Abstract
Prognosis and treatment options of hepatitis B virus‐related hepatocellular carcinoma (HBV‐HCC) are generally based on tumor burden and liver function. Yet, tumor growth and therapeutic resistance of HBV‐HCC are strongly influenced by intratumoral hypoxia and cells infiltrating the tumor microenvironment (TME). We, therefore, studied whether linking parameters associated with hypoxia and TME cells could have a better prediction of prognosis and therapeutic responses. Quantification of 109 hypoxia‐related genes and 64 TME cells was performed in 452 HBV‐HCC tumors. Prognostic hypoxia and TME cells signatures were determined based on Cox regression and meta‐analysis for generating the Hypoxia‐TME classifier. Thereafter, the prognosis, tumor, and immune characteristics as well as the benefit of therapies in Hypoxia‐TME defined subgroups were analyzed. Patients in the Hypoxialow/TMEhigh subgroup showed a better prognosis and therapeutic responses than any other subgroups, which can be well elucidated based on the differences in terms of immune‐related molecules, tumor somatic mutations, and cancer cellular signaling pathways. Notably, our analysis furthermore demonstrated the synergistic influence of hypoxia and TME on tumor metabolism and proliferation. Besides, the classifier allowed a further subdivision of patients with early‐ and late‐HCC stages. In addition, the Hypoxia‐TME classifier was validated in another independent HBV‐HCC cohort (n = 144) and several pan‐cancer cohorts. Overall, the Hypoxia‐TME classifier showed a pretreatment predictive value for prognosis and therapeutic responses, which might provide new directions for strategizing patients with optimal therapies.
Keywords: HBV‐related hepatocellular carcinoma, hypoxia, prognosis, therapeutic responses, tumor microenvironment
What's new?
Intratumoral hypoxia is associated with poor prognosis in hepatitis B virus (HBV)‐related hepatocellular carcinoma (HCC). Hence, an integrated hypoxia‐tumor microenvironment (TME) signature may benefit clinical classification of HBV‐related HCC. Here, the authors developed a hypoxia‐TME classifier based on comprehensive analysis of TME cell status and hypoxia‐related genes in HBV‐related HCC. Prognostic prediction of the classifier was confirmed in various HCC cohorts. Best prognosis and therapeutic responses were linked to low hypoxia/high TME scores. The findings highlight the significance of evaluating tumor‐specific biology based on hypoxia and TME cell signatures, contributing to new opportunities in prognostic and therapeutic assessment in HCC.
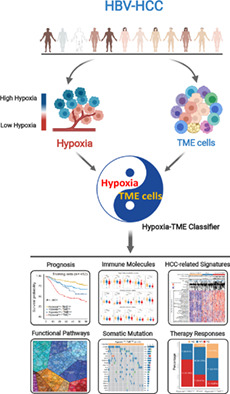
Abbreviations
- BCLC
Barcelona Clinic Liver Cancer
- DCs
dendritic cells
- FAO
fatty acid oxidation
- GSVA
Gene Set Variation Analysis
- HBV
hepatitis B virus
- HBV‐HCC
HBV‐related HCC
- HCC
hepatocellular carcinoma
- HIF‐1
hypoxia‐inducible factor 1
- ICI
immune checkpoint inhibitor
- KEGG
Kyoto Encyclopedia Genes and Genomes
- MDSCs
myeloid‐derived suppressor cells
- ssGSEA
single‐sample gene set enrichment analysis
- TMB
tumor mutational burden
- TME
tumor microenvironment
- Tregs
regulatory T cells
1. INTRODUCTION
Hepatocellular carcinoma (HCC) is the fourth leading cause of cancer death in the world, which also accounts for more than 85% of all primary liver malignancies. 1 Despite national vaccine programs and antiviral therapies, 2 hepatitis B virus (HBV) infection estimated to affect 292 million people globally, remains one of the largest causes for HCC. The prognosis of HCC remains poor with a 5‐year survival rate of approximately 18%. 3 The development of cancer immunotherapy provided a paradigm shift in the treatment of cancer, however, only a minority of patients benefit from it. 4 Thus, significant efforts are now being devoted to identifying prognostic and therapeutic biomarkers. Currently, the prognosis of resected HCC is evaluated mainly based on the clinical classification 5 and staging systems, 6 including tumor number/size, lymph nodes, metastases and liver function. Nonetheless, owing to the high tumor heterogeneity, 7 patients exhibit various clinical outcomes, even though presenting very similar clinical characteristics. One of the major proposed elucidation is that HCC is a quite heterogeneous tumor with different oncogenic pathways. 8 The above indicates that the current biomarkers explored are not satisfactory enough to accurately predict prognosis and strategize treatment options. 9
HBV plays a critical role in reprogramming the liver tumor microenvironment (TME), including the population of immune and nonimmune cells. 10 , 11 Given the presence of various cells within the TME proved essential for antitumor immune response, studying cell composition could bring not only prognostic information but also clues about the putative efficacy of immunotherapies. 12 , 13 However, previous studies were limited to a subset of 22 types of immune cells. 14 , 15 , 16 Beyond immune cells, also many nonimmune cells such as stromal cells in the TME interact with cancer cells, directly or indirectly promoting or inhibiting tumor growth and spread. 17 Consequently, we reasoned that quantification of the whole TME cellular landscape might provide a better metric for predicting prognosis and therapeutic responses.
Moreover, HCC is characterized as one of the most aggressive malignancies, giving rise to intratumoral hypoxia as a result of relatively poor blood supply coupled with rapid tumor growth. Hypoxia as an unfavorable determinant for angiogenesis, metastasis and altered metabolism in HCC, overexpression of hypoxia‐inducible factor 1 (HIF‐1) is strongly associated with poor prognosis. 18 Because hypoxia can facilitate the accumulation of poorly differentiated tumor cells. 19 Beyond the influence on tumor cells, hypoxia indeed enables several events in TME and affects many TME surrounding cells which play a crucial role in the tumorigenic process, thereby facilitating tumor aggressiveness and inhibition of antitumor responses. 20 Such as cells in a hypoxic state tend to switch to glycolytic metabolism, increasing the level of lactate which will in turn reinforce the acidification of the immune‐suppressive TME. 21 In addition, the hypoxic microenvironment will further facilitate protumoral immune cells infiltration markedly, such as M2 macrophages, 22 myeloid‐derived suppressor cells (MDSCs), 23 regulatory T cells (Tregs), 24 , 25 the three main immunosuppressive cells in TME. Furthermore, hypoxia also inhibits T cells‐mediated immune response strongly and drives immune escape in the TME. 26 , 27 Briefly, a strong cross‐interaction exists between the TME cells and intratumoral hypoxia.
To our knowledge, no study has been described concentrating on the HBV‐HCC tumor microenvironment based on combining hypoxia and comprehensive cellular landscape. Taking into account the effect of hypoxia modifiers 28 and immunotherapy in HCC, 29 an integrated Hypoxia‐TME signature might benefit the patients both for clinical classification and strategizing therapies. Therefore, in the present study, we aimed to systematically establish a Hypoxia‐TME classifier for prognostic and therapeutic responses prediction by incorporating hypoxia and tumor microenvironment cells. Our findings might contribute to a better understanding of tumor‐specific biology based on an integrated manner of hypoxic TME, which has important implications in clinical disease management.
