ABSTRACT
Convergence is the phenomenon whereby similar phenotypes evolve independently in different lineages. One example is resistance to toxins in animals. Toxins have evolved many times throughout the tree of life. They disrupt molecular and physiological pathways in target species, thereby incapacitating prey or deterring a predator. In response, molecular resistance has evolved in many species exposed to toxins to counteract their harmful effects. Here, we review current knowledge on the convergence of toxin resistance using examples from a wide range of toxin families. We explore the evolutionary processes and molecular adaptations driving toxin resistance. However, resistance adaptations may carry a fitness cost if they disrupt the normal physiology of the resistant animal. Therefore, there is a trade‐off between maintaining a functional molecular target and reducing toxin susceptibility. There are relatively few solutions that satisfy this trade‐off. As a result, we see a small set of molecular adaptations appearing repeatedly in diverse animal lineages, a phenomenon that is consistent with models of deterministic evolution. Convergence may also explain what has been called ‘autoresistance’. This is often thought to have evolved for self‐protection, but we argue instead that it may be a consequence of poisonous animals feeding on toxic prey. Toxin resistance provides a unique and compelling model system for studying the interplay between trophic interactions, selection pressures and the molecular mechanisms underlying evolutionary novelties.
Keywords: convergent evolution, toxin resistance, molecular adaptation, functional constraint, deterministic evolution, co‐evolutionary arms races
I. INTRODUCTION
Convergent evolution is the independent emergence of similar traits across different lineages (Storz, 2016). Toxins are key innovations that have evolved throughout the tree of life (Yamaguchi, Park & Inouye, 2011; Casewell et al., 2013). They act on specific molecular targets (e.g. receptors, ion‐channels, enzymes, and plasma membranes), causing a range of pathophysiological disruptions throughout the cardiovascular, circulatory and nervous systems (Fry et al., 2009; Casewell et al., 2013). Venom is a mixture of proteinaceous toxins exploited for a variety of functions, including prey capture and defence, ultimately resulting in severe pain, incapacitation or death. By definition, venoms are typically injected into a target system via a mechanical wound caused for example by fangs (e.g. snakes, centipedes and spiders), spines (e.g. fish), nematocysts (e.g. jellyfish and sea anemones) or stingers (bees, wasps and other arthropods). Poisons on the other hand tend to consist of small‐molecule toxins (e.g. low‐molecular‐weight alkaloid or steroidal‐based compounds) that are almost exclusively utilised to deter predators – often by causing rapid pain or paralysis upon contact or ingestion (Duran‐Riveroll & Cembella, 2017; Botelho et al., 2019).
Therefore, toxins can be considered ecologically functional traits that mediate antagonistic interactions between predator and prey, driven by natural selection. To counteract the deleterious effects of these toxins, some animals have evolved resistance. Toxin resistance is the increased ability of an animal to survive exposure to one or more toxins without being functionally affected. As a result, toxin resistance has evolved in at least three distinct ecological contexts (Fig. 1): predator resistance, where a predator is resistant to the toxins of its prey (Fig. 1A–C); prey resistance, where the prey is resistant to the toxins of a predator (Fig. 1D); or autoresistance, where an animal is resistant to its own toxins (Fig. 1E).
Fig. 1.
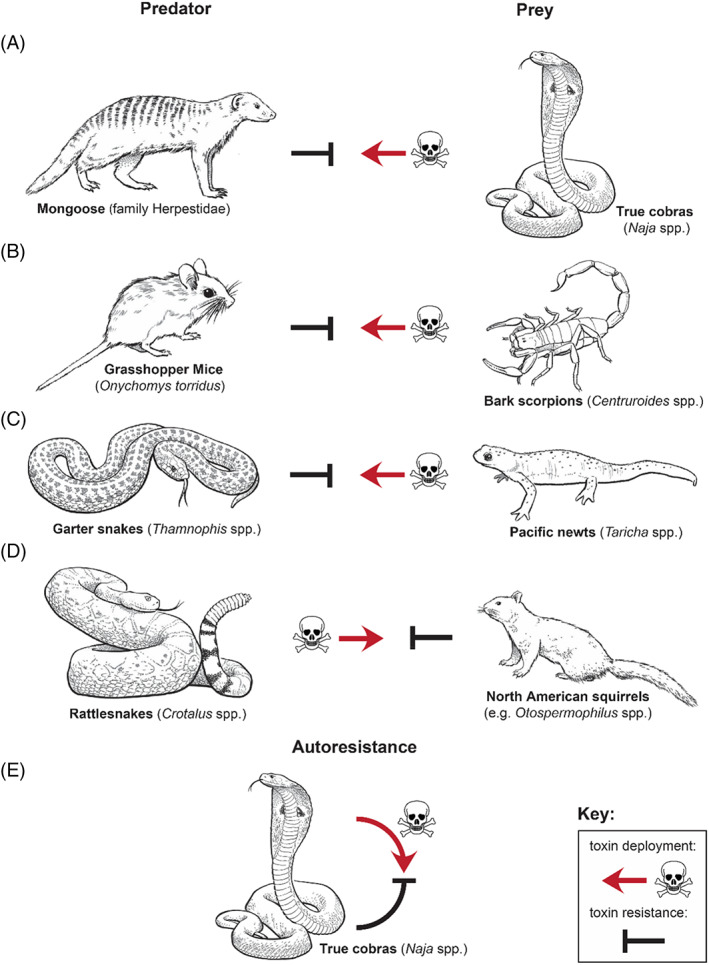
Well‐known examples of ecological contexts underpinning toxin resistance. (A–C) predator resistance, where a predator is resistant to the toxins of its prey. (A) The mongoose is known to predate on true cobras. (B) The grasshopper mouse preys on bark scorpions. (C) Garter snakes prey on toxic newts. (D) Prey resistance is resistance of a prey species to the toxins of a predator and is exemplified here by rattlesnakes preying on North American ground squirrels. (E) Autoresistance is where an animal is resistant to its own toxins. The example shown here is of true cobras that show resistance to cobra α‐neurotoxins.
Resistance particularly evolves in molecular targets involving key regulatory processes that are under strong selection pressures, i.e. that are crucial for the survival of an organism. There are several molecular mechanisms underlying toxin resistance [reviewed in Holding et al., 2016b and Arbuckle, Rodriguez de la Vega & Casewell, 2017]. First, target modification is a change in the gene sequences that encode receptors or circulating proteins to which specific toxins bind, resulting in a reduction of binding affinity of the toxin towards the target (Barchan et al., 1992; Geffeney et al., 2005; Jansa & Voss, 2011; Tarvin et al., 2017; Karageorgi et al., 2019). Second, off‐target repurposing is the molecular modification of a previously non‐target site that increases its toxin‐binding affinity so that the physiological effect induced by the toxin is altered, and the desired effect has thereby been repurposed (Rowe et al., 2013). Finally, toxin scavenging involves serum‐based components that patrol the circulatory system and inhibit the activity of enzymatic toxins (Perez, Pichyangkul & Garcia, 1979; Biardi et al., 2011; Gibbs et al., 2020).
Venom systems have been proposed as models for studying processes underlying evolutionary adaptations (Zancolli & Casewell, 2020), including convergent evolution (Casewell et al., 2019; Kazandjian et al., 2021; Walker et al., 2021). The well‐defined function (e.g. ecological interactions) and the discrete genotype–phenotype association (e.g. molecular adaptations reducing the binding affinity of a certain toxin) involving toxin resistance provide a compelling model system for studying evolutionary adaptations and drivers of fundamental processes in biology. Similar toxins are found in many different animals (Casewell et al., 2013; Schendel et al., 2019), and therefore they are involved in many different trophic interactions (i.e. ecological interplay between species related to diet) across many animal lineages. This provides a fascinating opportunity to study the extent of convergent evolution across unrelated taxa by incorporating the evolutionary drivers stimulating molecular adaptations leading to toxin resistance.
Here, we review current knowledge on the convergent evolution of toxin resistance, with a particular emphasis on its molecular basis and evolutionary drivers. We discuss the molecular mechanisms underpinning toxin resistance across a wide range of toxin families, with diverse evolutionary drivers. We then discuss several intriguing evolutionary aspects of toxin resistance, namely: (i) the evolutionary framework underlying the appearance of toxin resistance; (ii) the role of functional constraints on molecular targets leading to convergence; and (iii) a re‐examination of the evolutionary drivers of autoresistance in poisonous animals.
II. CONVERGENT MECHANISMS OF RESISTANCE
(1). Snake venom metalloproteinases
Snake venom metalloproteinases (SVMPs) are zinc‐dependent proteinases capable of exerting coagulopathic and haemorrhagic effects (Ferraz et al., 2019), and they are particularly abundant in viper (family Viperidae) venoms (Tasoulis & Isbister, 2017). Many animals have evolved an innate immune response mediated by SVMP inhibitors (SVMPIs). SVMPIs are acidic glycoproteins present in the circulatory system that neutralise the activity of SVMPs using a toxin‐scavenging mechanism. These ‘scavenging’ inhibitors display a tight‐binding reaction mechanism, mediated by the formation of non‐covalent interactions and ultimately preventing the pathophysiological effects of SVMPs (Valente et al., 2001). Although sharing similar functionality, SVMPIs are related to three different supergene families: (i) ficolin/opsonin p35 (Omori‐Satoh, Yamakawa & Mebs, 2000); (ii) immunoglobulin (Hood, Kronenberg & Hunkapiller, 1985); or (iii) cystatin (Rawlings & Barrett, 1990). SVMPIs evolved independently across many distinct mammal (class Mammalia) and snake (suborder Serpentes) lineages.
A well‐studied example of SVMP resistance can be seen in several North American squirrels (e.g. Otospermophilus spp., Ictidomys sp. and Sciurus sp.) that are sympatric (i.e. occurring in the same geographical area) with rattlesnakes (Crotalus spp.; Martinez et al., 1999; Biardi et al., 2011; Biardi & Coss, 2011; Pomento et al., 2016). This is in contrast to squirrels that are allopatric (i.e. occurring in distinct, non‐overlapping geographical areas) with rattlesnakes, which show less resistance (Poran, Coss & Benjamini, 1987; Holding, Biardi & Gibbs, 2016a; Pomento et al., 2016; Gibbs et al., 2020). Holding et al. (2016a) showed that local rattlesnake populations demonstrate higher SVMP activity in their venom; this higher activity was linked to the increased SVMPI activity in sympatric squirrels. The latter was not observed in allopatric populations. This suggests that rattlesnake venom has become adapted to maintain its selective advantage in overcoming squirrel resistance (Holding et al., 2016a). This highlights the evolutionary interplay between predator and prey, resulting in convergent, geographically restricted adaptations within and across different squirrel species.
Another example is the opossum family (Didelphidae), which is sympatric with pitvipers (subfamily Crotalinae). Interestingly, they have reciprocal trophic relationships: opossums predate upon pitvipers (Oliveira & Santori, 1999; Almeida‐Santos et al., 2000) and pitvipers prey on opossums (Voss, 2013). Many opossum species show resistance to injected pitviper venoms (Werner & Vick, 1977; Perales et al., 1994), a resistance mediated by serum SVMPIs. Evolutionary drivers underpinning resistance are likely to be species dependent; more data on trophic interactions will help address this issue (Voss, 2013).
Various other mammals, including several North American rodents (Sigmodon sp., Microtus sp. and Neotoma spp.), have evolved resistance to the SVMPs of sympatric pitviper species (Pichyangkul & Perez, 1981; de Wit, 1982; Garcia & Perez, 1984). Furthermore, some animals that prey on snakes, including the Indian grey mongoose (Herpestes edwardsii) and the European hedgehog (Erinaceus europaeus), also have serum SVMPIs. Most mammalian SVMPIs are related to the immunoglobulin family; however, erinacin, isolated from E. europaeus, is related to the ficolin/opsonin p35 family (Mebs et al., 1996; Omori‐Satoh et al., 2000). An overview of mammalian SVMPIs derived from the literature is provided as online supporting information in Table S1.
In addition to mammals, several snakes have also evolved serum SVMPIs that may confer resistance. In pitvipers this is likely an example of autoresistance, but in the eastern indigo snake (Drymarchon couperi) and Ryukyu odd‐tooth snake (Lycodon semicarinatus) it may rather be predator resistance because these species are known to prey on sympatric pitvipers (Tomihara et al., 1988; Goetz et al., 2019). Another snake with serum SVMPIs is the Burmese python (Python bivittatus; Duan et al., 2017), which likely evolved resistance in response to predation by venomous, snake‐eating snakes (Jones et al., 2020; Smith et al., 2021). While most research has focused on serum SVMPIs, some snakes also have SVMPIs in their venom and venom glands (Munekiyo & Mackessy, 2005; Yee et al., 2016; Valente et al., 2018). This could be an example of autoresistance, whereby the SVMPIs protect the secretory epithelium of the venom gland and venom components from harmful effects of endogenous SVMPs (Mackessy & Baxter, 2006; Valente et al., 2018). An overview of snake SVMPIs derived from the literature is provided in Table S2.
(2). Snake venom phospholipases A2
Phospholipases A2 (PLA2s) are esterolytic enzymes that can cause a variety of pathological effects including myotoxicity, neurotoxicity and haemotoxicity (Manjunatha Kini, 2003; Ferraz et al., 2019). They are major venom components across different snake lineages (Tasoulis & Isbister, 2017). Several animals have evolved an innate immune response mediated by PLA2 inhibitors (PLA2Is) that neutralise the activity of PLA2s using a toxin‐scavenging mechanism (Lizano, Domont & Perales, 2003). PLA2Is form stable complexes, and by mimicking PLA2‐acceptors prevent binding to the cell membrane, ultimately resulting in the inhibition of the pathological effects of PLA2s (Perales et al., 1995). PLA2Is are assigned to three structural classes (reviewed in Lizano et al., 2003): PLA2I‐α (C‐type lectin domain); PLA2I‐β (leucine‐rich repeats domain); and PLA2I‐γ (three‐finger domain). These PLA2Is evolved convergently in snakes and opossums.
Serum‐derived PLA2Is are predominantly found in venomous snakes, including species with an abundance of PLA2s in their venom (Tasoulis & Isbister, 2017). Interestingly, all elapid inhibitors are classified as PLA2I‐γ class, whereas viperid inhibitors are also in the PLA2I‐α and PLA2I‐β classes (see Table S3 for an overview of PLA2Is in snakes). PLA2I‐γ of some non‐venomous snakes may represent prey resistance that evolved in response to predation from venomous snakes (Thwin et al., 2000; Zhong & Huang, 2016; Fortes‐Dias et al., 2020; Rodrigues et al., 2020, 2021). PLA2Is have also been characterised in the venom and venom gland itself, possibly representing a form of autoresistance (Mackessy & Baxter, 2006; Valente et al., 2018).
The only PLA2Is known from mammals are those of opossums. A PLA2I has been isolated from the white‐eared opossum (Didelphis albiventris; Soares et al., 1997). PLA2Is are also known in the big‐eared opossum (D. aurita). Interestingly, they show homology with the immunoglobulin supergene family. This is significant because SVMPIs isolated from other didelphids share high sequence similarities with their PLA2I‐counterpart, suggesting that similar inhibitors act on distinct snake venom toxins (Rocha et al., 2002).
(3). Snake venom C‐type lectins
C‐type lectins (CTLs) are members of the lectin family and are predominantly found in the venom of vipers (Tasoulis & Isbister, 2017). CTLs bind to glycoprotein 1b and von Willebrand factor (vWF), thereby promoting abnormal platelet aggregation. This resistance convergently evolved within the opossum lineage, which are known to have trophic interactions with pitvipers (Oliveira & Santori, 1999; Almeida‐Santos et al., 2000; Voss, 2013). Several opossum species (Didelphis spp., Philander spp. and Lutreolina sp.) show modifications of the CTL‐binding site of the vWF protein (A1 domain) under positive selection (Jansa & Voss, 2011). The modified vWF protein has substitutions that change its hydrophobicity and net charge, which is hypothesised to inhibit the binding of CTLs (Jansa & Voss, 2011). This hypothesis was supported by functional in vitro experiments revealing significant decreases in platelet aggregation in opossum plasma exposed to CTLs (Drabeck et al., 2020). This is the first documented example of resistance‐associated adaptations in a non‐receptor protein targeted by venom.
