Abstract
Aims
To investigate the priming effects of sub‐inhibitory concentrations of biocides on antibiotic resistance in bacteria.
Methods and results
Escherichia coli, Pseudomonas aeruginosa and Staphylococcus aureus were exposed to sub‐inhibitory concentrations of biocides via a gradient plate method. Minimum inhibitory concentration (MIC) and antibiotic susceptibility were determined, and efflux pump inhibitors (thioridazine and chlorpromazine) were used to investigate antibiotic resistance mechanism(s). Escherichia coli displayed a twofold increase in MIC (32–64 mg l−1) to H2O2 which was stable after 15 passages, but lost after 6 weeks, and P. aeruginosa displayed a twofold increase in MIC (64–128 mg l−1) to BZK which was also stable for 15 passages. There were no other tolerances observed to biocides in E. coli, P. aeruginosa or S. aureus; however, stable cross‐resistance to antibiotics was observed in the absence of a stable increased tolerance to biocides. Sixfold increases in MIC to cephalothin and fourfold to ceftriaxone and ampicillin were observed in hydrogen peroxide primed E. coli. Chlorhexidine primed S. aureus showed a fourfold increase in MIC to oxacillin, and glutaraldehyde‐primed P. aeruginosa showed fourfold (sulphatriad) and eightfold (ciprofloxacin) increases in MIC. Thioridazine increased the susceptibility of E. coli to cephalothin and cefoxitin by fourfold and twofold, respectively, and both thioridazine and chlorpromazine increased the susceptibility S. aureus to oxacillin by eightfold and fourfold, respectively.
Conclusions
These findings demonstrate that sub‐inhibitory concentrations of biocides can prime bacteria to become resistant to antibiotics even in the absence of stable biocide tolerance and suggests activation of efflux mechanisms may be a contributory factor.
Significance and Impact of the Study
This study demonstrates the effects of low‐level exposure of biocides (priming) on antibiotic resistance even in the absence of obvious increased biocidal tolerance.
INTRODUCTION
The rise in multidrug‐resistant bacteria means that some conventional antimicrobials have become ineffective in the treatment of infections (Roca Subirà et al., 2012; Tuon et al., 2012; Walsh & Toleman, 2011). Biocides have been used for centuries to control infectious agents (Maillard, 2002; Morente et al., 2013; Russell, 2002). They are either applied as or added to formulated products that are used as disinfectants, preservatives, pesticides, antiseptics and even cosmetics (Gilbert & McBain, 2003; Knapp et al., 2015; Maillard, 2002). Public awareness to infection control has caused a rise in the use of biocide and biocidal products in the home environment. As a result of increased use of biocides, especially at sub‐inhibitory concentrations, there are concerns over how their selective pressure might potentially favour the development of less susceptible bacterial strains, as well as encourage the expression and dissemination of resistance mechanisms to biocides and other antimicrobial agents (Fraise, 2002; Knapp et al., 2015; Maillard et al., 2013; McBain & Gilbert, 2001; Pereira et al., 2021; SCENIHR, 2010). There are several laboratory studies demonstrating a link between exposure of bacteria to sub‐inhibitory concentrations and increased tolerance and resistance to biocides (Bock et al., 2016; Christensen et al., 2011; Escalada et al., 2005; Knapp et al., 2013; Walsh et al., 2003). Some studies show both increased tolerance to biocides plus antibiotic cross‐resistance in bacteria after exposure to sub‐inhibitory concentrations of biocides (Kurenbach et al., 2015; Wand et al., 2016). While exposure to biocides has been linked to reduced susceptibility and cross‐resistance to antibiotics (Braoudaki & Hilton, 2004; Chuanchuen et al., 2001; Slayden et al., 2000; Soumet et al., 2012; Tkachenko et al., 2007; Wand, 2017), it is paramount that we understand what happens in the absence of obvious increased biocidal tolerance after low‐level exposure; will there be a change in bacterial resistance to antibiotics in that instance?
Since biocides have several target sites within bacteria (Maillard, 2002; Russell, 2002), the most common mechanisms for cross‐resistance are via non‐specific processes such as efflux‐pumps (Bogomolnaya et al., 2013; Costa et al., 2013) and changes in properties of their cell wall, for example reduced permeability due to porin downregulation (Jaffe et al., 1982; Manzoor et al., 1999).
A review by Maillard et al. (2013) and a report from the scientific committee for emerging and newly identified health risks (SCENIHR, 2010) identified biocides as a risk due to selective pressure for less susceptible strains. Their findings highlighted a key gap in current knowledge in understanding the effect of low concentrations of biocides on bacterial cells, as well as the mechanisms involved in the development of resistance and cross‐resistance (Jaffe et al., 1982; Knapp et al., 2015; Maillard, 2002; SCENIHR, 2010). Because of these concerns, the European Union and the United States have proposed regulatory changes requiring manufacturers of biocidal products to provide data on the risks of resistance development in organisms targeted by biocidal products (Knapp et al., 2015; SCENIHR, 2010).
As previously demonstrated by several studies, prolonged exposure of bacteria to biocides under laboratory conditions can generate less susceptible mutants which can often display reduced susceptibility to various antibiotics (Fernández Márquez et al., 2017; Hardy et al., 2017; Karatzas et al., 2007, 2008; Randall et al., 2007; Whitehead et al., 2011).
The present study investigates the effect of biocide priming on bacterial resistance to antibiotics after continuous exposure of Escherichia coli, Staphylococcus aureus and Pseudomonas aeruginosa to low concentrations of hydrogen peroxide (H2O2), chlorhexidine (CHG), benzalkonium chloride (BZK) and glutaraldehyde (GTA). The three organisms used are all commonly associated with hospital‐acquired infections (HAIs) and the strains selected are those listed in the BS EN 1276:2009 disinfection test method. Escherichia coli is a Gram‐negative bacteria commonly transmitted to humans via the consumption of contaminated food and water (Kaper et al., 2004) and known to cause infections such as diarrhoea and haemorrhagic colitis to severe complications such as haemolytic anaemia and acute renal failure (Brzuszkiewicz et al., 2011). Outer membrane protein modification like the downregulation of porins and upregulation of efflux pumps such as AcrAB have been linked with increased antimicrobial resistance in E. coli (Ma et al., 1993, 1994). Staphylococcus aureus is a Gram‐positive, opportunistic pathogen responsible for both hospital‐ and community‐acquired infections (Boucher et al., 2010; Latimer et al., 2012). Upregulation of QacA‐D, E and H (Heir et al., 1998; Kazama et al., 1998; Rouch et al., 1990) and NorA, NorB and NorC efflux pumps (Truong‐Bolduc et al., 2006) have been linked to antimicrobial resistance in S. aureus. Pseudomonas aeruginosa is an opportunistic, intrinsically resistant, Gram‐negative bacteria that is responsible for a range of infections to the eyes and ears to serious complications in cystic fibrosis and bronchiectasis patients (Lambert, 2002; Soothill, 2013). Upregulation of efflux pumps such as MexAB, MexCD and MexEF and outer membrane protein modification have been linked to antimicrobial resistance in P. aeruginosa (Poole, 2001; Schweizer, 1998). This study aims to understand the impact of prolonged exposure of bacteria to low concentrations of biocide, and the possible cross‐resistance to antibiotics.
MATERIALS AND METHODS
Bacterial strains and storage
Escherichia coli ATCC 8739, P. aeruginosa ATCC 15442 and S. aureus ATCC 6538 strains were grown in tryptone soya broth (TSB) (Oxoid) at 37°C with shaking at 100 rev min−1 for 24 h and stored on protect beads (Scientific Laboratory Supplies Limited) at −80°C.
