Abstract
Endoplasmic reticulum (ER) stress and apoptosis play a critical role in liver injury. Endoplasmic reticulum oxidoreductase 1α (ERO1α) is an oxidase that exists in the luminal side of the ER membrane, participating in protein folding and secretion and inhibiting apoptosis, but the underlying mechanism on liver injury induced by homocysteine (Hcy) remains obscure. In this study, hyperhomocysteinemia (HHcy) mice model was established in cbs +/− mice by feeding a high‐methionine diet for 12 weeks; and cbs +/− mice fed with high‐methionine diet exhibited more severe liver injury compared to cbs +/+ mice. Mechanistically, we found that Hcy promoted ER stress and apoptosis of hepatocytes and thereby aggravated liver injury through inhibiting ERO1α expression; accordingly, overexpression of ERO1α remarkably alleviated ER stress and apoptosis of hepatocytes induced by Hcy. Epigenetic modification analysis revealed that Hcy significantly increased levels of DNA methylation and H3 lysine 9 dimethylation (H3K9me2) on ERO1α promoter, which attributed to upregulated DNA methyltransferase 1 (DNMT1) and G9a, respectively. Further study showed that DNMT1 and G9a cooperatively regulated ERO1α expression in hepatocytes exposed to Hcy. Taken together, our work demonstrates that Hcy activates ER stress and apoptosis of hepatocytes by downregulating ERO1α expression via cooperation between DNMT1 and G9a, which provides new insight into the mechanism of Hcy‐induced ER stress and apoptosis of hepatocytes in liver injury.
Keywords: apoptosis, DNMT1, endoplasmic reticulum stress, ERO1α, G9a, homocysteine
Abbreviations
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- CBS
cystathionine beta‐synthase
- ChIP
chromatin immunoprecipitation
- DMSO
dimethyl sulfoxide
- DNMT1
DNA methyltransferase 1
- DNMTs
DNA methyltransferases
- ER
endoplasmic reticulum
- ERO1α
endoplasmic reticulum oxidoreductase 1α
- H3K9me2
H3 lysine 9 dimethylation
- Hcy
homocysteine
- HE
hematoxylin‐eosin
- HHcy
hyperhomocysteinemia
- HMTs
histone methyltransferase
- NA
nanaomycin A
- nMS‐PCR
Nested methylation‐specific‐polymerase chain reaction
- PBS
phosphate buffer saline
- PCR
polymerization chain reaction
- PMSF
phenylmethanesulfonyl fluoride
- qRT‐PCR
quantitative real‐time PCR
- SAM
S‐adenosyl‐methionine
- SDS‐PAGE
sulfate‐polyacrylamide gel electrophoresis
- TEM
transmission electron microscopy
- TF‐3
theaflavin‐3
- UPR
unfolded protein response
1. INTRODUCTION
As a sulfur‐containing amino acid, homocysteine (Hcy) can induce oxidative stress and endoplasmic reticulum (ER) stress through methionine cycle‐related trans‐sulfuration pathway (Majumder et al., 2018). Abnormal level of Hcy often results in multiple severe diseases, including liver disease, atherosclerosis, and renal failure (Cheng et al., 2017; Medici et al., 2010; Xu et al., 2019). It has been reported that Hcy induces hepatic dysfunction through eliciting ER stress and activating the unfolded protein response (UPR) (Ai et al., 2017), and sustained ER stress and UPR activation promote cell apoptosis. However, the underlying mechanism that regarding the involvement of ER stress and apoptosis in Hcy‐induced liver injury remains unclear.
Endoplasmic reticulum oxidoreductase 1α (ERO1α) is a glycosylated flavoenzyme that is located on the luminal side of the ER membrane, which plays an important role in catalyzing the formation of protein disulfide bonds and ER redox homeostasis (Ramming & Appenzeller‐Herzog, 2012; Rao et al., 2015). Contribution of ERO1α to protein folding and restoring the homeostasis of cells under the condition of stress has been reported in pichia pastoris and proinsulin (Wright et al., 2013). Apart from its role in normal cellular functions, ERO1α oxidizes PDI to generate oxidative stress from molecular oxygen that forms H2O2, which results in a hyperoxidizing environment within the ER leading to increased cell death (Yan et al., 2020). Recent studies have shown that ERO1α plays an important role in ER stress‐induced CHOP‐dependent apoptosis in macrophages (Tanaka et al., 2016). Nevertheless, there was little available information about whether ERO1α involves in ER stress and apoptosis of liver injury induced by Hcy.
Besides the trans‐sulfuration pathway, Hcy also plays a role in the trans‐methylation cycle, in which a methyl group is transferred to macromolecules (Ehrlich, 2009). This process is related with epigenetic changes including DNA methylation and histone methylation. Generally, DNA methylation inhibits transcription, and loss of methylation (hypomethylation) activates gene expression (Schmitz et al., 2019). Increasing evidence have indicated that liver injury is either caused or impacted by abnormal methylation (Fromenty, 2020). In addition to DNA methylation, other study also reported that significantly less levels of H3K9 di‐methylation on LXRa and ERO1α gene promoter regions lead to dramatic elevation of lipogenesis and ER stress in the liver of mice with high‐fat diet (Shukla et al., 2015). In the cell interior, epigenetic modifications usually regulate gene expression through cooperative or antagonistic interactions with each other (Corso‐Diaz et al., 2018). It has been reported that p16 silencing is simultaneously characterized by DNA hypermethylation and H3K9 hypermethylation in gastric cancer (Bearzatto et al., 2002). When treated with 5‐Aza‐dC, an antagonist of DNA methylation, H3K9 dimethylation at the p16 hypermethylated promoter was also reduced (Zhang et al., 2012). Therefore, it is of significance to further elucidate the relationship between DNA methylation and histone methylation in regulating ERO1α expression during liver injury induced by Hcy.
In this study, we demonstrate that the abnormal expression of ERO1α in Hcy‐induced ER stress and apoptosis of hepatocytes is regulated by DNA methylation and H3K9me2. Our results reveal a novel molecular mechanism for the contribution of ERO1α to ER stress and apoptosis of hepatocytes induced by Hcy. Undoubtedly, a deep understanding of ERO1α‐regulating mechanism in Hcy‐induced liver injury will be beneficial for the potential ERO1α‐targeted clinical therapy of liver disease.
2. MATERIALS AND METHODS
2.1. Animals
The 8–10 weeks male cystathionine beta‐synthase (CBS) heterozygous knockout mice (cbs +/− ) were purchased from Jackson Laboratory, and housed in a controlled environment on 12:12 h light‐dark cycle. The mice were fed with regular mice chow and water ad libitum. To induce HHcy, all mice were fed chow diet plus 2.0% methionine for 12 weeks. The treatment and experimental protocol of mice followed the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978). All animal experiments procedures were performed according to guidelines approved by the Institutional Animal Care and Use Committee at the Ningxia medical university Laboratory Animal Center (Ethics approval number is NYDWZX‐2018‐083). Hematoxylin‐eosin (HE) staining was used to stain tissues in accordance with the instructions of the reagent manufacturer.
