Abstract
The β-lactamase-producing Asia-type plasmid pJD4 of Neisseria gonorrhoeae is a 7.4-kb, broad-host-range plasmid. It is part of a family of plasmids which are structurally related yet vary in size, found in both N. gonorrhoeae and Haemophilus ducreyi. Branch-point analysis by electron microscopy indicates that pJD4 carries three clustered but distinguishable origins of replication, which we named ori1, ori2, and ori3. Although pJD4 belongs to incompatibility (Inc) group W, it also carries a silent IncFII determinant which is expressed when ori2 and ori3 are absent. The Africa-type plasmid pJD5, a naturally occurring deletion derivative of pJD4, carries only ori1, belongs to the IncFII group, and, in contrast to pJD4, requires DNA polymerase I (Pol I) for replication. Plasmids constructed from pJD4 which lack ori1 but carry ori2 and ori3 do not require Pol I and are incompatible with IncW plasmids, suggesting that the ori2 or ori3 region contains the IncW determinant. We have cloned a replication initiation protein (RepB) that is necessary for ori2 and ori3 to function. This Rep protein is distinct from RepA, which is necessary for ori1. Thus, pJD4 is unique because it is the smallest plasmid characterized containing three origins of replication and two unique Rep proteins.
Plasmids of gram-negative bacteria containing three origins of replication are rare (1, 9, 23); to date, only three such naturally occurring plasmids have been described. The first plasmid discovered, F (94.5 kb), contains remnants of three independent replication regions, RepFIC, RepFIA, and RepFIB (3, 8). A 9-kb mini-F plasmid, containing the complete RepF1A region, includes seven genes encoding proteins involved in the replication and maintenance of the plasmid, including a single replication initiation protein (Rep) and two origins, oriV and oriS (8). Related plasmids lacking the RepF1A region require RepF1B, a less stable replicon (27). The RepF1C replicon was shown to have been rendered nonfunctional on the F plasmid by the natural insertion of the transposon Tn1000 (25, 40). This replicon, in members of the F plasmid family, resembles the replication region of the plasmid R1 (25). The IncN plasmid pCU1 contains three origins of replication (oriB, oriS, and oriV), all located on a 2-kb DNA fragment, which are driven by a single Rep protein (1, 38). The third and best-characterized plasmid with three origins (α, β, and γ) is the 38-kb plasmid RK6. This plasmid also carries genes for two replication proteins, pir (encoding the π protein for origins α and γ) and bis (Bis for origin β) (31). These origins and genes are located on a 4-kb DNA fragment (18, 31).
It appears that the penicillinase-producing plasmids of N. gonorrhoeae contain different and multiple origins of replication as well. These plasmids belong to a family of plasmids that are genetically related (33). Since the isolation of the first penicillinase-producing isolate in 1976, their worldwide spread and prevalence (i.e., over 50 to 80% of all gonococci isolated in some countries) have led to the demise of penicillin as a useful antibiotic for treating gonococcal infections (13). Penicillinase production in these plasmids is mediated by a TEM1-type β-lactamase encoded by the TnA transposon Tn2, which is truncated and includes 84% of tnpR, noncoding sequences, the entire bla gene, and the right inverted repeat (IR-R) (6, 15). Initially, a 3.8-kb BamHI-PvuII fragment of a 7.4-kb Asia-type plasmid was shown to be essential for replication (24). When this fragment was cloned into ColE1-type plasmids such as pBR322, pMB8, and pBluescript KS+, these vectors which normally depend on DNA polymerase I (Pol I) could be maintained in polA hosts, indicating that the replication was independent of polA (12). Subsequently, Yeung and Dillon (45) constructed nested deletions of the Asia-type plasmid (pJD4) and deduced that this plasmid had two replication origin regions, which they designated a and b. The b region was further characterized on plasmid pFA3, an Asia-type plasmid similar to pJD4, and included a putative replication protein (19). None of the replication regions have been additionally characterized since.
In the present study, we investigate further the properties of the origins of replication regions of the β-lactamase-producing plasmids of N. gonorrhoeae (45). Since broad-host-range gonococcal plasmids such as pJD4 have been shown to replicate in N. gonorrhoeae, Escherichia coli, Salmonella enterica serotype Minnesota, and Haemophilus influenzae (21), the present study focuses on origin usage in E. coli. Electron microscopy (EM) was used to confirm and locate origins in both naturally occurring and in vitro deletion derivatives of the Asia-type plasmid pJD4. The incompatibility of the origins to other enteric-derived incompatibility determinants was ascertained. Structural properties for each ori, as determined by DNA sequence analysis and cloning of putative Rep proteins, was determined by molecular and genetic approaches.
MATERIALS AND METHODS
E. coli strains and plasmids.
E. coli strains and the various plasmids used in this study are listed in Table 1. Bacterial cultures were maintained in brain heart infusion broth (Difco, Detroit, Mich.) containing 15% glycerol and stored at −70°C. E. coli strains were subcultured from frozen cells by growing them overnight on Luria-Bertani (LB) broth or agar (Difco) at 37°C containing ampicillin (100 μg/ml), chloramphenicol (50 μg/ml), or tetracycline (50 μg/ml) where appropriate. Individual colonies were purified by subculturing for 3 days prior to plasmid DNA isolation. Restriction endonuclease (R/E) analysis was performed as previously described (11) to ensure plasmid integrity before enriching for plasmid replicative molecules or for use in incompatibility studies. Plasmid maps are shown in Fig. 1, and details are based on our previously submitted (33) DNA sequences [GenBank accession U20374 (pJD4), U20375 (pJD5)] and work in this study.
TABLE 1.