2. METHODS
2.1. Data source
Gene datasets of HCC tumors were collected from five different public databases. Only patients with complete clinical information and positive HBV infection (n = 596) were enrolled for the present study in the end. The study included four cohorts with 452 HBV‐HCC tumors (GSE14520, GSE10143, ICGC‐LIRI‐JP, and Gao et al 30 ) and another independent validation cohort with 144 HBV‐HCC tumors from TCGA‐LIHC. In addition, single‐cell transcriptome data with cell annotations of 9 HCC tumors were collected from GSE125449 to visualize Hypoxia and TME scores in each cell. Besides, we also enrolled three HCV‐HCC cohorts, as well as 32 pan‐cancer cohorts to validate the general applicability of the Hypoxia‐TME classifier externally. Furthermore, to test the predictive value of therapeutic responses based on the classifier, a set of 67 HCC patients with Sorafenib treatment (GSE109211) and a set of 65 melanoma patients with MAGE‐A3 immunotherapy (GSE35640) from the Gene Expression Omnibus (GEO) databases were recruited. A detailed summary of these cohorts is summarized in Supplementary Table S1. For microarray data, the samples were downloaded and log‐transformed. As to the datasets in TCGA‐LIHC and ICGC‐LIRI‐JP, RNA sequencing data (FPKM value) were collected. Gene data from 32 pan‐cancer cohorts were downloaded from https://xenabrowser.net. Gene levels with detailed clinical information of the Gao et al cohort were collected directly from the supplementary files of the published paper. 30
2.2. Hypoxia‐related genes and TME cells quantification
A compendium of 109 hypoxia‐related genes constituting the HIF‐1 signaling pathway was obtained from the KEGG database (pathway “hsa04066”). Expression of all hypoxia‐related genes was extracted from all the above cohorts. For TME cells, we used a novel gene signature‐based method—xCell that integrates the advantages of gene set enrichment with deconvolution approaches. 31 This method allows the calculation of 64 TME cell types including immune and nonimmune cells based on transcriptomes of all collected tumor samples, and it outperforms other methods. 32 The enrichment scores calculated by xCell were utilized to represent the abundance of each TME cell type in each tumor sample of all cohorts.
2.3. Identification of overall prognostic hypoxia‐related genes and TME cells
The prognostic evaluation of hypoxia‐related genes and TME cells was determined by Cox proportional hazards regression analysis in each of the four different cohorts (Tables S2 and S3). To avoid bias across different source‐derived cohorts, we first evaluated each cohort independently and then integrated them using a fixed‐effects model by meta‐analysis for an overall prognostic value of hypoxia‐related genes and TME cells. The integrated analyses of multiple cohorts allow buffering of the bias and variability which often come from a single cohort. The overall prognostic value of hypoxia‐related genes and TME cells was described by hazard ratio (HR) and their 95% CI (confidence interval). In the end, 48 hypoxia‐related genes (Table S4) and 19 TME cells (Table S5) were identified to be statistically significantly associated with prognostic outcome in HBV‐related HCC (Cox regression, P < .05, and FDR < 0.05). For each prognostic factor, its hazard ratio (HR) and the standard estimates (SE) of HR in the meta‐analysis model were also calculated.
2.4. Establishment of Hypoxia score, TME score and Hypoxia‐TME classifier
The development of the Hypoxia score and TME score was based on the corresponding HR values with SE of 48 hypoxia‐related genes and 19 TME cells, respectively, which is an extension of a previous study. 33 The HR and corresponding SE of HR for each of the 48 hypoxia‐related genes decided their weight in the Hypoxia score. Simplistically, the Hypoxia score was given by
Similarly, the TME score was given by
where Gi or Cj are the abundances of gene or TME cell in each tumor sample. The normalized Z‐score for Hypoxia score and TME score was used for further analysis. Then Hypoxia and TME scores were integrated for the development of the Hypoxia‐TME classifier (Table S8). Thereafter, tumors were further divided into the following subgroups: Hypoxialow/TMEhigh, intermediate mixed (Hypoxialow/TMElow and Hypoxiahigh/TMEhigh) and Hypoxiahigh/TMElow based on the mean value of Hypoxia score and TME score in each cohort.
2.5. Single sample gene set enrichment analysis (ssGSEA) of other HCC‐related molecular classification signatures
To compare the Hypoxia‐TME classifier with other HCC‐related molecular classification gene signatures, the enrichment scores of 87 gene sets signatures that associated with HCC status collected in the Molecular Signatures Database (MSigDB) 34 (Table S14) were analyzed by the “GSVA” package in the R program. Next, a complex heatmap of these HCC‐related signatures was built in different Hypoxia‐TME subgroups to compare the consistency between the Hypoxia‐TME classifier and the 87 HCC‐related molecular classification signatures.
2.6. Tumor somatic mutation, functional annotation and KEGG pathway analysis
Somatic mutation data of TCGA‐LIHC and Gao et al cohorts were available in the TCGA‐LIHC database and the supplementary materials of the previous study, 30 respectively. The top 20 mutation genes were obtained and then compared between Hypoxia‐TME subgroups. Oncoprints for these genes were built by the R package “ComplexHeatmap.” Candidate genes with significant differences among Hypoxia‐TME subgroups were then extracted for further prognosis analysis. The tumor mutational burden (TMB) score of each tumor was also calculated by previously described methods. 35 Differentially expressed genes (DEGs) analysis was performed by the “limma” package in R. Besides, Proteomaps were developed by a web tool (https://bionic-vis.biologie.uni-greifswald.de/) based on the DEGs values. 36 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses for all cohorts were performed by the “clusterProfiler” package in R.
2.7. Statistical analysis
All statistical analysis was performed in R 3.6.2. Standard tests included the Student's t‐test, Wilcoxon rank‐sum test, and Fisher's exact test. The relationship between the Hypoxia score/TME score and other continuous variables was calculated by the Spearman method. The log‐rank test and Cox proportional hazard regression were used to explore related independent predictors of patients' prognosis. All reported P‐values were two‐sided with a significance level of .05.
3. RESULTS
3.1. A meta‐analysis of four different HBV‐HCC cohorts uncovering the prognostic value of Hypoxia and TME
To develop a method indicative of tumor hypoxic and TME cells status, a total of 596 HBV‐HCC tumors, including four cohorts (GSE14520, ICGC‐LIRI‐JP, GSE 10143, Gao et al, n = 452) and an independent validation cohort (TCGA‐LIHC, n = 144) were enrolled. The schematic diagram of the whole study is shown in Figure 1. For all patients, prognostic values of 109 hypoxia‐related genes and 64 TME cell types were analyzed in each of the four different cohorts independently (Tables S2 and S3). Then, a meta‐analysis as described in the methods section, was used to integrate the overall prognostic evaluation for each hypoxia‐related gene and TME cell. In the end, 48 hypoxia‐related genes and 19 TME cells were identified as independent prognostic factors (Cox proportional hazard model, P < .05, FDR < 0.05) based on 452 HBV‐HCC tumor samples (Tables S4 and S5). These prognostic genes/TME cells and their corresponding subpopulations (grouped based on their gene function or cell type) are shown in a network (Figure S1). Among them, most of the selected hypoxia‐related genes (31/48) were prognostically unfavorable. Of note, all the genes whose functions are related to reducing oxygen consumption are unfavorable factors, including, for example, HK2, LDHA, PGK1, ALDOA and so forth (Figure S1, Table S4). For the selected TME cells, 12/19 were prognostically favorable cells, including CD8+ effector memory T cells, B cells, plasma cells, NK cells and so forth. The remaining seven prognostically unfavorable cells included γδ T cells (Tgd), basophils, mast cells and so forth (Figure S1, Table S5). The intrinsic relationship between these hypoxia‐related genes and TME cells in each cohort was visualized in Figure S2.
FIGURE 1.