2.2. Cell culture
Hepatocyte cell line HL‐7702 was obtained from the Japanese Collection of Research Bioresources. The cells were cultured in 1640 plus 10% serum and 1% antibiotics. When the cells were 70% confluent, serum was deprived for 24 h to synchronize cells, cells were then cultured with 5% fetal calf serum before treatment with 0, 50, 100, 200, and 500 μM Hcy (Sigma‐Aldrich). To compensate for the short half‐life of Hcy, the compound was replenished every 12 h.
2.3. Detect the serum levels of Hcy, alanine aminotransferase (ALT), and aspartate aminotransferase (AST)
Blood samples of mice were centrifuged at 5000g for 15 min to separate serum samples. The serum Hcy, ALT, and AST levels were assayed by the Cobas e411 automatic biochemical analyzer (Roche Diagnostics).
2.4. Transmission electron microscopy (TEM)
Biopsied liver tissues were cut into pieces (1 × 1 × 5 mm) and fixed with glutaraldehyde in 0.1 M phosphate buffer saline (PBS) (pH 7.2) at 4°C for 2 h. After washing with PBS, samples were post‐fixed in 1% osmium tetroxide for 1 h at room temperature followed by dehydration with alcohol. Tissues were then embedded and sectioned with a diamond knife, stained with 1% uranyl acetate and 1% lead citrate, and examined with a transmission electron microscope (JME‐1220; JEOL) at an acceleration voltage of 80 kV.
2.5. MTT assay
To evaluate hepatocyte viability, we seeded hepatocytes in 96‐well plates and examined them via 3‐(4,5‐dimethyl‐2‐thiazolyl)‐2,5‐diphenyl‐2H‐tetrazolium bromide (MTT) assay (Sigma‐Aldrich). Hepatocytes were then cultured in 500 µg/ml MTT solution for 4 h. We then removed the MTT solution and added 100 µl of dimethyl sulfoxide (DMSO) per well into the 96‐well plates. After being vibrated for 60 s, the absorbance of 450 nm was monitored via a fully functional microporous plate detector (BioTek).
2.6. Analysis of apoptosis
Hepatocyte apoptosis was detected using a commercial PE Annexin V Apoptosis Detection Kit I (BD Pharmingen) according to the manufacturer's instructions. After Hcy treatment, the cells were washed with ice‐cold PBS, and incubated with fluoresce in conjugated 7‐AAD and PE for 15 min, cell apoptosis was analyzed using a fluorescence‐activated cell sorting (FACS) flow cytometer equipped with the FACS talion data management system and Cell Quest software (Becton Dickinson).
2.7. Quantitative real‐time polymerase chain reaction (qRT‐PCR)
The total RNA was isolated using Trizol reagent (Invitrogen). And the RNA was reverse‐transcribed by the Revert Aid first‐strand complementary DNA (cDNA) Synthesis Kit (Fermentas, Carlsbad, California, USA). The primer sequences are listed in Table 1. The qRT‐PCR was carried out by applying an FTC3000 real‐time PCR detection system (Funglyn Biotech Inc.). The 2‐ΔΔCt method was used to perform the gene messenger RNA (mRNA) quantifications, GAPDH was used as an internal control gene to normalize the amount of transcript in each sample to correct for differences in the cDNAs used.
Table 1.
Primer sequences for qRT‐PCR
| Gene | Species | Primer sequence (5′→3′) |
|---|---|---|
| ERO1α | Mouse | Forward: 5′‐CCTCCACTCGGAAGGACTATC‐3′ |
| Reverse: 5′‐TTGTGTGTTCGCCTCTTGAC‐3′ |
Abbreviation: qRT‐PCR, quantitative real‐time polymerase chain reaction.
2.8. Western blot
Liver tissues or hepatocytes were lysed in a cold lysis buffer (Keygene) supplemented with phenylmethanesulfonyl fluoride (PMSF) at 4°C for 30 min. After separation by 12% sulfate‐polyacrylamide gel electrophoresis, the proteins were transferred to polyvinylidene difluoride membrane. The membrane was blocked at room temperature in phosphate‐buffered saline with Tween 20 buffer with 5% nonfat milk followed by incubation with primary antibody, including anti‐ERO1α (ab172954, Abcam), anti‐GRP78 (ab21685, Abcam), anti‐PERK (ab156919, Abcam), anti‐ATF6 (ab203119, Abcam), and anti‐XBP‐1 (ab37152, Abcam), anti‐DNMT1 (ab13537, Abcam), anti‐G9a (#3306, Cell Signaling Technology), anti‐H3K9me2 (ab176882, Abcam), anti‐H3 (ab1791, Abcam) at 4°C overnight. Blotting of β‐actin (bsm‐33036M, BIOSS) was used as a loading control. The membrane was washed three times and incubated with horseradish peroxidase‐conjugated secondary antibody (ZB‐2306, ZSGB‐BIO) for 2 h. After washing three times, the target proteins were detected using enhanced chemiluminescence solution (Beyotime Institute of Biotechnology), and the relative protein levels were determined by band densitometry using a gel imaging system (Bio‐Rad Laboratories, Inc.).
2.9. RNA interference
The vectors expressing small interfering RNA (siRNA) for ERO1α, DNMT1, and G9a were purchased from Genechem Co. Ltd. (Genechem Co. Ltd.). Hepatocytes were transfected with siRNA constructs using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instruction.
2.10. Construction of recombinant plasmid
Total mRNA was purified from hepatocytes and converted to cDNA by reverse transcription, ERO1α, DNMT1, and G9a fragments were amplified by PCR and cloned into pEGFP‐N1. Hepatocytes were transfected with pEGFP‐N1‐pERO1α, pEGFP‐N1‐pDNMT1, pEGFP‐N1‐pG9a when they are 60%–80% confluent using Lipofectamine 2000 (Invitrogen). Cells were used for further experiments in 48 h after transfection.
2.11. Reporter gene construction and promoter activity study
Fragments of the ERO1α promoter, a 1700 bp fragment (−1/−1700), a 1100 bp fragment (−1/−1100), a 900 bp fragment (−1/−900) and 600 bp fragment (−1/−600) were inserted into the luciferase reporter plasmid pGL3‐basic (Promega). Promoter activity of ERO1α constructs was examined in cultured HEK293T cells by transient transfection and luciferase assay. The ratio of luciferase activity to Renilla luciferase in each transfection was used to normalize luciferase activity. The data presented were obtained from three independent experiments.
2.12. Nested methylation‐specific polymerase chain reaction (nMS‐PCR)
Genomic DNA was isolated from the liver tissues and hepatocytes by using the Wizard® Genomic DNA Purification Kit (Promega). DNA denaturation and bisulfite conversion processes are integrated into one‐step by EZ DNA Methylation‐Gold™ Kit (ZYMO RESEARCH). nMS‐PCR consists of two‐step PCR amplifications after a standard sodium bisulfite DNA modification. The first step of nMS‐PCR used an outer primer pair set that does not contain any CpG. The second‐step PCR was carried out with conventional PCR primers. Primers used for the nMS‐PCR assays are listed in Table 2. The PCR products were separated by 2% agarose gel containing ethidium bromide. DNA bands were visualized with ultraviolet light, and methylation was calculated by the formula: Methylation% = methylation/(methylation + unmethylation) × 100%.
Table 2.
