SUMMARY
The Oryza sativa (rice) carotenoid cleavage dioxygenase OsZAS was described to produce zaxinone, a plant growth‐promoting apocarotenoid. A zas mutant line showed reduced arbuscular mycorrhizal (AM) colonization, but the mechanisms underlying this behavior are unknown. Here, we investigated how OsZAS and exogenous zaxinone treatment regulate mycorrhization. Micromolar exogenous supply of zaxinone rescued root growth but not the mycorrhizal defects of the zas mutant, and even reduced mycorrhization in wild‐type and zas genotypes. The zas line did not display the increase in the level of strigolactones (SLs) that was observed in wild‐type plants at 7 days post‐inoculation with AM fungus. Moreover, exogenous treatment with the synthetic SL analog GR24 rescued the zas mutant mycorrhizal phenotype, indicating that the lower AM colonization rate of zas is caused by a deficiency in SLs at the early stages of the interaction, and indicating that during this phase OsZAS activity is required to induce SL production, possibly mediated by the Dwarf14‐Like (D14L) signaling pathway. OsZAS is expressed in arbuscule‐containing cells, and OsPT11prom::OsZAS transgenic lines, where OsZAS expression is driven by the OsPT11 promoter active in arbusculated cells, exhibit increased mycorrhization compared with the wild type. Overall, our results show that the genetic manipulation of OsZAS activity in planta leads to a different effect on AM symbiosis from that of exogenous zaxinone treatment, and demonstrate that OsZAS influences the extent of AM colonization, acting as a component of a regulatory network that involves SLs.
Keywords: apocarotenoids, arbuscular mycorrhizal symbiosis, GR24, in situ hybridization, OsPT11, Oryza sativa, strigolactones, zaxinone, zaxinone synthase
Significance Statement
OsZAS was described to produce zaxinone, a plant growth‐promoting apocarotenoid in Oryza sativa (rice). Here we demonstrate the importance of OsZAS to guarantee the correct extent of root colonization by arbuscular mycorrhizal (AM) fungi. We show that OsZAS controls the extent of AM colonization, acting as a component of a regulatory network, possibly mediated by the Dwarf14‐Like (D14L) signaling pathway, that involves SLs.
INTRODUCTION
Most terrestrial plants, including major crops, establish a root mutualistic association called arbuscular mycorrhizal (AM) symbiosis (Genre et al., 2020) with soil fungi belonging to Glomeromycotina (Spatafora et al., 2016). This evolutionarily ancient interaction implies a reciprocal delivery of nutrients: host plants receive mineral nutrients, mainly phosphorus (P), whereas AM fungi rely on plant‐derived fixed carbon (Rich et al., 2017). Additional benefits at organism and ecosystem levels make AM symbiosis a promising component of sustainable agricultural production (Chen et al., 2018; Rillig et al., 2019).
The establishment of AM symbiosis follows a finely tuned colonization pattern. The pre‐symbiotic phase is characterized by a molecular dialog involving the release of diffusible signals (Lanfranco, Fiorilli, & Gutjahr, 2018; Lanfranco, Fiorilli, Venice, & Bonfante, 2018) that leads to the activation of the so‐called common symbiosis signaling pathway (MacLean et al., 2017). Upon reaching the roots epidermis, the fungus develops adhesion structures called hyphopodia that enable the fungus to penetrate host tissues and proliferate via intercellular and/or intracellular routes. The symbiotic phase culminates when the fungal hyphae penetrate single cells of the inner cortical layer and form highly branched, tree‐shaped structures, called arbuscules. Arbuscules are always enveloped by a plant‐derived periarbuscular membrane (PAM) that forms an extensive interface for nutrient exchange (Gutjahr & Parniske, 2013). The PAM is indeed populated by a unique set of proteins, such as Pht1 phosphate (Pi) transporters that are responsible for the uptake of Pi delivered by the fungus (Harrison et al., 2002; Yang et al., 2012).
Phytohormones and other signaling molecules have been shown to play a role mainly in the control of the extent of fungal colonization of the root system (Müller & Harrison, 2019). Strigolactones (SLs), a group of carotenoid‐derived hormones, are the best‐known molecules active in early plant–AM fungal interaction (Lanfranco, Fiorilli, & Gutjahr, 2018; Lanfranco, Fiorilli, Venice, & Bonfante, 2018). SLs are produced by roots of Pi‐starved plants and exported to the rhizosphere, where they stimulate AM fungal metabolism, gene expression and hyphal branching, enhancing the chances of the fungus intercepting host plants (Akiyama et al., 2005; Besserer et al., 2006, 2008). However, the dynamics of SL production and their role during the later steps of AM colonization remain elusive.
The involvement of carotenoid metabolism in AM symbiosis is not restricted to SLs and to the early steps of colonization. Indeed, several lines of evidence suggest the initiation and the development of AM symbiosis are influenced by other apocarotenoids (Fiorilli et al., 2019 and reference therein). Among them, the well‐characterized plant hormone abscisic acid (ABA; C15) plays key roles in plant response to abiotic stress (Felemban et al., 2019; Peleg & Blumwald, 2011), regulates plant growth and development, and promotes pathogen defense responses (Ma et al., 2018; Ton et al., 2009) and mycorrhizal colonization (Charpentier et al., 2014; Herrera‐Medina et al., 2007; Martín‐Rodríguez et al., 2011). The role of ABA in AM symbiosis remains enigmatic: Solanum lycopersicum (tomato) ABA mutants showed reduced levels of AM colonization compared with the wild type; however, in Medicago truncatula, ABA treatment promotes AM colonization at low concentrations (Charpentier et al., 2014; Herrera‐Medina et al., 2007; Martín‐Rodríguez et al., 2011). Other works have highlighted an antagonistic interaction between ABA and other hormones involved in AM symbiosis, such as ethylene (Martín‐Rodríguez et al., 2011) and gibberellins (GAs) (Floss et al., 2013; Martín‐Rodríguez et al., 2016).
In addition, other specific classes of apocarotenoids, such as mycorradicins (C14) and blumenols (C13), are nowadays considered a signature for the establishment of AM symbiosis, as they are specifically accumulated in mycorrhizal plants (Hill et al., 2018; Moreno et al., 2021; Walter et al., 2007; Wang et al., 2018).
The formation of most of the plant apocarotenoid hormones and signaling molecules involves carotenoid cleavage dioxygenases (CCDs), an evolutionarily conserved family of non‐heme Fe2+‐dependent enzymes (Giuliano et al., 2003; Hou et al., 2016; Moise et al., 2005; Wang et al., 2021). The recent characterization of a member of the overlooked sixth CCD subfamily led to the identification of zaxinone (3‐OH‐all‐trans‐apo‐13‐carotenone), an important growth‐regulating apocarotenoid metabolite in plants (Ablazov et al., 2020; Wang et al., 2019). The enzyme responsible for its biosynthesis in Oryza sativa (rice), zaxinone synthase (ZAS), has a wide distribution in the plant kingdom although a homolog gene is absent in the genomes of non‐AM host species, such as Arabidopsis thaliana (Wang et al., 2019). A rice mutant (zas), defective in OsZAS, showed lower zaxinone content and higher levels of SLs in roots, as well as severely retarded root and shoot growth. Exogenous application of zaxinone not only rescued the zas root phenotype but also promoted root growth in wild‐type plants and reduced SL biosynthesis and exudation under Pi‐limited and non‐mycorrhizal conditions (Wang et al., 2019). Despite the increased SL content the rice zas mutant displayed a reduced, by half, level of AM colonization, compared with wild‐type plants. However, the mechanisms leading to the impaired mycorrhization of the mutant line are not known.
The aim of this study was to understand the role of OsZAS and its product zaxinone in the regulation of AM symbiosis. It has been shown that zaxinone has no effect on Gigaspora margarita spore germination (Wang et al., 2020), suggesting that perturbation of the fungal asymbiotic phase is unlikely. We therefore hypothesized that zaxinone controls the rate of colonization success through interactions with SLs and other hormones. To address these issues we investigated the phytohormone contents of wild‐type and zas genotypes; we performed different exogenous treatments with the aim to restore the expected colonization level in the zas mutant. In addition, we analyzed OsZAS gene expression at cellular resolution and we characterized transgenic lines in which the expression of OsZAS is driven by a promoter active in arbusculated cells (OsPT11prom::OsZAS lines). Our findings highlight that the SL profiles of wild‐type and zas genotypes depend on the plant developmental stage as well as the AM colonization process. In this context we demonstrate that OsZAS plays a regulatory role in SL production, possibly through D14‐Like signaling, during the early colonization process and, when expressed under the OsPT11 promoter, promotes fungal intraradical development.