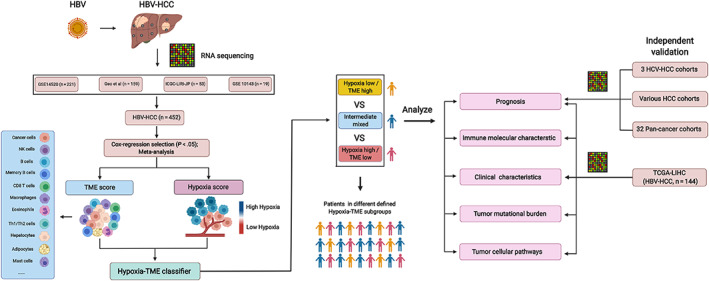
Schematic diagram depicting the establishment and validation of Hypoxia‐TME classifier. HCC patients with complete clinical information and positive HBV infection were enrolled from the four cohorts (GSE14520, ICGC‐LIHC‐JP, Gao et al, GSE 10143), which were used to establish the prognostic TME score and Hypoxia score, respectively. Cox regression analyses of 64 TME cells and 109 hypoxia‐related genes were performed in each of the four cohorts independently, and then meta‐analysis was used for the evaluation of the overall prognostic values of TME cells and genes. Briefly, 19 cell types within tumor microenvironment were selected for the development of TME score. As well, 48 hypoxia‐related genes were used for the establishment of Hypoxia score. The Hypoxia‐TME classifier which integrating the TME score and hypoxia score, divided all the patients into three different subgroups: Hypoxialow/TMEhigh, Hypoxiahigh/TMElow and the intermediate mixed subgroups. The difference in terms of prognosis, immune‐related molecules, clinical characteristics, tumor mutational burden and tumor cellular pathways were explored in different patients' subgroups based on the Hypoxia‐TME classifier. Furthermore, the classifier's performance was further validated in another independent HBV‐HCC cohort (TCGA‐LIHC), HCV‐HCC, various HCC cohorts and pan‐cancer patient cohorts, respectively. The figure was created with BioRender.com [Color figure can be viewed at wileyonlinelibrary.com]
We subsequently developed a Hypoxia score and TME score, respectively, based on the prognostic hypoxia and TME signatures by taking their contributions to risk prediction into account. According to the mean value of the Hypoxia score and TME score in each cohort, tumors were classified into two subgroups, respectively. Notably, the Hypoxia score was strongly positively correlated with prognostically unfavorable hypoxia‐related genes and negatively correlated with prognostically favorable genes (Figure 2A). While for the TME score, a strong positive correlation was observed between the TME score and prognostically favorable TME cells, and a negative correlation was found between the TME score and prognostically unfavorable TME cells (Figure 2B). Besides, it was observed that the patients with low Hypoxia score and high TME score showed a statistically longer survival compared to patients with a high Hypoxia score and low TME score (Figure 2C,D). And tumors with a high Hypoxia score were significantly enriched for the HIF‐1 pathway genes (Figures 2E and S3). Similarly, tumors with high TME scores were considerably enriched for the immune response pathway (Figures 2F and S4, Table S6). Notably, to further validate the Hypoxia and TME scores in the single‐cell transcriptomic landscape of liver cancer, we performed t‐distributed stochastic neighbor embedding (t‐SNE) analysis on single‐cell data of liver cancer biospecimens from nine patients (Figure 2G) and calculated Hypoxia and TME scores, respectively. It was observed that malignant cells and stromal cells exhibited higher Hypoxia scores than immune cells (Figure 2H,J). While immune cells such as monocytes/macrophages, CD8 T cells and plasma cells were found to have higher TME scores than any other stromal fibroblasts, malignant cells or endothelial cells (Figure 2I,K).
FIGURE 2.
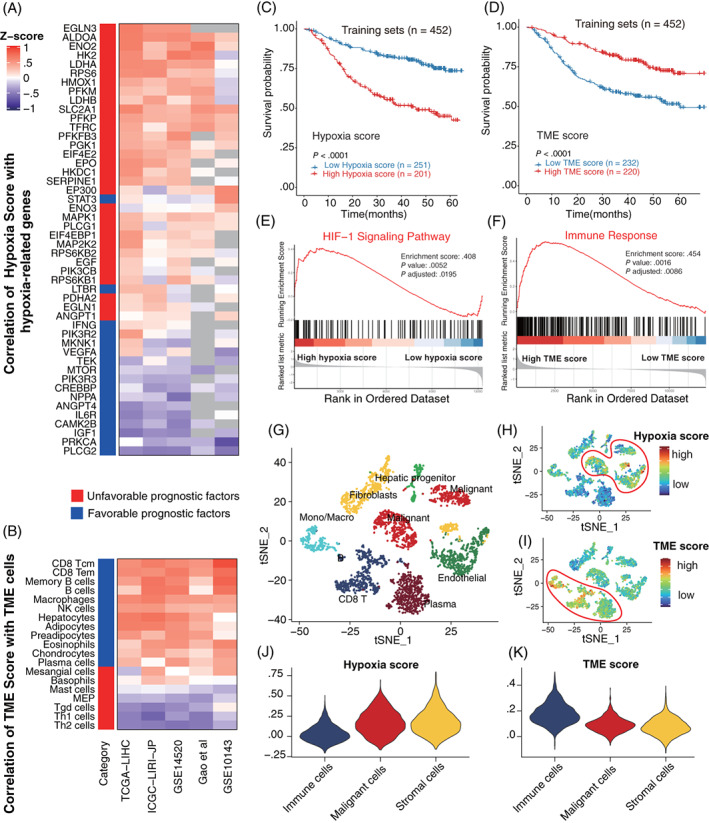
Development of the Hypoxia score and TME score based on hypoxia‐related genes and TME cells respectively. (A) Heat map showing the relationships between the Hypoxia score and 48 prognostic hypoxia‐related genes in five HBV‐HCC cohorts. Positive (red) and negative (purple) correlations are indicated. Gray means missing values in that cohort. (B) Heat map showing the relationships between the TME score and 19 prognostic TME cells in five HBV‐HCC cohorts. Positive (red) and negative (purple) correlations are indicated. (C) Kaplan‐Meier overall survival curves of four training cohorts in tumors with high Hypoxia score vs low Hypoxia score. (D) Kaplan‐Meier overall survival curves of four training cohorts in tumors with high TME score vs low TME score. (E) Gene Set Enrichment Analysis (GSEA) of 109 genes represent HIF‐1 signaling pathway reveals the association between Hypoxia score and HIF‐1 signaling pathway. High hypoxia score located in the left approaching the origin of the x‐axis, by contrast, low hypoxia score lay on the right of the x‐axis (GSE14520). (F) Gene Set Enrichment Analysis (GSEA) of 332 marker genes represent immune response display the association between TME score and immune response. High TME score located in the left approaching the origin of the x‐axis, by contrast, low TME score lay on the right of the x‐axis (GSE14520). (G) t‐SNE plot of all 3836 cells from 9 primary liver cancer patients. Cells were annotated based on known lineage‐specific marker genes as CD8 T cells, monocytes/macrophages, plasma cells, fibroblasts, malignant cells, endothelial cells, hepatic progenitor (GSE125449). (H) t‐SNE plot of all the single cells colored by Hypoxia score. (I) t‐SNE plot of all the single cells colored by TME score. (J) Comparisons of Hypoxia score among immune cells, malignant cells and stromal cells. (K) Comparisons of TME score among immune cells, malignant cells and stromal cells [Color figure can be viewed at wileyonlinelibrary.com]
3.2. Prognostic value of the established Hypoxia‐TME classifier
Based on the results above, we asked whether it would be possible to combine the Hypoxia score and TME score to characterize the hypoxic TME. Since a statistically significant negative correlation between Hypoxia score and TME score was not only found in most HBV‐HCC cohorts, but also in all the 32 pan‐cancer cohorts (Figure S5 and Table S7). Therefore, we combined the Hypoxia score with the TME score, and developed the Hypoxia‐TME classifier which resulted in dividing patients into four subgroups: Hypoxialow/TMEhigh, Hypoxialow/TMElow, Hypoxiahigh/TMEhigh, Hypoxiahigh/TMElow. The Hypoxia‐TME classifier showed a statistically different prognosis in HBV‐HCC cohorts (n = 452) (Figure 3A). It was found that both the Hypoxia score and TME score contribute significantly to the prognostic value. Patients in the Hypoxialow/TMEhigh subgroup were revealed to have the best prognosis compared to patients from the other three subgroups. The prognosis of patients in the Hypoxialow/TMElow and Hypoxiahigh/TMEhigh subgroups are less divergent, and the percentage of different disease stages in subgroups of Hypoxialow/TMElow and Hypoxiahigh/TMEhigh was very similar in the two largest cohorts (Figure S6). Thus, we combined these two subgroups as a mixed subgroup. Importantly, the performance of the Hypoxia‐TME classifier was further validated in another independent HBV‐HCC cohort (n = 144) (Figure 3B), HCV‐HCC cohort (n = 270) and various HCC patients (n = 1294) (Figure S7). Additionally, a correlation coefficient heatmap was generated to visualize a comprehensive internal and intergroup correlation among Hypoxia‐TME factors (hypoxia‐related genes and TME cells) based on the three subgroups, respectively. An internal positive relationship and an intergroup negative correlation were characterized in the Hypoxialow/TMEhigh subgroup and the Hypoxiahigh/TMElow subgroups (Figure 3C).
FIGURE 3.
