Abstract
We have characterized the effects of the T199S, T199A, and K70A mutations on the biochemical activity and in vivo functioning of Escherichia coli DnaK. Threonine-199 is the site of autophosphorylation of DnaK, and the lysine residue of bovine Hsc70 corresponding to K70 of DnaK has been shown to be essential for the hydrolysis of ATP. The dnaK alleles T199A and K70A are completely unable, and the T199S allele is only partially able, to complement the defects of a ΔdnaK mutant. The ATPase activities of the DnaK T199A and DnaK K70A proteins are nearly abolished, while the ATPase activity of the DnaK T199S protein has a steady-state rate similar to that of wild-type DnaK. The DnaK T199S protein also retains approximately 13% of the autophosphorylation activity of wild-type DnaK, while the autophosphorylation activities of the T199A and K70A derivatives are completely abolished. All four DnaK proteins bind a model peptide substrate, and the wild-type, T199A, and T199S DnaK proteins release the peptide with similar kinetics upon the addition of ATP. The DnaK K70A protein, in contrast, does not release the peptide upon the addition of ATP. ATP induces a conformational change in the wild-type, T199A, and T199S DnaK proteins but not in the DnaK K70A protein. The T199A and K70A mutations both disrupt the ATPase activity of DnaK but have profoundly different effects on the ATP-induced conformational change and peptide release activities of DnaK, implying that the two mutations affect different steps in the functional cycle of DnaK. The DnaK T199S protein represents a new class of DnaK mutant, one which has near-normal levels of ATPase activity and undergoes an ATP-induced conformational change that results in the release of peptide but which is not able to fully complement loss of DnaK function in the cell.
Hsp70 proteins are a highly conserved family of molecular chaperones, proteins that facilitate the folding of other proteins. Hsp70s are found in all prokaryotic cells and in most compartments of all eukaryotic cells (12). Escherichia coli has one primary member of the Hsp70 family, the DnaK protein. E. coli, like all organisms, responds to sudden increases in temperature by transiently increasing the level of expression of a group of proteins called heat shock proteins. Over 30 genes, including the dnaK gene, belong to the heat shock regulon of E. coli, which consists of genes that are transcriptionally activated by the heat shock factor ς32 (11). The processes in which DnaK functions include the folding of nascent polypeptides (21), protein degradation (40), and disassembly of protein complexes (42). As a molecular chaperone, DnaK binds polypeptides in unstable or unfolded states, protects them from aggregation and denaturation, and helps them along their folding pathway (5, 6).
All Hsp70 proteins have a weak ATPase activity that is functionally linked to the cycles of peptide binding and release that characterize the chaperone activity (25). The ATP binding and hydrolysis activity is associated with the amino-terminal 44-kDa tryptic fragment of the protein (14). This region is extremely conserved at the level of amino acid sequence from E. coli DnaK to human Hsp70. The tertiary structures of this domain are highly conserved as well, and the crystallographic structure of the amino-terminal ATPase domain of E. coli DnaK (20) is nearly identical to those of bovine Hsc70 (14) and human Hsp70 (39). The biochemical mechanisms involved in ATP binding and hydrolysis are believed to be very similar, if not identical, for all Hsp70 proteins (15).
Hsp70 proteins also have a weak, calcium-dependent autophosphorylation activity (44). The phosphorylated residue of E. coli DnaK is the threonine at position 199 (28), which is a completely conserved residue among Hsp70s and corresponds to residue threonine-204 of Hsc70. Replacement of threonine-199 of DnaK with alanine, valine, or aspartic acid results in a protein with no autophosphorylation activity and a greatly reduced ATPase activity (28). These DnaK mutants also fail to complement the loss of DnaK function in ΔdnaK cells (29). Structural studies of the 44-kDa amino-terminal fragment of Hsc70 showed that this conserved threonine residue is not essential for ATP hydrolysis (31). Lysine-71 of bovine Hsc70, on the other hand, was found to be an essential residue for the chemical hydrolysis of ATP by the 44-kDa amino-terminal fragment (30). This residue is also completely conserved in the Hsp70 family and corresponds to lysine-70 of DnaK. This lysine residue is the only residue to date to be identified as essential for ATP hydrolysis. ATP does not induce a conformational change in the 60-kDa amino-terminal fragment of bovine Hsc70 in which lysine-71 has been mutated to methionine (22) and does not induce the release of bound peptide by human cytosolic Hsp70 in which lysine-71 has been mutated to glutamate (35).
In this study, we carried out biochemical and functional characterizations of DnaK and DnaK derivatives with substitutions of residues threonine-199 and lysine-70. Based on our previous findings that nucleoside diphosphate kinase (NDP kinase) is present at very low levels in DnaK preparations and that its presence can result in inaccurate kinetic measurements of the DnaK ATPase activity (2), we made certain that our preparations were as free from NDP kinase as possible by purifying DnaK from ndk::km cells and using an extended purification protocol. We constructed and characterized a new DnaK derivative in which threonine-199 is replaced by serine in order to examine what effect a conservative substitution of this residue has on DnaK function. We also constructed and characterized a new DnaK derivative in which lysine-70 is replaced by alanine. We found that this K70A mutation results in a DnaK protein that has a defective ATPase activity and does not release peptide or undergo a conformational change upon the binding of ATP.
MATERIALS AND METHODS
Reagents and media.
N-{[2-(Iodoacetoxy)ethyl]-N-methyl}amino-7-nitrobenz-2-oxa-1,3-diazole (I-ANBD) was purchased from Molecular Probes (Eugene, Oreg.). ATP was purchased from Sigma Chemical Co. (St. Louis, Mo.). [α-32P]ATP and [γ-32P]ATP were purchased from DuPont NEN Life Science Products, Inc. (Boston, Mass.). Adenylyl-imidodiphosphate (AMP-PNP) was purchased from Boehringer Mannheim. Luria-Bertani (LB) liquid medium was as described previously (37). Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; tetracycline, 12.5 μg/ml; chloramphenicol, 30 μg/ml; and kanamycin, 50 μg/ml.
Strains and plasmids.
The E. coli strains and plasmids used in this work are listed with their relevant features in Table 1. Plasmids were transformed by the standard CaCl2-heat shock procedure (37). The E. coli strains NA7623 and JC7623 were generously provided to us by the laboratory of Masayori Inouye. Strain NA7623 has the gene encoding NDP kinase (ndk) disrupted and is otherwise isogenic to JC7623. The ndk gene of NA7623 is disrupted by a kanamycin resistance gene which is inserted into its EcoRI site as described previously (26). The ΔdnaK52 allele was transduced into both NA7623 and JC7623 by using P1(GW8306) lysate as described previously (29). The resulting strains were TB3200 (ndk::km ΔdnaK52) and TB3500 (ndk+ ΔdnaK52) and were transformed with the DnaK overexpression plasmid pJM6 (28) to create strains TB3220 (JC7623 ndk::km ΔdnaK52/pJM6) and TB3520 (JC7623 ndk+ ΔdnaK52/pJM6).
