Abstract
Adenosine bases of RNA can be transiently modified by the deposition of a methyl-group to form N6-methyladenosine (m6A). This adenosine-methylation is an ancient process and the enzymes involved are evolutionary highly conserved. A genetic screen designed to identify suppressors of late flowering transgenic Arabidopsis plants overexpressing the miP1a microProtein yielded a new allele of the FIONA1 (FIO1) m6A-methyltransferase. To characterize the early flowering phenotype of fio1 mutant plants we employed an integrative approach of mRNA-seq, Nanopore direct RNA-sequencing and meRIP-seq to identify differentially expressed transcripts as well as differentially methylated RNAs. We provide evidence that FIO1 is the elusive methyltransferase responsible for the 3’-end methylation of the FLOWERING LOCUS C (FLC) transcript. Furthermore, our genetic and biochemical data suggest that 3’-methylation stabilizes FLC mRNAs and non-methylated FLC is a target for rapid degradation.
Author summary
Plants constantly measure environmental parameters such as temperature, light-quality, -quantity and -duration, as well as other parameters to adjust their growth and development. The transition to flowering is a crucial transition during plant development because it impacts the number of fertile offspring a plant can produce. Thus, this transition is under tight control by various transcription factors and epigenetic mechanisms. In a genetic screen for early flowering mutants, we identified the gene encoding the FIONA1 methyltransferase to act as a repressor of flowering. Investigation of the changes of the transcriptome and m6A-RNA-methylome revealed a large number of differentially expressed genes and methylated RNAs. Among these RNAs is the also the mRNA of the floral repressor FLOWERING LOCUS C that shows loss of m6A methylation in the 3’UTR that causes destabilization of the FLC transcript.
Introduction
Modification of RNA is pervasive and found across the entire tree of life [1]. In mRNA, N6-methyladenosine (m6A) is the most abundant internal covalent modification. m6A methylation patterns in plant mRNA have been found to be conserved between distant ecotypes [2] suggesting ancient regulatory functions. Biochemical studies have revealed that the mammalian m6A -writer complex consists of METTL3, METTL14, and associated proteins, such as WTAP and the ubiquitin ligase HAKAI [3–5]. Besides METTL3 and METTL14, METTL16 is a U6 adenosine methyltransferase that has been implicated in controlling m6A -methylation of mRNAs in humans [6] and has been shown in worms to affect diet-induced splicing of mRNA transcripts [7]. In plants, the functions of homologs of METTL3 and METTL14, MTA and MTB, respectively, as m6A -methylation writers are well characterized [4, 5]. For example, it is known that loss of MTA causes embryonic arrest at the globular stage [4], demonstrating the biological importance of m6A. In addition to m6A -writers, m6A readers, i.e. RNA binding proteins with specificity for m6A, can recognize m6A marks and affect RNA stability, splicing and translation through an unknown molecular mechanism [8]. The analysis of an early flowering knock-down allele of the METTL16-homolog FIONA1, fio1-2, revealed changes in the m6A methylation status of many transcripts, several encoding flowering regulators including SUPPRESSOR OF OVEREXPRESSION OF CONSTANS (SOC1) [9]. Besides SOC1 mRNA, the mRNA of the flowering regulator FLOWERING LOCUS C (FLC) has also been shown to be modified by m6A -methylation [10]. The latter study showed that an R-loop forms at the FLC locus that is resolved by the RNA-binding proteins FCA and FY. In this process, FCA binds the FLC COOLAIR antisense transcript to facilitate m6A -methylation [10]. Interestingly, the authors also detected m6A -methylation of the 3’UTR of FLC mRNA that appeared to be installed independently of FCA.
Here, we isolated a novel allele of FIONA1 (FIO1) in a genetic screen for suppressors of the late flowering phenotype of plants overexpressing the miP1a microProtein [11]. We present evidence that FIO1 acts as an m6A -methyltransferase in Arabidopsis and is the functional homolog of human METTL16. Using a combination of mRNA-seq, meRIP-seq and Nanopore direct RNA-sequencing, we provide further evidence that FIO1 is the elusive 3’UTR methyltransferase of FLC. Moreover, our data shows that the largely pleiotropic phenotype of fio1 mutant plants is a result of massive transcriptome and RNA-methylome changes. In the case of FLC, FIO1 is needed to maintain 3’-end methylation. Abrogation of this methylation mark causes depletion of FLC mRNA.