ERO1α primer sequences for nMS‐PCR
| Gene | Species | Primer Sequence (5′→3′) |
|---|---|---|
| Outer primer | Mouse | Forward: 5′‐GTTTTGGGAATTGGAAGGATT‐3′ |
| Reverse: 5′‐AAAAAAAATAAAACAAATTATTAAACTTAC‐3′ | ||
| Methylation primer | Mouse | Forward: 5′‐ACTAACCTACAATAATACGATCGCA‐3′ |
| Reverse: 5′‐AGTTTGGGTAATATGG TGAAATTTC‐3′ | ||
| Unmethylation primer | Mouse | Forward: 5′‐ACTAACCTACAATAAT ACAATCACA‐3′ |
| Reverse: 5′‐AGTTTGGGTAATAT GGTGAAATTTC‐3′ |
Abbreviations: ERO1α, Endoplasmic reticulum oxidoreductase 1α; nMS‐PCR, nested methylation‐specific polymerase chain reaction.
2.13. MassARRAY methylation analysis
For every cleaved CpG site, the intensity of a pair of mass signals, one representing methylated and/or another one representing unmethylated DNA, was recorded and interpreted by MassARRAY EpiTYPER software. The relative methylation status was estimated by dividing the peak intensity, or area of the methylated DNA, by the sum of the intensities or areas of the methylated and unmethylated components. The ratios between methylated and unmethylated DNA were obtained as quantity of methylation for further analysis.
2.14. Chromatin immunoprecipitation (ChIP) assays
ChIP assays were performed according to the protocol of the manufacturer (Millipore). Antibodies against H3K9me2 and ERO1α (Abcam Biotech Inc.) were used for immunoprecipitate chromatin assay. Primers were designed to detect the proximal promoter region of ERO1α by PCR as described above. The Forward primer sequence: 5′‐CGCTGCTGGGCGCTC TTCCTGTCTC‐3′ and the reverse primer sequence: 5′‐AGAGTGGGAGCGGGTCAT‐3′. Nonimmune IgG was used as a control to measure nonspecific background intensity in immunoprecipitation.
2.15. Statistical analysis
GraphPad Prism 6.0 software was used for statistical analysis. All data are presented as mean ± SD from at least three independent experiments. Single parameters between two groups were compared by the paired Student's t test. One‐way analysis of variance was used to compare the means of multiple groups, followed by Newman–Keuls test. p < 0 .05 was considered statistically significant.
3. RESULTS
3.1. Hcy aggravates liver injury by promoting ER stress and hepatocytes apoptosis
To investigate the mechanism by which Hcy induces liver injury, cbs +/ − mice were fed with high‐methionine diet to establish an animal model of HHcy. Successful establishment of the model was confirmed by the elevation of serum Hcy levels in cbs +/ − mice (Figure 1a). As alanine aminotransferase (ALT) and aspartate aminotransferase (AST) are the most specific indicators of hepatocellular necrosis to evaluate the degree of liver injury in clinic (Torruellas et al., 2014), we compared serum levels of ALT and AST in cbs +/− and cbs +/+ mice fed with high‐methionine diet. As shown in Figure 1b, the levels of AST and ALT in serum of cbs +/− mice with HHcy were much higher than that in cbs +/+ mice. The H&E staining also showed a more considerable mass of fat vacuoles or deposition in hepatic tissues of cbs +/+ mice with HHcy, as well as ballooning and hydropic degeneration of hepatocytes (Figure 1c), indicating the causal effect of HHcy on liver injury. Since Hcy is a sulfur‐containing amino acid with reducing power, we next studied whether ER stress involves in HHcy‐induced liver injury. The ER status was observed using transmission electron microscopy (TEM), which showed an obvious swell and deregulation of ER with expansion of ER lumen, formation of vacuole, and ecclasis of ribosomes from rough ER (indicated by the red arrow), accompanied by serious pyknotic nucleus, in the liver tissues of cbs +/− mice with HHcy (Figure 1d). Consistently, ER stress‐associated proteins including GRP78, ATF6, PERK, and XBP‐1 were upregulated in the liver tissues from cbs +/ − mice with HHcy (Figure 1e). Next, hepatocytes were treated with different concentration of Hcy for further confirmation. As shown in Figure 1f,g, Hcy treatment significantly inhibited viability of hepatocytes while promoted cell apoptosis, especially at 100 μM. Simultaneously, protein levels of GRP78, ATF6, PERK, and XBP‐1 were obviously increased in hepatocytes with Hcy treatment (Figure 1h). Taken together, these results suggest that Hcy induces ER stress and apoptosis in hepatocytes to aggravate liver injury.
Figure 1.
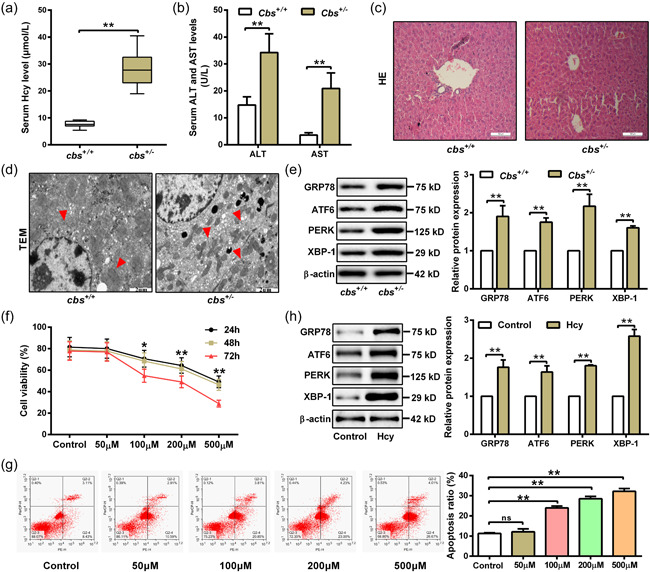
Homocysteine (Hcy) promotes endoplasmic reticulum (ER) stress and apoptosis of hepatocytes leading to liver injury. (a and b) Serum levels of Hcy, aspartate aminotransferase (AST) and alanine transaminase (ALT) in cbs +/+ and cbs +/ − mice fed with high‐methionine diet were measured by automatic biochemical analyzer (n = 6). (c) Liver injury was evaluated by hematoxylin and eosin (H&E) staining of liver slices from cbs +/+ and cbs +/ − mice. Scale bar, 100 μM. (d) Transmission electron microscopy (TEM) was performed to observe endoplasmic reticulum in liver slices from cbs +/+ and cbs +/‐ mice. Red arrows indicate hyperplasia of rough endoplasmic reticulum in hepatocytes. Scale bar, 2 μM. (e) Western blot was used to measure GRP78, ATF6, PERK, and XBP‐1 protein expression in liver tissue of cbs +/+ and cbs +/ − mice (n = 6). (f) Cell viability was detected by MTT assay in hepatocytes treated with Hcy at indicated concentrations (0, 50, 100, 200, and 500 μM) for 24, 48, and 72 h (n = 3). (g) Apoptosis rate was measured by flow cytometry after treating hepatocytes with different concentration of Hcy (0, 50, 100, 200, and 500 μM) for 48 h (n = 3). (h) The protein expression of GRP78, ATF6, PERK, and XBP‐1 were detected by western blot after treating hepatocytes with 100 μM Hcy for 48 h (n = 3). Data represents mean ± SD, **p <0 .01. MTT, 3‐(4,5‐dimethyl‐2‐thiazolyl)‐2,5‐diphenyl‐2H‐tetrazolium bromide; SD, standard deviation
3.2. Hcy promotes ER stress and apoptosis of hepatocytes through inhibiting ERO1α expression
As a pivotal ER sulfhydryl oxidase, ERO1α has been reported to be induced by reductive stress, such as hypoxia and dithiothreitol stimuli (Seervi et al., 2013). To explore whether ERO1α involves in Hcy‐induced ER stress and apoptosis in hepatocytes, we firstly detected the expression of ERO1α in liver tissues of cbs +/− and cbs +/+ mice fed with high‐methionine diet. The result showed that both mRNA and protein levels of ERO1α were obviously decreased in cbs +/‐ mice with HHcy (Figure 2a). Similar results were also observed in hepatocytes treated with Hcy (Figure 2b). Next, ERO1α was knocked down or overexpressed in hepatocytes to further confirm the role of ERO1α in ER stress and apoptosis induced by Hcy (Figure 2c,d). As presented in Figure 2e, knockdown of ERO1α remarkably increased the expression of ER stress‐associated proteins in hepatocytes treated with Hcy, including GRP78, ATF6, PERK, and XBP‐1. In contrary, overexpression of ERO1α could alleviate Hcy‐induced ER stress according to the alteration of ER stress‐associated proteins levels. Meanwhile, the results of flow cytometry showed that knockdown of ERO1α promoted hepatocyte apoptosis, while overexpression of ERO1α had an opposite effect (Figure 2f). These data indicate that Hcy induces ER stress and apoptosis of hepatocytes through downregulation of ERO1α.