Preparation of inoculum
Bacterial strains were grown on Mueller Hinton agar (MHA) (Oxoid) at 37°C for 24 h and a single colony was transferred to 10 ml sterile saline. The turbidity of the suspension was adjusted to the equivalence 0.5 McFarland (absorbance range between 0.08 and 0.13) at 625 nm line with the European Committee for Antimicrobial Susceptibility Testing (EUCAST, 2015) and of the European Society of Clinical Microbiology and Infectious Diseases [ESCMID], 2000).
Preparation of biocide working solutions
A twofold dilution series of biocides (mg l−1) was prepared in Mueller‐Hinton broth following the BS EN ISO 20776‐6 guidelines/protocol (BSI, 2006). Hydrogen peroxide (H2O2) concentration range 1–512 mg l−1 (Fisher Scientific Belgium), formulated chlorhexidine gluconate (CHG) concentration range 0.976–500 mg l−1 (Molnlycke Health Care Ltd), benzalkonium chloride (BZK) concentration range 1–512 mg l−1 and glutaraldehyde (GTA) concentration range 8–4096 mg l−1 (Sigma‐Aldrich).
Preparation of antibiotic working solutions
Antibiotic discs Mastring‐S (M13 and M14) were obtained from Mast Diagnostics UK. Antibiotics and efflux pump inhibitors (EPIs) were purchased from Sigma Aldrich and used at the following concentrations: oxacillin sodium salt concentration 0.0020–8 mg l−1, cefoxitin sodium salt 0.25–32 mg l−1, cephalothin sodium salt 0.25–32 mg l−1, ceftriaxone disodium salt hemi (heptahydrate) 0.016–2 mg l−1, ampicillin 0.25–32 mg l−1, sulfathiazole 37% v/v, sulfadiazine 37% v/v, sulfamerazine 26% v/v, ciprofloxacin sodium salt 0.125–2 mg l−1, thioridazine hydrochloride (TZ) 1.9–500 mg l−1 and chlorpromazine hydrochloride (CPZ) 4–256 mg l−1. Stock solutions were prepared by dissolving salts in sterile distilled water and working solutions were prepared in Mueller‐Hinton broth following the BS EN ISO 20776‐6 guidelines/protocol (BSI, 2006).
Adaptation of bacterial strains to biocides using gradient plate
A twofold dilution series of biocides were prepared in sterile distilled water following the EUCAST guidelines of ESCMID (EUCAST, 2015; ESCMID 2000). The required biocide (1 ml) was added to 19 ml sterile molten nutrient agar to give specific final concentrations (from a concentration just below the MIC of the biocide against tested bacteria strain). The molten agar and biocide mixture was poured into sterile Petri dishes and set at an angle, after which plates were placed on a flat surface and 20 ml sterile molten agar were poured over the first layer and allowed to set. Plates were left at 4°C for 24 h to allow diffusion of biocide. The 24 h bacterial cultures were streaked along the concentration gradient starting at the point of the gradient plate containing the lowest concentration of biocide. These streaked plates were incubated at 37°C for 24 h. Bacterial colonies that grew the furthest along the gradient towards the high concentration were used to streak a new gradient plate containing the next highest concentration, until no further increases in tolerance were observed, at which point the furthest‐growing colonies were selected and stored on protect beads at −80°C. For example, the H2O2 MIC against E. coli was 32 mg l−1; therefore, the bacteria was first streaked on a plate containing 16 mg l−1 H2O2 (concentration twofold below the MIC), then moved to 32, 64, 128 and 256 mg l−1 until no growth were observed on the 512 mg l−1 plate.
Stability of adaptive resistance
To confirm the stability of the primed strains produced, the gradient plate isolates were subcultured 15 times in 10 ml of TSB without biocide. MIC between parent and primed strains were compared after 1, 2, 10 and 15 subcultures, following EUCAST guidelines ESCMID (EUCAST, 2015; ESCMID 2000).
Determination of minimum inhibitory concentrations
The minimum inhibitory concentrations (MICs) were determined for each strain before and after exposure to the biocides. This was carried out using the colony suspension method of testing in accordance with BS EN ISO 20776‐1 guidelines/protocol (BSI, 2006). Briefly, 50 μl of the standardized inoculum was dispensed into each well of a 96‐well plate containing 50 μl of the appropriate concentration of biocides or antimicrobial agents to give a final inoculum concentration of 5 × 105 CFU per ml. The inoculated 96‐well plate was incubated at 37°C for 24 h. The lowest concentration of compound that prevented bacterial growth after 24 h of incubation was used to establish the MIC. Bacterial growth was determined in two ways: by visual observation of growth on the 96‐well plate and by measuring absorbance at 625 nm in a Spectramax plus plate reader. All experiments were carried out as biological and technical triplicates.
Antibiotic susceptibility testing using disc diffusion test
The antibiotic susceptibility of each bacterial strain was determined before and after biocide treatment with biocides using the disc diffusion method of antimicrobial susceptibility testing. This was done in accordance with the guidelines of European committee on antimicrobial susceptibility testing (EUCAST, 2015). Briefly, antibiotic discs were applied to surfaces of the inoculated plates within 15 min of inoculation. After 24 h of incubation at 37°C, zones of inhibition were measured with a digital Vernier calliper.
The effect of EPIs
Standardized bacterial suspensions of primed strains were prepared from 24 h culture as previously described. Suspensions were grown in 96‐well plates for 24 h at 37°C in concentrations of antibiotics they were resistant to, plus an EPI at half its MIC (a concentration that does not inhibit growth of strain). H2O2 primed E. coli (EcH2O), GTA primed P. aeruginosa (PaGTA) and BZK primed P. aeruginosa (PaBZK) were tested with TZ, and CHG primed S. aureus (SaCHG) was tested with CPZ and TZ. EcH2O, 40 mg l−1 TZ was tested with cephalothin and cefoxitin, PaGTA, 250 mg l−1 TZ was tested with ciprofloxacin and sulphatriad, PaBZK 250 mg l−1 TZ was tested with BZK, and SaCHG was tested with oxacillin with either 32 mg l−1 CPZ or 15.625 mg l−1 TZ. The lowest concentration of antibiotic plus EPI that prevented bacterial growth after 24 h of incubation was used to establish MIC.
Confirmation of strain identity
16S rRNA genes were amplified from genomic DNA of parent and biocide‐treated strains using Q5® Hot Start High‐Fidelity 2X Master Mix (New England Biolabs) and primer pairs 16S rRNA For 1 (5′‐AGAGTTTGATCCTGGCTCAG‐3′) and 16S rRNA Rev 1 (5′‐ACGGCTACCTTGTTACGACTT‐3′) for S. aureus, 16S rRNA For 2 (5′‐AGAGTTTGATCATGGCTCAG‐3′) and 16S rRNA Rev 2 (5′‐ACGGTTACCTTGTTACGACTT‐3′) for E. coli, and 16S rRNA For 2 and 16S rRNA Rev 1 for P. aeruginosa. PCR products were analysed on 0.8% (w/v) agarose gels and sequenced by automated DNA sequencing (Eurofins).
Fitness of primed strains
Overnight cultures of parent and primed bacterial strains were inoculated in antibiotic‐free Mueller Hinton broth (MHB) and absorbance at 600 nm was recorded every 10 min for 16 h. Resulting data and standard deviations were plotted. Each experiment was conducted in biological triplicates.
RESULTS
Effect of sub‐inhibitory concentrations of biocides
The MIC of four biocides against pre‐ and post‐primed strains of bacteria are summarized in Table 1. There was an initial increase in MIC to H2O2 from 32 mg l−1 for the E. coli parent strain to 64 mg l−1 after H2O2 priming (EcH2O2), which was stable after 15 passages but lost after storage for 6 weeks. The MIC of E. coli did not change after exposure to other biocides used in this study. Apart from a twofold increased MIC from 64 mg l−1 for the parent strain of P. aeruginosa to 128 mg l−1 (stable after 15 passages) after BZK priming (PaBZK), there was no other obvious increased tolerance observed to biocides tested in P. aeruginosa. No biocide tolerance was observed with S. aureus, but bacteria exposed to CHG (SaCHG) were included in further testing to determine whether there was any observed antibiotic cross‐resistance.