RESULTS AND DISCUSSION
The low colonization level of the zas mutant is rescued by SLs, but not by exogenous treatments with zaxinone
As the zas mutant displayed decreased mycorrhizal colonization (Wang et al., 2019), we tested whether this phenotype could be restored by an exogenous treatment with zaxinone. Therefore, we applied zaxinone at different concentrations (5, 0.5 and 0.05 μm) on 10‐day‐old wild‐type and zas mycorrhizal plants. Although the application of zaxinone successfully rescued most plant phenotypic defects in the mutant, i.e. crown root number, shoot length and biomass (Figure S1), it did not restore the expected AM colonization level, as shown by the quantitative reverse transcription polymerase chain reaction (qRT‐PCR) on plant AM marker genes OsPT11 (Figure 1a) and OsLysM, and the fungal 18S rRNA, and also by a morphological assessment (Figures S2, S3 and S4). In wild‐type plants the lowest zaxinone concentration (0.05 μm) had no impact on mycorrhization but the 0.5 or 5 μm concentrations strongly reduced the AM colonization (Figures 1a, S2, S3 and S4). We hypothesize that the reduced mycorrhization of wild‐type plants might be caused by the negative impact of exogenous zaxinone on SL biosynthesis (Wang et al., 2019) or to alterations to other plant hormones involved in AM symbiosis. As the terpenoid‐derived phytohormones ABA and GAs were shown to play a role in regulating the extent of the AM colonization (Liao et al., 2018; Pozo et al., 2015), we determined their profile in wild‐type and zas genotypes in non‐mycorrhizal and mycorrhizal conditions. In non‐mycorrhizal conditions, the zas mutant displayed a decrease in ABA level at 10 days post germination (10 dpg) and at 45 dpg, whereas we observed an increase in gibberellin (GA3, GA20, GA13 and GA29) content in at least one of the considered time points (Tables S1 and S2). In mycorrhizal plants, zas showed an increase in ABA and GA (particularly GA1 and GA20) content compared with the wild type (Tables S1 and S2). As it has been shown that biologically active GAs suppress arbuscule development and negatively affect the frequency of mycorrhization (Floss et al., 2013), we tested whether an increased level of GA could be responsible for the low level of mycorrhizal colonization in the zas mutant. We treated wild‐type and zas mycorrhizal plants with paclobutrazol (PAC), which reduces GA levels by inhibiting the ent‐kaurene oxidase/CYP701 (Rademacher, 2000). The effect of the PAC treatment was verified by the plant growth inhibition in both genotypes (Figure S5). In the zas mutants, PAC supply rescued neither the growth phenotype nor the mycorrhizal phenotype (Figures 1b, S5 and S6), indicating that the low level of mycorrhization of the mutant line was not cause by a perturbation in GA levels.
Figure 1.
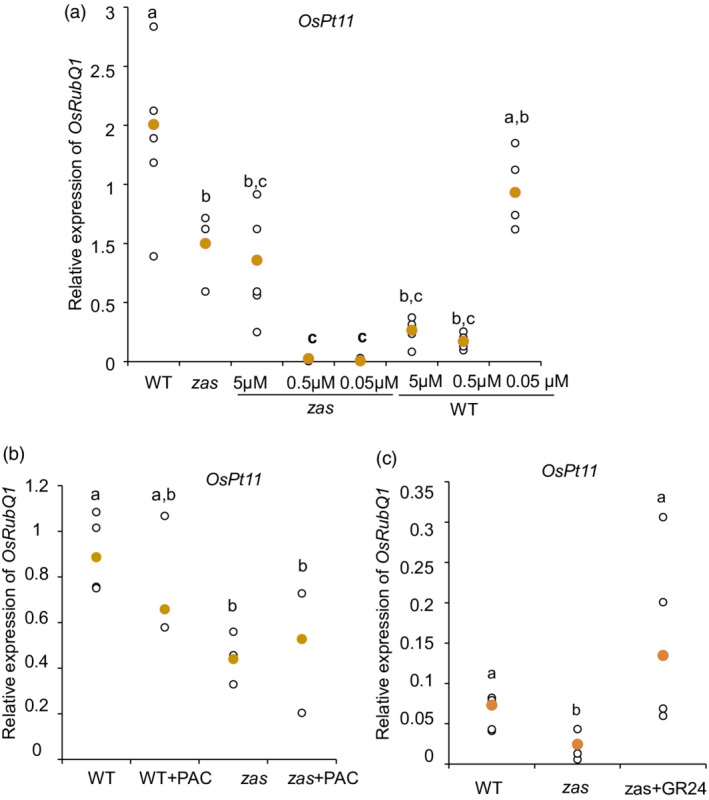
Mycorrhization level in wild‐type and zas mutant Oryza sativa (rice) plants grown under different treatments, evaluated on the abundance of phosphate transporter 11 gene (OsPT11) transcripts. (a) The relative expression levels of OsPT11 in mycorrhizal Nipponbare wild‐type (WT) and zas mutant (zas) roots, treated or not with different zaxinone concentrations (5, 0.5 and 0.05 μm). Mock treatment has the respective % of acetone used for 5‐μm treatments. (b) The relative expression levels of OsPT11 in mycorrhizal Nipponbare wild‐type (WT) and zas mutant (zas) roots, treated or not with paclobutrazol (PAC). (c) The relative expression level of OsPT11 in mycorrhizal roots of wild‐type (WT), zas and zas + GR24 plants. All plants were harvested at 35 days post inoculation (35 dpi) with Funneliformis mosseae. Zaxinone and PAC treatments were performed once a week directly in the nutrient solution, starting 10 days after mycorrhizal inoculation. GR24 (10 nm) treatment was applied once a week directly in the nutrient solution for the entire growing period. Ubiquitin was used as a reference gene. Individual data for each condition are shown as white dots and the median values are shown as yellow dots. For each experiment we considered at least n = 4 plants. Different letters represent statistically significant differences (P < 0.05, one‐way ANOVA; nsd, not statistically different). All experiments were repeated twice with equivalent results.
So far, the only well‐characterized hormones that promote the establishment of AM symbiosis are SLs, which are active at the early stage of AM interaction (Lanfranco, Fiorilli, Venice, & Bonfante, 2018).
We therefore treated the zas mutant with a racemic solution of GR24, an SL synthetic analog, and evaluated the AM colonization at 35 dpi. Notably, the GR24 treatment completely rescued the zas mutant mycorrhizal defect (Figures 1c and Figure S7), including the number of hyphopodia and arbuscules that were severely reduced in the untreated zas mutant compared with the wild type (Figures 1c and S7). These data suggest that the lower AM colonization rate of zas was linked to a deficiency in SLs at the early stage of the AM interaction. In addition, and notably, GR24 treatments did not rescue the zas root defects in non‐mycorrhizal (Figure S8) and mycorrhizal (Figure S9) conditions, which were by contrast rescued by the exogenous supply of zaxinone (Wang et al., 2019). These results indicate that the mycorrhizal and root defects of the zas mutant could be restored by distinct molecules: SLs and zaxinone, respectively, confirming the prominent role of zaxinone in controlling root development (Wang et al., 2019).
We therefore hypothesized a lower SL content in the zas mutant during the early stages of AM symbiosis. To investigate this hypothesis we quantified the 4DO content in both genotypes, along a time‐course experiment and during the colonization process. A different trend in 4DO content was observed in non‐mycorrhizal roots, with the 4DO content increasing along with the developmental stages (7, 21 and 35 dpi; Figure 2a). In mycorrhizal roots the highest 4DO content was observed at 7 dpi (Figure 2a), in agreement with the hypothesis that 4DO content facilitates host plant–AM fungus contact during the early stages (López‐Ráez et al., 2015), whereas at later stages (21 and 35 dpi) the 4DO content decreased, as previously observed in different plant species (Figure 2a; Lanfranco, Fiorilli, Venice, & Bonfante, 2018). Concerning the zas mutant, in non‐mycorrhizal roots the 4DO content increased over time and a higher 4DO content compared with the wild type was observed at 21 dpi, as described by Wang et al. (2019). Notably, this difference was not statistically significant at earlier (7 dpi) and later (35 dpi) developmental stages (Figure 2a). These data indicate that, under our growth conditions, the increase in SL content in the zas mutant varies depending on the developmental stage, which is a rather common phenomenon for plant hormones (Rizza & Jones, 2019). In mycorrhizal conditions the 4DO profile of the zas mutant was similar to that of wild‐type plants at 21 and 35 dpi, whereas at 7 dpi no 4DO increment was detected (Figure 2a), suggesting that OsZAS activity is involved in the increase in SLs required at this specific stage of the interaction. The different 4DO contents in wild‐type and zas mycorrhizal roots at 7 dpi was also supported by the upregulation of OsCCD8 (D10), a key SL biosynthetic gene, which was exclusively observed in wild‐type roots upon AM fungal inoculation (Figure S10). These results suggest the occurrence of a regulatory link between OsZAS function and SL production during the early colonization process. The increase of OsZAS mRNA abundance and zaxinone content observed at the early stage of AM colonization in wild‐type mycorrhizal plants (Wang et al., 2019) is also in line with this model.
Figure 2.