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| Strains | ||
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 deoC1 ptsF25 rpsR flbB301 | 10 |
| BB1553 | MC4100 ΔdnaK52 sidB1 | 8 |
| GW8301 | MC4100/pJM100 | 29 |
| GW8302 | BB1553/pJM100 | 29 |
| GW8303 | GW8301/pBR322 | 29 |
| GW8305 | GW8301/pJM41 | 29 |
| GW8306 | GW8302/pJM41 | 29 |
| GW8309 | GW8301/pJM43 | 29 |
| TB1009 | GW8301/pTB204 | This work |
| TB1031 | GW8301/pTB107 | This work |
| JC7623 | recB recC sbcB | 26 |
| NA7623 | JC7623 ndk::km | 26 |
| TB3200 | NA7623 ΔdnaK52 | This work |
| TB3500 | JC7623 ΔdnaK52 | This work |
| TB3220 | TB3200/pJM6 | This work |
| TB3520 | TB3500/pJM6 | This work |
| TB3221 | TB3200/pTB127 | This work |
| TB3222 | TB3200/pJM8 | This work |
| TB3223 | TB3200/pTB124 | This work |
| Plasmids | ||
| pBS | Stratagene | |
| pBR322 | New England Biolabs | |
| pJM100 | pACYC177 KmrlacIq | 29 |
| pJM41 | pBR322-PlacdnaK+dnaJ+ | 29 |
| pJM43 | pBR322-PlacdnaK(T199A) dnaJ+ | 29 |
| pTB204 | pBR322-PlacdnaK(T199S) dnaJ+ | This work |
| pTB107 | pBR322-PlacdnaK(K70A) dnaJ+ | This work |
| pJM6 | pBS-PlacdnaK+ | 28 |
| pJM8 | pBS-PlacdnaK(T199A) | 28 |
| pTB124 | pBS-PlacdnaK(T199S) | This work |
| pTB127 | pBS-PlacdnaK(K70A) | This work |
Plasmid constructions.
Mutagenesis was performed using the Sculptor in vitro mutagenesis system (Amersham) according to the standard protocol. All other DNA manipulations were performed as described previously (37). The plasmid pJM11 (29), which is a pBS derivative with the dnaK dnaJ operon under Plac control and a BamHI site introduced 230 bp upstream from the start codon, was used as a parent plasmid for all constructions. Site-directed mutagenesis was used to change pJM11 (pBS-PlacdnaK+ dnaJ+) to pTB104 [pBS-PlacdnaK(T199S) dnaJ+] and pTB107[pBS-PlacdnaK(K70A) dnaJ+]. HindIII digestion followed by religation removed the dnaJ gene from these plasmids to create pTB124 [pBS-PlacdnaK(T199S)] and pTB127 [pBS-PlacdnaK(K70A)]. The PlacdnaK(T199S) dnaJ+ BamHI fragment of plasmid pTB104 was subcloned into the pBR322 BamHI site to create plasmid pTB204 [pBR322-PlacdnaK(T199S) dnaJ+].
DnaK purification.
Strains TB3220, TB3221, TB3222, and TB3223 were used to produce DnaK+, DnaK K70A, DnaK T199A, and DnaK T199S, respectively, in an ndk::km background, and DnaK and DnaK mutant proteins were purified as previously described (2). Nucleotide was removed from DnaK, and the protein concentration was determined as previously described (2).
Synthesis of PepH-ANBD.
PepH (NRLLLCG) was synthesized by the Massachusetts Institute of Technology Biopolymers Laboratory using a peptide synthesizer (Applied Biosystems 430A). A 5.0-mg amount of PepH (6.3 μmol) in 0.5 ml of 5.0 mM Tris buffer (pH 8.0) was combined with 2.5 mg of I-ANBD (6.3 μmol) in 0.5 ml of acetonitrile and incubated at 37°C for 1 h. The modified PepH (PepH-ANBD) was purified by high-pressure liquid chromatography with a Waters Delta-pak C18 cartridge column. The column was run at 2 ml/min, and a linear gradient of 0 to 80% acetonitrile in 0.1% trifluoroacetic acid was run over 60 min. PepH-ANBD eluted from the column 6 min after unmodified PepH. The PepH-ANBD peak was collected and lyophilized.
ATPase assays.
Reaction mixtures (100 μl) contained modified ATPase buffer (50 mM HEPES-KOH [pH 7.6], 40 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 10% glycerol, 0.2 mg of ovalbumin per ml), [α-32P]ATP, and DnaK. Radiolabeled ATP stocks were made by adding radiolabeled ATP to unlabeled ATP to give a final activity of 82.6 μCi/ml. The ATP and ATPase buffer were mixed and preincubated at 30°C for 5 min, and hydrolysis was initiated by the addition of DnaK (t = 0). For each DnaK sample, a range of ATP concentrations of 6.35 to 510 nM was used. DnaK was present in each reaction mixture at a concentration of 7.24 nM. The reaction mixture was incubated in a water bath at 30°C, and the reaction was stopped at 5, 10, and 20 min by spotting 2 μl of the reaction mixture onto a polyethyleneimine-cellulose thin-layer chromatography (TLC) plate (J. T. Baker Inc., Phillipsburg, N.J.). Spotting the mixture on the TLC plate stopped the reaction immediately, as reactions quenched with 1 N HCl prior to spotting and reaction mixtures spotted without chemical quenching show the same extent of hydrolysis. The TLC plate was developed in 1 M formic acid–0.5 M LiCl, dried, and exposed to a Molecular Dynamics Storage Phosphor Screen. Data were obtained using a Molecular Dynamics PhosphorImager 445 Si and analyzed with ImageQuant 5.0. The amount of each radiolabeled species present was determined by volume integration. The data were corrected for the level of background hydrolysis (typically less than 1%). The velocity of the reaction was determined by multiplying the value for ADP/(ATP + ADP) by the starting amount of ATP in the reaction mixture and dividing by time.
Autophosphorylation assays.
Reaction mixtures (25 μl) contained buffer (40 mM HEPES-KOH [pH 7.6], 50 mM KCl, 10 mM CaCl2), [γ-32P]ATP (8.0 μM; 0.476 μCi/μl), and DnaK (0.929 μM). The reaction mixture was incubated at 37°C for 30 min. Protein was precipitated with 20% trichloroacetic acid–10 mM sodium pyrophosphate and washed with 20% trichloroacetic acid followed by acetone. The samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the gel was exposed to a Molecular Dynamics Storage Phosphor Screen. Data were obtained using a Molecular Dynamics PhosphorImager 445 Si and analyzed with ImageQuant 5.0. The amount of radiolabel in each DnaK band (if any) was determined by volume integration.