Bacterial strains and plasmids
| E. coli strain | Plasmid | GenBank accession no. | Description and source or reference |
|---|---|---|---|
| SR1758 | Δ(gal bio) thi-1 relA1 spoT1 (1) | ||
| SR1672 | ΔpolA Δ(gal bio) thi-1 relA1 spoT1 (1) | ||
| C600 | F−thi-1 leuB6 lacY1 tonA21 supE44 (Stratagene) | ||
| DH5α | F− φ80dlacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR-17 (rk− mk+) supE44 relA1 deoR Δ(lacZYA-argF)U169 (Stratagene) | ||
| BLR(DE3) | F−ompT hsdSB(rB−mB−) gal dcm Δ(srl-recA)306::Tn10 (DE3) (Novagen) | ||
| pBluescript KSII | X52328 | Stratagene | |
| pORF15 | pFP9 with 80% of ORF15 deleted (this study) | ||
| pBlueORF15 | pBluescript KSII containing ORF15 under T7 promoter (this study) | ||
| pACYC184 | X06403 | New England Biolabs | |
| C600-JD4 | pJD4 | U20374 | Asia plasmid (12) |
| C600-JD5 | pJD5 | U20375 | Africa plasmid (12) |
| C600-JD7 | pJD7 | U20419 | Toronto plasmid (12) |
| C600-JD9 | pJD9 | U20420 | In vitro deletion derivative of pJD4 (45) |
| C600-FP5 | pFP5 | Non-Tn2 region of pJD5 PCR amplified and ligated to chloramphenicol acetyltransferase gene (this study) | |
| C600-FP9 | pFP9 | Non-Tn2 region of pJD9 PCR amplified and ligated to chloramphenicol acetyltransferase gene (this study) | |
| C600-GF1 | pGF1 | U20421 | Nimes plasmid (20) |
| C600(AS84/417) | pAS84/417 | U20422 | New Zealand plasmid (4) |
| C600(GO4717) | pGO4717 | U55934 | Rio plasmid (41) |
| C600 | pULB2429 | IncU (8) | |
| pULB2433 | IncHI2 (8) | ||
| pULB2436 | IncHI1 (8) | ||
| pULB2440 | IncFIC (8) | ||
| pULB2426 | IncW (8) | ||
| pULB2424 | IncQ (8) | ||
| pULB2410 | IncY (8) | ||
| pULB2423 | IncL/M (8) | ||
| pULB2406 | IncB/O (8) | ||
| pULB2432 | IncN (8) | ||
| pULB2425 | IncT (8) | ||
| pULB 2404 | IncFIB (8) | ||
| pULB2439 | IncK (8) | ||
| pUBL2420 | IncP (8) |
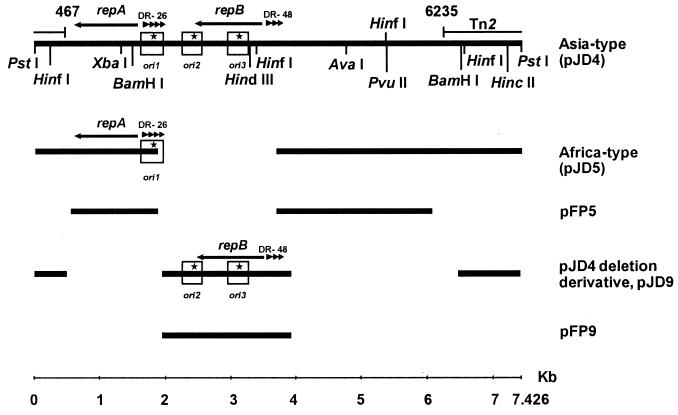
FIG. 1.
Linear map of plasmids used in this study. Spaces indicate DNA sequences not present in alignments or on plasmids. Replication initiation protein genes are indicated with arrows, direct repeats are indicated by triangles, and origin of replication regions are depicted as open boxes. The star indicates where DNA replication initiates based on branch-point analysis (Fig. 2 to 4). Plasmid pFP5 is identical to pJD5, and pFP9 is identical to pJD9 except that Tn2 sequences were replaced with a chloramphenicol acetyltransferase cassette in the FP series of plasmids.
Construction of pFP5 and pFP9.
Derivatives of pJD5 and pJD9 lacking Tn2 were constructed as follows: the chloramphenicol acetyltransferase cassette was amplified from pACYC184 using primers AC1(5′-CTAGCTGCAGGCCGCTGAACTGGTGTCCCTGTTGATA-3′; gonococcal uptake sequence in bold) and AC2 (5′-CGTGCTGCAGTTCTGCCATTCATCC-3′), with each primer including a PstI site (italic). The non-Tn2 region from pJD5 (3,888 bp) was amplified using the primers FP9, 5′-ATGTCTGCAGGCCGCTCTAACCGCT-3′, and FP10, 5′CAGTCTGCAGTCGCCGTTCTGGTTG-3′. Similarly, the 1,923-bp non-Tn2 region of pJD9 was amplified with primers FP7, 5′-CGCTCTGCAGCAACCGAAGCCGTTA-3′, and FP8, 5′-CACTCTGCAGCCGTCGGTACTCTCA-3′. PCR-generated products were digested with PstI, ligated, and then transformed into E. coli DH5α grown on LB containing chloramphenicol (11). PCR conditions reflected the thermal melting points of the primers, and amplification conditions can be supplied upon request.
Plasmid enrichment and isolation.
Replicative molecules were isolated as described previously (2), with the following modifications: cells were chilled in an ice bath for 30 min prior to plasmid extraction; after centrifugation of the lysate, the supernatant containing plasmid DNA was passed through four layers of gauze to remove bacterial debris; to precipitate the DNA (approximately 10 to 11 ml), 3.5 ml of 50% (wt/vol) polyethylene glycol (PEG) 8000 and 2 ml of 5 M NaCl were added. The solution was placed on ice for 60 min and centrifuged at 10 000 × g for 20 min, and then the pellet was resuspended in 2 ml of TE buffer (Tris-EDTA, pH 8.0). NaCl (5 M) was added to a final concentration of 100 mM, followed by the addition of an equal amount of 10 mM Tris-HCl (pH 8.0)-equilibrated phenol-chlorofoform-isoamyl alcohol (25:24:1). DNA was precipitated with 2 volumes of ice-cold ethanol, washed with 70% ethanol, and dissolved in 200 μl of TE buffer (pH 8.0) containing 1 μg/ml RNase A.
EM and branch-point analysis.
Plasmid DNA, prepared for EM as described above, was digested with PstI or HindIII. The PstI and HindIII sites are unique within Tn2 and is asymmetrically located with respect to the origins of replication regions (Fig. 1). Approximately 1 μg of linearized plasmid molecules was purified by phenol and chloroform extractions as described above and prepared for EM as described by Ferguson and Davis (16). Copper grids (300 and 400 mesh, coated with Formvar [0.3% in ethylene dichloride]) were used to lift the DNA from the cytochrome-DNA monolayer, followed by staining in uranyl acetate (10 μl of 21.2% uranyl acetate in 0.5 M HCl, made in 90% ethanol, to 10 ml of 90% ethanol). There was a 10-min interval between lifts to allow the molecules to diffuse to the surface, as the hyperphase becomes disturbed following each lift. Four lifts were performed per DNA sample.