Figure 2.
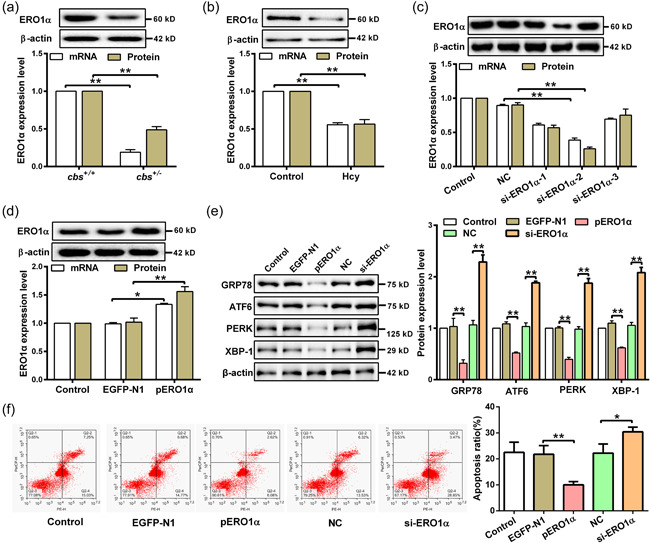
Hcy induces ER stress and apoptosis by downregulation of ERO1α in hepatocyte. (a and b) Western blot and qRT‐PCR were used to detect the mRNA and protein expression of ERO1α in the liver tissue of cbs +/+ and cbs +/ − mice (n = 6) and hepatocytes treated with 100 μM Hcy (n = 3). (c) The expression of ERO1α was examined by qRT‐PCR and western blot in hepatocytes transfected with siRNAs targeting ERO1α (si‐ERO1α‐1/2/3) (n = 3). (d) ERO1α expression in hepatocytes transfected with ERO1α‐overexpressed plasmids (pERO1α) or negative control vector (EGFP‐N1) was detected by qRT‐PCR and western blot (n = 3). (e) The protein expression of GRP78, ATF6, PERK, and XBP‐1 were measured by western blot in hepatocytes transfected with pERO1α or si‐ERO1α in the presence of Hcy (n = 3). (f) Flow cytometry was used to detect the apoptosis rate of hepatocytes transfected with pERO1α or si‐ERO1α in the presence of Hcy (n = 3). Data represents mean ± SD, *p < 0.05, **p < 0 .01. ER, endoplasmic reticulum; ERO1α, endoplasmic reticulum oxidoreductase 1α; Hcy, homocysteine; qRT‐PCR, quantitative real‐time polymerase chain reaction; SD, standard deviation
3.3. Hcy downregulates ERO1α through DNMT1‐mediated hypermethylation on ERO1α promoter
DNA methylation plays a key role in regulating gene expression. To get insight into the underlying mechanism of ERO1α downregulation in hepatocytes with Hcy treatment, we analyzed the sequence of ERO1α promoter with the MethPrimer. As shown in Figure 3a, three CpG island lying from −2000 to −1 at the promoter of ERO1α relative to the TSS was found, meaning that ERO1α promoter has the potential to be methylated, which may alter its transcriptional activity. To support this hypothesis, several fragments (−1 to −600, −1 to −900, −1 to −1100, −1 to −1700) of ERO1α promoter were inserted into the firefly luciferase vector pGL3 respectively, and luciferase activity assay was then performed to measure their transcriptional activity. As shown in Figure 3b, the region from −1 to −1100 of ERO1α promoter had the highest activity, suggesting that this region is predominant for maintaining high transcriptional activity of ERO1α. Subsequently, DNA methylation levels of ERO1α promoter were detected by nMS‐PCR. The results showed that methylation levels of ERO1α promoter in liver tissue of cbs +/ − mice with HHcy or hepatocytes treated with Hcy were noticeably increased as compared to their respective controls, implying that DNA methylation is responsible for regulation of ERO1α expression (Figure 3c,d). Given DNA methylation in mammals is catalyzed by DNA methyltransferases (DNMTs), including DNMT1, DNMT3a, and DNMT3b (Moore et al., 2013), we then treated hepatocytes with specific inhibitors (DC‐05, TF‐3, and NA) of DNMT1, DNMT3a, and DNMT3b to clarify which enzyme is responsible for Hcy‐induced transcriptional repression of ERO1α. Among them, we found that treatment of DC‐05, the DNMT1 inhibitor, could significantly upregulate ERO1α expression, while no differences of ERO1α expression were observed in hepatocytes treated with TF‐3 and NA (Figure 3e). This data suggests that DNMT1 might play a role in regulating ERO1α expression in response to Hcy. Correspondingly, DNMT1 expression was obviously enhanced in liver tissue of cbs +/ − mice with HHcy and hepatocytes treated with Hcy (Figure 3f,g). To determine whether DNMT1 involves in the downregulation of ERO1α induced by Hcy, the expression of ERO1α was measured after overexpression or silence of DNMT1 in hepatocytes (Figure 3h,i). As expected, overexpression of DNMT1 significantly increased the methylation level of ERO1α promoter while decreased its expression in hepatocytes treated with Hcy, and vice versa. (Figure 3j,k). These data support that DNMT1 is responsible for the hypermethylation of ERO1α promoter, which leads to suppression of ERO1α transcription in hepatocytes treated with Hcy.
Figure 3.
