Abstract
A sequenced collection of plasmid-borne random fusions of Escherichia coli DNA to a Photorhabdus luminescens luxCDABE reporter was used as a starting point to select a set of 689 nonredundant functional gene fusions. This group, called LuxArray 1.0, represented 27% of the predicted transcriptional units in E. coli. High-density printing of the LuxArray 1.0 reporter strains to membranes on agar plates was used for simultaneous reporter gene assays of gene expression. The cellular response to nalidixic acid perturbation was analyzed using this format. As expected, fusions to promoters of LexA-controlled SOS-responsive genes dinG, dinB, uvrA, and ydjM were found to be upregulated in the presence of nalidixic acid. In addition, six fusions to genes not previously known to be induced by nalidixic acid were also reproducibly upregulated. The responses of two of these, fusions to oraA and yigN, were induced in a LexA-dependent manner by both nalidixic acid and mitomycin C, identifying these as members of the LexA regulon. The responses of the other four were neither induced by mitomycin C nor dependent on lexA function. Thus, the promoters of ycgH, intG, rihC, and a putative operon consisting of lpxA, lpxB, rnhB, and dnaE were not generally DNA damage responsive and represent a more specific response to nalidixic acid. These results demonstrate that cellular arrays of reporter gene fusions are an important alternative to DNA arrays for genomewide transcriptional analyses.
Reporter gene fusions have been widely and successfully used to monitor gene expression in microbes, leading to important fundamental discoveries (35) and numerous applications (19). The facile methods for generating (4) and screening (10, 17, 51) such fusions have allowed genomewide surveys of gene expression in a response to a variety of stimuli. However, typically only the gene fusions having the desired property are identified in such surveys. With the availability of complete genomic sequences for many microbes, use of large sets of precisely defined reporter gene fusions is practical. Recently, defined sets of transcriptional gene fusions using gfp as a reporter in Saccharomyces cerevisiae (9) and luxCDABE (46) as a reporter in Escherichia coli have been described. The advantages of easily assayed reporter end products, such as fluorescence or light production, become increasingly critical for high-density analyses. For bacteria, bioluminescence as a reporter of gene expression is particularly useful because of the sensitivity and large dynamic range (5). Furthermore, use of the five-gene luxCDABE operon allows facile monitoring of kinetic responses because all the components necessary for light production are present in the cell, thus obviating the need for cell lysis and substrate addition (21).
Cellular arrays of reporter genes can be considered an alternative, complementary approach to other currently used genomewide transcriptional analyses such as those involving DNA arrays. Despite the success and broad use of DNA arrays, there are limitations, such as artifacts from RNA isolation (38) and cross hybridization (31). Here we describe a new solid-phase assay system consisting of 689 reporter strains, each containing a different, precisely sequenced fusion. The reporter strains were chosen from a collection of random, genomewide luxCDABE gene fusions in E. coli that had been sequenced to determine the identity and orientation of the genomic fragment upstream of the reporter genes; this collection includes representative members of several global stress response regulons and thus mirrors the E. coli transcriptional wiring diagram (46). The essence of this assay is to collect an image of the signal generated from reporter constructs arrayed in high density, such that the signal intensity can be subsequently quantified. Thus, bioluminescence from a high-density array of luxCDABE gene fusions in E. coli was measured by creating an image of the entire reporter array and quantitating the pixel density. Differences in the pixel density measurements in the presence or absence of a perturbation defined gene expression alterations.
The utility of this solid-phase cellular array was demonstrated by detection of responses to nalidixic acid, an inhibitor of DNA gyrase. Treatment of E. coli with this compound is known to result in selective transcriptional upregulation. The predominant response to nalidixic acid treatment is induction of the LexA-controlled SOS response (49). In addition to the SOS response, upregulation of the ς32-controlled heat shock response (18, 40, 45), increased activity of promoters responsive to DNA supercoiling (25, 37), increased expression of emr genes (20), and other responses (13, 36) are induced by nalidixic acid treatment. Accordingly, upregulation of gene fusions in two classes, fusions to genes of the LexA regulon and fusions to genes that are not part of the SOS response, was expected and was found. The genes included several novel nalidixic acid-upregulated genes, some of which were demonstrated to be members of the LexA regulon.
MATERIALS AND METHODS
E. coli strains, genetic nomenclature, growth medium, and chemicals.
The construction (42), sequencing, and characterization (46) of a collection of random, 1.8-kbp-average-size E. coli genomic fragments upstream of Photorhabdus luminescens luxCDABE in moderate-copy-number plasmid pDEW201 (44) have been described. The host strain for transformants in this collection is E. coli DPD1675 (ilvB2101 ara thi Δ[proAB-lac] tolC::miniTn10). The list of the 689 transcriptional fusions between predicted E. coli promoters and the luxCDABE operon (described in Results) is available upon request. Individual strains from this set are available to members of the scientific community for noncommercial purposes by contacting the corresponding author. Strain and plasmid names, in addition to the lux clone identification number, were assigned for strains containing gene fusions that were reproducibly upregulated by nalidixic acid, as shown in Table 1. Plasmid DNA isolated using a QiaPrep Spin kit (Qiagen Corp.) was used for transformation of E. coli DM800 (F− metA28 lacY1 or lacZ4 thi-1 xyl-5 or xyl-7 galK2 tsx-6) and otherwise isogenic lexA1 strain DM803 (24), selecting for ampicillin resistance.
TABLE 1.
E. coli strains containing nalidixic acid-upregulated gene fusions
| Strain | Lux identification | Plasmid | Gene fusion structure |
|---|---|---|---|
| DPD2354 | lux-a.pk033.c5 | pDEW630 | ′yniC ydjM-luxCDABE |
| DPD2248 | lux-a.pk024.f5 | pDEW235 | ′rhlE ybiA dinG′-luxCDABE |
| DPD2249 | lux-a.pk055.a3 | pDEW236 | fhiA′ mbhA dinB′-luxCDABE |
| DPD2253 | lux-a.pk001.b6 | pDEW240 | yjc′ yjcB ssb uvrA′-luxCDABE |
| DPD2345 | lux-a.pk015.d6 | pDEW621 | ′ycg ycgH′-luxCDABE |
| DPD2347 | lux-a.pk019.g1 | pDEW623 | yedL yedN yedM intG′-luxCDABE |
| DPD2362 | lux-a.pk061.c3 | pDEW638 | ′fabZ lpxA lpxB′-luxCDABE |
| DPD2361 | lux-a.pk058.f5 | pDEW637 | ′mltB ygaD recA oraA′-luxCDABE |
| DPD2353 | lux-a.pk031.e7 | pDEW629 | ′lytB rihC′-luxCDABE |
| DPD2358 | lux-a.pk046.f11 | pDEW634 | ′udp yigN′-luxCDABE |
Common, mnemonic names for E. coli genes were used. Where no common name had been given, the Rudd systematic nomenclature, found at http: //bmb.med.miami.edu/ecogene/ecoweb/ (34), was used.