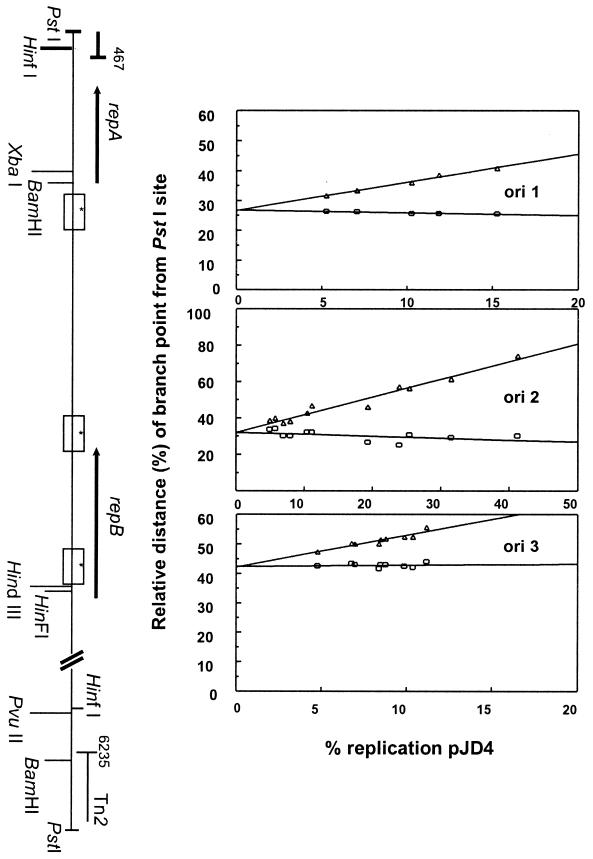
To enhance the contrast of DNA molecules, the samples were coated with a fine layer of metal particles by rotary shadowing using a Balzers vacuum evaporator as previously described (7). Evaporation was for 30 s, with the specimen platform rotating at 30 to 60 rpm, at an angle of 8.5°, using a 5-cm piece of Pt-Pd wire (80:20 ratio). Micrographs were taken using a Philips EM201 microscope. Replicating molecules were traced and measured for statistical and branch-point analysis as previously described (1, 7). We chose only those molecules that contained small, replicating bubbles (versus two extended branches) to ascertain the position of the origins. Molecules were measured from the end of each molecule (i.e., PstI site) to the near and far branches of the replication bubble and aligned so that the shortest, unreplicated arm was on the left. The relative distances to the nearest (i.e., PstI to beginning of bubble) and farthest (i.e., PstI to end of bubble) branch point are depicted by squares and triangles, respectively (see Fig. 2, 3, and 4). The lines were drawn by linear regression analysis of the points obtained, using Sigma Plot version 5.0
FIG. 2.
Plots of branch-point positions against percent replication of partially replicated plasmid molecules of pJD4 in E. coli C600, depicting the location of replication origins. Plots of branch-point positions against the percent plasmid replication of pJD4 digested with PstI are shown. Measurements of the distances from the PstI site to the branch points are relative to the linearized molecule digested with the same enzyme and are set at 100%. Each molecule has different symbols, representing the relative distances to the nearest (squares) and farthest (triangles) branch point from the PstI site. The lines were drawn using linear regression analysis. A schematic of the plasmid (compare to Fig. 1) is shown on the y axis for orientation purposes. Stars within the origins (boxes) indicate where the lines meet at the y axis, corresponding to the start of DNA replication.
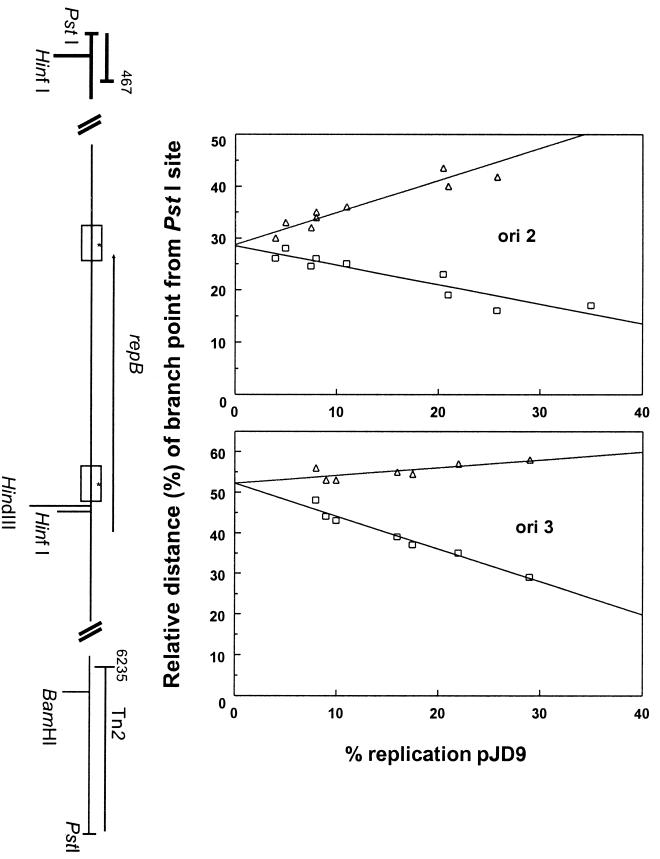
FIG. 3.
Plots of branch-point positions against percent replication of partially replicated plasmid molecules of pJD9 in E. coli C600, depicting the location of replication origins. See legend to Fig. 2 for details.
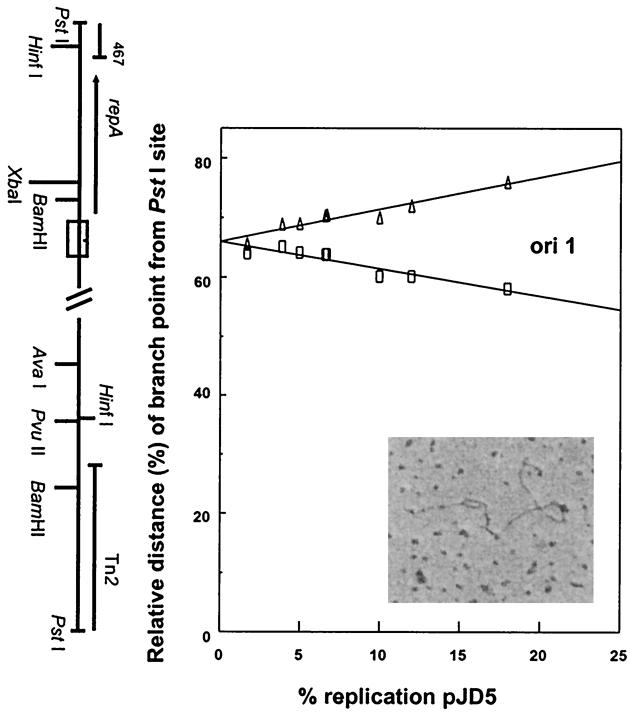
FIG. 4.
Plots of branch-point positions against percent replication of partially replicated plasmid molecules of pJD5 in E. coli C600, depicting the location of replication origins. (Inset) Electron micrograph showing a plasmid molecule with a replication “bubble.” See legend to Fig. 2 for details.
Test for DNA Pol I dependence.
Isogenic E. coli strains SR1758 (polA+) and SR1672 (polA) (1) were used to investigate the requirement of various gonococcal plasmids for PolA in replication. Plasmids were transformed into these hosts using electrotransformation (29) or calcium chloride methods (11). Transformants were purified by subculturing them for three consecutive days on selective medium. Plasmids were isolated and analyzed by R/E digestions to screen for possible structural changes (e.g., deletions, insertions, and rearrangements).
Incompatibility determination.