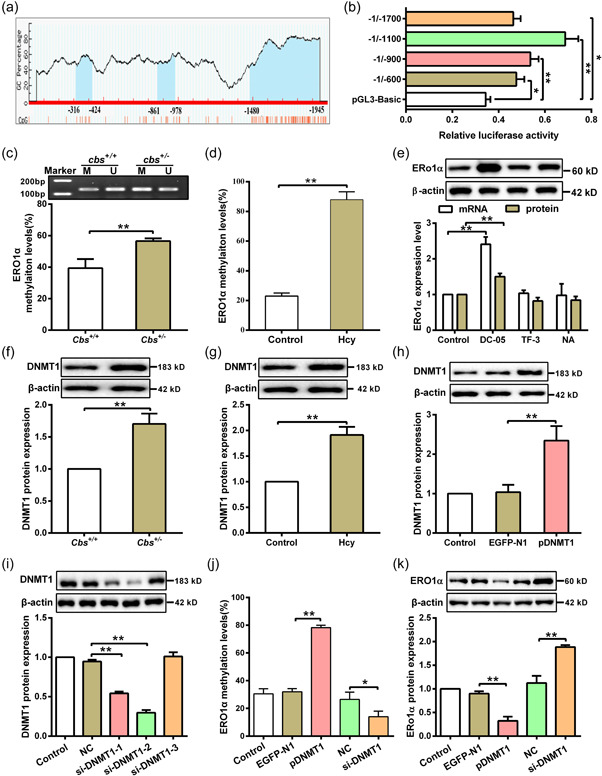
Hcy inhibits ERO1α expression through DNA methylation. (a) Bioinformatics was used to analyze the methylation sites of ERO1α promoter from −2000 to −1 region on the MethPrimer website (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi), and the predicted CpG islands were indicated by blue color. (b) Several fragments (−1 to −600, −1 to −900, −1 to −1100, −1 to −1700) of the ERO1α promoter were cloned into a pGL3‐basic reporter vector and cotransfected with a Renilla reporter plasmid in HEK293T cells, and the ERO1α promoter activity was analyzed by dual‐luciferase reporter assay (n = 3). (c) Methylation levels of ERO1α promoter in liver tissues from cbs +/ − and cbs +/+ mice were detected by nMS‐PCR after bisulfite modification of the DNA (n = 6). M: methylation; U: unmethylation. (d) MassARRAY methylation analysis was used to measure the methylation level of ERO1α promoter in hepatocytes cells treated with 100 μM Hcy (n = 3). (e) The mRNA and protein expression of ERO1α were determined by qRT‐PCR and western blot in hepatocytes treated with DNMT1, DNMT3a, and DNMT3b specific inhibitors (DC‐05, Theaflavin‐3 (TF‐3), and Nanaomycin A (NA), respectively (n = 3). (f) DNMT1 protein expression was analyzed by western blot in liver tissues of cbs +/ − and cbs +/+ mice (n = 6). (G) Protein expression of DNMT1 was analyzed by western blot in hepatocytes treated with Hcy (n = 3). (h) Protein expression of DNMT1 was analyzed in hepatocytes after transfection with negative control vector (EGFP‐N1) or DNMT1‐overexpressed plasmid (pDNMT1) by western blot (n = 3). (i) Western blot was used to detect DNMT1 protein expression in hepatocytes transfected with DNMT1 siRNAs (si‐DNMT1‐1/2/3) (n = 3). (j) ERO1α methylation level was measured with MassARRAY methylation analysis after hepatocytes transfected with pDNMT1 or si‐DNMT1 in the presence of Hcy (n = 3). (k) Protein expression of ERO1α was analyzed by western blot in Hcy‐treated hepatocytes after transfection with pDNMT1 or si‐DNMT1 (n = 3). Data represents mean ± SD, *p < 0.05, **p < 0 .01. DNMT1, DNA methyltransferase 1; ER, endoplasmic reticulum; ERO1α, endoplasmic reticulum oxidoreductase 1α; Hcy, homocysteine; nMS‐PCR, nested methylation‐specific polymerase chain reaction; qRT‐PCR, quantitative real‐time polymerase chain reaction; SD, standard deviation
3.4. Hcy inhibits ERO1α expression through G9a‐mediated H3K9me2 in hepatocytes
Studies have indicated that H3K9 di‐methylation (H3K9me2) is implicated in the promoter region of silenced gene, which can suppress gene expression (Shinkai & Tachibana, 2011). Therefore, we measured the level of H3K9me2 in liver tissue of our mice model and hepatocytes treated with Hcy. The results showed that the levels of H3K9me2 were increased in liver tissue of the cbs +/ − mice and hepatocytes treated with Hcy, as compared to their respective controls (Figure 4a,b). Consistent with western blot results, ChIP assay showed an elevated enrichment of H3K9me2 on ERO1α promoter in hepatocytes after Hcy treatment (Figure 4c). These data suggest that modification of H3K9me2 on ERO1α promoter is another potential mechanism contributing to Hcy‐induced suppression of ERO1α expression. Since G9a is the well‐characterized histone methyltransferase (HMTs) catalyzing H3K9me2 (Yu et al., 2013), we investigated the effect of Hcy on G9a expression in hepatocytes. The results showed that G9a protein expression was dramatically increased in liver tissue of cbs +/ − mice with HHcy and hepatocytes under Hcy treatment (Figure 4d,e). To further confirm the regulatory role of G9a on ERO1α expression under Hcy treatment, G9a was overexpressed or knocked down in hepatocytes (Figure 4f,g). The enrichment of H3K9me2 on ERO1α promoter was increased in Hcy treated hepatocytes with G9a overexpression, whereas the opposite results were found in hepatocytes with G9a knockdown (Figure 4h). Accordingly, overexpression of G9a inhibited the expression of ERO1α in Hcy‐treated hepatocytes, while knockdown of G9a promoted ERO1α expression in the cells (Figure 4i). These results suggest that G9a mediates H3K9me2 modification on ERO1α promoter leading to downregulation of ERO1α in the presence of Hcy.
Figure 4.
