Abstract
Mohs micrographic surgery (MMS) is considered the gold standard for difficult‐to‐treat malignant skin tumors, whose incidence is on the rise. Currently, there are no agreed upon classifiers to predict complex MMS procedures. Such classifiers could enable better patient scheduling, reduce staff burnout and improve patient education. Our goal was to create an accessible and interpretable classifier(s) that would predict complex MMS procedures. A retrospective study applying machine learning models to a dataset of 8644 MMS procedures to predict complex wound reconstruction and number of MMS procedure stages. Each procedure record contained preoperative data on patient demographics, estimated clinical tumor size prior to surgery (mean diameter), tumor characteristics and tumor location, and postoperative procedure outcomes included the wound reconstruction technique and the number of MMS stages performed in order to achieve tumor‐free margins. For the number of stages complexity classification model, the area under the receiver operating characteristic curve (AUROC) was 0.79 (good performance), with model accuracy of 77%, sensitivity of 71%, specificity of 77%, positive prediction value (PPV) of 14% and negative prediction value (NPV) of 98%. The results for the wound reconstruction complexity classification model were 0.84 for the AUROC (excellent performance), with model accuracy of 75%, sensitivity of 72%, specificity of 76%, PPV of 39% and NPV of 93%. The ML models we created predict the complexity of the components that comprise the MMS procedure. Using the accessible and interpretable tool we provide online, clinicians can improve the management and well‐being of their patients. Study limitation is that models are based on data generated from a single surgeon.
Keywords: artificial intelligence, machine learning, Mohs complexity, Mohs micrographic surgery, skin cancer
1. INTRODUCTION
Non‐melanoma skin cancer (NMSC) has the highest and an increasingly rising incidence of all cancers in Caucasians. 1 , 2 Despite this high incidence, NMSC mortality rate is relatively low 3 due to effective treatment and other medical factors.
Considered the gold standard, Mohs micrographic surgery (MMS), first described by Frederic Mohs in 1937, 4 offers precise surgical margin control, along with high cure rates for difficult‐to‐treat malignant skin tumors, while simultaneously providing maximum tissue preservation. 5 Unfortunately, due to the time and expense involved with this procedure, it is indicated only in patients with aggressive tumors or for those who are at risk of disfigurement or functional impairment. 6
While the benefits and indications for MMS are well established, the factors that can predict complicated and prolonged procedures have not been well studied. Sahai and Walling 7 studied the factors that influence the number of stages performed, defining procedures with more than three stages as complex, based on 77 complex cases and 154 control cases. These findings support previous studies, and while each study differs slightly in the definition of a complex procedure, all emphasize the influence of tumor recurrence, tumor location and tumor type on complexity. 8 , 9 , 10 , 11 Boyle et al. 12 studied the correlation of clinic–demographic features with the need for extensive reconstruction (tissue transfer—skin graft or flap). Their findings proposed that the patient's age, tumor location and history of previous reconstruction are the most important features that predict the need for advanced reconstruction. One might infer that the need for advanced reconstruction correlates with higher complexity of tumor excision; however, the two are not necessarily directly related.
The increase in demand for MMS requires the surgeon to better distinguish complex from simple procedures in order to maximize the utilization of time, trained MMS surgeons and operating room resources. In order to optimize efficiency, technical performance and resource allocations, Webb and Rivera published the first score for MMS complexity in 2012 (WAR score) based on 211 reported cases including preoperative assessment, complexity and the time each procedure required. 13 Based on classical statistics, the study's outcome included a form in which surgeons could calculate the WAR score and apply the authors' recommendations for patient scheduling and improved patient management.
