ABSTRACT
Bone regeneration involves skeletal stem/progenitor cells (SSPCs) recruited from bone marrow, periosteum, and adjacent skeletal muscle. To achieve bone reconstitution after injury, a coordinated cellular and molecular response is required from these cell populations. Here, we show that SSPCs from periosteum and skeletal muscle are enriched in osteochondral progenitors, and more efficiently contribute to endochondral ossification during fracture repair as compared to bone‐marrow stromal cells. Single‐cell RNA sequencing (RNAseq) analyses of periosteal cells reveal the cellular heterogeneity of periosteum at steady state and in response to bone fracture. Upon fracture, both periosteal and skeletal muscle SSPCs transition from a stem/progenitor to a fibrogenic state prior to chondrogenesis. This common activation pattern in periosteum and skeletal muscle SSPCs is mediated by bone morphogenetic protein (BMP) signaling. Functionally, Bmpr1a gene inactivation in platelet‐derived growth factor receptor alpha (Pdgfra)‐derived SSPCs impairs bone healing and decreases SSPC proliferation, migration, and osteochondral differentiation. These results uncover a coordinated molecular program driving SSPC activation in periosteum and skeletal muscle toward endochondral ossification during bone regeneration. © 2022 The Authors. Journal of Bone and Mineral Research published by Wiley Periodicals LLC on behalf of American Society for Bone and Mineral Research (ASBMR).
Keywords: BONE REPAIR, SINGLE CELL RNA SEQUENCING, PERIOSTEUM, SKELETAL STEM/PROGENITOR CELLS, BMP PATHWAY
Introduction
Tissue regeneration is mediated by resident stem/progenitor cells that are activated locally within injured tissues or organs. During bone regeneration, skeletal stem/progenitor cells (SSPCs) are mobilized not only from bone compartments; ie, bone marrow and periosteum, but also from adjacent skeletal muscle.( 1 , 2 , 3 , 4 , 5 , 6 , 7 , 8 , 9 , 10 ) These SSPC populations are diverse in their tissue origin and in their cellular composition, as revealed by single‐cell RNAseq (scRNAseq).( 2 , 11 , 12 , 13 , 14 , 15 , 16 ) Multiple markers have been used to identify SSPCs in bone marrow (ie, Mx1, Grem1, LepR, Nes, Cxcl12, Pdgfra, Gli1) or in periosteum (ie, Ctsk, Acta2),( 12 , 17 , 18 , 19 , 20 , 21 , 22 , 23 ) but none of these markers is restricted to a single tissue.( 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 )
Despite the cellular diversity of SSPCs, functional analyses in adult tissues indicate unique regenerative potential of SSPCs according to their origin. Bone marrow SSPCs are osteogenic, support hematopoiesis and osteoclast formation, play paracrine and immunomodulatory roles, but show limited capacity to form cartilage during bone repair( 19 , 24 , 25 ). SSPCs within periosteum contribute efficiently to both cartilage and bone, and SSPCs from skeletal muscle mainly participate in cartilage formation.( 1 , 2 , 10 ) Here, we assessed the activation pattern of SSPCs from periosteum and skeletal muscle after fracture to understand their role in the endochondral ossification process. Compared to bone marrow, SSPCs from periosteum and skeletal muscle are enriched in osteochondral progenitors and efficiently participate in endochondral ossification after transplantation. Using scRNAseq analyses, we uncover the composition of the periosteal cell populations at steady state and compare their response to fracture with that of skeletal muscle‐derived progenitors. We found that bone morphogenetic protein (BMP) signaling is activated in periosteum‐derived and skeletal muscle‐derived progenitors after fracture. We investigated the consequences of bmpr1a gene inactivation in periosteum and skeletal muscle SSPCs marked by platelet‐derived growth factor receptor alpha (Pdgfra) on proliferation, migration, and differentiation. These results reveal that independent of their tissue origin and heterogeneity, SSPCs from periosteum and skeletal muscle share a common molecular response after fracture to support endochondral ossification during bone repair.
Subjects and Methods
Mice
C57BL6/J, Prx1 Cre ,( 26 ) Prx1 CreERT (Stock number #029211),( 27 ) Pdgfra CreERT (Stock number # 018280),( 28 ) Rosa‐tdTomato‐EGFP (Rosa mTmG ) and Rosa LacZ were obtained from The Jackson Laboratory (Bar Harbor, ME, USA). Bmpr1a fl/fl mice were provided by Dr. Yuji Mishina (University of Michigan, Ann Arbor, MI, USA).( 29 , 30 ) All strains were maintained on a C57BL6/J background. Mice were bred and kept under controlled pathogen conditions in separated ventilated cages with controlled humidity and ambient temperature, with 12:12‐hour light:dark cycles and free access to food and water in the animal facilities of IMRB, Creteil, and Imagine Institute, Paris. All experiments were performed in compliance with procedures approved by the Paris Est Creteil and Paris University Ethical Committees. Both males and females were used in all experiments. For in vitro experiments, 4‐week‐old to 8‐week‐old mice were used, and for in vivo experiments 12‐week‐old to 14‐week‐old mice were used. No specific randomization methods were used. Sample labeling allowed blind analyses.
Tamoxifen injection
Tamoxifen (TMX; Sigma‐Aldrich, St. Louis, MO, USA; ref T5648) was dissolve at 10 mg/mL in corn oil (Sigma‐Aldrich; ref C8267) at 60°C for 1 hour. Mice received 300 μL per intraperitoneally injection. To maximize recombination efficiency with the Prx1 CreERT line, we tested two different tamoxifen induction protocols. Prx1 CreERT ;Rosa LacZ and Prx1 CreERT ;Bmpr1a fl/fl mice were injected three times a week from week 9 to week 11 post‐birth, the day before fracture, and days 1 and 3 following fracture (Fig. S7 A) or three times the week before fracture (Fig. S7 B). For phenotypic characterization, Pdgfra CreERT ;Bmpr1a fl/fl and Pdgfra CreERT ;Bmpr1a +/+ mice were injected once a week before fracture, the day before fracture, the day of fracture, and 1 day following fracture, to target stem/progenitor cells at the time of fracture (Fig. 6). Pdgfra CreERT ;Rosa mTmG ;Bmpr1a +/+ (Bmpr1a control ) and Pdgfra CreERT ;Rosa mTmG ;Bmpr1a fl/fl (Bmpr1a cKO ) mice used as donors for extensor digitorum lengus (EDL) or periosteum grafts and for in vitro experiments were injected 3 consecutive days the week before harvest and the day before harvest (Fig. 7).
Fig. 6.
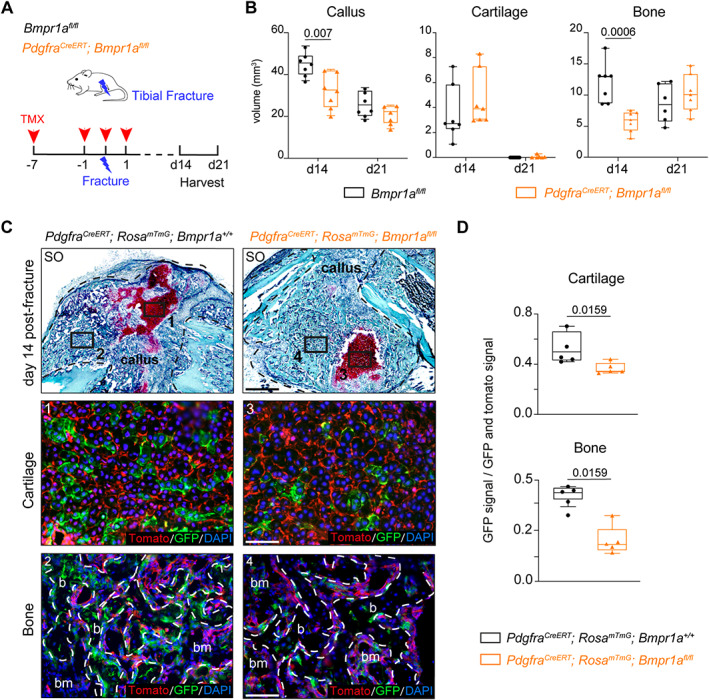
Loss of Bmpr1a in Pdgfra‐derived progenitors impairs bone healing. (A) Experimental design. Bmpr1a fl/fl (control) and Pdgfra CreERT ;Bmpr1a fl/fl mice were induced with TMX at days 7 and 1 before fracture, day 0 and day 1 post‐fracture. Tibial fractures were induced at d0 and calluses were harvested at d14 and d21 post‐fracture. (B) Histomorphometric quantification of callus, cartilage, and bone volumes at d14 and d21 post‐fracture in Bmpr1a fl/fl control and Pdgfra CreERT ;Bmpr1a fl/fl mutant mice (in black and orange, respectively) (n = 6–7 animals per group). (C) Longitudinal callus sections from Pdgfra CreERT ;Rosa mTmG ;Bmpr1a +/+ control (left) and Pdgfra CreERT ;Rosa mTmG ;Bmpr1a fl/fl mutant (right) mice at d14 post‐fracture stained with SO (callus delimited by a black dotted line). High magnifications of boxed areas of cartilage (1 and 3) and bone (2 and 4) show Pdgfra‐derived cells in cartilage and bone. (D) Quantification of GFP+ signal normalized on total GFP+ and Tomato+ signal in cartilage and bone. n = 4–5 animals per group, each dot represents a single animal. Exact p value calculated with two‐sided Mann‐Whitney test, values represent median and interquartile range. Scale bars: low magnification = 1 mm, high magnification = 200 μm. b = bone; bm = bone‐marrow; d0 = day 0; SO = Safranin O; TMX = tamoxifen.
Fig. 7.