λ sensitivity assays.
Strains GW8305, GW8309, TB1009, and TB1031 (Table 1) were transduced with the ΔdnaK52 allele as described previously (29). Overnight cultures of each of these strains were grown at 30°C in LB medium with ampicillin, kanamycin, and chloramphenicol and subcultured (1:20) into LB medium with ampicillin, kanamycin, and chloramphenicol plus 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), 0.2% maltose, and 8 mM MgSO4. Following a 2-h incubation at 30°C, 100 μl of each incubation mixture was mixed with 3 ml of λ top agar with 1 mM IPTG, 0.2% maltose, and 8 mM MgSO4. The top agar was poured onto LB agar plates and allowed to solidify. Serial dilutions of λcI− phage were spotted (2 μl each) onto the plates. The plates were incubated at 30°C overnight and inspected for clearing of cells on the bacterial lawn.
Fluorescence measurements.
Fluorescence measurements were performed using a FloroMax-2 spectrofluorimeter (ISA Jobin Yvon-Spex Instruments, S.A., Inc., Edison, N.J.). For PepH-ANBD binding studies, the excitation wavelength was set at 480 nm. For DnaK conformational analysis, the excitation wavelength was set at 295 nm. For both sets of experiments, the slit widths were set at 5 nm for excitation and 7 nm for emission. All experiments were carried out in ATPase buffer (40 mM HEPES-KOH [pH 7.6], 50 mM KCl, 11 mM magnesium acetate), and all measurements were taken at 22°C.
Peptide release kinetics.
The kinetics of PepH-ANBD release from DnaK were measured on a Applied Photosystems stopped-flow spectrofluorimeter with a dead time of 1 ms. Reactions were initiated by mixing equal volumes (≈70 μl). The excitation wavelength was set at 480 nm, and a 495-nm-cutoff filter was used. The slit widths were set at 0.5 mm for both excitation and emission. All measurements were taken at 22°C.
RESULTS
DnaK T199A, T199S, and K70A derivatives are unable to perform DnaK function in vivo.
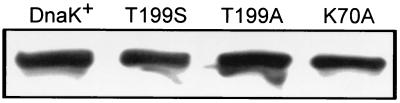
We constructed T199A, T199S, and K70A mutations of E. coli DnaK and measured the ability of each mutant to perform DnaK function in vivo. Cells containing a deletion of the dnaK gene display a number of aberrant phenotypes, including insensitivity to λ phage, inability to grow at temperatures of 42°C or higher, and extensive filamentation of the cells (7, 32). GW8301 cells were transformed with pBR322 plasmids carrying the dnaK or mutant dnaK gene under Plac control. The ΔdnaK52 allele was transduced into each of these strains, and the strains were grown in the presence of 0.5 mM IPTG to induce the expression of DnaK or the DnaK derivative. In order to ensure that the mutant derivatives of DnaK are present in cells at levels similar to that of wild-type DnaK, the amount of each DnaK derivative was visualized by Western blot analysis. Figure 1 shows that the steady-state amount of each DnaK derivative is approximately equal to the amount of wild-type DnaK in these cells. As summarized in Table 2, the previously described DnaK T199A (29) and DnaK K70A fail to provide DnaK function to the ΔdnaK52 mutant, while DnaK T199S only partially complements the loss of DnaK. Expression of DnaK+ complements the filamentation defect of ΔdnaK52 cells, while DnaK T199S, DnaK T199A, and DnaK K70A fail to complement the defect. DnaK T199S is able to partially complement the λ insensitivity and temperature sensitivity defects of ΔdnaK cells (Table 2). As judged by the number of PFU of λcI− phage required to cause detectable clearing of cells on a bacterial lawn, DnaK T199S-expressing cells are 100 times less sensitive to λ phage than DnaK+-expressing cells. Cells expressing DnaK T199S also show weak growth at 42°C. The DnaK T199A- and DnaK K70A-expressing ΔdnaK cells, by contrast, show no λ sensitivity or growth at 42°C.
FIG. 1.
Steady-state amount of DnaK protein in E. coli cells expressing dnaK mutations in a ΔdnaK52 background. The ΔdnaK52 allele was transduced into strains GW8305, GW8309, TB1009, and TB1031. LB broth (5 ml) containing 0.5 mM IPTG, ampicillin, chloramphenicol, and kanamycin was inoculated with single-colony picks from these transductions and incubated at 30°C until cells reached an A600 of 0.6. Cells were lysed by boiling in SDS-PAGE sample buffer, and equal amounts of protein from each cell lysate were loaded for SDS-PAGE. DnaK was detected by Western blot analysis using a polyclonal anti-DnaK antiserum and Western Light chemiluminescent detection (Tropix).
TABLE 2.
Phenotypes of E. coli cells expressing dnaK mutations in a ΔdnaK52 backgrounda
| DnaK | λ sensitivityb | Growth at 42°Cc | Cell morphologyd |
|---|---|---|---|
| Wild type | 106 | +++ | Unfilamented |
| T199S | 108 | + | Filamented |
| T199A | None | None | Filamented |
| K70A | None | None | Filamented |
The ΔdnaK52 allele was transduced into strains GW8305, GW8309, TB1009, and TB1031.
λ sensitivity assays were performed as described in the text, and λ sensitivity is expressed as the number of PFU of λcI− phage required to show any visible clearing of cells when spotted in a 2-μl drop on a growing lawn of cells.
LB broth (5 ml) containing 0.5 mM IPTG, ampicillin, chloramphenicol, and kanamycin was inoculated with single-colony picks from the transductions and incubated at 42°C. Following a 16-h incubation, the extent of growth in each culture was scored from no growth (0) to confluent (+++).
For cell morphology observations, strains expressing wild-type or mutant DnaK in a ΔdnaK background were grown at 30°C on LB plates containing 0.5 mM IPTG, ampicillin, chloramphenicol, and kanamycin. Multiple colonies were picked for each strain and observed microscopically. Microscopic analyses were done on a Zeiss Axioplan Universal microscope with Nomarski optics.
ATPase and autophosphorylation activities of DnaK mutants.
The DnaK proteins used in the following biochemical assays were all purified by an extended protocol used to ensure that all DnaK proteins were as free of NDP kinase activity as possible (2). All DnaK preparations used in the biochemical assays had an ADP kinase activity of less than 0.025 pmol of ADP phosphorylated per min per pmol of DnaK.