Machine learning (ML) has been successfully integrated into almost every field of medicine. What differentiates ML from traditional statistical tools is that an ML model does not need to be programmed with a set of rules; the model learns from examples provided, broken down into features and their matching labels. 14 Thus, it is an ideal tool for pattern recognition. ML has made a huge impact in the field of radiology, mainly due to the ability to evaluate a large workload of visual content. 15 , 16 This feature helped ease the transition of ML into the field of dermatology, as a diagnostic and classification aid for skin lesions. 17 In addition, ML has been used to review electronic health records (EHR), revealing patterns that standard correlations have missed. 18 The limitation of ML models is their lack of interpretability. Complex models are considered a “black box,” discouraging clinicians from incorporating ML models into daily practice. 19
In this study, we developed models for MMS complexity prediction utilizing the capabilities of ML. Similar work has been done by Tan et al., published in 2016; however, their model was used to predict the surgical complexity of periocular basal cell carcinomas and was based on a dataset of 156 procedures, 20 while we are basing our models on a dataset 50 times larger and aspiring to achieve robust models that are not based on prior histopathological knowledge.
2. MATERIAL AND METHODS
2.1. Patient population and tumor characteristics
The ML models were developed and tested on a dataset of 8644 MMS procedures, all meeting the appropriate Mohs use criteria, 21 performed by a single surgeon in an outpatient setting (from July 2010 through August 2021) as a retrospective study in order to identify complex MMS procedures.
Prior to each patient's surgery, a clinical lesion evaluation was performed and recorded. Each procedure record contained preoperative data on patient demographics, tumor size (mean diameter), tumor characteristics and tumor location; however, unlike previous studies, we intentionally did not include the tumor's histological subtype. The postoperative procedure outcomes measured included the wound reconstruction technique (primary closure, local flap or skin graft) and the number of MMS stages performed in order to achieve tumor‐free margins. The data went through a preprocessing pipeline; the features extracted for the models included age and estimated clinical tumor size prior to surgery – all expressed as numerical features. Tumor location, encoded with an anatomy map, and tumor topography (flat, elevated or mixed) were encoded from categorical features to numeric array features. Patient gender and the tumor features of pigmented, ulcerated, keratinized, scarred and regular borders were designated as categorical features.
Our goal was to predict complex MMS surgery; however, we could not find an accepted definition of what is considered complex. In terms of surgical difficulty, surgery duration and overall procedure time, we decided to divide procedure complexity into two ML prediction models. The first model predicted the complexity of tumor excision, which was translated into the number of stages the surgeon performs in order to achieve tumor‐free margins. The average number of stages reported typically varies between 1.5 and 2 stages per case. 22 , 23 We decided to categorize tumor excisions that required more than two stages as complex, which was how we recorded them. The second model predicted the complexity of surgical wound reconstruction. We labelled all non‐primary wound reconstruction procedures as complicated.
2.2. Machine learning model
In order to understand how the features contributed to the models’ output, we developed an algorithm. The algorithm was a modification of the marginal contribution feature importance (MCI) algorithm, a robust addition of the correlated features method that identifies important features. 24 (The MCI algorithm is described in the Supporting Information.) After receiving the modified MCI output, we excluded features with low contribution to model performance. Feature exclusion based on the modified MCI results would reduce noise, reduce the risk of overfitting and improve model prediction.
The dataset was shuffled and divided to train and test datasets, 90% of the data were allocated to training and 10% were allocated to testing. The models were created using the training dataset for a 10‐fold cross‐validation. We used a gradient boosting decision tree (GBDT) for classification models, constructed using XGBoost (version 1.2). 25 XGBoost is an optimized distributed gradient boosting library (open source) and the most common method for structured problems in Kaggle competitions. XGBoost shares qualities with other decision tree ensemble algorithms, such as the ability to handle a mixture of numerical and categorical data features. Moreover, XGBoost is a highly suitable tool for these models because it learns trees in a sequential order such that each additional tree is a gradient step with respect to a loss function, and has an extra randomization parameter used to decorrelate the individual trees. 26 (Additional information regarding model design can be found in the Supporting Information.) We selected the area under the receiver operating characteristic curve (AUROC) for the models' evaluation metric. ROC graphs are an extremely useful tool for evaluating classifiers; the AUROC is equivalent to the probability that the classifier will rank a randomly chosen positive instance higher than a randomly chosen negative instance. 27 To meet the reporting standard of ML predictive models in biomedical research, we reported accuracy, sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV). 28 All machine learning models were implemented using Python (The Python Software Foundation, Fredericksburg, VA, USA) (version 3.7).