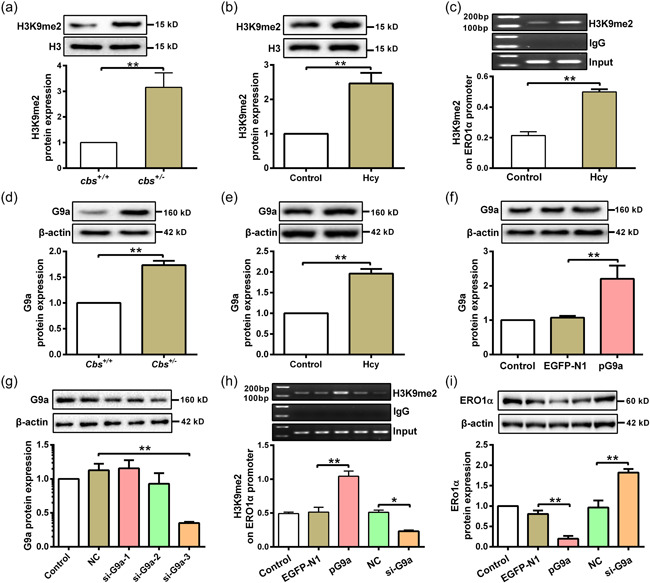
G9a mediates H3K9me2 at ERO1α promoter in hepatocytes with Hcy treatment. (a and b) The H3K9me2 levels in liver tissues of cbs +/ − mice (n = 6) and hepatocytes treated with Hcy (n = 3) were analyzed by western blot. (c) The enrichment of H3K9me2 on ERO1α promoter was assayed by ChIP assay in hepatocytes after Hcy treatment (n = 6). (d and e) G9a protein expression was detected by western blot in liver tissue of cbs +/ − mice (n = 6) and hepatocytes treated with Hcy (n = 3). (f) G9a protein expression was measured by western blot in hepatocytes transfected with G9a‐overexpressed plasmid (pG9a) (n = 3). (g) Protein expression of G9a was examined by western blot in hepatocytes after transfection with three G9a siRNAs (si‐G9a‐1/2/3) (n = 3). (h) ChIP assay for analysis of the enrichment of H3K9me2 on ERO1α promoter in hepatocytes after transfection with pG9a or si‐G9a in the presence of Hcy (n = 3). (i) Western blot was used to measure the protein expression of ERO1α in Hcy‐treated hepatocytes after transfection with pG9a and si‐G9a (n = 3). Data represents mean ± SD, *p < 0 .05, **p < 0 .01. ChIP, chromatin immunoprecipitation; ERO1α, endoplasmic reticulum oxidoreductase 1α; Hcy, homocysteine; siRNA, small interfering RNA
3.5. Hcy inhibits ERO1α expression via the cooperation of DNMT1 and G9a in the hepatocytes
Considering that both DNA methylation and histone methylation can regulate ERO1α expression, it is necessary to clarify the relationship between them. As shown in Figure 5a, DNA methylation level of ERO1α promoter was reduced in hepatocytes after transfection with si‐G9a, and knockdown of both DNMT1 and G9a expression further enhanced the inhibitory effect in comparison with single gene knockdown. Similar to the above results, knockdown of DNMT1 expression inhibited H3K9me2 enrichment on the ERO1α promoter, the co‐knockdown of DNMT1 and G9a further enhanced the inhibitory effect in comparison with either alone (Figure 5b). Moreover, silencing of both DNMT1 and G9a expression further increased the ERO1α protein expression (Figure 5c). To further confirm these observations, hepatocytes were treated with a G9a specific inhibitor Bix01294 (Bix) and DC‐05 in the presence of Hcy. Consistent with the previous results, co‐treatment of DC‐05 and Bix further inhibited DNA methylation level and H3K9me2 modification on the ERO1α promoter compared with individual treatment (Figure 5d,e). In addition, ERO1α protein expression was found to be increased in hepatocytes treated with DC‐05 or Bix, and combinational treatment of them further enhanced ERO1α expression (Figure 5f). In summary, these data demonstrate that the interaction between DNMT1 and G9a plays a vital role in regulation of ERO1α expression.
Figure 5.
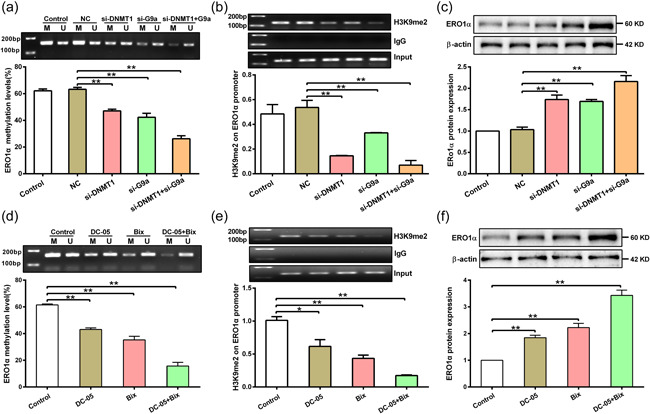
DNMT1 cooperates with G9a to regulate ERO1α expression in hepatocytes treated with Hcy. (a) DNA methylation level of ERO1α promoter was measured by nMS‐PCR in hepatocytes after transfection with siRNA of DNMT1 and G9a in the presence of Hcy (n = 3). (b) Enrichment of H3K9me2 on ERO1α promoter was detected by ChIP assay in hepatocytes after transfection with si‐DNMT1 and si‐G9a in the presence of Hcy (n = 3). (c) Protein expression of ERO1α was measured by western blot in hepatocytes after transfection with si‐DNMT1 and si‐G9a in the presence of Hcy (n = 3). (d) DNA methylation of ERO1α was detected by nMS‐PCR in hepatocytes after treatment with DC‐05 and Bix (Bix01294, G9a specific inhibitor) in the presence of Hcy (n = 3). (e) ChIP assay was used to detect the enrichment of H3K9me2 on ERO1α promoter in hepatocytes after treatment with DC‐05 and Bix in the presence of Hcy (n = 3). (f) Detection of ERO1α protein expression in hepatocytes treated with DC‐05 and Bix in the presence of Hcy (n = 3). Data represents mean ± SD, *p <0 .05, **p < 0.01. DNMT1, DNA methyltransferase 1; ChIP, chromatin immunoprecipitation; ERO1α, endoplasmic reticulum oxidoreductase 1α; Hcy, homocysteine; nMS‐PCR, nested methylation‐specific polymerase chain reaction
4. DISCUSSION
In general, aberrant levels of Hcy can influence the status of liver, in return, dysfunction of the liver alters methionine metabolism, resulting in elevated Hcy that is released into the plasma as well. Growing evidence support that elevated Hcy levels contributes to liver injury, and ER stress; and subsequent apoptosis is implicated in the pathogenesis of liver injury, which correlates with liver disease severity (Malhi & Kaufman, 2011). In this study, we confirmed that Hcy aggravates ER stress and apoptosis of hepatocytes, which might explain the pathogenic effects of Hcy on liver injury. However, the underlying mechanism of ER stress and subsequent apoptosis in Hcy‐mediated liver injury remains obscure.
Previous studies proved that ERO1α is a critical ER sulfhydryl oxidase that exists in the ER, which is ubiquitously expressed in numerous tissues and cell types; and the main function of ERO1α is to promote protein folding and secretion and inhibiting apoptosis (Zhang et al., 2014). ERO1α activity may be an important factor contributing to the large production of ROS in cells, as it has been previously found that ERO1α dysfunction may result in a rapid decrease in ER‑derived oxidative stress (Zhang et al., 2020). In this study, we found that overexpression of ERO1α reduced Hcy‐induced ER stress and apoptosis in hepatocytes, which in agreement with a previous report that ERO1α can inhibit ER stress. In the contrary, ERO1α overexpression restores galactosamine/LPS induced liver injury in CHOP‐deficient mice during acute liver failure (Wang et al., 2019). The difference between our findings and others may be attributed to the different disease models and different drug intervention factors. As the traditional metabolic pathway of Hcy is a trans‐sulfuration pathway, and ERO1α can form disulfide bonds, we speculated that Hcy may transfer sulfide groups to ERO1α to cause ER stress, which may be a novel mechanism for Hcy induced liver injury which is worth further study.
