. Data were fitted with the ‘One binding site model’ of ORIGIN.
Pull‐down assays
Overnight cultures of P. putida 1290 grown in M9 minimal medium supplemented with glucose as a carbon source was used to inoculate fresh medium to reach an OD660 of 0.075. After overnight growth, cultures were diluted to an OD600 of 0.075 in the same medium until an OD660 of 0.6. Subsequently, pellets were re‐suspended in buffer A containing 10 mM IAA and broken by French press treatment at a gauge pressure of 62.5 lb in−2. After centrifugation at 10 000 xg for 1 h, the supernatant was loaded onto a HisTrap column on which PcpI‐LBD had previously been immobilized. The column was washed with buffer A prior to protein elution using a 0–6 M guanidine hydrochloride gradient in buffer A. Finally, to release PcpI‐LBD or any other protein bound to the HisTrap column, a gradient of 10–500 mM imidazole in buffer A was applied. As a control, the P. putida 1290 supernatant was applied to a column that did not contain PcpI‐LBD. Bands of interest were excised from an SDS‐PAGE gel, digested with trypsin and analyzed by MALDI‐TOF mass spectrometry at the proteomics service of the Faculty of Pharmacy – Complutense University of Madrid (Spain). Protein identity was established using the MASCOT software.
Competitive root colonization assays
Maize seeds were sterilized and germinated as described previously (Matilla et al., 2007). Thereafter, germinated seeds were planted at the centre of a 50 ml Sterilin tubes containing 40 g of sterile washed silica sand. For the competitive root colonization assays, 100 μl of a 107 CFU ml−1 1:1 mixture of the wild type P. putida 1290 and a pcpI mutant were inoculated at the edge of each Sterilin tube. Subsequently, plants were maintained at 24°C with a daily light period of 16 h. After 10 days, bacterial cells were recovered from the rhizosphere or from 1 mm of the main root apex, as described previously (Matilla et al., 2007). Serial dilutions were plated in minimal medium‐agar and minimal medium‐agar supplemented with 50 μg ml−1 of kanamycin to select the pcpI mutant strain.
Collection of maize root exudates
The collection of maize root exudates was carried out as previously indicated (Lopez‐Farfan et al., 2019). Briefly, maize seeds were sterilized and germinated as described previously (Matilla et al., 2007). Sixteen germinated seeds were transferred into an axenic system with 450 ml of sterile water and allowed to grow at room temperature. After 8 days, the water containing root exudates was collected and vacuum filtrated (0.45 μm cut‐off). An aliquot was taken and plated onto solid LB media to check for contamination. Maize root exudates were aliquoted, freeze‐dried and stored at −80°C. Before use, the lyophilized exudates were re‐suspended in chemotaxis medium and filter‐sterilized.
Growth experiments
Pseudomonas putida 1290 strains were grown overnight in an M9 minimal medium containing 15 mM glucose. Cultures were washed twice with M9 salts medium and then diluted to an OD600 of 0.02 in M9 containing 5 mM glucose (positive control) and medium supplemented with 5 mM IAA, benzoate, 3‐MBA and salicylate as carbon sources. Two‐hundred microliters of these cultures were transferred to microwell plates and growth (OD600) at 30°C was followed over time using Bioscreen Microbiological Growth Analyser (Oy Growth Curves Ab, Helsinki, Finland).
Supporting information
Appendix S1: Supporting Information.
Acknowledgements
This study was supported through grants from the CSIC to M.A.M. (PIE‐202040I003), from the Spanish Ministry for Science and Innovation/Agencia Estatal de Investigación 10.13039/501100011033 (PID2019‐103972GA‐I00 to M.A.M. and grant PID2020‐112612GB‐I00 to T.K.) and the Junta de Andalucía (grant P18‐FR‐1621 to T.K.). A.R. was supported by the Ramon y Cajal R&D&i Programme (RYC2019‐026481‐I) from the Spanish Ministry for Science and Innovation/Agencia Estatal de Investigación/10.13039/501100011033 y FSE ‘El FSE invierte en tu futuro’.