Luria-Bertani (LB) medium (23) was used for all experiments and was supplemented with 100 μg of ampicillin/ml. A stock solution of 20 mg of nalidixic acid (Sigma Chemical Co.)/ml in 1 M NaOH was diluted to appropriate concentrations into LB medium. Likewise, a stock solution of 1 mg of mitomycin C (Sigma Chemical Co.)/ml in water was used.
Preparation of reporter arrays.
Duplicate cultures of the E. coli strains in the LuxArray were grown overnight at 37°C in 40 μl of LB medium supplemented with 100 μg of ampicillin/ml in a set of 16 96-well plates. These cultures were used as the cell source to manufacture the arrays. Porous nylon membranes (8 by 12 cm; Biodyne B; Nunc) were sterilized by UV illumination for 10 min and then placed in contact with solid LB growth media in a culture dish (50 ml; OmniTray; Nunc) that had been prewarmed to 37°C. Printing of 1,536 spots in four-by-four subarrays was accomplished using a BioMek 2000 (Beckman Coulter) equipped with a high-density replication tool. Sterilization between transfers was accomplished by soaking the pins successively in 0.2% sodium dodecyl sulfate in water, sterile water, and 70% ethanol. After sterilization, the pins were air dried prior to the next transfer. The E. coli reporter strains and control strains were printed in triplicate for each treatment.
Following the printing, the arrays were incubated for 6 h at 37°C to allow the cells to become actively growing and to increase the bioluminescent signal. Then the membranes were moved to new, prewarmed plates containing either LB media or LB media supplemented with 5 μg of nalidixic acid/ml. These plates were replaced at 37°C to continue growing. Sixteen-bit gray-scale tagged-image file format (TIFF) images were collected for each array every 2 h from 0 to 8 h after relocation using a cooled charge-coupled device camera (FluorChem 8000; f0.85 lens; AlphaInnotech) with constant focal plane, magnification, and integration time (2 min) empirically determined to maintain the signal within saturation limits. An additional image was collected after overnight growth of all plates but was not analyzed.
Data analysis from reporter arrays.
Spot intensity of each image was determined using ArrayVision (Imaging Research, Toronto, Canada), and the resultant pixel density measurements were imported into a template with identifiers for each spot. The background signal, which results from cross illumination of neighboring spots, was estimated by finding the median of the 24 spots containing a strain with the parental plasmid on each of the triplicate arrays followed by calculation of the average of the three medians. The background signal at each of the time points was subtracted from each reporter strain measurement at the corresponding time point. All negative numbers were converted to zero. The average signal for each of the triplicate spots was calculated.
Data normalization to correct for growth during the course of the experiments with the control and nalidixic acid-treated arrays was based on the assumption that the bioluminescence increases over time for the vast majority of the reporter strains were proportional to the increases in cell density. Likewise, decreases in cellular metabolism resulting in reduced bioluminescence in the chemically treated plate would be generally reflected as decreased signals from the vast majority of the reporter strains. Thus, the sum of the averaged, background-subtracted signals of the array for each treatment at each time point was assumed to correlate with total cell density and overall bioluminescent activity in the array. A normalization factor was calculated as follows: normalization factor = total array signal (time zero, LB control)/total array signal (time x, condition y). Each measurement was multiplied by this normalization factor to yield a normalized signal. The ratio of each nalidixic acid-treated spot to the corresponding control spot was calculated using this normalized data.
Kinetic analysis of bioluminescence using liquid cultures.
Kinetic analyses of bioluminescent response to chemicals was done as previously described (42, 43), except that a Luminoskan Ascent microplate luminometer (Labsystems) was used to measure light production at 37°C. Briefly, fresh overnight cultures for each strain grown in LB medium supplemented with ampicillin were diluted into LB medium and grown to mid-exponential phase at 37°C. These actively growing cultures were divided at the initiation of chemical treatment in 100 μl (total volume) in the wells of white, 96-well microplates (Microlite; Dynex). Pipetting the cell cultures was done rapidly at room temperature, after which the microplate was placed immediately into a prewarmed luminometer. Nonetheless, some cooling of the cell cultures occurred during pipetting, leading to an initial rise in the bioluminescence of both the control and chemically treated cultures as they warmed toward the temperature optimum of the Lux proteins. A typical liquid culture experiment used seven concentrations of the chemical of interest such that the range included a sublethal dose at which stress responses were induced without severe toxicity that would inhibit the light production reactions (41). Nevertheless, some growth inhibition may occur, and thus the response ratio, which is the bioluminescence of the chemically treated culture divided by the bioluminescence of the control at each time point (41), represents a minimum estimate of gene expression responses because it does not include a correction for reduced cell density in the presence of the added chemical.
RESULTS
Selection of a maximal nonredundant set of lux gene fusions.
A random luxCDABE gene fusion collection of 4,988 plasmid-borne gene fusions, each with boundary information relative to the E. coli genome and the orientation relative to the lux operon (46) was the starting point from which an optimum subset was selected. A functional construct was defined as one consisting of a genomic fragment encompassing a promoter adjacent to the promoterless lux operon in an orientation that causes transcription initiated at the promoter to proceed into and through the lux operon. Therefore, to identify the functional subset of the collection, Perl scripts were used to computationally filter the list of gene fusions first for functionality and second for redundancy. A list of definitions of documented and predicted operons (39) was used to define genomic coordinates of the operons as the translational start codon position of the first open reading frame (ORF) in the operon and the translational stop codon position of the last ORF in the operon. Additional information included the strand on which the operon is contained (direction of transcription) and gene names. The lux gene fusions were filtered computationally using the following criteria to select functional transcriptional fusions: (i) the genomic fragment must start more than 50 bp upstream of the start codon of the first ORF and end anywhere between the start codon of the first ORF in the operon and the stop codon of the last ORF in the operon, thereby eliminating the occurrence of a transcriptional stop signal in the construct between the promoter and the lux operon; (ii) the promoter contained in the genomic fragment must be orientated such that it directs transcription into the lux operon. Finally, when more then one gene fusion fit the criteria for a single operon, only the construct containing the genomic fragment representing the greatest amount of upstream sequence was retained, thereby eliminating redundancy. The resulting 689 selected gene fusions represent 27% of the 2,584 known and predicted transcriptional units in the E. coli genome (3, 39). The working hypothesis based on the predicted operon structures is that bioluminescence from cells containing each of the selected gene fusions reports on the expression of the operon to which luxCDABE is joined.
Individual cultures of strains containing the 689 identified gene fusions were rearrayed from the 90 original culture plates to create a set of 16 96-well microplates containing each of the identified fusions in duplicate. Also included were multiple replicates of two control strains, one bearing a lacZYA promoter fusion and another containing the parental plasmid, pDEW201. These 16 plates were used to generate the high-density cellular arrays designated LuxArray 1.0, as described in Materials and Methods, for use in subsequent analyses. The identity and location of each culture in the resultant array are available upon request.
Initial characterization of the LuxArray.