(4). Snake venom three‐finger toxins
Three‐finger toxins (3FTX) are one of the most abundant non‐enzymatic toxin families in elapid (family Elapidae) and some colubrid (family Colubridae) venoms (Tasoulis & Isbister, 2017; Modahl & Mackessy, 2019), and their principal effects are cytotoxicity and neurotoxicity. The basal activity is post‐synaptic neurotoxicity through antagonistic binding to the muscle‐type nicotinic acetylcholine receptor (nAChR), causing muscular paralysis and respiratory failure (Barchan et al., 1995; Takacs, Wilhelmsen & Sorota, 2004). These neurotoxic effects are caused by α‐neurotoxins, which primarily bind to Loop C of the ligand‐binding domain (α1‐subunit) of the nAChR. Furthermore, they also show interaction with the Cys Loop, Loop F, and neighbouring delta and gamma subunits (Rahman et al., 2020). Resistance to α‐neurotoxins is underpinned by different molecular modifications of the ligand‐binding domain of the nAChR, causing a significant reduction in their binding affinity (Barchan et al., 1995; Kachalsky et al., 1995; Asher et al., 1998; Takacs, Wilhelmsen & Sorota, 2001; Takacs et al., 2004; Dellisanti et al., 2007; Rahman et al., 2020; Harris & Fry, 2021; Jones et al., 2021). These resistance adaptations can be generally categorised as one of four different amino acid substitutions. First, asparagine resistance is characterised by a change to asparagine, resulting in a glycosylation motif (187–189NVT or 189–191NXS/T). As a result, the N‐glycosylation of asparagine sterically hinders the binding of α‐neurotoxins (Asher et al., 1998; Takacs et al., 2001, 2004; Rahman et al., 2020). Secondly, arginine resistance is characterised by a replacement to arginine (187R). This change sterically hinders the binding of α‐neurotoxins (Rahman et al., 2020). However, since arginine is a positively charged amino acid, it has also been suggested to affect toxin binding by means of electrostatic repulsion of the positively charged α‐neurotoxins (Dellisanti et al., 2007; Drabeck, Dean & Jansa, 2015). Lysine resistance is characterised by a replacement to lysine (191K, 195K and/or 196K). This change to the positively charged amino acid affects the binding by means of electrostatic repulsion of α‐neurotoxins (Harris & Fry, 2021). Finally, proline resistance is characterised by a replacement from proline to either a leucine or histidine (194L, 197L/H). These replacements change the structural conformation of the ligand‐binding domain, and therefore affect the binding of α‐neurotoxins (Kachalsky et al., 1995). An overview of the key sites and mutations that confer α‐neurotoxin resistance is provided in Table S4. α‐Neurotoxin resistance has evolved convergently across a broad diversity of vertebrates (Fig. 2).
Fig. 2.
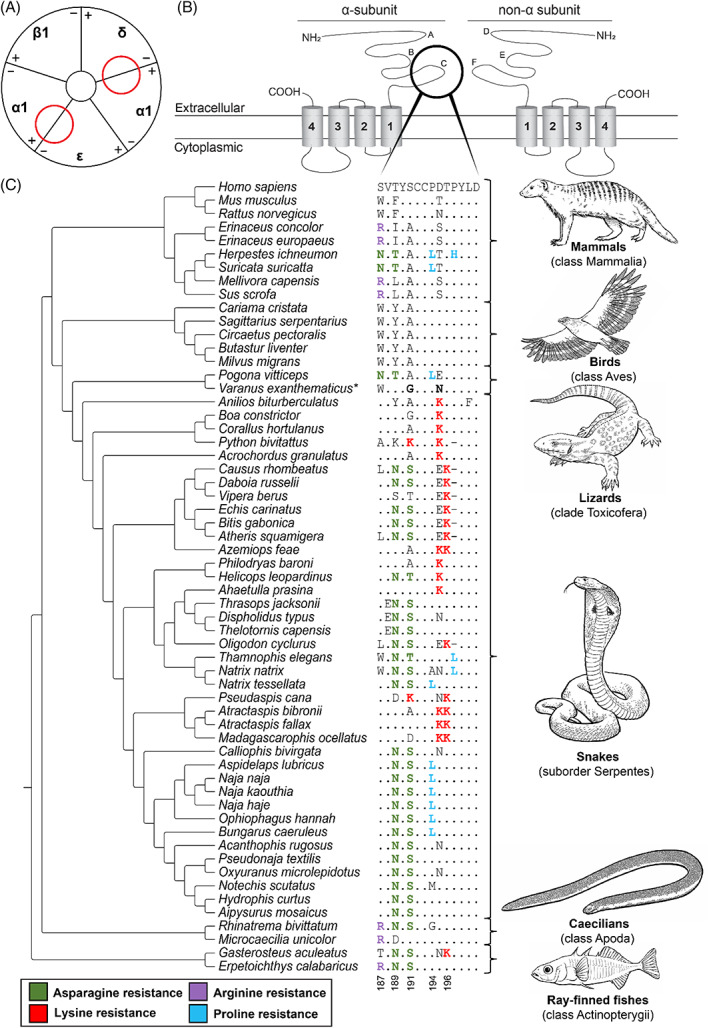
Convergent evolution of α‐neurotoxin resistance in animals. (A) Schematic representation (based on Kini, 2019) of the α‐1 muscle‐type nicotinic acetylcholine receptor (nAChR). Red circles indicate the position of the ligand‐binding domain of α‐neurotoxins in the nAChR. (B) Protein topology of an α‐subunit and a non‐α‐subunit of the muscle‐type nAChR. A–F indicate the loop structures at the extracellular domain in the respective subunits (Rahman et al., 2020). The black circle indicates the C‐loop involved in α‐neurotoxin binding. (C) Sequence alignment of the α1‐nAChR ligand‐binding domain. The reference amino acid sequence is from humans (Homo sapiens) and differences from this sequence are displayed for all other species. Substitutions associated with resistance are highlighted in coloured font. The asterisk (*) in Varanus exanthematicus indicates that the two substitutions shown are associated with reduced binding affinity (Jones, Harris & Fry, 2021). Tree topology based on Khan et al. (2020). Sequence accession numbers are provided in Table S5.
A classic example of α‐neurotoxin resistance is seen in mongooses (family Herpestidae). For example, the Egyptian mongoose (Herpestes ichneumon) and the meerkat (Suricata suricatta) both possess a combination of asparagine resistance (187–189NVT) and proline resistance (194L; Fig. 2; Barchan et al., 1992; Kachalsky et al., 1995; Asher et al., 1998; Khan et al., 2020). Furthermore, the Egyptian mongoose also shows an additional proline replacement (197H; Fig. 2). These adaptations presumably evolved in response to predation on venomous snakes including true cobras (Naja spp.; Stuart, 1983; Struhsaker & McKey, 1975). However, more studies are needed to characterise the extent of trophic interactions between these mammals and snakes in the wild. Other resistant mammals include the honey badger (Mellivora capensis), hedgehogs (Erinaceus europaeus and E. concolor), and the wild boar (Sus scrofa), all of which show arginine resistance (187R; Fig. 2; Barchan et al., 1995; Drabeck et al., 2015; Harris & Fry, 2021). The honey badger predates upon venomous snakes, including the Cape cobra (Naja nivea), which has α‐neurotoxin in its venom (Begg et al., 2003). Wild boars and hedgehogs are also known occasionally to prey on snakes (Reeve, 1994; Tanaka & Mori, 2000; Jolley et al., 2010; Wilcox, 2015). Interestingly, a recent study of primates highlighted that some groups that are sympatric with α‐neurotoxic snakes had a reduced susceptibility towards α‐neurotoxins of true cobras (Naja spp.). Members of the subfamily Homininae (Homo, Pan and Gorilla spp.) showed the lowest degree of susceptibility compared to other primates (Harris, Nekaris & Fry, 2021). In principle, this pattern could be explained in terms of the long history of interactions between primates and venomous snakes (Isbell, 2009; Kazandjian et al., 2021).
Many birds prey on venomous snakes, including snake specialists such as the secretary bird (Sagittarius serpentarius), snake eagles (Circaetus spp.), and seriemas (family Cariamidea; Redford & Peters, 1986; Portugal et al., 2016; Mori, Vyas & Upadhyay, 2017). Birds do not show any known resistance‐related modifications associated with α‐neurotoxins (Fig. 2); Khan et al., 2020). To explain this apparent paradox, we propose that a set of morphological exaptations and behavioural traits in snake‐eating birds prevent envenomation in the first place (Fig. 3). Some examples of morphological exaptations are plumage and leg scales that may provide a physical barrier against snakebite envenomation (Lucas & Stettenheim, 1972). Additionally, bird legs mainly contain tendons and lack highly vascular tissue such as skeletal muscle; this may limit the uptake of venom if the bird is bitten. In particular, the secretary bird attacks snakes aggressively, directing kicks to the head and neck (Portugal et al., 2016). Its elongated tibiotarsus and tarsometatarsus may facilitate a powerful kick (Portugal et al., 2016). Birds of prey, many of which are snake‐eaters, have high visual acuity and ambush hunting strategies that may minimise the risk of snakebite (Potier et al., 2020). The red‐legged seriema (Cariama cristata) uses its beak to grasp prey behind the neck and then shakes the prey violently so as to fracture its spine (Silva et al., 2016). In summ, these bird‐specific morphological and behavioural traits might explain why molecular adaptations conferring resistance have not evolved among them, in contrast to other snake‐eating lineages [e.g. mongoose, honey badger (Drabeck et al., 2015; Khan et al., 2020)].
Fig. 3.
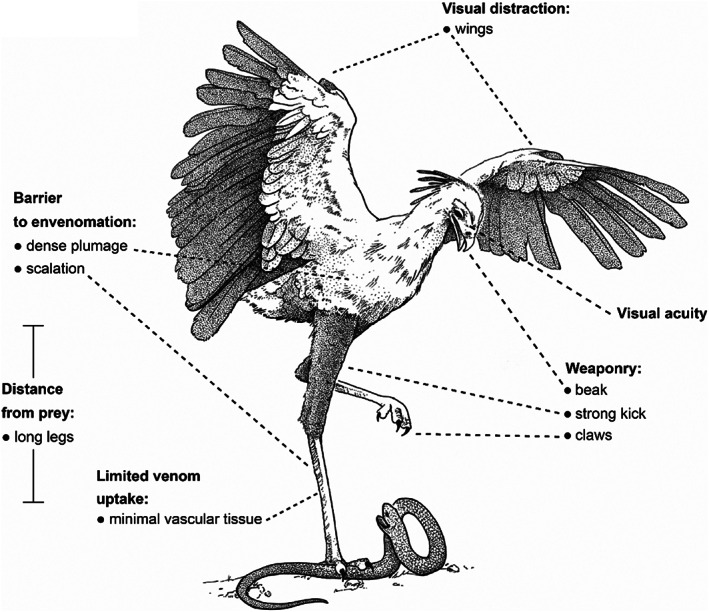
Morphological exaptations and behavioural traits proposed to negate selection pressures for evolving molecular resistance in snake‐eating birds such as the secretary bird (Sagittarius serpentarius). This figure represents multiple examples of traits unique to birds, and particularly to snake‐eating birds, that might contribute to the prevention of snakebite envenomation. Drawing from an original photograph by Jason Shallcross, with permission.
Resistance‐related mutations have been documented in lizards (clade Toxicofera) that are potentially vulnerable to predation by sympatric, neurotoxic snakes, such as the central bearded dragon (Pogona vitticeps; 187–189NYT, 194L) and the savannah monitor [Varanus exanthematicus; 191G and 195N; Fig. 2 (Khan et al., 2020; Jones et al., 2021)]. However, resistance has not been documented in monitor lizards (Varanus spp.) that have been suggested to prey on neurotoxic snakes (Jones et al., 2021). Several studies hypothesised that morphological exaptations (thick, osteodermic scales) and prey‐handling behaviour negated selection pressure for molecular resistance in these lizards (Jones et al., 2021; Youngman, Llinas & Fry, 2021). The evolution of such strategies to avoid envenomation is comparable to what we propose for snake‐eating birds (Fig. 3).
α‐neurotoxin resistance is particularly widespread in snakes, which have convergently evolved asparagine resistance (189–191NXS/T), lysine resistance (191K, 195K and/or 196K) and/or, proline resistance (194L, 197L) to elapid α‐neurotoxins (Fig. 2; Khan et al., 2020; Harris & Fry, 2021). Resistance to these toxins in snakes is particularly interesting because – uniquely among vertebrates – snakes show all three ecological functions of resistance, namely, predator resistance, prey resistance and autoresistance. The driver of this trait is either due to selection pressure from sympatric venomous snakes or autoresistance. Elapids, including true cobras (Naja spp.) and the king cobra (Ophiophagus hannah), prey on other snakes, and this includes cannibalism (Shine et al., 2007; Layloo, Smith & Maritz, 2017; Maritz, Alexander & Maritz, 2019; Jones et al., 2020), and numerous species are specialised snake predators, which may have been the ancestral condition of the clade (Shine, 1991; Kgaditse, 2016). It has been suggested that predation from snake‐eating elapids may have contributed to the evolution of resistance in multiple non‐elapid snake lineages, an assertion that is supported by ecological observations of predation events and diet studies (Alexander & Maritz, 2010; Maritz et al., 2019; Jones et al., 2020). Notably, the European viper (Vipera berus) shows a reversal of the asparagine‐resistance genotype (Fig. 2), secondary to vipers radiating into geographic areas lacking sympatric neurotoxic snakes; this may indicate that resistance mutations carry a fitness cost in species that no longer encounter α‐neurotoxins (Khan et al., 2020). On the other hand, this resistance may also prevent self‐envenomation (e.g. accidental occasions when a venomous snake bites itself) in snakes with α‐neurotoxins. All elapid snakes sequenced to date share the N‐glycosylation form of resistance to α‐neurotoxins found in their own venom (Fig. 2; Khan et al., 2020). Resistance to α‐neurotoxins is less common in non‐elapid neurotoxic snakes – but is ubiquitous within the elapids (Khan et al., 2020; Harris & Fry, 2021), perhaps suggesting a strong selection pressure for the evolution of autoresistance.
Less well‐documented examples have been observed in both caecilians (clade Apoda) and ray‐finned fishes (class Actinopterygii). Two caecilian species have convergently evolved resistance elements: the tiny Cayenne caecilian (Microcaecilia unicolor; 187R) and the two‐lined caecilian (Rhinatrema bivittatum; 189–191NYS and 187R; Fig. 2; Khan et al., 2020). Both species are sympatric with caecilian‐eating coral snakes (Micrurus spp.), which could explain this resistance (Martins & Ermelinda Oliveira, 1998). Additionally, asparagine resistance (189–191NYS) has evolved in the three‐spined stickleback (Gasterosteus aculeatus) and the reedfish (Erpetoichthys calabaricus; Fig. 2; Khan et al., 2020). The three‐spined stickleback additionally evolved lysine resistance (196K) whereas the reedfish additionally shows arginine resistance (187R; Fig. 2). The ecological role of these modifications, if any, is unknown. A highly speculative possibility is that it could have evolved against anatoxin‐a (which is an α‐neurotoxin) secreted by freshwater cyanobacteria (Aráoz, Molgó & Tandeau de Marsac, 2010).
(5). Pain‐inducing scorpion toxins
The venom of bark scorpions (Centruroides spp.) is potentially lethal, and their venom rapidly induces intense pain, presumably to deter attackers. The pain is caused by the activation of voltage‐gated Na+ channels (Nav 1.7), which are responsible for transmitting pain signals (nociceptive action potentials) to the central nervous system (Rowe et al., 2011, 2013). The grasshopper mouse (Onychomys torridus) preys on arthropods including bark scorpions. It shows toxin resistance towards bark scorpion venom characterised by a diminished pain response (Rowe & Rowe, 2008; Rowe et al., 2013). This resistance is underpinned by two substitutions: glutamine (859Q) and glutamic acid (862E) in the Nav 1.8 channel (Fig. 4; Rowe et al., 2013). Interestingly, these substitutions do not occur in the original targeted Na+ channel (Nav 1.7) but in the previously non‐target Na+ channel (Nav 1.8), an example of off‐target repurposing (Rowe et al., 2013). The negatively charged glutamic acid facilitates binding of the toxin to Nav 1.8 channels, inhibiting the transmission of pain signals by the neuron, inducing an analgesic effect (Rowe et al., 2013). The pallid bat (Antrozous pallidus) also preys on bark scorpions (Bell, 1982; Johnston & Fenton, 2001; Lenhart, Mata‐Silva & Johnson, 2010) and shows resistance to the venom of the Arizona bark scorpion (Centruroides sculpturatus; Hopp et al., 2017). However, the unknown mechanism of resistance appears to be different; the pallid bat does not possess the resistant genotype observed in the grasshopper mouse (Fig. 4; Hopp et al., 2017). This highlights that similar selection pressures do not always stimulate convergence at the molecular level.
Fig. 4.