References
- Abril, M.A. , Michan, C. , Timmis, K.N. , and Ramos, J.L. (1989) Regulator and enzyme specificities of the TOL plasmid‐encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J Bacteriol 171: 6782–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addi, T. , Poitevin, S. , McKay, N. , El Mecherfi, K.E. , Kheroua, O. , Jourde‐Chiche, N. , et al. (2019) Mechanisms of tissue factor induction by the uremic toxin indole‐3 acetic acid through aryl hydrocarbon receptor/nuclear factor‐kappa B signaling pathway in human endothelial cells. Arch Toxicol 93: 121–136. [DOI] [PubMed] [Google Scholar]
- Aklujkar, M. , Risso, C. , Smith, J. , Beaulieu, D. , Dubay, R. , Giloteaux, L. , et al. (2014) Anaerobic degradation of aromatic amino acids by the hyperthermophilic archaeon Ferroglobus placidus . Microbiology 160: 2694–2709. [DOI] [PubMed] [Google Scholar]
- Allard‐Massicotte, R. , Tessier, L. , Lecuyer, F. , Lakshmanan, V. , Lucier, J.F. , Garneau, D. , et al. (2016) Bacillus subtilis early colonization of Arabidopsis thaliana roots involves multiple chemotaxis receptors. mBio 7: e01664‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez‐Ortega, C. , and Harwood, C.S. (2007) Identification of a malate chemoreceptor in Pseudomonas aeruginosa by screening for chemotaxis defects in an energy taxis‐deficient mutant. Appl Environ Microbiol 73: 7793–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin, S.A. , Hmelo, L.R. , van Tol, H.M. , Durham, B.P. , Carlson, L.T. , Heal, K.R. , et al. (2015) Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522: 98–101. [DOI] [PubMed] [Google Scholar]
- Antunez‐Lamas, M. , Cabrera, E. , Lopez‐Solanilla, E. , Solano, R. , Gonzalez‐Melendi, P. , Chico, J.M. , et al. (2009) Bacterial chemoattraction towards jasmonate plays a role in the entry of Dickeya dadantii through wounded tissues. Mol Microbiol 74: 662–671. [DOI] [PubMed] [Google Scholar]
- Bains, M. , Fernandez, L. , and Hancock, R.E. (2012) Phosphate starvation promotes swarming motility and cytotoxicity of Pseudomonas aeruginosa . Appl Environ Microbiol 78: 6762–6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker, P.A.H.M. , Ran, L. , and Mercado‐Blanco, J. (2014) Rhizobacterial salicylate production provokes headaches! Plant Soil 382: 1–16. [Google Scholar]
- Bi, S. , and Sourjik, V. (2018) Stimulus sensing and signal processing in bacterial chemotaxis. Curr Opin Microbiol 45: 22–29. [DOI] [PubMed] [Google Scholar]
- Bogaert, K.A. , Blommaert, L. , Ljung, K. , Beeckman, T. , and De Clerck, O. (2019) Auxin function in the Brown alga Dictyota dichotoma . Plant Physiol 179: 280–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl, M.T. , and Lindow, S.E. (1998) Contribution of indole‐3‐acetic acid production to the epiphytic fitness of Erwinia herbicola . Appl Environ Microbiol 64: 3256–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerna‐Vargas, J.P. , Santamaría‐Hernando, S. , Matilla, M.A. , Rodríguez‐Herva, J.J. , Daddaoua, A. , Rodríguez‐Palenzuela, P. , et al. (2019) Chemoperception of specific amino acids controls Phytopathogenicity in Pseudomonas syringae pv. tomato. mBio 10: e01868‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, A.I. , Dolben, E.F. , Okegbe, C. , Harty, C.E. , Golub, Y. , Thao, S. , et al. (2014) Candida albicans ethanol stimulates Pseudomonas aeruginosa WspR‐controlled biofilm formation as part of a cyclic relationship involving phenazines. PLoS Pathog 10: e1004480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin, R. , Ni, B. , Laganenka, L. , and Sourjik, V. (2021) Multiple functions of flagellar motility and chemotaxis in bacterial physiology. FEMS Microbiol Rev 45: fuab038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, K.D. , Lacal, J. , and Ottemann, K.M. (2014) Internal sense of direction: sensing and signaling from cytoplasmic chemoreceptors. Microbiol Mol Biol Rev 78: 672–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton, K.K. , and Scharf, B.E. (2021) Rhizobial chemoattractants, the taste and preferences of legume symbionts. Front Plant Sci 12: 686465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral‐Lugo, A. , De la Torre, J. , Matilla, M.A. , Fernández, M. , Morel, B. , Espinosa‐Urgel, M. , and Krell, T. (2016) Assessment of the contribution of chemoreceptor‐based signalling to biofilm formation. Environ Microbiol 18: 3355–3372. [DOI] [PubMed] [Google Scholar]