Interestingly, Hcy in the liver is methylated to methionine by methionine synthase (requiring folate) and by betaine‐homocysteine methyltransferase or converted to cystathionine by CBS. As a component of the methionine recycle system, Hcy is involved in one‐carbon methyl group‐transfer metabolism and acts as a methyl donor when it is converted to S‐adenosyl‐methionine (SAM), in which a methyl group is transferred to not only DNA, but also histone and other proteins (Kinoshita et al., 2013). DNA methylation is one of the best‐known epigenetic factors which is catalyzed by DNMTs. The reaction involves the transfer of a methyl group to the 5′ position of a cytosine ring of CpG dinucleotides (Lee et al., 2020). In this study, we observed that DNMT1 upregulation is responsible for the DNA hypermethylation on ERO1α promoter, which leads to suppression of ERO1α in ER stress and apoptosis of hepatocytes induced by Hcy. These data suggest that DNA methylation of ERO1α promoter may be an important regulation mechanism of ERO1α transcription, which is consistent with that alteration of DNA hypermethylation levels at the promoter region leading to gene silencing.
In addition to DNA methylation, gene expression also can be regulated by histone methylation at lysine residues (Yan et al., 2003). The most extensively studied histone methylation event is the methylation at lysine 9 of histone 3 (H3K9), which is generally linked to gene silencing and chromatin condensation (Fujioka et al., 2020). More and more studies indicate that histone methylation is involved in the liver physiology and disease (Price et al., 2020). To know the role of H3K9me2 in the regulation of ERO1α expression, we detected the H3K9me2 levels and found that the enrichment of H3K9me2 levels on ERO1α promoter was increased after Hcy treatment, which attributes to the enhanced expression of histone lysine methyltransferase enzyme G9a. Since both DNA methylation and histone methylation can regulate ERO1α expression, it is of interest to clarify the relationship between them. Some experiments supported that DNA methylation guides histone modifications, and DNA methylation can lead to gene silencing by either preventing or promoting the recruitment of regulatory proteins to DNA, it can provide binding sites for methyl‐binding domain proteins, which can mediate gene repression through interactions with histone lysine methyltransferases (Johnson et al., 2007, Xu et al., 2016). We also observed that DNA methylation and histone methylation cooperatively regulate ERO1α expression via DNMT1 and G9a in hepatocytes induced by Hcy. Our results were similar with the findings that histone acetylation/methylation interacts with DNA methylation at the GATA4 promoter during differentiation of mesenchymal stem cells into cardiomyocyte‐like cells (Markmee et al., 2019). All these data suggest that the interaction between DNA methylation and histone methylation might plays a vital role in Hcy‐induced ER stress and apoptosis of hepatocytes.
In summary, dynamic expression and epigenetic modification of ERO1α have been investigated in Hcy‐induced liver injury. We demonstrated that the interaction between DNA hypermethylation and H3K9me2 at ERO1α promoter is induced by Hcy, leading to the silencing of ERO1α expression and promoting hepatic ER stress and apoptosis. These results may shed light on a new approach to prevent Hcy‐induced liver injury.
AUTHOR CONTRIBUTIONS
Jiangyong Shen, Yun Jiao, Huiping Zhang, and Yideng Jiang conceived and designed research. Jiangyong Shen and Yun Jiao drafted manuscript. Jiangyong Shen, Ning Ding, Lin Xie, and Shengchao Ma performed experiments. Yun Jiao, Hui Zhang, and Anning Yang analyzed the data.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
ACKNOWLEDGMENTS
This study was supported by grants from the National Natural Science Foundation of China [82060110, 81870332, 81860555, 81870225, and 81760139]. Key Research and Development Projects in Ningxia Province (2020BFH02003, 2020BEG03005, 2019BFG02004, and 2018BEG02004). Natural Science Foundation of Ningxia Province (2020AAC02021, 2020AAC02038, and 2021AAC03337).
Shen, J. , Jiao, Y. , Ding, N. , Xie, L. , Ma, S. , Zhang, H. , Yang, A. , Zhang, H. , & Jiang, Y. (2022). Homocysteine facilitates endoplasmic reticulum stress and apoptosis of hepatocytes by suppressing ERO1α expression via cooperation between DNMT1 and G9a. Cell Biology International, 46, 1236–1248. 10.1002/cbin.11805
Jiangyong Shen and Yun Jiao contributed equally to this work.
Contributor Information
Huiping Zhang, Email: zhp19760820@163.com.
Yideng Jiang, Email: jydeng@nxmu.edu.cn.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Ai, Y. , Sun, Z. , Peng, C. , Liu, L. , Xiao, X. , & Li, J. (2017). Homocysteine induces hepatic steatosis involving ER stress response in high methionine diet‐fed mice. Nutrients, 9, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearzatto, A. , Conte, D. , Frattini, M. , Zaffaroni, N. , Andriani, F. , Balestra, D. , Tavecchio, L. , Daidone, M. G. , & Sozzi, G. (2002). p16(INK4A) hypermethylation detected by fluorescent methylation‐specific PCR in plasmas from non‐small cell lung cancer. Clinical Cancer Research, 8, 3782–3787. [PubMed] [Google Scholar]
- Cheng, Y. , Kong, F. Z. , Dong, X. F. , Xu, Q. R. , Gui, Q. , Wang, W. , Feng, H. X. , Luo, W. F. , Gao, Z. E. , & Wu, G. H. (2017). Influence of renal function on the association between homocysteine level and risk of ischemic stroke. American Journal of Translational Research, 9, 4553–4563. [PMC free article] [PubMed] [Google Scholar]
- Corso‐Diaz, X. , Jaeger, C. , Chaitankar, V. , & Swaroop, A. (2018). Epigenetic control of gene regulation during development and disease: A view from the retina. Progress in Retina and Eye Research, 65, 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich, M. (2009). DNA hypomethylation in cancer cells. Epigenomics, 1, 239–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromenty, B. (2020). Alteration of mitochondrial DNA homeostasis in drug‐induced liver injury. Food and Chemical Toxicology, 135, 110916. [DOI] [PubMed] [Google Scholar]
- Fujioka, M. , Suzuki, S. , Gi, M. , Kakehashi, A. , Oishi, Y. , Okuno, T. , Yukimatsu, N. , & Wanibuchi, H. (2020). Dimethylarsinic acid (DMA) enhanced lung carcinogenesis via histone H3K9 modification in a transplacental mouse model. Archives of Toxicology, 94, 927–937. [DOI] [PubMed] [Google Scholar]
