Abstract
The nitrate reductase of the hyperthermophilic archaeon Pyrobaculum aerophilum was purified 137-fold from the cytoplasmic membrane. Based on sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis, the enzyme complex consists of three subunits with apparent molecular weights of 130,000, 52,000, and 32,000. The enzyme contained molybdenum (0.8-mol/mol complex), iron (15.4-mol/mol complex) and cytochrome b (0.49-mol/mol complex) as cofactors. The P. aerophilum nitrate reductase distinguishes itself from nitrate reductases of mesophilic bacteria and archaea by its very high specific activity using reduced benzyl viologen as the electron donor (Vmax with nitrate, 1,162 s−1 (326 U/mg); Vmax with chlorate, 1,348 s−1 (378 U/mg) [assayed at 75°C]). The Km values for nitrate and chlorate were 58 and 140 μM, respectively. Azide was a competitive inhibitor and cyanide was a noncompetitive inhibitor of the nitrate reductase activity. The temperature optimum for activity was >95°C. When incubated at 100°C, the purified nitrate reductase had a half-life of 1.5 h. This study constitutes the first description of a nitrate reductase from a hyperthermophilic archaeon.
Nitrate serves as electron acceptor to many prokaryotic microbes that thrive under anaerobic conditions. Nitrate respiration occurs via two independent pathways, the denitrification pathway and the ammonification pathway (3). Nitrate is reduced sequentially to dinitrogen gas in the denitrification pathway, while ammonium is the product of the two-step ammonification pathway. The first reaction, in which nitrate is reduced to nitrite via the membrane-bound nitrate reductase, is identical in both pathways (3, 21). In general, the dissimilatory nitrate reductase is conserved among bacteria and archaea that have been investigated thus far. The enzyme has been extensively studied in mesophilic nitrate reducing bacteria such as the ammonifier Escherichia coli and the denitrifiers Paracoccus denitrificans, Pseudomonas stuzeri, Pseudomonas denitrificans, and others (5, 6, 11, 12, 16). The E. coli NarGHI enzyme is one of the best-characterized enzymes (3). The enzyme complex consists of three subunits (11, 16). The α subunit (NarG) has an Mr of 145,000 and contains a molybdopterin cofactor at its active site, where nitrate is reduced to nitrite. The β subunit (NarH) has an Mr of 58,000 and is the location of one [3Fe-4S] center and three [4Fe-4S] centers. Both the α and β subunits are attached to the cytoplasmic membrane by the 25,000-Da γ subunit (NarI). This polypeptide contains cytochrome b and functions to oxidize the menaquinol or ubiquinol of the quinone pool. Electrons are transferred from the quinol pool via the β subunit to the α subunit active site (3). Recently, nitrate reductases from several archaeal species have been described. While Haloferax volcanii contains a heterotrimeric enzyme complex similar to the bacterial dissimilatory nitrate reductases, the nitrate reductase from Haloferax denitrificans was purified as a heterodimeric enzyme possibly lacking the membrane anchor subunit (4, 9). In contrast, Haloarcula marismortui contains a homotetrameric nitrate reductase (20).
The archaeon Pyrobaculum aerophilum is the only hyperthermophilic denitrifier that has been characterized thus far (18). P. aerophilum was isolated from a hot spring in Italy and grows optimally at 100°C (18). Based on 16S rRNA gene sequence analysis, P. aerophilum was placed in the order Thermoproteales within the Crenarchaeota branch of the phylogenetic tree (18). In contrast to its strictly anaerobic, sulfur-respiring relatives, P. aerophilum can respire aerobically with 0.6 to 1% oxygen present in the gas phase. Alternatively, P. aerophilum can grow anaerobically by dissimilatory nitrate reduction, a catabolic feature not found among its close relatives (1, 18). Previous studies have established that growth of P. aerophilum is strictly dependent on the availability of tungstate in the culture medium (1). Tungstate, however, has been demonstrated to inactivate molybdoenzymes such as the nitrate reductase and formate dehydrogenase in mesophilic bacteria (8). In contrast to E. coli, where nitrate reductase enzyme function was abolished by the addition of tungstate to the medium, the enzyme was still active in P. aerophilum cultured with elevated tungstate concentrations (1). Thus, P. aerophilum appears to be adapted to an extremely thermophilic environment that typically contains elevated levels of tungstate (1). In this study we report the purification and characterization of the thermostable nitrate reductase from P. aerophilum.