- Corral‐Lugo, A. , Matilla, M.A. , Martín‐Mora, D. , Silva Jiménez, H. , Mesa Torres, N. , Kato, J. , et al. (2018) High‐affinity chemotaxis to histamine mediated by the TlpQ chemoreceptor of the human pathogen Pseudomonas aeruginosa . mBio 9: e01894‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cserzo, M. , Wallin, E. , Simon, I. , von Heijne, G. , and Elofsson, A. (1997) Prediction of transmembrane alpha‐helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng 10: 673–676. [DOI] [PubMed] [Google Scholar]
- da Silva Lima, L. , Olivares, F.L. , Rodrigues de Oliveira, R. , Vega, M.R.G. , Aguiar, N.O. , and Canellas, L.P. (2014) Root exudate profiling of maize seedlings inoculated with Herbaspirillum seropedicae and humic acids. Chem Biol Technol Agric 1: 23. [Google Scholar]
- Djami‐Tchatchou, A.‐T. , Li, Z.A. , Stodghill, P. , Filiatrault, M.J. , and Kunkel, B.N. (2021) Identification of IAA‐regulated genes in Pseudomonas syringae pv. tomato strain DC3000. J Bacteriol 204: e0038021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati, A.J. , Lee, H.I. , Leveau, J.H. , and Chang, W.S. (2013) Effects of indole‐3‐acetic acid on the transcriptional activities and stress tolerance of Bradyrhizobium japonicum . PLoS One 8: e76559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duca, D. , Lorv, J. , Patten, C.L. , Rose, D. , and Glick, B.R. (2014) Indole‐3‐acetic acid in plant‐microbe interactions. Antonie Van Leeuwenhoek 106: 85–125. [DOI] [PubMed] [Google Scholar]
- Duca, D.R. , and Glick, B.R. (2020) Indole‐3‐acetic acid biosynthesis and its regulation in plant‐associated bacteria. Appl Microbiol Biotechnol 104: 8607–8619. [DOI] [PubMed] [Google Scholar]
- Ebenau‐Jehle, C. , Thomas, M. , Scharf, G. , Kockelkorn, D. , Knapp, B. , Schühle, K. , et al. (2012) Anaerobic metabolism of indoleacetate. J Bacteriol 194: 2894–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbourne, L.D.H. , Tetu, S.G. , Hassan, K.A. , and Paulsen, I.T. (2017) TransportDB 2.0: a database for exploring membrane transporters in sequenced genomes from all domains of life. Nucleic Acids Res 45: D320–D324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, B. , Carvalhais, L.C. , Becker, A. , Fedoseyenko, D. , von Wiren, N. , and Borriss, R. (2012) Transcriptomic profiling of Bacillus amyloliquefaciens FZB42 in response to maize root exudates. BMC Microbiol 12: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, H. , Zhang, N. , Fu, R. , Liu, Y. , Krell, T. , Du, W. , et al. (2019) Recognition of dominant attractants by key chemoreceptors mediates recruitment of plant growth‐promoting rhizobacteria. Environ Microbiol 21: 402–415. [DOI] [PubMed] [Google Scholar]
- Fernandez, M. , Matilla, M.A. , Ortega, A. , and Krell, T. (2017) Metabolic value chemoattractants are preferentially recognized at broad ligand range chemoreceptor of Pseudomonas putida KT2440. Front Microbiol 8: 990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández, M. , Morel, B. , Corral‐Lugo, A. , and Krell, T. (2016) Identification of a chemoreceptor that specifically mediates chemotaxis toward metabolizable purine derivatives. Mol Microbiol 99: 34–42. [DOI] [PubMed] [Google Scholar]
- Finkel, O.M. , Salas‐González, I. , Castrillo, G. , Conway, J.M. , Law, T.F. , Teixeira, P.J.P.L. , et al. (2020) A single bacterial genus maintains root growth in a complex microbiome. Nature 587: 103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick, C.R. , Salas‐González, I. , Conway, J.M. , Finkel, O.M. , Gilbert, S. , Russ, D. , et al. (2020) The plant microbiome: from ecology to reductionism and beyond. Annu Rev Microbiol 74: 81–100. [DOI] [PubMed] [Google Scholar]
- Fu, S.F. , Wei, J.Y. , Chen, H.W. , Liu, Y.Y. , Lu, H.Y. , and Chou, J.Y. (2015) Indole‐3‐acetic acid: a widespread physiological code in interactions of fungi with other organisms. Plant Signal Behav 10: e1048052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher, N.B. , Holliday, P.M. , Klem, E. , Cann, M.J. , and Wolfgang, M.C. (2010) The Pseudomonas aeruginosa Chp chemosensory system regulates intracellular cAMP levels by modulating adenylate cyclase activity. Mol Microbiol 76: 889–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallei, M. , Luschnig, C. , and Friml, J. (2020) Auxin signalling in growth: Schrödinger's cat out of the bag. Curr Opin Plant Biol 53: 43–49. [DOI] [PubMed] [Google Scholar]