TABLE 1.
MIC of biocides against pre‐ and post‐primed strains of bacteria
| Strain | MIC (mg l−1), n = 3 | |||
|---|---|---|---|---|
| Hydrogen peroxide | Glutaraldehyde | Benzalkonium chloride | Chlorhexidine | |
| E. coli | 32 | 1024 | 16 | 15.6 |
| EcH2O2 | 64 a | 1024 | 16 | 15.6 |
| S. aureus | 4 | 512 | 4 | 7.8 |
| SaCHG | 4 | 512 | 4 | 7.8 |
| P. aeruginosa | 32 | 1024 | 64 | 31.3 |
| PaGTA | 32 | 1024 | — | 31.3 |
| PaBZK | — | — | 128 b | — |
Note: EcH2O2, H2O2 primed E. coli; SaCHG, CHG primed S. aureus; PaGTA, GTA primed P. aeruginosa; and PaBZK, BZK primed P. aeruginosa.
EcH2O2 tolerance to H2O2 was unstable.
Only in PaBZK was there a stable twofold increase to tolerance BZK.
The results of antibiotic susceptibility tests for parent and primed strains of bacteria are summarized in Table 2. Two sample t‐tests assuming unequal variances (p = 0.05) were used to compared the data and statistically significant changes were observed in the following five cases. Changes in zones of inhibition were observed in EcH2O2, where there was no zone of inhibition around cephalothin or ampicillin discs compared to the parent strain E. coli strain, where 10.43 mm (cephalothin) and 18.88 mm (ampicillin) zones of inhibition where observed. EUCAST (2020) zone diameter breakpoint for ampicillin and Enterobacterales reports resistance at <14 mm, indicating a change in clinical susceptibility; the corresponding data for cephalothin are not available. For SaCHG, there was no zone of inhibition around oxacillin discs compared to 22.28 mm for the parent strain (EUCAST breakpoint data not available) and for PaGTA and PaBZK there was no zone of inhibition around sulphatriad discs compared to 15.11 mm for the parent strain (EUCAST breakpoint data not available).
TABLE 2.
Antibiotic susceptibility profiles of pre‐ and post‐primed bacteria strains
| Antibiotics (μg) | Zone of inhibition (mm) (SE) | ||||||
|---|---|---|---|---|---|---|---|
| EcATCC | EcH2O2 | SaATCC | SaCHG | PaATCC | PaGTA | PaBZK | |
| Ampicillin (10) | 18.88 a (1.50) | NZ a | 31.90 (0.05) | 31.81 (0.14) | NZ | NZ | NZ |
| Cephalothin (5) | 10.43 a (0.24) | NZ a | 31.67 (0.21) | 31.84 (0.16) | NZ | NZ | NZ |
| Colistin (25) | 14.41 (0.24) | 14.37 (0.23) | NZ | NZ | 15.92 (0.59) | 15.87 (0.59) | 13.86 (0.39) |
| Gentamicin (10) | 21.67 (0.22) | 21.90 (0.46) | 22.82 (0.084) | 22.76 (0.12) | 15.96 (0.82) | 15.62 (0.58) | 13.23 (3.29) |
| Streptomycin (10) | 18.18 (0.94) | 17.98 (0.83) | 19.11 (0.06) | 18.78 (0.21) | 10.00 (0.18) | 9.75 (0.37) | 7.20 (3.60) |
| Sulphatriad (200) | 31.26 (0.62) | 31.20 (0.56) | 26.13 (0.85) | 26.14 (0.83) | 15.11 a (0.58) | NZ a | NZ a |
| Tetracycline (25) | 21.99 (1.27) | 21.93 (1.27) | 23.16 (0.37) | 23.09 (0.37) | 8.779 (0.05) | 8.78 (0.07) | 10.00 (0.34) |
| Cotrimoxazole (25) | 27.46 (1.02) | 28.11 (0.77) | 20.35 (0.19) | 20.57 (0.13) | NZ | NZ | NZ |
| Chloramphenicol (25) | 20.42 (0.33) | 21.37 (0.33) | 20.03 (0.21) | 19.97 (0.22) | NZ | NZ | NZ |
| Erythromycin (5) | NZ | NZ | 17.12 (0.42) | 17.08 (0.38) | NZ | NZ | NZ |
| Fusidic Acid (10) | NZ | NZ | 26.71 (0.12) | 26.50 (0.22) | NZ | NZ | NZ |
| Oxacillin (5) | NZ | NZ | 22.28 a (0.44) | NZ a | NZ | NZ | NZ |
| Novabiocin (5) | NZ | NZ | 22.39 (0.07) | 22.35 (0.02) | NZ | NZ | NZ |
| Penicillin G 1 unit | NZ | NZ | 21.26 (0.24) | 21.23 (0.22) | NZ | NZ | NZ |
| Streptomycin (10) | 18.55 (0.99) | 19.04 (1.21) | 16.13 (0.67) | 16.09 (0.61) | 8.96 (1.26) | 8.96 (1.25) | 10.87 (0.34) |
| Tetracycline (25) | 20.77 (0.32) | 20.94 (0.15) | 21.06 (0.26) | 20.94 (0.21) | NZ | NZ | 7.82 (0.27) |
Note: NZ, no zone (6 mm disc diameter). Standard error shown in parentheses (n = 3). EcH2O2, H2O2 primed E. coli; SaCHG, CHG primed S. aureus; PaGTA, GTA primed P. aeruginosa; and PaBZK, BZK primed P. aeruginosa.
Changes in zones of inhibition where observed compared to parent strain.
To investigate the resistance more closely, the MIC of selected antibiotics against biocide primed strains of bacteria was determined (summarized in Table 3); the strain and antibiotic combinations tested were based on the results obtained from antibiotic disc diffusion tests. Due to the increased resistance of EcH2O2 to cephalothin (a first‐generation cephalosporin), both cefoxitin and ceftriaxone, second‐ and third‐generation cephalosporins respectively, were included in the MIC testing to see whether the cross‐resistance extended to newer antibiotics. There was an eightfold increased MIC for EcH2O2 to cephalothin from 4 mg l−1 for the parent strain to 32 mg l−1 for EcH2O2, a fourfold increase in MIC for EcH2O2 to cefoxitin to 16 mg l−1 compared to 4 mg l−1 in the parent strain and a twofold increase to ceftriaxone (parent 0.0625 mg l−1 and primed strain 0.125 mg l−1) and ampicillin (parent 2 mg l−1 and primed 4 mg l−1). The MIC breakpoint (EUCAST, 2020) for Enterobacterales reports resistance at >2 mg l−1 with ceftriaxone indicating no change in clinical susceptibility, data for cephalothin and cefoxitin are not available. A fourfold increased MIC for oxacillin was observed in SaCHG (2 mg l−1 compared to parent strain S. aureus 0.5 mg l−1; breakpoint data not available). In PaGTA, an eightfold increased MIC to ciprofloxacin was observed (1 mg l−1 compared to 0.125 mg l−1 for the parent P. aeruginosa strain) and a fourfold increased MIC to sulphatriad was also recorded. The MIC breakpoint (EUCAST, 2020) for Pseudomonas spp. and ciprofloxacin reports resistance at >0.5 mg l−1 indicating clinically significant resistance in PaGTA (data for sulphatriad are not available).
TABLE 3.
MICs of selected antibiotics against parent and primed strains of bacteria
| Strains | MIC (mg l−1) | ||||||
|---|---|---|---|---|---|---|---|
| Cephalothin | Cefoxitin | Ceftriaxone | Ampicillin | Oxacillin | Sulphatriad | Ciprofloxacin | |
| E. coli | 4 | 4 | 0.0625 | 2 | — | — | — |
| EcH2O2 | 32 a | 16 a | 0.125 a | 4 a | — | — | — |
| S. aureus | — | — | — | — | 0.5 | — | — |
| SaCHG | — | — | — | — | 2 a | — | — |
| P. aeruginosa | — | — | — | — | — | 256 | 0.125 |
| PaGTA | — | — | — | — | — | 1024 a | 1 a |
Note: EcH2O2, H2O2 primed E. coli; SaCHG, CHG primed S. aureus; PaGTA, GTA primed P. aeruginosa. Standard error shown in parentheses (n = 3).