2.3. Interpretability
We want clinicians to use our models, while at the same time, we want them to understand how the models arrive at their predictions.
For each model prediction, we provide a SHapley Additive exPlanations (SHAP) waterfall plot. SHAP builds model explanations by asking the same question for every prediction and feature: “How does prediction i change when feature j is removed from the model?”. 29 , 30 The so‐called SHAP values are the answers. They quantify the magnitude and direction (positive or negative) of a feature's effect on a prediction. Waterfall plots are designed to display explanations for individual predictions. The bottom of a waterfall plot displays the expected value of the model output. Each subsequent row up the vertical axis shows how the positive (red) or negative (blue) contribution of each feature moves the value from the expected model output over the background dataset to the model output for this prediction. 31 The models' output is presented in logarithm of odds (log‐odds) units, and with a simple conversion, the models can represent the probability of a complicated MMS procedure. (Additional information regarding SHAP can be found in the Supporting Information.)
3. RESULTS
The study cohort included 8644 procedures, the average patient age was 69 ± 13 years and 55% of patients were males. The vast majority of tumor excisions were conducted in up to two stages and underwent primary wound reconstruction (94% and 84% respectively). The most prevalent tumor locations were on the nose (22%) (which was divided into the sub‐locations of the columella, dorsum, nostril and tip), cheek (18%), forehead (12%), scalp (9%), neck (8%) and ears (8%) (Figure 1). The average estimated clinical tumor size prior to surgery was 12 ± 6 mm. Other tumor characteristics varied among patients; however, 67% of tumors had regular borders that were sharply demarcated during preoperative clinical or dermoscopic evaluation. The complete list of feature descriptions and values is available in Table 1.
FIGURE 1.
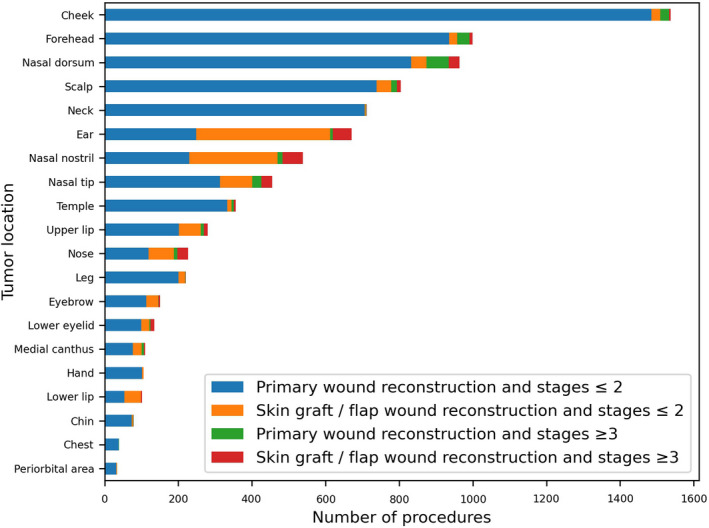
Top 20 tumor locations and procedure outcome labelling
TABLE 1.
Participant demographics, preoperative tumor evaluation characteristics and surgical outcomes
| Feature | Total |
|---|---|
| Demographics | (n = 8644) |
| Age, years, mean ± SD | 69 ± 13 |
| Gender, male, n (%) | 4725 (55) |
| Tumor size, mm, mean ± SD | 12 ± 6 |
| Tumor characteristics | |
| Pigmented, n (%) | 739 (8.5) |
| Ulcerated, n (%) | 1478 (17) |
| Keratinized, n (%) | 1477 (17.1) |
| Scarred, n (%) | 712 (8.2) |
| Regular borders, n (%) | 5776 (66.8) |
| Tumor topography | |
| Flat, n (%) | 1207 (14) |
| Elevated, n (%) | 4241 (49) |
| Mixed, n (%) | 3196 (37) |
| Procedure outcome | |
| MMS stages > 2, n (%) | 479 (5.5) |
| Skin graft/flap wound reconstruction, n (%) | 1379 (16) |
Abbreviation: MMS, Mohs micrographic surgery.