- Garcia, V. , Reyes‐Darias, J.A. , Martin‐Mora, D. , Morel, B. , Matilla, M.A. , and Krell, T. (2015) Identification of a chemoreceptor for C2 and C3 carboxylic acids. Appl Environ Microbiol 81: 5449–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavira, J.A. , Gumerov, V.M. , Rico‐Jiménez, M. , Petukh, M. , Upadhyay, A.A. , Ortega, A. , et al. (2020) How bacterial chemoreceptors evolve novel ligand specificities. mBio 11: e03066‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhut, I.V. , Slezak, B.L. , and Leveau, J.H.J. (2018) iac gene expression in the indole‐3‐acetic acid‐degrading soil bacterium Enterobacter soli LF7. Appl Environ Microbiol 84: e01057‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde, M. , Englert, D.L. , Schrock, S. , Cohn, W.B. , Vogt, C. , Wood, T.K. , et al. (2011) Chemotaxis to the quorum‐sensing signal AI‐2 requires the Tsr chemoreceptor and the periplasmic LsrB AI‐2‐binding protein. J Bacteriol 193: 768–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herud‐Sikimić, O. , Stiel, A.C. , Kolb, M. , Shanmugaratnam, S. , Berendzen, K.W. , Feldhaus, C. , et al. (2021) A biosensor for the direct visualization of auxin. Nature 592: 768–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hida, A. , Oku, S. , Kawasaki, T. , Nakashimada, Y. , Tajima, T. , and Kato, J. (2015) Identification of the mcpA and mcpM genes, encoding methyl‐accepting proteins involved in amino acid and l‐malate chemotaxis, and involvement of McpM‐mediated chemotaxis in plant infection by Ralstonia pseudosolanacearum (formerly Ralstonia solanacearum p). Appl Environ Microbiol 81: 7420–7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hida, A. , Oku, S. , Miura, M. , Matsuda, H. , Tajima, T. , and Kato, J. (2020) Characterization of methyl‐accepting chemotaxis proteins (MCPs) for amino acids in plant‐growth‐promoting rhizobacterium Pseudomonas protegens CHA0 and enhancement of amino acid chemotaxis by MCP genes overexpression. Biosci Biotechnol Biochem 84: 1948–1957. [DOI] [PubMed] [Google Scholar]
- Hong, C.S. , Kuroda, A. , Ikeda, T. , Takiguchi, N. , Ohtake, H. , and Kato, J. (2004a) The aerotaxis transducer gene aer, but not aer‐2, is transcriptionally regulated by the anaerobic regulator ANR in Pseudomonas aeruginosa . J Biosci Bioeng 97: 184–190. [DOI] [PubMed] [Google Scholar]
- Hong, C.S. , Shitashiro, M. , Kuroda, A. , Ikeda, T. , Takiguchi, N. , Ohtake, H. , and Kato, J. (2004b) Chemotaxis proteins and transducers for aerotaxis in Pseudomonas aeruginosa . FEMS Microbiol Lett 231: 247–252. [DOI] [PubMed] [Google Scholar]
- Huang, W.E. , Huang, L. , Preston, G.M. , Naylor, M. , Carr, J.P. , Li, Y. , et al. (2006) Quantitative in situ assay of salicylic acid in tobacco leaves using a genetically modified biosensor strain of Acinetobacter sp. ADP1. Plant J 46: 1073–1083. [DOI] [PubMed] [Google Scholar]
- Jahn, L. , Hofmann, U. , and Ludwig‐Müller, J. (2021) Indole‐3‐acetic acid is synthesized by the endophyte Cyanodermella asteris via a tryptophan‐dependent and ‐independent way and mediates the interaction with a non‐host plant. Int J Mol Sci 22: 2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansari, V.H. , Potharla, V.Y. , Riddell, G.T. , and Bardy, S.L. (2016) Twitching motility and cAMP levels: signal transduction through a single methyl‐accepting chemotaxis protein. FEMS Microbiol Lett 363: fnw119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, P. , Garcia, B.J. , Furches, A. , Tuskan, G.A. , and Jacobson, D. (2019) Plant host‐associated mechanisms for microbial selection. Front Plant Sci 10: 862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, L.A. , Mezulis, S. , Yates, C.M. , Wass, M.N. , and Sternberg, M.J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.E. , Shitashiro, M. , Kuroda, A. , Takiguchi, N. , and Kato, J. (2007) Ethylene chemotaxis in Pseudomonas aeruginosa and other Pseudomonas species. Microbes Environ 22: 186–189. [Google Scholar]
- Knights, H.E. , Jorrin, B. , Haskett, T.L. , and Poole, P.S. (2021) Deciphering bacterial mechanisms of root colonization. Environ Microbiol Rep 13: 428–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel, B.N. , and Harper, C.P. (2018) The roles of auxin during interactions between bacterial plant pathogens and their hosts. J Exp Bot 69: 245–254. [DOI] [PubMed] [Google Scholar]