MATERIALS AND METHODS
Growth conditions.
P. aerophilum was grown anaerobically at 95°C in a marine medium containing yeast extract (0.05%), peptone (0.05%), nitrate (10 mM), molybdate (0.5 μM), and tungstate (0.4 μM) as previously described (1). For routine cell transfers, P. aerophilum was cultured in 50 ml of medium in 125-ml anaerobic serum vials (1). For large-scale cultivation, P. aerophilum was grown in 70 liters of culture medium in a 100-liter glass-lined fermentor (Pfaudler). Cultures were harvested in late exponential phase by concentration with a hollow-fiber filter (A/G Technology) and subsequent centrifugation for 20 min at 16,000 × g. The pelleted cells were stored at −20°C. Typical cell yields were 35 g (wet weight) of cells per 70 liter of culture volume. Cells that were used for the preparation of the membrane fraction for absorption spectroscopy were grown in the absence of molybdenum and with 4.5 μM tungstate.
Purification of the nitrate reductase.
Enzyme purification was performed under aerobic conditions and at room temperature unless otherwise indicated. Frozen cells were resuspended in 20 mM sodium phosphate buffer (pH 7.0) and broken by sonication (1). The membrane fraction was collected by ultracentrifugation at 100,000 × g for 60 min at 4°C. The membrane pellet was then resuspended in 20 mM sodium phosphate (pH 7.0). Membrane-bound proteins were solubilized by extracting the membrane fraction with 2% Triton X-100 in 20 mM sodium phosphate (pH 7.0) at a ratio of 6 mg of Triton X-100 per mg of protein. The mixture was stirred for 45 min and centrifuged at 100,000 × g for 60 min at 4°C to remove the insoluble fraction. The supernatant was applied to a 5-ml Hi-Trap Q Sepharose column (Pharmacia Biotech) equilibrated with 20 mM sodium phosphate (pH 7.0) containing 0.1% Triton X-100. After the column was washed with 15 volumes of the same buffer, nitrate reductase activity was detected in the flowthrough fractions. The flowthrough fractions were loaded onto a 5-ml hydroxyapatite column (Bio-Rad) equilibrated with 10 mM sodium phosphate buffer (pH 7.2) containing 0.1% Triton X-100. A 10 to 200 mM linear sodium phosphate gradient was applied, and nitrate reductase activity was recovered in fractions containing about 130 mM sodium phosphate. Fractions containing nitrate reductase activity were combined and concentrated with Centricon 100 concentrators (Amicon). The buffer of the concentrated sample was exchanged, using a PD-10 Sephadex G-25 M column (Pharmacia) for the buffer used for the subsequent chromatography column (i.e., 20 mM Tricine [pH 8.0] plus 0.1% Triton X-100). The final chromatography step was performed with a MonoQ HR5/5 column (Pharmacia), which was developed with a linear gradient of NaCl from 0 to 200 mM. Nitrate reductase eluted at about 100 mM NaCl. The purified enzyme was concentrated and stored at 4°C or, for long-term storage, at −70°C.
Enzyme assay.
Nitrate reductase activity was determined with reduced benzyl viologen as the electron donor in an anaerobic cuvette assay as previously described (1). The assay was performed at 75°C unless otherwise indicated. One unit of enzyme activity is defined as 1 μmol of nitrate reduced per min.
Characterization of the pH and temperature optima.
The pH and temperature optima for nitrate reductase activity were measured at the pH values and temperatures indicated in the figures.
Determination of temperature stability.
The temperature stability of the P. aerophilum nitrate reductase was determined by incubating the enzyme (0.2 mg of protein/ml) in 10 mM Tricine (pH 8.0)–0.1. M NaCl–0.1% Triton X-100 at the indicated temperatures for more than 30 h. Throughout the experiment, samples were removed and assayed for nitrate reductase activity at 75°C.