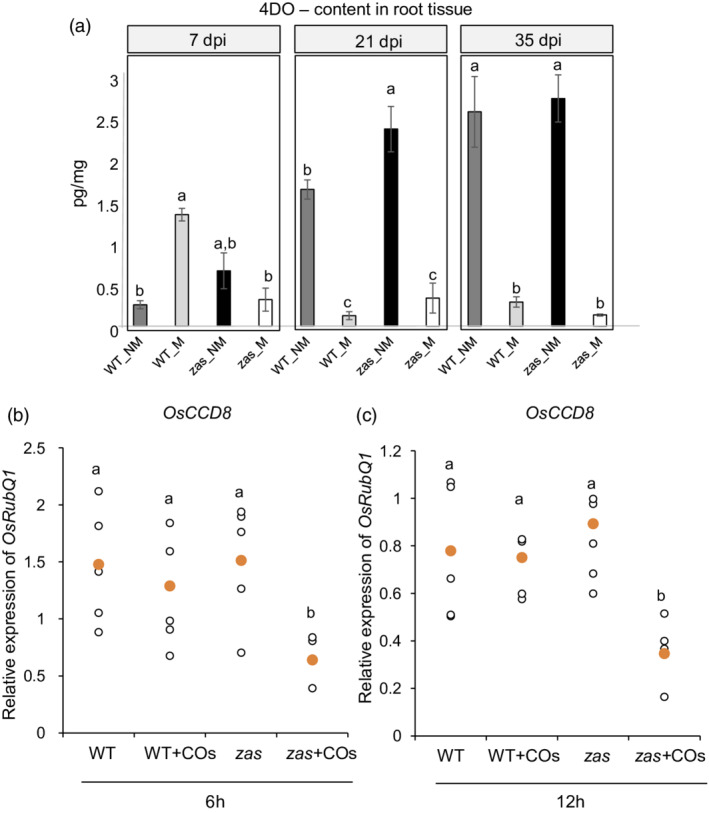
Strigolactone (SL) content and effect of treatment with short‐chain chito‐oligosaccharides (COs) on OsCCD8 in wild‐type and zas Oryza sativa (rice) plants. (a) SL, 4‐deoxyorobanchol (4DO), quantification in wild‐type and zas mutant roots in mycorrhizal and non‐mycorrhizal conditions. Roots from mycorrhizal (MYC) and non‐mycorrhizal (NM) plants were collected at 7, 21 and 35 days post fungal inoculation (dpi). Three independent biological replicates (each replicate is a pool of 40 plants) have been considered for the analysis. Different letters represent statistically significant differences within time points (P < 0.05, one‐way ANOVA). (b, c) The relative expression level of OsCCD8 in non‐mycorrhizal roots of wild‐type (WT) and zas plants treated (+COs) or not with COs. All plants were harvested at 7 days of growth: (b) after 6 hours post‐COs treatment (hpt); (c) and after 12 hpt. Ubiquitin was used as a reference gene (n = 5 plants). Different letters represent statistically significant differences (P < 0.05, one‐way ANOVA).
The increase of SLs induced by the presence of AM fungus seems, therefore, to be dependent on a fully functional OsZAS. To follow this hypothesis we investigated the impact of short‐chain chito‐oligosaccharides (COs), the early signaling molecules released by AM fungi that are known to trigger symbiotic responses in the host (Genre et al., 2013; Volpe et al., 2020), on SL biosynthetic gene expression in the wild type and the zas mutant. We monitored the expression of OsCCD8 (Figure 2b,c) and OsMAX1‐1400 at 6 h (Figure S10b) and 12 h after treatment with COs (hpt) (Figure S10). As previously reported in other host plants (Giovannetti et al., 2015), no differences were observed in wild‐type plants treated with COs, whereas, interestingly, both SL biosynthetic genes were downregulated in the zas mutant after the treatment with COs. This accords with a reduced accumulation of SLs in the zas mutant at 7 dpi compared with the wild type in mycorrhizal conditions (Figure 2a). Altogether, these data indicate that at the early stage of the AM interaction, OsZAS regulates SLs biosynthesis during the plant–fungus molecular dialog.
The cooperation between SLs and zaxinone biosynthesis during the mycorrhizal colonization of rice is also revealed by recent findings on the signaling pathway mediated by the α/β‐fold hydrolase Dwarf14‐Like (D14L) (Choi et al., 2020), which has been demonstrated to be indispensable for the establishment of AM symbiosis in rice (Gutjahr et al., 2015). It has been shown that D14L signaling positively regulates SL biosynthesis, and therefore AM symbiosis, by eliminating the negative regulator SMAX1 (Choi et al., 2020). Notably, the removal of SMAX1 leads to the upregulation not only of genes involved in SL biosynthesis (i.e. D10 and D17), but also several genes evolutionarily conserved in AM hosts, including OsZAS. Therefore, OsZAS transcription appears to depend on the activation of the D14L signaling pathway, which is also required to induce SL biosynthetic genes. To support the connection between OsZAS and D14L and SMAX1, we investigated the gene expression level of OsD14L and OsSMAX1 in wild‐type and zas genotypes and observed a downregulation of both genes in the mutant compared with wild‐type plants (Figure 3). Intriguingly, these data suggest that the low colonization level of zas could also be related to a downregulation of the D14L signaling pathway, which also negatively impacts SL biosynthesis.
Figure 3.
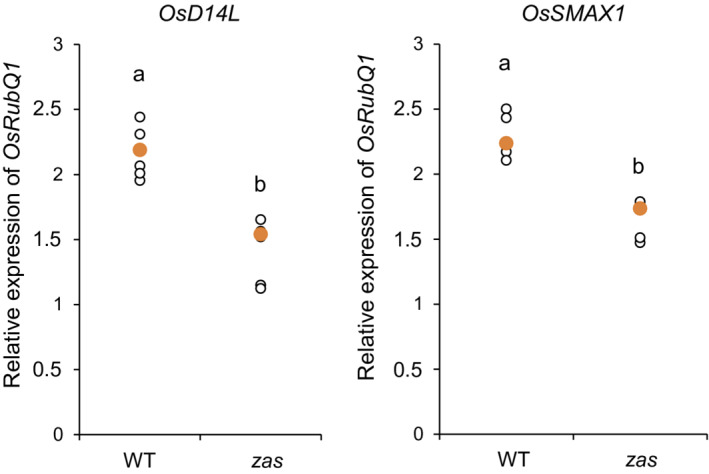
Expression levels of the Dwarf14‐Like (D14L) signaling pathway in wild‐type and zas mutant Oryza sativa (rice) plants. The relative expression levels of (a) D14L and (b) OsSMAX1 in non‐mycorrhizal Nipponbare wild‐type (WT) and zas mutant (zas) roots at 7 days post growth (dpg). Ubiquitin was used as a reference gene (n = 5 plants). Different letters represent statistically significant differences (P < 0.05, one‐way ANOVA).
Taken as a whole, our data indicate that OsZAS takes part in the mechanisms underpinning the early symbiotic programs that are instrumental in achieving normal mycorrhization levels, influencing both SLs and D14‐L signaling pathways.
OsZAS mRNA is localized in the arbusculated cells
We showed that OsZAS gene expression is induced in rice roots upon mycorrhizal colonization at 7 and 35 dpi (Wang et al., 2019). With the aim to gain data on OsZAS spatial expression, in situ hybridization assays were performed on roots of 35‐dpi plants, which correspond to mature mycorrhizas. OsZAS mRNA accumulated in cells with fully developed arbuscules where a strong chromogenic signal was observed (Figure 4a,b). By contrast, no signal was detected in non‐colonized cells from mycorrhizal roots (Figure 4a,b) or in sections from mycorrhizal roots hybridized with the OsZAS sense probe (Figure 4c,d). Although it was expected to detect OsZAS mRNA in non‐mycorrhizal roots, no hybridization signal was observed (Figure 4e,f). We hypothesize that in non‐mycorrhizal cortical cells the level of OsZAS mRNA is relatively low, making it undetectable by in situ hybridization; an alternative explanation is that OsZAS expression is associated with other parts of the root. OsZAS spatial expression in mycorrhizal roots is consistent with transcript accumulation observed in the late stages of mycorrhization (Wang et al., 2019) and suggests an involvement of OsZAS in the functioning of arbusculated cells.
Figure 4.