- Kunkel, B.N. , and Johnson, J.M.B. (2021) Auxin plays multiple roles during plant‐pathogen interactions. Cold Spring Harb Perspect Biol 13: a040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacal, J. , Alfonso, C. , Liu, X. , Parales, R.E. , Morel, B. , Conejero‐Lara, F. , et al. (2010a) Identification of a chemoreceptor for tricarboxylic acid cycle intermediates: differential chemotactic response towards receptor ligands. J Biol Chem 285: 23126–23136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacal, J. , García‐Fontana, C. , Muñoz‐Martínez, F. , Ramos, J.‐L. , and Krell, T. (2010b) Sensing of environmental signals: classification of chemoreceptors according to the size of their ligand binding regions. Environ Microbiol 12: 2873–2884. [DOI] [PubMed] [Google Scholar]
- Laganenka, L. , Colin, R. , and Sourjik, V. (2016) Chemotaxis towards autoinducer 2 mediates autoaggregation in Escherichia coli . Nat Commun 7: 13979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird, T.S. , Flores, N. , and Leveau, J.H.J. (2020) Bacterial catabolism of indole‐3‐acetic acid. Appl Microbiol Biotechnol 104: 9535–9550. [DOI] [PubMed] [Google Scholar]
- Laird, T.S. , and Leveau, J.H.J. (2019) Finished genome sequence of the indole‐3‐acetic acid‐catabolizing bacterium Pseudomonas putida 1290. Microbiol Resour Announc 8: e00519‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveau, J.H. , and Gerards, S. (2008) Discovery of a bacterial gene cluster for catabolism of the plant hormone indole 3‐acetic acid. FEMS Microbiol Ecol 65: 238–250. [DOI] [PubMed] [Google Scholar]
- Leveau, J.H. , and Lindow, S.E. (2005) Utilization of the plant hormone indole‐3‐acetic acid for growth by Pseudomonas putida strain 1290. Appl Environ Microbiol 71: 2365–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Wang, D. , Hu, F. , Li, H. , Ma, L. , and Xu, L. (2016) Exogenous IAA treatment enhances phytoremediation of soil contaminated with phenanthrene by promoting soil enzyme activity and increasing microbial biomass. Environ Sci Pollut Res Int 23: 10656–10664. [DOI] [PubMed] [Google Scholar]
- Liu, X. , Wood, P.L. , Parales, J.V. , and Parales, R.E. (2009) Chemotaxis to pyrimidines and identification of a cytosine chemoreceptor in Pseudomonas putida . J Bacteriol 191: 2909–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.‐Y. , Chen, H.‐W. , and Chou, J.‐Y. (2016) Variation in indole‐3‐acetic acid production by wild Saccharomyces cerevisiae and S. paradoxus strains from diverse ecological sources and its effect on growth. PLoS One 11: e0160524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, J.G. , and Sourjik, V. (2018) Chemotaxis of Escherichia coli to major hormones and polyamines present in human gut. ISME J 12: 2736–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Farfan, D. , Reyes‐Darias, J.A. , Matilla, M.A. , and Krell, T. (2019) Concentration dependent effect of plant root exudates on the chemosensory systems of Pseudomonas putida KT2440. Front Microbiol 10: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe‐Power, T.M. , Jacobs, J.M. , Ailloud, F. , Fochs, B. , Prior, P. , and Allen, C. (2016) Degradation of the plant defense signal salicylic acid protects Ralstonia solanacearum from toxicity and enhances virulence on tobacco. mBio 7: e00656‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, Y. , Zhao, K. , Baker, A.E. , Kuchma, S.L. , Coggan, K.A. , Wolfgang, M.C. , et al. (2015) A hierarchical cascade of second messengers regulates Pseudomonas aeruginosa surface behaviors. mBio 6: e02456‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein, R.Q. , Joachimiak, A. , Sprinzl, M. , and Sigler, P.B. (1987) The structural basis for the interaction between L‐tryptophan and the Escherichia coli trp aporepressor. J Biol Chem 262: 4922–4927. [PubMed] [Google Scholar]
- Marmorstein, R.Q. , and Sigler, P.B. (1989) Stereochemical effects of L‐tryptophan and its analogues on trp repressor's affinity for operator‐DNA. J Biol Chem 264: 9149–9154. [PubMed] [Google Scholar]
- Martin‐Mora, D. , Fernandez, M. , Velando, F. , Ortega, A. , Gavira, J.A. , Matilla, M.A. , and Krell, T. (2018a) Functional annotation of bacterial signal transduction systems: progress and challenges. Int J Mol Sci 19: 3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin‐Mora, D. , Ortega, A. , Perez‐Maldonado, F.J. , Krell, T. , and Matilla, M.A. (2018b) The activity of the C4‐dicarboxylic acid chemoreceptor of Pseudomonas aeruginosa is controlled by chemoattractants and antagonists. Sci Rep 8: 2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla, M.A. , Daddaoua, A. , Chini, A. , Morel, B. , and Krell, T. (2018) An auxin controls bacterial antibiotics production. Nucleic Acids Res 46: 11229–11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla, M.A. , Espinosa‐Urgel, M. , Rodriguez‐Herva, J.J. , Ramos, J.L. , and Ramos‐Gonzalez, M.I. (2007) Genomic analysis reveals the major driving forces of bacterial life in the rhizosphere. Genome Biol 8: R179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla, M.A. , and Krell, T. (2018) The effect of bacterial chemotaxis on host infection and pathogenicity. FEMS Microbiol Rev 42: fux052. [DOI] [PubMed] [Google Scholar]