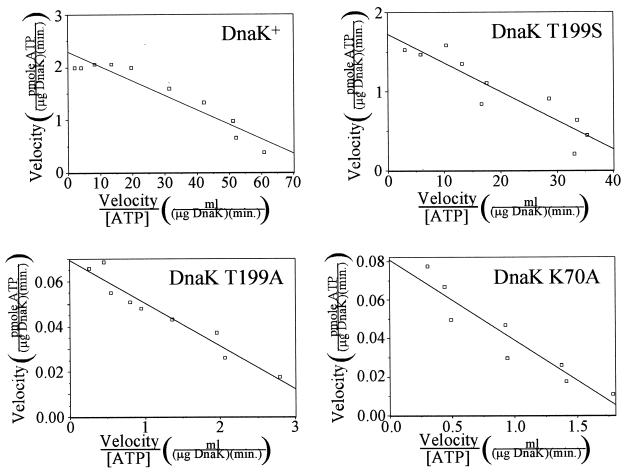
ATPase assays were performed with wild-type DnaK as well as with derivatives in which threonine-199 replaced with alanine (T199A) or serine (T199S) or in which lysine-70 was replaced with alanine (K70A). A low concentration of DnaK (7.24 nM) was used in the assays to avoid the phenomenon of mutual depletion kinetics (9, 19), which would result in the determination of erroneously high Km values. For each reaction, the kinetics of ADP formation were monitored and were shown to be linear and without initial burst activity (2). The DnaK was incubated with ATP at concentrations ranging from 6.35 to 510 nM, the velocity of ATP hydrolysis was determined for each reaction, and Eadie-Hofstee plots were made of the data (Fig. 2). Table 3 shows the values for Vmax, kcat, and Km for the ATPase activities of DnaK and the T199S, T199A, and K70A derivatives determined from the Eadie-Hofstee plots. The amount of calcium-dependent autophosphorylation was also determined for each protein and expressed as a fraction of the amount of autophosphorylation that occurs in wild-type DnaK. The turnover rate determined for wild-type DnaK (0.158 min−1) is similar to some recently reported values (13, 38) and a fewfold higher than some other recently reported values (9, 23, 24, 27). The Km value for wild-type DnaK (27.5 nM) correlates well with those determined in other studies in which mutual depletion kinetics were avoided (9). The Km values of the three mutants are all within twofold of the value for wild-type DnaK. However, the turnover rate for the T199A mutant is only 3% of that of the wild type and is similar to the previously reported value (28). Mutation of lysine-70, reported to be absolutely required for ATP hydrolysis by the amino-terminal ATPase domain of Hsc70 (30), results in a DnaK protein with a significantly lowered but not abolished ATPase activity. The kcat of DnaK K70A is 3.5% of that of wild-type DnaK. Also significant is the finding that most of the wild-type ATPase activity is preserved when residue threonine-199, which is completely conserved in the Hsp70 family, is replaced by serine. This mutant is also the only one with a residual autophosphorylation activity, measured to be 12.9% of that of wild-type DnaK. The K70A mutant, in which the site of autophosphorylation (threonine-199) is preserved, has no autophosphorylation activity. The ability of each DnaK derivative to bind ATP was confirmed by incubating the protein with [α-32P]ATP and separating free nucleotide from DnaK and DnaK-ATP complexes by rapid gel filtration chromatography. Wild-type DnaK as well as the T199A, T199S, and K70A derivatives bound ATP near saturation with ATP concentrations of as low as 1 μM (data not shown).
FIG. 2.
ATPase and autophosphorylation activities of DnaK and DnaK derivatives free of NDP kinase activity. Eadie-Hofstee plots of ATPase activity for DnaK and DnaK derivatives purified by the extended protocol are shown. DnaK (final concentration, 7.24 nM) was incubated with [α-32P]ATP (final concentration, 6.35 to 510 nM) in modified ATPase buffer at 30°C. The amounts of ADP and total nucleotide were determined following various incubation times as described in Materials and Methods, and the velocity of the ATPase reaction for each [ATP] was calculated. The line drawn on each graph is a best-fit line determined from a least-squares linear regression analysis of the data points.
TABLE 3.
ATPase activity parameters and extent of autophosphorylation for DnaK and DnaK derivatives
| DnaK | ATPase activitya
|
Extent of autophosphorylationb | ||
|---|---|---|---|---|
| Vmax (pmol of ATP/μg of DnaK/min) | kcat (min−1) | Km (nM) | ||
| Wild type | 2.29 | 0.158 | 27.5 | 1.00 |
| T199S | 1.73 | 0.120 | 36.5 | 0.129 |
| T199A | 0.0696 | 0.00481 | 19.1 | 0 |
| K70A | 0.0808 | 0.00558 | 41.8 | 0 |
The Vmax, Kcat, and Km values were derived from the y intercept and slope of the Eadie-Hofstee plots (Fig. 1).
The extent of autophosphorylation was determined as described in Materials and Methods. The extent of autophosphorylation of wild-type DnaK was set as 1.00.
ATP does not induce DnaK K70A to release peptide.
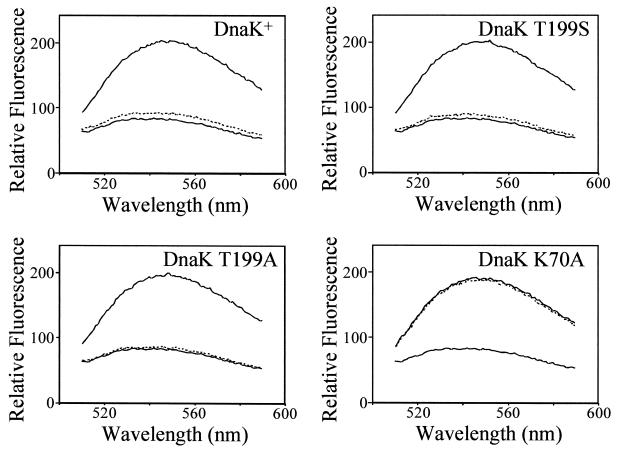
Short peptides can be used to study the interaction of Hsp70 proteins with polypeptide substrate (16). We used a derivative (called PepH [NRLLLCG]) of the 7-amino-acid peptide NR (NRLLLSG), which has been shown to bind to DnaK with high affinity (18), as a peptide substrate for DnaK. A cysteine-specific fluorescent probe, I-ANBD, was covalently attached to PepH, allowing it to be used for kinetic binding studies. The intensity of the fluorescent signal of PepH-ANBD increases when it binds to DnaK, allowing for a direct assay of the extent of binding (43). DnaK and the T199S, T199A, and K70A derivatives were incubated with PepH-ANBD, and binding was measured using a fluorescence spectrometer. Figure 3 shows that when PepH-ANBD is mixed with DnaK or any of the three derivatives, the emission spectrum signal has a greater intensity than the emission spectrum of an equal amount of PepH-ANBD alone, showing that PepH-ANBD binds to all of the proteins. The addition of ATP results in the rapid release of peptide by DnaK (38). ATP was added to the various DnaK proteins with bound PepH-ANBD. The addition of ATP results in the reduction of fluorescence signal by PepH-ANBD almost to the level of PepH-ANBD alone for wild-type DnaK, DnaK T199S, and DnaK T199A but results in very little change in the fluorescence signal of PepH-ANBD bound to DnaK K70A. These results indicate that the addition of ATP results in the nearly complete dissociation of PepH-ANBD from DnaK, DnaK T199S, and DnaK T199A but does not result in the dissociation of PepH-ANBD from DnaK K70A.