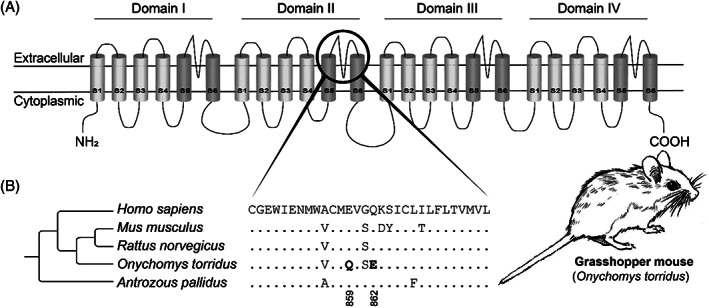
Resistance against pain‐inducing scorpion venom in grasshopper mouse (Onychomys torridus). (A) Protein topology of voltage‐gated Na + channel (Nav 1.8). The black circle indicates the outer pore associated with scorpion‐venom binding in the Nav 1.8 channel. Structure based on Shen et al. (2017). (B) Partial sequence alignment of the outer pore of the α‐subunit of domain II of the Nav 1.8 channel. The reference amino acid sequence is from humans (Homo sapiens) and differences from this sequence are displayed for all other species. Substitutions associated with resistance are highlighted in bold. Tree topology based on TimeTree.org (Kumar et al., 2017). For sequence accession numbers, see Table S6.
(6). Guanidinium toxins
Guanidinium toxins, including tetrodotoxin (TTX) and saxitoxin (STX), are alkaloids that bind to the outer pores of voltage‐gated Na+ channels (Nav) on excitable tissues, causing muscle paralysis and even death (Duran‐Riveroll & Cembella, 2017). Animals that deploy these toxins for defence presumably sequester them from their diet or produce them by means of symbiotic microorganisms (Hwang et al., 1989; Duran‐Riveroll & Cembella, 2017). Guanidinium toxins are synthesised by several species of bacteria and then enter food webs, where they are assimilated by a wide variety of organisms (Miyazawa & Noguchi, 2001). Resistance against TTX and STX is attributed to amino acid substitutions in the α‐subunit of skeletal muscle‐type (Nav 1.4) and neuronal‐type (Nav 1.6 and Nav 1.7) voltage‐gated Na+ channels (Soong & Venkatesh, 2006). These substitutions alter TTX binding by changing the outer pore structures and/or by altering the electrostatic interaction between TTX and the pore residues (Geffeney et al., 2005; Feldman et al., 2012). Guanidinium toxin resistance has evolved independently in phylogenetically distinct animal lineages (Fig. 5).
Fig. 5.
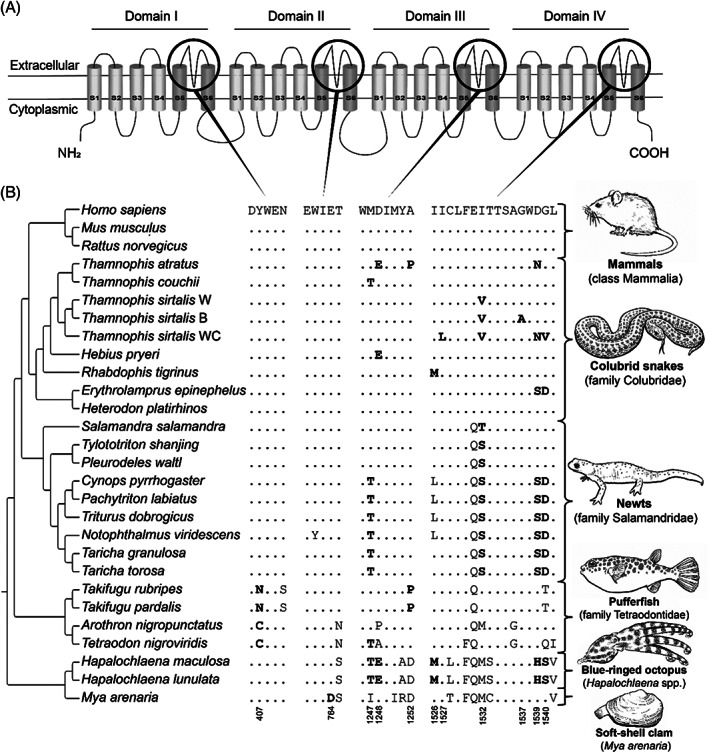
Convergent evolution of guanidinium toxin resistance in animals. (A) Protein topology of the voltage‐gated Na+ channel, Nav 1.4. The black circles indicate the outer pores involved in guanidinium toxin binding. Structure based on Shen et al. (2017). (B) Partial sequence alignments of the outer pores of the voltage‐gated Na + channel Nav 1.4. The reference amino acid sequence is from humans (Homo sapiens) and differences from this sequence are displayed for all other species. Substitutions associated with resistance are highlighted in bold, and their respective amino‐acid positions are numbered based on Nav 1.4 from Homo sapiens. Tree topology based on TimeTree.org (Kumar et al., 2017) and taxon‐specific phylogenies (Geffeney et al., 2005; Feldman et al., 2012; Hanifin & Gilly, 2015). Key for the Thamnophis sirtalis populations: B, Benton County; W, Warrenton; WC, Willow Creek. For sequence accession numbers, see Table S7.
Resistance against TTX has evolved convergently in at least six colubrid snake lineages (Feldman et al., 2012). TTX‐resistance is underpinned by numerous substitutions that decrease the binding affinity of TTX with the Nav 1.4 channel (Fig. 5). Among these colubrid snakes, North American garter snakes (Thamnophis spp.) are one of the best‐studied examples of TTX resistance (Brodie, 1990; Geffeney, Brodie & Ruben, 2002; Brodie III et al., 2005; Geffeney et al., 2005; Feldman et al., 2009, 2010). Interestingly, resistance has evolved independently among species as well as within one species, as observed in distinct populations of the common garter snake (T. sirtalis; Fig. 5; Geffeney et al., 2002, 2005), highlighting the ecological importance of this resistance trait. Common garter snakes also evolved TTX resistance in two additional Nav paralogs: Nav 1.6 and Nav 1.7 (McGlothlin et al., 2014). These paralogs are exclusively expressed in the peripheral nervous system, which is frequently exposed to TTX. By contrast, the Nav paralogs in the central nervous system (Nav 1.1–1.3), which are not exposed to TTX, lack any resistance modifications (McGlothlin et al., 2014). The resistant Nav paralogs (Nav 1.6 and Nav 1.7) were present in the common ancestor of all snakes – which initially allowed predation on toxic prey. Subsequently, this facilitated the evolution of resistance at the skeletal muscle Nav 1.4 channel (McGlothlin et al., 2016). This resistance in garter snakes is tightly linked to predation on Pacific newts (Taricha spp.) that deploy TTX on their skin for defence against predators (Brodie III & Brodie Jr., 1999; Williams, Brodie & Brodie, 2004; Hanifin & Gilly, 2015). Additionally, several geographically widespread colubrids (e.g. Hebius sp., Rhabdophis sp., and Erythrolamprus sp.) have also evolved substitutions conferring TTX‐resistance (Feldman et al., 2012), facilitating predation on distinct TTX‐bearing amphibians (Fig. 5; Feldman et al., 2012). However, the eastern hognose snake (Heterodon platirhinos) displays high levels of TTX resistance, but lacks these Nav channel substitutions, suggesting a different mechanism of adaptation that remains to be elucidated (Fig. 5; Feldman et al., 2016).
Another example of TTX resistance is provided by newts (family Salamandridae). One or more substitutions associated with TTX resistance evolved among these amphibians, providing them with the ability to accumulate TTX from dietary sources or (symbiotic) microorganisms and to exploit it for defensive purposes (Hanifin & Gilly, 2015; Vaelli et al., 2020). Interestingly, all species show a single substitution (1532T/S) associated with TTX resistance in the Nav 1.4 channel (Fig. 5; Hanifin & Gilly, 2015). However, some newts show three additional substitutions (1247T, 1539S, and 1540D), conferring an increased level of TTX resistance and additionally deploying higher concentrations of TTX, and they are therefore more toxic to adversaries (Fig. 5; Hanifin & Gilly, 2015). Interestingly, a recent study revealed that TTX resistance in newts is not exclusively restricted to the Nav 1.4 channel but extends across the entire Nav gene family, which is driven by positive selection, relaxed constraints and gene conversion events (Gendreau et al., 2021).
The pufferfishes (family Tetraodontidae) are another group of animals that show TTX resistance. Different substitutions (407N/C, 1247T, and 1252P) evolved in Nav 1.4 channels conferring resistance to TTX (Fig. 5; Yotsu‐Yamashita et al., 2000; Venkatesh et al., 2005; Jost et al., 2008). Pufferfish exploit their resistant genotype for different ecological functions, enabling them to prey on TTX‐bearing species such as gastropods and echinoderms (Noguchi, Arakawa & Takatani, 2006b). Pufferfish thus accumulate high concentrations of dietary TTX in their organs, including liver, skin and ovaries (Noguchi et al., 2006b). Notably, captive pufferfish kept on a TTX‐free diet are not toxic, but when fed a TTX‐containing diet, they started to accumulate the toxins (Noguchi, Arakawa & Takatani, 2006a; Noguchi et al., 2006b). These accumulated TTXs are primarily exploited to deter predators of this otherwise innocuous fish. When threatened, pufferfish inflate their body, erecting spines and releasing TTX from the skin, thereby deterring predators (Kodama, Ogata & Sato, 1985).
TTX resistance has also evolved in some invertebrates. The greater blue‐ringed octopus (Hapalochlaena lunulata) and southern blue‐ringed octopus (H. maculosa) show five amino acid substitutions (1247T, 1248E, 1526M, 1539H, 1540S) associated with TTX resistance (Fig. 5; Geffeney et al., 2019; Whitelaw et al., 2020). Produced by symbiotic bacteria, this toxin is sequestered by these octopuses in their posterior salivary glands (Sheumack et al., 1978), and in their skin and other organs, so TTX is assumed to play a role in both prey capture as well as predator deterrence (Yotsu‐Yamashita, Mebs & Flachsenberger, 2007; Williams & Caldwell, 2009). Another example is the soft‐shell clam (Mya arenaria), which has evolved resistance towards STX, a toxin produced by algal blooms of dinoflagellates (Alexandrium spp.; Bricelj et al., 2005; Phillips et al., 2018). Softshell clam populations frequently exposed to blooms have evolved resistance underpinned by an aspartic acid substitution (764D; Fig. 5; Bricelj et al., 2005; Phillips et al., 2018). This resistance increases their capacity to accumulate these toxins, thereby enhancing the risk of paralytic shellfish poisoning after human consumption (Bricelj et al., 2005).
Another relatively underexplored strategy associated with guanidinium toxin resistance involves toxin‐binding proteins. The best‐studied example is saxiphilin, which is a soluble, well‐characterised STX‐binding protein (Yen et al., 2019). Saxiphilin has particularly been studied in frogs (Mahar et al., 1991; Yen et al., 2019), but STX‐binding activity has also been detected in other amphibians, reptiles, fish and some arthropods (Llewellyn, Bell & Moczydlowski, 1997). By contrast, STX‐binding activity has not been detected in any mammal or bird (Llewellyn et al., 1997). Other soluble guanidinium toxin‐binding proteins have been identified in pufferfish (Yotsu‐Yamashita et al., 2000, 2010), crabs (Lin et al., 2015) and gastropods (Hwang et al., 2007; Takati et al., 2007). Toxin‐binding proteins have been identified in plasma, haemolymph, and a diverse range of tissues [e.g., liver, stomach, kidney and heart (Mahar et al., 1991; Llewellyn et al., 1997; Yotsu‐Yamashita et al., 2010)]. Remarkably, such toxin‐binding proteins not only interact with STX and/or TTX but also with some other small‐molecule neurotoxins [e.g. batrachotoxin and decahydroquinoline (Mahar et al., 1991; Llewellyn et al., 1997; Abderemane‐Ali et al., 2021; O'Connell et al., 2021)]. Ultimately, toxin‐binding proteins such as saxiphilin can provide resistance against guanidinium toxins and are proposed to play a role in sequestration mechanisms that may facilitate autoresistance.
(7). Batrachotoxins
Batrachotoxin (BTX) is a steroidal alkaloid that targets voltage‐gated Na+ channels (Nav), causing irreversible depolarisation of muscles and nerves leading to paralysis, cardiac arrest, and other harmful effects (Albuquerque, Daly & Witkop, 1971). Resistance against BTX has evolved in poison dart frogs (family Dendrobatidae) and a few passerine bird species (e.g. Pitohui spp.).
Poison dart frogs are known for sequestering a variety of toxins including BTX (Daly, 1995). These frogs selectively sequester such toxins from their diet (Daly et al., 1994; Clark et al., 2005; Saporito et al., 2007b), and these are then used for chemical defence that is generally accompanied by vivid aposematic warning patterns (Summers & Clough, 2001). The golden poison dart frog (Phyllobates terribilis), which contains high BTX concentrations in its tissues, shows BTX resistance (Daly et al., 1980). Partial sequencing of the skeletal muscle Nav 1.4 channel suggested convergence of several substitutions conferring BTX resistance in poison frogs (Tarvin et al., 2016). Wang & Wang (2017) showed that one of these substitutions reduces BTX sensitivity and suggested that a single amino acid replacement confers BTX resistance. However, a more recent study failed to support the idea that Nav 1.4 channel mutations confer resistance in poison dart frogs (Abderemane‐Ali et al., 2021). It has been hypothesised that these frogs may have autoresistance based on toxin‐binding proteins (Abderemane‐Ali et al., 2021).
Certain passerine birds native to New Guinea (Pitohui spp. and Ifrita kowaldi) are known to sequester dietary BTXs (Dumbacher et al., 1992, 2004; Dumbacher, Spande & Daly, 2000). BTXs have been identified in significant concentrations across several organs (e.g. heart, skeletal muscle and liver), but the highest abundance is present in their skin and feathers (Dumbacher, Menon & Daly, 2009). Therefore, BTX is likely utilised for chemical defence against ectoparasites and/or predators (Dumbacher, 1999; Dumbacher et al., 2000). Despite the high concentrations found in these passerine birds, there are no resistance‐related modifications in the Nav channels (Nav 1.4 and Nav 1.5, respectively), which could suggest a comparable strategy using toxin‐binding proteins as proposed in poison dart frogs (Abderemane‐Ali et al., 2021).
(8). Cardiac glycosides
Cardiac glycosides (e.g. cardenolides and bufadienolides) are steroidal compounds that cause cardiotoxicity by inhibiting the sodium–potassium pump (Na+/K+‐ATPase; Schoner, 2002). A variety of animals and plants exploit cardiac glycosides as defensive poisons to deter potential predators (Botelho et al., 2019). As a response, many animals have evolved resistance underpinned by substitutions in the H1–H2 extracellular domain of the Na+/K+‐ATPase (Ujvari et al., 2015; Karageorgi et al., 2019). These substitutions are predominantly characterised by the replacement of neutral amino acids with charged amino acids, causing a reduction in binding affinity of cardiac glycosides (Ujvari et al., 2015). Cardiac glycoside resistance has evolved across the animal kingdom (Fig. 6).
Fig. 6.
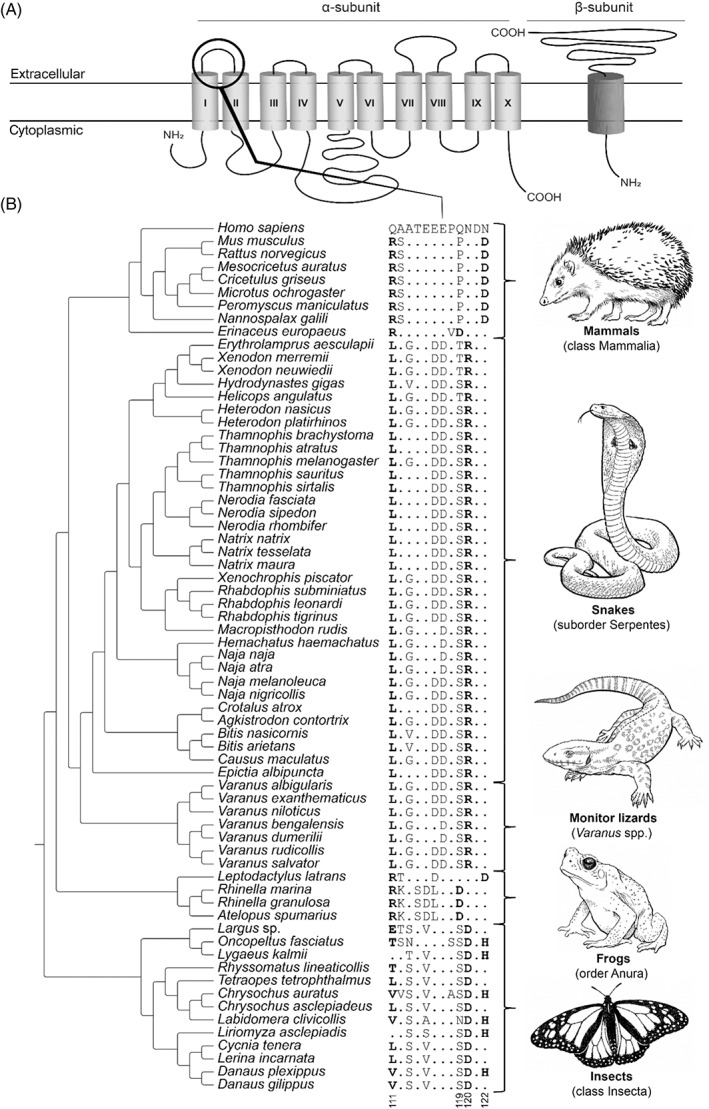
Convergent evolution of cardiac glycoside resistance in animals. (A) Protein topology of the sodium–potassium pump (Na+/K+‐ATPase). The black circle indicates the H1–H2 extracellular loop that is involved in cardiac glycoside binding. Structure based on Bagrov, Shapiro & Fedorova (2009). (B) Sequence alignment of the H1–H2 extracellular domain of Na+/K+‐ATPase. The reference amino acid sequence is from humans (Homo sapiens) and differences from this sequence are displayed for all other species. Substitutions associated with resistance are highlighted in bold. Tree topology based on TimeTree.org (Kumar et al., 2017) and taxon‐specific phylogenies (Dobler et al., 2012; Ujvari et al., 2015; Mohammadi et al., 2016). For sequence accession numbers, see Table S8.