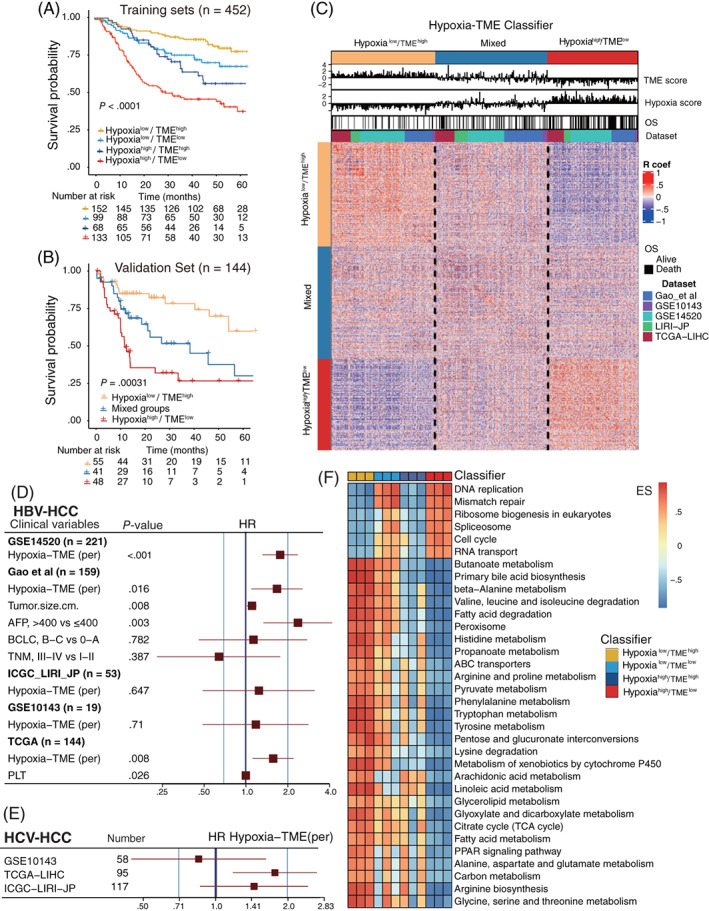
Prognostic value and tumor cellular signaling pathways analysis based on Hypoxia‐TME classifier. (A) Kaplan‐Meier overall survival curves of four HBV‐HCC training cohorts (n = 452) stratified into four different subgroups based on the Hypoxia‐TME classifier (Hypoxialow/TMEhigh, Hypoxialow/TMElow, Hypoxiahigh/TMEhigh and Hypoxiahigh/TMElow). Log‐rank test, P < .001. (B) Kaplan‐Meier overall survival curves of the independent validation HBV‐HCC cohort (n = 144) stratified into three different subgroups based upon the Hypoxia‐TME classifier (Hypoxialow/TMEhigh, mixed and Hypoxiahigh/TMElow). Log‐rank test, P < .001. (C) Heat map showing the correlations matrix among all Hypoxia‐TME signatures in three different subgroups based upon Hypoxia‐TME classifier. Positive (red) and negative (purple) correlations are indicated. (D) Multivariate cox analysis of the Hypoxia‐TME classifier in five HBV‐HCC cohorts. (E) Cox analysis of the Hypoxia‐TME classifier in three HCV‐HCC cohorts. (F) Compare tumor proliferative signaling pathways and tumor metabolism‐related pathways among four Hypoxia‐TME subgroups based on mRNA expression levels (GSE14520, Gao et al, TCGA cohorts): higher enrichment scores (red), lower enrichment scores (blue) [Color figure can be viewed at wileyonlinelibrary.com]
To portray a detailed prognostic value of the Hypoxia‐TME classifier, we performed the univariate and multivariate Cox regression analysis in all five HBV‐HCC cohorts and three HCV‐HCC cohorts, respectively. The Hypoxia‐TME classifier was significantly related to overall survival in training cohorts with enough patients and one validation HBV‐HCC cohort (Figure 3D, Table S9). Also, the classifier showed a statistically significant prognostic value in one out of three HCV‐HCC cohorts (Figure 3E). Likewise, the prognostic value of the Hypoxia‐TME classifier could be validated in 10/32 pan‐cancer cohorts (Figure S8, Table S10).
3.3. Differences in cellular signaling pathways among different Hypoxia‐TME subgroups
Considering the significant prognostic differences based on Hypoxia‐TME classifier, an intratumor cellular signaling pathways analysis was performed based on the Hypoxia‐TME subgroups in three cohorts with enough genes for pathway analysis (Figure 3F). The detailed enrichment scores of these pathways among Hypoxia‐TME subgroups are listed in Tables S11–S13. Tumors in the subgroups exhibited different patterns in terms of tumor proliferation and cancer metabolism related genes expression. Intriguing, it was observed that tumors in subgroups with low Hypoxia score (Hypoxialow/TMEhigh and Hypoxialow/TMElow) showed a much higher enrichment of amino acid metabolism, fatty acid metabolism, carbon metabolism and ATP‐binding cassette (ABC) transporters related genes expression than tumors with a high Hypoxia score (Hypoxiahigh/TMEhigh and Hypoxiahigh/TMElow). Furthermore, tumors in subgroups with a low TME score (Hypoxialow/TMElow and Hypoxiahigh/TMElow) displayed a much higher enrichment of DNA replication, mismatch repair, cell cycle and RNA transport related genes expression than tumors with a high TME score (Hypoxialow/TMEhigh and Hypoxiahigh/TMEhigh). Which suggest a stronger antitumor immune response and less tumor growth in patients with high TME scores. The above furthermore demonstrated the synergistic influence of hypoxia and TME on tumor metabolism and cancer cells proliferation, which implies the significance of an integrated analysis of hypoxic TME.
In addition, we constructed proteomaps to better visualize the cellular signaling pathway differences between Hypoxialow/TMEhigh and Hypoxiahigh/TMElow subgroups. Tumors in the Hypoxialow/TMEhigh subgroup showed higher enrichment of complement cascades 37 and PPAR‐related proteins, 38 all of which play essential roles in enhanced host immune response. While tumors in the Hypoxiahigh/TMElow subgroup showed higher enrichment of cell cycle, DNA replication and transcription factors (Figure S9). The above results further indicate the physiological significance of the intergated Hypoxia score (hypoxic status) and TME score (immune responses) in the tumor microenvironment, which might help to elucidate the prognostic differences between different subgroups based on tumor biology.
3.4. Association between Hypoxia‐TME classifier with clinical features and other HCC‐related molecular classification signatures
We next investigated clinical features and other HCC‐related signatures based on the Hypoxia‐TME classifier. Patients with advanced HCC and larger tumor size were found slightly more in the Hypoxiahigh/TMElow subgroup compared to other subgroups. Yet, AFP, multinodular and cirrhosis do not differ among different Hypoxia‐TME subgroups (Figure 4A, up panel). To further compare the Hypoxia‐TME classifier with other HCC‐related molecular classification signatures as documented previously, 34 we analyzed the previous HCC‐related molecular classification signatures as comprehensively as possible (Table S14). The heatmap revealed that the Hypoxia‐TME classifier was very consistent with 57 gene sets signatures from 24 different studies 34 , 39 in predicting the prognosis of HCC (Figure 4A, bottom panel, all P < .01). Patients in the Hypoxialow/TMEhigh subgroup showed low enrichment of unfavorable prognosis related signatures, while these signatures were highly enriched in patients within the Hypoxiahigh/TMElow subgroup. Similarly, favorable prognostic signatures were greatly enriched in the Hypoxialow/TMEhigh subgroup, while much less expressed in the Hypoxiahigh/TMElow subgroup. The mixed subgroups also showed a mixture pattern of these signatures. Besides, the Hypoxia‐TME classifier could significantly distinguish the overall survival of the HBV‐HCC patients (Figure 4B). Figure 4C shows that the Hypoxia‐TME classifier could predict overall survival at 1, 2, 3 and 5 years with a range of AUCs from 0.68 to 0.73. By comparison with the present clinical staging systems under the time‐dependent ROC for the incidence of overall survival, the Hypoxia‐TME classifier showed higher efficacy than BCLC staging classification and similar to the performance of TNM staging system (Figure 4D). Notably, our analysis furthermore demonstrated that the Hypoxia‐TME classifier allowed a further subdivision of patients both with early HCC stage (TNM I‐II) and late HCC stage (TNM III‐IV). The survival of patients in the late‐stage HCC subgroup with Hypoxialow/TMEhigh was significantly different from the late‐stage Hypoxiahigh/TMElow subgroup but similar to the survival of patients in the early‐stage HCC subgroup with Hypoxiahigh/TMElow (Figure 4E). Thus, the Hypoxia‐TME classifier in combination with the TNM classification seems to provide a more accurate predictive value than the TNM classifier alone.