Increase in MIC compared to parent strain.
The effect of efflux pump inhibitors
The MICs of cephalothin and cefoxitin against EcH2O2 were reduced in the presence of the efflux pump inhibitor TZ (from 32 to 8 mg l−1 for cephalothin and from 16 to 8 mg l−1 for cefoxitin; Table 4). The MIC of oxacillin against SaCHG was reduced from 2 to 0.25 mg l−1 in the presence of TZ and to 0.5 mg l−1 in the presence of CPZ (Table 5). The effect of TZ on the MIC of ciprofloxacin and sulphatriad against PaGTA could not be determined due to the turbidity, caused by insolubility, observed when 250 mg l−1 of TZ (a concentration that does not inhibit growth) was combined with either ciprofloxacin or sulphatriad. To further determine the effect of TZ in the presence of either antibiotics, contents of the previously observed turbid wells were spread onto agar and further incubated at 37°C for 24 h. Reduced growth on agar of PaGTA was observed from wells containing ciprofloxacin + TZ and sulphatriad + TZ, compared to wells with only ciprofloxacin, TZ or sulphatriad (data not shown). The same observation was seen in the case of PaBZK, where no growth on agar was observed from wells containing 128 mg l−1 BZK + TZ, compared to significant growth from wells with only BZK or TZ (data not shown).
TABLE 4.
The effect of TZ on the tolerance of cephalothin and cefoxitin in EcH2O2
| Strain | MIC (mg l−1) | ||||
|---|---|---|---|---|---|
| TZ | Cephalothin | + TZ | Cefoxitin | + TZ | |
| EcH2O2 | 128 | 32 | 8a | 16 | 8b |
Note: EcH2O2, hydrogen peroxide primed E. coli. TZ, thioridazine.
Reduced MIC for aCephalothin and bcefoxitin was observed in the presence of 40 mg l−1 thioridazine.
TABLE 5.
The effects of TZ and CPZ on oxacillin in SaCHG
| Strain | MIC (mg l−1), mode n = 3 | ||||
|---|---|---|---|---|---|
| TZ | CPZ | Ox | + ½ TZ | + ½ CPZ | |
| SaCHG | 31.25 | 64 | 2 | 0.25 | 0.5 |
Note: Reduced MIC to oxacillin was observed in the presence of TZ and CPZ. SaCHG, CHG primed S. aureus; TZ, thioridazine; CPZ, chlorpromazine. Standard error shown in parentheses (n = 3).
Confirmation of strain identity
16S rRNA sequencing confirmed all parent (wild type) and biocide primed strains used/generated in this study were at least 99.9% identical to the type strains; E. coli ATCC 8739, S. aureus ATCC 6538 and P. aeruginosa ATCC 15442 (data not shown).
Fitness of primed strains
Results of 16‐h growth curves of parent and primed strains are shown in Figure 1. There was no difference in the length of the lag phase between the E. coli parent strain and primed isolate, but growth was decreased from 4 h onwards (Figure 1a). For S. aureus, the CHG primed isolate had an extended lag phase compared to the parent strain, but overall growth was the same from 6 h onwards (Figure 1b). Interestingly, both the P. aeruginosa BZK and GTA isolates had an extended lag phase and decreased overall growth when compared to the parent strain (Figure 1c). This effect was more pronounced with the BZK isolate.
FIGURE 1.
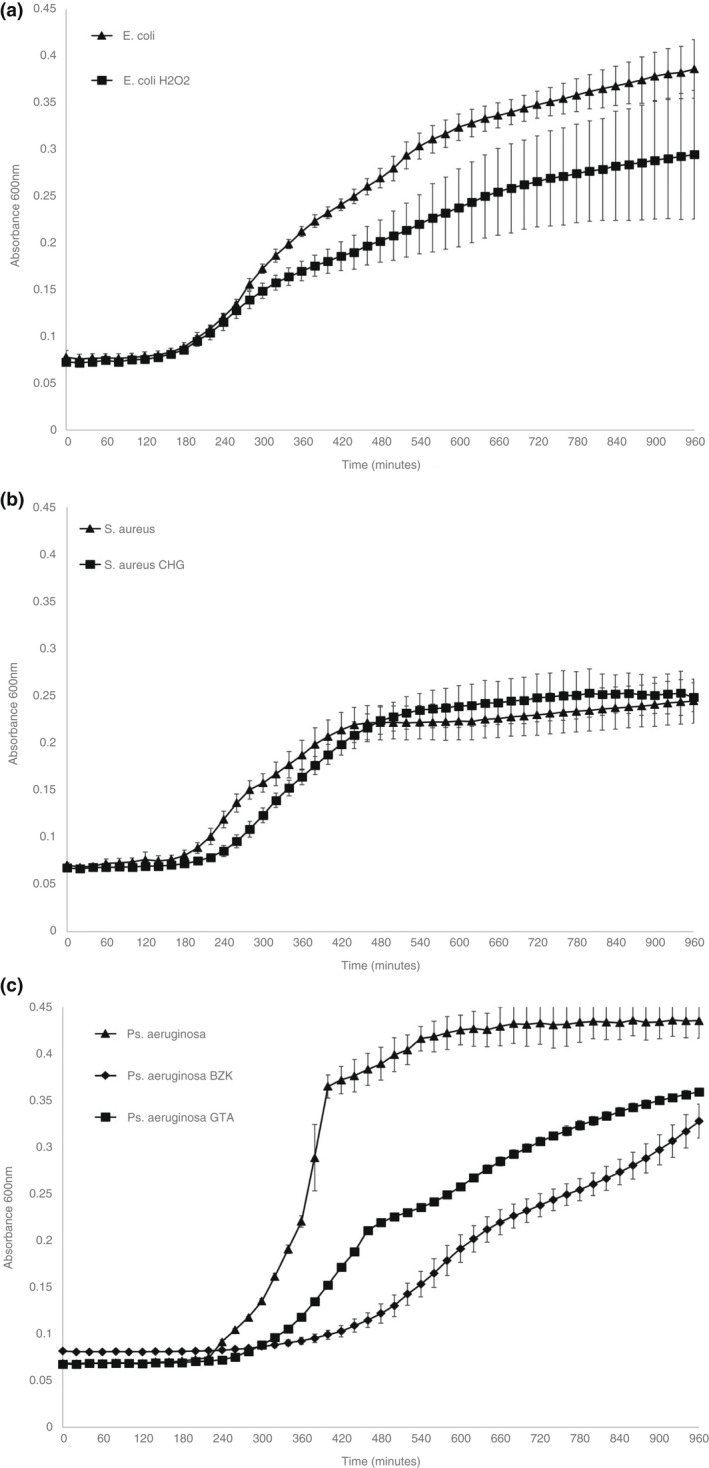
Growth curves of E. coli (a), S. aureus (b) and Ps. aeruginosa (c) primed and parent strains in antibiotic‐free Mueller Hinton broth, absorbance at 600 nm recorded for 16 h (n = 3, error bars denote standard deviation)
DISCUSSION
The goal of the present study was to test whether biocide priming has an effect on bacterial resistance to antibiotics. Low concentrations of H2O2 have previously been shown to promote tolerance to the biocide (Bogomolnaya et al., 2013; Imlay & Linn, 1987) as was initially seen in this study with E. coli. In contrast to previous studies, here the increased tolerance to H2O2 was short lived. Stable cross‐resistance of EcH2O2 to cephalothin, cefoxitin, ceftriaxone and ampicillin antibiotics was observed, similar to the findings of Pereira et al. (2021). Previous studies show H2O2 activates the SoxRS and the OxyR regulons (Aslund et al., 1999; Manchado et al., 2000) both of which control the expression of over 100 oxidative stress response genes (Blanchard et al., 2007, 2012; Demple, 1991), including acrAB, which encode a multidrug efflux pump (Dukan et al., 1996; Storz & Imlay, 1999). Although this might explain the cross‐tolerance to antibiotics after H2O2 priming observed here, further work is required to see if either the SoxRS or OxyR regulons are activated in the present study, by comparing the expression levels of genes involved in both parent and primed strains. This could help to establish the mechanism(s) behind the differences seen between this study and others. Specifically, the ability of biocides to prime for antibiotic resistance without the usual observation of stable biocidal tolerance. Exposure of S. aureus to CHG did not reduce its susceptibility to the biocide, contrasting the findings of Hardy et al. where prolonged exposure of S. aureus isolates to CHG led to reduced susceptibility (Hardy et al., 2017). Here, cross‐resistance to oxacillin was observed with no corresponding tolerance to the biocide.