Protein determination.
The protein concentration was determined by the modified Lowry procedure for membrane-bound and detergent-containing protein samples (Sigma). Bovine serum albumin was used as the standard.
Absorption spectroscopy.
The oxidized and dithionite-reduced absorption spectra of the purified enzyme were recorded on a Beckman DU 640 spectrophotometer as previously described (17). The amount of cytochrome b present in the enzyme preparation was based on an extinction coefficient for cytochrome b of 20 mM−1 cm−1 at 556 nm (13). The absorption spectrum of the P. aerophilum membrane fraction was recorded by an SLM Aminco DW 2000 spectrophotometer using cuvettes with a 1-mm path length. After the spectrum of the oxidized sample was recorded, anaerobic conditions were established by three cycles of evacuation and flushing with N2 gas. The sample was subsequently reduced with 1 mM sodium dithionite, and the spectrum was recorded after 3 and 6 min at room temperature to ensure complete reduction of the cytochrome-containing membrane proteins. The oxidation of the nitrate reductase cytochrome b was recorded immediately after addition of sodium chlorate at 3.3 mM (final concentration).
Molecular weight determination.
The apparent molecular weight of the purified, denatured nitrate reductase was determined from the mobility of the protein in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12.5% polyacrylamide). The molecular weight standards were bovine serum albumin (M, 66,000), chicken egg albumin (M, 45,000), glyceraldehyde-3-phosphate-dehydrogenase (M, 36,000), bovine carbonic anhydrase (M, 29,000), bovine pancreas trypsinogen (M, 24,000), soybean trypsin inhibitor (M, 20,000), and bovine milk α-lactalbumin (M, 14,200) (Sigma).
Metal analysis.
The molybdenum, tungsten, and iron content was determined using inductively coupled plasma mass spectroscopy performed by the Soil and Plant Analysis Laboratory, University of Wisconsin—Madison/Extension, Madison, Wis.
Gel electrophoresis.
Protein samples were diluted 1:2 with 10 mM Tris-HCl (pH 8.0)–5% SDS–10% β-mercaptoethanol–0.02% bromphenol blue and incubated for 30 min at 100°C. The protein samples (1 μg) were then separated on an SDS–12.5% polyacrylamide gel (Phastsystem; Pharmacia). Precast polyacrylamide gels and SDS buffer strips were purchased from Pharmacia.
Chemicals.
Benzyl viologen, Lubrol, Thesit, and Brij 35 were obtained from Sigma Chemical Co. Triton X-100 was purchased from Boehringer. All other chemicals were of the highest purity commercially available.
RESULTS
Purification of the nitrate reductase.
The nitrate reductase from P. aerophilum was purified from cells that had been grown anaerobically in a nitrate-containing medium supplemented with molybdate and a limiting amount of tungstate (i.e., 0.4 μM). The latter metal oxyanion concentration had been shown previously to promote high nitrate reductase levels (1). Since nitrate reductase activity had been previously localized to the membrane fraction, membranes were extracted using several different detergents. About 100% extraction of nitrate reductase activity from the cytoplasmic membrane was accomplished with 2% Triton X-100 at a ratio of 6 mg of Triton X-100 per mg of protein. A 5% solution of the nonionic detergent Thesit extracted 60% of the enzyme activity, whereas neither 5% Lubrol nor 5% Brij 35 extracted any nitrate reductase activity (data not shown). The detergents used did not aversely affect the nitrate reductase activity.
Nitrate reductase was purified from the Triton X-100 extract by Q-Sepharose, hydroxyapatite, and Mono Q ion-exchange chromatography (Table 1). The enzyme separated as a broad band on the last column. Only the peak fractions were combined to give the final preparation. This, however, resulted in a low yield (2%). After the last purification step, the nitrate reductase enzyme was enriched 137-fold with respect to the membranes.
TABLE 1.