- Matilla, M.A. , Martín‐Mora, D. , Gavira, J.A. , and Krell, T. (2021a) Pseudomonas aeruginosa as a model to study chemosensory pathway signaling. Microbiol Mol Biol Rev 85: e00151‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla, M.A. , Ortega, Á. , and Krell, T. (2021b) The role of solute binding proteins in signal transduction. Comput Struct Biotechnol J 19: 1786–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla, M.A. , Velando, F. , Martín‐Mora, D. , Monteagudo‐Cascales, E. , and Krell, T. (2022) A catalogue of signal molecules that interact with sensor kinases, chemoreceptors and transcriptional regulators. FEMS Microbiol Rev 46: fuab043. [DOI] [PubMed] [Google Scholar]
- Miethke, M. , and Marahiel, M.A. (2007) Siderophore‐based iron acquisition and pathogen control. Microbiol Mol Biol Rev 71: 413–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, A.K. , and Baek, K.‐H. (2021) Salicylic acid biosynthesis and metabolism: a divergent pathway for plants and bacteria. Biomolecules 11: 705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, R. , Kohn, S. , Hwang, S.H. , Hassett, D.J. , and Sauer, K. (2006) BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa . J Bacteriol 188: 7335–7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal, A.L. , Ahmad, S. , Gordon‐Weeks, R. , and Ton, J. (2012) Benzoxazinoids in root exudates of maize attract Pseudomonas putida to the rhizosphere. PLoS One 7: e35498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro, R. , Raucci, S. , Michel, A.H. , Stumpe, M. , Osuna, G.M.G. , Jaquenoud, M. , et al. (2021) Indole‐3‐acetic acid is a physiological inhibitor of TORC1 in yeast. PLoS Genet 17: e1009414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor, J.R. , Kuwada, N.J. , Huangyutitham, V. , Wiggins, P.A. , and Harwood, C.S. (2012) Surface sensing and lateral subcellular localization of WspA, the receptor in a chemosensory‐like system leading to c‐di‐GMP production. Mol Microbiol 86: 720–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtaka, K. , Hori, K. , Kanno, Y. , Seo, M. , and Ohta, H. (2017) Primitive auxin response without TIR1 and Aux/IAA in the charophyte alga Klebsormidium nitens . Plant Physiol 174: 1621–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku, S. , Komatsu, A. , Nakashimada, Y. , Tajima, T. , and Kato, J. (2014) Identification of Pseudomonas fluorescens chemotaxis sensory proteins for malate, succinate, and fumarate, and their involvement in root colonization. Microbes Environ 29: 413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku, S. , Komatsu, A. , Tajima, T. , Nakashimada, Y. , and Kato, J. (2012) Identification of chemotaxis sensory proteins for amino acids in Pseudomonas fluorescens Pf0‐1 and their involvement in chemotaxis to tomato root exudate and root colonization. Microbes Environ 27: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira, D.L. , Pugine, S.M. , Ferreira, M.S. , Lins, P.G. , Costa, E.J. , and de Melo, M.P. (2007) Influence of indole acetic acid on antioxidant levels and enzyme activities of glucose metabolism in rat liver. Cell Biochem Funct 25: 195–201. [DOI] [PubMed] [Google Scholar]
- O'Neal, L. , Vo, L. , and Alexandre, G. (2020) Specific root exudate compounds sensed by dedicated chemoreceptors shape Azospirillum brasilense chemotaxis in the rhizosphere. Appl Environ Microbiol 86: e01026‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega, A. , Zhulin, I.B. , and Krell, T. (2017) Sensory repertoire of bacterial chemoreceptors. Microbiol Mol Biol Rev 81: e00033‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parales, R.E. , Luu, R.A. , Chen, G.Y. , Liu, X. , Wu, V. , Lin, P. , et al. (2013) Pseudomonas putida F1 has multiple chemoreceptors with overlapping specificity for organic acids. Microbiology 159: 