FIGURE 4.
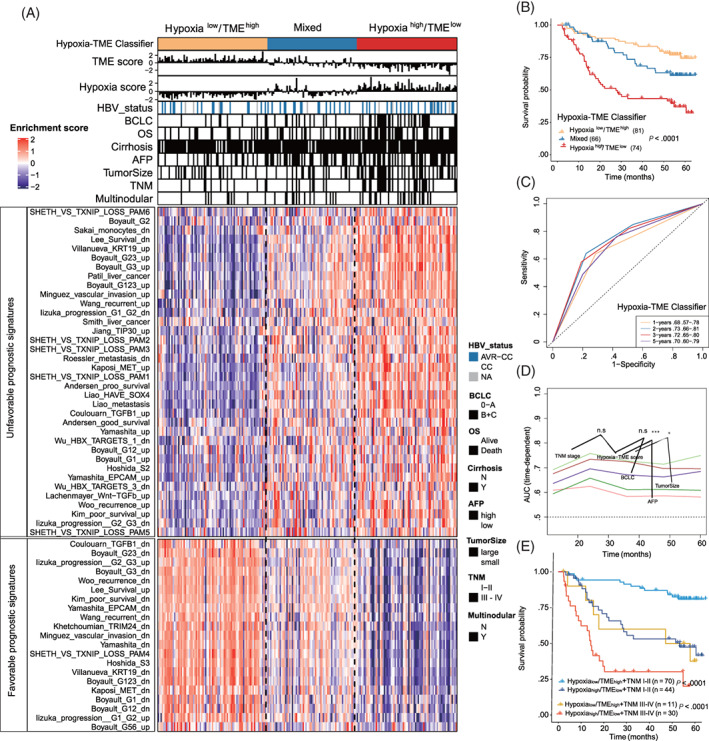
Association between Hypoxia‐TME classifier and other HCC‐related molecular signatures. (A) The association between Hypoxia‐TME classifier and clinical characteristics/other HCC‐related molecular signatures. AVR‐CC, active viral replication chronic carrier; CC, chronic carrier; AFP (high) means >400 ng/mL; AFP (low) means <400 ng/mL; tumor size (large) >5 cm, tumor size (small) <5 cm. Higher enrichment scores (red), lower enrichment scores (purple) (GSE14520). (B) Kaplan‐Meier overall survival curves of an HBV‐HCC cohort stratified into three different subgroups based on the Hypoxia‐TME classifier. Log‐rank test, P < .001. (C) ROC curves for the 1‐, 2‐, 3‐, 5‐year survival according to the Hypoxia‐TME classifier. (D) Comparisons of Hypoxia‐TME classifier with other clinical prognostic staging systems/biomarkers under the time‐dependent ROC for the incidence of overall survival. TNM staging system (light green), Hypoxia‐TME classifier (brown), BCLC staging system (purple), tumor size (dark green), AFP (brick‐red). (E) Kaplan‐Meier overall survival curves of HBV‐HCC patients, divided by the combination of Hypoxia‐TME classifier and TNM staging system. Log‐rank test, P < .001 [Color figure can be viewed at wileyonlinelibrary.com]
3.5. Distinct immune response profile in tumors among Hypoxia‐TME subgroups
We then further investigated the immune response associated genes among different subgroups from several aspects: major histocompatibility complex (MHC), inhibitory immune markers, activation immune markers, anti‐inflammatory markers, and pH regulation marker (Figure 5). It was noted that the HypoxialowTMEhigh subgroup in general had a higher expression of all MHC, most of the inhibitory immune marker (except TIM‐1) and all activation immune markers compared to the intermediate mixed and HypoxiahighTMElow subgroups. Besides, the HypoxialowTMEhigh subgroup demonstrated a lower expression of TGFB1(anti‐inflammatory gene) than the other two subgroups. While the HypoxiahighTMElow subgroup was observed to have significantly higher CA9 expression which are responsible for pH regulation. More detailed results about immune‐related genes are also depicted in a heatmap (Figure S10).
FIGURE 5.

Comparison of immune‐related markers in three subgroups based upon Hypoxia‐TME classifier. Comparison of the expression of immune‐related genes in defined three subgroups based on Hypoxia‐TME classifier. Box and whisker plots showing normalized expression of mRNA for selected markers. The Hypoxialow/TMEhigh, intermediate mixed and Hypoxiahigh/TMElow subgroups are represented as yellow, blue and red, respectively (GSE14520) [Color figure can be viewed at wileyonlinelibrary.com]
3.6. Differential patterns of tumor somatic mutations in patients among Hypoxia‐TME subgroups
We next investigated the tumor somatic alterations among different Hypoxia‐TME subgroups. The top 20 variant mutations in the TCGA‐LIHC cohort were identified (Figure 6A). Among the top 20 mutations, 12 out of 20 mutations showed a higher level in the Hypoxiahigh/TMElow subgroup compared to the Hypoxialow/TMEhigh subgroup. Especially, a TP53 mutation was significantly higher in the Hypoxiahigh/TMElow subgroup. A more pronounced result was further validated in another cohort (Figure S11). As well, the average lower expression of TP53 in the Hypoxialow/TMEhigh subgroup and improved survival in patients without TP53 mutation are consistent with previous studies (Figure 6C,D). 40 , 41 Notably, within the subgroup of patients with TP53 mutations, the Hypoxia‐TME classifier could identify patients with better prognosis (Figure 6E). Besides, as tumor mutational burden is an emerging therapeutic measurement of sensitivity to immunotherapy, we also calculated the TMB score of each tumor. No significant difference was found among Hypoxia‐TME subgroups (Figure 6B). These results might indicate that the Hypoxia‐TME classifier is more sensitive than TMB score to distinguish patients and it can more finely identify better prognosis in patients with P53 mutations.
FIGURE 6.