Purification of the P. aerophilum nitrate reductasea
| Purification step | Total amt of protein (mg) | Total activity (U) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Membrane fraction | 78 | 191 | 2.4 | 100 | 1 |
| Triton X-100 extract | 33 | 193 | 7.9 | 100 | 3 |
| Q-Sepharose | 5.5 | 134 | 24.5 | 69 | 10 |
| Hydroxyapatite | 0.8 | 77 | 100 | 40 | 42 |
| Mono-Q | 0.01 | 3 | 326 | 1.6 | 137 |
The activity was measured at 75°C. One unit of enzyme activity is defined as 1 μmol of nitrate reduced per min.
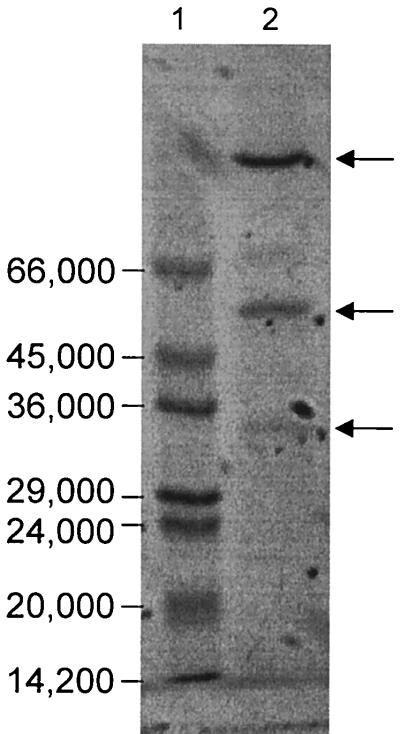
The active nitrate reductase at the final purification step consisted of three polypeptides with apparent molecular weights of 130,000, 52,000, and 32,000 as determined by SDS-PAGE (Fig. 1). The purity of the enzyme preparation was estimated to be about 80% as judged from SDS-PAGE. Based on the purity of the preparation and using the enrichment factor of 137, it was calculated that the nitrate reductase constitutes about 0.6% of the membrane-bound protein.
FIG. 1.
SDS-PAGE analysis of the purified nitrate reductase from P. aerophilum. Lanes: 1, molecular weight markers (see Materials and Methods); 2, 1 μg of nitrate reductase.
Cofactor composition.
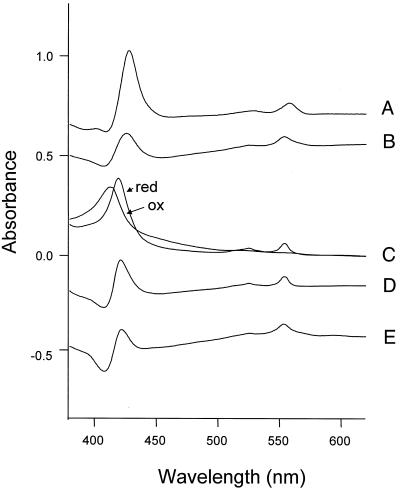
The purified nitrate reductase exhibited a visible absorbance spectrum characteristic of cytochrome b (Fig. 2, traces C and D), with absorbance maxima of the reduced enzyme at 420 and 555 nm for the γ and α band, respectively. Based on the calculated M, of 214,000 for the three-subunit complex (see below), the amount of cytochrome b was estimated to be 0.49 mol/mol of enzyme complex. Nitrate or the alternative substrate chlorate can reoxidize the reduced cytochrome b of nitrate reductases. This can be demonstrated with the nitrate reductase present in the membrane fraction of the denitrifying bacterium Paracoccus denitrificans by adding chlorate to dithionite reduced membranes (Fig. 2, trace A). Unlike nitrite, the product of nitrate reduction, chlorite, the product of chlorate reduction, cannot be further reduced by other cytochrome-containing enzymes of the denitrification pathway and will therefore not interfere with the difference spectrum. In Paracoccus denitrificans, the absorbance maxima for the nitrate reductase cytochrome b in the reduced-minus-oxidized difference spectrum are identical to those for the purified enzyme (7). The same experiment performed with membranes of P. aerophilum identifies a similar chlorate-oxidizable cytochrome b (Fig. 2, trace B). The spectrum of the chlorate-oxidizable cytochrome b in the membranes is very similar to that of the chlorate-oxidizable cytochrome b in the purified Nar (trace E). The spectrum is also similar to that of the spectrum shown in trace D, which was generated from the absorption spectra of purified, air-oxidized, and dithionite reduced nitrate reductase. This is conclusive for the existence of a chlorate-oxidizable cytochrome b in the P. aerophilum nitrate reductase.