1086–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parales, R.E. , Nesteryuk, V. , Hughes, J.G. , Luu, R.A. , and Ditty, J.L. (2014) Cytosine chemoreceptor McpC in Pseudomonas putida F1 also detects nicotinic acid. Microbiology 160: 2661–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson, J.S. , Hazelbauer, G.L. , and Falke, J.J. (2015) Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol 23: 257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascale, A. , Proietti, S. , Pantelides, I.S. , and Stringlis, I.A. (2020) Modulation of the root microbiome by plant molecules: the basis for targeted disease suppression and plant growth promotion. Front Plant Sci 10: 1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasupuleti, S. , Sule, N. , Cohn, W.B. , MacKenzie, D.S. , Jayaraman, A. , and Manson, M.D. (2014) Chemotaxis of Escherichia coli to norepinephrine (NE) requires conversion of NE to 3,4‐dihydroxymandelic acid. J Bacteriol 196: 3992–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Y. , Yang, J. , Li, X. , and Zhang, Y. (2021) Salicylic acid: biosynthesis and signaling. Annu Rev Plant Biol 72: 761–791. [DOI] [PubMed] [Google Scholar]
- Petersson, S.V. , Johansson, A.I. , Kowalczyk, M. , Makoveychuk, A. , Wang, J.Y. , Moritz, T. , et al. (2009) An auxin gradient and maximum in the Arabidopsis root apex shown by high‐resolution cell‐specific analysis of IAA distribution and synthesis. Plant Cell 21: 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova, O.E. , and Sauer, K. (2012a) Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc Natl Acad Sci U S A 109: 16690–16695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova, O.E. , and Sauer, K. (2012b) PAS domain residues and prosthetic group involved in BdlA‐dependent dispersion response by Pseudomonas aeruginosa biofilms. J Bacteriol 194: 5817–5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda‐Molina, E. , Reyes‐Darias, J.‐A. , Lacal, J. , Ramos, J.L. , García‐Ruiz, J.M. , Gavira, J.A. , and Krell, T. (2012) Evidence for chemoreceptors with bimodular ligand‐binding regions harboring two signal‐binding sites. Proc Natl Acad Sci U S A 109: 18926–18931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, C.T.D. , Lee, I.R. , and Gustafson, J.E. (2000) The effects of salicylate on bacteria. Int J Biochem Cell Biol 32: 1029–1043. [DOI] [PubMed] [Google Scholar]
- Raina, J.‐B. , Fernandez, V. , Lambert, B. , Stocker, R. , and Seymour, J.R. (2019) The role of microbial motility and chemotaxis in symbiosis. Nat Rev Microbiol 17: 284–294. [DOI] [PubMed] [Google Scholar]
- Ratzinger, A. , Riediger, N. , von Tiedemann, A. , and Karlovsky, P. (2009) Salicylic acid and salicylic acid glucoside in xylem sap of Brassica napus infected with Verticillium longisporum . J Plant Res 122: 571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes‐Darias, J.A. , Yang, Y. , Sourjik, V. , and Krell, T. (2015) Correlation between signal input and output in PctA and PctB amino acid chemoreceptor of Pseudomonas aeruginosa . Mol Microbiol 96: 513–525. [DOI] [PubMed] [Google Scholar]
- Rico‐Jimenez, M. , Munoz‐Martinez, F. , Garcia‐Fontana, C. , Fernandez, M. , Morel, B. , Ortega, A. , et al. (2013) Paralogous chemoreceptors mediate chemotaxis towards protein amino acids and the non‐protein amino acid gamma‐aminobutyrate (GABA). Mol Microbiol 88: 1230–1243. [DOI] [PubMed] [Google Scholar]
- Rico‐Jimenez, M. , Reyes‐Darias, J.A. , Ortega, A. , Diez Pena, A.I. , Morel, B. , and Krell, T. (2016) Two different mechanisms mediate chemotaxis to inorganic phosphate in Pseudomonas aeruginosa . Sci Rep 6: 28967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio‐Alvarez, I. , Munoz‐Gomez, C. , Navas‐Vasquez, M. , Martinez‐Garcia, P.M. , Antunez‐Lamas, M. , Rodriguez‐Palenzuela, P. , and Lopez‐Solanilla, E. (2015) Role of Dickeya dadantii 3937 chemoreceptors in the entry to Arabidopsis leaves through wounds. Mol Plant Pathol 16: 685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampedro, I. , Parales, R.E. , Krell, T. , and Hill, J.E. (2015) Pseudomonas chemotaxis. FEMS Microbiol Rev 39: 17–46. [DOI] [PubMed] [Google Scholar]
- Sanchis‐López, C. , Cerna‐Vargas, J.P. , Santamaría‐Hernando, S. , Ramos, C. , Krell, T. , Rodríguez‐Palenzuela, P. , et al. (2021) Prevalence and specificity of chemoreceptor profiles in plant‐associated bacteria. mSystems 6: e0095121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarand, I. , Osterberg, S. , Holmqvist, S. , Holmfeldt, P. , Skarfstad, E. , Parales, R.E. , and Shingler, V. (2008) Metabolism‐dependent taxis towards (methyl)phenols is coupled through the most abundant of three polar localized Aer‐like proteins of Pseudomonas putida . Environ Microbiol 10: 1320–1334. [DOI] [PubMed] [Google Scholar]