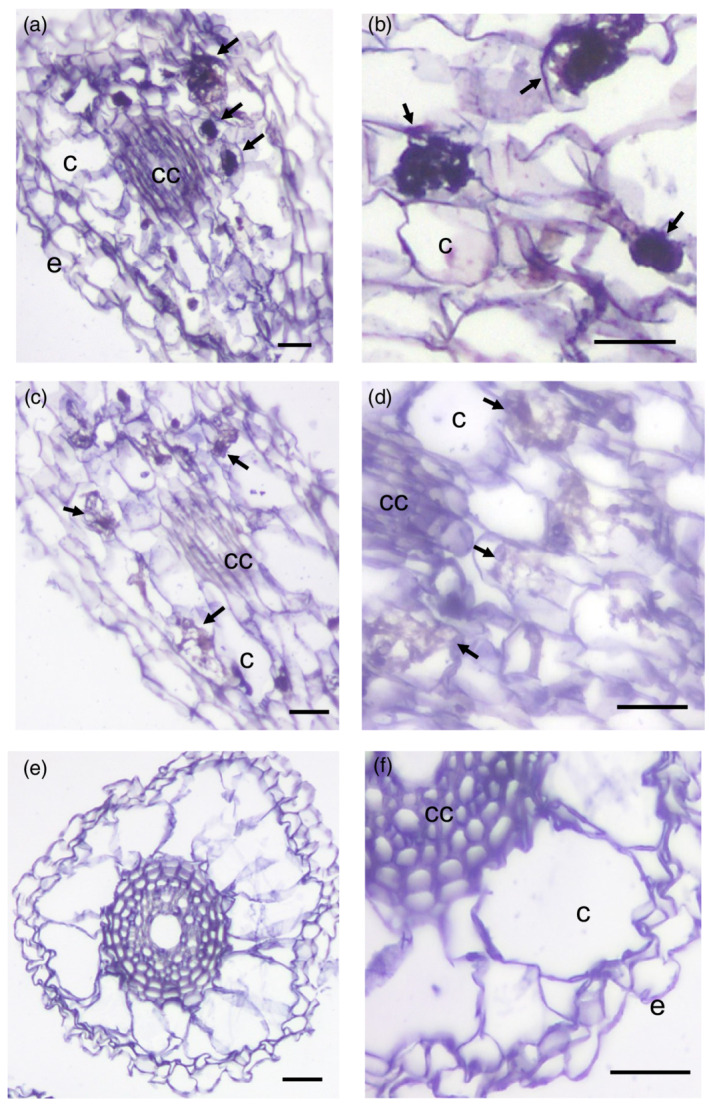
Spatial expression of OsZAS. Localization of OsZAS mRNA in sections from differentiated regions of inoculated (a–d) and non‐inoculated (e, f) roots of Oryza sativa (rice) by cold in situ hybridization. (a, b) Section of mycorrhizal roots treated with OsZAS antisense probe. A strong chromogenic signal, which mirrors the presence of the OsZAS transcripts, is visible in arbuscule‐containing cells (arrows). (c, d) Section of mycorrhizal roots treated with OsZAS sense probe, where a hybridization signal is not evident. Arrows indicate the arbuscule‐containing cells that are not labeled. (e, f). Only a very low background is present in uninoculated root segments. c, non‐colonized cortical cells; cc, central cylinder; e, epidermal cells. Scale bars: 50 μm.
OsPT11prom:: OsZAS lines show higher root colonization and normal arbuscule morphology
As in situ hybridization experiments revealed that OsZAS mRNA accumulates in arbusculated cells, we then investigated whether the OsZAS expression level has an impact on the intraradical phase and, in particular, on arbuscule formation and development. We thus generated rice transgenic lines, called OsPT11prom::OsZAS (Figure S11), where OsZAS expression is driven by the OsPT11 promoter that is active in arbuscule‐containing cells (Paszkowski et al., 2002). Two independent, hygromycin‐selected lines, PT11prom:OsZAS_6 (PT11p::zas6) and PT11prom:OsZAS_18 (PT11p::zas18), were identified by PCR (Figure S11). Both lines were then phenotyped in non‐mycorrhizal (Figure S12) and mycorrhizal conditions (Figure S13). The two lines under non‐mycorrhizal conditions showed an increased crown root length compared with the wild type (Figure S12). This phenotype was similar to that observed in wild‐type plants treated with exogenous zaxinone (Wang et al., 2019). As it has been shown that in the legumes Medicago truncatula and Lotus japonicus the OsPT11 homologs are also expressed in root tips when plants are grown under Pi‐limiting conditions (Volpe et al., 2016), we verified whether OsPT11 was also expressed in rice root apexes. Indeed, OsPT11 transcripts were detected in root tips of both the wild type and PT11p::zas6 (Figure S14); notably, we also found that also OsZAS is expressed in root apexes and, as expected, transcripts were more abundant in the transgenic line compared with the wild type (Figure S14).
These findings clearly show that in non‐mycorrhizal roots the OsPT11 promoter is active in root apices, and that the OsZAS gene is also expressed in this root tissue. This spatial expression, together with the fact that OsZAS expression is induced by Pi starvation (Wang et al., 2019), suggests that OsZAS may be involved in Pi sensing, as has been proposed for the OsPT11 homologs in legumes (Volpe et al., 2016). The recent discovery that promoters of both OsZAS and OsPT11 genes carry the conserved Pi starvation‐responsive motif P1BS, and are transactivated by the central regulator of Pi signaling, PHR2, has strengthened the idea that these genes have been co‐opted for the Pi sensing pathway and the establishment of AM symbiosis (Das et al., 2021; Shi et al., 2021).
Moreover, the enhanced growth observed in OsPT11prom::OsZAS non‐mycorrhizal roots could be the result of localized OsZAS upregulation. Indeed, a higher zaxinone content was detected in roots of the non‐mycorrhizal PT11p::zas6 line, whereas the PT11p::zas18 line showed a higher but not statistically different level compared with the wild type (Figure 5a). At the same growth stage non‐mycorrhizal wild‐type and OsPT11prom::OsZAS plants showed similar levels of SLs in the roots (Figure 5b). Remarkably, a higher trend of 4DO level was detected in root exudates of both transgenic lines compared with the wild type (Figure 5c).
Figure 5.
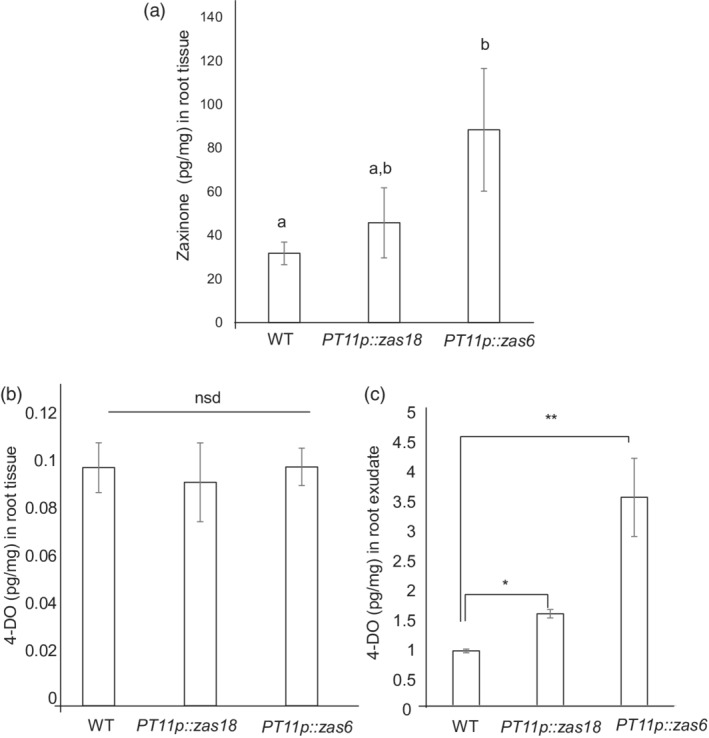
Zaxinone and 4‐deoxyorobanchol (4DO) quantification in OsPT11prom::OsZAS (PT11p::ZAS) lines of Oryza sativa (rice). Zaxinone (a) and 4DO content in root tissue (b) and in root exudate (c) were quantified in wild‐type (WT) and OsPT11prom::OsZAS (PT11p::ZAS) lines in non‐mycorrhizal conditions at 21 days post‐germination. Means and standard errors of four biological replicates are shown. Different letters indicate significant differences (*P < 0.05, **P < 0.01).
It has been shown that hyphopodium formation is severely attenuated in SL‐deficient d17 (CCD7) d10 (CCD8) rice double mutants, suggesting that a continuous requirement of SLs is essential for hyphopodia formation and the promotion of secondary infection (Kobae et al., 2018). In accordance with these data, OsPT11prom::OsZAS lines showed an increased AM colonization level in terms of mycorrhization frequency, number of arbuscules and number of hyphopodia, compared with the wild type (Figure 6a–c), whereas the arbuscule morphology was unaltered (Figure 6e). The morphological results were also confirmed by gene expression analyses in the plant OsPT11 (Figure 6d).
Figure 6.