FIG. 3.
Peptide binding and release by DnaK and DnaK derivatives. DnaK or mutant DnaK protein (2.0 μM) was incubated with PepH-ANBD (1.0 μM) for 30 min at 30°C in ATPase buffer. The sample was split into two aliquots, and ATP (50 μM) was added to one of the aliquots. The emission spectrum of each mixture was collected as described in Materials and Methods. For comparison, the emission spectrum of PepH-ANBD (1.0 μM) alone was collected. Shown are the spectra for DnaK and mutant DnaK proteins, as follows: DnaK plus PepH-ANBD (upper solid line), DnaK plus PepH-ANBD plus ATP (dashed line), and PepH-ANBD alone (lower solid line).
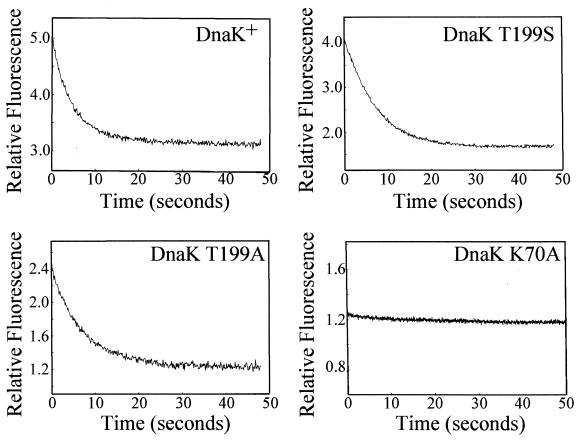
The kinetics of the release of PepH-ANBD by DnaK upon the addition of ATP were also measured. The reaction was initiated in a stopped-flow device by rapidly mixing ATP with prebound complexes of DnaK and PepH-ANBD. Figure 4 shows traces of the fluorescence signal over time for DnaK+, DnaK T199S, DnaK T199A, and DnaK K70A with PepH-ANBD bound following rapid mixing with ATP. The decreasing fluorescence signal shows that DnaK+, DnaK T199S, and DnaK T199A all release PepH-ANBD upon the addition of ATP with relatively rapid and similar kinetics. The fluorescence signal for PepH-ANBD bound to DnaK K70A, by contrast, remains flat, indicating that ATP does not induce DnaK K70A to release peptide.
FIG. 4.
Kinetics of peptide release by DnaK and DnaK derivatives. DnaK or mutant DnaK protein (1 μM) was incubated with PepH-ANBD (0.5 μM) in ATPase buffer for 30 min at 30°C and mixed in a stopped-flow apparatus with an equal volume of ATP (2 μM) in ATPase buffer. The excitation wavelength used was 480 nm, and a 495-nm-cutoff filter was used. For details of fluorescence measurements, see Materials and Methods. All reaction traces, except for that of DnaK K70A, could be fit to a single-exponential decay function.
ATP does not induce DnaK K70A to undergo a conformational change.
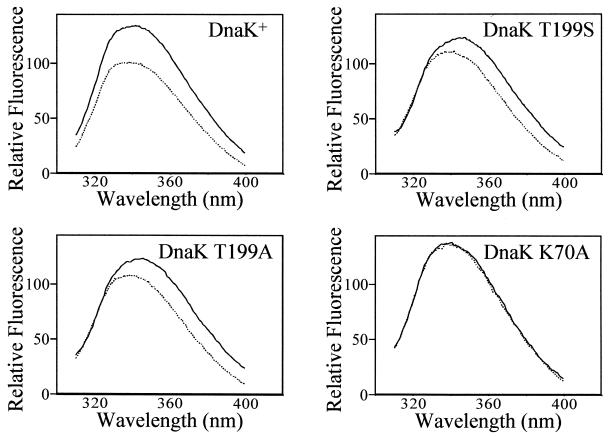
The binding of ATP induces a global conformational change in DnaK which results in the release of bound peptide (25). DnaK contains a single tryptophan residue (tryptophan-102), the fluorescence of which changes upon the addition of ATP to DnaK (1). This fluorescence shift is the result of the global conformational change that occurs when DnaK binds ATP and can be used to observe the conformational changes of DnaK. The fluorescence spectra of DnaK, DnaK T199S, DnaK T199A, and DnaK K70A were collected in the presence and absence of ATP by exciting the samples with light at a wavelength of 295 nm in a fluorescence spectrometer. Figure 5 shows that for DnaK, DnaK T199S, and DnaK T199A, the fluorescence spectrum in the presence of 50 μM ATP is reduced in intensity and slightly shifted to shorter wavelengths compared to the fluorescence spectrum without ATP added. These results indicate that ATP induces a conformational change in these three proteins. The fluorescence spectra of DnaK K70A, however, are the same in both the presence and absence of ATP (Fig. 5). There was no change in the spectrum even following prolonged incubation of DnaK K70A with ATP (45 min at room temperature). These results indicate that ATP does not induce a conformational change in DnaK K70A, at least not a change similar to the one that occurs in wild-type DnaK, and results in a change in the local environment of tryptophan-102 and its fluorescence spectrum. Therefore, ATP induces DnaK K70A neither to undergo a conformational change nor to release peptide.
FIG. 5.
ATP-induced conformational change of DnaK and DnaK derivatives. The fluorescence of the single tryptophan residue in DnaK and mutant DnaK proteins was measured in the absence (solid line) or presence (dashed line) of ATP. The fluorescence spectrum of DnaK and mutant DnaK protein (2.0 μM) with or without ATP (50 μM) in ATPase buffer was collected as described in Materials and Methods.
DISCUSSION
We have biochemically characterized E. coli DnaK and derivatives with alterations at the threonine-199 and lysine-70 positions. The DnaK proteins used in this study were purified from ndk::km cells in order to prevent inaccuracies in the biochemical measurements due to the presence of NDP kinase in the preparations (2). The preparations were also treated to remove bound nucleotide using a procedure which we found further reduces the amount of residual copurifying NDP kinase activity (2). All four DnaK preparations used in this study had measurable, but very small, amounts of ADP kinase activity, ranging from 0.015 to 0.025 pmol of ADP phosphorylated/min/pmol of DnaK. This level of ADP kinase activity would be caused by approximately 5 × 10−7 pmol of NDP kinase per pmol of DnaK.