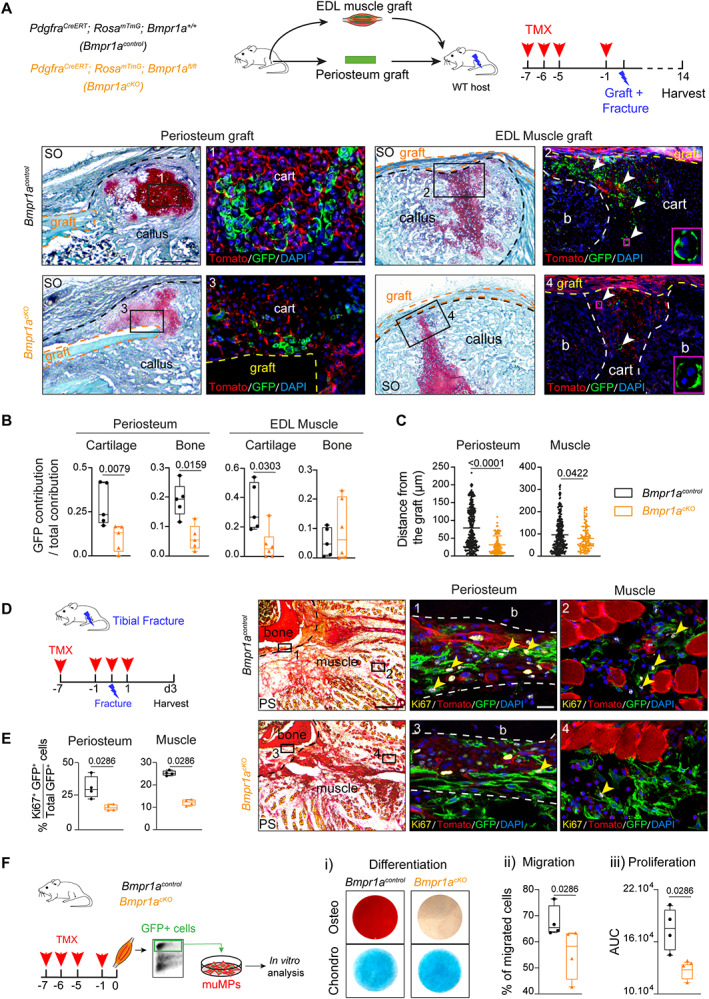
Bmpr1a inactivation in Pdgfra‐derived cells affects periosteum and muscle progenitors during bone repair. (A) Top, Experimental design of periosteum or EDL grafts from Pdgfra CreERT ;Rosa mTmG ;Bmpr1a +/+ (Bmpr1a control ) or Pdgfra CreERT ; Rosa mTmG ;Bmpr1a fl/fl (Bmpr1a cKO ) mice transplanted at the fracture site of wild‐type hosts. Donor mice were induced with TMX at d7, d6, d5, and d1 prior to fracture. Fractures were induced at d0, and samples were collected at d14 post‐fracture. Bottom, Longitudinal callus sections stained with SO at d14 post‐fracture and periosteum (left) or EDL muscle (right) graft from Bmpr1a control (top) and Bmpr1a cKO (bottom) mice (callus delimited by a black dotted line, graft delimited by an orange/yellow dotted line). High magnifications of boxed areas of cartilage show reduced contribution to cartilage of Pdgfra‐derived cells from Bmpr1a cKO periosteal and EDL muscle grafts compared to Bmpr1a control . Magenta boxed areas show GFP+ hypertrophic chondrocytes from EDL muscle grafts. (B) Quantification of GFP+ contribution normalized on total cellular contribution to cartilage and bone of periosteum and EDL muscle grafts. n = 5–6 animals per group, each dot represents a single animal. (C) Distance between GFP+ chondrocytes and the border of periosteum or EDL grafts (delimited by an orange dotted line) from Bmpr1a control and Bmpr1a cKO donors. Control periosteum: n = 284 cells, mutant periosteum: n = 99 cells, control EDL: n = 302 cells, and mutant EDL: n = 120 cells. Five animals were used per group. Each dot represents an individual cell. (D) Left, Experimental design. Bmpr1a control and Bmpr1a cKO mice were induced with TMX at d7 and d1 before fracture, d0 and d1 post‐fracture. Samples were harvested at d3 post‐fracture. Right, Transverse sections of fracture site at d3 post‐fracture were stained with PS and adjacent sections were immunostained for Ki67 (Ki67 cells in yellow, pointed with yellow arrowhead). High magnifications of boxed areas in activated periosteum (delimited by a dotted line) and skeletal muscle adjacent to fracture site show less Ki67+/GFP+ cells in Bmpr1a cKO mice compared to Bmpr1a control mice. (E) Quantification of GFP + Ki67+ over the total GFP+ cells in activated periosteum and skeletal muscle at d3 post‐fracture in tamoxifen induced Bmpr1a control and Bmpr1a cKO mice. n = 3–4 animals per group, each dot represents a single animal. (F) Left, Experimental design of in vitro cell differentiation, migration, and proliferation assays. Bmpr1a control and Bmpr1a cKO mice were induced with tamoxifen at d7, d6, d5, and d1 before experiment. Pdgfra‐derived GFP+ cells were collected from skeletal muscles surrounding the tibia, sorted based on GFP expression and cultured in vitro prior analyses. (i) Representative images of osteogenic (top, osteo) and chondrogenic (bottom, chondro) differentiation assays of GFP+ cells isolated from Bmpr1a control and Bmpr1a cKO mice, (ii) Percentage of migrating cells assessed by in vitro transwell migration assay, (iii) AUC of proliferation. n = 3 independent primary cultures per group, each dot represents a primary culture. Statistical analyses: Exact p value calculated with two‐sided Mann‐Whitney test; values represent median and interquartile range. Scale bars: low magnification = 200 μm, high magnification = 50 μm. AUC = area under the curve; b = bone; bm = bone‐marrow; cart = cartilage; chondro = chondrogenic; d0 = day 0; osteo = osteogenic; PS = Picrosirius; SO = Safranin O; TMX = tamoxifen.
Non‐stabilized tibial fracture
Mice were anesthetized with an intraperitoneal injection of Ketamine (50 mg/mL) and Medetomidine (1 mg/kg) and received a subcutaneous injection of Buprenorphine (0.1 mg/kg) for analgesia. The right leg was shaved and cleaned using Vetidine soap and solution (Vetoquinol, Lure Cedex, France; ref VET 001). The tibial surface was exposed, and the tibia was cut in the mid‐diaphysis to create the fracture. At the end of the procedure, the skin was sutured using non‐resorbable sutures (Harvard Apparatus, Holliston, MA, USA; ref 72‐3318). Mice were revived with an intraperitoneal injection of atipamezole (1 mg/mL) and kept on heated plate. Two additional doses of analgesia were administrated within 24 hours post‐surgery.
Isolation and primary culture of muscle mesenchymal progenitors, periosteal cells, and bone marrow stromal cells
Primary culture of skeletal muscle mesenchymal progenitors (muMPs) was performed as described.( 2 ) Briefly, 4‐week‐old to 8‐week‐old mice were euthanized and hindlimbs were harvested. After removing skin and fascia, skeletal muscles surrounding the tibia were dissected. Only the middle part of the muscle tissue free of tendon was used for cell isolation. In a Petri dish with Dulbecco's modified Eagle medium (DMEM) (Life Technologies, Grand Island, NY, USA; ref 21063029) skeletal muscles were minced with scissors. Skeletal muscles were then digested in digesting medium composed of DMEM with 1% Trypsin (Life Technologies; ref 210234) and 1% collagenase D (Roche Diagnostics, Mannheim, Germany; ref 11088866001) and incubated at 37°C for 2 hours. Every 20 minutes individualized cells were removed and transferred into growth media on ice: α minimum essential medium (α‐MEM) (Life Technologies; ref 32561029) with 1% penicillin–streptomycin (P/S) (Life Technologies; ref 15140122), 20% lot‐selected non‐heat‐inactivated fetal bovine serum (FBS) (Life Technologies; ref 10270106) and 10 ng/mL basic fibroblast growth factor (bFGF; R&D Systems, Minneapolis, MN, USA; ref 3139‐FB‐025/CF) and digesting medium was renewed. This step was repeated until all skeletal muscle tissue was digested. After the digestion, cells were filtered sequentially through 100‐μm (Dutscher, Bernolsheim, France; ref 352360) and 40‐μm filters (Dutscher; ref 352340), centrifuged 10 minutes at 300 g, resuspended in growth medium and placed in culture in growth medium.
Primary cultures of periosteal cells (PCs) and bone marrow stromal cells (BMSCs) were performed as described.( 10 , 31 ) Briefly, 4‐week‐old to 8‐week‐old mice were euthanized, and femurs and tibias were dissected to remove fat, adjacent skeletal muscles, and tendons. Epiphyses were then cut and removed. For BMSC culture, bone marrow was flushed out, collected in growth medium, and centrifuged 10 minutes at 300 g. Bone marrow cells were resuspended in growth medium and placed in culture. Growth medium was changed every day for 3 days to eliminate floating cells, and then every 3 days. For PC culture, flushed femurs and tibias were placed in 6‐cm culture plates and covered with a drop of growth medium to allow PCs to migrate out of the bone explants. When PCs reached confluence, bone explants were removed and cells at passage 0 (P0) were directly subjected to scRNAseq analyses or expanded for subsequent analyses.
Cell sorting
For cell transplantation experiments, PCs, BMSCs, and muMPs were trypsinized (Life Technologies; ref 25200056) and resuspended in growth medium. After 10 minutes of centrifugation at 300 g, cells were resuspended in sorting medium containing α‐MEM with 1% P/S and 2% lot‐selected non‐heat‐inactivated FBS. Sytox blue (1/1000; Thermo Fisher Scientific, Waltham, MA, USA; ref S34857) was added just before sorting to stain dead cells. Sytox Blue‐/Prx1‐derived green fluorescent protein–positive (GFP+) cells were sorted from Prx1 Cre ;Rosa mTmG mice. For scRNAseq of muMPs, Sytox Blue‐/Prx1‐derived GFP+ cells were sorted after muscle digestion as described in 'Isolation of primary culture of muscle mesenchymal progenitors' section. For in vitro experiments, SytoxBlue‐/Pdgfra‐derived GFP+ cells were sorted directly after muscle digestion from Bmpr1a control and Bmpr1a cKO mice. Equivalent percentage of GFP+ cells in the muscle of Bmpr1a control and Bmpr1a cKO mice was observed (13.6 ± 1.2% and 13.9 ± 4.7%, respectively). Cell sorting was performed on BD FACS Aria II SORP (BD Biosciences, San Jose, CA, USA) for scRNAseq and cell transplantation experiments and Aria Fusion (BD Biosciences) for in vitro experiments.