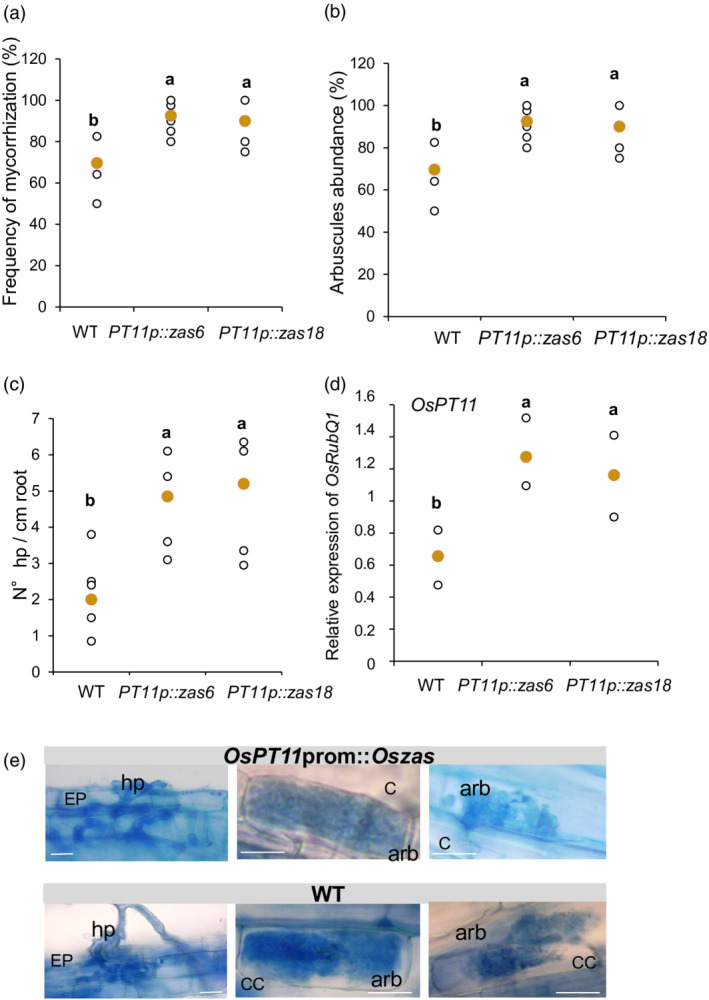
Molecular and phenotypic evaluation of mycorrhizal colonization in OsPT11prom::OsZAS lines of Oryza sativa (rice). After blue cotton staining: (a) the frequency of mycorrhizal (F%) colonization; (b) arbuscule abundance (A%); and (c) number of hyphopodia per cm of roots were evaluated in the wild‐type (WT) and OsPT11prom::OsZAS lines (PT11p::zas6; PT11p::zas18) (n = 5 plants). (d) The relative expression levels of OsPT11 in mycorrhizal wild‐type (WT) and OsPT11prom::OsZAS lines (PT11p::zas6; PT11p::zas18). Ubiquitin was used as a reference gene (n = 4 plants). (e) Root epidermal cells (EP) and cortical cells (CC) from wild‐type (WT) and OsPT11prom::OsZAS lines where hyphopodia (hp) and arbuscules (arb) are shown, respectively; the blue color indicates the cotton blue staining. Scale bars: 80 μm. All plants were harvested 21 days post inoculation with Funneliformis mosseae. Individual data for each condition are shown as white dots and median values are shown as yellow dots. Different letters represent statistically significant differences (P < 0.05, one‐way ANOVA).
The data obtained from OsPT11prom::OsZAS lines confirmed the role of OsZAS in promoting the AM colonization level, probably by inducing SL biosynthesis, which triggers hyphopodia formation that in turn promotes arbuscule formation; they also provide evidence that localized OsZAS overexpression in arbusculated cells does not have an impact on intracellular fungal morphology (Figure 6e).
CONCLUSION
Overall, the data we present here demonstrate the importance of OsZAS to guarantee the correct extent of AM root colonization. In the early stages of the AM interaction, OsZAS modulates AM colonization and exerts its function as a component of a regulatory network that involves SLs and D14L pathways. The overexpression of OsZAS in arbusculated cells (OsPT11prom::OsZAS lines) leads to increased mycorrhization, including an increased abundance of hyphopodia and arbuscules (Figure 7), that could be related to the higher content of SLs in the OsPT11prom::OsZAS root exudates.
Figure 7.
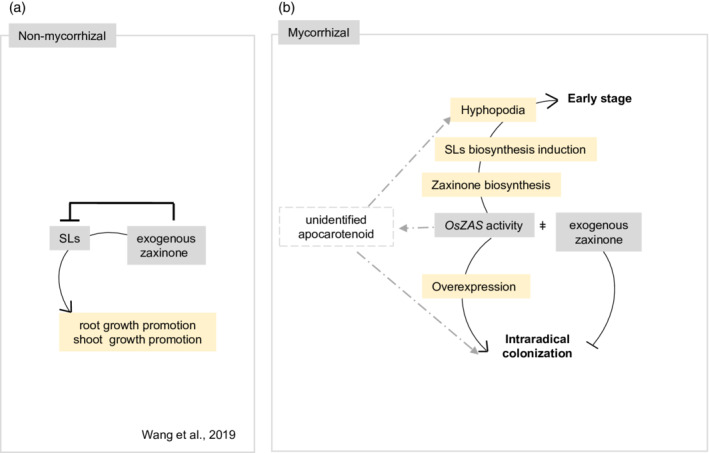
Schematic model for OsZAS and exogenous zaxinone regulation in wild‐type Oryza sativa (rice) plants grown in non‐mycorrhizal (a) and mycorrhizal (b) conditions. (a) In low‐Pi conditions, exogenous zaxinone treatment negatively regulates rice SL biosynthesis and release. The zaxinone root and shoot growth promotion requires functional strigolactone (SL) biosynthesis (Wang et al., 2019). (b) OsZAS activity increases zaxinone content and promotes the biosynthesis of SLs and hyphopodia formation in the early stage of mycorrhizal colonization. Overexpression of OsZAS under the OsPT11 promoter increases the intraradical colonization. By contrast, exogenous application of zaxinone negatively impact AM colonization. The discrepancy between the impacts on AM symbiosis of exogenous and endogenous (by OsZAS localized overexpression) accumulation of zaxinone suggests that, besides zaxinone, OsZAS can form in planta a yet unidentified apocarotenoid required for optimal mycorrhization (dashed arrow and lines). Positive and negative effects are illustrated by arrows and blunt‐ended bars, respectively.
Our results show that the genetic manipulation of OsZAS activity in planta leads to a different effect on the AM symbiosis from that of an exogenous zaxinone treatment. Although we found a clear positive correlation between the expression level of OsZAS and the extent of colonization, exogenous zaxinone repressed the AM symbiosis, probably through the strong negative impact of a continuous application of this compound on SL biosynthesis (Wang et al., 2019; Figure 7). This highlights that appropriate levels of this apocarotenoid are needed to assist root colonization by AM fungi and that OsZAS activity is involved in a complex network that could not be mimicked by an exogenous supply of its product. One could also speculate that, besides zaxinone, OsZAS can form in planta a yet unidentified apocarotenoid required for optimal mycorrhization.
EXPERIMENTAL PROCEDURES
Plant and fungal material
For all the experiments, seeds of the Nipponbare wild‐type cultivar, the mutants zas (Wang et al., 2019) and d17 (Butt et al., 2018), and two independent OsPT11prom::Oszas lines (see Supplemental methods) cv. Nipponbare were germinated as described by Fiorilli et al. (2015). Mycorrhizal plants were colonized with Funneliformis mosseae (BEG 12; MycAgroLab, http://www.mycagrolab.com) using a fungal inoculum mixed (25%) with sterile quartz sand. Plants were grown and watered as described by Vallino et al. (2014). Mycorrhizal roots were stained with cotton blue and the level of mycorrhizal colonization was assessed according to the method described by Trouvelot et al. (1986). Hyphopodia were counted manually in each root section.
In situ hybridization and detection
To generate the probe for in situ hybridization, a primer pair (Table S3) was used to amplify an Oszas sequence of 470 bp. The amplicon was cloned in the sense and antisense orientation into the pCR2.1‐TOPO (TA Cloning®; ThermoFisher Scientific, https://www.thermofisher.com) with respect to the T7 promoter. Digoxigenin‐labeled RNA probes were synthesized from PCR fragments with T7 or SP6 RNA polymerase, as described by Balestrini et al. (1997). Root segments of 1 cm in length were fixed in 4% paraformaldehyde in PBS overnight at 4°C. Samples were then dehydrated in an ethanol series, embedded in Paraplast Plus and sectioned to 8‐μm thickness using a rotary microtome. In situ hybridization and color development were performed as described by Balestrini et al. (1997) (see Supplemental methods). Sections were observed under a light microscope (Primo Star Zeiss; Carl Zeiss, https://www.zeiss.com) with a Leica DFC425 digital camera (Leica Microsystems, https://www.leica‐microsystems.com). The experiment was repeated twice with equivalent results.
Phytohormone quantification
For the quantification of targeted plant hormones and related compounds, 20‐mg (fresh weight) portions of separately harvested roots were frozen in liquid nitrogen. Concentration levels of endogenous phytohormones (ABA and GAs) were determined in four biological replicates according to the modified method described by Šimura et al. (2018) (see Supplemental methods).
To measure SLs and zaxinone contents, the protocol described by Wang et al. (2019) was followed for different stages of mycorrhizal symbiosis in a time‐course experiment: plants inoculated or not with F. mosseae were sampled at 7, 20 and 35 dpi. To measure SL content in root exudate of OsPT11prom::Oszas lines, the protocol described by Wang et al. (2019) was followed.