We found that wild-type DnaK and DnaK T199S have ATPase activities with rates about 30 times higher than those of DnaK T199A and DnaK K70A. The kcat values for the ATPase reaction were found to be 0.158, 0.120, 0.00481, and 0.00558 min−1 for DnaK+, DnaK T199S, DnaK T199A, and DnaK K70A, respectively. Because the background amount of ADP kinase activity was nearly the same in all four DnaK preparations, these kcat values represent real differences in the ATPase activities of DnaK+ and DnaK T199S from those of DnaK T199A and DnaK K70A. We recognize that the low ATPase rates of the DnaK T199A and DnaK K70A proteins might actually represent the amount of background hydrolysis due to the small amount of NDP kinase in the preparations. Therefore, the ATPase activities of the DnaK T199A and K70A proteins may actually have kcat values lower than those we assigned and could be 0. However, since the amount of residual NDP kinase activity varied by less than twofold between the DnaK+, DnaK T199S, DnaK T199A, and DnaK K70A preparations, the amount of NDP kinase-dependent ATPase activity in the DnaK+ and DnaK T199S preparations would not be expected to be significantly greater than that in the DnaK T199A and DnaK K70A preparations. For this reason, the true kcat values for the ATPase activities of DnaK+ and DnaK T199S are at most ≈0.006 min−1 lower than those we report in Table 3.
The kcat value that we determined for the ATPase activity of wild-type DnaK (0.158 min−1) correlates well with previously reported values of 0.180 min−1 (33) and 0.130 min−1 (13, 38) but is severalfold higher than other reported values (9, 27, 36) and nearly 10-fold higher than that in one report (34). The reason for these differences is not known. It is possible that despite our effort to obtain pure samples of DnaK, our preparations contain peptide fragments that stimulate the ATPase activity of DnaK. It is also possible that some fraction of the DnaK in preparations from other labs is inactive. The DnaK T199A mutant has previously been studied, and the kcat value determined by us for the ATPase activity of this protein (0.00481 min−1) agrees with previously published values of 0.007 min−1 (28) and 0.011 min−1 (33). The K71A mutation in the 44-kDa amino-terminal ATPase fragment of Hsc70 was found to have no measurable ATPase activity (30). The small, but measurable, ATPase activity in our DnaK K70A preparation could be due to a difference between full-length protein and the 44-kDa amino-terminal fragment, a difference between DnaK and bovine Hsc70, or the total absence of NDP kinase in the Hsc70 preparation. The kcat value reported for the ATPase activity of the human cytosolic Hsp70 K71E mutant of 0.0048 min−1 (35) is similar to the value we obtained for the DnaK K70A mutant of 0.00558 min−1. The complete lack of autophosphorylation by the DnaK T199A mutant that we observed has been previously reported (28). The lack of autophosphorylation by the DnaK K70A mutant is not surprising considering that the autophosphorylation activity is considered to be a side reaction of ATP hydrolysis (15) and the mutant has very little ATPase activity.
It is interesting that the DnaK T199S mutant retains most of the ATPase activity of wild-type DnaK but is not able to fully function as DnaK in the cell, as shown by the low sensitivity to λ phage, slow growth at 42°C, and filamentation of ΔdnaK cells expressing DnaK T199S. These phenotypes represent a true defect of function of the DnaK T199S protein, as the amount of DnaK T199S protein in these cells is approximately equal to the amount of wild-type DnaK in homologous cells (Fig. 1). Furthermore, the DnaK T199S protein also undergoes a conformational change and releases peptide when mixed with ATP, so it does not appear to be defective in the coupling of the ATPase and peptide release activities. Threonine-199 of DnaK is completely conserved among Hsp70 proteins. While it was initially thought that phosphorylation of this threonine might play an important role in the reaction pathway of ATP hydrolysis, this notion was disproved by structural studies of mutants altered at the threonine-204 (homologous to threonine-199 of DnaK) position of Hsc70 (31). The Hsp70 protein immunoglobin-binding protein (BiP) with a serine substituted for the homologous threonine residue, threonine-229, was found to be autophosphorylated at a level of 11.6% of that of wild-type BiP (17), similar to our finding that the DnaK T199S mutant is autophosphorylated at a level of 12.9% of that of wild-type DnaK. However, the velocity of the ATPase activity of the BiP T229S mutant was found to be only 21.7% of that of wild-type BiP, and the BiP T299S mutant was not measured for ATP-dependent peptide release and conformational change or the ability of the protein to function in vivo (17). The reason that the DnaK T199S protein is unable to fully perform DnaK function in the cell is not known, but it represents a new class of DnaK mutant: one with a functional ATPase activity coupled to a functional peptide release activity but unable to complement the ΔdnaK52 mutation. Previously defined classes of DnaK mutants include those with defective ATPase activities but functional peptide release activities, such as DnaK T199A (33), and those with functional ATPase activities but defective peptide release activities, such as DnaK E171A (4).
The DnaK K70A derivative belongs to a class of mutant not yet described specifically for DnaK proteins but previously described for other Hsp70 proteins: those with both defective ATPase activities and defective ATP-induced peptide release activities. The K71E derivative of human cytosolic Hsp70 (35) and the K71M derivative of the 60-kDa fragment of Hsc70 (22) also have both defective ATPase and defective ATP-induced peptide release activities. DnaK K70A binds ATP and Hsp70 K71E binds ATP, albeit with lower affinity than wild-type Hsp70 (35), showing that the lack of ATP hydrolysis is due to a true catalytic defect and not simply to the lack of ATP binding. Structural studies of lysine-71 mutations in the 44-kDa amino-terminal ATPase fragment of bovine Hsc70 indicated that lysine-71 participates in the catalysis of ATP hydrolysis by stabilizing an H2O molecule or OH− ion for a nucleophilic attack on the gamma-phosphate of the bound ATP (30). The results of the Hsc70 study identified lysine-71 as the only residue yet known to be essential for the hydrolysis of ATP. Our results indicate that mutation of the homologous lysine in DnaK, lysine-70, results in a protein that does not undergo a conformational change or release peptide in the presence of ATP. Two possible explanations for this behavior are (i) that the K70A mutant does not bind ATP with the correct geometry required to elicit a conformational change and (ii) that the K70A mutant, because it is unable to hydrolyze ATP, does not undergo a conformational change in the presence of ATP.
DnaK and other Hsp70 proteins undergo a conformational change which results in the release of bound polypeptide. It is widely believed that this conformational change is elicited by ATP binding and not the hydrolysis of ATP (3). This conclusion was initially drawn to explain the observation that the DnaK T199A mutant releases bound peptide upon the addition of ATP (33). It was assumed that the T199A derivative of DnaK is defective in ATP hydrolysis, and it was thus concluded that ATP hydrolysis is not required for peptide release. Previously, it had been concluded that ATP hydrolysis is required for DnaK to undergo a conformational change and release bound peptide, since nonhydrolyzable analogs of ATP do not induce these activities of DnaK (25). The model that ATP binding, and not ATP hydrolysis, triggers the conformational change of DnaK that results in peptide release is compatible with the observation that AMP-PNP, AMP-PCP, and ATP-γS do not elicit such a conformational change in DnaK if these nonhydrolyzable analogs do not bind DnaK in the same manner as ATP. Similarly, to be compatible with this model, the assumption must be made that ATP does not induce a conformational change in DnaK K70A, not because this mutant protein is defective in ATP hydrolysis but because either ATP does not bind to it in the same manner that ATP binds to wild-type DnaK or the K70A mutation renders the protein unable to undergo a conformational change.