FIG. 2.
Optical difference spectra of membranes from Paracoccus denitrificans and P. aerophilum and of purified P. aerophilum nitrate reductase. The traces contain the following spectra: A, difference spectrum of Paracoccus denitrificans membranes (dithionite reduced minus chlorate [3.3 mM] oxidized); B, difference spectrum of P. aerophilum membranes (dithionite reduced minus chlorate [3.3 mM] oxidized); C, absolute spectra of oxidized (ox) and reduced (red) purified P. aerophilum nitrate reductase; D, difference spectrum of dithionite-reduced minus air-oxidized purified P. aerophilum nitrate reductase (generated from trace C); E, difference spectrum of dithionite-reduced minus chlorate (3.3 mM)-oxidized purified P. aerophilum nitrate reductase.
To determine whether this nitrate reductase contains molybdenum or tungsten as a cofactor, the molybdenum content of the purified nitrate reductase was analyzed by inductively coupled plasma mass spectroscopy. The enzyme contained 3.6 nmol of molybdenum and 0.27 nmol of tungsten per mg of protein. Based on the Mr of 214,000 for the nitrate reductase complex and not normalizing for the protein purity, 0.8 mol of molybdenum and 0.06 mol of tungsten per mol of complex can be calculated. Thus, the P. aerophilum nitrate reductase is a molybdenum-containing enzyme similar to the enzymes purified from mesophilic bacteria. Iron was present at 15.4 mol/mol of enzyme (72 nmol/mg of protein), consistent with the notion that the enzyme contains one [3Fe-4S] cluster and three [4Fe-4S] clusters, similar to mesophilic nitrate reductases (3, 21).
Kinetic properties.
Using reduced benzyl viologen as the electron donor and either nitrate or chlorate as the electron acceptor, the kinetic properties of the purified nitrate reductase were determined. The Km value for nitrate was 58 μM, with a Vmax of 1162 s−1 (326 U/mg). The affinity of nitrate reductase was weaker when chlorate was used as a substrate, i.e., a Km of 140 μM; the Vmax with chlorate was measured to be 1,348 s−1 (378 U/mg).
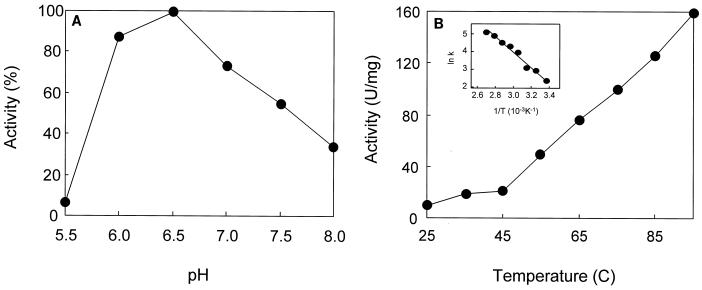
Both azide and cyanide, known inhibitors of dissimilatory nitrate reductases in mesophilic microbes, were tested for their effect on P. aerophilum nitrate reductase activity (21). Azide was determined to be a competitive inhibitor of the P. aerophilum nitrate reductase with a Ki value of 31 μM. Cyanide was a noncompetitive inhibitor, which reduced the Vmax of the enzyme by threefold (i.e., to 108 U/mg). The pH optimum of nitrate reductase was determined to be pH 6.5 (Fig. 3A).
FIG. 3.
Determination of the pH (A) and temperature (B) optima of the P. aerophilum nitrate reductase. The inset in panel B shows the Arrhenius plot, which was calculated using the data from panel B. The symbol k is the rate constant for nitrate reduction, and T is the temperature. The activity was determined as described in Materials and Methods.
Temperature optimum and stability.