Plant treatments
For zaxinone treatment, a set of wild‐type and zas mycorrhizal plants were watered twice a week, once by applying 5, 0.5 or 0.05 μm of the compound in the nutrient solution, starting at 10 dpi to avoid a decrease of SL content during the early phase of AM symbiosis. For SL treatment, 10 nm of the SL analog GR24 (racemic solution) was applied once a week on non‐mycorrhizal and mycorrhizal wild‐type and zas plants. Both zaxinone and GR24 were dissolved in acetone. For treatment with paclobutrazol (PAC), an inhibitor of GA biosynthesis, 10 μm PAC was applied 10 days after AM fungal inoculation once a week for a total period of 4 weeks.
For the chitooligosaccharides (COs; CO4–CO5) treatment, rice seeds of wild‐type and zas mutant plants were germinated in pots containing sand and incubated for 10 days in a growth chamber under a 14‐h light (23°C)/10‐h dark (21°C) photoperiod. Seedlings were transferred to 5‐ml Eppendorf tubes (https://www.eppendorf.com) and were grown hydroponically in a modified Long Ashton (LA) solution containing 3.2 μm Na2HPO4·12H2O. A set of wild‐type (WT + CO) and zas (zas + CO) plants were treated with a concentration of 10−5 M (Carotenuto et al., 2017) of COs mix, previously withthe protocol from Crosino et al. (2021), for 6 and 12 h, and then roots were collected for gene expression analysis.
Nucleic acid extraction and cDNA synthesis
Total RNA was extracted from rice roots using the Plant RNeasy Kit (Qiagen, https://www.qiagen.com). Samples were treated with TURBO™ DNase (Ambion, now ThermoFisher Scientific, https://www.thermofisher.com). The RNA samples were routinely checked for DNA contamination using PCR analysis, using primers for OsRubQ1 (Güimil et al., 2005). For single‐strand cDNA synthesis, about 1000 ng of total RNA was reverse‐transcribed using Super‐Script II (Invitrogen, now ThermoFisher Scientific, https://www.thermofisher.com).
Real‐time quantitative RT‐PCR
Quantitative RT‐PCR (qRT‐PCR) was performed using a Rotor‐Gene Q 5plex HRM Platform (Qiagen). Each PCR reaction was carried out as described by Fiorilli et al. (2015). All reactions were performed on at least four biological and two technical replicates. The transcript levels of rice OsPT11 (Güimil et al., 2005), OsLysM (Fiorilli et al., 2015), OsCCD8 and OsMAX1 (Wang et al., 2019), and OsD14L (Gutjahr et al., 2015) and OsSMAX1 (Choi et al., 2020) and fungal housekeeping Fm18S (Balestrini et al., 2007) were normalized using the OsRubQ1 housekeeping gene (Table S3).
Statistics
Statistical tests were carried out through one‐way analysis of variance (one‐way ANOVA) and Tukey's post hoc test, using a probability level of P < 0.05. All statistical elaborations were performed using past 2.16 (Hammer et al., 2001).
AUTHOR CONTRIBUTIONS
VF, SA‐B, PB and LL designed the investigation. CV and VF performed the cellular and molecular experiments concerning mycorrhization. RB contributed with the in situ hybridization. JYW carried out the quantification of zaxinone and SLs. IH and AS generated the transgenic lines. IP, DT and ON conducted the quantification of hormones. All authors contributed to the results and discussion, and VF, SA‐B, PB and LL wrote the article.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest associated with this work.
Supporting information
Figure S1. Phenotypic evaluation of sand‐grown Oryza sativa (rice) wild‐type (WT) and zas mycorrhizal plants treated with 5 μm zaxinone.
Figure S2. Analysis of arbuscular mycorrhizal level in wild‐type (WT) and zas mutant plants under 5 μm zaxinone treatment.
Figure S3. Analysis of arbuscular mycorrhizal level in wild‐type (WT) and zas mutant plants under 0.5 μm zaxinone treatment.
Figure S4. Analysis of arbuscular mycorrhizal level in wild‐type (WT) and zas mutant plants under 0.05 μm zaxinone treatment.
Figure S5. Phenotypic evaluation of sand‐grown Oryza sativa (rice) wild‐type (WT) and zas plants treated or not with 10 μm paclobutrazol (PAC).
Figure S6. Analysis of arbuscular mycorrhizal level in wild‐type (WT) and zas mutant plants under 10 μm paclobutrazol (PAC) treatment.
Figure S7. Analysis of arbuscular mycorrhizal level in wild‐type (WT) and zas mutant plants under 10 nm GR24 treatment.
Figure S8. Effect of GR24 treatment on the shoot and root phenotypes of wild‐type (WT) and zas mutant plants grown in non‐mycorrhizal conditions.
Figure S9. Effect of GR24 treatment on the shoot and root phenotypes of wild‐type (WT) and zas mutant plants in mycorrhizal conditions.
Figure S10. Relative expression level of SL biosynthesis genes (OsCCD8 and OsMAX1) in wild‐type (WT) and zas mutant plants in the early stage of the AM interaction; the number of hyphopodia per cm of root evaluated in WT, zas and zas + GR24 plants at 35 dpi; and relative expression level of OsMAX1‐1400 in non‐mycorrhizal roots of wild type (WT) and zas mutant plants treated (+COs) or not with COs.
Figure S11. Molecular analysis of the OsPT11prom:OsZAS transgenic Oryza sativa (rice) lines.
Figure S12. Phenotypic evaluation of Oryza sativa (rice) wild‐type (WT) and OsPT11prom::OsZAS lines grown in sand in non‐mycorrhizal conditions.
Figure S13. Phenotypic evaluation of Oryza sativa (rice) wild‐type (WT) and OsPT11prom::OsZAS lines in mycorrhizal conditions grown in sand at 21 days post‐inoculation.
Figure S14. Gel electrophoresis of RT‐PCR products obtained from RNA of root apexes of non‐mycorrhizal wild‐type (WT) and OsPT11prom::OsZAS(Pt11_6) line samples using specific primers.
Table S1. ABA quantification (pmol/gFW) in wild‐type (WT) and zas non‐mycorrhizal and mycorrhizal roots in a time‐course experiment.
Table S2. Gibberellin quantification (pmol/gFW) in wild‐type (WT) and zas non‐mycorrhizal and mycorrhizal roots.
Table S3. Primer sequences used in this study.
Supplemental methods
ACKNOWLEDGEMENTS
This work was supported by baseline funding of the University of Turin (LL and VF) and the Competitive Research Grant (CRG2017) given to SA‐B and LL from King Abdullah University of Science and Technology, by the Ministry of Education, Youth and Sports of the Czech Republic (European Regional Development Fund‐Project ‘Centre for Experimental Plant Biology’ no. CZ.02.1.01/0.0/0.0/16_019/0000738), and by the Internal Grant Agency of Palacký University (IGA_PrF_2021_011) to ON. The authors thank Dr Mara Novero and Dr Mara Politi for their technical assistance and Prof. Andrea Genre for providing chito‐olisaccharides. Open Access Funding provided by Universita degli Studi di Torino within the CRUI‐CARE Agreement.
Contributor Information
Salim Al‐Babili, Email: salim.babili@kaust.edu.sa.
Luisa Lanfranco, Email: luisa.lanfranco@unito.it.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding authors, upon reasonable request.