Flow cytometry analyses
For flow cytometry analyses, PCs, BMSCs and muMPs at passage 0–1 were trypsinized and resuspended in growth medium. After 10 minutes of centrifugation at 300 g, cells were resuspended in sorting medium and counted. A total of 500,000 cells were incubated with 50 μL of BD Horizon Brilliant Stain Buffer (BD Biosciences; ref 563794), CD45‐BV650 (Clone 30‐F11, 1/300; BD Biosciences; ref 563410), TER‐119‐BV650 (Clone TER‐119, 1/300; BD Biosciences; ref 747739), CD51‐BV711 (Clone RMV‐7, 1/300; BD Biosciences; ref 740755), TIE2‐APC (1/300; BioLegend, San Diego, CA, USA; ref 124009), 6C3‐PECy7 (1/300; Ozyme, Saint‐Cyr‐l'École, France; ref BLE108313), THY.2‐BV786 (Clone 53–2.1, 1/300; BD Biosciences; ref 564365), CD105‐BV421 (Clone MJ7/18, 1/300; BD Biosciences; ref 562760), CD200‐BV605 (Clone OX‐90, 1/300; BD Biosciences; ref 745255), PDGFRα‐PECy7 (Clone APA5, 1/300; eBioscience, Santa Clara, CA, USA; ref 25‐1401‐80), SCA1‐APC (Clone REA422, 1/200; Miltenyi Biotec, Bergisch Gladbach, Germany; ref 103‐123‐848), CD29‐APC (Clone HMβ1‐1, 1/400; Miltenyi Biotech; ref 130‐102‐557), or PDGFRα‐BV711 (Clone APA5, 1/200; BD Biosciences; ref 740740) for 30 minutes on ice and protected from light. Cells were then washed by adding 1 mL of sorting medium and centrifuged for 10 minutes at 300 g. Supernatant was discarded and cell pellets were resuspended in 200 μL of sorting medium. Compensation beads (Thermo Fischer Scientific; ref 01‐2222‐42) were used for initial compensation set up and fluorescence minus one (FMO) controls were used for the gating. Analyses were performed on BD LSR Fortessa SORP (BD Biosciences) and results were analyzed using FlowJo software, version 10.2 (FlowJo, LLC, Ashland, OR, USA). The gating strategy used for the analyses is available in Fig. S1 A–C.
In vitro differentiation
In vitro differentiation was performed as described.( 31 ) Skeletal muscle from tamoxifen induced Bmpr1a control and Bmpr1a cKO mice were digested and Pdgfra‐derived muMPs were sorted based on GFP+ expression. After cell sorting, Pdgfra‐derived muMPs were expanded in six‐well plates for in vitro differentiation. For osteogenic differentiation, cells at confluence were cultured in osteogenic medium containing αMEM supplemented with 10% lot‐selected non‐heat‐inactivated FBS, 0.1μM dexamethasone (Sigma‐Aldrich; ref D2915), 0.2mM L‐ascorbic acid (Sigma‐Aldrich; ref A8960) and 10mM glycerol 2‐phosphate disodium salt hydrate (Sigma‐Aldrich; ref G9422). Medium was changed every 3 days for 3 weeks. Mineralized particles were stained with 0.2% Alizarin red staining (Sigma‐Aldrich; ref A5533). For chondrogenic differentiation, 1.5 × 105 cells were plated as micromass in 200 μL of growth media for 2 hours. Then, growth medium was replaced by chondrogenic medium composed of DMEM with 10% lot‐selected non‐heat‐inactivated FBS, 0.1μM dexamethasone, 100 μg/mL sodium pyruvate (Sigma‐Aldrich; ref P5280), 40 μg/mL L‐proline (Sigma‐Aldrich; ref P0380), 50 μg/mL L‐ascorbic acid, 50 mg/mL Insulin‐Transferrin‐Selenium (Sigma‐Aldrich; ref I1884), and 10 ng/mL transforming growth factor β1 (TGFβ1) (Sigma‐Aldrich; ref T7039). Proteoglycans were stained with Alcian blue (Sigma‐Aldrich; ref A5268). All pictures were obtained with a Leica DM IRB light microscope (Leica Microsystems, Inc., Buffalo Grove, IL, USA).
In vitro cell migration
A total of 100,000 cells resuspended in 200 μL of medium supplemented with 0.2% bovine serum albumin (Sigma‐Aldrich; ref A2153) were seeded in the upper chamber of an 8‐μm pore transwell plate (Thermo Fischer Scientific; ref 141082) and incubated with 800 μL of α‐MEM at 1% P/S and 10% FBS in the lower chamber for 15 hours. Anti‐mitotic Cytosine β‐D‐arabinofuranoside hydrochloride (Sigma‐Aldrich; ref C6645) was added to synchronize cells. Cells were fixed with 4% paraformaldehyde (PFA) and nuclei were stained with 4′,6‐diamidino‐2‐phenylindole (DAPI). Pictures were taken using EVOS Cell Imaging Systems (Thermo Fischer Scientific). Non‐migrated cells were then removed from the upper side of the membrane with a cotton swap (VWR, Leicestershire, UK; ref PURJ896‐PC) and migrated cells on the bottom side of the membrane were counted. DAPI+ nuclei were counted using ImageJ software (NIH, Bethesda, MD, USA; https://imagej.nih.gov/ij/).
In vitro cell proliferation
A total of 20,000 GFP+ sorted cells from Bmpr1a control or Bmpr1a cKO mice were plated in 12‐well plates. Each sample was analyzed in duplicate. Cells were then manually counted at d2, d4 and d6 after plating. Growth curve was then generated and area under the curve (AUC) was used to determine cell proliferation using GraphPad Prism (GraphPad Software, Inc., La Jolla, CA, USA).
Genotyping of Bmpr1a mutant cells
GFP+ sorted cells from Bmpr1a control or Bmpr1a cKO mice were lysed in NaOH 50mM at 95°C for 10 minutes. The solution was then equilibrated with 23% Tris–HCl. Bmpr1a polymerase chain reaction (PCR) was performed with GoTaq G2 Hot Start Green Master Mix (Promega, San Luis Obispo, CA, USA; ref M7423) using the following primers: Fx2: 5′‐GCA GCT GCT GCT GCA GCC TCC‐3′, Fx4: 5′‐TGG CTA CAA TTT GTC TCA TGC‐3′, Fx1:5′‐GGT TTG GAT CTT AAC CTT AGG‐3′, according to Mishina and colleagues.( 30 ) PCR products were then run on 4% agarose gel.
Tissue and cell transplantation
Tissue and cell transplantations were performed as described.( 2 , 10 , 31 , 32 ) Tibial fracture was induced in the host mice as described in 'Non‐stabilized tibial fracture' section. For EDL muscle transplantation, EDL muscle was dissected from tendon to tendon and grafted adjacent to the fracture site. EDL graft was positioned on the anterior surface of the tibia, free of endogenous skeletal muscle, and sutured to the host patellar and peroneus muscle tendons with nonresorbable sutures (Fine Science Tools, Foster, CA, USA; ref 12051‐08). The skin was sutured and the mice revived. For periosteum transplantation, the tibia of donor mice was collected and a fragment of cortical bone of approximately 2 mm in length and 1 mm in width was cut in the anterior‐proximal area of each tibia. The endosteum and bone marrow were removed from the graft. Host mice were prepared by creating a cortical defect on the anterior‐proximal surface of the tibia adjacent to a fracture. The graft was placed in the cortical defect. The muscle was sutured over the defect to hold the graft in place, and wounds were closed.
For cell transplantation, PCs, BMSCs, and muMPs at passage 1 were trypsinized, washed and resuspended in 1 mL of sorting medium. GFP+ cells were sorted as described in 'Cell sorting' section. The viability of MuMPs, PCs, and BMSCs was 99.8 ± 0.2%, 99.5 ± 0.2%, and 97.1 ± 2.3%, respectively. A total of 150,000 sorted cells were embedded in Tisseel Prima fibrin gel, composed of fibrinogen and thrombin (Baxter, Deerfield, IL, USA; ref 3400894252443), according to the manufacturer's instructions. Briefly, 1 × 105 cells were resuspended in 15 μL of fibrin (diluted at 1/4), 15 μL of thrombin (diluted at 1/4) was added and cells were placed on ice for at least 15 minutes to let the matrix polymerize. The cell pellet was transplanted at the fracture site and the wound was closed.
Sample processing, histology, and histomorphometric analyses
For Ki67 and phoshoSmad1/5/9 immunostaining, calluses were fixed in 4% PFA (Euromedex, Souffelweyersheim, France; ref 15714) for 4 hours and decalcified in 19% ethylenediamine tetraacetic acid (EDTA) (Euromedex; EU00084) for 3 to 4 days at 4°C under agitation in the dark. All other fractured tibias were harvested, fixed in 4% PFA for 24 hours and decalcified in 19% EDTA for 3 weeks at 4°C under agitation in the dark. Samples were embedded in paraffin or in optimal cutting temperature compound (OCT) (MM France, Brignais, France; ref F/62550–1). The entire callus was sectioned, and all consecutive sections were collected. After deparaffinization in NeoClear® (VWR; ref 1098435000) for 2 × 5 minutes, sections were rehydrated and rinsed in PBS for 5 minutes. Frozen sections were dried at room temperature for 30 minutes in the dark and rehydrated in PBS for 10 minutes. After staining, sections were dehydrated in series of graded alcohols and incubated in NeoClear® for 10 minutes. Slides were mounted with NeoMount® mounting medium (VWR; ref 1090160100).
Safranin‐O staining
Sections were stained with Weigert's solution for 5 minutes, rinsed in tap running water for 3 minutes and stained with 0.02% Fast Green for 30s (Sigma‐Aldrich; ref F7252), followed by 1% acetic acid for 30s and Safranin'O (SO) solution for 45 minutes (Sigma‐Aldrich; ref S2255).
Masson's trichrome (TC) staining
Sections were stained with Harris hematoxylin (dilution ½) for 5 minutes (MM France; ref F/C0283), rinsed in running tap water 5 minutes, stained with Mallory red for 10 minutes, rinsed for 5 minutes, and then incubated with phosphomolybdic acid 1% for 10 minutes (Sigma‐Aldrich; ref HT153). Collagen fibers were stained inu light green for 20 minutes (VWR; ref 720‐0335) and fixed in 1% acetic acid.
Picrosirius staining
Sections were stained with Picrosirius solution (PS) (0.1 g of Direct Red 80; Sigma‐Aldrich; ref 43665‐25G; diluted into 100 mL of saturated solution of picric acid; Sigma‐Aldrich; ref 80456) for 2 hours at room temperature, protected from light.
For histomorphometric analyses, every tenth slide throughout the entire callus was stained with SO, TC, or counterstained with DAPI to visualize fluorescent GFP and Tomato signals. Images were captured using a Zeiss Imager D1 AX10 light microscope (Carl Zeiss Microscopy GmbH, Jena, Germany). Areas of callus, cartilage, bone, and GFP or Tomato signal were determined using ZEN software v1.1.2.0 (Carl Zeiss Microscopy GmbH) and volumes were calculated via the following formula:
𝑉𝑜𝑙u𝑚𝑒=
where Ai and Ai+1 were the areas of callus, cartilage, bone, or fluorescent signal in sequential sections, h was the distance between Ai and Ai+1 and equal to 300 μm, n was the total number of sections analyzed in the sample.