Incompatibility tests, which include establishment, maintenance, and segregation tests, were performed in E. coli DH5α as described previously (3). The establishment test ascertains the ability of a plasmid to establish itself and to replicate in the presence of a resident plasmid. Cells carrying the resident plasmid were electroporated with incoming plasmid DNA, and selection was made for one or both plasmids. The maintenance test determines the ability of two plasmids to coexist once they have been established within a strain. Cells containing both plasmids were grown overnight in the presence of both antibiotics (approximately stationary phase) before being diluted 106-fold into LB broth without antibiotics. Cells were grown in antibiotic-free LB broth for 3 days (106-fold dilution every 12 h), and subsequently appropriate dilutions of the culture were spread onto LB agar containing antibiotics for the donor, resident, or both plasmids. The segregation test was performed to confirm the existence of weakly compatible plasmids (i.e., when initial transformants isolated carried both plasmids). Colonies carrying both plasmids were grown in antibiotic-free medium without selection for 48 to 72 h in LB broth, and cell dilutions were plated on nonselective agar. After 24 h of incubation, colonies were subcultured with sterile toothpicks on LB agar containing antibiotics selecting for the incoming, resident, or both plasmids. These experiments were repeated three to five times, after which plasmid preparations and DNA R/E analysis on representative transformants were performed to confirm the presence of the plasmid(s), as well as to examine the possibility of DNA rearrangements.
Cloning of RepB (ORF15).
We ascertained that RepB was equivalent to ORF15 which we initially identified from DNA sequence analysis of pJD4 (33). The repB gene was PCR amplified from pJD9 using a Perkin-Elmer 9600 thermal cycler with primers ORF15(5′-GCGCGAGCTCTGTTTTTTTATTGACC-3′) and ORF15COM (5′-GGCGTCTAGAATTTTTCTGTCTCTG-3′) (SacI site italic; XbaI site boldface). PCR amplification conditions are available upon request. Amplified DNA (1,101 bp, including 81 bp upstream of start codon) was analyzed using agarose gel electrophoresis followed by ethidium bromide staining. Gels were photographed using the Gel Print 2000i digital system (Bio Photonics Corp., Ann Arbor, Mich.). Prior to restriction endonuclease digestion of ORF15 amplicons, samples were passed through a Qiagen QIAQuick PCR purification column to remove PCR components. The amplicon was digested with SacI and XbaI, ligated to SacI- and XbaI-digested pBluescript KS+ II, and transformed into E. coli C600, DH5α, or BLR(DE3) with selection on ampicillin-containing medium. The plasmid content of selected transformants was verified by R/E analysis. The recombinant plasmid selected for further analysis was named pBlueORF15.
Deletion of RepB from pFP9.
Plasmid pFP9 was linearized using the unique HindIII site found within the 5′ end of RepB (1,020 bp, 339 amino acids [aa], 39,448 Da; HindIII at position 198) and made blunt-ended using T4 DNA polymerase, creating a frameshift that introduces stop codons throughout ORF15. DNA sequence analysis revealed a truncated (200 bp) form of ORF15 which would produce a much shorter protein (67aa) with a putative molecular mass of 7,593 Da. This mutated plasmid (pORF15) was religated and introduced into E. coli BLR(DE3) which contained pBlueORF15, thereby providing ORF15 in trans following induction with 1 mM IPTG (isopropylthiogalactopyranoside). Transformants were recovered on medium containing chloramphenicol, ampicillin, and IPTG and analyzed for their plasmid content by sequencing and R/E analysis.
In vitro transcription-translation reactions.
The E. coli S30 Extract System for Linear DNA Templates (Promega) was used with PCR-amplified DNA from pJD4 using primers FP7/FP8 and FP9/FP10 according to the manufacturer's instruction. [35S]methionine (15 mCi ml−1) (Amersham Canada) and 1 μl of RNA Guard (Pharmacia Biotech Inc.) were also added to each reaction. Products were separated on denaturing 12% polyacrylamide–sodium dodecyl sulfate (SDS) gels. Gels were dried in a model 583 gel dryer (Bio-Rad), and Kodak X-OMAT AR films were exposed overnight to the dried gel and developed.
DNA sequencing and analysis.
DNA sequencing was performed at the University of Ottawa Biotechnology Research Institute (UOBRI, Ottawa, Canada) using an Applied Biosystems 373A DNA sequencer (Applied Biosystems Canada, Mississauga, Canada), and the PRISM Ready Reaction DyeDeoxy Terminator sequencing kit (Applied Biosystems) in conjunction with Centri-Sep spin columns (Princeton Separations, Philadelphia, N.J.). Primers were designed using the Primer Designer software package (Scientific and Educational Software, Durham, N.C.) and purchased from the UOBRI. Analysis of DNA, RNA, and protein sequences was performed using the PC-Gene software (Intelligenetics, Mountain View, Calif.).
RESULTS
Gonococcal β-lactamase-producing plasmids have multiple origins of replication.
Branch-point analysis revealed the presence of three clustered yet spatially distinguishable origins of replication on pJD4, corresponding to initiation coordinates 1867, 2400, and 3100 (based on where the lines meet using regression analysis; see stars on Fig. 2). We have named these origins ori1, ori2, and ori3. Molecules corresponding to a complete length expected upon digestion of pJD4 with PstI were used to localize these origins. Molecules observed under the electron microscope contained only one bubble, as multiple bubbles were never observed. Bubbles observed represent double-stranded DNA that is newly synthesized (7). EM and branch-point analyses now confirm our previous studies, which suggested that pJD4 contained two replication regions, a and b (45), in that the previously identified a region is shown to contain ori2 and ori3, whereas region b contains ori1. Statistical inference from experimental observations (Fig. 2) indicates that in an E. coli host, ori1 was used 20% of the time and replicated bidirectionally and ori2 was used 44% of the time with bidirectional replication. ori3 was used 36% of the time and appeared to replicate unidirectionally.
The Asia-type plasmid pJD4 is 7.4 kb in size and, due to its size, presented some challenge with respect to the collection of linear molecules of expected length containing small, replicating bubbles. We were able to separate ori2 and ori3 from ori1 using pJD9, an in vitro deletion derivative of pJD4 (45). In pJD9 (Fig. 3), the ori1 and ori2 mapped to positions 2424 and 3200 (using pJD4 coordinates), respectively. According to the molecules obtained, the origins were used equally, with replication proceeding bidirectionally. Based on DNA sequence analysis (33), these origins correspond to ori2 and ori3 of pJD4, respectively (Fig. 1). The naturally occurring Africa-type plasmid in which ori2 and ori3 would be absent due to a deletion in the plasmid (33), typified by pJD5, was found to have one bidirectional origin of replication (Fig. 4). The origin starts at position 1867 (using pJD4 coordinates) and is the same as ori1 of pJD4, based on DNA sequencing studies (33) (Fig. 1). Additional experiments using the single-cutting restriction endonuclease enzyme HindIII and Xba confirmed the three origin locations (data not shown).
Gonococcal plasmids have at least two incompatibility determinants.