The P. aerophilum nitrate reductase exhibited its highest activity at or above 95°C (Fig. 3B), which is slightly below the optimal growth temperature of P. aerophilum (i.e., 100°C). Noteworthy, at 37 to 45°C, nitrate reductase still exhibited 10% of its optimal activity. The Arrhenius plot indicates an activation energy of 36.6 kJ/mol for nitrate reduction (Fig. 3B, inset).
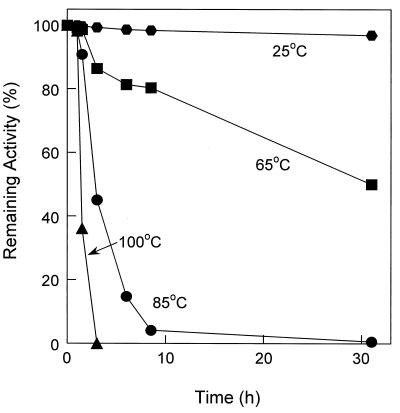
After incubation of purified nitrate reductase at 100°C, its activity declined with a half-life of 1.5 h (Fig. 4). In contrast, the half-life of nitrate reductase activity within the cell membranes was 6 h (data not shown), suggesting that the lipid environment stabilized the enzyme. At incubation temperatures of 85 and 65°C, the purified enzyme exhibited a t1/2 of 16 and 32 h, respectively (Fig. 4). The enzyme was stable at room temperature for many weeks, with negligible loss of activity.
FIG. 4.
Determination of the thermostability of the P. aerophilum nitrate reductase enzyme. The nitrate reductase was incubated at various temperatures. Samples were removed and analyzed for nitrate reductase activity as described in Materials and Methods.
DISCUSSION
Hyperthermophiles, such as the denitrifying P. aerophilum, are naturally exposed to high levels of tungsten, a heavy metal that is abundant in high-temperature environments (14). In E. coli, anaerobic growth with nitrate is abolished when tungstate is present in the medium. Growth inhibition is based on the inactivation of molybdenum-containing enzymes such as nitrate reductase by the incorporation of tungsten in place of molybdenum into molybdenum cofactor of the enzyme (8). Previous studies have shown that the hyperthermophile P. aerophilum is well adapted to a high-tungsten environment in that it has a strict requirement for this heavy metal for its anaerobic growth mode on nitrate (1). However, also in this organism, nitrate reductase activity is decreased when the tungstate concentration in the environment is increased, suggesting that this enzyme might contain molybdenum as a cofactor (1). In contrast to other mesophilic nitrate reducers, P. aerophilum growth with nitrate is not abolished at high tungstate concentrations.
The purpose of this study was to characterize the nitrate reductase from this organism and to determine the nature of its cofactors. This report is the first description of a thermostable nitrate reductase from a hyperthermophilic archaeon. The enzyme was purified from cells that had been grown anaerobically with nitrate and in the presence of low levels of tungstate and molybdenum. These growth conditions had been demonstrated previously to promote the highest nitrate reductase activity (1). We demonstrate here that the P. aerophilum nitrate reductase is also a molybdenum-containing enzyme (0.8 mol of molybdenum/mol of enzyme) with similar subunit composition to that found for mesophilic nitrate reducers (3, 21). In addition, cytochrome b is an integral cofactor of the P. aerophilum nitrate reductase, which can be reoxidized by its substrate in a similar fashion to that shown for the mesophilic bacterium Paracoccus denitrificans (Fig. 2).
The P. aerophilum nitrate reductase distinguishes itself from mesophilic membrane-bound nitrate reductases by its unusually high specific activity of 326 U/mg as measured at 75°C with reduced benzyl viologen as the electron donor (Table 1). Considering that the activity at 75°C is 62% of that at 95°C, nitrate reductase activity under physiological conditions would be about 526 U/mg (turnover of 1,875 s−1). This is about 7 to 40 times higher than the activity of nitrate reductases from mesophilic bacteria and archaea measured at their respective optimal temperatures. Typically, enzymes isolated from hyperthermophiles, such as the NAD-dependent glutamate dehydrogenase from the related Pyrobaculum islandicum, have comparable specific activities to those of enzymes from their mesophilic counterparts (15). Like other mesophilic dissimilatory nitrate reductases, the P. aerophilum enzyme can also utilize chlorate as a substrate at a rate slightly higher than the nitrate reduction rate. Interestingly, the apparent affinities for both nitrate and chlorate are 4- to 60-fold higher than those reported for other dissimilatory nitrate reductases (e.g., Km of 0.2 to 3 mM) (21). It is possible that this highly active nitrate reductase may be an adaptation to counteract inhibition by tungsten under physiological growth conditions: P. aerophilum needs to support growth by nitrate respiration even when the tungsten concentration in the environment is high.