The results from the present study show an increased MIC to BZK in PaBZK in comparison to the parent P. aeruginosa strain. Activation of multidrug efflux systems such as the RND‐type MexCD‐OprJ was previously linked to the use of sub‐inhibitory concentrations of membrane damaging biocides including BZK and CHG (Fraud et al., 2008; Morita et al., 2003). Previous adaptation of P. aeruginosa to sub‐inhibitory concentrations of BZK was attributed to increased efflux (Loughlin et al., 2002; McCay et al., 2010). In their work, McCay et al. (2010) observed cross‐resistance with ciprofloxacin in BZK primed P. aeruginosa grown in continuous culture, in contrast to this study whereby no cross‐resistance to antibiotics, including ciprofloxacin, was seen. This could be attributed to the different methodology used when priming strains as here a serial batch method, gradient plating, was used in comparison to the continuous culture method employed by McCay et al. (2010). Our findings are consistent with those of Loughlin et al., where cross‐resistance was observed with BZK and other quaternary ammonium compounds after serial batch culture (Loughlin et al., 2002). The varied outcome in these studies shows how different growth conditions may influence adaptation and selection of bacteria to antimicrobials.
There are several reports of bacterial resistance to GTA (Kampf et al., 2013; Kirschke et al., 2003; Simoes et al., 2006; Simões et al., 2011; Svetlikova et al., 2009; Tschudin‐Sutter et al., 2011), which was not observed in this study. Cross‐resistance after priming with GTA to two unrelated antibiotics, ciprofloxacin and sulphatriad, was however observed. A study using Pseudomonas fluorescens biofilms showed that exposure of the bacteria to GTA significantly induced expression of two genes encoding multidrug efflux pumps, PFLU2929 and PFLU3876, which appear to be orthologs of OprN and PA5159 in P. aeruginosa (Vikram et al., 2015). The cross‐resistance observed with unrelated antibiotics in this study might suggest efflux is involved, as seen previously (Ferreira et al., 2011; Maseda et al., 2009; Poole, 2002; Sanchez et al., 2005).
Phenothiazines such as TZ and CPZ have been shown to potentiate the effect of antimicrobials against bacteria (Viveiros & Amaral, 2001; Wainwright et al., 1998), eliminate antibiotic resistant plasmids (Evdokimova et al., 1997; Radhakrishnan et al., 1999) and inhibit bacterial efflux pumps (Costa et al., 2013; Machado et al., 2018; Ordway et al., 2003; Viveiros & Amaral, 2001). The most well‐studied efflux pump inhibitor is CPZ, but both CPZ and TZ have the same antimicrobial properties against efflux and phagocytosed bacteria (Machado et al., 2018; Ordway, Viveiros, Leandro, Arroz, & Amaral, 2002; Ordway, Viveiros, Leandro, Arroz, Molnar, et al., 2002). Here we tested the effect of TZ at sub‐inhibitory concentration on EcH2O2 in the presence of varying concentrations of cephalothin and cefoxitin, to which the primed strain had previously shown cross‐resistance. TZ greatly increased the susceptibility of EcH2O2 to cephalothin and cefoxitin, suggesting that efflux mechanisms may be involved in the observed cross‐resistance. Our results corroborate the findings of Amaral et al. where CPZ reduced the MIC of ceftazidime and ceftriaxone against E. coli from 1.0 to 0.08 mg l−1 and 0.07 mg l−1, respectively (Amaral et al., 1992). These findings suggest that increased efflux might be a contributory factor in the cross‐resistance observed with EcH2O2.
When the effect of both TZ and CPZ on MIC levels of oxacillin on SaCHG was evaluated, the results revealed a significant reduction of MIC in the presence of CPZ and TZ. These results suggest that efflux mechanisms may be contributing to the cross‐resistance observed. Furthermore, this agrees with the findings of Kristiansen et al. (2006) and Costa et al. (2013).
Although efflux contributes highly to antimicrobial resistance in P. aeruginosa, reduced influx/impermeability has also been shown to contribute (Li et al., 2000), for example mutation in or loss of the transmembrane porin OprD is significant in resistance to carbapenems (Bradford et al., 1999). Thus, a combination of increased efflux and decreased influx may contribute to antimicrobial resistance in P. aeruginosa resistance, even though it was not fully demonstrated with the methods used here.
The increased susceptibility seen with some biocide primed strains to antibiotics in the presence of EPIs suggests efflux as a contributory mechanism to the cross‐resistance observed in this study. The next step will be to employ the use of q qPCR to compare gene expression in parent and primed strains of bacteria.
The growth curve results comparing both parent and primed strains indicated that the resistance phenotype did have an impact on overall growth in the case of E. coli, the length of the lag phase with S. aureus and both overall growth and lag phase with P. aeruginosa. This suggests that although there may be potential fitness cost associated with these phenotypic adaptations in terms of initial or overall speed of replication, this is not impacting on the conservation of the resistance after repeated subculture under laboratory conditions. This agrees with other findings, where generally most mutations had an impact on fitness (Melnyk et al., 2015). It is not unexpected to observe this with biocide induced cross‐resistance, as biocides generally have broader modes of action in terms of cellular targets.
Results from this study clearly demonstrate that continuous exposure to sub‐inhibitory concentrations of biocides far below the recommended in‐use concentration can, under laboratory conditions, prime bacteria to become resistant to antibiotics even in the absence of increased tolerance to the biocides. This raises the important question of whether this phenomenon is occurring in clinical settings and contributing to dissemination to antimicrobial resistance.
CONFLICT OF INTEREST
None declared.