There are no established plasmid incompatibility groups among the gonococcal plasmids. The IncP plasmid pUB307 was shown to mobilize ampicillin resistance gonococcal plasmids from E. coli to N. gonorrhoeae (36), yet no naturally occurring IncP plasmid or IncP DNA element has yet to be isolated in a gonococcus. However, plasmids having homology to the IncQ groups, such as RSF1010, have been found in commensal Neisseria spp. and Neisseria meningitidis (14, 37, 39). Therefore, it was of interest to ascertain whether the various origins of replication on pJD4 were incompatible with characterized enteric plasmids (8). These experiments were necessarily completed in E. coli because most origins do not replicate in an N. gonorrhoeae background. Plasmids pJD9/pFP9 and pJD5/pFP5 were tested for their incompatibility with plasmid constructs containing known incompatibility determinants as described in the Materials and Methods section. These plasmid pairs were used because they contain the same origins but different antibiotic resistance markers for selection (i.e., penicillin on pJD9 and pJD5, chloramphenicol on their derivatives). All tests revealed that plasmids pJD9 and pFP9, containing ori2/ori3, belonged to the IncW group (Table 2), whereas pJD5 and pFP5 (ori1-containing plasmids) belonged to the IncFII group (Table 2).
TABLE 2.
Incompatibility results for gonococcal plasmids pJD5/pFP5 (ori1) and pJD9/pFP9 (ori2/ori3)
| Incompatibility group | Resident/incoming plasmida
|
Incompatibilityb | |
|---|---|---|---|
| pJD9/pFP9 | pJD5/pFP5 | ||
| IncW | X | X | Yes with pJD9/pFP9 |
| IncL/M | X | X | No |
| IncB/O | X | X | No |
| IncH12 | X | X | No |
| IncF1 | X | X | No |
| IncY | X | X | No |
| IncN | X | X | No |
| IncT | X | X | No |
| IncQ | X | X | No |
| IncV | X | X | No |
| IncFIIA | X | X | Yes with pJD5/pFP5 |
| IncII | X | X | No |
| IncFIB | X | X | No |
| IncU | X | X | No |
Experiments included resident as well as incoming plasmids. X denotes that the plasmid was used as the resident or incoming plasmid when tested against the plasmid of a known incompatibility group.
Yes indicates that plasmids cannot be maintained in the same cell (i.e., they are incompatible). No indicates that plasmids can be maintained in the same cell.
Incompatibility studies suggested that, since plasmid pJD4 contains all three origins, it should express both Inc phenotypes (W and FII). This was determined to be the case. However, as might be expected, only one incompatibility determinant was expressed at a time. For example, plasmid pJD4 was incompatible with the IncW plasmids when incompatibility tests were performed but expressed IncFII incompatibility, characteristic of ori1, in establishment experiments in which pFP5 (ori1, IncFII) was resident and pJD4 was the incoming plasmid (data not shown). However, when pFP5 was the incoming plasmid, both plasmids were stably maintained within the cell, indicating that ori2/3 (IncW) was being used by pJD4. These data indicate that, once established in E. coli, pJD4 replicates preferentially from ori2/3. This observation was also confirmed by the small number of ori1 bubbles (versus ori2 and ori3) obtained in EM experiments (Fig. 2).
Novel Rep protein is required for the replication of pFP9 (ori2 and ori3).
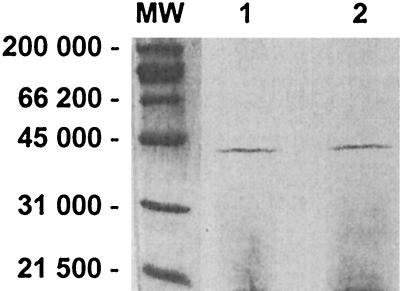
The above experiments suggested the existence of a replication protein preferentially expressed by pJD4 which is used for ori2/ori3. To look for this protein, we used in vitro transcription-translation studies to ascertain whether the 1.9-kb DNA fragment with ori2/ori3 of plasmid pFP9, which excluded the ampicillin resistance determinant, contained open reading frames (ORFs) encoding proteins that might be involved with replication or maintenance functions. Bioinformatics evidence also suggested that one ORF, which we called ORF15, might be implicated in the replication process (34) (data not shown). Results from these experiments revealed a single protein of approximately 39 kDa (Fig. 5, lane 2). Evidence that this protein, named RepB (1,020 bp, 339 aa, 39,449 Da) (33), might be the Rep protein for ori2 and ori3 included the failure to obtain a self-replicating plasmid (pORF15) construct when RepB was truncated by exonuclease digestion (or end-filling) at its unique HindIII site. However, when RepB was provided in trans to rescue plasmid pORF15 (pFP9 with a truncated repB), the plasmid was able to replicate (data not shown). When pORF15 was isolated and transformed into a cell not supplying RepB, it was unable to replicate, further indicating a role for RepB in the replication process. Thus, RepB is a novel replication initiation protein for the β-lactamase-producing plasmids which functions in trans. We have named it RepB to distinguishing it from a previously characterized RepA protein, and the location of repB is indicated in Fig. 1 (19).
FIG. 5.
In vitro transcription-translation. DNA was amplified from pJD5 using primers FP9 and FP10 (lane 1) or from pJD9 using primers FP7 and FP8 (lane 2). Based on computer analysis, the RepA protein (lane 1) is 38.6 kDa and RepB (lane 2) is 39.4 kDa. Lane MW, size standards (in daltons).
RepA is necessary for ori1, not required for ori2/3, and expendable for pJD4 (ori1/2/3).
Other investigators first described a protein (RepA) which was involved with the replication of ori1 on a fragment of the Asia-type plasmid pFA3, since removal of repA abolished replication of a DNA fragment containing repA and ori1 (19). We confirmed this observation using plasmids pJD4 (ori1/2/3) and pJD5 (ori1). Functional replication origin regions of plasmids pJD5 (and pJD4) contain repA (Fig. 1), as identified by in vitro transcription-translation experiments and DNA sequence analysis (19, 33, 44) (Fig. 5, lane 1). Inactivation of RepA in plasmid pJD5 by end-filling the unique XbaI site near the 5′ end of repA or the unique BamHI site in pFP5 (Fig. 1) resulted in a plasmid which could not replicate autonomously (data not shown). To date, we have been unable to rescue a DNA sequence containing the ori1 region by supplying the RepA protein in trans, suggesting that the repA gene is required in cis for ori1 to function.
As pJD4 contains both repA and repB, attempts to knock out either gene were successful only for the repA gene (data not shown). That is, all constructs containing an intact repB and truncated repA gene replicated autonomously, whereas no self-replicating plasmids with an intact repA and truncated repB gene were obtained in an E. coli background. Thus, RepA is dispensable for replication of pJD4 in E. coli and required only for replication of DNA sequences containing only ori1.
Gonococcal origins of replication are similar in their structural organization.