As is typical for enzymes of hyperthermophiles, the P. aerophilum nitrate reductase is most active at or above 95°C. The enzyme appears to be stabilized by its membrane environment, since detergent extraction results in a fourfold loss of the thermostability of nitrate reductase activity.
Denitrification in the archaea is in general not well understood. Archaeal denitrifiers that have been isolated thus far include P. aerophilum, the hyperthermophilic Ferroglobus placidus, and the several Haloferax species (1, 2, 10, 18–20, 21). Thus far, five nitrate reductases, including the enzyme described in this study and one copper-containing nitrite reductase, have been reported (4, 9, 20). In subunit composition, the P. aerophilum nitrate reductase appears to be most similar to the Haloferax volcanii enzyme, which also consists of three subunits with molecular masses of 100, 60, and 31 kDa (4). Like the P. aerophilum enzyme, the H. volcanii nitrate reductase is membrane bound. In contrast, soluble nitrate reductases were purified from Haloferax denitrificans, Haloferax mediterranei, and Haloarcula marismortui (2, 9, 20). It is possible that an additional protein exists that associates these soluble nitrate reductases with the cytoplasmic membrane. The Haloarcula marismortui enzyme appears to differ from all the other enzymes in that it comprises a homotetrameric complex composed of a 63-kDa polypeptide (20). All nitrate reductases from halophilic archaea except the enzyme from H. denitrificans require high salt concentrations for activity and structural stability (4). It is interesting that the presence of increasing salt concentrations shifts the temperature optimum for activity of the H. mediterranei enzyme to a higher value (2). Thus, temperature optima for nitrate reduction of 59 and 89°C were assayed in the presence of 0.6 and 3.2 M NaCl, respectively.
In conclusion, the nitrate reductase isolated from P. aerophilum is a molybdenum-containing enzyme with unusually high specific activity. From an evolutionary point of view, the enzyme from P. aerophilum is the oldest nitrate reductase characterized thus far and argues for the presence of a heterotrimeric enzyme in the last common-ancestor group of microbes.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Science Foundation (MCB-9631006). Some of the recent work was also supported by grant MCB-0091351 from the National Science Foundation.
REFERENCES
- 1.Afshar S, Kim C, Monbouquette H G, Schröder I. Effect of tungstate on nitrate reduction by the hyperthermophilic archaeon Pyrobaculum aerophilum. Appl Environ Microbiol. 1998;64:3004–3008. doi: 10.1128/aem.64.8.3004-3008.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez-Ossorio M, Muriana F J G, de la Rosa F F, Relimpio A M. Purification and characterization of nitrate reductase from the halophile archaebacterium Haloferax mediterranei. Z Naturforsch. 1992;47:670–676. doi: 10.1042/bj2780149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berks B C, Ferguson S J, Moir J W B, Richardson D J. Enzymes and associated electron transport systems that catalyze the respiratory reduction of nitrogen oxides and oxyanions. Biochim Biophys Acta. 1995;1232:97–173. doi: 10.1016/0005-2728(95)00092-5. [DOI] [PubMed] [Google Scholar]
- 4.Bickel-Sankötter S, Ufer M. Properties of a dissimilatory nitrate reductase from the halophilic archaeon Haloferax volcanii. Z Naturforsch. 1995;50:365–372. [Google Scholar]