Results
FIONA1 acts as a floral repressor that functions partially independent of the photoperiod pathway
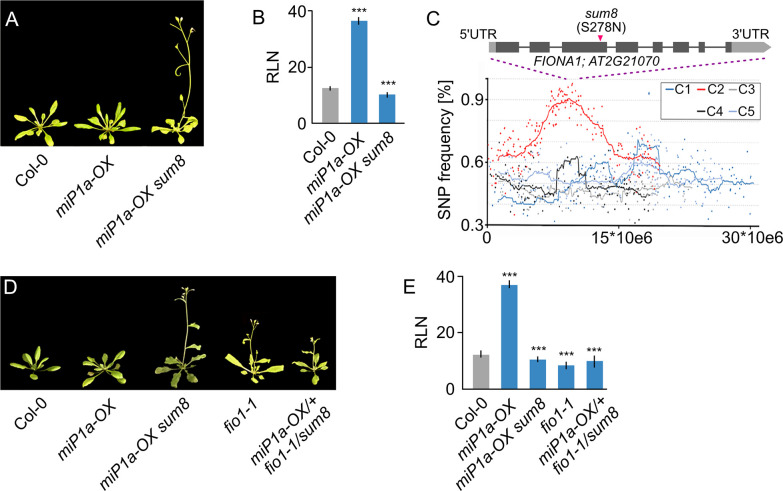
The miP1a/miP1b microProteins act as suppressors of flowering by interacting with a TOPLESS-containing repressor complex [11, 12]. To identify factors that are required for the repressor complex to suppress flowering, we performed a genetic screen with transgenic miP1a-OX (35S::MIP1A) plants. We identified a set of suppressor of miP1a (sum) mutants, that, despite high levels of miP1a protein, flowered early under inductive long day conditions [12]. One of the suppressors, sum8, we describe here, showed accelerated flowering compared to the non-mutagenized miP1a-OX parental plant (Fig 1A and 1B). To identify the causal mutation in the sum8 background, we crossed miP1a-OX sum8 plants to Col-0 wildtype, self-pollinated the offspring and selected a pool of 20 BASTA-resistant suppressor mutants of the following generation. Pooled DNA of the sum8 suppressor mutant and the parental line was then analyzed by genome re-sequencing. In total, we detected 685 EMS-induced SNPs with a frequency enrichment in the middle of chromosome 2 (Fig 1C). At the summit region of the enrichment peak we identified a point mutation in the FIONA1 (FIO1) gene which converted serine 278 into an asparagine (S278N). To verify that the mutation in FIO1 is causal for the early flowering phenotype, we obtained a second EMS allele (fio1-1) that had been described earlier [13] and crossed it with miP1a-OX sum8 plants. The resultant nullizygote offspring (miP1a-OX/+ fio1-1/sum8) flowered early (Fig 1D and 1E), supporting that the mutation in FIO1 is indeed causal for the flowering phenotype. The fio1-1 allele is a splice site mutation that results in the loss of five amino acids while sum8 is a point mutation. To obtain an additional FIO1 allele, we used a CRISPR approach with multiple sgRNAs and obtained the new fio1-5 allele. Like fio1-1 and sum8, fio1-5 also showed early flowering in long day conditions (S1A and S1B Fig). The fio1-5 deletion occurred close to a splice site and caused the loss of amino acids 53–64 and the conversion of amino acid residues 66–72 (S1C Fig).
Fig 1. Identification of the flowering repressor FIONA1 by whole-genome re-sequencing.
(A) Phenotype of the sum8 (fio1) mutant in the miP1a-OX background compared to the Col-0 wildtype grown in LD conditions. (B) Determination of flowering by counting the number of rosette leaves (RLN = rosette leaf number) at the bolting stage in LD. Plotted are average leaf number +/- SD, ***p = <0.001, N = 10. (C) Mapping-by-sequencing of the sum8 suppressor mutation. Plotted are SNP frequencies of a pool of segregating F2 plants. Increased SNP frequencies were observed in chromosome 2 and the FIO1 locus is at the summit of the plot. (D) Genetic complementation experiment proving that the sum8 mutation affects FIO1. Shown are the flowering phenotypes of plants grown in LD conditions. (E) Determination of flowering by counting the number of rosette leaves (RLN = rosette leaf number) at the bolting stage in LD. Plotted are average leaf number +/- SD, ***p = <0.001, N = 10.