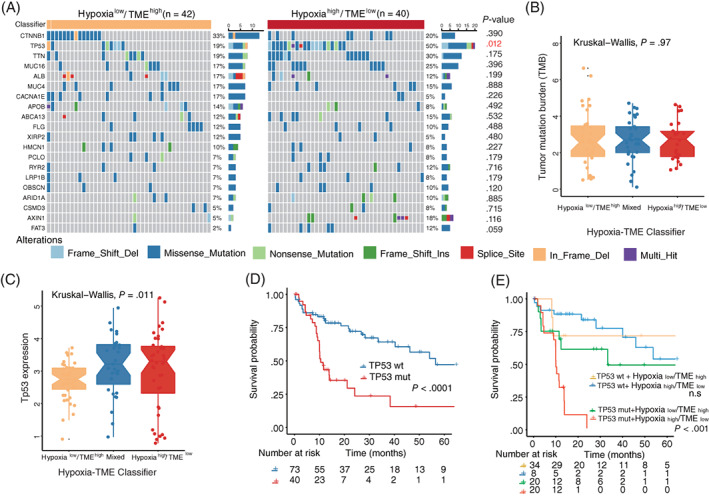
Association between tumor somatic mutations and Hypoxia‐TME classifier. (A) The OncoPrint was constructed by the top 20 mutation genes between Hypoxialow/TMEhigh and Hypoxiahigh/TMElow subgroups. Each liver tumor from an individual patient was represented in each column (TCGA‐LIHC). (B) Comparison of tumor mutational burden among defined subgroups based on Hypoxia‐TME classifier. (C) Comparison of TP53 expression among defined subgroups according to Hypoxia‐TME classifier. (D) Kaplan‐Meier overall survival curves of HBV‐HCC patients with or without TP53 gene mutation. (E) Kaplan‐Meier overall survival curves of HBV‐HCC patients divided by TP53 mutation status and Hypoxia‐TME classifier [Color figure can be viewed at wileyonlinelibrary.com]
3.7. Prediction of therapies responses based on Hypoxia‐TME classifier
We next tested whether the Hypoxia‐TME classifier could be used to predict clinical responses in patients undergoing therapies. Sequencing data from tumor samples (biopsies) was collected before sorafenib and MAGE‐A3 immunotherapy, respectively. Then we evaluated the predictive ability of the Hypoxia‐TME classifier in therapeutic responses, in patients treated with the multityrosine kinase inhibitor (mTKI) sorafenib, which exhibits both antiproliferative and antiangiogenic activity. 8 Patients in sorafenib responder group showed significantly lower Hypoxia score and higher TME score (Figure 7A,B). Compared to patients with Hypoxiahigh/TMElow, patients with Hypoxialow/TMEhigh had a much higher percentage (59.3%) of sorafenib responders, while only 13.6% and 11.1% sorafenib responders in the mixed and Hypoxiahigh/TMElow subgroups, respectively (Figure 7C). Similarly, patients with metastatic melanoma responding to MAGE‐A3 immunotherapy showed statistically lower Hypoxia score and higher TME score, respectively (Figure 7D,E). The highest percentage (63.16%) of patients with a therapeutic response were found in the Hypoxialow/TMEhigh subgroup, while the Hypoxiahigh/TMElow subgroup was only 14.29% (Figure 7F). Additionally, the Proteomap was used to intuitively reveal the potential mechanism of Hypoxia‐TME classifier predicting therapy responses in patients undergoing MAGE‐A3 immunotherapy. Interestingly, the pattern of Proteomap in the Hypoxialow/TMEhigh and MAGE‐A3 responder are quite similar. And high similarity between Hypoxiahigh/TMElow subgroup and those MAGE‐A3 nonresponder was also observed (Figure 7G,H). To summarize, these results might suggest that the pretreatment Hypoxia‐TME signature can depict the tumor immune microenvironment thus benefit the prediction of patient's therapy responses.
FIGURE 7.
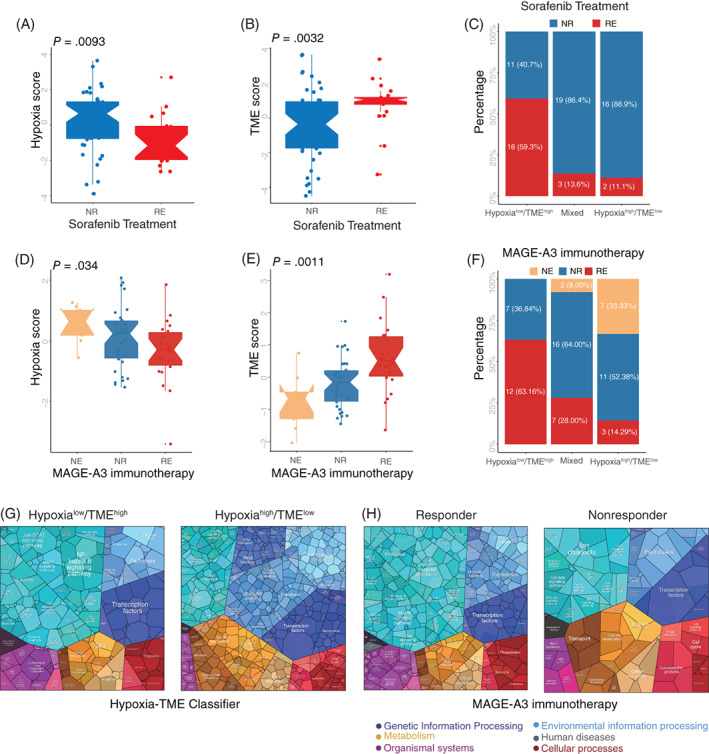
Therapy responses prediction based on Hypoxia‐TME classifier. (A) Comparison of Hypoxia scores between sorafenib responder and nonresponder. NR (nonresponder), RE (responder). (B) Comparison of TME scores between sorafenib responder and nonresponder. (C) The different percentages of sorafenib responder among subgroups based on Hypoxia‐TME classifier. (D) Comparison of Hypoxia scores among patients with different MAGE‐A3 immunotherapy response status. NE (not evaluable), NR (nonresponder), RE (responder). (E) Comparison of TME scores among patients with different MAGE‐A3 immunotherapy response status. (F) The different percentage of MAGE‐A3 immunotherapy responder among subgroups based on Hypoxia‐TME classifier. (G) Functional analysis in Hypoxialow/TMEhigh (left) and Hypoxiahigh/TMElow (right) of patients under MAGE‐A3 immunotherapy illustrated using Proteomaps. Each small polygon corresponds to a single KEGG pathway, and the size correlates with the ratio between the subgroups. (H) Functional analysis in responder (left) and nonresponder (right) of patients under MAGE‐A3 immunotherapy illustrated using Proteomaps. Each small polygon corresponds to a single KEGG pathway, and the size correlates with the ratio between the subgroups [Color figure can be viewed at wileyonlinelibrary.com]
4. DISCUSSION
The explosion of research on hypoxia and TME strengthens our understanding of the importance of a hypoxic TME in prognosis and therapies of cancer patients. 20 , 42 However, few studies integrated hypoxia and TME signatures for prediction of prognosis and therapy responses. When considering targeting hypoxia 18 , 43 combined with immunotherapy for the treatments of HCC, 29 , 44 signatures based on the combination of hypoxia and TME might enable both clinical classification and optimizing therapy strategies. In the present study, we systematically utilized large‐scale HBV‐HCC cohorts to assess the integrated value of hypoxic TME for prognostic and therapeutic prediction based on the Hypoxia‐TME classifier.
Patients in the Hypoxialow/TMEhigh subgroup showed the best prognosis and clinical responses to treatments. Besides, the prognostic prediction ability of the classifier was further confirmed in an independent HBV‐HCC cohort, an HCV‐HCC cohort, various HCC cohorts (including nonviral HCC) and 10/32 pan‐cancer cohorts, indicating its broad applicability in cancer patients. This might suggest certain common features existing in host antitumor immune response within the hypoxic TME.
Interestingly, within the selected 48 hypoxia‐related genes, 12 genes cataloged into a functional group related to reduction of oxygen consumption were all unfavorable prognostic factors. This could be an adaptive reaction of tumor exposed to a reduced‐oxygen environment. When total oxygen is limited, HIF‐1‐dependent blocking of oxygen utilization causes a drop in mitochondrial oxygen consumption and results in a relative increase in intracellular oxygen tension in human carcinoma cells. 45 This, in turn can decrease tumor cells death and favors tumor survival, explains the 12 genes related to oxygen consumption reduction as poor prognostic factors. Of the 19 TME cell types, γδ T cells, Th1 and Th2 cells play an unfavorable role in the HBV‐HCC cohorts studied. Notably, prognosis based on γδ T cells seems very divergent among different cancer types. 46 Yet, in line with our results, Kong et al demonstrated that γδT cells enhance MDSCs infiltration in liver, leading to MDSC‐mediated CD8+ T cell exhaustion. While furthermore, γδT cell deficiency led to a break in HBV‐induced tolerance and subsequent recovery of hepatic HBV‐specific CD8+ T cells. 47 This could explain the unfavorable prognostic role of γδT cells in HBV‐HCC. Moreover, the levels of Th1 cell densities in HCC tissues predicted poor survival, 48 and Th2 cells are also related to HCC tumor growth or metastasis. 49 As for myeloid cells, mast cells were determined an unfavorable prognostic factor that accumulation of tumor‐infiltrating mast cells predicts poor survival in several cancers including liver cancer. 50 Apart from lymphocytes and myeloid cells, some nonimmune cells within the tumor were also selected as prognostic factors, confirming our initial hypothesis that nonimmune cells are also associated with the prognosis of cancer patients. 51
Patients with high Hypoxia score showed lower metabolism genes enrichment, demonstrating the phenomenon of “hypoxic hypometabolism” in the tumor microenvironment of HCC, which was defined as a drop in metabolic rate during hypoxia. 52 Additionally, the negative correlation between Hypoxia score and TME score in different HBV‐HCC and 32 pan‐cancer cohorts might indicate hypoxia can inhibit the antitumor immune response. 53 These results all suggested the tumor biological significance for establishing a Hypoxia‐TME classifier. Besides, active DNA replication and mismatch repair together with the downregulated fatty acid oxidation (FAO) in the Hypoxiahigh/TMElow subgroup is also in agreement with recent studies showing that FAO is downregulated in multiple tumor types and activation of FAO may inhibit cancer cell proliferation. 54 The above results might elucidate the mechanism behind Hypoxia‐TME classifier predicting prognosis and therapy responses.