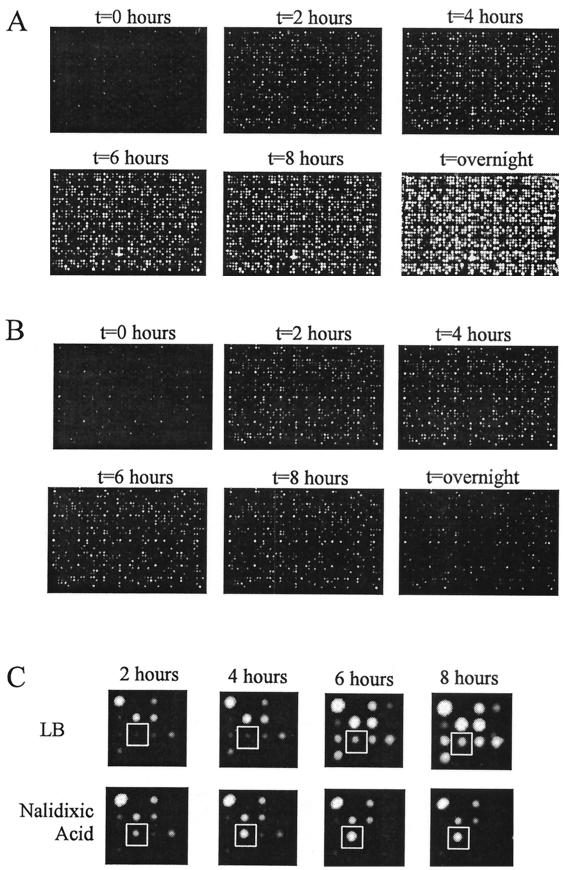
The bioluminescence over time from a LuxArray consisting of 1,536 spots arrayed at a density of 16 spots/cm2 of medium is shown in Fig. 1A. Each array in Fig. 1 was a side-by-side replicate, such that the left half was identical to the right half. The intensity of bioluminescence at each location reflected the relative activity level of each individual promoter controlling expression of the luxCDABE reporter. The increased bioluminescent signal at each spot location with time was, in general, due primarily to cell growth.
FIG. 1.
Images of duplicate LuxArray 1.0 cellular bioluminescent reporter arrays. Following the spotting of the E. coli strains containing reporter gene fusions onto membranes on LB agar plates and growth for 6 h, the membranes were moved to LB medium plates (A) or LB medium plates containing 5 μg of nalidixic acid/ml (B). The images were taken as described in Materials and Methods immediately after moving the membrane and subsequently at 2, 4, 6, and 8 h and after overnight incubation. A magnification of the 16 spots in the D-4 primary location from panels A and B is shown in panel C. The spot at the secondary location of row 3, column 2, containing cells with upregulated gene fusion dinB-luxCDABE is boxed.
The growth rate of individual cultures spotted in the array was evaluated to differentiate between strain-dependent and system-dependent sources of signal variability. Cellular arrays were generated in triplicate on different days using LB agar media containing 10 μg of tetrazolium blue/ml. The product generated when live cells reduce tetrazolium blue is an insoluble blue precipitate. This greatly increased the contrast between the cells and the media, simplifying direct imaging of the cells by normal light. For each of the triplicate experiments, each spot was visually scored to determine the size of the growth generated during 8 h of incubation at 37°C (data not shown). Variability was clearly strain specific and consistent from day to day. As the majority of proposed analyses are relative measurements and as interstrain comparisons are unlikely, this type of growth variability does not affect the applicability or robustness of the overall assay system. No further attempts to determine the source of the variability were made; however the variability can be assumed to be a result of the plasmid constructs carried by the individual strains.
Identification of nalidixic acid-responsive gene fusions using LuxArray.
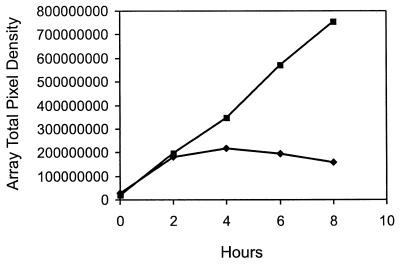
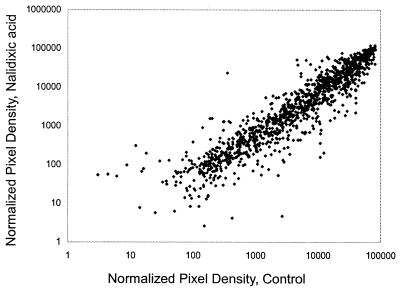
The antibiotic nalidixic acid, an inhibitor of DNA gyrase known to be an effective inducer of the SOS DNA damage stress response (49), was used to test the utility of this array for detecting changes in gene expression after perturbation. Figure 1B displays the bioluminescence over time of the LuxArray treated with 5 μg of nalidixic acid/ml. In contrast to that of the control array (Fig. 1A), the bioluminescence levels of the majority of the spots did not increase substantially after 2 h of nalidixic acid treatment (Fig. 1B). This general inhibition of bioluminescence increase was due to growth inhibition or metabolic inhibition, which lowers cellular ATP levels and reducing power required for the bioluminescence reactions. This inhibition of bioluminescence increase was also evident from the sum of all bioluminescent signals in the array plotted as a function of time (Fig. 2). Thus, each bioluminescence measurement was normalized to correct for the lower cellular density in the nalidixic acid-treated array (see Materials and Methods). A scatter plot of this normalized data comparing the nalidixic acid-treated spots to untreated spots at the 4-h time point is shown in Fig. 3. It is evident that the signals from vast majority of the spots in the array were not affected by nalidixic acid treatment but that spots with increased signal in response to nalidixic acid, as well as those with decreased signal, were present. Also evident from this scatter plot is that there was a wide range of signal strengths and that the data points for low signals showed more scatter than those for high signals.
FIG. 2.
Total light production from LuxArray 1.0 over time. The averaged pixel density of each spot on triplicate membranes was summed over the entire array and plotted at each time point for each condition. Squares, control LB plates; diamonds, LB plates containing 5 μg of nalidixic acid/ml.
FIG. 3.
Scatter plot of showing the relationship of normalized signal intensity with and without nalidixic acid treatment at the 4-h time point. The data from all signals greater than 1.0 for both treatments are plotted.
To identify nalidixic acid-induced gene expression responses, ratios of the normalized pixel intensity data in the presence of nalidixic acid to that in its absence were calculated for each of the duplicate spots formed by independent cultures. Putative nalidixic acid-upregulated gene fusions were selected as those for which these ratios in both independent, duplicate cultures were at least 2.0 at both the 2- and 4-h time points. Twelve gene fusions were identified by these criteria (Table 2). An example of the original image of one of the nalidixic acid-upregulated gene fusions is shown in Fig. 1C.
TABLE 2.