DNA sequence comparisons between ori1 and ori2/ori3 regions on pJD4 and its related plasmids pJD9 (ori2/3) and pJD5 (ori1) indicated structural similarities between them (Fig. 6). Both origins appear to belong to the iteron family, as they both contained iterons (DR-48 for ori2/3 and DR-26 for ori1) and replication initiation proteins (Fig. 6). While repA is distinct from the actual denaturation site as mapped by EM, repB spans all of ori3 and part of ori2 (Fig. 1). DNA sequence analysis of the promoter regions for repA and repB revealed that the putative transcription signals (−10 and −35) were located within or adjacent to the direct repeats DR-26 and DR-48 (Fig. 6), respectively. Interestingly, the two iterons DR-26 and DR-48 were 36% homologous (Fig. 6C), having an 8-nucleotide conserved sequence motif. Sequence analysis demonstrated that RepA (339 aa, 39,448 Da) and RepB (328 aa, 38,636 Da) were 61% identical at the nucleotide level and 57% homologous in their amino acid sequence.
FIG. 6.
Promoter regions for repA (A) and repB (B). Start sites for Rep proteins, putative ribosome-binding sites (RBS), −10 and −35 regions (thick and thin boxes, respectively), and direct repeat (long arrows) are indicated. Mismatches to E. coli consensus −10 and −35 regions are indicated with asterisks. (C) Sequence alignment between DR-26, DR-48, and iterons of plasmid pSa. Underlined sequences indicate identical matches between DR-26 and DR-48, and the boxed sequences demonstrate the conserved sequence between DR-26, DR-48, and pSa.
Requirements for PolA.
Iteron and RNA-regulated plasmids differ in their dependence for DNA Pol I (25). Since the gonococcal penicillinase plasmids appeared to have an iteron-based type of organization (Fig. 6), the whole family of gonococcal β-lactamase-producing plasmids were tested for their ability to replicate and maintain themselves in an E. coli polA deletion mutant (Table 3). Interestingly, only plasmid pJD5 and its insertion derivative pGF1 (Nîmes-type), which both carry only ori1, were unable to grow in a polA host.
TABLE 3.
Dependence of gonococcal β-lactamase-producing plasmids on DNA Pol I
| Plasmid (type) | Establisment and maintenancea
|
|
|---|---|---|
| SR1672 (polA) | SR1758 (polA+) | |
| pJD4 (Asia) | + | + |
| pAS84/417 (New Zealand) | + | + |
| pJD7 (Toronto) | + | + |
| pGO4117 (Rio) | + | + |
| pJD9/pFP9 | + | + |
| pJD5 (Africa) | − | + |
| pGF1 (Nîmes) | − | + |
| pBluescript KS+ | − | + |
| pBlu-BamHIb | + | + |
| pBR322 | − | + |
Symbols: +, plasmid was able to maintain itself in the host; −, plasmid was not able to establish and maintain itself in the host.
The large BamHI fragment of pJD4 was inserted into the BamHI site of pBluescript KS.
DISCUSSION
Plasmids containing three origins of replication are rare and generally large (1, 9, 23). Using an E. coli background, we show that the gonococcal Asia-type plasmid pJD4 is the smallest described plasmid to date containing three distinct origins of replication (ori1, ori2, and ori3). The naturally occurring Africa-type plasmid, typified by pJD5, contains a single origin of replication corresponding to ori1 of pJD4. Current attempts to transform gonococci with plasmids containing ori2/ori3 have been unsuccessful, indicating that ori2 and ori3 may not function in a gonococcal background. The host range of an Asia-type plasmid (ori1/2/3) was shown to include gonococci, members of the family Enterobacteriaceae, and Haemophilus influenzae, but not Acinetobacter calcoaceticus or Pseudomonas aeruginosa (21). However, we have been able to transform P. aeruginosa with pFP9 (ori2/3) but not pFP5 (ori1), confirming that ori1 has a different host range than plasmids containing ori2/ori3 (Pagotto and Dillon, unpublished data). Previous work demonstrated that the 3.8-kb BamHI-PvuII fragment was essential for plasmid replication in E. coli (24). This study has shown that this 3.8-kb DNA fragment contains ori2/ori3 and RepB. Plasmid constructs containing ori1 and repA have been shown to be functional in an E. coli background (19; this study). In addition, we have shown recently that plasmids containing ori1 and repA are functional in both gonococcal and Haemophilus backgrounds (34).
The Africa-type plasmid pJD5, containing ori1, demonstrated different replication characteristics than gonococcal plasmids containing ori2/ori3. Plasmid R1, used to classify the incompatibility of pJD5/pFP5 in this study, belongs to the IncFII plasmid family. It does not have iterons for the Rep protein and is classified as an RNA-regulated plasmid (22). In plasmid R1, translation of the RepA initiation protein is controlled by the antisense molecule copA (28, 43). However, it should be noted that plasmids containing iterons have not been shown to possess antisense molecules associated with replication, copy number, or incompatibility properties (10, 22, 26). Thus, the regulation of repA in ori1-containing gonococcal plasmids and the role of the iterons in the replication process remain to be determined.
Plasmids pJD4 and pJD9/FP9 were shown to be incompatible with the IncW plasmid pUB2426, which contains a 1.5-kb fragment of DNA from plasmid pSa (8). This 1.5-kb DNA fragment contains oriV, three iterons, and a major part of the repA gene. The RepA protein of pSa is proposed to bind the iterons and thus control its own rate of synthesis (32). The iterons were also proposed to be the incompatibility determinant for this plasmid (32). Alignment between the consensus sequence of the pSa iterons and DR-48 revealed that they were 41% identical and contained 4 conserved nucleotides (Fig. 6C). This suggests that the iteron DR-48 (and those from pSa) would bind to RepB.
Of the iteron-regulated plasmids described to date, only some have experimentally shown definitive binding of the iterons to the Rep protein. Other plasmids have been classified as iteron-regulated based on the genetic organization and the presence of iterons (22). Based on available information, we propose that pJD4/pJD9 is incompatible with the IncW plasmid pUB2426 because RepB from pJD4/pJD9 might be interacting with the iterons on pUB2426, thereby disrupting replication from either ori2 or ori3. At least two possibilities exist for the role of these iterons. In the first, the iterons from pUB2426 bind and titrate away RepB, thereby reducing replication from ori2/ori3 (referred to as the titration model [5, 43]). In the second, the iterons from pUB2426 bind to the iterons in ori2/ori3-containing plasmids and cause a pairing of gonococcal-pUB2426 iterons and shutting off of replication through the coupling of all plasmid molecules within the cell (referred to as the handcuffing or coupling model [10, 35]). Which of these situations is occurring is not known at this time, and we have not ruled out a novel mechanism to explain gonococcal plasmid replication. While there are some similarities between the gonococcal iterons DR-48 and DR-26 and those of pSa, comparisons of iterons of various plasmids revealed no consensus sequence and showed variation in both number and size (22).