For transplanted cell and tissue contribution to cartilage and bone, GFP and Tomato signal surface were quantified throughout the entire callus on sections adjacent to SO and TC using a Zeiss Imager D1 AX10 light microscope and ZEN software. Volume of fluorescent signal was calculated as described for histomorphometric analyses above. For Bmpr1 control and Bmpr1a cKO fracture calluses, the volume of GFP and Tomato signals were quantified on three sections 300 μm apart in the central part of the callus. GFP+ signal in each area was normalized over the total fluorescence signal (sum of GFP and Tomato fluorescent signals).
Immunofluorescence
GFP and Tomato signals were detected without immunofluorescence staining. Cryosections were dried at room temperature for 30 minutes, rehydrated in PBS for 10 minutes, and then mounted with Fluoromount (eBioscience, Santa Clara, CA, USA; ref 495952).
For Ki67 immunostaining, tissue sections were incubated in PBS supplemented with 5% normal goat serum for 30 minutes and with rabbit anti‐mouse Ki67 antibody (dilution 1/200; Abcam, Cambridge, MA, USA; ref ab15580) overnight at 4°C. Secondary goat anti‐rabbit AF647 antibody (Life Technologies; ref A‐21245) was incubated for 1 hour at room temperature and slides were mounted with Fluoromount (ref 495952, eBioscience). For phospho‐Smad1/5/9 immunostaining, citrate buffer antigen retrieval was used for 20 minutes at 95°C followed by 20 minutes at 4°C. Sections were incubated in PBS supplemented with 5% normal goat serum for 30 minutes, before incubation with rabbit anti‐mouse phoshoSmad1/5/9 antibody (dilution 1/200; Cell Signaling Technology, Danvers, MA, USA; ref 13820 T) overnight at 4°C, and with secondary goat anti‐rabbit AF647 antibody for 1 hour at room temperature. Slides were mounted with Fluoromount and images were acquired using a Zeiss LSM 800 confocal microscope.
X‐Gal staining
Samples were harvested, fixed in 0.2% glutaraldehyde solution overnight at 4°C, cryoprotected in 30% sucrose solution and embedded in OCT. Sections were dried at room temperature for 30 minutes, rehydrated in PBS for 5 minutes and post‐fixed in 0.2% glutaraldehyde solution for 15 minutes at room temperature. Slides were then washed 3 × 15 minutes in the washing buffer containing 1M MgCl2 (Sigma‐Aldrich; ref M8266), 1% Na‐deoxycholate (Sigma‐Aldrich; ref D6750), 2% NP40 (ref 74385, diluted in H2O) in PBS. Sections were incubated overnight at 37°C in a humidified chamber in X‐Gal solution containing X‐Gal (Thermo Fisher Scientific; ref R0404; 50 mg/mL in DMSO), 1X potassium ferrocyanide, 1X potassium ferrocyanide, 1M Tris (pH 7.3–7.4) diluted in washing buffer. Sections were washed in PBS 3 × 5 minutes, counterstained with 1% eosin for 2 minutes, dehydrated, and mounted with NeoMount® mounting medium.
Quantification of cell migration and proliferation in vivo
In vivo cell migration was determined using ImageJ by measuring the minimal distance between each GFP+ cell in the callus and the border of the tissue graft. Every 30th section was used throughout the entire callus.
In vivo cell proliferation in the activated periosteum and skeletal muscle was measured by manual counting of Ki67 + GFP+ and total GFP+ in three independent regions per section. The results represent the mean of three different sections per sample.
Microarray analyses
For microarray analyses, datasets from Lu and colleagues( 33 ) were reanalyzed. Wild‐type fractured hindlimbs between knee and ankle were collected free of skin followed by RNA extraction at day 2 (n = 3) and day 7 (n = 4) and from uninjured limbs (n = 4) using Trizol (Invitrogen, Carlsbad, CA, USA). Microarrays were obtained using Agilent Mouse single‐color 4 × 44 K arrays (Agilent Technologies, Santa Clara, CA, USA). Microarray feature extraction was performed using Agilent's Extraction 9.1 (Agilent Technologies).( 33 ) Gene set enrichment analysis (GSEA) was performed using all normalized probes on “curated gene set” and “ontology gene set” collections of the Molecular Signatures Database v7 with gene sets between 15 and 5000 genes, 1000 permutations, FDR <0.25 and p value <0.05. Enrichment map was performed with Cytoscape software v3.8.1. using GSEA results with FDR cutoff <0.25, p value <0.05 and filtered by gene expression. Only clusters with more that five terms were retained for the analyses.
Single‐cell RNAseq analyses
For skeletal muscle–derived muMPs, datasets from Julien and colleagues( 2 ) were reanalyzed. Briefly, Prx1‐derived skeletal muscle cells were isolated directly by enzymatic and mechanical digestion of skeletal muscles surrounding the tibia from Prx1 Cre ;Rosa mTmG uninjured mice, and at day 3 and day 5 post–tibial fracture. Two mice were used per sample and only skeletal muscles adjacent to the fracture site were dissected. For PC scRNAseq, uninjured cells were isolated from three mice (ie, six uninjured tibias) by explant culture, and d3 post‐fracture cells were isolated from five mice (ie, five injured tibias) as described.( 10 ) PCs were then subjected to scRNAseq at passage 0 without further in vitro cell expansion. No specific analysis of the between‐animal variance was conducted due to the design of the experiment. All scRNAseq libraries were generated using Chromium Single Cell 3′Library & Gel Bead Kit v.2 (10X Genomics, San Francisco, CA, USA; ref PN‐120237) according to the manufacturer's protocol. Libraries were sequenced on NovaSeq 600 (Illumina, San Diego, CA, USA) with 26 cycles of read 1, eight cycles of i7 index, and 98 cycles of read 2. FastQ files from the scRNA 10X libraries were processed using the Cell Ranger Count pipeline with its default parameters (v5.0.1). Reads were aligned against the mm10 reference genome customized by adding GFP sequence.
Data analyses
Seurat v4.0.1 and RStudio v1.3.1073 were used for analysis of scRNA‐seq data.( 34 , 35 )
As described in Julien and colleagues,( 2 ) for muMPs, cells expressing between 350 and 8000 genes and expressing less that 20% of mitochondrial genes were retained for analysis, genes expressed in less than three cells were excluded from the analysis. For PCs, cells expressing between 100 and 8000 genes and expressing less that 10% of mitochondrial genes were retained for analysis. Genes expressed in less than three cells were excluded from the analysis. After quality control, 4458 uninjured PCs, 15726 PCs at d3, 4013 uninjured muMPs, 5313 muMPs at d3, and 1449 muMPs at d5 post‐fracture were retained for the analysis.
Normalization was performed using sctransform pipeline to integrate datasets and raw counts were log normalized and scaled for gene expression. All datasets were regressed on mitochondrial content. Clustering was performed using the first 20 principal components and a resolution of 0.6 for muMPs, and the first 25 principal components and a resolution of 0.5 for PCs. The number of principal components was determined using the ElbowPlot function of Seurat package. Differentially expressed genes were determined using Wilcoxon rank sum test with p value <0.05. Gene ontology (GO) analyses were done using differentially upregulated genes and implemented in Enrichr interface (https://amp.pharm.mssm.edu/Enrichr/).( 36 ) GO functions including less than five genes and with adjusted p value >0.05 were excluded. GO functions were classified into manually annotated general functions and the number of GO terms per general function was plotted.
Monocle analysis
Monocle3 v0.2.3.0 was used for pseudotime analysis.( 37 ) Sctransform normalized data were used as matrix to perform pseudotime analyses. Starting points correspond to the highest expression of stem/progenitor genes (Cd34, Ly6a) and the lowest expression of chondrogenic genes (Sox9/Acan). Pseudotime values were then added as metadata into Seurat object and pseudotime was plotted as feature using Scatterplot function.
Cell cycle analysis
Cell Cycle Regression vignette from Seurat package was used to study cell cycle.
Lineage analysis
Signature score was calculated for each cell as arithmetic mean of the expression of the indicated genes (Table S1) and implemented as metadata in Seurat object.
Statistical analyses
Data are presented as mean ± standard deviation (SD) and were obtained from at least two independent experiments; n represents the number of samples used for the analysis. Two‐sided Mann‐Whitney test was used to compare two groups. For comparison of more than three groups, one‐way or two‐way analysis of variance (ANOVA) was used. One‐way ANOVA was followed by two‐sided Mann‐Whitney test and two‐way ANOVA was followed by Tukey test or Šídák's multiple comparison test as indicated in the legend. Statistical analyses were done using GraphPad Prism v6.0a. Differences were considered significant for p value <0.05. Statistical analyses are provided in the Table S2.
Results
Periosteum and skeletal muscle are the sources of osteochondral progenitors
Prx1‐derived SSPCs form cartilage and bone during bone regeneration, and are localized within periosteum, bone marrow and skeletal muscle.( 2 , 3 , 4 , 10 , 23 ) To directly compare the expression profiles of SSPCs, we performed flow cytometry analyses of sorted Prx1‐derived cultured cells isolated from bone marrow (BMSCs), periosteum (PCs), and skeletal muscle (muMPs) from Prx1 Cre ;Rosa mTmG uninjured hindlimbs (Fig. 1A ). We used the strategy proposed by Chan and colleagues,( 38 ) a combination of cell surface markers that identify skeletal stem/progenitor, osteochondral, and stromal subpopulations from the entire bone. Hematopoietic and endothelial cells were excluded from the analyses and skeletal stem/progenitors were defined as ITGAV+/THY1−/6C3− cells, osteochondral progenitors as ITGAV+/THY1+/6C3− cells and stromal cells as ITGAV+/THY1−/6C3+ cells (Fig. 1B ). We evaluated the proportions of each subpopulation isolated from bone marrow, periosteum, and skeletal muscle. We observed that BMSCs exhibit reduced proportions of non‐hematopoietic and non‐endothelial cells and increased proportions of ITGAV+/THY1−/6C3− cells compared to PCs and muMPs. However, PCs and muMPs were enriched in ITGAV+/THY1+/6C3− cells and were mostly Prx1‐derived (Figs. 1C and S1). In vivo transplantation of Prx1‐derived BMSCs, PCs, and muMPs at the fracture site showed that PCs have a higher capacity to integrate into the callus compared to BMSCs and muMPs, and both PCs and muMPs have an enhanced chondrogenic potential compared to BMSCs. Further, PCs were bi‐potent because they also formed bone at d14 post‐transplantation (Fig. 1D,E ).
Fig. 1.