The modified MCI results for the MMS stages model emphasized tumor characteristics as the most important feature, followed by tumor location and tumor topography. However, relative to these features, the contributions of gender and tumor size features were one order of magnitude smaller. Moreover, the age feature was two orders of magnitude smaller (Table 2). For the wound reconstruction model, the order of features by importance of contribution was tumor location, tumor characteristics, tumor topography and tumor size, while age and gender features were one order of magnitude smaller.
TABLE 2.
Complexity classification models: the marginal contribution of each feature in the model
| Feature | Stage complexity classification model's modified MCI (AUROC) | Wound reconstruction complexity classification model's modified MCI (AUROC) |
|---|---|---|
| Tumor location | 0.2073 | 0.3592 |
| Gender | 0.0125 | 0.0214 |
| Age | 0.0086 | 0.0510 |
| Tumor size | 0.0274 | 0.1212 |
| Tumor topography | 0.1897 | 0.1244 |
| Tumor characteristics | 0.2611 | 0.1614 |
Abbreviations: AUROC, Area under the receiver operating characteristic curve; MCI, Marginal contribution feature importance.
For the stages complexity classification model, the average 10‐fold cross‐validation AUROC was 0.75 ± 0.03 (Figure 2A). The test set AUROC was 0.79 (Figure 3). We created a confusion matrix, and the model's accuracy was 77%, with a sensitivity of 71%, specificity of 77%, PPV of 14% and NPV of 98%. The results for the wound reconstruction complexity classification model were 0.86 ± 0.02 (Figure 2B) for the average 10‐fold AUROC, with a test set AUROC of 0.84 (Figure 3), model accuracy of 75%, sensitivity of 72%, specificity of 76%, PPV of 39% and NPV of 93%.
FIGURE 2.
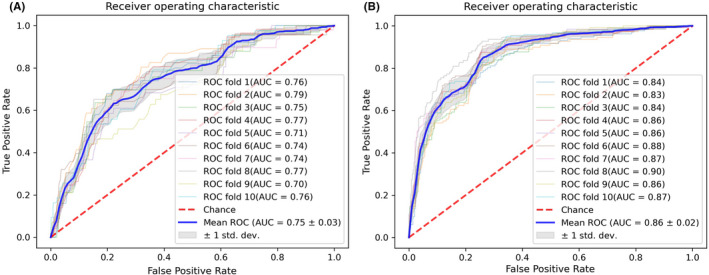
Receiver operating characteristic curves. (A) ROC for MMS stages complexity model prediction. (B) ROC for wound reconstruction complexity model prediction
FIGURE 3.
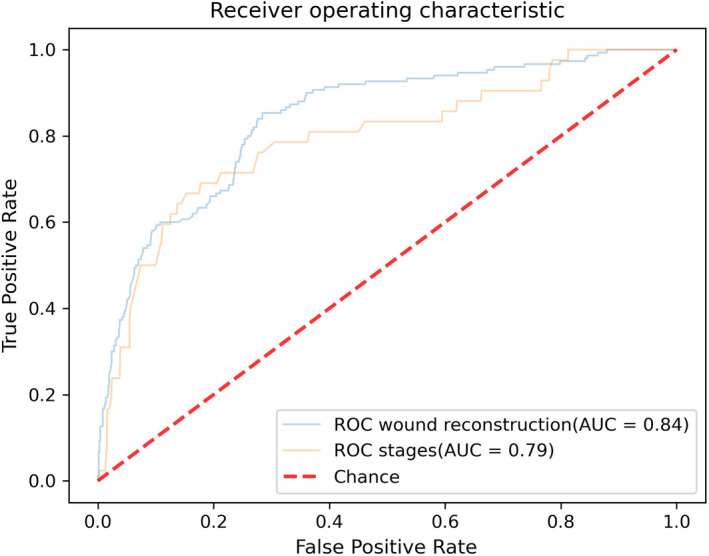
Test dataset receiver operating characteristic curves
In our pursuit of model accessibility, we created a web application (www.MMS‐Prediction.com) 32 , in which clinicians can enter tumor features and receive the complexity prediction, prediction probability and SHAP waterfall plot.