Adkin, P. , Hitchcock, A. , Smith, L.J. & Walsh, S.E. (2022) Priming with biocides: A pathway to antibiotic resistance?. Journal of Applied Microbiology, 133, 830–841. Available from: 10.1111/jam.15564
REFERENCES
- Amaral, L. , Kristiansen, J. & Lorian, V. (1992) Synergic effect of chlorpromazine on the activity of some antibiotics. J Antimicrob Chemother, 4, 556–558. [DOI] [PubMed] [Google Scholar]
- Aslund, F. , Zheng, M. , Beckwith, J. & Storz, G. (1999) Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol‐disulfide status. Proc Natl Acad Sci USA, 11, 6161–6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard, J.L. , Wholey, W. , Conlon, E.M. & Pomposiello, P.J. (2007) Rapid changes in gene expression dynamics in response to superoxide reveal SoxRS‐dependent and independent transcriptional networks. PLoS One, 2, e1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard, J.L. , Wholey, W. , Conlon, E.M. & Pomposiello, P.J. (2012) Correction: rapid changes in gene expression dynamics in response to superoxide reveal SoxRS‐dependent and independent transcriptional networks. PLoS One, 7, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock, L. , Wand, M. & Sutton, J. (2016) Varying activity of chlorhexidine‐based disinfectants against Klebsiella pneumoniae clinical isolates and adapted strains. J Hosp Infect, 93, 42–48. [DOI] [PubMed] [Google Scholar]
- Bogomolnaya, L.M. , Andrews, K.D. , Talamantes, M. , Maple, A. , Ragoza, Y. , Vazquez‐Torres, A. et al. (2013) The ABC‐type efflux pump MacAB protects Salmonella enterica serovar typhimurium from oxidative stress. mBio, 4, e00630‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher, H. , Miller, L.G. & Razonable, R.R. (2010) Serious infections caused by methicillin‐resistant Staphylococcus aureus . Clin Infect Dis, 51, 183–S197. [DOI] [PubMed] [Google Scholar]
- Bradford, P.A. , Petersen, P.J. , Fingerman, I.M. & White, D.G. (1999) Characterization of expanded‐spectrum cephalosporin resistance in E. coli isolates associated with bovine calf diarrhoeal disease. J Antimicrob Chemother, 44, 607–610. [DOI] [PubMed] [Google Scholar]
- Braoudaki, M. & Hilton, A.C. (2004) Adaptive resistance to biocides in Salmonella enterica and Escherichia coli O157 and cross‐resistance to antimicrobial agents. J Clin Microbiol, 42, 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzuszkiewicz, E. , Thürmer, A. , Schuldes, J. , Leimbach, A. , Liesegang, H. , Meyer, F.D. et al. (2011) Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: Entero‐Aggregative‐Haemorrhagic Escherichia coli (EAHEC). Arch Microbiol, 193, 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BSI 2006, EN ISO 20776‐1 (2006) Clinical laboratory testing and in vitro diagnostic test systems. Susceptibility testing of infectious agents and evaluation of performance of antimicrobial susceptibility test devices. Part 1. Reference method for testing the in vitro activity of antimicrobial agents against rapidly growing aerobic bacteria involved in infectious diseases, BSI, London United Kingdom.
- Christensen, E.G. , Gram, L. & Kastbjerg, V.G. (2011) Sublethal triclosan exposure decreases susceptibility to gentamicin and other aminoglycosides in Listeria monocytogenes . Antimicrob Agents Chemother, 55, 4064–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuanchuen, R. , Beinlich, K. , Hoang, T.T. , Becher, A. , Karkhoff‐Schweizer, R.R. & Schweizer, H.P. (2001) Cross‐resistance between triclosan and antibiotics in Pseudomonas aeruginosa is mediated by multidrug efflux pumps: exposure of a susceptible mutant strain to triclosan selects nfxB mutants overexpressing MexCD‐OprJ. Antimicrob Agents Chemother, 45, 428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, S.S. , Junqueira, E. , Palma, C. , Viveiros, M. , Melo‐Cristino, J. , Amaral, L. et al. (2013) Resistance to antimicrobials mediated by efflux pumps in Staphylococcus aureus . Antibiotics, 2, 83–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demple, B. (1991) Regulation of bacterial oxidative stress genes. Annu Rev Genet, 25, 315–337. [DOI] [PubMed] [Google Scholar]
- Dukan, S. , Dadon, S. , Smulski, D.R. & Belkin, S. (1996) Hypochlorous acid activates the heat shock and soxRS systems of Escherichia coli . Appl Environ Microbiol, 62, 4003–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalada, M.G. , Harwood, J. , Maillard, J. & Ochs, D. (2005) Triclosan inhibition of fatty acid synthesis and its effect on growth of Escherichia coli and Pseudomonas aeruginosa . J Antimicrob Chemother, 55, 879–882. [DOI] [PubMed] [Google Scholar]
- EUCAST (2015) EUCAST disk diffusion method, version 5.0. European Society of Clinical Microbiology and Infectious Diseases, 1–21.
- EUCAST (2020) European Committee on Antimicrobial Susceptibility Testing: Breakpoint Tables for Interpretation of MICs and Zone Diameters. European Society of Clinical Microbiology and Infectious Diseases
- European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) . (2000) Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by agar dilution. Clin Microbiol Infect, 6, 509–515. [DOI] [PubMed] [Google Scholar]
- Evdokimova, O.V. , Smirnov, I.V. , Artem’eva, N.A. & Rozhkova, E.A. (1997) Effect of promethazine hydrochloride (pipolphen) on the stability of R plasmid resistance in Escherichia coli . Antibiot. Khimioter, 42, 8–11. [PubMed] [Google Scholar]
- Fernández Márquez, M.L. , Burgos, M.J.G. , Pulido, R.P. , Gálvez, A. & López, R.L. (2017) Biocide tolerance and antibiotic resistance in Salmonella isolates from hen eggshells. Foodborne Pathog Dis, 14, 89–95. [DOI] [PubMed] [Google Scholar]
- Ferreira, C. , Pereira, A. , Pereira, M. , Melo, L. & Simões, M. (2011) Physiological changes induced by the quaternary ammonium compound benzyldimethyldodecylammonium chloride on Pseudomonas fluorescens . J Antimicrob Chemother, 66, 1036–1043. [DOI] [PubMed] [Google Scholar]
- Fraise, A. (2002) Biocide abuse and antimicrobial resistance – a cause for concern? J Antimicrob Chemother, 49, 11–12. [DOI] [PubMed] [Google Scholar]
- Fraud, S. , Campigotto, A.J. , Chen, Z. & Poole, K. (2008) MexCD‐OprJ multidrug efflux system of Pseudomonas aeruginosa: involvement in chlorhexidine resistance and induction by membrane‐damaging agents dependent upon the AlgU stress response sigma factor. Antimicrob Agents Chemother, 52, 4478–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, P. & McBain, A.J. (2003) Potential impact of increased use of biocides in consumer products on prevalence of antibiotic resistance. Clin Microbiol Rev, 16, 189–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy, K. , Sunnucks, K. , Gil, H. , Shabir, S. , Trampari, E. , Hawkey, P. et al. (2017) Increased usage of antiseptics is associated with reduced susceptibility in clinical isolates of S. aureus . mBio, 9, e00894–e00818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heir, E. , Sundheim, G. & Holck, A.L. (1998) The Staphylococcus qacH gene product: a new member of the SMR family encoding multidrug resistance. FEMS Microbiol Lett, 163, 49–56. [DOI] [PubMed] [Google Scholar]