In addition, several other HCC‐related molecular classification signatures could be classified into clearly distinguishable subgroups according to the Hypoxia‐TME classifier. Which suggests the Hypoxia‐TME classifier correlates well with other HCC‐related molecular classification signatures. It may be explained by the existence of an overlapped or associated cellular pathways or genes between the hypoxia‐immune signatures and other HCC‐related signatures. Furthermore, an intriguing result showed that both activating and inhibitory immune markers were, in general, highly expressed in the Hypoxialow/TMEhigh subgroup, suggesting that a stronger antitumor immune response would likely be restored through immune checkpoint blockade in the Hypoxialow/TMEhigh subgroup. This imply the Hypoxia‐TME classifier could be applied for preimmunotherapy stratification of cancer patients. Additionally, statistically higher CA9 expression in Hypoxiahigh/TMElow subgroup indicates an acidic extracellular milieu favoring tumor growth, poor tumor differentiation and development, 55 , 56 which further exhibits the predictive ability of this classifier.
Apart from the above results, the observed different patterns between subgroups in terms of tumor somatic genome alterations revealed that the mRNA‐based Hypoxia‐TME classifier also reflects DNA molecular heterogeneity in tumor. Our results further support the observations that CCTNB1 mutation is associated with favorable prognosis and low‐stage HCC 57 and TP53 mutation is an indicator for poor prognosis. 40 Sorafenib treatment showed a significant recurrence‐free survival improvement in responders than nonresponders, although sorafenib responders status was predicted due to the original clinical trial limitation. 58 In the present research, more sorafenib responder patients existed in Hypoxialow/TMEhigh subgroup partially showed the classifier is predictive for therapeutic responses and better survival. Due to the unavailability of large‐scale HCC sequencing data with clinical information, immunotherapy and prognosis of patients. Also, in order to investigate the classifier's applicability in a different cancer setting, melanoma patients treated with MAGE‐A3 immunotherapy were enrolled. An obvious higher percentage (63.16%) of MAGE‐A3 immunotherapy responders within the Hypoxialow/TMEhigh subgroup validated its ability to predict therapeutic effect. The similarity of proteomap patterns between Hypoxialow/TMEhigh and MAGE‐A3 immunotherapy‐responder subgroups might reveal certain commonality in a determinative interplay between patients' immune system and cancer cells, which further indicated the therapeutic predictive value of Hypoxia‐TME classifier.
We acknowledge certain limitations to our study. First, the Hypoxia‐TME signatures which we based on gene expression requires more validation in accordance with immunofluorescence or flow cytometry of tumor samples (biopsies). Second, due to the limitation of public datasets, an in‐house cohort should be performed to further evaluate the classifier's performance. To conclude, portraying the integrated hypoxia and cellular landscape signatures within the tumor microenvironment, benefits the prediction of the prognosis and therapy responses. It might be a potential method for prognosis estimation and stratification of patients for clinical disease management in the future.
AUTHOR CONTRIBUTIONS
The work reported in the paper has been performed by the authors, unless clearly specified in the text. Conception and design: Shipeng Chen, Yuzhen Gao, Toos Daemen; Development of methodology: Yuzhen Gao, Shipeng Chen, Ying Wang; Acquisition of data: Shipeng Chen, Yuzhen Gao, Ying Wang; Analysis: Yuzhen Gao, Shipeng Chen; Interpretation of results: Shipeng Chen, Yuzhen Gao, Toos Daemen; Writing, revision and review of the manuscript: Shipeng Chen, Yuzhen Gao, Ying Wang, Toos Daemen; Study supervision: Toos Daemen.
CONFLICT OF INTEREST
The authors declare no potential conflicts of interests.
ETHICS STATEMENT
All the patients' data involved in our study was collected from public research databases. The application of the public data is already properly anonymized and informed consent was also obtained at the time of the original data collection.
Supporting information
Supplementary Figures Supporting information
Supplementary Tables Supporting information
ACKNOWLEDGMENTS
The authors acknowledge the China Scholarship Council (CSC) and Graduate School of Medical Sciences (GSMS) of the University of Groningen for a scholarship for S. Chen.
Chen S, Gao Y, Wang Y, Daemen T. The combined signatures of hypoxia and cellular landscape provides a prognostic and therapeutic biomarker in hepatitis B virus‐related hepatocellular carcinoma. Int J Cancer. 2022;151(5):809‐824. doi: 10.1002/ijc.34045
Shipeng Chen and Yuzhen Gao contributed equally to this work.
Funding information Graduate School of Medical Sciences (GSMS) of the University of Groningen; China Scholarship Council
DATA AVAILABILITY STATEMENT
Only publicly available data were used in our study, and data sources and handling of these data are described in the Materials and Methods and in the Supplementary Tables. Further information is available from the corresponding author upon request.
REFERENCES
- 1. Villanueva A. Hepatocellular Carcinoma. N Engl J Med. 2019;380:1450‐1462. [DOI] [PubMed] [Google Scholar]
- 2. Polaris Observatory Collaborators . Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol Hepatol. 2018;3:383‐403. [DOI] [PubMed] [Google Scholar]
- 3. Jemal A, Ward EM, Johnson CJ, et al. Annual report to the nation on the status of cancer, 1975–2014, featuring survival. J Natl Cancer Inst. 2017;109:djx030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pinter M, Scheiner B, Peck‐Radosavljevic M. Immunotherapy for advanced hepatocellular carcinoma: a focus on special subgroups. Gut. 2021;70:204‐214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Satala CB, Jung I, Kobori L, et al. Benefits of the 8th American Joint Committee on Cancer System for Hepatocellular Carcinoma Staging. J Gastrointest Cancer. 2021;52:243‐248. [DOI] [PubMed] [Google Scholar]
- 6. Llovet JM, Brú C, Bruix J. Prognosis of hepatocellular carcinoma: the BCLC staging classification. Semin Liver Dis. 1999;19:329‐337. [DOI] [PubMed] [Google Scholar]
- 7. Hung MH, Wang XW. Molecular Alterations and Heterogeneity in Hepatocellular Carcinoma. Cham: Humana Press; 2019:293‐316. [PubMed] [Google Scholar]
- 8. Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15:599‐616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pinto Marques H, Gomes da Silva S, De Martin E, Agopian VG, Martins PN. Emerging biomarkers in HCC patients: current status. Int J Surg. 2020;82:70‐76. [DOI] [PubMed] [Google Scholar]
- 10. Yang P, Markowitz GJ, Wang XF. The hepatitis B virus‐associated tumor microenvironment in hepatocellular carcinoma. Natl Sci Rev. 2014;1:396‐412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lim CJ, Lee YH, Pan L, et al. Multidimensional analyses reveal distinct immune microenvironment in hepatitis B virus‐related hepatocellular carcinoma. Gut. 2019;68:916‐927. [DOI] [PubMed] [Google Scholar]
- 12. Kurebayashi Y, Ojima H, Tsujikawa H, et al. Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification. Hepatology. 2018;68:1025‐1041. [DOI] [PubMed] [Google Scholar]
- 13. Miao YR, Zhang Q, Lei Q, et al. ImmuCellAI: a unique method for comprehensive T‐cell subsets abundance prediction and its application in cancer immunotherapy. Adv Sci. 2020;7:1902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782‐795. [DOI] [PubMed] [Google Scholar]
- 15. Charoentong P, Finotello F, Angelova M, et al. Pan‐cancer immunogenomic analyses reveal genotype‐immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248‐262. [DOI] [PubMed] [Google Scholar]
- 16. Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Valkenburg KC, De Groot AE, Pienta KJ. Targeting the tumour stroma to improve cancer therapy. Nat Rev Clin Oncol. 2018;15:366‐381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nat Rev Cancer. 2011;11:393‐410. [DOI] [PubMed] [Google Scholar]