4. LIMITATIONS
ML models create algorithms that are designed based on examples rather than a set of rules; thus, the study's limitations are derived from and driven by the example dataset. Our dataset was based on the practice of a single centre and a single surgeon (over a period of a decade), which limits the diversity of examples. The absence of histological subtypes, which might seem like a limitation, was intentionally omitted from the algorithm developed. This is because the models in this study are intended to be used as decision‐making tools during the preoperative clinic visit and for planning before the histological subtype is known.
5. DISCUSSION
The increasing rise in NMSC translates to an increasing demand for MMS, and although healthcare systems perform differently in each country, optimizing treatment management is a necessity worldwide, as is reducing physician burnout rate. 33
As a community of researchers, we should always strive towards excellence, and even a slight improvement could serve as a stepping stone for future progress. The WAR score was the first to classify complicated MMS procedures by measuring procedure duration. Tan et al. demonstrated the opportunities that lie in utilizing ML algorithms, 20 providing us with a proof of concept. In our research, we aim to upscale, learning from the successes and limitations of previous studies and creating a more meaningful and accessible tool.
The question of what is complex MMS was a significant part of the study design. Unlike the WAR score, which was constructed based on data from an outpatient setting, we wished to create more robust models that would fit the complexities of inpatient settings as well, ones that would extend beyond predicting procedure duration—and be applicable to different surgeon expertise levels, not only post‐fellowship MMS surgeon levels. Since we decided to divide the complexity model into two separate models, prediction of wound reconstruction complexity and the prediction of tumor excision complexity, according to the number of stages needed, these predictions enable each physician to schedule and manage their patients based on his or her own skill and operating environment, which dictates procedure duration.
Both prediction models did not underperform compared to Tan et al. 20 models, even though when performing tasks with much more difficult classifications, both models were able to identify more than 75% of the complicated procedures and predict uncomplicated procedures with high certainty, almost guaranteed in the stages model, with an NPV of 98%. Regarding the relatively low PPV values, as a tool, the models' predictions are not intended to replace the surgeon's clinical evaluation, and as with other AI tools in medicine, serve as recommendations to be considered. Moreover, our models' predictions are accompanied by a prediction probability and SHAP plot that display how each feature contributed to the prediction. This additional information will enable the surgeon to understand the model prediction and decide what actions to take.
Another interesting finding is the lack of contribution of demographic data to the models' performance, which means no identifying details could compromise patient privacy, which simplifies ethics committee approvals.
Although our dataset is based on the experience of a single surgeon, the vast number of procedures (covering a 10‐year period) and their supporting data partially compensate for such a limitation. By shuffling the dataset, we trained the model using examples that reflected various experience levels, expressed in example labels, to adapt the model to novice and experienced MMS surgeons, which contributed to model robustness. Moreover, the features used for model prediction included features such as tumor location and size, which require no special expertise. Other features reflect accepted clinical evaluation standards taught in dermatology textbooks.
In some cases, following clinical evaluation, surgeons have no doubt regarding the nature of the lesion. 34 Thus, performing a biopsy will not only further burden the patient but also delay treatment. Moreover, currently, non‐invasive tools for lesion classification are starting to deliver a high level of certainty and may perhaps replace lesion biopsy in the future. 17 , 35
When we decided to intentionally exclude lesion histopathology as a feature, we knew this decision would not be accepted easily. We wanted to create both a robust tool that could be applied when the histopathology is not available, and a flexible tool that would integrate into future machine learning pipelines, which would not necessarily include lesion histopathology reports. For these important reasons, we decided to exclude histopathology from the model.
The purpose of this study was not only to report our findings but also to create and share an accessible and interpretable tool that would aid our fellow clinicians in making better decisions. The use of the modified MCI will assist in understanding how the models were constructed, while the SHAP plot will simplify the prediction for physicians and patients, showing the prediction visually, shedding some light on what the “black box” is. By creating an open web application, we have accomplished the desired leap from bench to bedside.