REFERENCES
- Ablazov, A. , Mi, J. , Jamil, M. , Jia, K.P. , Wang, J.Y. , Feng, Q. et al. (2020) The apocarotenoid zaxinone is a positive regulator of strigolactone and abscisic acid biosynthesis in arabidopsis roots. Frontiers in Plant Science, 11, 578. 10.3389/fpls.2020.00578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama, K. , Matsuzaki, K. & Hayashi, H. (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature, 435, 824–827. [DOI] [PubMed] [Google Scholar]
- Balestrini, R. , Estanyol, M.J. , Puigdoménech, P. & Bonfante, P. (1997) Hydroxyproline‐rich glycoprotein mRNA accumulation in maize root cells colonized by an arbuscular mycorrhizal fungus as revealed by in situ hybridization. Protoplasma, 198, 36–42. [Google Scholar]
- Balestrini, R. , Gómez‐Ariza, J. , Lanfranco, L. & Bonfante, P. (2007) Laser microdissection reveals that transcripts for five plant and one fungal phosphate transporter genes are contemporaneously present in arbusculated cells. Molecular Plant‐Microbe Interactions, 20, 1055–1062. [DOI] [PubMed] [Google Scholar]
- Besserer, A. , Bécard, G. , Jauneau, A. , Roux, C. & Séjalon‐Delmas, N. (2008) GR24, a synthetic analogue of strigolactones, stimulates the mitosis and growth of the arbuscular mycorrhizal fungus Gigaspora rosea by boosting its energy metabolism. Plant Physiology, 148, 402–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besserer, A. , Puech‐Pagès, V. , Kiefer, P. , Gomez‐Roldan, V. , Jauneau, A. , Roy, S. et al. (2006) Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biology, 4, e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt, H. , Jamil, M. , Wang, J.Y. , Al‐Babili, S. & Mahfouz, M. (2018) Engineering plant architecture via CRISPR/Cas9‐mediated alteration of strigolactone biosynthesis. BMC Plant Biology, 18, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carotenuto, G. , Chabaud, M. , Miyata, K. , Capozzi, M. , Takeda, N. , Kaku, H. , et al. (2017) The rice LysM receptor-like kinasesOsCERK1 is required for the perception of short-chain chitin oligomers in arbuscularmycorrhizal signaling. New Phytologist, 214, 1440–1446. [DOI] [PubMed] [Google Scholar]
- Charpentier, M. , Sun, J. , Wen, J. , Mysore, K.S. & Oldroyd, G.E.D. (2014) Abscisic acid promotion of arbuscular mycorrhizal colonization requires a component of the PROTEIN PHOSPHATASE 2A complex. Plant Physiology, 166, 2077–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M. , Arato, M. , Borghi, L. , Nouri, E. & Reinhardt, D. (2018) Beneficial services of arbuscular mycorrhizal fungi – from ecology to application. Frontiers in Plant Science, 9, 1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J. , Lee, T. , Cho, J. , Servante, E.K. , Pucker, B. , Summers, W. et al. (2020) The negative regulator SMAX1 controls mycorrhizal symbiosis and strigolactone biosynthesis in rice. Nature Communications, 11, 2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosino, A. , Moscato, E. , Blangetti, M. , Carotenuto, G. , Spina, F. , Bordignon, S. et al. (2021) Extraction of short chain chitooligosaccharides from fungal biomass and their use as promoters of arbuscular mycorrhizal symbiosis. Scientific Reports, 11, 3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, D. , Paries, M. , Hobecker, K. , Gigl, H. , Dawid, C. , Lam, H.M. et al. (2021) Phosphate starvation response enables arbuscular mycorrhiza symbiosis. bioRxiv preprint 10.1101/2021.11.05.467437 [DOI] [PMC free article] [PubMed]
- Felemban, A. , Braguy, J. , Zurbriggen, M.D. & Al‐Babili, S. (2019) Apocarotenoids Involved in Plant Development and Stress Response. Frontiers in Plant Science, 10, 1168. 10.3389/fpls.2019.01168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorilli, V. , Vallino, M. , Biselli, C. , Faccio, A. , Bagnaresi, P. & Bonfante, P. (2015) Host and non‐host roots in rice: cellular and molecular approaches reveal differential responses to arbuscular mycorrhizal fungi. Frontiers in Plant Science, 6, 636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorilli, V. , Wang, J.Y. , Bonfante, P. , Lanfranco, L. & Al‐Babili, S. (2019) Apocarotenoids: old and new mediators of the arbuscular mycorrhizal symbiosis. Frontiers in Plant Science, 10, 1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floss, D.S. , Levy, J.G. , Levesque‐Tremblay, V. , Pumplin, N. & Harrison, M.J. (2013) DELLA proteins regulate arbuscule formation in arbuscular mycorrhizal symbiosis. Proceeding of the National Academy of Science U S A, 110, E5025–E5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre, A. , Chabaud, M. , Balzergue, C. , Puech‐Pagès, V. , Novero, M. , Rey, T. et al. (2013) Short‐chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytologist, 198, 190–202. [DOI] [PubMed] [Google Scholar]
- Genre, A. , Lanfranco, L. , Perotto, S. & Bonfante, P. (2020) Unique and common traits in mycorrhizal symbioses. Nature Review Microbiology, 18, 649–660. [DOI] [PubMed] [Google Scholar]
- Giovannetti, M. , Mari, A. , Novero, M. & Bonfante, P. (2015) Early Lotus japonicus root transcriptomic responses to symbiotic and pathogenic fungal exudates. Frontiers in Plant Science, 6, 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano, G. , Al‐Babili, S. & von Lintig, J. (2003) Carotenoid oxygenases: cleave it or leave it. Trends in Plant Science, 8, 145–149. [DOI] [PubMed] [Google Scholar]
- Güimil, S. , Chang, H.S. , Zhu, T. , Sesma, A. , Osbourn, A. , Roux, C. et al. (2005) Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proceeding of the National Academy of Science U S A, 102, 8066–8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr, C. , Gobbato, E. , Choi, J. , Riemann, M. , Johnston, M.G. , Summers, W. et al. (2015) Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science, 350, 1521–1524. [DOI] [PubMed] [Google Scholar]
- Gutjahr, C. & Parniske, M. (2013) Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annual Review of Cell and Developmental Biology, 29, 593–617. [DOI] [PubMed] [Google Scholar]
- Hammer, Ø. , Harper, D.A.T. & Ryan, P.D. (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica, 4, 9. [Google Scholar]
- Harrison, M.J. , Dewbre, G.R. & Liu, J. (2002) A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. The Plant Cell, 14, 2413–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera‐Medina, M.J. , Steinkellner, S. , Vierheilig, H. , Ocampo Bote, J.A. & García Garrido, J.M. (2007) Abscisic acid determines arbuscule development and functionality in the tomato arbuscular mycorrhiza. New Phytologist, 175, 554–564. [DOI] [PubMed] [Google Scholar]
- Hill, E.M. , Robinson, L.A. , Abdul‐Sada, A. , Vanbergen, A.J. , Hodge, A. & Hartley, S.E. (2018) Arbuscular mycorrhizal fungi and plant chemical defence: effects of colonisation on aboveground and belowground metabolomes. Journal of Chemical Ecology., 44, 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, X. , Rivers, J. , León, P. , McQuinn, R.P. & Pogson, B.J. (2016) Synthesis and function of apocarotenoid signals in plants. Trends in Plant Science, 21, 792–803. [DOI] [PubMed] [Google Scholar]
- Kobae, Y. , Kameoka, H. , Sugimura, Y. , Saito, K. , Ohtomo, R. , Fujiwara, T. et al. (2018) Strigolactone biosynthesis genes of rice are required for the punctual entry of arbuscular mycorrhizal fungi into the roots. Plant Cell Physiology, 59, 544–553. [DOI] [PubMed] [Google Scholar]
- Lanfranco, L. , Fiorilli, V. & Gutjahr, C. (2018) Partner communication and role of nutrients in the arbuscular mycorrhizal symbiosis. New Phytologist, 220, 1031–1046. [DOI] [PubMed] [Google Scholar]
- Lanfranco, L. , Fiorilli, V. , Venice, F. & Bonfante, P. (2018) Strigolactones cross the kingdoms: plants, fungi, and bacteria in the arbuscular mycorrhizal symbiosis. Jornal of Experimental Botany, 69, 2175–2188. [DOI] [PubMed] [Google Scholar]
- Liao, D. , Wang, S. , Cui, M. , Liu, J. , Chen, A. & Xu, G. (2018) Phytohormones regulate the development of arbuscular mycorrhizal symbiosis. International Journal of Molecular Sciences, 19, 3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Ráez, J.A. , Fernández, I. , García, J.M. , Berrio, E. , Bonfante, P. , Walter, M.H. et al. (2015) Differential spatio‐temporal expression of carotenoid cleavage dioxygenases regulates apocarotenoid fluxes during AM symbiosis. Plant Science, 230, 59–69. [DOI] [PubMed] [Google Scholar]