- Imlay, J.A. & Linn, S. (1987) Mutagenesis and stress responses induced in Escherichia coli by hydrogen peroxide. J Bacteriol, 169, 2967–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe, A. , Chabbert, Y.A. & Semonin, O. (1982) Role of porin proteins OmpF and OmpC in the permeation of beta‐lactams. Antimicrob Agents Chemother, 22, 942–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampf, G. , Ostermeyer, C. , Tschudin‐Sutter, S. & Widmer, A. (2013) Resistance or adaptation? How susceptible is a glutaraldehyde‐resistant Pseudomonas aeruginosa isolate in the absence of selection pressure? J Hosp Infect, 84, 316–318. [DOI] [PubMed] [Google Scholar]
- Kaper, J.B. , Nataro, J.P. & Mobley, H.L. (2004) Pathogenic Escherichia coli . Nat Rev Microbiol, 2, 123–140. [DOI] [PubMed] [Google Scholar]
- Karatzas, K.A. , Webber, M.A. , Jorgensen, F. , Woodward, M.J. , Piddock, L.J. & Humphrey, T.J. (2007) Prolonged treatment of Salmonella enterica serovar Typhimurium with commercial disinfectants selects for multiple antibiotic resistance, increased efflux and reduced invasiveness. J Antimicrob Chemother, 60, 947–955. [DOI] [PubMed] [Google Scholar]
- Karatzas, K.A. , Randall, L.P. , Webber, M. , Piddock, L.J. , Humphrey, T.J. , Woodward, M.J. et al. (2008) Phenotypic and proteomic characterization of multiply antibiotic‐resistant variants of Salmonella enterica serovar Typhimurium selected following exposure to disinfectants. Appl Environ Microbiol, 74, 1508–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama, H. , Hamashima, H. , Sasatsu, M. & Arai, T. (1998) Distribution of the antiseptic‐resistance gene qacE Δ 1 in Gram‐positive bacteria. FEMS Microbiol Lett, 165, 295–299. [DOI] [PubMed] [Google Scholar]
- Kirschke, D.L. , Jones, T.F. , Craig, A.S. , Chu, P.S. , Mayernick, G.G. , Patel, J.A. et al. (2003) Pseudomonas aeruginosa and Serratia marcescens contamination associated with a manufacturing defect in bronchoscopes. N Engl J Med, 348, 214–220. [DOI] [PubMed] [Google Scholar]
- Knapp, L. , Rushton, L. , Stapleton, H. , Sass, A. , Stewart, S. , Amezquita, A. et al. (2013) The effect of cationic microbicide exposure against Burkholderia cepacia complex (Bcc); the use of Burkholderia lata strain 383 as a model bacterium. J Appl Microbiol, 115, 1117–1126. [DOI] [PubMed] [Google Scholar]
- Knapp, L. , Amezquita, A. , McClure, P. , Stewart, S. & Maillard, J.Y. (2015) Development of a protocol for predicting bacterial resistance to microbicides. Appl Environ Microbiol, 81, 2652–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen, M.M. , Leandro, C. , Ordway, D. , Martins, M. , Viveiros, M. , Pacheco, T. et al. (2006) Thioridazine reduces resistance of methicillin‐resistant Staphylococcus aureus by inhibiting a reserpine‐sensitive efflux pump. In vivo (Athens, Greece), 20, 361–366. [PubMed] [Google Scholar]
- Kurenbach, B. , Marjoshi, D. , Amabile‐Cuevas, C.F. , Ferguson, G.C. , Godsoe, W. , Gibson, P. et al. (2015) Sublethal exposure to commercial formulations of the herbicides dicamba, 2,4‐dichlorophenoxyacetic acid, and glyphosate cause changes in antibiotic susceptibility in Escherichia coli and Salmonella enterica serovar Typhimurium. mBio, 6, e00009–e00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert, P.A. (2002) Mechanisms of antibiotic resistance in Pseudomonas aeruginosa . J R Soc Med, 95, 22–26. [PMC free article] [PubMed] [Google Scholar]
- Latimer, J. , Forbes, S. & McBain, A.J. (2012) Attenuated virulence and biofilm formation in Staphylococcus aureus following sublethal exposure to triclosan. Antimicrob Agents Chemother, 56, 3092–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Zhang, L. & Poole, K. (2000) Interplay between the MexA‐MexB‐OprM multidrug efflux system and the outer membrane barrier in the multiple antibiotic resistance of Pseudomonas aeruginosa . J Antimicrob Chemother, 45, 433–436. [DOI] [PubMed] [Google Scholar]
- Loughlin, M. , Jones, M. & Lambert, P. (2002) Pseudomonas aeruginosa cells adapted to benzalkonium chloride show resistance to other membrane‐active agents but not to clinically relevant antibiotics. J Antimicrob Chemother, 49, 631–639. [DOI] [PubMed] [Google Scholar]
- Ma, D. , Cook, D.N. , Alberti, M. , Pon, N. , Nikaido, H. & Hearst, J. (1993) Molecular cloning and characterization of acrA and acrE genes of Escherichia coli . J Bacteriol, 175, 6299–6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, D. , Cook, D.N. , Hearst, J.E. & Nikaido, H. (1994) Efflux pumps and drug resistance in Gram‐negative bacteria. Trends Microbiol, 2, 489–493. [DOI] [PubMed] [Google Scholar]
- Machado, D. , Girardini, M. , Viveiros, M. & Pieroni, M. (2018) Challenging the drug‐likeness dogma for new drug discovery in tuberculosis. Front Microbiol, 9, 1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard, J. (2002) Bacterial target sites for biocide action. J Appl Microbiol, 92, 16S–27S. [PubMed] [Google Scholar]
- Maillard, J. , Bloomfield, S. , Coelho, J.R. , Collier, P. , Cookson, B. , Fanning, S. et al. (2013) Does microbicide use in consumer products promote antimicrobial resistance? A critical review and recommendations for a cohesive approach to risk assessment. Microb Drug Resist, 19, 344–354. [DOI] [PubMed] [Google Scholar]
- Manchado, M. , Michan, C. & Pueyo, C. (2000) Hydrogen peroxide activates the SoxRS regulon in vivo. J Bacteriol, 182, 6842–6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzoor, S. , Lambert, P. , Griffiths, P. , Gill, M. & Fraise, A. (1999) Reduced glutaraldehyde susceptibility in Mycobacterium chelonae associated with altered cell wall polysaccharides. J Antimicrob Chemother, 43, 759–765. [DOI] [PubMed] [Google Scholar]
- Maseda, H. , Hashida, Y. , Konaka, R. , Shirai, A. & Kourai, H. (2009) Mutational upregulation of a resistance‐nodulation‐cell division‐type multidrug efflux pump, SdeAB, upon exposure to a biocide, cetylpyridinium chloride, and antibiotic resistance in Serratia marcescens . Antimicrob Agents Chemother, 53, 5230–5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mc Cay, P.H. , Ocampo‐Sosa, A.A. & Fleming, G.T. (2010) Effect of sub inhibitory concentrations of benzalkonium chloride on the competitiveness of Pseudomonas aeruginosa grown in continuous culture. Microbiology, 156, 30–38. [DOI] [PubMed] [Google Scholar]
- McBain, A.J. & Gilbert, P. (2001) Biocide tolerance and the harbingers of doom. Int Biodeterior Biodegrad, 47, 55–61. [Google Scholar]
- Melnyk, A.H. , Wong, A. & Kassen, R. (2015) The fitness costs of antibiotic resistance mutations. Evol Appl, 8, 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morente, E.O. , Fernández‐Fuentes, M.A. , Burgos, M.J.G. , Abriouel, H. , Pulido, R.P. & Galvez, A. (2013) Biocide tolerance in bacteria. Int J Food Microbiol, 162, 13–25. [DOI] [PubMed] [Google Scholar]
- Morita, Y. , Murata, T. , Mima, T. , Shiota, S. , Kuroda, T. , Mizushima, T. et al. (2003) Induction of mexCD‐oprJ operon for a multidrug efflux pump by disinfectants in wild‐type Pseudomonas aeruginosa PAO1. J Antimicrob Chemother, 51, 991–994. [DOI] [PubMed] [Google Scholar]
- Ordway, D. , Viveiros, M. , Leandro, C. , Arroz, M.J. & Amaral, L. (2002a) Intracellular activity of clinical concentrations of phenothiazines including thioridiazine against phagocytosed Staphylococcus aureus . Int J Antimicrob Agents, 20, 34–43. [DOI] [PubMed] [Google Scholar]
- Ordway, D. , Viveiros, M. , Leandro, C. , Arroz, M.J. , Molnar, J. , Kristiansen, J.E. et al. (2002b) Chlorpromazine has intracellular killing activity against phagocytosed Staphylococcus aureus at clinical concentrations. J Inf Chemother, 8, 227–231. [DOI] [PubMed] [Google Scholar]