Based on the results obtained in this study, the following model is proposed for replication of β-lactamase-producing plasmids of N. gonorrhoeae. RepA is essential for ori1 and controls its own rate of synthesis by binding to iteron DR-26, which contains its promoter region. Binding of RepA to the iterons would also create a structural distortion in the DNA, allowing for the melting of the two strands and for the loading of DNA replication proteins at ori. For ori2 and ori3, RepB acts as the replication initiation protein and most likely controls its own rate of synthesis by binding to iteron DR-48. Binding of RepB to the iterons would recruit the replication machinery to either ori2 or ori3 in a similar manner to RepA (ori1) and as described for iteron-containing plasmids (22, 26). Exactly how the binding of RepB causes either ori2 or ori3 to be used preferentially is unknown. It is most likely that ori2 and ori3 mimic the situation described with plasmid R6K, in which the gamma origin is rarely used but is required in cis for the alpha and beta origins to be used (30).
In conclusion, this is the first report of a naturally occurring plasmid of N. gonorrhoeae displaying three functional origins of replication when studied in an enteric background such as E. coli. The plasmid also encodes two functionally distinct replication initiation proteins and belongs to two incompatibility groups. We are continuing to investigate the phenomenon of multiple origins of replication and their differential usage with the gonococcal plasmids as a model system. Future work will include studies involving replication of the gonococcal β-lactamase-producing plasmids in some of their other hosts, including Neisseria, Pseudomonas, and Haemophilus spp.
ACKNOWLEDGMENTS
This work was funded by start-up funds from the University of Ottawa to J.R.D. F. Pagotto acknowledges the Ontario Graduate Scholarship and the FCAR (Fonds pour la Formation de Chercheurs et l'Aide à la Recherche) doctoral awards for partial financial assistance.
We are grateful to Stéphane Bernatchez for performing the in vitro transcription-translation experiments; Roberto Catana for performing incompatibility studies; S. Banerjee (Institute of Biochemistry, Carleton University, Ottawa, Canada) for technical advice on branch point analysis; and D. Brown for providing electron microscopy facilities (Department of Biology, University of Ottawa, Ottawa, Canada). W. Maas (New York University School of Medicine, New York) supplied the set of incompatibility plasmids, and V. N. Iyer (formerly of Carleton University) provided polA+ and polA strains. We thank B. Valentine (Department of Biology, University of Ottawa, Ottawa, Canada) for technical discussions regarding sample preparations for EM and S. Ramirez and J. Szeto for their comments on various drafts of the manuscript.
REFERENCES
- 1.Banerjee S K, Luck B T, Kim H Y, Iyer V N. Three clustered origins of replication in a promiscuous-plasmid replicon and their differential use in a PolA+ strain and a ΔPolA strain of Escherichia coli K-12. J Bacteriol. 1992;174:8139–8143. doi: 10.1128/jb.174.24.8139-8143.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee S K, Iyer V N. Quick and easy spreading technique for electron microscopy of DNA. BioTechniques. 1995;18:946–947. [PubMed] [Google Scholar]
- 3.Bergquist P L. Incompatibility. In: Hardy K G, editor. Plasmids: a practical approach. Washington, D.C.: IRL Press; 1988. pp. 37–78. [Google Scholar]
- 4.Brett M. A novel gonococcal β-lactamase plasmid. J Antimicrob Chemother. 1989;23:653–654. doi: 10.1093/jac/23.4.653. [DOI] [PubMed] [Google Scholar]
- 5.Chattoraj D K. Control of plasmid DNA replication by iterons: no longer paradoxical. Mol Microbiol. 2000;37:467–476. doi: 10.1046/j.1365-2958.2000.01986.x. [DOI] [PubMed] [Google Scholar]
- 6.Chen S T, Clowes R C. Nucleotide sequence comparisons of plasmids pHD131: pJB1, pFA3, pFA7, and β-lactamase expression in Escherichia coli, Haemophilus influenzae, and Neisseria gonorrhoeae. J Bacteriol. 1987;169:3124–3130. doi: 10.1128/jb.169.7.3124-3130.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coggins L W. Preparation of nucleic acids for electron microscopy. In: Sommerville J, Scheer U, editors. Electron microscopy in molecular biology: a practical approach. Oxford, England: IRL Press; 1987. pp. 1–29. [Google Scholar]
- 8.Couturier M, Bex F, Bergquist P L, Maas W K. Identification and classification of bacterial plasmids. Microbiol Rev. 1988;52:375–395. doi: 10.1128/mr.52.3.375-395.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crosa J H. Three origins of replication are active in vivo in the R-plasmid RSF1040. J Biol Chem. 1980;255:11075–11077. [PubMed] [Google Scholar]
- 10.del Solar G, Giraldo R, Ruiz-Echevarria M J, Espinosa M, Diaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dillon J A, Nasim A, Nestmann E R. Recombinant DNA Methodology. New York, N.Y: John Wiley and Sons; 1985. [Google Scholar]
- 12.Dillon J A R, Yeung K-H. β-Lactamase plasmids and chromosomally mediated antibiotic resistance in pathogenic Neisseria species. Clin Microbiol Rev. 1989;2:S125–S133. doi: 10.1128/cmr.2.suppl.s125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dillon J R, Pagotto F J. Importance of drug resistance in gonococci: from mechanisms to monitoring. Curr Opin Infect Dis. 1999;12:35–40. doi: 10.1097/00001432-199902000-00007. [DOI] [PubMed] [Google Scholar]
- 14.Facinelli B, Varaldo P E. Plasmid-mediated sulfonamide resistance in Neisseria meningitidis. Antimicrob Agents Chemother. 1987;31:1642–1643. doi: 10.1128/aac.31.10.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fayet O, Froment Y, Piffaretti J C. β-Lactamase-specifying plasmids isolated from Neisseria gonorrhoeae have retained an intact right part of a Tn3-like transposon. J Bacteriol. 1982;149:136–144. doi: 10.1128/jb.149.1.136-144.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferguson J, Davis R W. Quantitative electron microscopy of nucleic acids. In: Koehler J K, editor. Advanced techniques in biological electron microscopy II. New York, N.Y: Springer-Verlag; 1978. pp. 123–171. [Google Scholar]
- 17.Filutowicz M, McEachern M J, Helinski D R. Positive and negative roles of an initiator protein at an origin of replication. Proc Natl Acad Sci USA. 1986;83:9645–9649. doi: 10.1073/pnas.83.24.9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filutowicz M, Rakowski S A. Regulatory implications of protein assemblies at the (origin of plasmid R6K — a review. Gene. 1998;223:195–204. doi: 10.1016/s0378-1119(98)00367-9. [DOI] [PubMed] [Google Scholar]