- Sasse, J. , Martinoia, E. , and Northen, T. (2018) Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci 23: 25–41. [DOI] [PubMed] [Google Scholar]
- Schweinitzer, T. , and Josenhans, C. (2010) Bacterial energy taxis: a global strategy? Arch Microbiol 192: 507–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, J.C. , Greenhut, I.V. , and Leveau, J.H. (2013) Functional characterization of the bacterial iac genes for degradation of the plant hormone indole‐3‐acetic acid. J Chem Ecol 39: 942–951. [DOI] [PubMed] [Google Scholar]
- Silver, N. , Best, S. , Jiang, J. , and Thein, S.L. (2006) Selection of housekeeping genes for gene expression studies in human reticulocytes using real‐time PCR. BMC Mol Biol 7: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith‐Becker, J. , Marois, E. , Huguet, E.J. , Midland, S.L. , Sims, J.J. , and Keen, N.T. (1998) Accumulation of salicylic acid and 4‐hydroxybenzoic acid in phloem fluids of cucumber during systemic acquired resistance is preceded by a transient increase in phenylalanine ammonia‐lyase activity in petioles and stems. Plant Physiol 116: 231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soby, S. , Kirkpatrick, B. , and Kosuge, T. (1991) Chemotaxis of Pseudomonas syringae subsp. savastanoi virulence mutants. Appl Environ Microbiol 57: 2918–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik, V. , and Berg, H.C. (2002) Receptor sensitivity in bacterial chemotaxis. Proc Natl Acad Sci U S A 99: 123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaepen, S. , Das, F. , Luyten, E. , Michiels, J. , and Vanderleyden, J. (2009) Indole‐3‐acetic acid‐regulated genes in Rhizobium etli CNPAF512. FEMS Microbiol Lett 291: 195–200. [DOI] [PubMed] [Google Scholar]
- Spaepen, S. , and Vanderleyden, J. (2011) Auxin and plant‐microbe interactions. Cold Spring Harb Perspect Biol 3: a001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunchai, M. , Hida, A. , Oku, S. , Tajima, T. , and Kato, J. (2021) Chemotactic disruption as a method to control bacterial wilt caused by Ralstonia pseudosolanacearum . Biosci Biotechnol Biochem 85: 697–702. [DOI] [PubMed] [Google Scholar]
- Upadhyay, A.A. , Fleetwood, A.D. , Adebali, O. , Finn, R.D. , and Zhulin, I.B. (2016) Cache domains that are homologous to, but different from PAS domains comprise the largest superfamily of extracellular sensors in prokaryotes. PLoS Comput Biol 12: e1004862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetting, M.W. , Al‐Obaidi, N. , Zhao, S. , San Francisco, B. , Kim, J. , Wichelecki, D.J. , et al. (2015) Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes. Biochemistry 54: 909–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives‐Peris, V. , de Ollas, C. , Gómez‐Cadenas, A. , and Pérez‐Clemente, R.M. (2020) Root exudates: from plant to rhizosphere and beyond. Plant Cell Rep 39: 3–17. [DOI] [PubMed] [Google Scholar]
- Wu, H. , Kato, J. , Kuroda, A. , Ikeda, T. , Takiguchi, N. , and Ohtake, H. (2000) Identification and characterization of two chemotactic transducers for inorganic phosphate in Pseudomonas aeruginosa . J Bacteriol 182: 3400–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Q. , Pande, G.S.J. , Wang, Z. , Lin, B. , Rubin, R.A. , Vora, G.J. , and Defoirdt, T. (2017) Indole signalling and (micro)algal auxins decrease the virulence of Vibrio campbellii, a major pathogen of aquatic organisms. Environ Microbiol 19: 1987–2004. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Li, S. , Liu, X. , Wang, Z. , Jiang, M. , Wang, R. , et al. (2020) Sensing of autoinducer‐2 by functionally distinct receptors in prokaryotes. Nat Commun 11: 5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Gardina, P.J. , Kuebler, A.S. , Kang, H.S. , Christopher, J.A. , and Manson, M.D. (1999) Model of maltose‐binding protein/chemoreceptor complex supports intrasubunit signaling mechanism. Proc Natl Acad Sci U S A 96: 939–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. (2018) Essential roles of local auxin biosynthesis in plant development and in adaptation to environmental changes. Annu Rev Plant Biol 69: 417–435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting Information.