The threonine-204 residue of bovine Hsc70, which is homologous to threonine-199 of E. coli DnaK, was found not to be essential for the breaking of the β-γ phosphate bond of ATP (31). It is possible that the threonine-199 residue of DnaK is required for some other step in the functional cycle of DnaK. The complete disruption of this putative step could significantly lower the steady-state rate of ATP hydrolysis as seen with the T199A mutation, while a partial disruption of this step could lead to a nonfunctional chaperone with a near-normal steady-state rate of ATP hydrolysis as seen with the T199S mutation. Since the DnaK T199A and DnaK T199S mutant proteins would be able to break the β-γ phosphate bond of ATP, it is possible that this step, and not the binding of ATP by DnaK, induces the conformational change that results in the release of bound peptide substrate. ATP binds to DnaK in a process involving at least two steps: the rapid formation of a weak complex followed by a slower isomerization that is kinetically correlated to the conformational change of DnaK, resulting in peptide release (41). The explanation that this second step of ATP binding actually represents the reversible breaking of the β-γ phosphate bond of ATP and a step that is defective in the K70A mutant DnaK would be consistent with the observations that the DnaK K70A mutant does not undergo an ATP-induced conformational change and that nonhydrolyzable analogs of ATP do not induce a conformational change in wild-type DnaK. If this was true, then ATP hydrolysis, and not the binding of ATP alone, would be required to induce the conformational change in DnaK that results in peptide release.
ACKNOWLEDGMENTS
We are grateful to Alok Srivastava for his help with the Sauer lab's stopped-flow apparatus and the Baker lab's spectrofluorimeter. We thank Christophe Herman for constructing the pJM11-PlacdnaK(K70A) dnaJ+ plasmid. We thank Bob Sauer for the use of his stopped-flow spectrofluorimeter and Tania Baker for the use of her phosphorimager and spectrofluorimeter. We thank the members of our laboratory for helpful discussions.
This work was supported by Public Health Service grant CA21615 from the National Institutes of Health.
REFERENCES
- 1.Banecki B, Zylicz M, Bertoli E, Tanfani F. Structural and functional relationships in DnaK and DnaK756 heat-shock Proteins from Escherichia coli. J Biol Chem. 1992;267:25051–25058. [PubMed] [Google Scholar]
- 2.Barthel T K, Walker G C. Inferences concerning the ATPase properties of DnaK and other HSP70s are affected by the ADP kinase activity of copurifying nucleoside-diphosphate kinase. J Biol Chem. 1999;274:36670–36678. doi: 10.1074/jbc.274.51.36670. [DOI] [PubMed] [Google Scholar]
- 3.Buchberger A, Reinstein J, Bukau B. The DnaK chaperone system: mechanism and comparison with other Hsp70 systems. In: Bukau B, editor. Molecular chaperones and folding catalysts: regulation, cellular function, and mechanisms. Amsterdam, The Netherlands: Harwood Academic Publishers; 1999. pp. 609–635. [Google Scholar]
- 4.Buchberger A, Valencia A, McMacken R, Sander C, Bukau B. The chaperone function of DnaK requires the coupling of ATPase activity with substrate binding through residue E171. EMBO J. 1994;13:1687–1695. doi: 10.1002/j.1460-2075.1994.tb06433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukau B, editor. Molecular chaperones and folding catalysts: regulation, cellular function and mechanisms. Amsterdam, The Netherlands: Harwood Academic Publishers; 1999. [Google Scholar]
- 6.Bukau B, Horwich A L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 7.Bukau B, Walker G C. Cellular defects caused by deletion of the Escherichia coli dnaK gene indicate roles for heat shock protein in normal metabolism. J Bacteriol. 1989;171:2337–2346. doi: 10.1128/jb.171.5.2337-2346.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukau B, Walker G C. Mutations altering heat shock specific subunit of RNA polymerase suppress major cellular defects of E. coli mutants lacking the DnaK chaperone. EMBO J. 1990;9:4027–4036. doi: 10.1002/j.1460-2075.1990.tb07624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burkholder W F, Panagiotidis C A, Silverstein S J, Cegielska A, Gottesman M E, Gaitanaris G A. Isolation and characterization of an Escherichia coli DnaK mutant with impaired ATPase activity. J Mol Biol. 1994;242:364–377. doi: 10.1006/jmbi.1994.1587. [DOI] [PubMed] [Google Scholar]
- 10.Casadaban M J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 11.Connolly L, Yura T, Gross C A. Autoregulation of the heat shock response in prokaryotes. In: Bukau B, editor. Molecular chaperones and folding catalysts: regulation, cellular function and mechanisms. Amsterdam, The Netherlands: Harwood Academic Publishers; 1999. [Google Scholar]
- 12.Craig E A, Weissman J S, Horwich A L. Heat shock proteins and molecular chaperones: mediators of protein conformation and turnover in the cell. Cell. 1994;78:365–372. doi: 10.1016/0092-8674(94)90416-2. [DOI] [PubMed] [Google Scholar]
- 13.de Crouy-Chanel A, Kohiyama M, Richarme G. Specificity of DnaK for arginine/lysine and effect of DnaJ on the amino acid specificity of DnaK. J Biol Chem. 1996;271:15486–15490. doi: 10.1074/jbc.271.26.15486. [DOI] [PubMed] [Google Scholar]
- 14.Flaherty K M, DeLuca-Flaherty C, McKay D B. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature. 1990;346:623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- 15.Flaherty K M, Wilbanks S M, Deluca-Flaherty C, McKay D B. Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment. J Biol Chem. 1994;269:12899–12907. [PubMed] [Google Scholar]
- 16.Flynn G C, Chappell T G, Rothman J E. Peptide binding and release by proteins implicated as catalysts of protein assembly. Science. 1989;245:385–390. doi: 10.1126/science.2756425. [DOI] [PubMed] [Google Scholar]
- 17.Gaut J R, Hendershot L M. The immunoglobulin-binding protein in vitro autophosphorylation site maps to a threonine within the ATP binding cleft but is not a detectable site of in vivo phosphorylation. J Biol Chem. 1993;268:12691–12698. [PubMed] [Google Scholar]