- 5.Blasco F, Iobbi C, Ratouchniak J, Bonnefoy V, Chippaux M. Nitrate reductases of Escherichia coli: sequence of the second nitrate reductase and comparison to that encoded by the narGHIJ operon. Mol Gen Genet. 1990;222:104–111. doi: 10.1007/BF00283030. [DOI] [PubMed] [Google Scholar]
- 6.Bluemle S, Zumft W G. Respiratory nitrate reductase from denitrifying Pseudomonas stuzeri, purification, properties and target of proteolysis. Biochim Biophys Acta. 1991;1057:102–108. [Google Scholar]
- 7.Craske A L, Ferguson S J. The respiratory nitrate reductase from Paracoccus denitrificans. Eur J Biochem. 1986;158:429–436. doi: 10.1111/j.1432-1033.1986.tb09771.x. [DOI] [PubMed] [Google Scholar]
- 8.Enoch H G, Lester R L. Effects of molybdate, tungstate, and selenium compounds on formate dehydrogenase and other enzyme systems in Escherichia coli. J Bacteriol. 1972;110:1032–1040. doi: 10.1128/jb.110.3.1032-1040.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hochstein L I, Lang F. Purification and properties of a dissimilatory nitrate reductase from Haloferax denitrificans. Arch Biochem Biophys. 1991;288:380–385. doi: 10.1016/0003-9861(91)90210-a. [DOI] [PubMed] [Google Scholar]
- 10.Inatomi K L, Hochstein L I. The purification and properties of a copper nitrite reductase from Haloferax denitrificans. Curr Microbiol. 1996;32:72–76. [Google Scholar]
- 11.Iobbi-Nivol C, Santini C, Blasco F, Giordano G. Purification and further characterization of the second nitrate reductase of Escherichia coli K12. Eur J Biochem. 1990;188:679–687. doi: 10.1111/j.1432-1033.1990.tb15450.x. [DOI] [PubMed] [Google Scholar]
- 12.Ishizuka M, Toraya T, Fukui S. Purification, properties and limited proteolysis of nitrate reductase from Pseudomonas denitrificans. Biochim Biophys Acta. 1984;786:133–143. [Google Scholar]
- 13.Jones C W, Poole R K. The analysis of cytochromes. Methods Microbiol. 1985;18:285–327. [Google Scholar]
- 14.Klenitz A, Adams M W W. Tungsten in biological systems. FEMS Microbiol Rev. 1996;18:5–63. doi: 10.1016/0168-6445(95)00025-9. [DOI] [PubMed] [Google Scholar]
- 15.Kujo C, Ohshima T. Enzymological characteristics of the hyperthermostable NAD-dependent glutamate dehydrogenase from the archaeon Pyrobaculum islandicum and effects of denaturants and organic solvents. Appl Environ Microbiol. 1998;64:2152–2157. doi: 10.1128/aem.64.6.2152-2157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morpeth F F, Boxer D H. Kinetic analysis of respiratory nitrate reductase from Escherichia coli K12. Biochem. 1985;24:40–46. doi: 10.1021/bi00322a007. [DOI] [PubMed] [Google Scholar]
- 17.Schröder I, Rech S, Krafft T, Macy J M. Purification and characterization of the selenate reductase from Thauera selenatis. J Biol Chem. 1997;272:23765–23768. doi: 10.1074/jbc.272.38.23765. [DOI] [PubMed] [Google Scholar]
- 18.Völki P, Huber R, Drobner E, Rachel R, Burggraf S, Trincone A, Stetter K O. Pyrobaculum aerophilum sp. nov., a novel nitrate reducing hyperthermophilic archaeum. Appl Environ Microbiol. 1993;59:2918–2926. doi: 10.1128/aem.59.9.2918-2926.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vorholt J A, Hafenbradl D, Stetter K O, Thauer R K. Pathways of autotrophic CO2 fixation and of dissimilatory nitrate reduction to N2O in Ferroglobus placidus. Arch Microbiol. 1997;167:19–23. doi: 10.1007/s002030050411. [DOI] [PubMed] [Google Scholar]
- 20.Yoshimatsu K, Sakurai T, Fujiwara T. Purification and characterization of dissimilatory nitrate reductase from a denitrifying halophilic archaeon, Haloarcula marismortui. FEBS Lett. 2000;470:216–220. doi: 10.1016/s0014-5793(00)01321-1. [DOI] [PubMed] [Google Scholar]
- 21.Zumft W G. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]