Nalidixic acid responses on solida and in liquid media
| Fusion to gene or operon | Lux identification | Solid response ratio at:
|
Liquid maximal response concn (μg/ml) | Liquid response ratio at 2 h | Liquid concn range >1.5-fold upb (μg/ml) | |||
|---|---|---|---|---|---|---|---|---|
| 2 h | 4 h | 6 h | 8 h | |||||
| Known SOS regulon members | ||||||||
| ydjM | lux-a.pk033.c5 | 3.7 | 4.3 | 3.2 | 2.6 | 5 | 3.7 | 1.2–80 |
| 2.1 | 3.1 | 3.3 | 2.5 | |||||
| dinG ybiB | lux-a.pk024.f5 | 3.4 | 4.2 | 3.1 | 2.6 | 5 | 4.2 | 1.2–40 |
| 2.5 | 3.7 | 3.5 | 2.9 | |||||
| dinB | lux-a.pk055.a3 | 3.1 | 5.1 | 4.2 | 3.1 | 5 | 7.6 | 1.2–80 |
| 5.5 | 11.0 | 8.1 | 6.3 | |||||
| uvrA | lux-a.pk001.b6 | 2.5 | 2.3 | 1.6 | 1.0 | ndc | nd | nd |
| 2.9 | 2.4 | 1.1 | 0.6 | |||||
| Nalidixic acid upregulated on solid LuxArray and in liquid culture | ||||||||
| ycgH | lux-a.pk015.d6 | 2.3 | 6.4 | 2.7 | 0.6 | 20 | 2.7 | 1.2–40 |
| 2.2 | 4.1 | 1.9 | 0.8 | |||||
| intG | lux-a.pk019.g1 | 2.5 | 7.7 | 9.7 | 7.1 | 10 | 2.3 | 1.2–40 |
| 4.4 | 8.2 | 15.5 | 10.2 | |||||
| lpxA lpxB rnhB dnaE | lux-a.pk061.c3 | 3.5 | 3.1 | 2.8 | 1.7 | 2.5 | 1.8 | 1.2–5.0 |
| 2.7 | 3.5 | 3.0 | 1.9 | |||||
| oraA | lux-a.pk058.f5 | 5.1 | 7.9 | 5.9 | 3.9 | 10 | 7.2 | 1.2–80 |
| 4.7 | 7.7 | 5.6 | 3.8 | |||||
| rihC | lux-a.pk031.e7 | 2.1 | 2.0 | 1.3 | 1.0 | 2.5 | 1.9 | 1.2–5.0 |
| 2.0 | 2.1 | 1.8 | 1.5 | |||||
| yigN | lux-a.pk046.f11 | 2.1 | 2.5 | 0.3 | 0.1 | 1.2 | 2.5 | 1.2–20 |
| 7.1 | 4.3 | 2.5 | 2.2 | |||||
| Nalidixic acid upregulation not reproduced in liquid culture | ||||||||
| frvR frvX frvB frvA | lux-a.pk046.e6 | 2.8 | 2.3 | 0.8 | 0.5 | 20 | 1.5 | None |
| 2.1 | 2.3 | 2.0 | 1.7 | |||||
| yfhJ fdx hscA yfhE | lux-a.pk0019.g2 | 2.1 | 2.0 | 1.7 | 1.4 | 1.2 | 1.0 | None |
| 2.1 | 2.2 | 1.5 | 1.3 | |||||
The responses in the solid LuxArray experiment are given for each of the two independent spots.
Nalidixic acid concentration ranges in cultures with response ratios of 1.5 or greater.
nd, not done..
These upregulated gene fusions included four characterized members of the LexA regulon, ydjM, dinG, dinB, and uvrA (11, 50). In addition, eight fusions to promoters not previously known to be upregulated by nalidixic acid treatment were included. The DNA sequence of plasmid DNA isolated from each of these 12 cultures confirmed the identity of each inserted DNA. The LuxArray contains a total of seven gene fusions to LexA-regulated operons, all of which would be expected to be upregulated by nalidixic acid. Three were, thus, scored falsely as negatives. An examination of the data showed that two of these false negatives were reproducibly upregulated at modest levels: a fusion to uvrD was upregulated by 1.6- and 1.9-fold after 4 h of nalidixic acid treatment, while one to ruvA was upregulated 1.7- and 2.1-fold at that time point. The third had inconsistent responses; the ratios of bioluminescence from the nalidixic acid-treated culture to that in the untreated control for the dinF-lux fusion were found to be 4.2 in one of the duplicate spots and 0.7 in the other.
Validation of nalidixic acid-upregulated gene fusions with cultures in liquid medium.
Each of the eight newly identified nalidixic acid-upregulated gene fusions and three of the known SOS gene fusions were tested in liquid medium using a range of nalidixic acid concentrations (80 μg/ml in twofold dilutions to 1.2 μg/ml). Table 2 shows the results expressed as ratios of the signal from the nalidixic acid-treated cultures to that from the untreated control at 2 h without correction for growth inhibition caused by nalidixic acid. The concentration that yielded the maximal response and the range of nalidixic acid concentrations that resulted in response ratios of 1.5 or greater are also given. By the criteria of a maximal response ratio of at least 1.8 and of more than one concentration tested yielding response ratios of at least 1.5, six of the eight putative novel nalidixic acid-upregulated gene fusions were reproduced in liquid medium.
Mitomycin C responses and effect of a noninducible lexA allele.
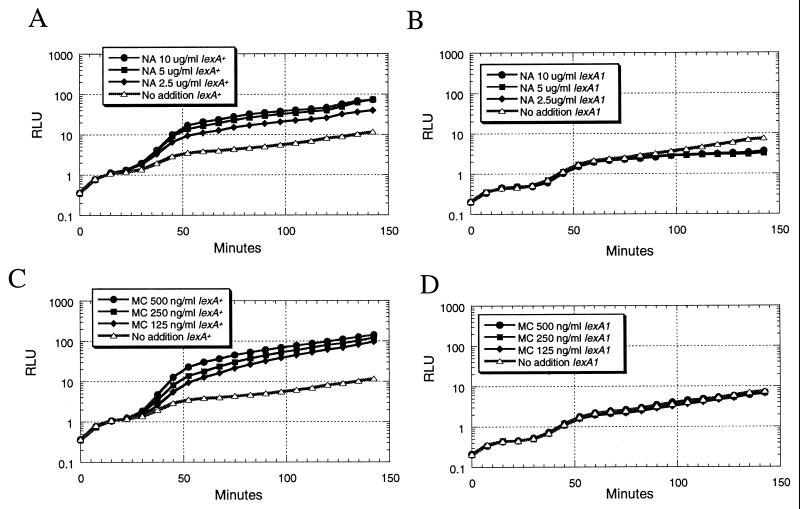
Mitomycin C, a compound that damages DNA by intercalation and covalent modification, was used to determine if these newly discovered nalidixic acid-upregulated gene fusions were generally responsive to DNA damage. In addition, the effect of a lexA1 mutation, which renders the LexA repressor noncleavable by the RecA coprotease (24), was tested. The expectation was that expression of SOS-responsive gene fusions controlled by LexA-regulated promoters would be induced by both nalidixic acid and mitomycin C in a manner that is dependent on normal LexA function. As shown in Fig. 4, the expression of the gene fusion to yigN was markedly induced by both nalidixic acid and mitomycin C in the lexA+ host. The kinetics of this response, consisting of a 20-min lag time, where the bioluminescence of the nalidixic acid-treated cultures was not different from that of the untreated control, followed by a dose-dependent nalidixic acid-mediated increase in light production, were similar to the those of the response of other LexA-regulated gene fusions (46). This upregulation of gene expression was completely eliminated when the same plasmid was put into an otherwise isogenic lexA1 host strain (Fig. 4). Likewise, the expression of a gene fusion to oraA was also induced by both mitomycin C and nalidixic acid only in the lexA+ host (Table 3). In contrast, four other nalidixic acid-responsive gene fusions were not upregulated by mitomycin C in the lexA+ host, suggesting that they are not generally DNA damage responsive but rather are more specifically responsive to nalidixic acid. Consistent with this observation, the upregulation by nalidixic acid, which was weaker in general than the LexA-controlled responses, was still present in the lexA1 mutant host (Table 3). Unexpectedly, the bioluminescence of these four gene fusions was slightly upregulated in response to mitomycin C in the lexA1 mutant host. Also shown in Table 3 are the levels of light production in the absence of chemical treatment. These levels, which reflect promoter strength, were largely unaffected by the lexA1 mutation. As expected, expression of ςS-controlled gene fusion yciG-lux (42), which was not expected to respond to DNA damage, was not induced by either nalidixic acid or mitomycin C (Table 3).