Cardiac glycoside resistance evolved at least twice in mammals (Fig. 6). The European hedgehog (Erinaceus europaeus) shows two substitutions (111R and 119D), facilitating predation on cardiac glycoside‐containing toads, and could also be associated with the hedgehog's habit of anointing itself with the toad toxins (Brodie, 1977; Ewert & Traud, 1979; Ujvari et al., 2015). Also, some rodents (Muroidea) show resistance characterised by two amino acid substitutions (111R and 122D) – most likely associated with feeding on bufonid toads, insects or plants that contain cardiac glycosides (Ujvari et al., 2015).
Reptiles of the order Squamata have convergently evolved resistance mediated by identical substitutions in their Na+/K+‐ATPase (111L and 120R; Fig. 6; Ujvari et al., 2015; Mohammadi et al., 2016, 2018). This resistant genotype is widespread across the snake phylogeny, and likely contributes to their ability to prey on toads (Ujvari et al., 2015; Mohammadi et al., 2016). Interestingly, keelback snakes (Rhabdophis spp.) not only consume toxin‐bearing amphibians, but they also sequester these toxins in their nuchal glands for antipredator defence (Hutchinson et al., 2007; Mori et al., 2012). Additionally, varanid lizards (Varanus spp.) native to Africa and Asia that occasionally feed on toxin‐bearing toads also have evolved this resistant genotype (Ujvari et al., 2013). However, their Australian congeners, which originally did not share their habitat with bufonid toads, were found to have a reversal of this genotype (Ujvari et al., 2013, 2015). This lack of resistance in Australian varanid lizards resulted in major population declines after the introduction of the invasive cane toad (Rhinella marina), on which they occasionally predate (Madsen & Ujvari, 2009; Jolly, Shine & Greenlees, 2015).
Amphibians of the order Anura have convergently evolved resistance towards cardiac glycosides on two occasions (Ujvari et al., 2015). Bufonid toads (family Bufonidae) evolved resistance under strong positive selection based on two substitutions in Na+/K+‐ATPase (111R and 119D; Fig. 6) – presumably enabling the synthesis of their cardiac glycosides which they exploit to deter aggressors (Moore et al., 2009; Ujvari et al., 2015). By contrast, the South American spotted grassfrog (Leptodactylus latrans) evolved two substitutions in Na+/K+‐ATPase (111R and 122D; Fig. 6), allowing them to feed upon otherwise chemically protected bufonid prey (Ujvari et al., 2015; Mohammadi et al., 2021).
Another example of resistance in invertebrates is provided by insects (class Insecta). At least 21 lineages of insects have independently evolved the ability to feed on plants that contain cardiac glycosides and to sequester those toxins (Karageorgi et al., 2019). These feeding habits are facilitated by toxin resistance underpinned by combinations of two or three substitutions at residues 111, 120 and/or 122 in Na+/K+‐ATPase (111E, 111T, 111L, 111V, 120D, and/or 122H; Fig. 6; Holzinger, Frick & Wink, 1992; Dobler et al., 2012; Zhen et al., 2012; Ujvari et al., 2015; Karageorgi et al., 2019).
(9). Epibatidine
Epibatidine is an alkaloid toxin that targets neural‐type nicotinic acetylcholine receptors (nAChRs), causing muscle paralysis (Tarvin et al., 2017). This rarely observed form of resistance has evolved independently in distinct poison dart frog lineages. Poison dart frogs are known to exploit multiple toxins, including epibatidine, for chemical defence (Daly, 1995). Three distinct dendrobatid frog clades (Oophaga spp., Ameerega spp., and Epipedobates spp.) have convergently evolved resistance mediated by a single amino acid replacement to a cysteine in the β2 subunit of neural‐type nAChR (108C; Fig. 7; Tarvin et al., 2017). The cysteine residue has a sulphur‐containing side chain that is bulkier compared to the naïve serine residue. This substitution occurs at a key position for epibatidine binding and is therefore hypothesised to alter the epibatidine–receptor interaction (Tarvin et al., 2017). Additionally, another amino acid replacement to a valine (118V) in the Ameerega lineage has also been shown to reduce epibatidine binding (Fig. 7; Tarvin et al., 2017). The resistant phenotype might allow storage of epibatidine in the granular skin glands, which subsequently can be used for deterring adversaries.
Fig. 7.
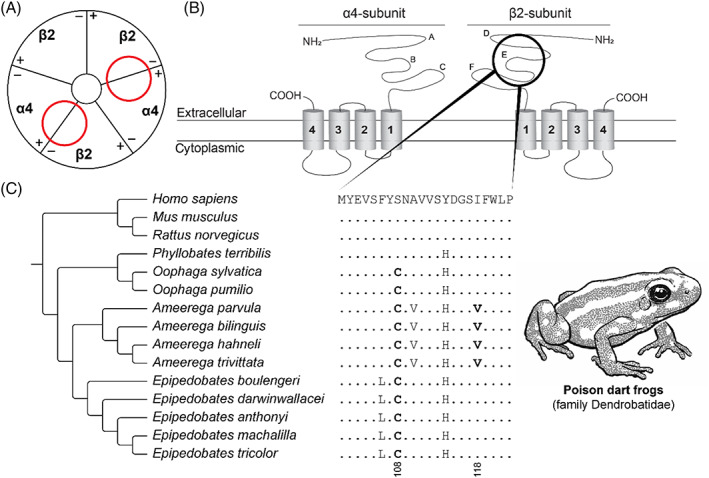
Convergent evolution of epibatidine resistance in poison dart frogs. (A) Schematic representation (based on Kini, 2019) of the neural‐type nicotinic acetylcholine receptor [nAChR; (α4)2(β2)3]. Red circles indicate the ligand‐binding domain of epibatidine in the nAChR. (B) Protein topology of the α4‐subunit and the β2‐subunit of the neural‐type nAChR. A–F indicate the loop structures at the extracellular domain in the respective subunits (Rahman et al., 2020). The black circle indicates the E‐loop involving the ligand‐binding domain of epibatidine. (C) Sequence alignment of the β2‐nAChR ligand‐binding domain. The reference amino acid sequence is from humans (Homo sapiens) and differences from this sequence are displayed for all other species. Substitutions associated with resistance are highlighted in bold. Tree topology based on Tarvin et al. (2017). For sequence accession numbers, see Table S9.
III. EVOLUTIONARY IMPLICATIONS OF TOXIN RESISTANCE
(1). When does toxin resistance evolve?
Prey and predator inevitably exert reciprocal selection pressures on each other. A prey species is under selection to avoid capture, whereas a predator is under selection to acquire the energy resources contained in the prey. In any predator–prey relationship involving a poisonous or venomous participant, this will translate into selection to evade these toxic armaments. Given sufficient reciprocal selection, this can in turn trigger an evolutionary response in the toxic participant to maintain the effectiveness of its weaponry, potentially leading to an evolutionary arms race.
The intensity and symmetry of selective forces between prey and predator are highly variable, depending on the importance of the prey species as a resource to the predator, and the importance of the predator as a cause of loss in fitness to the prey. For example, as we discussed above, some animals show a reversal of their resistant genotype in the absence of their toxic counterparts (Ujvari et al., 2015; Khan et al., 2020).
Life‐history theory predicts that toxin resistance is most likely to evolve when the poisonous or venomous opponent exerts strong selection, whether as prey or as predator. In predators of toxic prey, resistance is most likely to evolve when the predator is under strong selection to exploit an abundant but toxic food source. Examples include many reptiles that prey on toxic amphibians (Feldman et al., 2012; Ujvari et al., 2015), mammalian mesopredators feeding on venomous snakes (Drabeck et al., 2015, 2020) and grasshopper mice eating bark scorpions (Rowe et al., 2013). In prey species subject to predation by a venomous predator, prey resistance will most likely evolve if the predator is an important overall cause of mortality, e.g. sea kraits (Laticauda spp.), a lineage unrelated to other sea snakes, preying on moray eels (Gymnothorax spp. Heatwole & Poran, 1995) and rattlesnakes preying on North American ground squirrels and other rodents (de Wit, 1982; Holding et al., 2016a; Gibbs et al., 2020). In the latter example, reciprocal adaptation has been demonstrated, as rattlesnakes match their venom phenotype to the resistance profile of local prey to retain a selective advantage (Holding et al., 2016a; Margres et al., 2017), contrary to the predictions of the ‘Life‐Dinner Principle’ (Dawkins et al., 1979). That principle implies asymmetric selection, such that the prey is under greater selection pressure to escape capture, compared to the selection pressure on the predator to secure a meal.
By contrast, life‐history theory predicts that resistance is unlikely to evolve when selection pressure is low, for example: (i) when predation by a venomous predator is a relatively unimportant selective force for the prey because of the scarcity of encounters; (ii) a short temporal window of exposure exists (Marques et al., 2012); or (iii) when behavioural avoidance of toxic prey is more advantageous than evolving resistance (Smith, 1977; Brodie III, 1993; Portugal et al., 2016); see also Fig. 3).
Finally, it is also possible that resistance is most likely to evolve in situations where incremental increases in resistance confer an increasing selective advantage. Relatively low‐level resistance could be adaptive where prey toxicity varies geographically (Feldman et al., 2012). It could also be adaptive where different life stages differ in their toxin content, as appears to be the case in cane toads (Rhinella marina), for example (Hayes et al., 2009). Finally, low‐level resistance could be adaptive where partial failure of predatory envenomation is common, as seen in venomous snakes (Whitford et al., 2019). In summary, resistance is seen in many diverse ecological contexts and can be interpreted under a range of evolutionary scenarios. Despite the complex routes towards resistance, a few outcomes are repeatedly seen in unrelated lineages.
(2). Competing selection pressures and convergent evolution
Evolutionary trade‐offs usually come with a fitness disadvantage (Brodie III & Brodie Jr, 1999; Blanchard & Moreau, 2017; Hague et al., 2018). It is important that resistance modifications do not disrupt the physiology of the resistant animal. Therefore, there is a trade‐off between a functional target (e.g. binding site of the endogenous ligand) and the modifications enhancing toxin resistance. The emergence of similar adaptations is likely mediated by constraints on a functional target when subjected to similar selection pressures. Some examples of this can be seen across the animal kingdom.
Poison dart frog clades convergently evolved an identical substitution conferring epibatidine‐resistance, causing a decrease in acetylcholine sensitivity. As a result, this was then compensated by additional substitutions to maintain the receptor function (Tarvin et al., 2017). A similar phenomenon can be observed in α‐neurotoxin resistance. Multiple substitutions convergently evolved to reduce α‐neurotoxin binding but without compromising the amino acid residues vital for acetylcholine binding (Barchan et al., 1992; Khan et al., 2020). Similar convergent adaptations are found in multiple distinct colubrid snakes showing tetrodotoxin resistance. This trait is mediated by a functional trade‐off between ion channel function and tetrodotoxin insensitivity (Lee et al., 2011; Feldman et al., 2012). Cardiac glycoside resistance consistently evolved many times by two or three substitutions (respectively positioned at 111, 119, 120 or 122) – suggesting that these widespread genotypes also are constrained (Dobler et al., 2012; Ujvari et al., 2015; Karageorgi et al., 2019).
In summary, toxin resistance shows fascinating examples of non‐random and deterministic evolution mediated by constraints on sequence plasticity, while retaining receptor functionality. Thus, there may be a limited number of functional amino acid substitutions that reduce the binding affinity of toxins, even when different species are under similar selection pressures. This limited number of functional solutions available for adaptive evolution results in repeated funnelling of the same molecular pathway, leading to convergence.
(3). Origins of autoresistance in poisonous animals
Some animals are resistant to their own toxins, referred to as autoresistance. Venomous animals likely evolved resistance both for the prevention of self‐envenomation (autoresistance) and for defence against other venomous animals (see Section II.(4)). However, we suggest that this is a much more complicated evolutionary scenario in the case of poisons (e.g. tetrodotoxin, cardiac glycosides, batrachotoxin, epibatidine) – which has already been partially touched upon in previous literature (Saporito et al., 2012; Santos, Tarvin & O'Connell, 2016). We propose a scenario in which there was a three‐step evolution of resistance across phylogenetically distinct poisonous animals: (i) predator resistance, followed by (ii) sequestration of the toxin by the predator, and finally (iii) exploitation of the toxin for defence (Fig. 8).
Fig. 8.
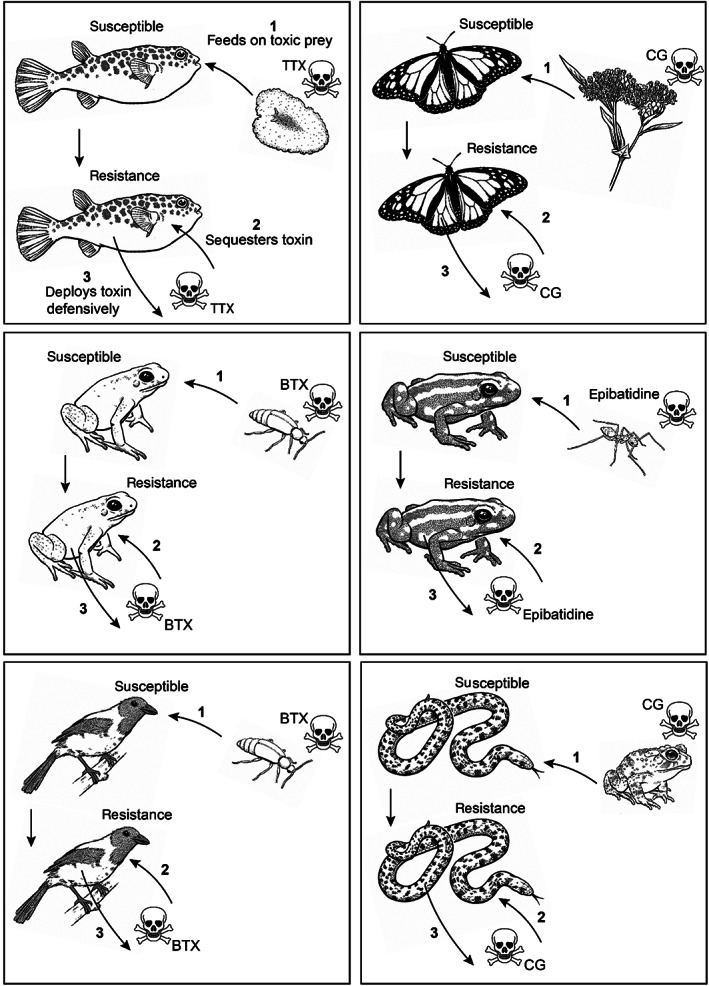
Hypothesised convergent evolutionary scenarios for autoresistance in poisonous animals. It is generally assumed that autoresistance is a self‐protection phenomenon. Here, we propose a three‐step evolution scenario for the origins of autoresistance: (1) predator resistance, followed by (2) sequestration of the toxin by the predator, and (3) exploitation of the toxin for defence. As this figure indicates, a similar three‐step process can be seen in diverse lineages, suggesting evolutionary convergence. The displayed examples include (A) pufferfish (family Tetraodontidae) feeding on TTX‐bearing flatworms, gastropods and echinoderms, (B) herbivorous insects feeding on CG‐containing plants, (C, D) poison dart frogs (family Dendrobatidae) feeding on toxic arthropods, (E) pitohui birds (Pitohui spp.) feeding on (among others) BTX‐bearing melyrid beetles, and (F) keelback snakes (Rhabdophis spp.) feeding on CG‐bearing anuran amphibians. BTX, batrachotoxin; CG, cardiac glycosides; TTX, tetrodotoxin.