- 18.Gragerov A, Gottesman M E. Different peptide binding specificities of hsp70 family members. J Mol Biol. 1994;241:133–135. doi: 10.1006/jmbi.1994.1482. [DOI] [PubMed] [Google Scholar]
- 19.Griffiths J R. Steady-state enzyme kinetics in mutual depletion systems. Biochem Rev. 1979;7:15–25. doi: 10.1042/bst0070429. [DOI] [PubMed] [Google Scholar]
- 20.Harrison C J, Hayer-Hartl M, Di Liberto M, Hartl F-U, Kuriyan J. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science. 1997;276:431–435. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- 21.Hendrick J P, Hartl F-U. Molecular chaperone functions of heat-shock proteins. Annu Rev Biochem. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- 22.Johnson E R, McKay D B. Mapping the role of active site residues for transducing an ATP-induced conformational change in the bovine 70-kDa heat shock cognate protein. Biochemistry. 1999;38:10823–10830. doi: 10.1021/bi990816g. [DOI] [PubMed] [Google Scholar]
- 23.Jordan R, McMacken R. Modulation of the ATPase activity of the molecular chaperone DnaK by peptides and the DnaJ and GrpE heat shock proteins. J Biol Chem. 1995;270:4563–4569. doi: 10.1074/jbc.270.9.4563. [DOI] [PubMed] [Google Scholar]
- 24.Karzai A W, McMacken R. A bipartite signaling mechanism involved in DnaJ-mediated activation of the Escherichia coli DnaK protein. J Biol Chem. 1996;271:11236–11246. doi: 10.1074/jbc.271.19.11236. [DOI] [PubMed] [Google Scholar]
- 25.Liberek K, Skowyra D, Zylicz M, Johnson C, Georgopoulos C. The Escherichia coli DnaK chaperone, the 70-kDa heat shock protein eukaryotic equivalent, changes conformation upon ATP hydrolysis, thus triggering its dissociation from a bound target protein. J Biol Chem. 1991;266:14491–14496. [PubMed] [Google Scholar]
- 26.Lu Q, Zhang X, Almaula N, Mathews C K, Inouye M. The gene for nucleoside diphosphate kinase functions as a mutator gene in Escherichia coli. J Mol Biol. 1995;254:337–341. doi: 10.1006/jmbi.1995.0620. [DOI] [PubMed] [Google Scholar]
- 27.McCarty J S, Buchberger A, Reinstein J, Bukau B. The role of ATP in the functional cycle of the DnaK chaperone Syst. J Mol Biol. 1995;249:126–137. doi: 10.1006/jmbi.1995.0284. [DOI] [PubMed] [Google Scholar]
- 28.McCarty J S, Walker G C. DnaK as a thermometer: threonine-199 is site of autophosphorylation and is critical for ATPase activity. Proc Natl Acad Sci USA. 1991;88:9513–9517. doi: 10.1073/pnas.88.21.9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarty J S, Walker G C. DnaK mutants defective in ATPase activity are defective in negative regulation of the heat shock response: expression of mutant DnaK proteins results in filamentation. J Bacteriol. 1994;176:764–780. doi: 10.1128/jb.176.3.764-780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Brien M C, Flaherty K M, McKay D B. Lysine 71 of the chaperone protein Hsc70 is essential for ATP hydrolysis. J Biol Chem. 1996;271:15874–15878. doi: 10.1074/jbc.271.27.15874. [DOI] [PubMed] [Google Scholar]
- 31.O'Brien M C, McKay D B. Threonine 204 of the chaperone protein Hsc70 influences the structure of the active site, but is not essential for ATP hydrolysis. J Biol Chem. 1993;268:24323–24329. [PubMed] [Google Scholar]
- 32.Paek K-H, Walker G C. Escherichia coli dnaK null mutants are inviable at high temperature. J Bacteriol. 1987;169:283–290. doi: 10.1128/jb.169.1.283-290.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palleros D R, Reid K L, Shi L, Welch W J, Fink A L. ATP-induced protein-Hsp70 complex dissociation requires K+ but not ATP hydrolysis. Nature. 1993;365:664–666. doi: 10.1038/365664a0. [DOI] [PubMed] [Google Scholar]
- 34.Pierpaoli E V, Sandmeier E, Baici A, Schànfeld H-J, Gisler S, Christen P. The power stroke of the DnaK/DnaJ/GrpE molecular chaperone Syst. J Mol Biol. 1997;269:757–768. doi: 10.1006/jmbi.1997.1072. [DOI] [PubMed] [Google Scholar]
- 35.Rajapandi T, Wu C, Eisenberg E, Greene L. Characterization of D10S and K71E mutants of human cytosolic Hsp70. Biochemistry. 1998;37:7244–7250. doi: 10.1021/bi972252r. [DOI] [PubMed] [Google Scholar]
- 36.Russell R, Jordan R, McMacken R. Kinetic characterization of the ATPase cycle of the DnaK molecular chaperone. Biochemistry. 1998;37:596–607. doi: 10.1021/bi972025p. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Schmid D, Baici A, Gehring H, Christen P. Kinetics of molecular chaperone action. Science. 1994;263:971–973. doi: 10.1126/science.8310296. [DOI] [PubMed] [Google Scholar]
- 39.Sriram M, Osipiuk J, Freeman B C, Morimoto R I, Joachimiak A. Human Hsp70 molecular chaperone binds two calcium ions within the ATPase domain. Structure. 1997;5:403–414. doi: 10.1016/s0969-2126(97)00197-4. [DOI] [PubMed] [Google Scholar]
- 40.Straus D, Walter W, Gross C A. DnaK, DnaJ, and GrpE heat shock proteins negatively regulate heat shock gene expression by controlling the synthesis and stability of ς32. Genes Dev. 1990;4:2202–2209. doi: 10.1101/gad.4.12a.2202. [DOI] [PubMed] [Google Scholar]
- 41.Theyssen H, Schuster H-P, Packschies L, Bukau B, Reinstein J. The second step of ATP binding to DnaK induces peptide release. J Mol Biol. 1996;263:657–670. doi: 10.1006/jmbi.1996.0606. [DOI] [PubMed] [Google Scholar]
- 42.Wickner S, Hoskins J, McKenney K. Monomerization of RepA dimers by heat shock proteins activates binding to DNA replication origin. Proc Natl Acad Sci USA. 1991;88:7903–7907. doi: 10.1073/pnas.88.18.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J, Walker G C. Interactions of peptides with DnaK and C-terminal DnaK fragments studied using fluorescent and radioactive peptides. Arch Biochem Biophys. 1998;356:177–186. doi: 10.1006/abbi.1998.0784. [DOI] [PubMed] [Google Scholar]
- 44.Zylicz M, LeBowitz J H, McMacken R, Georgopoulos C. The dnaK protein of Escherichia coli possesses an ATPase and autophosphorylating activity and is essential in an in vitro DNA replication system. Proc Natl Acad Sci USA. 1983;80:6431–6435. doi: 10.1073/pnas.80.21.6431. [DOI] [PMC free article] [PubMed] [Google Scholar]