FIG. 4.
Response of the yigN-luxCDABE gene fusion to nalidixic acid (NA) and mitomycin C (MC) in lexA+ and lexA1 host strains. Actively growing cultures in LB medium were mixed with chemicals at time zero, and light production was measured in a Luminoskan Ascent microplate luminometer. (A and C) Plasmid pDEW634, containing the yigN-luxCDABE gene fusion, in E. coli strain DM800. (B and D) Plasmid pDEW634 in E. coli strain DM803. RLU, relative light units.
TABLE 3.
Nalidixic acid and mitomycin C responses in lexA+ and lexA1 host strains
| Fusion to gene or operon | Host straina | Response ratio for:
|
||
|---|---|---|---|---|
| Nalidixic acid (2 h, 10 μg/ml) | Mitomycin C (2 h, 250 ng/ml) | RLU at 2 h, no chemical addedb | ||
| Known LexA regulon members | ||||
| ydjM | lexA+ | 5.2 | 9.0 | 19.58 |
| lexA1 | 0.4 | 0.9 | 12.91 | |
| dinG ybiB | lexA+ | 4.0 | 5.0 | 15.88 |
| lexA1 | 1.0 | 1.1 | 12.96 | |
| Nalidixic acid upregulated on the solid LuxArray and in liquid culturec | ||||
| oraA | lexA+ | 11.1 | 15.3 | 2.69 |
| lexA1 | 1.1 | 1.2 | 2.18 | |
| yigN | lexA+ | 5.7 | 9.8 | 8.08 |
| lexA1 | 0.6 | 0.9 | 5.30 | |
| ycgH | lexA+ | 3.2 | 1.1 | 0.90 |
| lexA1 | 2.3 | 1.7 | 0.64 | |
| intG | lexA+ | 3.2 | 1.2 | 0.08 |
| lexA1 | 1.6 | 2.5 | 0.13 | |
| lpxA lpxB rnhB dnaE | lexA+ | 2.9 | 1.1 | 0.29 |
| lexA1 | 2.4 | 1.3 | 0.29 | |
| rihC | lexA+ | 1.5 | 1.0 | 8.81 |
| lexA1 | 1.2 | 1.0 | 7.28 | |
| Non-DNA damage-responsive control | ||||
| yciG | lexA+ | 0.3 | 0.6 | 0.31 |
| lexA1 | 0.7 | 0.6 | 0.33 | |
The lexA+ host strain was E. coli DM800, while the lexA1 host strain was E. coli DM803.
The light production in the absence of perturbation reflects the relative basal levels of promoter activity. RLU, relative light units.
Fusions to oraA and yigN were LexA dependent and generally DNA damage responsive, while those to ycgH, intG, the lpxA lpxB rnhB dnaE operon, and rihC were nalidixic acid upregulated and not generally DNA damage inducible.
DISCUSSION
A high-density cellular array of transcriptional reporter gene fusions.
LuxArray 1.0 consists of a set of 689 E. coli strains, each containing a reporter gene fusion to a different E. coli promoter element. Overall, 27% of the known or predicted transcriptional units in E. coli are represented in this array. It has previously been shown that several major stress-responsive regulons are each represented by one or more gene fusions in this collection (46). Thus, it is reasonable to assume that activation of most other major regulatory circuits will be reported by gene fusions in this array. Although the LuxArray does not afford a complete analysis of transcriptional alterations in E. coli, it provides a representative view of transcriptional changes because the set of strains originated in a sequenced collection of random gene fusions. Given that a larger collection of random gene fusions was culled using a compilation of known and predicted promoters in E. coli (39) as a reference and that predicting promoters and operons is not yet entirely accurate (15), the LuxArray may include some gene fusions to promoters that are not active despite predictions (46) and may have missed some promoters that were not predicted yet that are functional. Nevertheless, LuxArray 1.0 provides a genomewide collection that will yield a much more detailed and representative transcriptional pattern in response to perturbation than previously used panels of selected stress-responsive gene fusions (2, 27, 45).
The solid format incorporating growth on the surface of the membrane allowed the reporter assay to be moved between chemical perturbations as required. Experimental protocols often involve perturbations that prohibit long-term exposure due to cell death or other irreversible effects. The ability to move the entire array to new growth conditions allows a great variety of experimental schemes such as pulse or pulse-chase exposures, reversibility, and short-term kinetics studies. Furthermore, the solid format with a printing density of 16 spots/cm2 permitted all members of LuxArray 1.0 to be arrayed in duplicate on a single 8- by 12-cm membrane. Additional experiments (data not shown) demonstrated that up to 64 spots/cm2 could be printed and resolved. Discrete areas of growth did not overlap even at these high densities due to the self-limiting nature of nutrient availability in the media. At this maximal density, a single 8- by 12-cm array could contain gene fusions to all the transcriptional units in the E. coli genome, in duplicate, with capacity left for controls. However, the cross illumination from one spot to another during quantitation of pixel density during image analysis may limit the practical printing density.
Other methods for bioluminescence detection of multiple samples, such as those involving microplates, reduce cross illumination by using specifically designed microplates and microplate luminometers (48). An advantage of a cellular array of reporter genes is the flexibility to use other formats for signal detection. Accordingly, a liquid format for the LuxArray, in which the set of reporter strains are grown and assayed in a series of 96-well microplates, has also been implemented (T. K. Van Dyk, unpublished data). Although this format requires a greater number of liquid-handling manipulations than does the solid format, it allows the LuxArray analysis to be conducted using cultures that are more uniformly in a given growth stage. A further advantage of a cellular array is that facile follow-up experimentation with individual strains from the array to validate initial results and further characterize the responses is possible, as the work described here and elsewhere (46) demonstrates. Moreover, because the gene fusions in LuxArray 1.0 are plasmid borne, testing regulation of gene expression can be readily accomplished by isolation of plasmid DNA and transformation of regulatory mutant host strains (this work; 42, 46).