Over the course of evolution, predation on a toxic species leads to frequent exposure to one or more specific toxins through generalised trophic interactions. In most cases, naïve predators feeding on highly toxic prey (such as TTX‐containing newts) are rapidly eliminated, with negative selection on the wild type thus favouring toxic prey avoidance. However, if variants that are capable of tolerating potent toxins exist in the population, then positive selection should favour the resistant phenotype, as this allows the predator to capitalise on abundant, often underutilised prey species. This then provides an evolutionary selection pressure on evolving, and maybe in some cases even maintaining, a less‐susceptible genotype. Interestingly, several animals (e.g. poison dart frogs and pufferfish) have been shown to be toxic only after the ingestion of a toxic diet, indicating that the toxins originated exogenously (Noguchi et al., 2006a; Saporito et al., 2007a; Yotsu‐Yamashita et al., 2012). Subsequently, the resistant phenotype increases the accumulation capacity of the toxin compared to non‐resistant animals (for example, as observed in clams; Bricelj et al., 2005). This phenomenon is not likely to occur in predators of venomous animals due to the proteinaceous nature of venom toxins that are easily metabolised after ingestion. By contrast, poisons (e.g. alkaloid or steroidal‐based toxins) are less easily metabolised and thus accumulate in the body. Ultimately, this enabled the exploitation of the accumulated toxins for defensive purposes in poisonous animals (reviewed in Savitzky et al., 2012). Therefore, we hypothesise that autoresistance primarily evolved as predator resistance rather than as an evolutionary driver itself, suggesting that multiple widespread taxa convergently evolved this three‐step evolution of resistance (Fig. 8).
IV. CONCLUSIONS
(1) Toxin resistance is an adaptive response seen at many trophic levels, underscoring how relatively simple adaptations can lead to solutions to complicated problems. This review has shown that molecular adaptations conferring toxin resistance have evolved repeatedly in diverse animal lineages, highlighting how different selection pressures can result in convergence at the molecular level.
(2) Convergent evolution involving toxin resistance can be explained by functional constraints. These constraints are mediated by a trade‐off between maintaining a functional molecular target and reducing toxin susceptibility. This trade‐off limits the functional solutions available for adaptive evolution.
(3) We propose a novel scenario for the evolution of ‘autoresistance’ in poisonous animals. We suggest that autoresistance did not evolve primarily as a form of self‐protection, but as a consequence of those animals feeding on toxic prey. This would imply that multiple diverse taxa convergently evolved this scenario.
(4) Similar selection pressures do not always lead to convergent molecular adaptations (as shown in certain bird, mammal, and reptile species). Molecular adaptations are only one of the ways in which organisms deal with toxins. We propose that some animals have elaborated or exploited existing behavioural or morphological traits, which may be exaptations, as alternative strategies to prevent intoxication in the first place.
(5) Toxin resistance is a phenomenon existing at the crossroads between molecular evolution, selection pressures and ecological interactions. The emergence of new or improved research technologies (e.g. ‐omics, functional assays and genetic modification techniques), combined with more robust ecological models, will provide opportunities to study novel and unexplored forms of resistance, as well as fundamental knowledge on how animals cope with direct or indirect exposure to toxic molecules in their environment. Toxin resistance is a compelling and multidisciplinary model system for studying evolutionary novelties with relevance in many branches of biology.
Supporting information
Table S1. Snake venom metalloproteinase inhibitors (SVMPIs) from mammalian serum, plasma or muscle tissue.
Table S2. Snake venom metalloproteinase inhibitors (SVMPIs) derived from snake serum, plasma or liver tissue.
Table S3. Phospholipase A2 inhibitors (PLA2Is) from snake serum, plasma or liver.
Table S4. Overview of the key sites on the muscle‐type nicotinic acetylcholine receptor (nAChR) and mutations that confer α‐neurotoxin resistance.
Table S5. Accession numbers of sequences included in Fig. 2.
Table S6. Accession numbers of sequences included in Fig. 4.
Table S7. Accession numbers of sequences included in Fig. 5.
Table S8. Accession numbers of sequences included in Fig. 6.
Table S9. Accession numbers of sequences included in Fig. 7.
V. ACKNOWLEDGEMENTS
M. A. K. and M. K. R. were supported by Elise Mathilde Fonds of Leids Universiteits Fonds (grant number 6113/21‐6‐16), Academy Ecology Funds of Royal Netherlands Academy of Arts and Sciences (grant number 713/18015), Higher Education Commission, Islamabad, Pakistan; R. J. H. was supported by the University of Queensland International PhD scholarship fund; and R. M. K. was supported by Ministry of Education, Singapore (grant number MOE2017‐T2‐1‐045). All line drawings of animals and plants were created by Sven Ballinger.
Jory van Thiel and Muzaffar A. Khan contributed equally to this work.
Contributor Information
Jory van Thiel, Email: joryvthiel@gmail.com.
Michael K. Richardson, Email: m.k.richardson@biology.leidenuniv.nl.
REFERENCES
References marked with asterisk have been cited within the supporting information
- Abderemane‐Ali, F. , Rossen, N. D. , Kobiela, M. E. , Craig, R. A. II , Garrison, C. E. , Chen, Z. , Colleran, C. M. , O'Connell, L. A. , Du Bois, J. , Dumbacher, J. P. & Minor, D. L. Jr. (2021). Evidence that toxin resistance in poison birds and frogs is not rooted in sodium channel mutations and may rely on “toxin sponge” proteins. Journal of General Physiology 153, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuquerque, E. X. , Daly, J. W. & Witkop, B. (1971). Batrachotoxin: chemistry and pharmacology. Science 172, 995–1002. [DOI] [PubMed] [Google Scholar]
- Alexander, G. J. & Maritz, B. (2010). Bitis arietans arietans: partial resistance to Naja venom. African Herp News 50, 34–36. [Google Scholar]
- Almeida‐Santos, S. M. , Antoniazzi, M. M. , Sant'Anna, O. A. & Jared, C. (2000). Predation by the opossum Didelphis marsupialis on the rattlesnake Crotalus durissus . Current Herpetology 19, 1–9. [Google Scholar]
- Aráoz, R. , Molgó, J. & Tandeau de Marsac, N. (2010). Neurotoxic cyanobacterial toxins. Toxicon 56, 813–828. [DOI] [PubMed] [Google Scholar]
- Arbuckle, K. , Rodriguez de la Vega, R. C. & Casewell, N. R. (2017). Coevolution takes the sting out of it: evolutionary biology and mechanisms of toxin resistance in animals. Toxicon 140, 118–131. [DOI] [PubMed] [Google Scholar]
- Asher, O. , Lupu‐Meiri, M. , Jensen, B. S. , Paperna, T. , Fuchs, S. & Oron, Y. (1998). Functional characterization of mongoose nicotinic acetylcholine receptor alpha‐subunit: resistance to alpha‐bungarotoxin and high sensitivity to acetylcholine. FEBS Letters 431, 411–414. [DOI] [PubMed] [Google Scholar]
- Bagrov, A. Y. , Shapiro, J. I. & Fedorova, O. V. (2009). Endogenous cardiotonic steroids: physiology, pharmacology, and novel therapeutic targets. Pharmacological Reviews 61, 9–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchan, D. , Kachalsky, S. , Neumann, D. , Vogel, Z. , Ovadia, M. , Kochva, E. & Fuchs, S. (1992). How the mongoose can fight the snake: the binding site of the mongoose acetylcholine receptor. Proceedings of the National Academy of Sciences of the United States of America 89, 7717–7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchan, D. , Ovadia, M. , Kochva, E. & Fuchs, S. (1995). The binding site of the nicotinic acetylcholine receptor in animal species resistant to α‐bungarotoxin. Biochemistry 34, 9172–9176. [DOI] [PubMed] [Google Scholar]
- Begg, C. M. , Begg, K. S. , Du Toit, J. T. & Mills, M. G. L. (2003). Sexual and seasonal variation in the diet and foraging behaviour of a sexually dimorphic carnivore, the honey badger (Mellivora capensis). Journal of Zoology 260, 301–316. [Google Scholar]
- Bell, G. (1982). Behavioral and ecological aspects of gleaning by a desert insectivorous bat Antrozous pallidus (Chiroptera: Vespertilionidae). Behavioral Ecology and Sociobiology 10, 217–223. [Google Scholar]
- Biardi, J. E. & Coss, R. G. (2011). Rock squirrel (Spermophilus variegatus) blood sera affects proteolytic and hemolytic activities of rattlesnake venoms. Toxicon 57, 323–331. [DOI] [PubMed] [Google Scholar]
- Biardi, J. , Ho, C. , Marcinczyk, J. & Nambiar, K. (2011). Isolation and identification of a snake venom metalloproteinase inhibitor from California ground squirrel (Spermophilus beecheyi) blood sera. Toxicon 58, 486–493. [DOI] [PubMed] [Google Scholar]
- Blanchard, B. D. & Moreau, C. S. (2017). Defensive traits exhibit an evolutionary trade‐off and drive diversification in ants. Evolution 71, 315–328. [DOI] [PubMed] [Google Scholar]
- * Borkow, G. , Gutierrez, J. M. & Ovadia, M. (1994). A potent antihemorrhagin in the serum of the non‐poisonous water snake Natrix tessellata: isolation, characterization and mechanism of neutralization. Biochimica et Biophysica Acta 1201, 482–490. [DOI] [PubMed] [Google Scholar]
- * Borkow, G. , Gutierrez, J. M. & Ovadia, M. (1995). Isolation, characterization and mode of neutralization of a potent antihemorrhagic factor from the serum of the snake Bothrops asper . Biochimica et Biophysica Acta 1245, 232–238. [DOI] [PubMed] [Google Scholar]
- Botelho, A. F. M. , Pierezan, F. , Soto‐Blanco, B. & Melo, M. M. (2019). A review of cardiac glycosides: structure, toxicokinetics, clinical signs, diagnosis and antineoplastic potential. Toxicon 158, 63–68. [DOI] [PubMed] [Google Scholar]
- Bricelj, V. M. , Connell, L. , Konoki, K. , Macquarrie, S. P. , Scheuer, T. , Catterall, W. A. & Trainer, V. L. (2005). Sodium channel mutation leading to saxitoxin resistance in clams increases risk of PSP. Nature 434, 763–767. [DOI] [PubMed] [Google Scholar]
- Brodie, E. D. (1977). Hedgehogs use toad venom in their own defence. Nature 268, 627–628. [Google Scholar]
- Brodie, E. D. (1990). Tetrodotoxin resistance in garter snakes: an evolutionary response of predators to dangerous prey. Evolution 44, 651–659. [DOI] [PubMed] [Google Scholar]
- Brodie, E. D. III (1993). Differential avoidance of coral snake banded patterns by free‐ranging avian predators in Costa Rica. Evolution 47, 227–235. [DOI] [PubMed] [Google Scholar]
- Brodie, E. D. III & Brodie, E. D. Jr. (1999). Costs of exploiting poisonous prey: evolutionary trade‐offs in a predator‐prey arms race. Evolution 53, 626–631. [DOI] [PubMed] [Google Scholar]
- Brodie, E. D. III & Brodie, E. D. Jr. (1999). Predator–prey arms races: asymmetrical selection on predators and prey may be reduced when prey are dangerous. Bioscience 49, 557–568. [Google Scholar]
- Brodie, E. D. III , Feldman, C. R. , Hanifin, C. T. , Motychak, J. E. , Mulcahy, D. G. , Williams, B. L. & Brodie, E. D. Jr. (2005). Parallel arms races between garter snakes and newts involving tetrodotoxin as the phenotypic interface of coevolution. Journal of Chemical Ecology 31, 343–356. [DOI] [PubMed] [Google Scholar]
- Casewell, N. R. , Wüster, W. , Vonk, F. J. , Harrison, R. A. & Fry, B. G. (2013). Complex cocktails: the evolutionary novelty of venoms. Trends in Ecology & Evolution 28, 219–229. [DOI] [PubMed] [Google Scholar]
- Casewell, N. R. , Petras, D. , Card, D. C. , Suranse, V. , Mychajliw, A. M. , Richards, D. , Koludarov, I. , Albulescu, L.‐O. , Slagboom, J. , Hempel, B.‐F. , Ngum, N. M. , Kennerley, R. J. , Brocca, J. L. , Whiteley, G. , Harrison, R. A. , Bolton, F. M. S. , et al. (2019). Solenodon genome reveals convergent evolution of venom in eulipotyphlan mammals. Proceedings of the National Academy of Sciences of the United States of America 116, 25745–25755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Catanese, J. J. & Kress, L. F. (1992). Isolation from opossum serum of a metalloproteinase inhibitor homologous to human alpha 1B‐glycoprotein. Biochemistry 31, 410–418. [DOI] [PubMed] [Google Scholar]
- Clark, V. C. , Raxworthy, C. J. , Rakotomalala, V. , Sierwald, P. & Fisher, B. L. (2005). Convergent evolution of chemical defense in poison frogs and arthropod prey between Madagascar and the Neotropics. Proceedings of the National Academy of Sciences of the United States of America 102, 11617–11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly, J. W. (1995). The chemistry of poisons in amphibian skin. Proceedings of the National Academy of Sciences of the United States of America 92, 9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly, J. W. , Myers, C. W. , Warnick, J. E. & Albuquerque, E. X. (1980). Levels of batrachotoxin and lack of sensitivity to its action in poison‐dart frogs (Phyllobates). Science 208, 1383–1385. [DOI] [PubMed] [Google Scholar]
- Daly, J. W. , Secunda, S. I. , Garraffo, H. M. , Spande, T. F. , Wisnieski, A. & Cover, J. F. (1994). An uptake system for dietary alkaloids in poison frogs (Dendrobatidae). Toxicon 32, 657–663. [DOI] [PubMed] [Google Scholar]
- Dawkins, R. , Krebs, J. R. , Maynard Smith, J. & Holliday, R. (1979). Arms races between and within species. Proceedings of the Royal Society of London. Series B. Biological Sciences 205, 489–511. [DOI] [PubMed] [Google Scholar]
- Dellisanti, C. , Yao, Y. , Stroud, J. C. , Wang, Z.‐Z. & Chen, L. (2007). Structural determinants for α‐neurotoxin sensitivity in muscle nAChR and their implications for the gating mechanism. Channels 1, 234–237. [DOI] [PubMed] [Google Scholar]
- Dobler, S. , Dalla, S. , Wagschal, V. & Agrawal, A. A. (2012). Community‐wide convergent evolution in insect adaptation to toxic cardenolides by substitutions in the Na,K‐ATPase. Proceedings of the National Academy of Sciences of the United States of America 109, 13040–13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabeck, D. H. , Dean, A. M. & Jansa, S. A. (2015). Why the honey badger don't care: convergent evolution of venom‐targeted nicotinic acetylcholine receptors in mammals that survive venomous snake bites. Toxicon 99, 68–72. [DOI] [PubMed] [Google Scholar]