- Johnson, L. M. , Bostick, M. , Zhang, X. , Kraft, E. , Henderson, I. , Callis, J. , & Jacobsen, S. E. (2007). The SRA methyl‐cytosine‐binding domain links DNA and histone methylation. Current Biology, 17, 379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, M. , Numata, S. , Tajima, A. , Shimodera, S. , Imoto, I. , & Ohmori, T. (2013). Plasma total homocysteine is associated with DNA methylation in patients with schizophrenia. Epigenetics, 8, 584–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C. J. , Ahn, H. , Jeong, D. , Pak, M. , Moon, J. H. , & Kim, S. (2020). Impact of mutations in DNA methylation modification genes on genome‐wide methylation landscapes and downstream gene activations in pan‐cancer. BMC Medical Genomics, 13, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder, A. , Singh, M. , Behera, J. , Theilen, N. T. , George, A. K. , Tyagi, N. , Metreveli, N. , & Tyagi, S. C. (2018). Hydrogen sulfide alleviates hyperhomocysteinemia‐mediated skeletal muscle atrophy via mitigation of oxidative and endoplasmic reticulum stress injury. American Journal of Physiology: Cell Physiology, 315, C609–C622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhi, H. , & Kaufman, R. J. (2011). Endoplasmic reticulum stress in liver disease. Journal of Hepatology, 54, 795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmee, R. , Aungsuchawan, S. , Pothacharoen, P. , Tancharoen, W. , Narakornsak, S. , Laowanitwattana, T. , Bumroongkit, K. , Puaninta, C. , & Pangjaidee, N. (2019). Effect of ascorbic acid on differentiation of human amniotic fluid mesenchymal stem cells into cardiomyocyte‐like cells. Heliyon, 5, e02018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medici, V. , Peerson, J. M. , Stabler, S. P. , French, S. W. , Gregory, J. F., 3rd , Virata, M. C. , Albanese, A. , Bowlus, C. L. , Devaraj, S. , Panacek, E. A. , Rahim, N. , Richards, J. R. , Rossaro, L. , & Halsted, C. H. (2010). Impaired homocysteine transsulfuration is an indicator of alcoholic liver disease. Journal of Hepatology, 53, 551–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, L. D. , Le, T. , & Fan, G. (2013). DNA methylation and its basic function. Neuropsychopharmacology, 38, 23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, A. J. , Manjegowda, M. C. , Kain, J. , Anandh, S. , & Bochkis, I. M. H. 3 (2020). Setdb1, and Kap1 mark H3K9me3/H3K14ac bivalent regions in young and aged liver. Aging Cell, 19, e13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramming, T. , & Appenzeller‐Herzog, C. (2012). The physiological functions of mammalian endoplasmic oxidoreductin 1: On disulfides and more. Antioxidants & redox signaling, 16, 1109–1118. [DOI] [PubMed] [Google Scholar]
- Rao, J. , Zhang, C. , Wang, P. , Lu, L. , Qian, X. , Qin, J. , Pan, X. , Li, G. , Wang, X. , & Zhang, F. (2015). C/EBP homologous protein (CHOP) contributes to hepatocyte death via the promotion of ERO1alpha signalling in acute liver failure. Biochemical Journal, 466, 369–378. [DOI] [PubMed] [Google Scholar]
- Schmitz, R. J. , Lewis, Z. A. , & Goll, M. G. (2019). DNA methylation: Shared and divergent features across eukaryotes. Trends in Genetics, 35, 818–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seervi, M. , Sobhan, P. K. , Joseph, J. , Ann Mathew, K. , & Santhoshkumar, T. R. (2013). ERO1alpha‐dependent endoplasmic reticulum‐mitochondrial calcium flux contributes to ER stress and mitochondrial permeabilization by procaspase‐activating compound‐1 (PAC‐1). Cell Death & Disease, 4, e968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai, Y. , & Tachibana, M. (2011). H3K9 methyltransferase G9a and the related molecule GLP. Genes and Development, 25, 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla, S. D. , Aroor, A. R. , Restrepo, R. , Kharbanda, K. K. , & Ibdah, J. A. (2015). In vivo acute on chronic ethanol effects in liver: A mouse model exhibiting exacerbated injury, altered metabolic and epigenetic responses. Biomolecules, 5, 3280–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, T. , Kutomi, G. , Kajiwara, T. , Kukita, K. , Kochin, V. , Kanaseki, T. , Tsukahara, T. , Hirohashi, Y. , Torigoe, T. , Okamoto, Y. , Hirata, K. , Sato, N. , & Tamura, Y. (2016). Cancer‐associated oxidoreductase ERO1‐alpha drives the production of VEGF via oxidative protein folding and regulating the mRNA level. British Journal of Cancer, 114, 1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torruellas, C. , French, S. W. , & Medici, V. (2014). Diagnosis of alcoholic liver disease. World Journal of Gastroenterology, 20, 11684–11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Chen, L. , Zhang, X. , Xu, L. , Xie, B. , Shi, H. , Duan, Z. , Zhang, H. , & Ren, F. (2019). Kaempferol protects mice from d‐GalN/LPS‐induced acute liver failure by regulating the ER stress‐Grp78‐CHOP signaling pathway. Biomedicine and Pharmacotherapy, 111, 468–475. [DOI] [PubMed] [Google Scholar]
- Wright, J. , Birk, J. , Haataja, L. , Liu, M. , Ramming, T. , Weiss, M. A. , Appenzeller‐Herzog, C. , & Arvan, P. (2013). Endoplasmic reticulum oxidoreductin‐1α (Ero1α) improves folding and secretion of mutant proinsulin and limits mutant proinsulin‐induced endoplasmic reticulum stress. The. Journal of Biological Chemistry, 288, 31010–31018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H. , Yi, Q. , Yang, C. , Wang, Y. , Tian, J. , & Zhu, J. (2016). Histone modifications interact with DNA methylation at the GATA4 promoter during differentiation of mesenchymal stem cells into cardiomyocyte‐like cells. Cell Proliferation, 49, 315–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L. , Hao, H. , Hao, Y. , Wei, G. , Li, G. , Ma, P. , Xu, L. , Ding, N. , Ma, S. , Chen, A. F. , & Jiang, Y. (2019). Aberrant MFN2 transcription facilitates homocysteine‐induced VSMCs proliferation via the increased binding of c‐Myc to DNMT1 in atherosclerosis. Journal of Cellular and Molecular Medicine, 23, 4611–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Y. , Kluz, T. , Zhang, P. , Chen, H. B. , & Costa, M. (2003). Analysis of specific lysine histone H3 and H4 acetylation and methylation status in clones of cells with a gene silenced by nickel exposure. Toxicology and Applied Pharmacology, 190, 272–277. [DOI] [PubMed] [Google Scholar]
- Yan, Y. , Wu, X. , Wang, P. , Zhang, S. , Sun, L. , Zhao, Y. , Zeng, G. , Liu, B. , Xu, G. , Liu, H. , Wang, L. , Wang, X. , & Jiang, C. (2020). Homocysteine promotes hepatic steatosis by activating the adipocyte lipolysis in a HIF1α‐ERO1α‐dependent oxidative stress manner. Redox Biology, 37, 101742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H. , Lin, Q. , Wang, Y. , He, Y. , Fu, S. , Jiang, H. , Yu, Y. , Sun, S. , Chen, Y. , Shou, J. , & Li, H. (2013). Inhibition of H3K9 methyltransferases G9a/GLP prevents ototoxicity and ongoing hair cell death. Cell Death & Disease, 4, e506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B. , Xiang, S. , Zhong, Q. , Yin, Y. , Gu, L. , & Deng, D. (2012). The p16‐specific reactivation and inhibition of cell migration through demethylation of CpG islands by engineered transcription factors. Human Gene Therapy, 23, 1071–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Yang, J. , Lin, C. , Liu, W. , Huo, Y. , Yang, M. , Jiang, S. H. , Sun, Y. , & Hua, R. (2020). Endoplasmic reticulum stress‐dependent expression of ERO1L promotes aerobic glycolysis in Pancreatic Cancer. Theranostics, 10, 8400–8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Niu, Y. , Zhu, L. , Fang, J. , Wang, X. , Wang, L. , & Wang, C. C. (2014). Different interaction modes for protein‐disulfide isomerase (PDI) as an efficient regulator and a specific substrate of endoplasmic reticulum oxidoreductin‐1alpha (Ero1alpha). Journal of Biological Chemistry, 289, 31188–31199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