Bioluminescent reporter gene fusions have typically been used to detect induction of gene expression in response to a perturbation. Determining repression has been more difficult because perturbations that affect the metabolic capability of the cell result in decreased bioluminescence due to decreased growth or production of ATP and reducing power required to produce light. Thus, decreased promoter activity cannot be readily separated from this general “lights out” response for an individual reporter gene fusion. However, use of a large set of gene fusions, such as in the LuxArray, allows normalization of the signals to correct for decreased bioluminescence unrelated to promoter activity levels. As shown in Fig. 3, a subset of gene fusions that appear to be downregulated upon nalidixic acid treatment as well as those that appear to be upregulated can be distinguished following normalization of the data. The putative downregulated gene fusions were not further considered; rather the upregulated gene fusions were examined.
Responses to nalidixic acid perturbation.
Twelve putative upregulated gene fusions were identified following perturbation of the LuxArray with nalidixic acid treatment. Ten of these were confirmed by demonstrated upregulation in response to nalidixic acid in liquid medium (Table 2) (46). Two with differing responses on solid medium and in liquid medium may be false positives or may represent responses that only occur when the cultures are grown on solid medium, as has been observed for aluminum-activated gene expression in E. coli (14). Another formal possibility is that overexpression of a full-length ORF contained in the plasmid with the gene fusion conferred increased resistance to nalidixic acid. Thus growth to a greater extent than that for the majority of the spots in the array could give the appearance of a greater signal after normalization to correct for growth inhibition. This explanation is ruled out for the 10 gene fusions that were demonstrated to be upregulated in liquid medium where the bioluminescent signal was greater in the presence of nalidixic acid than in its absence without correction for growth inhibition. Of the 10 gene fusions that yielded upregulation by nalidixic acid in both solid and liquid formats, 4 were fusions to known LexA-regulated genes. Also included in the LuxArray were three additional gene fusions to known members of the LexA regulon that did not meet the established criteria to be scored as upregulated. This rate of false negatives, 43%, is similar to the rate from a DNA array analysis where 7 of 19 SOS genes, 37%, were not scored as upregulated following mitomycin C treatment (46). Improvement in these rates of false negatives by optimization of conditions is possible and desirable for both methods. Even so, since all methods are likely to have associated errors, the importance of alternative, independent methods of genomewide gene expression for increased reliability of transcriptional response analyses is underscored.
The six reproducibly upregulated gene fusions that were not previously known to be part of the LexA-controlled SOS response were also not previously known to be induced by nalidixic acid. These were categorized by responses to mitomycin C and the effect of a mutation that eliminates the ability of the LexA protein to be cleaved in response to DNA damage.
New LexA-regulated SOS genes.
Gene fusions to two genes not previously known to be members of the LexA-dependent SOS regulon were found to be upregulated by two DNA-damaging agents with different mechanisms of action in a LexA-dependent fashion (Table 3 and Fig. 4). These results suggest that yigN and oraA are new members of this regulon. In agreement with the results from the LuxArray analysis reported here, a very recent publication reports lexA-dependent upregulation of yigN and oraA transcription in response to UV light treatment as determined by DNA array (7). Conversely, these results conflict with another recent report in which a LexA binding site upstream of yigN was identified but no increase in mRNA formation upon mitomycin C treatment or regulation by LexA was observed (11). However, the substantial, lexA-dependent upregulation of a luxCDABE gene fusion to yigN observed in response to both mitomycin C treatment and nalidixic acid treatment (Fig. 4) as well as the data from Courcelle et al. (7) suggest that this gene, which encodes a conserved protein with two modules, both related to transcriptional repressors (32, 33), is in the LexA-controlled SOS regulon.
In contrast to what was found for yigN, there is no predicted LexA binding site immediately upstream of oraA (11). This gene, also known as recX, encodes a putative regulatory protein conserved in gram-negative and gram-positive bacteria and is often located downstream of recA (8). Cotranscription of recA and recX is found in Pseudomonas aeruginosa (16), Mycobacterium smegmatis (28), and Streptomyces lividans, where expression of the recA-recX transcript is induced by DNA damage (47). In Xanthomonas campestris pv. citri, recA and recX are thought to be transcribed from their own promoters but expression of both is induced by DNA damage (53). Thus, despite the prediction that recA and oraA constitute independent transcriptional units in E. coli (39), it is likely that the LexA-regulated recA promoter present in the oraA-luxCDABE gene fusion plasmid (Table 1) controls transcription of oraA. Again, the LuxArray result is in agreement with those found by DNA array analysis following UV treatment (7) and suggests that oraA is another member of the LexA-controlled SOS response.
Novel nalidixic acid-upregulated genes that are not generally DNA damage inducible.
Four gene fusions demonstrated a specificity of response to nalidixic acid compared with mitomycin C, thus suggesting that these are not part of the DNA damage-responsive SOS regulon. None of the promoters for these four transcriptional units are known to be controlled by ς32, nor were any of the known ς32-controlled gene fusions in the LuxArray found to be upregulated by nalidixic acid treatment. Thus, this format of gene expression analysis does not report on the induction of the heat shock response by nalidixic acid, which has a magnitude lower than that of the SOS response (40, 45). These four nalidixic acid-specific gene fusions are likely responding to other signals, such as levels of DNA supercoiling (22), which are decreased in plasmid DNA upon inhibition of DNA gyrase with nalidixic acid (13, 26). However, currently available information about expression of these transcriptional units sheds little light on possible mechanisms of nalidixic acid-mediated regulation.
Expression of rihC, which was formerly named yaaF and which encodes a ribonucleoside hydrolase, is under catabolite repression (29). The relatively high levels of expression of the luxCDABE gene fusion to rihC determined by measuring unstressed bioluminescent light production in a rich medium lacking glucose (Table 3) are consistent with this regulation. However, the observed response induced by nalidixic acid is not likely to be related to catabolite repression and thus remains to be defined. The lpx/dnaE gene cluster in E. coli has multiple promoters (30). Thus, the selected gene fusion, lux-a.pk061.c3, may contain such an internal promoter driving expression of luxCDABE. Our results that demonstrate low-level basal activity of this fusion (Table 3) are consistent with a minor role of this promoter in expression of these essential genes. The regulation of this promoter is not known; thus the mechanism of nalidixic acid upregulation remains unknown. The other two gene fusions in this class are to genes lacking known function or regulation. Thus, the mechanisms of activation of ycgH and intG by nalidixic acid are unknown. Interestingly, UV-induced DNA damage upregulated transcription of ycgH but not of intG, rihC, or a putative operon consisting of lpxA, lpxB, rnhB, and dnaE (7), thus suggesting that at least two different regulatory mechanisms are involved.
Although the mechanism of regulation for this class of nalidixic acid-responsive gene fusions is not known, their activation is a useful empirical signature of the nalidixic acid mode of action. Upregulation of these four gene fusions by nalidixic acid but not mitomycin C provides a characteristic fingerprint of the transcriptional responses induced by these compounds, which cause DNA damage by different mechanisms. Recently, such characteristic gene expression signatures in Haemophilus influenzae have been also demonstrated for two DNA gyrase inhibitors, novobiocin and ciprofloxacin, with differing mechanisms of inhibition (12).
Comparison of reporter gene array and DNA array technologies.