- Ma, Y. , Cao, J. , He, J. , Chen, Q. , Li, X. & Yang, Y. (2018) Molecular mechanism for the regulation of ABA homeostasis during plant development and stress responses. International Journal of Molecular Sciences, 19, 3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean, A.M. , Bravo, A. & Harrison, M.J. (2017) Plant signaling and metabolic pathways enabling arbuscular mycorrhizal symbiosis. The Plant Cell, 29, 2319–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín‐Rodríguez, J.A. , Huertas, R. , Ho‐Plágaro, T. , Ocampo, J.A. , Turečková, V. , Tarkowská, D. et al. (2016) Gibberellin–abscisic acid balances during arbuscular mycorrhiza formation in tomato. Frontiers of Plant Science, 7, 1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín‐Rodríguez, J.A. , León‐Morcillo, R. , Vierheilig, H. , Ocampo, J.A. , Ludwig‐Müller, J. & García‐Garrido, J.M. (2011) Ethylene‐dependent/ethylene‐independent ABA regulation of tomato plants colonized by arbuscular mycorrhiza fungi. New Phytologist, 190, 193–205. [DOI] [PubMed] [Google Scholar]
- Moise, A. , Vonlintig, J. & Palczewski, K. (2005) Related enzymes solve evolutionarily recurrent problems in the metabolism of carotenoids. Trends in Plant Science, 10, 178–186. [DOI] [PubMed] [Google Scholar]
- Moreno, J.C. , Mi, J. , Alagoz, Y. & Al‐Babili, S. (2021) Plant apocarotenoids: from retrograde signaling to interspecific communication. The Plant Journal, 105, 351–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, L.M. & Harrison, M.J. (2019) Phytohormones, miRNAs, and peptide signals integrate plant phosphorus status with arbuscular mycorrhizal symbiosis. Current Opinion in Plant Biology, 50, 132–139. [DOI] [PubMed] [Google Scholar]
- Paszkowski, U. , Kroken, S. , Roux, C. & Briggs, S.P. (2002) Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proceeding of the National Academy of Science U S A, 99, 13324–13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg, Z. & Blumwald, E. (2011) Hormone balance and abiotic stress tolerance in crop plants. Current Opinion in Plant Biology, 14, 290–295. [DOI] [PubMed] [Google Scholar]
- Pozo, M.J. , López‐Ráez, J.A. , Azcón‐Aguilar, C. & García‐Garrido, J.M. (2015) Phytohormones as integrators of environmental signals in the regulation of mycorrhizal symbioses. New Phytologist, 205, 1431–1436. [DOI] [PubMed] [Google Scholar]
- Rademacher, W. (2000) Growth retardants: effects on gibberellin biosynthesis and other metabolic pathways. Annual Review of Plant Physiology and Plant Molecular Biology, 51, 501–531. [DOI] [PubMed] [Google Scholar]
- Rich, M.K. , Nouri, E. , Courty, P.E. & Reinhardt, D. (2017) Diet of arbuscular mycorrhizal fungi: bread and butter? Trends in Plant Science, 22, 652–660. [DOI] [PubMed] [Google Scholar]
- Rillig, M. , Aguilar‐Trigueros, C.A. , Camenzind, T. , Cavagnaro, T.R. , Degrune, F. , Hohmann, P. et al. (2019) Why farmers should manage the arbuscular mycorrhizal symbiosis. New Phytologist, 222, 1171–1175. [DOI] [PubMed] [Google Scholar]
- Rizza, A. & Jones, A.M. (2019) The makings of a gradient: spatiotemporal distribution of gibberellins in plant development. Current Opinion in Plant Biology, 47, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J. , Zhao, B. , Zheng, S. , Zhang, X. , Wang, X. , Dong, W. et al. (2021) A phosphate starvation response‐centered network regulates mycorrhizal symbiosis. Cell 28, 184(22), 5527‐5540.e18. [DOI] [PubMed] [Google Scholar]
- Šimura, J. , Antoniadi, I. , Široká, J. , Tarkowská, D. , Strnad, M. , Ljung, K. et al. (2018) Plant hormonomics: multiple phytohormone profiling by targeted metabolomics. Plant Physiology, 177, 476–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatafora, J.W. , Chang, Y. , Benny, G.L. , Lazarus, K. , Smith, M.E. , Berbee, M.L. et al. (2016) A phylum‐level phylogenetic classification of zygomycete fungi based on genome‐scale data. Mycologia, 108, 1028–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton, J. , Flors, V. & Mauch‐Mani, B. (2009) The multifaceted role of ABA in disease resistance. Trends in Plant Science, 14, 310–317. [DOI] [PubMed] [Google Scholar]
- Trouvelot, A. , Kough, J. & Gianinazzi‐Pearson, V. (1986) Evaluation of VA infection levels in root systems. Research for estimation methods having a functional significance. In: Gianinazzi‐Pearson, V. & Gianinazzi, S. (Eds.) Physiological and Genetical Aspects of Mycorrhizae. France: INRAPress, pp. 217–221. [Google Scholar]
- Vallino, M. , Fiorilli, V. & Bonfante, P. (2014) Rice flooding negatively impacts root branching and arbuscular mycorrhizal colonization, but not fungal viability. Plant Cell Environment, 37, 557–572. [DOI] [PubMed] [Google Scholar]
- Volpe, V. , Giovannetti, M. , Sun, X.G. , Fiorilli, V. & Bonfante, P. (2016) The phosphate transporters LjPT4 and MtPT4 mediate early root responses to phosphate status in non mycorrhizal roots: Characterization of AM‐induced Pi transporters. Plant Cell Environment, 39, 660–671. [DOI] [PubMed] [Google Scholar]
- Volpe, V. , Carotenuto, G. , Berzero, C. , Cagnina, L. , Puech-Pagès, V. & Genre, A. (2020) Short chainchito-oligosaccharides promote arbuscular mycorrhizal colonization in Medicagotruncatula. Carbohydrate Polymers, 229. 115505 [DOI] [PubMed] [Google Scholar]
- Walter, M.H. , Floß, D.S. , Hans, J. , Fester, T. & Strack, D. (2007) Apocarotenoid biosynthesis in arbuscular mycorrhizal roots: contributions from methylerythritol phosphate pathway isogenes and tools for its manipulation. Phytochemistry, 68, 130–138. [DOI] [PubMed] [Google Scholar]
- Wang, J.Y. , Haider, I. , Jamil, M. , Fiorilli, V. , Saito, Y. , Mi, J. et al. (2019) The apocarotenoid metabolite zaxinone regulates growth and strigolactone biosynthesis in rice. Nature Communications, 10, 810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.Y. , Jamil, M. , Lin, P.Y. , Ota, T. , Fiorilli, V. , Novero, M. et al. (2020) Efficient mimics for elucidating zaxinone biology and promoting agricultural applications. Molecular Plant, 13, 1654–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.Y. , Lin, P.Y. & Al‐Babili, S. (2021) On the biosynthesis and evolution of apocarotenoid plant growth regulators. Seminars in Cell and Devlopmental Biology, 109, 3–11. [DOI] [PubMed] [Google Scholar]
- Wang, M. , Schäfer, M. , Li, D. , Halitschke, R. , Dong, C. , McGale, E. et al. (2018) Blumenols as shoot markers of root symbiosis with arbuscular mycorrhizal fungi. eLife, 7, e37093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S.Y. , Grønlund, M. , Jakobsen, I. , Grotemeyer, M.S. , Rentsch, D. , Miyao, A. et al. (2012) Non redundant regulation of rice arbuscular mycorrhizal symbiosis by two members of the PHOSPHATE TRANSPORTER1 gene family. The Plant Cell, 24, 4236–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Phenotypic evaluation of sand‐grown Oryza sativa (rice) wild‐type (WT) and zas mycorrhizal plants treated with 5 μm zaxinone.
Figure S2. Analysis of arbuscular mycorrhizal level in wild‐type (WT) and zas mutant plants under 5 μm zaxinone treatment.
Figure S3. Analysis of arbuscular mycorrhizal level in wild‐type (WT) and zas mutant plants under 0.5 μm zaxinone treatment.
Figure S4. Analysis of arbuscular mycorrhizal level in wild‐type (WT) and zas mutant plants under 0.05 μm zaxinone treatment.
Figure S5. Phenotypic evaluation of sand‐grown Oryza sativa (rice) wild‐type (WT) and zas plants treated or not with 10 μm paclobutrazol (PAC).
Figure S6. Analysis of arbuscular mycorrhizal level in wild‐type (WT) and zas mutant plants under 10 μm paclobutrazol (PAC) treatment.
Figure S7. Analysis of arbuscular mycorrhizal level in wild‐type (WT) and zas mutant plants under 10 nm GR24 treatment.
Figure S8. Effect of GR24 treatment on the shoot and root phenotypes of wild‐type (WT) and zas mutant plants grown in non‐mycorrhizal conditions.
Figure S9. Effect of GR24 treatment on the shoot and root phenotypes of wild‐type (WT) and zas mutant plants in mycorrhizal conditions.
Figure S10. Relative expression level of SL biosynthesis genes (OsCCD8 and OsMAX1) in wild‐type (WT) and zas mutant plants in the early stage of the AM interaction; the number of hyphopodia per cm of root evaluated in WT, zas and zas + GR24 plants at 35 dpi; and relative expression level of OsMAX1‐1400 in non‐mycorrhizal roots of wild type (WT) and zas mutant plants treated (+COs) or not with COs.
Figure S11. Molecular analysis of the OsPT11prom:OsZAS transgenic Oryza sativa (rice) lines.
Figure S12. Phenotypic evaluation of Oryza sativa (rice) wild‐type (WT) and OsPT11prom::OsZAS lines grown in sand in non‐mycorrhizal conditions.
Figure S13. Phenotypic evaluation of Oryza sativa (rice) wild‐type (WT) and OsPT11prom::OsZAS lines in mycorrhizal conditions grown in sand at 21 days post‐inoculation.
Figure S14. Gel electrophoresis of RT‐PCR products obtained from RNA of root apexes of non‐mycorrhizal wild‐type (WT) and OsPT11prom::OsZAS(Pt11_6) line samples using specific primers.
Table S1. ABA quantification (pmol/gFW) in wild‐type (WT) and zas non‐mycorrhizal and mycorrhizal roots in a time‐course experiment.
Table S2. Gibberellin quantification (pmol/gFW) in wild‐type (WT) and zas non‐mycorrhizal and mycorrhizal roots.
Table S3. Primer sequences used in this study.
Supplemental methods
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors, upon reasonable request.