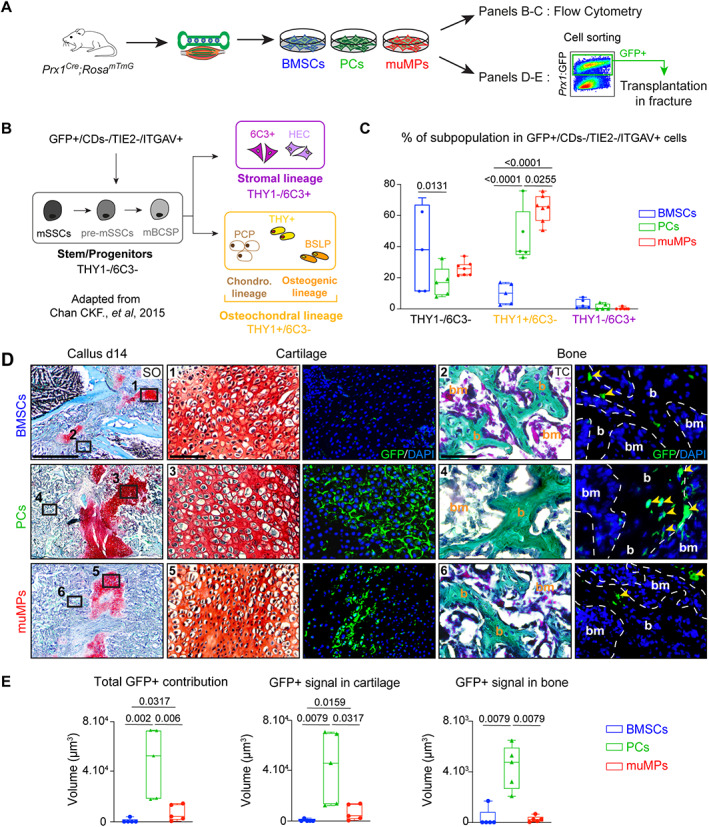
Periosteum‐derived and skeletal muscle‐derived cells are enriched in osteochondral progenitors and contribute efficiently to endochondral ossification during bone repair. (A) Experimental design. BMSCs, PCs, and skeletal muMPs were isolated from Prx1 Cre ;Rosa mTmG mice and cultured for one passage before flow cytometry analyses or cell sorting based on GFP expression prior to cell transplantation at the fracture site. (B) Schematic representation of hierarchical organization of skeletal stem/progenitor cells adapted from.( 38 ) Prx1‐derived GFP+ cells were gated first; hematopoietic and endothelial cells were excluded (CDs‐ = TER119−/CD45−/TIE2−) and ITGAV+ cells were included in the analysis. THY1 and 6C3 markers expression allow the identification of stem/progenitors (THY1−/6C3−), osteochondral (THY1+/6C3−) and stromal (THY1−/6C3+) subpopulations. (C) Percentage of skeletal stem/progenitor, osteochondral, and stromal cells in Prx1‐derived GFP+ BMSCs, PCs, and muMPs (n = 5–7 cell cultures per group). (D) Left, Longitudinal sections of fracture callus at d14 post‐fracture stained by SO. Middle‐right, High magnifications of boxed areas in cartilage and bone stained by SO and TC, respectively, and adjacent sections counterstained with DAPI (bone is delimited by a white dotted line). Boxed areas 1 and 2 showing limited contribution of BMSCs to cartilage and bone, boxed areas 3 and 4 showing robust contribution of PCs to cartilage and bone, and boxed areas 5 and 6 showing contribution of muMPs to cartilage and limited contribution to bone. Yellow arrowheads indicate contribution of transplanted cells to bone. (E) Histomorphometric quantification of total GFP+ signal and GFP+ signal in cartilage and bone, respectively. (C) Each dot represents an independent cell culture; (E) each dot represents a single animal. Values represent the median and interquartile range. (C) Exact p value calculated by two‐way ANOVA followed by Tukey test. (E) n = 5 per group, exact p value calculated with one‐way ANOVA followed by two‐sided Mann‐Whitney test. Scale bars: SO low magnification= 1 mm, cartilage high magnification = 200 μm, bone high magnification = 50 μm. b = bone; bm = bone marrow; BMSC = bone marrow stromal cell; muMP = muscle mesenchymal progenitor; PC = periosteal cell; SO = Safranin O; TC = trichrome.
Heterogeneity of periosteal cells at steady state
Periosteum is a well‐established source of SSPCs during bone repair( 1 , 5 , 6 , 10 , 32 ). To characterize the cellular composition of the periosteum, we performed scRNAseq analyses of primary PCs at P0 isolated from Prx1 Cre ;Rosa mTmG uninjured hindlimbs (Fig. 2A ). We identified nine clusters, grouped into three cell populations: stem/progenitors, encompassing clusters 0 to 5, expressing Prrx1, Pdgfra, Ly6a, Cd34, and Acta2; macrophages, corresponding to clusters 6 and 7, both expressing Ptprc, Adgre1, and Csfr1, and osteoclasts, ie, cluster 8, expressing Acp5 and Ocstamp (Table S3). Prx1‐driven GFP expression was detected almost exclusively in the stem/progenitor population (Fig. 2B,C ). Within the stem/progenitor population, we observed regionalized expression of markers. Ly6a and Pdgfra were highly expressed in clusters 0, 1, 2, and 3, whereas Acta2 was mainly expressed in clusters 0, 2, 4, and 5. In addition, we detected the expression of markers commonly defined as tenocyte (Scx, Tnmd) and pericyte (Mylk, Cspg4) markers, revealing heterogeneity within the periosteal stem/progenitor population (Figs. 2D and S2).
Fig. 2.
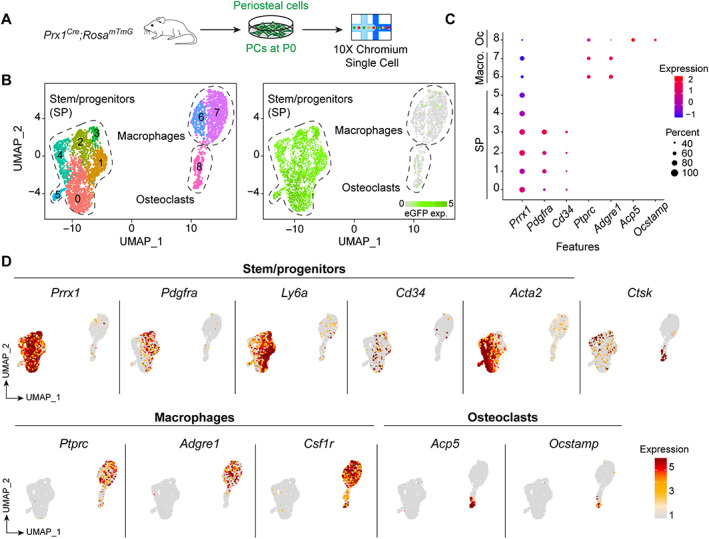
Single‐cell RNAseq of periosteal cells at steady state. (A) Experimental design of scRNAseq of PCs at steady state. PCs were isolated from uninjured tibia of Prx1 Cre ;Rosa mTmG mice by explant culture without expansion and subjected to scRNAseq analyses at P0. (B) Left: clusterization of PCs. Right: Feature plot of eGFP expression. (C) Expression of markers used to define SP, Macro, and Oc clusters. (D) Representation of SP, Macro, and OC marker expression. Macro = macrophage; Oc = osteoclast; P0 = passage 0; PC = periosteal cell; SP = stem/progenitor.
Periosteal cell response to bone injury at single‐cell resolution
To characterize the cellular response of PCs to fracture, we performed scRNAseq analyses of PCs isolated at d3 post‐fracture from Prx1 Cre ;Rosa mTmG mice, and integrated uninjured and d3 post‐fracture datasets (Fig. 3A ). In addition to the stem/progenitor, macrophage and osteoclast populations described at steady state (Fig. 2), we identified neutrophils expressing Ngp and Elane, fibrochondro progenitors (FCPs) expressing Col2a1 and Postn, and fibroblasts expressing S100a4 and Tpm2 (Figs. 3B–D and S3 A,B, Table S3). Prx1‐driven GFP expression was mainly detected in the non‐hematopoietic populations, ie, stem/progenitor, FCP, and fibroblast populations (Fig. S3 C). We then focused our analysis on these non‐hematopoietic populations that form cartilage and bone in the callus. To assess the fate of the non‐hematopoietic periosteal cells in response to fracture, we plotted the lineage score of stem/progenitor, fibrogenic (extracellular matrix [ECM]‐producing cells) and chondrogenic gene expression. The FCP cluster, found exclusively at d3 post‐fracture, was the only cluster containing cells with stem/progenitor, fibrogenic, and chondrogenic signatures (Fig. 3E–G ). GO analysis of upregulated genes in the FCP cluster showed a high number of biological functions related to stem/progenitor cell activation upon injury (proliferation and migration categories) and ossification (bone development and ECM categories). Detailed analyses of GO terms highlighted an overrepresentation of GO terms related to skeletal development, cartilage/chondrocyte and ossification/mineralization, as well as signaling pathways related to cartilage/bone formation such as TGFβ and BMP (Fig. 3H ). These results suggest that the FCP cluster contains the PC population specifically activated upon injury.
Fig. 3.
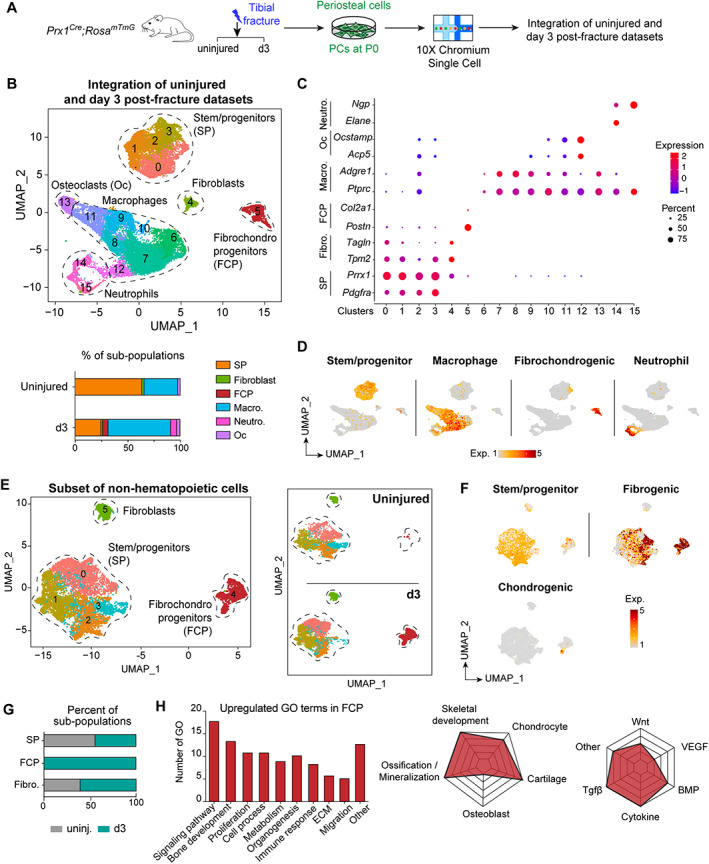
Single‐cell RNAseq of periosteal cells post‐fracture. (A) Experimental design of scRNAseq. PCs were isolated from Prx1 Cre ;Rosa mTmG mice by explant culture from uninjured tibia and from fractured tibia at d3 post‐fracture and used at P0 for scRNAseq. (B) Top, Clusterization of integrated uninjured and d3 post‐fracture datasets define six subpopulations (delimited by a black dotted line and named). Bottom, Percentage of subpopulations per sample. (C) Dot plot of markers used to define cell populations. (D) Feature plot of stem/progenitor, macrophage, fibrochondrogenic, and neutrophil lineage scores. (E) Left, UMAP projection of uninjured and d3 post‐fracture datasets subclusterized for non‐hematopoietic cells. Right, Split UMAP visualization of non‐hematopoietic cells from uninjured and d3 post‐fracture datasets. (F) Feature plot of stem/progenitor, fibrogenic, and chondrogenic lineage scores in the non‐hematopoietic cells. (G) Percentage of subpopulations per sample. (H) Left, GO analyses of upregulated genes in FCP subpopulation. Middle, Radar chart of skeletal related functions. Right, signaling pathways enriched in GO analyses. d3 = day 3; FCP = fibrochondro progenitor; Fibro. = fibroblast; GO = gene ontology; Macro. = macrophage; Neutro. = neutrophil; Oc = osteoclast; PC = periosteal cell; SP = stem/progenitor; UMAP = uniform manifold approximation and projection.