The results presented here demonstrate that a cellular array of reporter gene fusions can be used in a fashion analogous to that for DNA array hybridization assays to monitor transcriptional changes. The application of independent methods for genomewide transcriptional analysis is useful because there are advantages and disadvantages of each. An advantage of the current DNA array technology is the availability of comprehensive analysis of essentially all ORFs in several microorganisms (6, 31, 52, 54) due to the relative simplicity of their construction. Of course, comprehensive reporter arrays are also possible to construct by sequencing a larger collection of random gene fusions or by PCR amplification of promoter regions and subsequent cloning. However, not all applications require a comprehensive analysis. Thus, transcriptional fingerprints that distinguish chemicals with different modes of action are found with a representative subset of the genome, as shown here. DNA arrays are likely to be more useful than reporter gene arrays for genomewide transcription analyses of mutant strains because of the inconvenience in transferring a large set of reporter constructs to a mutant host. Implementation of both DNA arrays and reporter gene arrays requires a substantial amount of equipment; however, the automated liquid handling and camera required for the LuxArray are not specialized and thus do not need to be dedicated solely to this technology.
Cellular arrays of reporter genes are an important addition to methods of genomewide transcriptional analyses because they offer an alternative, independent method that overcomes some limitations in DNA array technology. One such limitation is the ability to distinguish the expression of closely related genes due to cross hybridization (31). Reporter arrays with a separate construct for each promoter do not face this limitation. Furthermore, analysis of RNA molecules with differential stability by DNA arrays can be problematic because of the RNA isolation steps required (1). The cellular array analysis that reports on promoter activity should not have this limitation, as all strains utilize the identical mRNA. Additionally, the requirement for RNA isolation and hybridization for each time point limits the feasibility of detailed kinetics time course analyses with DNA arrays. Thus, a significant advantage in using a bioluminescent reporter array is that kinetics analyses are readily accomplished by collecting as many images as desired of a single array over time.
ACKNOWLEDGMENTS
This work was supported by the DuPont Company Central Research and Development Department.
We thank Mario Chen and Michael Ramaker for assistance with Perl scripts and rearraying TCL code, Mary Jane Reeve for instruction on luminometer methods, Robert LaRossa for encouragement and critical reading of the manuscript, and Brooks Low for the gift of E. coli strains DM800 and DM803.
REFERENCES
- 1.Arfin S M, Long A D, Ito E T, Tolleri L, Riehle M M, Paegle E S, Hatfield G W. Global gene expression profiling in Escherichia coli K12: the effects of integration host factor. J Biol Chem. 2000;275:29672–29684. doi: 10.1074/jbc.M002247200. [DOI] [PubMed] [Google Scholar]
- 2.Belkin S, Smulski D R, Dadon S, Vollmer A C, Van Dyk T K, LaRossa R A. A panel of stress-responsive luminous bacteria for the detection of selected classes of toxicants. Water Res. 1997;31:3009–3016. [Google Scholar]
- 3.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 4.Casadaban M J, Cohen S N. Lactose genes fused to exogenous promoters in one step using a new-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci USA. 1980;76:4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatterjee J, Meighen E A. Biotechnological applications of bacterial bioluminescence (lux) genes. Photochem Photobiol. 1995;62:641–650. [Google Scholar]
- 6.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown P O, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 7.Courcelle J, Khodursky A, Peter B, Brown P O, Hanawalt P C. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Mot R, Schoofs G, Vanderleyden J. A putative regulatory gene downstream of recA is conserved in gram-negative and gram-positive bacteria. Nucleic Acids Res. 1994;22:1313–1314. doi: 10.1093/nar/22.7.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimster-Denk D, Rine J, Phillips J, Scherer S, Cundiff P, DeBord K, Gilliland D, Hickman S, Jarvis A, Tong L, Ashby M. Comprehensive evaluation of isoprenoid biosynthesis regulation in Saccharomyces cerevisiae utilizing the Genome Reporter Matrix. J Lipid Res. 1999;40:850–860. [PubMed] [Google Scholar]
- 10.Engebrecht J, Simon M, Silverman M. Measuring gene expression with light. Science. 1985;227:1345–1347. doi: 10.1126/science.2983423. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez De Henestrosa A R, Ogi T, Aoyagi S, Chafin D, Hayes J J, Ohmori H, Woodgate R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- 12.Gmuender H, Kuratli K, Di Padova K, Gray C, Keck W, Evers S. Gene expression changes triggered by exposure of Haemophilus influenzae to novobiocin or ciprofloxacin: combined transcription and translation analysis. Genome Res. 2001;11:28–42. doi: 10.1101/gr.157701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomez-Eichelmann M C. Effect of nalidixic acid and novobiocin on pBR322 genetic expression in Escherichia coli minicells. J Bacteriol. 1981;148:745–752. doi: 10.1128/jb.148.3.745-752.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guzzo A, Diorio C, DuBow M S. Transcription of the Escherichia coli fliC gene is regulated by metal ions. Appl Environ Microbiol. 1991;57:2255–2259. doi: 10.1128/aem.57.8.2255-2259.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henaut A, Danchin A. Analysis and predictions from Escherichia coli sequences, or E. coli in silico. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 2047–2066. [Google Scholar]
- 16.Horn J M, Ohman D E. Transcriptional and translational analyses of recA mutant alleles in Pseudomonas aeruginosa. J Bacteriol. 1988;170:1637–1650. doi: 10.1128/jb.170.4.1637-1650.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenyon C J, Walker G C. DNA damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc Natl Acad Sci USA. 1980;77:2819–2823. doi: 10.1073/pnas.77.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krueger J H, Walker G C. groEL and dnaK genes of Escherichia coli are induced by UV irradiation and nalidixic acid in an htpR+-dependent fashion. Proc Natl Acad Sci USA. 1984;81:1499–1503. doi: 10.1073/pnas.81.5.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LaRossa R A, Van Dyk T K. Applications of stress responses for environmental monitoring and molecular toxicology. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: ASM Press; 2000. pp. 455–468. [Google Scholar]