We are aware of the general opinion that senior surgeons have plenty of experience in identifying complicated procedures and do not need support tools; nonetheless, as clinicians, we are taught to appreciate the benefit of a second opinion, especially in difficult cases. Moreover, these tools could be aids for junior surgeons, giving them access to knowledge and experience. We believe another benefit of such a model is that patients would also appreciate knowing that a sophisticated model helped support and confirm the physician's experience—even for senior surgeons. For example, this tool would help the surgeon explain to the patient, “Based on procedures just like yours, we know with x% certainty that the procedure will not require complicated wound reconstruction.”
The fact that the large majority of MMS procedures conclude with patients being tumor free gives us the opportunity to test and integrate into practice new cutting‐edge ML models. We believe that this research, to our knowledge, is the first of its kind, which uses ML to classify complicated MMS procedures based on a vast dataset, will inspire fellow researchers to further integrate ML models into surgical dermatology clinical practice, leading to improvement in all aspects of treatment and resulting in better quality of life for patients and clinicians.
The next evolution of this study is to integrate our prediction models with deep learning models, such as those that were developed in the past for tumor classification. 17 This will create a tool that will be able to predict MMS complexity from a tumor's image. Nonetheless, machine learning models should constantly improve by including more diverse data and performing retraining, by more data analysis and by exploring new models.
A subsequent study could compare the accuracy of the tool across numerous surgeons with various years of experience.
The insights from this research serve as a tool that will:
Enable better planning of patient and staff scheduling by predicting long and complex procedures
Reduce surgeon overload and burnout by creating balanced work plans, spreading out complex procedures and adjusting them to surgeon ability in training environments, thus improving surgery outcomes
Perhaps most important, improve our understanding of planned procedures, so we can better express them to patients, assisting in patient education, managing expectations and reducing preoperative anxiety levels. 36
6. CONCLUSION
The ML models we created predict the complexity of the components that comprise the MMS procedure. Using the accessible and interpretable tool we provide online, clinicians can improve the management and well‐being of their patients.
ACKNOWLEDGEMENT
We would like to thank our patients, as we continue to strive for delivering better treatment. We also wish to thank Prof. Ran Gilad‐Bachrach from the faculty of Bio‐Medical Engineering, Tel‐Aviv University, Israel, for his guidance and support in the field of artificial intelligence.
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
AUTHOR CONTRIBUTIONS
Gon Shoham designed the research study, analysed the data, designed AI models and wrote the manuscript. Ariel Berl wrote the manuscript. Ofir Shir‐az wrote the manuscript. Sharon Shabo wrote the manuscript. Avshalom Shalom designed the research study, performed the research and wrote the manuscript.
Supporting information
Supplementary Material
Shoham G, Berl A, Shir‐az O, Shabo S, Shalom A. Predicting Mohs surgery complexity by applying machine learning to patient demographics and tumor characteristics. Exp Dermatol. 2022;31:1029–1035. doi: 10.1111/exd.14550
Gon Shoham, Ariel Berl, Ofir Shir‐az, Sharon Shabo, and Avshalom Shalom contributed equally to this research.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Non‐melanoma skin cancer incidence statistics . Cancer Research UK. Published July 2, 2018. Accessed February 1, 2021. https://www.cancerresearchuk.org/health‐professional/cancer‐statistics/statistics‐by‐cancer‐type/non‐melanoma‐skin‐cancer/incidence [Google Scholar]