To better understand how PCs are activated upon fracture, we performed pseudotime analyses of FCPs as they correspond to cells engaging in chondrogenesis (Fig. 4A ). The starting point of the pseudotime trajectory was defined by the least differentiated cells expressing the highest level of stem/progenitor genes (Cd34/Ly6a) and the lowest level of chondrogenic genes ( Sox9/Acan ). The pseudotime trajectory progressed through one main branch where PCs downregulated stem/progenitor genes and upregulated fibrogenic genes prior to engaging in chondrogenesis (Figs. 4B and S3 D). Genes associated with cell migration were increased in parallel with fibrogenic genes, whereas genes associated with proliferation were upregulated in parallel with chondrogenic genes (Fig. 4C ).
Fig. 4.
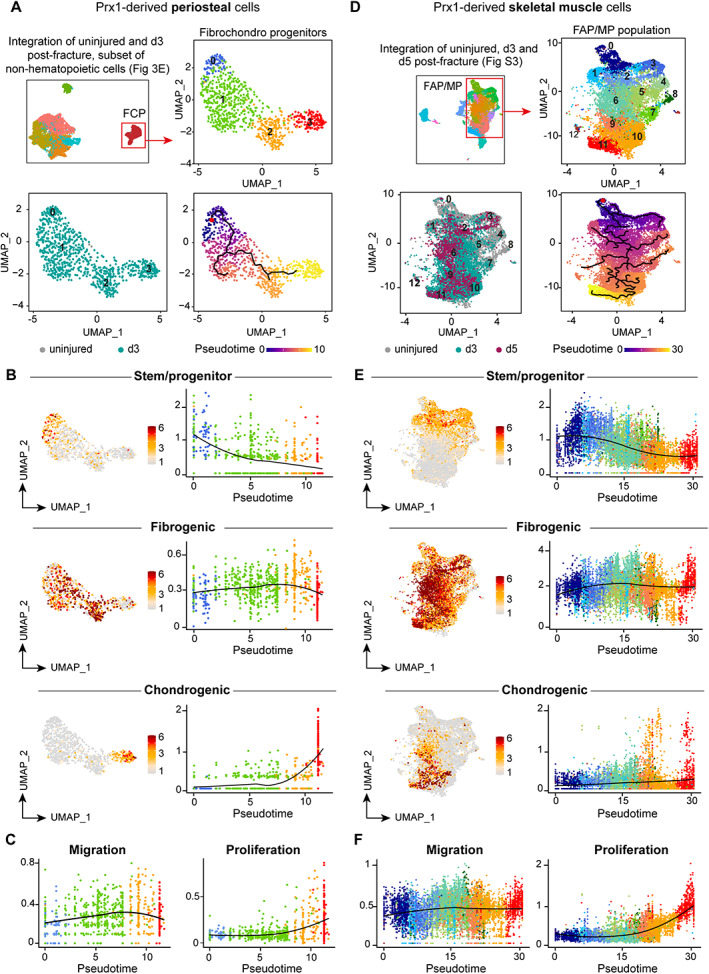
Single‐cell RNAseq reveals similar activation patterns of periosteum and muscle progenitors after bone fracture. (A) Top left, UMAP projection of subclusterization of non‐hematopoietic PCs as in Fig. 3E. Top right, UMAP projection of FCPs used for the subsequent analyses. Bottom left, UMAP visualization of sample origin of FCPs (uninjured in gray and d3 post‐fracture in green). Bottom right, Pseudotime trajectory analysis of periosteum‐derived FCPs. (B) Feature plot and scatter plot of stem/progenitor (top), fibrogenic (middle), and chondrogenic (bottom) lineage scores along pseudotime in periosteum derived FCPs. (C) Scatter plot of migration and proliferation lineage scores along pseudotime in FCPs. (D) Top left, UMAP projection of Prx1‐derived skeletal muscle cells from uninjured tissue and from d3 and d5 post‐fracture samples as in Fig. S4. Top right, UMAP projection of subclusterization of Prx1‐derived skeletal muscle FAP/MP used for the subsequent analyses. Bottom left, UMAP visualization of sample origin of Prx1‐derived skeletal muscle FAP/MP (uninjured in gray, d3 post‐fracture in green and d5 post‐fracture in purple). Bottom right, Pseudotime trajectory analysis of Prx1‐derived skeletal muscle FAP/MP. (E) Feature plot and scatter plot of stem/progenitor (top), fibrogenic (middle), and chondrogenic (bottom) lineage scores along pseudotime in FAP/MP. (F) Scatter plot of migration and proliferation lineage scores along pseudotime in FAP/MP. Color scheme used in B and C corresponds to the color of clusters used in A, and color scheme used in and E and F corresponds to the color of clusters used in D. d3 = day 3; FAP = fibroadipogenic progenitor; FCP = fibrochondro progenitor; MP = mesenchymal progenitor; PC = periosteal cell; UMAP = uniform manifold approximation and projection.
We previously reported the response to bone repair of Prx1‐derived skeletal muscle progenitors using scRNAseq( 2 ) (Fig. S4 A–D). Within Prx1‐derived skeletal muscle cells, the fibroadipogenic progenitor/mesenchymal progenitor (FAP/MP) population was the most responsive to injury( 2 ) (Table S3). Activated muMPs displayed the same cellular response to bone injury as activated PCs by losing their stem/progenitor identity, engaging in fibrogenesis, prior to engaging in chondrogenesis and proliferating (Fig. 4D–F ). However, in addition to the main trajectory, we observed multiple branches in the fibrogenic state for activated muMPs, suggesting that activated muMPs may not all engage in chondrogenesis and contain a subset of SSPCs (Fig. S4 E). Chondrogenic‐related and migration‐related genes were not highly expressed by d5 post‐fracture in activated muMPs because they were still localized in the skeletal muscle tissue prior to their migration into the fracture callus and only started expressing chondrogenic and migrating programs.
PC and muMP response to bone injury is mediated by BMP signaling
We then assessed the similarities in the molecular programs driving PC and muMP activation. Microarray datasets of uninjured, d2 and d7 post‐fracture hindlimbs were reanalyzed.( 33 ) We observed an overrepresentation of signaling pathways related to immune response such as Toll‐like receptors (Toll‐like R), chemokines, tumor necrosis factor (TNF), and interleukins, as well as Hedgehog (HH), platelet‐derived growth factor receptor (PDGFR), epidermal growth factor (EGF), TGF, and BMP pathways at d2 post‐fracture compared to uninjured samples (Fig. 5A,B ). This correlated with the enrichment in GO terms related to immune response (Fig. S5 A,B). At d7 post‐fracture, GO terms related to immune response and HH, PDGFR, and EGF signaling pathways were downregulated, but not GO terms related to BMP signaling. In parallel, GO terms related to metabolism, signal transduction, neurotrophin, and Wnt signaling pathways were upregulated (Fig. 5B ). This signature was associated with GO terms related to morphogenesis, skeletal development, and ECM secretion (Fig. S5 C,D). BMP signaling, defined as the mean expression of BMP receptors and effectors, was overexpressed at d2 and d7 post‐fracture, suggesting a role in the early stage of repair (Fig. 5C ). In the PC and muMP scRNAseq datasets from Fig. 4, we observed that BMP signaling was upregulated in FCPs at d3 and in activated muMPs at d3 and d5 post‐fracture (Figs. 5D,E and S6). Immunostaining for phoshoSMAD1/5/9 confirmed that the BMP pathway is active in Prx1‐derived periosteal and skeletal muscle cells at d3 post‐fracture in vivo (Fig. 5F ).
Fig. 5.