- Drabeck, D. H. , Rucavado, A. , Hingst‐Zaher, E. , Cruz, Y. P. , Dean, A. M. & Jansa, S. A. (2020). Resistance of South American opossums to vWF‐binding venom C‐type lectins. Toxicon 178, 92–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, J. , Sanggaard, K. W. , Schauser, L. , Lauridsen, S. E. , Enghild, J. J. , Schierup, M. H. & Wang, T. (2017). Transcriptome analysis of the response of Burmese python to digestion. GigaScience 6, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumbacher, J. P. (1999). Evolution of toxicity in pitohuis: I. Effects of homobatrachotoxin on chewing lice (order Phthiraptera). The Auk 116, 957–963. [Google Scholar]
- Dumbacher, J. P. , Beehler, B. M. , Spande, T. F. , Garraffo, H. M. & Daly, J. W. (1992). Homobatrachotoxin in the genus Pitohui: chemical defense in birds? Science 258, 799–801. [DOI] [PubMed] [Google Scholar]
- Dumbacher, J. P. , Spande, T. F. & Daly, J. W. (2000). Batrachotoxin alkaloids from passerine birds: a second toxic bird genus (Ifrita kowaldi) from New Guinea. Proceedings of the National Academy of Sciences of the United States of America 97, 12970–12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumbacher, J. P. , Wako, A. , Derrickson, S. R. , Samuelson, A. , Spande, T. F. & Daly, J. W. (2004). Melyrid beetles (Choresine): a putative source for the batrachotoxin alkaloids found in poison‐dart frogs and toxic passerine birds. Proceedings of the National Academy of Sciences of the United States of America 101, 15857–15860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumbacher, J. P. , Menon, G. K. & Daly, J. W. (2009). Skin as a toxin storage organ in the endemic New Guinean genus Pitohui . The Auk 126, 520–530. [Google Scholar]
- Duran‐Riveroll, L. M. & Cembella, A. D. (2017). Guanidinium toxins and their interactions with voltage‐gated sodium ion channels. Marine Drugs 15, 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Estevão‐Costa, M. I. , Rocha, B. C. , de Alvarenga Mudado, M. , Redondo, R. , Franco, G. R. & Fortes‐Dias, C. L. (2008). Prospection, structural analysis and phylogenetic relationships of endogenous γ‐phospholipase A2 inhibitors in Brazilian Bothrops snakes (Viperidae, Crotalinae). Toxicon 52, 122–129. [DOI] [PubMed] [Google Scholar]
- Ewert, J. P. & Traud, R. (1979). Releasing stimuli for antipredator behaviour in the Common Toad Bufo bufo (L.). Behaviour 68, 170–180. [Google Scholar]
- * Farah, M. F. , One, M. , Novello, J. C. , Toyama, M. H. , Perales, J. , Moussatche, H. , Domont, G. B. , Oliveira, B. & Marangoni, S. (1996). Isolation of protein factors from opossum (Didelphis albiventris) serum which protect against Bothrops jararaca venom. Toxicon 34, 1067–1071. [DOI] [PubMed] [Google Scholar]
- Feldman, C. R. , Brodie, E. D. Jr. , Brodie, E. D. III & Pfrender, M. E. (2009). The evolutionary origins of beneficial alleles during the repeated adaptation of garter snakes to deadly prey. Proceedings of the National Academy of Sciences of the United States of America 106, 13415–13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, C. R. , Brodie, E. D. , Brodie, E. D. & Pfrender, M. E. (2010). Genetic architecture of a feeding adaptation: garter snake (Thamnophis) resistance to tetrodotoxin bearing prey. Proceedings of the Royal Society B: Biological Sciences 277, 3317–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, C. R. , Brodie, E. D. , Brodie, E. D. & Pfrender, M. E. (2012). Constraint shapes convergence in tetrodotoxin‐resistant sodium channels of snakes. Proceedings of the National Academy of Sciences of the United States of America 109, 4556–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, C. R. , Durso, A. M. , Hanifin, C. T. , Pfrender, M. E. , Ducey, P. K. , Stokes, A. N. , Barnett, K. E. , Brodie, E. D. III & Brodie, E. D. Jr. (2016). Is there more than one way to skin a newt? Convergent toxin resistance in snakes is not due to a common genetic mechanism. Heredity 116, 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraz, C. R. , Arrahman, A. , Xie, C. , Casewell, N. R. , Lewis, R. J. , Kool, J. & Cardoso, F. C. (2019). Multifunctional toxins in snake venoms and therapeutic implications: from pain to hemorrhage and necrosis. Frontiers in Ecology and Evolution 7, 1–19. [Google Scholar]
- * Fortes‐Dias, C. L. , Fonseca, B. C. B. , Kochva, E. & Diniz, C. R. (1991). Purification and properties of an antivenom factor from the plasma of the south American rattlesnake (Crotalus durissus terrificus). Toxicon 29, 997–1008. [DOI] [PubMed] [Google Scholar]
- * Fortes‐Dias, C. L. , Barcellos, C. J. & Estevão‐Costa, M. I. (2003). Molecular cloning of a γ‐phospholipase A2 inhibitor from Lachesis muta muta (the bushmaster snake). Toxicon 41, 909–917. [DOI] [PubMed] [Google Scholar]
- Fortes‐Dias, C. L. , Macedo, D. H. F. , Barbosa, R. P. , Souza‐Silva, G. & Ortolani, P. L. (2020). Identification and characterization of the first endogenous phospholipase A2 inhibitor from a non‐venomous tropical snake, boa constrictor (Serpentes: Boidae). Journal of Venomous Animals and Toxins Including Tropical Diseases 26, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry, B. G. , Roelants, K. , Champagne, D. E. , Scheib, H. , Tyndall, J. D. , King, G. F. , Nevalainen, T. J. , Norman, J. A. , Lewis, R. J. , Norton, R. S. , Renjifo, C. & de la Vega, R. C. (2009). The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annual Review of Genomics and Human Genetics 10, 483–511. [DOI] [PubMed] [Google Scholar]
- Garcia, V. E. & Perez, J. C. (1984). The purification and characterization of an antihemorrhagic factor in woodrat (Neotoma micropus) serum. Toxicon 22, 129–138. [DOI] [PubMed] [Google Scholar]
- Geffeney, S. , Brodie, E. D. & Ruben, P. C. (2002). Mechanisms of adaptation in a predator–prey arms race: TTX‐resistant sodium channels. Science 297, 1336–1339. [DOI] [PubMed] [Google Scholar]
- Geffeney, S. L. , Fujimoto, E. , Brodie, E. D. , Brodie, E. D. & Ruben, P. C. (2005). Evolutionary diversification of TTX‐resistant sodium channels in a predator–prey interaction. Nature 434, 759–763. [DOI] [PubMed] [Google Scholar]
- Geffeney, S. L. , Williams, B. L. , Rosenthal, J. J. C. , Birk, M. A. , Felkins, J. , Wisell, C. M. , Curry, E. R. & Hanifin, C. T. (2019). Convergent and parallel evolution in a voltage‐gated sodium channel underlies TTX‐resistance in the greater blue‐ringed octopus: Hapalochlaena lunulata . Toxicon 170, 77–84. [DOI] [PubMed] [Google Scholar]
- Gendreau, K. L. , Hornsby, A. D. , Hague, M. T. J. & McGlothlin, J. W. (2021). Gene conversion facilitates the adaptive evolution of self‐resistance in highly toxic newts. Molecular Biology and Evolution 38, 4077–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs, H. L. , Sanz, L. , Perez, A. , Ochoa, A. , Hassinger, A. T. B. , Holding, M. L. & Calvete, J. J. (2020). The molecular basis of venom resistance in a rattlesnake‐squirrel predator‐prey system. Molecular Ecology 29, 2871–2888. [DOI] [PubMed] [Google Scholar]
- Goetz, S. M. , Piccolomini, S. , Hoffman, M. , Bogan, J. , Holding, M. L. , Mendonça, M. T. & Steen, D. A. (2019). Serum‐based inhibition of pitviper venom by eastern indigo snakes (Drymarchon couperi). Biology Open 8, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hague, M. T. J. , Toledo, G. , Geffeney, S. L. , Hanifin, C. T. , Brodie, E. D. Jr. & Brodie, E. D. III (2018). Large‐effect mutations generate trade‐off between predatory and locomotor ability during arms race coevolution with deadly prey. Evolution Letters 2, 406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Hains, P. G. & Broady, K. W. (2000). Purification and inhibitory profile of phospholipase A2 inhibitors from Australian elapid sera. Biochemical Journal 346, 139–146. [PMC free article] [PubMed] [Google Scholar]
- Hanifin, C. T. & Gilly, W. F. (2015). Evolutionary history of a complex adaptation: tetrodotoxin resistance in salamanders. Evolution 69, 232–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, R. J. & Fry, B. G. (2021). Electrostatic resistance to alpha‐neurotoxins conferred by charge reversal mutations in nicotinic acetylcholine receptors. Proceedings of the Royal Society B: Biological Sciences 288, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, R. J. , Nekaris, K. A.‐I. & Fry, B. G. (2021). Monkeying around with venom: an increased resistance to α‐neurotoxins supports an evolutionary arms race between afro‐Asian primates and sympatric cobras. BMC Biology 19, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, R. A. , Crossland, M. R. , Hagman, M. , Capon, R. J. & Shine, R. (2009). Ontogenetic variation in the chemical defenses of cane toads (Bufo marinus): toxin profiles and effects on predators. Journal of Chemical Ecology 35, 391–399. [DOI] [PubMed] [Google Scholar]
- Heatwole, H. & Poran, N. S. (1995). Resistances of sympatric and allopatric eels to sea snake venoms. Copeia 1995, 136–147. [Google Scholar]
- Holding, M. L. , Biardi, J. E. & Gibbs, H. L. (2016a). Coevolution of venom function and venom resistance in a rattlesnake predator and its squirrel prey. Proceedings of the Royal Society B: Biological Sciences 283, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holding, M. L. , Drabeck, D. H. , Jansa, S. A. & Gibbs, H. L. (2016b). Venom resistance as a model for understanding the molecular basis of complex coevolutionary adaptations. Integrative and Comparative Biology 56, 1032–1043. [DOI] [PubMed] [Google Scholar]
- Holzinger, F. , Frick, C. & Wink, M. (1992). Molecular basis for the insensitivity of the monarch (Danaus plexippus) to cardiac glycosides. FEBS Letters 314, 477–480. [DOI] [PubMed] [Google Scholar]
- Hood, L. , Kronenberg, M. & Hunkapiller, T. (1985). T cell antigen receptors and the immunoglobulin supergene family. Cell 40, 225–229. [DOI] [PubMed] [Google Scholar]
- Hopp, B. H. , Arvidson, R. S. , Adams, M. E. & Razak, K. A. (2017). Arizona bark scorpion venom resistance in the pallid bat, Antrozous pallidus . PLoS One 12, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Huang, K.‐F. , Chow, L.‐P. & Chiou, S.‐H. (1999). Isolation and characterization of a novel proteinase inhibitor from the snake serum of Taiwan habu (Trimeresurus mucrosquamatus). Biochemical and Biophysical Research Communications 263, 610–616. [DOI] [PubMed] [Google Scholar]
- Hutchinson, D. A. , Mori, A. , Savitzky, A. H. , Burghardt, G. M. , Wu, X. , Meinwald, J. & Schroeder, F. C. (2007). Dietary sequestration of defensive steroids in nuchal glands of the Asian snake Rhabdophis tigrinus . Proceedings of the National Academy of Sciences of the United States of America 104, 2265–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, D. F. , Arakawa, O. , Saito, T. , Noguchi, T. , Simidu, U. , Tsukamoto, K. , Shida, Y. & Hashimoto, K. (1989). Tetrodotoxin‐producing bacteria from the blue‐ringed octopus Octopus maculosus . Marine Biology 100, 327–332. [Google Scholar]
- Hwang, P.‐A. , Tsai, Y.‐H. , Lin, H.‐P. & Hwang, D.‐F. (2007). Tetrodotoxin‐binding proteins isolated from five species of toxic gastropods. Food Chemistry 103, 1153–1158. [Google Scholar]
- Isbell, L. A. (2009). The Fruit, the Tree, and the Serpent. Harvard University Press, Cambridge, MA. [Google Scholar]
- Jansa, S. A. & Voss, R. S. (2011). Adaptive evolution of the venom‐targeted vWF protein in opossums that eat pitvipers. PLoS One 6, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, D. S. & Fenton, M. B. (2001). Individual and population‐level variability in diets of pallid bats (Antrozous Pallidus). Journal of Mammalogy 82, 362–373. [Google Scholar]
- Jolley, D. B. , Ditchkoff, S. S. , Sparklin, B. D. , Hanson, L. B. , Mitchell, M. S. & Grand, J. B. (2010). Estimate of herpetofauna depredation by a population of wild pigs. Journal of Mammalogy 91, 519–524. [Google Scholar]
- Jolly, C. J. , Shine, R. & Greenlees, M. J. (2015). The impact of invasive cane toads on native wildlife in southern Australia. Ecology and Evolution 5, 3879–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, M. D. , Crane, M. S. , Silva, I. M. S. , Artchawakom, T. , Waengsothorn, S. , Suwanwaree, P. , Strine, C. T. & Goode, M. (2020). Supposed snake specialist consumes monitor lizards: diet and trophic implications of king cobra feeding ecology. Ecology 101, 1–4. [DOI] [PubMed] [Google Scholar]
- Jones, L. , Harris, R. J. & Fry, B. G. (2021). Not goanna get me: mutations in the savannah monitor lizard (Varanus exanthematicus) nicotinic acetylcholine receptor confer reduced susceptibility to sympatric cobra venoms. Neurotoxicity Research 39, 1116–1122. [DOI] [PubMed] [Google Scholar]
- Jost, M. C. , Hillis, D. M. , Lu, Y. , Kyle, J. W. , Fozzard, H. A. & Zakon, H. H. (2008). Toxin‐resistant sodium channels: parallel adaptive evolution across a complete gene family. Molecular Biology and Evolution 25, 1016–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Jurgilas, P. B. , Neves‐Ferreira, A. G. , Domont, G. B. & Perales, J. (2003). PO41, a snake venom metalloproteinase inhibitor isolated from philander opossum serum. Toxicon 42, 621–628. [DOI] [PubMed] [Google Scholar]
- Kachalsky, S. G. , Jensen, B. S. , Barchan, D. & Fuchs, S. (1995). Two subsites in the binding domain of the acetylcholine receptor: an aromatic subsite and a proline subsite. Proceedings of the National Academy of Sciences of the United States of America 92, 10801–10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karageorgi, M. , Groen, S. C. , Sumbul, F. , Pelaez, J. N. , Verster, K. I. , Aguilar, J. M. , Hastings, A. P. , Bernstein, S. L. , Matsunaga, T. , Astourian, M. , Guerra, G. , Rico, F. , Dobler, S. , Agrawal, A. A. & Whiteman, N. K. (2019). Genome editing retraces the evolution of toxin resistance in the monarch butterfly. Nature 574, 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazandjian, T. D. , Petras, D. , Robinson, S. D. , van Thiel, J. , Greene, H. W. , Arbuckle, K. , Barlow, A. , Carter, D. A. , Wouters, R. M. , Whiteley, G. , Wagstaff, S. C. , Arias, A. S. , Albulescu, L. O. , Plettenberg Laing, A. , Hall, C. , Heap, A. , et al. (2021). Convergent evolution of pain‐inducing defensive venom components in spitting cobras. Science 371, 386–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kgaditse, M. M. (2016). The evolution and diversification of diet in elapids . Doctoral dissertation: University of the Witwatersrand.