- 2. Skin cancer . Accessed February 1, 2021. https://www.aad.org/media/stats‐skin‐cancer
- 3. Fitzmaurice C, Abate D, Abbasi N, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability‐adjusted life‐years for 29 cancer groups, 1990 to 2017: a systematic analysis for the global burden of disease study. JAMA Oncol. 2019;5(12):1749‐1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mohs FE. Chemosurgery: a microscopically controlled CONTROLLED method of cancer excision. Arch Surg. 1941;42(2):279‐295. doi: 10.1001/archsurg.1941.01210080079004 [DOI] [Google Scholar]
- 5. Pennington BE, Leffell DJ. Mohs micrographic surgery: established uses and emerging trends. Oncology (Williston Park). 2005;19(9):1165‐1171; discussion 1171‐1172, 1175. [PubMed] [Google Scholar]
- 6. Neville JA, Welch E, Leffell DJ. Management of nonmelanoma skin cancer in 2007. Nat Clin Pract Oncol. 2007;4(8):462‐469. doi: 10.1038/ncponc0883 [DOI] [PubMed] [Google Scholar]
- 7. Sahai S, Walling HW. Factors predictive of complex Mohs surgery cases. J Dermatol Treat. 2012;23(6):421‐427. doi: 10.3109/09546634.2011.579083 [DOI] [PubMed] [Google Scholar]
- 8. Batra RS, Kelley LC. Predictors of extensive subclinical spread in nonmelanoma skin cancer treated with Mohs micrographic surgery. Arch. Dermatol. 2002;138(8):1043‐1051. [DOI] [PubMed] [Google Scholar]
- 9. Alam M, Berg D, Bhatia A, et al. Association between number of stages in Mohs micrographic surgery and surgeon‐, patient‐, and tumor‐specific features: a cross‐sectional study of practice patterns of 20 early‐ and mid‐career Mohs surgeons. Dermatol Surg. 2010;36(12):1915‐1920. doi: 10.1111/j.1524-4725.2010.01758.x [DOI] [PubMed] [Google Scholar]
- 10. Hendrix JD, Parlette HL. Duplicitous growth of infiltrative basal cell carcinoma. Dermatol Surg. 1996;22(6):535‐539. doi: 10.1111/j.1524-4725.1996.tb00370.x [DOI] [PubMed] [Google Scholar]
- 11. Orengo IF, Salasche SJ, Fewkes J, Khan J, Thornby J, Rubin F. Correlation of histologic subtypes of primary basal cell carcinoma and number of Mohs stages required to achieve a tumor‐free plane. J Am Acad Dermatol. 1997;37(3):395‐397. [DOI] [PubMed] [Google Scholar]
- 12. Boyle K, Newlands SD, Wagner RF, Resto VA. Predictors of reconstruction with Mohs removal of nonmelanoma skin cancers. Laryngoscope. 2008;118(6):975‐980. doi: 10.1097/MLG.0b013e31816a8cf2 [DOI] [PubMed] [Google Scholar]
- 13. Rivera AE, Webb JM, Cleaver LJ. The Webb and Rivera (WAR) score: a preoperative Mohs surgery assessment tool. Arch Dermatol. 2012;148(2):206‐210. doi: 10.1001/archdermatol.2011.1352 [DOI] [PubMed] [Google Scholar]
- 14. Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med. 2019;380(14):1347‐1358. doi: 10.1056/NEJMra1814259 [DOI] [PubMed] [Google Scholar]
- 15. Choy G, Khalilzadeh O, Michalski M, et al. Current applications and future impact of machine learning in radiology. Radiology. 2018;288(2):318‐328. doi: 10.1148/radiol.2018171820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang S, Summers RM. Machine learning and radiology. Med Image Anal. 2012;16(5):933‐951. doi: 10.1016/j.media.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Esteva A, Kuprel B, Novoa RA, et al. Dermatologist‐level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115‐118. doi: 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thomsen K, Iversen L, Titlestad TL, Winther O. Systematic review of machine learning for diagnosis and prognosis in dermatology. Null. 2020;31(5):496‐510. doi: 10.1080/09546634.2019.1682500 [DOI] [PubMed] [Google Scholar]
- 19. Lee EY, Maloney NJ, Cheng K, Bach DQ. Machine learning for precision dermatology: advances, opportunities, and outlook. J Am Acad Dermatol. 2020. doi:84(5), 1458‐1459. 10.1016/j.jaad.2020.06.1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tan E, Lin F, Sheck L, Salmon P, Ng S. A practical decision‐tree model to predict complexity of reconstructive surgery after periocular basal cell carcinoma excision. J Eur Acad Dermatol Venereol. 2017;31(4):717‐723. doi: 10.1111/jdv.14012 [DOI] [PubMed] [Google Scholar]