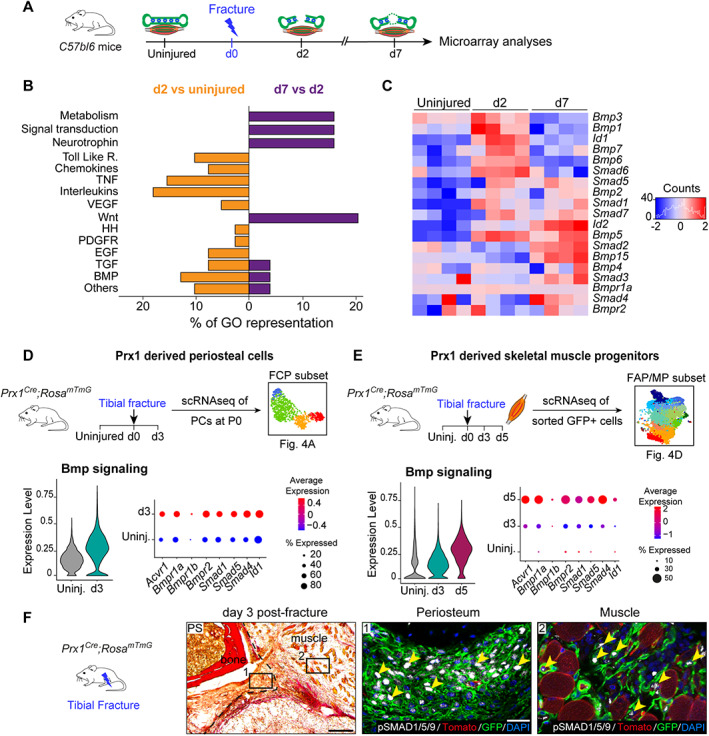
BMP signaling is upregulated in injury‐activated periosteum and muscle progenitors. (A) Experimental design of microarray analyses.( 33 ) Tibia and adjacent skeletal muscle were harvested from uninjured hindlimbs at d2 and d7 post‐fracture and used for microarray analyses. (B) Representation of upregulated signaling pathways from Gene Ontology analyses between d2 versus uninjured (in orange, left) and between d7 versus d2 (in purple, right). (C) Heat map of BMP signaling components from microarray dataset. (D,E) Experimental design of scRNAseq of Prx1‐derived periosteal cells and skeletal muscle progenitors. Violin plot of BMP signaling pathway expression (left) and detailed visualization of receptor (Acvr1, Bmpr1a, Bmpr1b, Bmpr2), co‐factor (Smad1, Smad4, Smad5), and target (Id1) gene expression (right) in Prx1‐derived FCPs (D) and skeletal muscle FAP/MP (E). (F) Transverse section of d3 post‐fracture Prx1 Cre ;Rosa mTmG hindlimb stained with PS showing activated periosteum (delimited by a black dotted line). High magnifications of boxed areas from adjacent section counterstained with DAPI show phosphoSMAD1/5/9 positive nuclei (white, pointed with yellow arrowhead) in GFP+ cells (in green) within periosteum (middle), and adjacent skeletal muscle (right). Scale bars: low magnification = 200 μm, high magnification = 50 μm. bm = bone‐marrow; d2 = day 2; PS = Picrosirius.
BMP signaling is required for SSPC activation upon fracture
To functionally evaluate the role of BMP signaling in SSPCs during bone regeneration, we performed genetic inactivation of the Bmpr1a gene.( 39 ) Due to the low efficiency of the Prx1 CreERT mouse model (Fig. S7), we used the Pdgfra CreERT line to target periosteal and skeletal muscle progenitors expressing Pdgfra (Figs. 2 and 3C , and Fig. S4 and S8A,B ).( 2 , 10 , 14 ) We evaluated a 65.9% Cre recombination efficiency in Pdgfra CreERT ;Rosa mTmG mice and confirmed the presence of Pdgfra‐derived skeletal progenitors contributing to fracture healing in both periosteum and skeletal muscle (Fig. S8C–E). Analysis of bone repair in tamoxifen‐induced Pdgfra CreERT ; Bmpr1a fl/fl mice showed decreased callus and bone volumes at d14 post‐fracture compared to Bmpr1a fl/fl controls and reduced contribution of Bmpr1a‐deficient Pdgfra‐derived cells within cartilage and bone as compared to control cells (Fig. 6A‐D ). However, the presence of non‐recombined cells using this CRE model suggests that these cells may partially compensate for the phenotype using a systemic induction strategy.
To specifically trace Bmpr1a‐deficient cells from periosteum and skeletal muscle, we used periosteum and EDL muscle graft transplantation. Grafts from tamoxifen‐induced Pdgfra CreERT ;Rosa mTmG ; Bmpr1a fl/fl (Bmpr1a cKO ) and Pdgfra CreERT ;Rosa mTmG ;Bmpr1a +/+ (Bmpr1a control ) mice were transplanted at the fracture site of wild‐type hosts. We observed a reduced contribution to cartilage of Bmpr1a‐deficient cells from periosteum and skeletal muscle, and reduced contribution to bone, of Bmpr1a‐deficient cells from periosteum, as compared to controls (Fig. 7A,B ). To determine whether abnormal cell migration could account for the impaired cellular contribution of periosteum and skeletal muscle to callus formation, we measured the distance between the graft and the Pdgfra‐derived migrating cells within cartilage. We observed that Bmpr1a‐deficient cells remained closer to the graft compared to control cells (Fig. 7C ). We then assessed cell proliferation in vivo using Ki67 immunostaining on Bmpr1a cKO and Bmpr1a control calluses at d3 post‐fracture. The percentage of proliferative Pdgfra‐derived cells over the total Pdgfra‐derived population was significantly reduced in the activated periosteum and adjacent skeletal muscle of Bmpr1a cKO mutant mice compared to control mice at day 3 post‐fracture, suggesting that a decrease in the proliferation of skeletal muscle and periosteum progenitors in the absence of BMP signaling could also impact the phenotype (Fig. 7D,E ). In vitro differentiation assays showed an abolition of the osteogenic potential of Bmpr1a‐deficient cells, whereas early chondrogenic differentiation was not affected. In vitro migration and proliferation capacities of Bmpr1a‐deficient cells were impaired, confirming in vivo observations (Fig. 7F ). We verified Cre‐mediated recombination in Pdgfra‐derived cells by Bmpr1a genotyping and confirmed that Bmpr1a deletion did not affect the cellular identity of Pdgfra‐derived cells by flow cytometry (Fig. S9). Altogether, these results highlight the role of BMP signaling as a common mediator of PC and muMP activation during the early stages of bone repair.
Discussion
Multiple markers have been proposed as specific for SSPC subpopulations in bone, but the diversity of these markers makes it challenging to investigate SSPC functions in skeletal growth, repair, and aging.( 9 , 12 , 14 , 18 , 19 , 20 , 21 , 22 , 23 , 39 ) In addition, during bone regeneration, SSPCs originate not only from bone compartments; ie, bone marrow and periosteum, but also from adjacent skeletal muscle.( 1 , 2 , 3 , 4 , 6 , 10 , 40 ) In skeletal muscle, the SSPC population overlaps with the non‐myogenic cell population commonly termed “mesenchymal” cells.( 2 ) These “mesenchymal” cells have fibrogenic, adipogenic, and osteochondrogenic potentials in muscle regeneration or pathological conditions.( 41 , 42 ) More knowledge is needed to improve the nomenclature and better describe “mesenchymal” cells that share common cell surface markers yet may have distinct embryonic origins and functions in different tissues.( 43 , 44 ) In this study we set out to compare SSPCs in bone marrow, periosteum, and skeletal muscle. In these three tissues, we identified skeletal stem/progenitor, osteochondral, and stromal cell populations expressing combinations of cell surface markers, as defined by Chan and colleagues.( 38 ) Furthermore, our flow cytometry results indicate that periosteum‐derived and skeletal muscle‐derived cells are enriched in osteochondral progenitors, which may correlate with their in vivo differentiation potential. Transplanted PCs show efficient chondrogenic and osteogenic potential in vivo, whereas muMPs are mainly chondrogenic, and BMSCs have a limited osteochondral potential. These results reinforce that the periosteum is the major source of SSPCs for bone repair.
We then focused on periosteum‐derived cells, which remain poorly characterized, and report here the cellular heterogeneity of periosteum‐derived cells by scRNAseq at steady state and in response to fracture. None of the clusters identified by scRNAseq coincided with populations defined by the flow cytometry panel. Given the cooperation between PCs and muMPs during endochondral ossification, we compared their cellular and molecular response to fracture at the single cell level. We identified a similar response to bone fracture toward chondrogenesis through pseudotime analyses. After bone fracture, both activated muMPs and PCs leave their stem/progenitor state to undergo fibrogenesis prior to chondrogenesis. Thus, SSPCs from periosteum and skeletal muscle follow a common molecular program during the initiation of the endochondral ossification process. Single‐cell analyses of freshly isolated SSPCs and from later time points post‐injury will be helpful in the future to further characterize the cellular heterogeneity of SSCPs and the in vivo differentiation potential of various SSPC subpopulations. In particular, given the bipotentiality of SSPCs in periosteum for osteogenesis and chondrogenesis, whether this is due to the presence of a single lineage or multiple lineages within periosteum remains to be clarified.
We show that the shared molecular response within periosteum and skeletal muscle is mediated by early activation of the BMP pathway, known to play a central role during skeletal development, bone homeostasis, and fracture healing.( 45 ) Genes associated with the BMP signaling pathway are expressed at early stages of bone repair from 3 days post‐fracture and regulate chondrogenesis and osteogenesis.( 46 , 47 , 48 , 49 , 50 , 51 ) Here, we combined lineage tracing, conditional knockout of Bmpr1a gene in Pdgfra‐derived progenitors and tissue transplantation to assess the role of BMP signaling in SSPCs from periosteum and skeletal muscle. Notably, due to the current limitations in easily discriminating SSPCs derived from periosteum and skeletal muscle in vivo, which would entail the analysis of a combination of several markers and their expression levels, cell or tissue transplantation at the injury site remains an efficient way to discriminate the regenerative potential of SSCPs from different tissues. Using these approaches, we show that the contribution to endochondral ossification of Bmpr1a deficient SSPCs from periosteum and skeletal muscle is impaired. Loss of BMP signaling in SSPCs leads to a decrease in proliferation, migration, and chondrogenic and osteogenic differentiation of periosteum and skeletal muscle SSPCs in vitro and in vivo. These functional analyses confirm that BMP signaling is a critical mediator for the activation of periosteum and skeletal muscle SSPCs during bone regeneration. Although there are reports showing that PDGFRβ or CCL5‐CCR5 signaling pathways are also essential( 52 , 53 ), little is known about the molecular regulation of SSPC activation. More work is required to provide insight and understanding of the mechanisms driving SSPC activation and their interactions with the bone injury environment.
Author Contributions
Anais Julien: Data curation; formal analysis; methodology; writing – original draft. Simon Perrin: Formal analysis; methodology; writing – review and editing. Ester Martinez‐Sarra: Formal analysis; writing – review and editing. Anuya Kanagalingam: Formal analysis. Caroline Carvalho: Formal analysis. Marine Luka: Methodology. Mickaël Menager: Funding acquisition; methodology; resources. Celine Colnot: Conceptualization; funding acquisition; supervision; writing – original draft.
Authors’ Roles
AJ, SP, and EMS performed the experiments and analyzed the data with the help of AK and CCarvalho. ML performed scRNAseq libraries and MM gave advice. AJ, SP, and CColnot designed the experiments. AJ and CColnot wrote the manuscript. SP and EMS reviewed the manuscript. CColnot conceived the project and supervised the study.
Conflicts of Interest
Authors declare no competing interests.
Data Availability
scRNAseq data from uninjured and d3 post‐fracture PCs have been deposited in the Gene Expression Omnibus (GEO) database under the accession number GSE195940. scRNAseq data from Prx1‐derived skeletal muscle cells are available through the accession number GSE164573. Microarray data are available upon request.