- Ordway, D. , Viveiros, M. , Leandro, C. , Bettencourt, R. , Almeida, J. , Martins, M. et al. (2003) Clinical concentrations of thioridazine kill intracellular multidrug‐resistant Mycobacterium tuberculosis . Antimicrob Agents Chemother, 47, 917–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, B.M.P. , Wang, X. & Tagkopoulos, I. (2021) Biocide‐induced emergence of antibiotic resistance in Escherichia coli . Front Microbiol, 12, 640923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole, K. (2001) Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J Mol Microbiol Biotech, 3, 255–264. [PubMed] [Google Scholar]
- Poole, K. (2002) Mechanisms of bacterial biocide and antibiotic resistance. J Appl Microbiol, 92, 55S–64S. [PubMed] [Google Scholar]
- Radhakrishnan, V. , Ganguly, K. , Ganguly, M. , Dastidar, S.G. & Chakrabarty, A. (1999) Potentiality of tricyclic compound thioridazine as an effective antibacterial and antiplasmid agent. Indian J Exp Biol, 37, 671–675. [PubMed] [Google Scholar]
- Randall, L. , Cooles, S. , Coldham, N. , Penuela, E. , Mott, A. , Woodward, M.J. et al. (2007) Commonly used farm disinfectants can select for mutant Salmonella enterica serovar Typhimurium with decreased susceptibility to biocides and antibiotics without compromising virulence. J Antimicrob Chemother, 60, 1273–1280. [DOI] [PubMed] [Google Scholar]
- Roca Subirà, I. , Espinal, P. , Vila‐Farrés, X. & Vila Estapé, J. (2012) The Acinetobacter baumannii oxymoron: commensal hospital dweller turned pan‐drug‐resistant menace. Front Microbiol, 3, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouch, D. , Cram, D. , Berardino, D. , Littlejohn, T. & Skurray, R. (1990) Efflux‐mediated antiseptic resistance gene qacA from Staphylococcus aureus: common ancestry with tetracycline‐and sugar‐transport proteins. Mol Microbiol, 4, 2051–2062. [DOI] [PubMed] [Google Scholar]
- Russell, A. (2002) Introduction of biocides into clinical practice and the impact on antibiotic‐resistant bacteria. J Appl Microbiol, 92, 121S–135S. [PubMed] [Google Scholar]
- Sanchez, P. , Moreno, E. & Martinez, J.L. (2005) The biocide triclosan selects Stenotrophomonas maltophilia mutants that overproduce the SmeDEF multidrug efflux pump. Antimicrob Agents Chemother, 49, 781–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCENIHR . (2010) Research strategy to address the knowledge gaps on the antimicrobial resistance effects of biocides. Brussels, Belgium: European Commission. [Google Scholar]
- Schweizer, H.P. (1998) Intrinsic resistance to inhibitors of fatty acid biosynthesis in Pseudomonas aeruginosa is due to efflux: application of a novel technique for generation of unmarked chromosomal mutations for the study of efflux systems. Antimicrob Agents Chemother, 42, 394–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes, M. , Pereira, M.O. , Machado, I. , Simões, L.C. & Vieira, M. (2006) Comparative antibacterial potential of selected aldehyde‐based biocides and surfactants against planktonic Pseudomonas fluorescens . J Ind Microbiol Biotechnol, 33, 741–749. [DOI] [PubMed] [Google Scholar]
- Simões, L.C. , Lemos, M. , Araújo, P. , Pereira, A.M. & Simões, M. (2011) The effects of glutaraldehyde on the control of single and dual biofilms of Bacillus cereus and Pseudomonas fluorescens . Biofouling, 27, 337–346. [DOI] [PubMed] [Google Scholar]
- Slayden, R.A. , Lee, R.E. & Barry, C.E. (2000) Isoniazid affects multiple components of the type II fatty acid synthase system of Mycobacterium tuberculosis . Mol Microbiol, 38, 514–525. [DOI] [PubMed] [Google Scholar]
- Soothill, J. (2013) Use of bacteriophages in the treatment of Pseudomonas aeruginosa infections. Expert Rev Anti‐Infect Ther, 11, 909–915. [DOI] [PubMed] [Google Scholar]
- Soumet, C. , Fourreau, E. , Legrandois, P. & Maris, P. (2012) Resistance to phenicol compounds following adaptation to quaternary ammonium compounds in Escherichia coli . Vet Microbiol, 158, 147–152. [DOI] [PubMed] [Google Scholar]
- Storz, G. & Imlay, J.A. (1999) Oxidative stress. Curr Opin Microbiol, 2, 188–194. [DOI] [PubMed] [Google Scholar]
- Svetlikova, Z. , Skovierova, H. , Niederweis, M. , Gaillard, J.L. , McDonnell, G. & Jackson, M. (2009) Role of porins in the susceptibility of Mycobacterium smegmatis and Mycobacterium chelonae to aldehyde‐based disinfectants and drugs. Antimicrob Agents Chemother, 53, 4015–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkachenko, O. , Shepard, J. , Aris, V.M. , Joy, A. , Bello, A. , Londono, I. et al. (2007) A triclosan‐ciprofloxacin cross‐resistant mutant strain of Staphylococcus aureus displays an alteration in the expression of several cell membrane structural and functional genes. Res Microbiol, 158, 651–658. [DOI] [PubMed] [Google Scholar]
- Truong‐Bolduc, Q.C. , Strahilevitz, J. & Hooper, D.C. (2006) NorC, a new efflux pump regulated by MgrA of Staphylococcus aureus . Antimicrob Agents Chemother, 50, 1104–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschudin‐Sutter, S. , Frei, R. , Kampf, G. , Tamm, M. , Pflimlin, E. , Battegay, M. et al. (2011) Emergence of glutaraldehyde‐resistant Pseudomonas aeruginosa . Infect Control Hosp Epidemiol, 32, 1173–1178. [DOI] [PubMed] [Google Scholar]
- Tuon, F.F. , Gortz, L.W. & Rocha, J.L. (2012) Risk factors for pan‐resistant Pseudomonas aeruginosa bacteremia and the adequacy of antibiotic therapy. Braz J Infect Dis, 16, 351–356. [DOI] [PubMed] [Google Scholar]
- Vikram, A. , Bomberger, J.M. & Bibby, K.J. (2015) Efflux as a glutaraldehyde resistance mechanism in Pseudomonas fluorescens and Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother, 59, 3433–3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viveiros, M. & Amaral, L. (2001) Enhancement of antibiotic activity against poly‐drug resistant Mycobacterium tuberculosis by phenothiazines. Int J Antimicrob Agents, 17, 225–228. [DOI] [PubMed] [Google Scholar]
- Wainwright, M. , Phoenix, D. , Laycock, S. , Wareing, D. & Wright, P. (1998) Photobactericidal activity of phenothiazinium dyes against methicillin‐resistant strains of Staphylococcus aureus . FEMS Microbiol Lett, 160, 177–181. [DOI] [PubMed] [Google Scholar]
- Walsh, T.R. & Toleman, M.A. (2011) The emergence of pan‐resistant Gram‐negative pathogens merits a rapid global political response. J Antimicrob Chemother, 67, 1–3. [DOI] [PubMed] [Google Scholar]
- Walsh, S.E. , Maillard, J. , Russell, A. , Catrenich, C. , Charbonneau, D. & Bartolo, R. (2003) Development of bacterial resistance to several biocides and effects on antibiotic susceptibility. J Hosp Infect, 55, 98–107. [DOI] [PubMed] [Google Scholar]
- Wand, M.E. (2017) Bacterial resistance to hospital disinfection. In Modeling the transmission and prevention of infectious disease Andersen, pp. 19–54. Springer, Switzerland. [Google Scholar]
- Wand, M.E. , Bock, L.J. , Bonney, L.C. & Sutton, J.M. (2016) Mechanisms of increased resistance to chlorhexidine and cross‐resistance to colistin following exposure of Klebsiella pneumoniae clinical isolates to chlorhexidine. Antimicrob Agents Chemother, 61, e01162‐16. 10.1128/AAC.01162-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead, R.N. , Overton, T.W. , Kemp, C.L. & Webber, M.A. (2011) Exposure of Salmonella enterica serovar Typhimurium to high level biocide challenge can select multidrug resistant mutants in a single step. PLoS One, 6, e22833. [DOI] [PMC free article] [PubMed] [Google Scholar]