- Khan, M. A. , Dashevsky, D. , Kerkkamp, H. , Kordis, D. , de Bakker, M. A. G. , Wouters, R. , van Thiel, J. , Op den Brouw, B. , Vonk, F. , Kini, R. M. , Nazir, J. , Fry, B. G. & Richardson, M. K. (2020). Widespread evolution of molecular resistance to snake venom alpha‐neurotoxins in vertebrates. Toxins 12, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kini, R. M. (2019). Toxins for decoding interface selectivity in nicotinic acetylcholine receptors. Biochemical Journal 476, 1515–1520. [DOI] [PubMed] [Google Scholar]
- Kodama, M. , Ogata, T. & Sato, S. (1985). External secretion of tetrodotoxin from puffer fishes stimulated by electric shock. Marine Biology 87, 199–202. [Google Scholar]
- * Kogaki, H. , Inoue, S. , Ikeda, K. , Samejima, Y. , Omori‐Satoh, T. & Hamaguchi, K. (1989). Isolation and fundamental properties of a phospholipase A2 inhibitor from the blood plasma of Trimeresurus flavoviridis . The Journal of Biochemistry 106, 966–971. [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. , Suleski, M. & Hedges, S. B. (2017). TimeTree: a resource for timelines, timetrees, and divergence times. Molecular Biology and Evolution 34, 1812–1819. [DOI] [PubMed] [Google Scholar]
- Layloo, I. , Smith, C. & Maritz, B. (2017). Diet and feeding in the cape cobra, Naja nivea . African Journal of Herpetology 66, 147–153. [Google Scholar]
- Lee, C. H. , Jones, D. K. , Ahern, C. , Sarhan, M. F. & Ruben, P. C. (2011). Biophysical costs associated with tetrodotoxin resistance in the sodium channel pore of the garter snake, Thamnophis sirtalis . Journal of Comparative Physiology A 197, 33–43. [DOI] [PubMed] [Google Scholar]
- Lenhart, P. A. , Mata‐Silva, V. & Johnson, J. D. (2010). Foods of the pallid bat, Antrozous pallidus (Chiroptera: Vespertilionidae), in the Chihuahuan Desert of Western Texas. The Southwestern Naturalist 55, 110–115. [Google Scholar]
- Lin, H. , Zhang, C. , Liao, J. , Yang, F. , Zhong, S. , Jiang, P. , Chen, X. & Nagashima, Y. (2015). Neutralizing effect of hemolymph from the shore crab, Thalamita crenata, on paralytic shellfish toxins. Toxicon 99, 51–57. [DOI] [PubMed] [Google Scholar]
- * Lizano, S. , Lomonte, B. , Fox, J. W. & Gutiérrez, J. M. (1997). Biochemical characterization and pharmacological properties of a phospholipase A2 myotoxin inhibitor from the plasma of the snake Bothrops asper . The Biochemical Journal 326, 853–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Lizano, S. , Angulo, Y. , Lomonte, B. , Fox, J. W. , Lambeau, G. , Lazdunski, M. & Gutiérrez, J. M. (2000). Two phospholipase A2 inhibitors from the plasma of Cerrophidion (Bothrops) godmani which selectively inhibit two different group‐II phospholipase A2 myotoxins from its own venom: isolation, molecular cloning and biological properties. The Biochemical Journal 346, 631–639. [PMC free article] [PubMed] [Google Scholar]
- Lizano, S. , Domont, G. & Perales, J. (2003). Natural phospholipase A2 myotoxin inhibitor proteins from snakes, mammals and plants. Toxicon 42, 963–977. [DOI] [PubMed] [Google Scholar]
- Llewellyn, L. E. , Bell, P. M. & Moczydlowski, E. G. (1997). Phylogenetic survey of soluble saxitoxin‐binding activity in pursuit of the function and molecular evolution of saxiphilin, a relative of transferrin. Proceedings of the Royal Society of London. Series B: Biological Sciences 264, 891–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas, A. & Stettenheim, P. (1972). Avian Anatomy‐Integument. US Department of Agriculture Handbook, Washington, DC. [Google Scholar]
- Mackessy, S. P. & Baxter, L. M. (2006). Bioweapons synthesis and storage: the venom gland of front‐fanged snakes. Zoologischer Anzeiger—A Journal of Comparative Zoology 245, 147–159. [Google Scholar]
- Madsen, T. & Ujvari, B. (2009). Increased mortality of naive varanid lizards after the invasion of non‐native cane toads (Bufo marinus). Herpetological Conservation and Biology 4, 248–251. [Google Scholar]
- Mahar, J. , Lukács, G. L. , Li, L. , Hall, S. & Moczydlowski, E. (1991). Pharmacological and biochemical properties of saxiphilin, a soluble saxitoxin‐binding protein from the bullfrog (Rana catesbeiana). Toxicon 29, 53–71. [DOI] [PubMed] [Google Scholar]
- Manjunatha Kini, R. (2003). Excitement ahead: structure, function and mechanism of snake venom phospholipase A2 enzymes. Toxicon 42, 827–840. [DOI] [PubMed] [Google Scholar]
- Margres, M. J. , Wray, K. P. , Hassinger, A. T. B. , Ward, M. J. , McGivern, J. J. , Moriarty Lemmon, E. , Lemmon, A. R. & Rokyta, D. R. (2017). Quantity, not quality: rapid adaptation in a polygenic trait proceeded exclusively through expression differentiation. Molecular Biology and Evolution 34, 3099–3110. [DOI] [PubMed] [Google Scholar]
- Maritz, B. , Alexander, G. J. & Maritz, R. A. (2019). The underappreciated extent of cannibalism and ophiophagy in African cobras. Ecology 100, 1–4. [DOI] [PubMed] [Google Scholar]
- Marques, O. A. V. , Martins, M. , Develey, P. F. , Macarrão, A. & Sazima, I. (2012). The golden lancehead Bothrops insularis (Serpentes: Viperidae) relies on two seasonally plentiful bird species visiting its Island habitat. Journal of Natural History 46, 885–895. [Google Scholar]
- Martinez, R. R. , Pérez, J. C. , Sánchez, E. E. & Campos, R. (1999). The antihemorrhagic factor of the Mexican ground squirrel (Spermophilus mexicanus). Toxicon 37, 949–954. [DOI] [PubMed] [Google Scholar]
- Martins, M. & Ermelinda Oliveira, M. (1998). Natural history of snakes in forests of the Manaus region, Central Amazonia, Brazil. Herpetological Natural History 6, 78–150. [Google Scholar]
- McGlothlin, J. W. , Chuckalovcak, J. P. , Janes, D. E. , Edwards, S. V. , Feldman, C. R. , Brodie, E. D. Jr. , Pfrender, M. E. & Brodie, E. D. III (2014). Parallel evolution of tetrodotoxin resistance in three voltage‐gated sodium channel genes in the garter snake Thamnophis sirtalis . Molecular Biology and Evolution 31, 2836–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlothlin, J. W. , Kobiela, M. E. , Feldman, C. R. , Castoe, T. A. , Geffeney, S. L. , Hanifin, C. T. , Toledo, G. , Vonk, F. J. , Richardson, M. K. , Brodie, E. D. Jr. , Pfrender, M. E. & Brodie, E. D. III (2016). Historical contingency in a multigene family facilitates adaptive evolution of toxin resistance. Current Biology 26, 1616–1621. [DOI] [PubMed] [Google Scholar]
- Mebs, D. , Omori‐Satoh, T. , Yamakawa, Y. & Nagaoka, Y. (1996). Erinacin, an antihaemorrhagic factor from the European hedgehog, Erinaceus europaeus . Toxicon 34, 1313–1316. [DOI] [PubMed] [Google Scholar]
- Miyazawa, K. & Noguchi, T. (2001). Distribution and origin of tetrodotoxin. Journal of Toxicology: Toxin Reviews 20, 11–33. [Google Scholar]
- Modahl, C. M. & Mackessy, S. P. (2019). Venoms of rear‐fanged snakes: new proteins and novel activities. Frontiers in Ecology and Evolution 7, 1–18. [Google Scholar]
- Mohammadi, S. , Gompert, Z. , Gonzalez, J. , Takeuchi, H. , Mori, A. & Savitzky, A. H. (2016). Toxin‐resistant isoforms of Na+/K+‐ATPase in snakes do not closely track dietary specialization on toads. Proceedings of the Royal Society B: Biological Sciences 283, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi, S. , Petschenka, G. , French, S. S. , Mori, A. & Savitzky, A. H. (2018). Snakes exhibit tissue‐specific variation in cardiotonic steroid sensitivity of Na+/K+‐ATPase. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology 217, 21–26. [DOI] [PubMed] [Google Scholar]
- Mohammadi, S. , Yang, L. , Harpak, A. , Herrera‐Álvarez, S. , del Pilar Rodríguez‐Ordoñez, M. , Peng, J. , Zhang, K. , Storz, J. F. , Dobler, S. , Crawford, A. J. & Andolfatto, P. (2021). Concerted evolution reveals co‐adapted amino acid substitutions in Na+K+‐ATPase of frogs that prey on toxic toads. Current Biology 31, 2530–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, D. J. , Halliday, D. C. , Rowell, D. M. , Robinson, A. J. & Keogh, J. S. (2009). Positive Darwinian selection results in resistance to cardioactive toxins in true toads (Anura: Bufonidae). Biology Letters 5, 513–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, A. , Burghardt, G. M. , Savitzky, A. H. , Roberts, K. A. , Hutchinson, D. A. & Goris, R. C. (2012). Nuchal glands: a novel defensive system in snakes. Chemoecology 22, 187–198. [Google Scholar]
- Mori, D. , Vyas, R. & Upadhyay, K. (2017). Breeding biology of the short‐toed Snake Eagle Circaetus gallicus . Indian Birds 12, 149–156. [Google Scholar]
- Munekiyo, S. M. & Mackessy, S. P. (2005). Presence of peptide inhibitors in rattlesnake venoms and their effects on endogenous metalloproteases. Toxicon 45, 255–263. [DOI] [PubMed] [Google Scholar]
- * Neves‐Ferreira, A. G. , Perales, J. , Fox, J. W. , Shannon, J. D. , Makino, D. L. , Garratt, R. C. & Domont, G. B. (2002). Structural and functional analyses of DM43, a snake venom metalloproteinase inhibitor from Didelphis marsupialis serum. Journal of Biological Chemistry 277, 13129–13137. [DOI] [PubMed] [Google Scholar]
- Noguchi, T. , Arakawa, O. & Takatani, T. (2006a). Toxicity of pufferfish Takifugu rubripes cultured in netcages at sea or aquaria on land. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics 1, 153–157. [DOI] [PubMed] [Google Scholar]
- Noguchi, T. , Arakawa, O. & Takatani, T. (2006b). TTX accumulation in pufferfish. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics 1, 145–152. [DOI] [PubMed] [Google Scholar]
- O'Connell, L. A. , LS50: Integrated Science Laboratory Course , O'Connell, J. D. , Paulo, J. A. , Trauger, S. A. , Gygi, S. P. & Murray, A. W. (2021). Rapid toxin sequestration modifies poison frog physiology. Journal of Experimental Biology 224, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Ohkura, N. , Inoue, S. , Ikeda, K. & Hayashi, K. (1994). Isolation and characterization of a phospholipase A2 inhibitor from the blood plasma of the Thailand cobra Naja naja kaouthia . Biochemical and Biophysical Research Communications 200, 784–788. [DOI] [PubMed] [Google Scholar]
- * Ohkura, N. , Okuhara, H. , Inoue, S. , Ikeda, K. & Hayashi, K. (1997). Purification and characterization of three distinct types of phospholipase A2 inhibitors from the blood plasma of the Chinese mamushi, Agkistrodon blomhoffii siniticus . The Biochemical Journal 325, 527–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Ohkura, N. , Kitahara, Y. , Inoue, S. , Ikeda, K. & Hayashi, K. (1999). Isolation and amino acid sequence of a phospholipase A2 inhibitor from the blood plasma of the sea krait, Laticauda semifasciata . The Journal of Biochemistry 125, 375–382. [DOI] [PubMed] [Google Scholar]
- Oliveira, M. E. & Santori, R. T. (1999). Predatory behavior of the opossum Didelphis albiventris on the pitviper Bothrops jararaca . Studies on Neotropical Fauna and Environment 34, 72–75. [Google Scholar]
- * Omori‐Satoh, T. , Sadahiro, S. , Ohsaka, A. & Murata, R. (1972). Purification and characterization of an antihemorrhagic factor in the serum of Trimeresurus flavoviridis, a crotalid. Biochimica et Biophysica Acta 285, 414–426. [DOI] [PubMed] [Google Scholar]
- Omori‐Satoh, T. , Yamakawa, Y. & Mebs, D. (2000). The antihemorrhagic factor, erinacin, from the European hedgehog (Erinaceus europaeus), a metalloprotease inhibitor of large molecular size possessing ficolin/opsonin P35 lectin domains. Toxicon 38, 1561–1580. [DOI] [PubMed] [Google Scholar]
- * Ovadia, M. (1978). Purification and characterization of an antihemorrhagic factor from the serum of the snake Vipera palaestinae . Toxicon 16, 661–672. [DOI] [PubMed] [Google Scholar]
- * Perales, J. , Munoz, R. & Moussatché, H. (1986). Isolation and partial characterization of a protein fraction from the opossum (Didelphis marsupialis) serum, with protecting property against the Bothrops jararaca snake venom. Anais da Academia Brasileira de Ciências 58, 155–162. [PubMed] [Google Scholar]
- Perales, J. , Moussatche, H. , Marangoni, S. , Oliveira, B. & Domont, G. B. (1994). Isolation and partial characterization of an anti‐bothropic complex from the serum of South American Didelphidae. Toxicon 32, 1237–1249. [DOI] [PubMed] [Google Scholar]
- Perales, J. , Villela, C. , Domont, G. B. , Choumet, V. , Saliou, B. , Moussatché, H. , Bon, C. & Faure, G. (1995). Molecular structure and mechanism of action of the crotoxin inhibitor from Crotalus durissus terrificus serum. European Journal of Biochemistry 227, 19–26. [DOI] [PubMed] [Google Scholar]
- Perez, J. C. , Pichyangkul, S. & Garcia, V. E. (1979). The resistance of three species of warm‐blooded animals to western diamondback rattlesnake (Crotalus atrox) venom. Toxicon 17, 601–607. [DOI] [PubMed] [Google Scholar]
- Phillips, J. M. , Bricelj, V. M. , Mitch, M. , Cerrato, R. M. , MacQuarrie, S. & Connell, L. B. (2018). Biogeography of resistance to paralytic shellfish toxins in softshell clam, Mya arenaria (L.), populations along the Atlantic coast of North America. Aquatic Toxicology 202, 196–206. [DOI] [PubMed] [Google Scholar]
- Pichyangkul, S. & Perez, J. C. (1981). Purification and characterization of a naturally occurring antihemorrhagic factor in the serum of the hispid cotton rat (Sigmodon hispidus). Toxicon 19, 205–215. [DOI] [PubMed] [Google Scholar]
- Pomento, A. M. , Perry, B. W. , Denton, R. D. , Gibbs, H. L. & Holding, M. L. (2016). No safety in the trees: local and species‐level adaptation of an arboreal squirrel to the venom of sympatric rattlesnakes. Toxicon 118, 149–155. [DOI] [PubMed] [Google Scholar]
- Poran, N. S. , Coss, R. G. & Benjamini, E. (1987). Resistance of California ground squirrels (Spermophilus beecheyi) to the venom of the northern Pacific rattlesnake (Crotalus viridis oreganus): a study of adaptive variation. Toxicon 25, 767–777. [DOI] [PubMed] [Google Scholar]
- Portugal, S. J. , Murn, C. P. , Sparkes, E. L. & Daley, M. A. (2016). The fast and forceful kicking strike of the secretary bird. Current Biology 26, 58–59. [DOI] [PubMed] [Google Scholar]
- Potier, S. , Lieuvin, M. , Pfaff, M. & Kelber, A. (2020). How fast can raptors see? Journal of Experimental Biology 223, 1–7. [DOI] [PubMed] [Google Scholar]
- * Qi, Z.‐Q. , Yonaha, K. , Tomihara, Y. & Toyama, S. (1995). Isolation of peptides homologous to domains of human α 1B‐glycoprotein from a mongoose antihemorrhagic factor. Toxicon 33, 241–245. [DOI] [PubMed] [Google Scholar]
- Rahman, M. M. , Teng, J. , Worrell, B. T. , Noviello, C. M. , Lee, M. , Karlin, A. , Stowell, M. H. B. & Hibbs, R. E. (2020). Structure of the native muscle‐type nicotinic receptor and inhibition by snake venom toxins. Neuron 106, 952–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings, N. D. & Barrett, A. J. (1990). Evolution of proteins of the cystatin superfamily. Journal of Molecular Evolution 30, 60–71. [DOI] [PubMed] [Google Scholar]
- Redford, K. H. & Peters, G. (1986). Notes on the biology and song of the red‐legged seriema (Cariama cristata). Journal of Field Ornithology 57, 261–269. [Google Scholar]
- Reeve, N. (1994). Hedgehogs. T. & A.D. Poyser, London. [Google Scholar]
- Rocha, S. L. , Lomonte, B. , Neves‐Ferreira, A. G. , Trugilho, M. R. , Junqueira‐de‐Azevedo Ide, L. , Ho, P. L. , Domont, G. B. , Gutierrez, J. M. & Perales, J. (2002). Functional analysis of DM64, an antimyotoxic protein with immunoglobulin‐like structure from Didelphis marsupialis serum. European Journal of Biochemistry 269, 6052–6062. [DOI] [PubMed] [Google Scholar]
- Rodrigues, C. F. B. , Serino‐Silva, C. , Morais‐Zani, K. D. , Kavazoi, V. K. , Carvalho, M. P. N. , Grego, K. F. , Chiarelli, T. , Tashima, A. K. , Toyama, M. H. & Tanaka‐Azevedo, A. M. (2020). BoaγPLI: structural and functional characterization of the gamma phospholipase A2 plasma inhibitor from the non‐venomous Brazilian snake boa constrictor . PLoS One 15, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues, C. F. B. , Zdenek, C. N. , Serino‐Silva, C. , de Morais‐Zani, K. , Grego, K. F. , Bénard‐Valle, M. , Neri‐Castro, E. , Alagón, A. , Tanaka‐Azevedo, A. M. & Fry, B. G. (2021). BoaγPLI from boa constrictor blood is a broad‐spectrum inhibitor of venom PLA2 pathophysiological actions. Journal of Chemical Ecology 47, 907–914. [DOI] [PubMed] [Google Scholar]
- Rowe, A. H. & Rowe, M. P. (2008). Physiological resistance of grasshopper mice (Onychomys spp.) to Arizona bark scorpion (Centruroides exilicauda) venom. Toxicon 52, 597–605. [DOI] [PubMed] [Google Scholar]