- 20.Lomovskaya O, Lewis K, Matin A. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J Bacteriol. 1995;177:2328–2344. doi: 10.1128/jb.177.9.2328-2334.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meighen E A. Molecular biology of bacterial bioluminescence. Microbiol Rev. 1991;55:123–142. doi: 10.1128/mr.55.1.123-142.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menzel R, Gellert M. Modulation of transcription by DNA supercoiling: a deletion analysis of the Escherichia coli gyrA and gyrB promoters. Proc Natl Acad Sci USA. 1987;84:4185–4189. doi: 10.1073/pnas.84.12.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 24.Mount D W, Low K B, Edmiston S J. Dominant mutations (lex) in Escherichia coli K-12 which affect radiation sensitivity and frequency of ultraviolet light-induced mutation. J Bacteriol. 1972;112:886–893. doi: 10.1128/jb.112.2.886-893.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neumann S, Quinones A. Discoordinate gene expression of gyrA and gyrB in response to DNA gyrase inhibition in Escherichia coli. J Basic Microbiol. 1997;37:53–69. doi: 10.1002/jobm.3620370109. [DOI] [PubMed] [Google Scholar]
- 26.Ohtsuka Y, Mizushima T, Miki T, Sekimizu K. Transient DNA relaxation in Escherichia coli induced by nalidixic acid. Biol Pharm Bull. 1997;20:467–470. doi: 10.1248/bpb.20.467. [DOI] [PubMed] [Google Scholar]
- 27.Orser C S, Foong F C F, Capaldi S R, Nalezny J, MacKay W, Benjamin M, Farr S B. Use of prokaryotic stress promoters as indicators of the mechanisms of chemical toxicity. In Vitro Toxicol. 1995;8:71–85. [Google Scholar]
- 28.Papavinasasundaram K, Movahedzadeh F, Keer J, Stoker N, Colston M, Davis E. Mycobacterial recA is cotranscribed with a potential regulatory gene called recX. Mol Microbiol. 1997;24:141–153. doi: 10.1046/j.1365-2958.1997.3441697.x. [DOI] [PubMed] [Google Scholar]
- 29.Petersen C, Moller L B. The RihA, RihB, and RihC ribonucleoside hydrolases of Escherichia coli. Substrate specificity, gene expression, and regulation. J Biol Chem. 2001;276:884–894. doi: 10.1074/jbc.M008300200. [DOI] [PubMed] [Google Scholar]
- 30.Raetz C R. Bacterial lipopolysaccarides: a remarkable family of bioactive macroamphiphiles. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: ASM Press; 1996. [Google Scholar]
- 31.Richmond C S, Glasner J D, Mau R, Jin H, Blattner F R. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 1999;27:3821–3835. doi: 10.1093/nar/27.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riley M, Labedan B. Protein evolution viewed through Escherichia coli protein sequences: introducing the notion of a structural segment of homology, the module. J Mol Biol. 1997;268:857–868. doi: 10.1006/jmbi.1997.1003. [DOI] [PubMed] [Google Scholar]
- 33.Riley M, Serres M H. Interim report on genomics of Escherichia coli. Annu Rev Microbiol. 2000;54:341–411. doi: 10.1146/annurev.micro.54.1.341. [DOI] [PubMed] [Google Scholar]
- 34.Rudd K E. EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res. 2000;28:60–64. doi: 10.1093/nar/28.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silhavy T J. Gene fusions. J Bacteriol. 2000;182:5935–5938. doi: 10.1128/jb.182.21.5935-5938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sitjes J, Ysern P, Barbe J, Llagostera M. Induction of ribonucleoside diphosphate reductase gene transcription by chemicals in Escherichia coli. Mutagenesis. 1992;7:47–49. doi: 10.1093/mutage/7.1.47. [DOI] [PubMed] [Google Scholar]
- 37.Sumantran V N, Tranguch A J, Datta P. Increased expression of biodegradative threonine dehydratase of Escherichia coli by DNA gyrase inhibitors. FEMS Microbiol Lett. 1989;53:37–40. doi: 10.1016/0378-1097(89)90362-5. [DOI] [PubMed] [Google Scholar]
- 38.Tao H, Bausch C, Richmond C, Blattner F R, Conway T. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J Bacteriol. 1999;181:6425–6440. doi: 10.1128/jb.181.20.6425-6440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thieffry D, Salgado H, Huerta A M, Collado-Vides J. Prediction of transcriptional regulatory sites in the complete genome sequence of Escherichia coli K-12. Bioinformatics. 1998;14:391–400. doi: 10.1093/bioinformatics/14.5.391. [DOI] [PubMed] [Google Scholar]
- 40.VanBogelen R A, Kelley P M, Neidhardt F C. Differential induction of heat shock, SOS, and oxidation stress regulons and accumulation of nucleotides in Escherichia coli. J Bacteriol. 1987;169:26–32. doi: 10.1128/jb.169.1.26-32.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Dyk T K. Stress detection using bioluminescent reporters of the heat shock response. Methods Mol Biol. 1998;102:153–160. doi: 10.1385/0-89603-520-4:153. [DOI] [PubMed] [Google Scholar]
- 42.Van Dyk T K, Ayers B L, Morgan R W, LaRossa R A. Constricted flux through the branched-chain amino acid biosynthetic enzyme acetolactate synthase triggers elevated expression of genes regulated by rpoS and internal acidification. J Bacteriol. 1998;180:785–792. doi: 10.1128/jb.180.4.785-792.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Dyk T K, Majarian W R, Konstantinov K B, Young R M, Dhurjati P S, LaRossa R A. Rapid and sensitive pollutant detection by induction of heat shock gene-bioluminescence gene fusions. Appl Environ Microbiol. 1994;60:1414–1420. doi: 10.1128/aem.60.5.1414-1420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Dyk T K, Rosson R A. Photorhabdus luminescens luxCDABE promoter probe vectors. Methods Mol Biol. 1998;102:85–95. doi: 10.1385/0-89603-520-4:85. [DOI] [PubMed] [Google Scholar]
- 45.Van Dyk T K, Smulski D R, Elsemore D A, LaRossa R A, Morgan R W. A panel of bioluminescent biosensors for characterization of chemically-induced bacterial stress responses. ACS Symp Ser. 2000;762:167–184. [Google Scholar]
- 46.Van Dyk T K, Wei Y, Hanafey M K, Dolan M, Reeve M J G, Rafalski J A, Rothman-Denes L B, LaRossa R A. A genomic approach to gene fusion technology. Proc Natl Acad Sci USA. 2001;98:2555–2560. doi: 10.1073/pnas.041620498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vierling S, Weber T, Wohlleben W, Muth G. Transcriptional and mutational analyses of the Streptomyces lividans recX gene and its interference with RecA activity. J Bacteriol. 2000;182:4005–4011. doi: 10.1128/jb.182.14.4005-4011.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vollmer A C. Visualization of bioluminescence. Methods Mol Biol. 1998;102:21–31. doi: 10.1385/0-89603-520-4:21. [DOI] [PubMed] [Google Scholar]
- 49.Walker G C. The SOS response of Escherichia coli. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 1400–1416. [Google Scholar]
- 50.Walker G C, Smith B T, Sutton M D. The SOS response to DNA damage. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: ASM Press; 2000. pp. 131–144. [Google Scholar]
- 51.Wanner B L, McSharry R. Phosphate-controlled gene expression in Escherichia coli using Mud1-directed lacZ fusions. J Mol Biol. 1982;158:347–363. doi: 10.1016/0022-2836(82)90202-9. [DOI] [PubMed] [Google Scholar]
- 52.Wilson M, DeRisi J, Kristensen H-H, Imboden P, Rane S, Brown P O, Schoolnik G K. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc Natl Acad Sci USA. 1999;96:12833–12838. doi: 10.1073/pnas.96.22.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang M K, Chou M E. Molecular characterization and expression of the recX gene of Xanthomonas campestris pv. citri. Curr Microbiol. 2001;42:257–263. doi: 10.1007/s002840110214. [DOI] [PubMed] [Google Scholar]
- 54.Ye R W, Tao W, Bedzyk L, Young T, Chen M, Li L. Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J Bacteriol. 2000;182:4458–4465. doi: 10.1128/jb.182.16.4458-4465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]