- 21. Connolly SM, Baker DR, Coldiron BM, et al. AAD/ACMS/ASDSA/ASMS 2012 appropriate use criteria for Mohs micrographic surgery: a report of the American Academy of Dermatology, American College of Mohs Surgery, American Society for Dermatologic Surgery Association, and the American Society for Mohs Surgery. J Am Acad Dermatol. 2012;67(4):531‐550. doi: 10.1016/j.jaad.2012.06.009 [DOI] [PubMed] [Google Scholar]
- 22. Krishnan A, Xu T, Hutfless S, et al. Outlier practice patterns in Mohs micrographic surgery: defining the problem and a proposed solution. JAMA Dermatol. 2017;153(6):565‐570. doi: 10.1001/jamadermatol.2017.1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Donaldson MR, Coldiron BM. Mohs micrographic surgery utilization in the medicare population, 2009. Dermatol Surg. 2012;38(9):1427‐1434. doi: 10.1111/j.1524-4725.2012.02464.x [DOI] [PubMed] [Google Scholar]
- 24. Catav A, Fu B, Zoabi Y, et al. Marginal contribution feature importance—an axiomatic approach for explaining data. In: International Conference on Machine Learning. PMLR; 2021:1324‐1335. Accessed July 25, 2021. http://proceedings.mlr.press/v139/catav21a.html [PMC free article] [PubMed] [Google Scholar]
- 25. Chen T, Guestrin C. XGBoost: a scalable tree boosting system. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. Published online August 13, 2016:785‐794. doi: 10.1145/2939672.2939785 [DOI] [Google Scholar]
- 26. Nielsen Didrik. Tree boosting with xgboost‐why does xgboost win "every" machine learning competition?. MS thesis. NTNU, 2016. Published online 2016.
- 27. Fawcett T. An introduction to ROC analysis. Pattern Recogn Lett. 2006;27(8):861‐874. doi: 10.1016/j.patrec.2005.10.010 [DOI] [Google Scholar]
- 28. Luo W, Phung D, Tran T, et al. Guidelines for developing and reporting machine learning predictive models in biomedical research: a multidisciplinary view. Journal of Medical Internet Research. 2016;18(12):e323. doi: 10.2196/jmir.5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shapley LS. A value for n‐person games. Contrib Theory Games. 1953;2(28):307‐317. [Google Scholar]
- 30. Lundberg S, Lee SI. A unified approach to interpreting model predictions. arXiv:170507874 [cs, stat]. Published online November 24, 2017. Accessed January 25, 2021. http://arxiv.org/abs/1705.07874
- 31. Waterfall plot—SHAP latest documentation . Accessed February 24, 2021. https://shap.readthedocs.io/en/latest/example_notebooks/api_examples/plots/waterfall.html
- 32. Shoham G. MMS complexity prediction. MMS—prediction. Published February 2021. Accessed February 18, 2021. https://www.mms‐prediction.com
- 33. Lam C, Kim Y, Cruz M, Vidimos AT, Billingsley EM, Miller JJ. Burnout and resiliency in Mohs surgeons: a survey study. Int J Womens Dermatol. Published online January 18, 2021. doi: 2021, 7(3), 319‐322. 10.1016/j.ijwd.2021.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mogensen M, Jemec GBE. Diagnosis of nonmelanoma skin cancer/keratinocyte carcinoma: a review of diagnostic accuracy of nonmelanoma skin cancer diagnostic tests and technologies. Dermatol Surg. 2007;33(10):1158‐1174. doi: 10.1111/j.1524-4725.2007.33251.x [DOI] [PubMed] [Google Scholar]
- 35. Abhishek K, Kawahara J, Hamarneh G. Predicting the clinical management of skin lesions using deep learning. Sci Rep. 2021;11(1):7769. doi: 10.1038/s41598-021-87064-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hawkins SD, Koch SB, Williford PM, Feldman SR, Pearce DJ. Web app– and text message‐based patient education in Mohs micrographic surgery—a randomized controlled trial. Dermatol Surg. 2018;44(7):924‐932. doi: 10.1097/DSS.0000000000001489 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