- Rowe, A. H. , Xiao, Y. , Scales, J. , Linse, K. D. , Rowe, M. P. , Cummins, T. R. & Zakon, H. H. (2011). Isolation and characterization of CvIV4: a pain inducing alpha‐scorpion toxin. PLoS One 6, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe, A. H. , Xiao, Y. , Rowe, M. P. , Cummins, T. R. & Zakon, H. H. (2013). Voltage‐gated sodium channel in grasshopper mice defends against bark scorpion toxin. Science 342, 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos, J. C. , Tarvin, R. D. & O'Connell, L. A. (2016). A review of chemical defense in poison frogs (Dendrobatidae): ecology, pharmacokinetics, and autoresistance. In Chemical Signals in Vertebrates 13 (eds Schulte B. A., Goodwin T. E. and Ferkin M. H.), pp. 305–337. Springer International Publishing, Cham. [Google Scholar]
- Saporito, R. A. , Donnelly, M. A. , Jain, P. , Martin Garraffo, H. , Spande, T. F. & Daly, J. W. (2007a). Spatial and temporal patterns of alkaloid variation in the poison frog Oophaga pumilio in Costa Rica and Panama over 30 years. Toxicon 50, 757–778. [DOI] [PubMed] [Google Scholar]
- Saporito, R. A. , Donnelly, M. A. , Norton, R. A. , Garraffo, H. M. , Spande, T. F. & Daly, J. W. (2007b). Oribatid mites as a major dietary source for alkaloids in poison frogs. Proceedings of the National Academy of Sciences of the United States of America 104, 8885–8890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saporito, R. A. , Donnelly, M. A. , Spande, T. F. & Garraffo, H. M. (2012). A review of chemical ecology in poison frogs. Chemoecology 22, 159–168. [Google Scholar]
- Savitzky, A. H. , Mori, A. , Hutchinson, D. A. , Saporito, R. A. , Burghardt, G. M. , Lillywhite, H. B. & Meinwald, J. (2012). Sequestered defensive toxins in tetrapod vertebrates: principles, patterns, and prospects for future studies. Chemoecology 22, 141–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schendel, V. , Rash, L. D. , Jenner, R. A. & Undheim, E. A. B. (2019). The diversity of venom: the importance of behavior and venom system morphology in understanding its ecology and evolution. Toxins 11, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoner, W. (2002). Endogenous cardiac glycosides, a new class of steroid hormones. European Journal of Biochemistry 269, 2440–2448. [DOI] [PubMed] [Google Scholar]
- Shen, H. , Zhou, Q. , Pan, X. , Li, Z. , Wu, J. & Yan, N. (2017). Structure of a eukaryotic voltage‐gated sodium channel at near‐atomic resolution. Science 355, 1–12. [DOI] [PubMed] [Google Scholar]
- Sheumack, D. D. , Howden, M. E. , Spence, I. & Quinn, R. J. (1978). Maculotoxin: a neurotoxin from the venom glands of the octopus Hapalochlaena maculosa identified as tetrodotoxin. Science 199, 188–189. [DOI] [PubMed] [Google Scholar]
- Shine, R. (1991). Australian Snakes: A Natural History. Reed, Sydney. [Google Scholar]
- Shine, R. , Branch, W. R. , Webb, J. K. , Harlow, P. S. , Shine, T. & Keogh, J. S. (2007). Ecology of cobras from Southern Africa. Journal of Zoology 272, 183–193. [Google Scholar]
- * Shioi, N. , Deshimaru, M. & Terada, S. (2014). Structural analysis and characterization of new small serum proteins from the serum of a venomous snake (Gloydius blomhoffii). Bioscience, Biotechnology, and Biochemistry 78, 410–419. [DOI] [PubMed] [Google Scholar]
- Silva, A. N. , Nunes, R. , Estrela, D. C. , Malafaia, G. & Castro, A. L. S. (2016). Behavioral repertoire of the poorly known red‐legged seriema, Cariama cristata (Cariamiformes: Cariamidae). Revista Brasileira de Ornitologia 24, 73–79. [Google Scholar]
- Smith, S. M. (1977). Coral‐snake pattern recognition and stimulus generalisation by naive great kiskadees (Aves: Tyrannidae). Nature 265, 535–536. [Google Scholar]
- Smith, S. N. , Jones, M. D. , Marshall, B. M. , Waengsothorn, S. , Gale, G. A. & Strine, C. T. (2021). Native Burmese pythons exhibit site fidelity and preference for aquatic habitats in an agricultural mosaic. Scientific Reports 11, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares, A. M. , Rodrigues, V. M. , Borges, M. H. , Andriao‐Escarso, S. H. , Cunha, O. A. , Homsi‐Brandeburgo, M. I. & Giglio, J. R. (1997). Inhibition of proteases, myotoxins and phospholipases A2 from Bothrops venoms by the heteromeric protein complex of Didelphis albiventris opossum serum. IUBMB Life 43, 1091–1099. [DOI] [PubMed] [Google Scholar]
- * Soares, A. M. , Marcussi, S. , Stábeli, R. G. , França, S. C. , Giglio, J. R. , Ward, R. J. & Arantes, E. C. (2003). Structural and functional analysis of BmjMIP, a phospholipase A2 myotoxin inhibitor protein from Bothrops moojeni snake plasma. Biochemical and Biophysical Research Communications 302, 193–200. [DOI] [PubMed] [Google Scholar]
- Soong, T. W. & Venkatesh, B. (2006). Adaptive evolution of tetrodotoxin resistance in animals. Trends in Genetics 22, 621–626. [DOI] [PubMed] [Google Scholar]
- Storz, J. F. (2016). Causes of molecular convergence and parallelism in protein evolution. Nature Reviews Genetics 17, 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhsaker, T. T. & McKey, D. (1975). Two cusimanse mongooses attack a black cobra. Journal of Mammalogy 56, 721–722. [Google Scholar]
- Stuart, C. T. (1983). Food of the large grey mongoose Herpestes ichneumon in the south‐west Cape Province. South African Journal of Zoology 18, 401–403. [Google Scholar]
- Summers, K. & Clough, M. E. (2001). The evolution of coloration and toxicity in the poison frog family (Dendrobatidae). Proceedings of the National Academy of Sciences of the United States of America 98, 6227–6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takacs, Z. , Wilhelmsen, K. C. & Sorota, S. (2001). Snake α‐neurotoxin binding site on the Egyptian cobra (Naja haje) nicotinic acetylcholine receptor is conserved. Molecular Biology and Evolution 18, 1800–1809. [DOI] [PubMed] [Google Scholar]
- Takacs, Z. , Wilhelmsen, K. C. & Sorota, S. (2004). Cobra (Naja spp.) nicotinic acetylcholine receptor exhibits resistance to Erabu Sea snake (Laticauda semifasciata) short‐chain alpha‐neurotoxin. Journal of Molecular Evolution 58, 516–526. [DOI] [PubMed] [Google Scholar]
- Takati, N. , Mountassif, D. , Taleb, H. , Lee, K. & Blaghen, M. (2007). Purification and partial characterization of paralytic shellfish poison‐binding protein from Acanthocardia tuberculatum . Toxicon 50, 311–321. [DOI] [PubMed] [Google Scholar]
- Tanaka, K. & Mori, A. (2000). Literature survey on predators of snakes in Japan. Current Herpetology 19, 97–111. [Google Scholar]
- * Tanizaki, M. M. , Kawasaki, H. , Suzuki, K. & Mandelbaum, F. R. (1991). Purification of a proteinase inhibitor from the plasma of Bothrops jararaca (jararaca). Toxicon 29, 673–681. [DOI] [PubMed] [Google Scholar]
- Tarvin, R. D. , Santos, J. C. , O'Connell, L. A. , Zakon, H. H. & Cannatella, D. C. (2016). Convergent substitutions in a sodium channel suggest multiple origins of toxin resistance in poison frogs. Molecular Biology and Evolution 33, 1068–1080. [DOI] [PubMed] [Google Scholar]
- Tarvin, R. D. , Borghese, C. M. , Sachs, W. , Santos, J. C. , Lu, Y. , O'Connell, L. A. , Cannatella, D. C. , Harris, R. A. & Zakon, H. H. (2017). Interacting amino acid replacements allow poison frogs to evolve epibatidine resistance. Science 357, 1261–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasoulis, T. & Isbister, G. K. (2017). A review and database of snake venom proteomes. Toxins 9, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thwin, M. M. , Gopalakrishnakone, P. , Kini, R. M. , Armugam, A. & Jeyaseelan, K. (2000). Recombinant antitoxic and antiinflammatory factor from the nonvenomous snake Python reticulatus: phospholipase A2 inhibition and venom neutralizing potential. Biochemistry 39, 9604–9611. [DOI] [PubMed] [Google Scholar]
- * Tomihara, Y. , Yonaha, K. , Nozaki, M. , Yamakawa, M. , Kamura, T. & Toyama, S. (1987). Purification of three antihemorrhagic factors from the serum of a mongoose (Herpestes edwardsii). Toxicon 25, 685–689. [DOI] [PubMed] [Google Scholar]
- Tomihara, Y. , Yonaha, K. , Nozaki, M. , Yamakawa, M. , Kawamura, Y. , Kamura, T. & Toyama, S. (1988). Purification of an antihemorrhagic factor from the serum of the non‐venomous snake Dinodon semicarinatus . Toxicon 26, 420–423. [DOI] [PubMed] [Google Scholar]
- Ujvari, B. , Mun, H. C. , Conigrave, A. D. , Bray, A. , Osterkamp, J. , Halling, P. & Madsen, T. (2013). Isolation breeds naivety: Island living robs Australian varanid lizards of toad‐toxin immunity via four‐base‐pair mutation. Evolution 67, 289–294. [DOI] [PubMed] [Google Scholar]
- Ujvari, B. , Casewell, N. R. , Sunagar, K. , Arbuckle, K. , Wüster, W. , Lo, N. , O'Meally, D. , Beckmann, C. , King, G. F. , Deplazes, E. & Madsen, T. (2015). Widespread convergence in toxin resistance by predictable molecular evolution. Proceedings of the National Academy of Sciences of the United States of America 112, 11911–11916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaelli, P. M. , Theis, K. R. , Williams, J. E. , O'Connell, L. A. , Foster, J. A. & Eisthen, H. L. (2020). The skin microbiome facilitates adaptive tetrodotoxin production in poisonous newts. eLife 9, 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente, R. H. , Dragulev, B. , Perales, J. , Fox, J. W. & Domont, G. B. (2001). BJ46a, a snake venom metalloproteinase inhibitor. Isolation, characterization, cloning and insights into its mechanism of action. European Journal of Biochemistry 268, 3042–3052. [DOI] [PubMed] [Google Scholar]
- Valente, R. H. , Luna, M. S. , de Oliveira, U. C. , Nishiyama‐Junior, M. Y. , Junqueira‐de‐Azevedo, I. L. , Portes‐Junior, J. A. , Clissa, P. B. , Viana, L. G. , Sanches, L. , Moura‐da‐Silva, A. M. , Perales, J. & Yamanouye, N. (2018). Bothrops jararaca accessory venom gland is an ancillary source of toxins to the snake. Journal of Proteomics 177, 137–147. [DOI] [PubMed] [Google Scholar]
- Venkatesh, B. , Lu, S. Q. , Dandona, N. , See, S. L. , Brenner, S. & Soong, T. W. (2005). Genetic basis of tetrodotoxin resistance in pufferfishes. Current Biology 15, 2069–2072. [DOI] [PubMed] [Google Scholar]
- Voss, R. S. (2013). Opossums (Mammalia: Didelphidae) in the diets of Neotropical pitvipers (Serpentes: Crotalinae): evidence for alternative coevolutionary outcomes? Toxicon 66, 1–6. [DOI] [PubMed] [Google Scholar]
- Walker, A. A. , Robinson, S. D. , Paluzzi, J.‐P. V. , Merritt, D. J. , Nixon, S. A. , Schroeder, C. I. , Jin, J. , Goudarzi, M. H. , Kotze, A. C. , Dekan, Z. , Sombke, A. , Alewood, P. F. , Fry, B. G. , Epstein, M. E. , Vetter, I. & King, G. F. (2021). Production, composition, and mode of action of the painful defensive venom produced by a limacodid caterpillar, Doratifera vulnerans . Proceedings of the National Academy of Sciences of the United States of America 118, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S.‐Y. & Wang, G. K. (2017). Single rat muscle Na+ channel mutation confers batrachotoxin autoresistance found in poison‐dart frog Phyllobates terribilis . Proceedings of the National Academy of Sciences of the United States of America 114, 10491–10496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, R. M. & Vick, J. A. (1977). Resistance of the opossum (Didelphis virginiana) to envenomation by snakes of the family Crotalidae. Toxicon 15, 29–32. [DOI] [PubMed] [Google Scholar]
- Whitelaw, B. L. , Cooke, I. R. , Finn, J. , da Fonseca, R. R. , Ritschard, E. A. , Gilbert, M. T. P. , Simakov, O. & Strugnell, J. M. (2020). Adaptive venom evolution and toxicity in octopods is driven by extensive novel gene formation, expansion, and loss. Gigascience 9, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford, M. D. , Freymiller, G. A. , Higham, T. E. & Clark, R. W. (2019). Determinants of predation success: how to survive an attack from a rattlesnake. Functional Ecology 33, 1099–1109. [Google Scholar]
- Wilcox, J. (2015). Implications of predation by wild pigs on native vertebrates: a case study. California Fish and Game 101, 72–77. [Google Scholar]
- Williams, B. L. & Caldwell, R. L. (2009). Intra‐organismal distribution of tetrodotoxin in two species of blue‐ringed octopuses (Hapalochlaena fasciata and H. lunulata). Toxicon 54, 345–353. [DOI] [PubMed] [Google Scholar]
- Williams, B. L. , Brodie, E. D. & Brodie, E. D. (2004). A resistant predator and its toxic prey: persistence of newt toxin leads to poisonous (not venomous) snakes. Journal of Chemical Ecology 30, 1901–1919. [DOI] [PubMed] [Google Scholar]
- de Wit, C. A. (1982). Resistance of the prairie vole (Microtus ochrogaster) and the woodrat (Neotoma floridana), in Kansas, to venom of the Osage copperhead (Agkistrodon contortrix phaeogaster). Toxicon 20, 709–714. [DOI] [PubMed] [Google Scholar]
- * de Wit, C. A. & Westrom, B. R. (1987). Venom resistance in the hedgehog, Erinaceus europaeus: purification and identification of macroglobulin inhibitors as plasma antihemorrhagic factors. Toxicon 25, 315–323. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, Y. , Park, J. H. & Inouye, M. (2011). Toxin‐antitoxin systems in bacteria and archaea. Annual Review of Genetics 45, 61–79. [DOI] [PubMed] [Google Scholar]
- Yee, K. T. , Pitts, M. , Tongyoo, P. , Rojnuckarin, P. & Wilkinson, M. C. (2016). Snake venom metalloproteinases and their peptide inhibitors from Myanmar russell's viper venom. Toxins 9, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen, T.‐J. , Lolicato, M. , Thomas‐Tran, R. , Bois, J. D. & Minor, D. L. (2019). Structure of the saxiphilin:saxitoxin (STX) complex reveals a convergent molecular recognition strategy for paralytic toxins. Science Advances 5, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yotsu‐Yamashita, M. , Nishimori, K. , Nitanai, Y. , Isemura, M. , Sugimoto, A. & Yasumoto, T. (2000). Binding properties of (3)H‐PbTx‐3 and (3)H‐saxitoxin to brain membranes and to skeletal muscle membranes of puffer fish Fugu pardalis and the primary structure of a voltage‐gated Na(+) channel alpha‐subunit (fMNa1) from skeletal muscle of F. pardalis . Biochemical and Biophysical Research Communications 267, 403–412. [DOI] [PubMed] [Google Scholar]
- Yotsu‐Yamashita, M. , Mebs, D. & Flachsenberger, W. (2007). Distribution of tetrodotoxin in the body of the blue‐ringed octopus (Hapalochlaena maculosa). Toxicon 49, 410–412. [DOI] [PubMed] [Google Scholar]
- Yotsu‐Yamashita, M. , Yamaki, H. , Okoshi, N. & Araki, N. (2010). Distribution of homologous proteins to puffer fish saxitoxin and tetrodotoxin binding protein in the plasma of puffer fish and among the tissues of Fugu pardalis examined by western blot analysis. Toxicon 55, 1119–1124. [DOI] [PubMed] [Google Scholar]
- Yotsu‐Yamashita, M. , Gilhen, J. , Russell, R. W. , Krysko, K. L. , Melaun, C. , Kurz, A. , Kauferstein, S. , Kordis, D. & Mebs, D. (2012). Variability of tetrodotoxin and of its analogues in the red‐spotted newt, Notophthalmus viridescens (Amphibia: Urodela: Salamandridae). Toxicon 59, 257–264. [DOI] [PubMed] [Google Scholar]
- Youngman, N. J. , Llinas, J. & Fry, B. G. (2021). Evidence for resistance to coagulotoxic effects of Australian elapid snake venoms by sympatric prey (blue tongue skinks) but not by predators (monitor lizards). Toxins 13, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zancolli, G. & Casewell, N. R. (2020). Venom systems as models for studying the origin and regulation of evolutionary novelties. Molecular Biology and Evolution 37, 2777–2790. [DOI] [PubMed] [Google Scholar]
- Zhen, Y. , Aardema, M. L. , Medina, E. M. , Schumer, M. & Andolfatto, P. (2012). Parallel molecular evolution in an herbivore community. Science 337, 1634–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, L. & Huang, C. (2016). Isolation and biochemical characterization of a gamma‐type phospholipase A2 inhibitor from Macropisthodon rudis snake serum. Toxicon 122, 1–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Snake venom metalloproteinase inhibitors (SVMPIs) from mammalian serum, plasma or muscle tissue.
Table S2. Snake venom metalloproteinase inhibitors (SVMPIs) derived from snake serum, plasma or liver tissue.
Table S3. Phospholipase A2 inhibitors (PLA2Is) from snake serum, plasma or liver.
Table S4. Overview of the key sites on the muscle‐type nicotinic acetylcholine receptor (nAChR) and mutations that confer α‐neurotoxin resistance.
Table S5. Accession numbers of sequences included in Fig. 2.
Table S6. Accession numbers of sequences included in Fig. 4.
Table S7. Accession numbers of sequences included in Fig. 5.
Table S8. Accession numbers of sequences included in Fig. 6.
Table S9. Accession numbers of sequences included in Fig. 7.