Supporting information
Appendix S1 Supplementary Information
Acknowledgments
This work was supported by Foundation de l'Avenir, ANR‐18‐CE14‐0033, NIAMS R01 AR072707 to CColnot, INSERM ATIP‐Avenir to MM. AJ and SP were supported by a PhD fellowship from Paris University. The LabTech Single‐Cell@Imagine is supported by the Paris Region and the “Investissements d'avenir” program 764 through the 2019 ATF funding – S same Filires PIA (Grant N 3877871) and the Agence National de la Recherche as part of the “Investment for the Future” program (Institut Hospitalo‐Universitaire Imagine, grant ANR‐10‐IAHU‐01). We thank C. Alms, R. Prota, A. Lermant, H. Mouigni and A. Henry for technical assistance. Flow cytometry and cell sorting were performed at the Flow Cytometry platforms of Imagine Institute and IMRB. Single‐cell RNAseq were performed at the Genomic and Bioinformatic platforms of Imagine Institute.
References
- 1. Colnot C. Skeletal cell fate decisions within periosteum and bone marrow during bone regeneration. J Bone Miner Res. 2009;24(2):274‐282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Julien A, Kanagalingam A, Martínez‐Sarrà E, et al. Direct contribution of skeletal muscle mesenchymal progenitors to bone repair. Nat Commun. 2021;12(1):1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bianco P, Cao X, Frenette PS, et al. The meaning, the sense and the significance: translating the science of mesenchymal stem cells into medicine. Nat Med. 2013;19(1):35‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Crane JL, Cao X. Bone marrow mesenchymal stem cells and TGF‐β signaling in bone remodeling. J Clin Invest. 2014;124(2):466‐472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roberts SJ, van Gastel N, Carmeliet G, Luyten FP. Uncovering the periosteum for skeletal regeneration: the stem cell that lies beneath. Bone. 2015;70:10‐18. [DOI] [PubMed] [Google Scholar]
- 6. Ferretti C, Mattioli‐Belmonte M. Periosteum derived stem cells for regenerative medicine proposals: boosting current knowledge. World J Stem Cells. 2014;6(3):266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shen B, Tasdogan A, Ubellacker JM, et al. A mechanosensitive peri‐arteriolar niche for osteogenesis and lymphopoiesis. Nature. 2021;591(7850):438‐444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sacchetti B, Funari A, Michienzi S, et al. Self‐renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131(2):324‐336. [DOI] [PubMed] [Google Scholar]
- 9. Matsushita Y, Ono W, Ono N. Skeletal stem cells for bone development and repair: diversity matters. Curr Osteoporos Rep. 2020;18(3):189‐198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Duchamp De Lageneste O, Julien A, Abou‐Khalil R, et al. Periosteum contains skeletal stem cells with high bone regenerative potential controlled by Periostin. Nat Commun. 2018;9(1):773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baccin C, Al‐Sabah J, Velten L, et al. Combined single‐cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nat Cell Biol. 2020;22(1):38‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Debnath S, Yallowitz AR, McCormick J, et al. Discovery of a periosteal stem cell mediating intramembranous bone formation. Nature. 2018;562(7725):133‐139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matsushita Y, Nagata M, Kozloff KM, et al. A Wnt‐mediated transformation of the bone marrow stromal cell identity orchestrates skeletal regeneration. Nat Commun. 2020;11(1):1‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matthews BG, Novak S, Sbrana FV, et al. Heterogeneity of murine periosteum progenitors involved in fracture healing. Elife. 2021;10:e58534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sivaraj KK, Jeong H‐W, Dharmalingam B, et al. Regional specialization and fate specification of bone stromal cells in skeletal development. Cell Rep. 2021;36(2):109352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tikhonova AN, Dolgalev I, Hu H, et al. The bone marrow microenvironment at single‐cell resolution. Nature. 2019;569(7755):222‐228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grcevic D, Pejda S, Matthews BG, et al. In vivo fate mapping identifies mesenchymal progenitor cells. Stem Cells. 2012;30(2):187‐196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Greenbaum A, Hsu YMS, Day RB, et al. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem‐cell maintenance. Nature. 2013;495(7440):227‐230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Méndez‐Ferrer S, Michurina TV, Ferraro F, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466(7308):829‐834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Park D, Spencer JA, Koh BI, et al. Endogenous bone marrow MSCs are dynamic, fate‐restricted participants in bone maintenance and regeneration. Cell Stem Cell. 2012;10(3):259‐272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shi Y, He G, Lee W‐C, McKenzie JA, Silva MJ, Long F. Gli1 identifies osteogenic progenitors for bone formation and fracture repair. Nat Commun. 2017;8(1):1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Worthley DL, Churchill M, Compton JT, et al. Gremlin 1 identifies a skeletal stem cell with bone, cartilage, and reticular stromal potential. Cell. 2015;160(1‐2):269‐284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou BO, Yue R, Murphy MM, Peyer JG, Morrison SJ. Leptin‐receptor‐expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15(2):154‐168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Calvi LM, Adams GB, Weibrecht KW, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425(6960):841‐846. [DOI] [PubMed] [Google Scholar]
- 25. Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505(7483):327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ. Expression of Cre recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33(2):77‐80. [DOI] [PubMed] [Google Scholar]
- 27. Kawanami A, Matsushita T, Chan YY, Murakami S. Mice expressing GFP and CreER in osteochondro progenitor cells in the periosteum. Biochem Biophys Res Commun. 2009;386(3):477‐482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kang SH, Fukaya M, Yang JK, Rothstein JD, Bergles DE. NG2+ CNS glial progenitors remain committed to the oligodendrocyte lineage in postnatal life and following neurodegeneration. Neuron. 2010;68(4):668‐681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mishina Y, Suzuki A, Ueno N, Behringer RR. Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes Dev. 1995;9(24):3027‐3037. [DOI] [PubMed] [Google Scholar]
- 30. Mishina Y, Hanks MC, Miura S, Tallquist MD, Behringer RR. Generation of Bmpr/Alk3 conditional knockout mice. Genesis. 2002;32(2):69‐72. [DOI] [PubMed] [Google Scholar]
- 31. Perrin S, Julien A, Duchamp de Lageneste O, Abou‐Khalil R, Colnot C. Mouse periosteal cell culture, in vitro differentiation, and in vivo transplantation in tibial fractures. Bio Protoc. 2021;11(15):e4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Julien A, Perrin S, Duchamp de Lageneste O, et al. FGFR3 in periosteal cells drives cartilage‐to‐bone transformation in bone repair. Stem Cell Reports. 2020;15(4):955‐967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lu C, Saless N, Hu D, et al. Mechanical stability affects angiogenesis during early fracture healing. J Orthop Trauma. 2011;25(8):494‐499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single‐cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411‐420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single‐cell data. Cell. 2019;177(7):1888‐1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90‐W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cao J, Spielmann M, Qiu X, et al. The single‐cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496‐502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chan CKF, Seo EY, Chen JY, et al. Identification and specification of the mouse skeletal stem cell. Cell. 2015;160(1‐2):285‐298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mizoguchi T, Ono N. The diverse origin of bone‐forming osteoblasts. J Bone Miner Res. 2021;36(8):1432‐1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Abou‐Khalil R, Yang F, Lieu S, et al. Role of muscle stem cells during skeletal regeneration. Stem Cells. 2015;33(5):1501‐1511. [DOI] [PubMed] [Google Scholar]
- 41. Theret M, Rossi FMV, Contreras O. Evolving roles of muscle‐resident fibro‐adipogenic progenitors in health, regeneration, neuromuscular disorders, and aging. Front Physiol. 2021;20(12):481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lees‐Shepard JB, Yamamoto M, Biswas AA, et al. Activin‐dependent signaling in fibro/adipogenic progenitors causes fibrodysplasia ossificans progressiva. Nat Commun. 2018;9(1):1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee JH, Tammela T, Hofree M, et al. Anatomically and functionally distinct lung mesenchymal populations marked by Lgr5 and Lgr6. Cell. 2017;170(6):1149‐1163.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stzepourginski I, Nigro G, Jacob JM, et al. CD34+ mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proc Natl Acad Sci U S A. 2017;114(4):E506‐E513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Salazar VS, Gamer LW, Rosen V. BMP signalling in skeletal development, disease and repair. Nat Rev Endocrinol. 2016;12:203‐221. [DOI] [PubMed] [Google Scholar]
- 46. Tsuji K, Bandyopadhyay A, Harfe BD, et al. BMP2 activity, although dispensable for bone formation, is required for the initiation of fracture healing. Nat Genet. 2006;38(12):1424‐1429. [DOI] [PubMed] [Google Scholar]
- 47. Yoon BS, Ovchinnikov DA, Yoshii I, Mishina Y, Behringer RR, Lyons KM. Bmpr1a and Bmpr1b have overlapping functions and are essential for chondrogenesis in vivo. Proc Natl Acad Sci U S A. 2005;102(14):5062‐5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Morgan EF, Pittman J, DeGiacomo A, et al. BMPR1A antagonist differentially affects cartilage and bone formation during fracture healing. J Orthop Res. 2016;34(12):2096‐2105. [DOI] [PubMed] [Google Scholar]
- 49. van Gastel N, Stegen S, Stockmans I, et al. Expansion of murine periosteal progenitor cells with fibroblast growth factor 2 reveals an intrinsic endochondral ossification program mediated by bone morphogenetic protein 2. Stem Cells. 2014;32(9):2407‐2418. [DOI] [PubMed] [Google Scholar]
- 50. Chappuis V, Gamer L, Cox K, Lowery JW, Bosshardt DD, Rosen V. Periosteal BMP2 activity drives bone graft healing. Bone. 2012;51(4):800‐809. [DOI] [PubMed] [Google Scholar]
- 51. Yu YY, Lieu S, Lu C, Miclau T, Marcucio RS, Colnot C. Immunolocalization of BMPs, BMP antagonists, receptors, and effectors during fracture repair. Bone. 2010;46(3):841‐851. [DOI] [PubMed] [Google Scholar]
- 52. Böhm AM, Dirckx N, Tower RJ, et al. Activation of skeletal stem and progenitor cells for bone regeneration is driven by PDGFRβ signaling. Dev Cell. 2019;51(2):236‐254.e12. [DOI] [PubMed] [Google Scholar]
- 53. Ortinau LC, Wang H, Lei K, et al. Identification of functionally distinct Mx1+αSMA+ periosteal skeletal stem cells. Cell Stem Cell. 2019;25(6):784‐796.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Supplementary Information
Data Availability Statement
scRNAseq data from uninjured and d3 post‐fracture PCs have been deposited in the Gene Expression Omnibus (GEO) database under the accession number GSE195940. scRNAseq data from Prx1‐derived skeletal muscle cells are available through the accession number GSE164573. Microarray data are available upon request.