- 19. Kim Y, Lin Q, Glazer P, Yun Z. Hypoxic tumor microenvironment and cancer cell differentiation. Curr Mol Med. 2009;9:425‐434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Petrova V, Annicchiarico‐Petruzzelli M, Melino G, Amelio I. The hypoxic tumour microenvironment. Oncogenesis. 2018;7:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bader JE, Voss K, Rathmell JC. Targeting metabolism to improve the tumor microenvironment for cancer immunotherapy. Mol Cell. 2020;78:1019‐1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Colegio OR, Chu NQ, Szabo AL, et al. Functional polarization of tumour‐associated macrophages by tumour‐derived lactic acid. Nature. 2014;513:559‐563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chiu DKC, Tse APW, Xu IMJ, et al. Hypoxia inducible factor HIF‐1 promotes myeloid‐derived suppressor cells accumulation through ENTPD2/CD39L1 in hepatocellular carcinoma. Nat Commun. 2017;8:517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Clambey ET, McNamee EN, Westrich JA, et al. Hypoxia‐inducible factor‐1 alpha‐dependent induction of FoxP3 drives regulatory T‐cell abundance and function during inflammatory hypoxia of the mucosa. Proc Natl Acad Sci U S A. 2012;109:E2784‐E2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dang EV, Barbi J, Yang HY, et al. Control of TH17/Treg balance by hypoxia‐inducible factor 1. Cell. 2011;146:772‐784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sitkovsky M, Lukashev D. Regulation of immune cells by local‐tissue oxygen tension: HIF1α and adenosine receptors. Nat Rev Immunol. 2005;5:712‐721. [DOI] [PubMed] [Google Scholar]
- 27. Vito A, El‐Sayes N, Mossman K. Hypoxia‐driven immune escape in the tumor microenvironment. Cell. 2020;9:992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ye Y, Hu Q, Chen H, et al. Characterization of hypoxia‐associated molecular features to aid hypoxia‐targeted therapy. Nat Metab. 2019;1:431‐444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Iñarrairaegui M, Melero I, Sangro B. Immunotherapy of hepatocellular carcinoma: facts and hopes. Clin Cancer Res. 2018;24:1518‐1524. [DOI] [PubMed] [Google Scholar]
- 30. Gao Q, Zhu H, Dong L, et al. Integrated Proteogenomic characterization of HBV‐related hepatocellular carcinoma. Cell. 2019;179:1240. [DOI] [PubMed] [Google Scholar]
- 31. Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sturm G, Finotello F, Petitprez F, et al. Comprehensive evaluation of transcriptome‐based cell‐type quantification methods for immuno‐oncology. Bioinformatics. 2019;35:i436‐i445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hao D, Liu J, Chen M, et al. Immunogenomic analyses of advanced serous ovarian cancer reveal immune score is a strong prognostic factor and an indicator of chemosensitivity. Clin Cancer Res. 2018;24:3560‐3571. [DOI] [PubMed] [Google Scholar]
- 34. Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database Hallmark gene set collection. Cell Syst. 2015;1:417‐425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meléndez B, Van Campenhout C, Rorive S, Remmelink M, Salmon I, D'Haene N. Methods of measurement for tumor mutational burden in tumor tissue. Transl Lung Cancer Res. 2018;7:661‐667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liebermeister W, Noor E, Flamholz A, Davidi D, Bernhardt J, Milo R. Visual account of protein investment in cellular functions. Proc Natl Acad Sci U S A. 2014;111:8488‐8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang R, Liu Q, Li T, Liao Q, Zhao Y. Role of the complement system in the tumor microenvironment. Cancer Cell Int. 2019;19:300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chowdhury PS, Chamoto K, Kumar A, Honjo T. PPAR‐induced fatty acid oxidation in T cells increases the number of tumor‐reactive CD8+ T cells and facilitates anti‐PD‐1 therapy. Cancer Immunol Res. 2018;6:1375‐1387. [DOI] [PubMed] [Google Scholar]
- 39. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545‐15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Villanueva A, Hoshida Y. Depicting the role of TP53 in hepatocellular carcinoma progression. J Hepatol. 2011;55:724‐725. [DOI] [PubMed] [Google Scholar]
- 41. Zhan P, Ji Y‐N. Prognostic significance of TP53 expression for patients with hepatocellular carcinoma: a meta‐analysis. Hepatobiliary Surg Nutr. 2014;3:11‐117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lequeux A, Noman MZ, Xiao M, et al. Impact of hypoxic tumor microenvironment and tumor cell plasticity on the expression of immune checkpoints. Cancer Lett. 2019;458:13‐20. [DOI] [PubMed] [Google Scholar]
- 43. Wu XZ, Xie GR, Chen D. Hypoxia and hepatocellular carcinoma: the therapeutic target for hepatocellular carcinoma. J Gastroenterol Hepatol. 2007;22:1178‐1182. [DOI] [PubMed] [Google Scholar]
- 44. Greten TF, Sangro B. Targets for immunotherapy of liver cancer. J Hepatol. 2018;68:157‐166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF‐1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187‐197. [DOI] [PubMed] [Google Scholar]
- 46. Gentles AJ, Newman AM, Liu CL, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938‐945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kong X, Sun R, Chen Y, Wei H, Tian Z. γδT cells drive myeloid‐derived suppressor cell–mediated CD8+ T cell exhaustion in hepatitis B virus–induced Immunotolerance. J Immunol. 2014;193:1645‐1653. [DOI] [PubMed] [Google Scholar]
- 48. Yan J, Liu XL, Xiao G, et al. Prevalence and clinical relevance of T‐helper cells, Th17 and Th1, in hepatitis B virus‐related hepatocellular carcinoma. PLoS One. 2014;9:e96080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee HL, Jang JW, Lee SW, et al. Inflammatory cytokines and change of Th1/Th2 balance as prognostic indicators for hepatocellular carcinoma in patients treated with transarterial chemoembolization. Sci Rep. 2019;9:3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Suzuki S, Ichikawa Y, Nakagawa K, et al. High infiltration of mast cells positive to tryptase predicts worse outcome following resection of colorectal liver metastases. BMC Cancer. 2015;15:840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li W, Han J, Yuan K, Wu H. Integrated tumor stromal features of hepatocellular carcinoma reveals two distinct subtypes with prognostic/predictive significance. Aging (Albany NY). 2019;11:4478‐4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gu C, Jun JC. Does hypoxia decrease the metabolic rate? Front Endocrinol (Lausanne). 2018;9:668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Noman MZ, Hasmim M, Messai Y, et al. Hypoxia: a key player in antitumor immune response. A review in the theme: cellular responses to hypoxia. Am J Physiol Cell Physiol. 2015;309:C569‐C579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Aiderus A, Black MA, Dunbier AK. Fatty acid oxidation is associated with proliferation and prognosis in breast and other cancers. BMC Cancer. 2018;18:805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Swietach P, Vaughan‐Jones RD, Harris AL. Regulation of tumor pH and the role of carbonic anhydrase 9. Cancer Metastasis Rev. 2007;26:299‐310. [DOI] [PubMed] [Google Scholar]
- 56. Amann T, Maegdefrau U, Hartmann A, et al. GLUT1 expression is increased in hepatocellular carcinoma and promotes tumorigenesis. Am J Pathol. 2009;174:1544‐1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hsu HC, Jeng YM, Mao TL, Chu JS, Lai PL, Peng SY. β‐Catenin mutations are associated with a subset of low‐stage hepatocellular carcinoma negative for hepatitis B virus and with favorable prognosis. Am J Pathol. 2000;157:763‐770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pinyol R, Montal R, Bassaganyas L, et al. Molecular predictors of prevention of recurrence in HCC with sorafenib as adjuvant treatment and prognostic factors in the phase 3 STORM trial. Gut. 2019;68:1065‐1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures Supporting information
Supplementary Tables Supporting information
Data Availability Statement
Only publicly available data were used in our study, and data sources and handling of these data are described in the Materials and Methods and in the Supplementary Tables. Further information is available from the corresponding author upon request.