- 19.Gilbride K A, Brunton J L. Identification and characterization of a new replication region in the Neisseria gonorrhoeae β-lactamase plasmid pFA3. J Bacteriol. 1990;172:2439–2446. doi: 10.1128/jb.172.5.2439-2446.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gouby A, Bourg G, Ramuz M. Previously undescribed 6.6-kilobase R plasmid in penicillinase-producing Neisseria gonorrhoeae. Antimicrob Agents Chemother. 1986;29:1095–1097. doi: 10.1128/aac.29.6.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guiney D G, Ito J I. Transfer of the gonococcal penicillinase plasmid: mobilization in Escherichia coli by IncP plasmids and isolation as a DNA-protein relaxation complex. J Bacteriol. 1982;150:298–302. doi: 10.1128/jb.150.1.298-302.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helinski D R, Toukdarian A E, Novick R P. Replication control and other stable maintenance mechanisms of plasmids. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Brooks Low K, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2295–2324. [Google Scholar]
- 23.Inuzuka N, Inuzuka M, Helinski D R. Activity in vitro of three replication origins of the antibiotic resistance plasmid RSF1040. J Biol Chem. 1980;255:11041–11074. [PubMed] [Google Scholar]
- 24.Johnson S R. Cloning of the replication region of pGR9091: an R factor of Neisseria gonorrhoeae. In: Schoolnik G K, Knapp J S, McCuchan J A, Morse S A, editors. The pathogenic neisseriae. Washington, D.C.: American Society for Microbiology; 1985. pp. 222–228. [Google Scholar]
- 25.Kornberg A, Baker T A. DNA replication. W. H. New York, N.Y: Freeman and Company; 1992. [Google Scholar]
- 26.Kues U, Stahl U. Replication of plasmids in gram-negative bacteria. Microbiol Rev. 1989;53:491–516. doi: 10.1128/mr.53.4.491-516.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lane D, Gardner R. Second EcoRI fragment of F capable of self-replication. J Bacteriol. 1979;139:141–151. doi: 10.1128/jb.139.1.141-151.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Light J, Molin S. Post-transcriptional control of expression of the repA gene of plasmid R1 mediated by a small RNA molecule. EMBO J. 1983;2:93–98. doi: 10.1002/j.1460-2075.1983.tb01387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller J F. Bacterial transformation by electroporation. Methods Enzymol. 1994;235:375–385. doi: 10.1016/0076-6879(94)35156-2. [DOI] [PubMed] [Google Scholar]
- 30.Mukherjee S, Erickson H, Bastia D. Enhancer-origin interaction in plasmid R6K involves a DNA loop mediated by initiator protein. Cell. 1988;52:375–383. doi: 10.1016/s0092-8674(88)80030-8. [DOI] [PubMed] [Google Scholar]
- 31.Mukhopadhyay P, Filutowicz M, Helinski D R. Replication from one of the three origins of the plasmid R6K requires coupled expression of two plasmid-encoded proteins. J Biol Chem. 1986;261:9534–9539. [PubMed] [Google Scholar]
- 32.Okumura M S, Kado C E. The region essential for efficient autonomous replication of pSa in Escherichia coli. Mol Gen Genet. 1992;235:55–63. doi: 10.1007/BF00286181. [DOI] [PubMed] [Google Scholar]
- 33.Pagotto F, Ng L-K, Aman T, Brett M, Yeung K, Dillon J R. Structural analysis of the family of penicillinase-producing plasmids of Neisseria gonorrhoeae based on DNA sequencing. Plasmid. 2000;43:24–34. doi: 10.1006/plas.1999.1431. [DOI] [PubMed] [Google Scholar]
- 34.Pagotto F, Saliminia H, Totten P, Dillon J R. Stable shuttle vectors for Neisseria gonorrhoeae, Haemophilus spp. and other bacteria based on a single origin of replication. Gene. 2000b;244:13–19. doi: 10.1016/s0378-1119(99)00557-0. [DOI] [PubMed] [Google Scholar]
- 35.Pal S K, Chattoraj D K. P1 plasmid replication: initiator sequestration is inadequate to explain control by initiator-binding sites. J Bacteriol. 1988;170:3554–3560. doi: 10.1128/jb.170.8.3554-3560.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piffaretti J C, Arini A, Frey J. pUB307 mobilizes resistance plasmids from Escherichia coli into Neisseria gonorrhoeae. Mol Gen Genet. 1988;212:215–218. doi: 10.1007/BF00334687. [DOI] [PubMed] [Google Scholar]
- 37.Pintado C, Salvador C, Rotger R, Nombela C. Multiresistance plasmid from commensal Neisseria strains. Antimicrob Agents Chemother. 1985;27:120–124. doi: 10.1128/aac.27.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajendra Krishnan B, Iyer V N. IncN plasmid replicon: a deletion and subcloning analysis. J Mol Biol. 1990;213:777–788. doi: 10.1016/S0022-2836(05)80263-3. [DOI] [PubMed] [Google Scholar]
- 39.Rotger R, Rubio F, Nombela C. A multi-resistance plasmid isolated from commensal Neisseria species is closely related to the enterobacterial plasmid RSF1010. J Gen Microbiol. 1986;132:2491–2496. doi: 10.1099/00221287-132-9-2491. [DOI] [PubMed] [Google Scholar]
- 40.Saadi S, Maas W, Hill D F, Bergquist P. Nucleotide sequence analysis of RepFIC, a basic replicon present in IncFI plasmids p307 and F, and its relation to the RepA replicon of IncFII plasmids. J Bacteriol. 1986;169:1836–1846. doi: 10.1128/jb.169.5.1836-1846.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Embden A J, Dessens-Kroom D A, Van Klingeren B. A new β-lactamase plasmid in Neisseria gonorrhoeae. J Antimicrob Chemother. 1985;15:247–350. doi: 10.1093/jac/15.2.247. [DOI] [PubMed] [Google Scholar]
- 42.Wickner S H, Chattoraj D K. Replication of mini-P1 plasmid DNA in vitro requires two initiation proteins, encoded by the repA gene of phage P1 and the dnaA gene of Escherichia coli. Proc Natl Acad Sci USA. 1987;84:3668–3672. doi: 10.1073/pnas.84.11.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Womble D D, Dong X, Wu R P, Luckow V A, Martinez A, Rownd R H. IncFII plasmid incompatibility product and its target are both RNA transcripts. J Bacteriol. 1984;160:28–35. doi: 10.1128/jb.160.1.28-35.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeung K-H, Dillon J A. In vitro transcription/translation products and molecular characterization of naturally occurring and in vitro deletion derivatives of the 7.2-Kilobase plasmid of Neisseria gonorrhoeae. In: Schoolnik G K, editor. Proceedings of the 4th International Symposium of Pathogenic Neisseriae. Washington, D.C.: American Society for Microbiology; 1985. pp. 209–215. [Google Scholar]
- 45.Yeung K-H, Dillon J A. Construction of miniplasmids from the 7.2-kb and 5.1-kb penicillinase-producing plasmids of Neisseria gonorrhoeae reveals two replication origins. Plasmid. 1988;20:232–240. doi: 10.1016/0147-619x(88)90029-7. [DOI] [PubMed] [Google Scholar]