FIO1 is related to the human METTL16 protein
FIO1 is a nuclear localized protein containing a DUF890 domain, making it a member of the METTL16-like protein family that is comprised of, among others, the human and mouse METTL16 and the C. elegans METT-10 proteins. Animals carrying loss-of-function alleles of METT-10/METTL16 have been described to show severe developmental defects, and sometimes, lethality [14, 15]. The latter finding raised the question of whether we were dealing with complete loss-of-function or reduced function alleles of FIO1. All mutants had either smaller deletions or single amino acid changes suggesting they could be weak, reduced function alleles.
To gain further insights into the alleles that we had obtained, we created a homology model of the FIO1 methyltransferase (MTase) domain and compared it against the crystal structure of the human homologue, METTL16. In the case of the sum8 mutation (S278N, S2 Fig), we found that the sidechain of S278 normally forms hydrogen bonds with the nitrogen on the W330 within the protein core. Upon mutating the serine to an asparagine, we expect that the larger asparagine sidechain cannot be accommodated in the protein interior, leading to disrupted domain fold and function. The fio1-1 mutation involves the deletion of five amino acids 145–149 in the FIO1 protein (S2 Fig) which includes the disruption of a potential hydrogen bond between the sidechains of Q82 and T147 and the loss of a flexible loop connecting an alpha helix and a beta sheet. The fio1-5 mutation involves the large deletion of amino acids 57–68 and the non-conservative mutation of residues 53–56 and 69–72 (S2 Fig). Both fio1-1 and fio1-5 involve the large-scale disruption of hydrophobic and hydrogen bonding interactions and are likely to result in misfolded or aggregated protein. Thus, it is highly likely that all three mutations (sum8, fio1-1 and fio1-5) disrupt the methyltransferase function of FIO1.
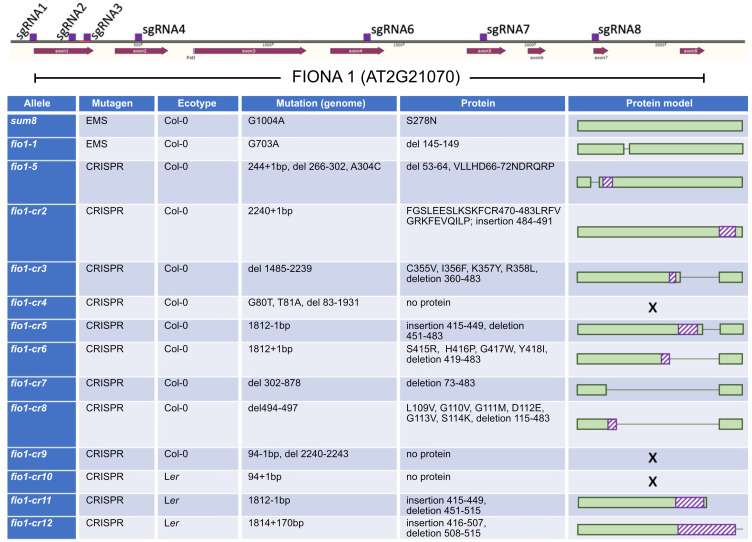
To validate the findings of the protein modeling we employed a second CRISPR mutagenesis approach and designed eight sgRNAs spanning the entire FIO1 locus and transformed these in bulk to obtain larger structural mutations (Fig 2). We identified 11 new FIO1 alleles, of which several had large structural deletions. Three of these alleles (fio1-cr4, fio1-cr9, fio1-cr10) had frame-shift mutations that would not lead to the production of functional proteins. All new alleles were early flowering (S3 Fig) but viable and produced fertile offspring. Taken together, these results show that the loss of METTL16 function is not lethal in plants but affects the transition to flowering.
Fig 2. Overview of fiona1 mutant plants analyzed and generated in this study.
Gene model depicting the FIO1 locus (exons in dark red and location of sgRNAs in purple). All sgRNAs were transformed in bulk and from all early flowering individuals the FIO1 gene was sequenced to determine the nature of CRISPR-induced mutations.
The loss of FIO1 function results in a pleiotropic phenotype
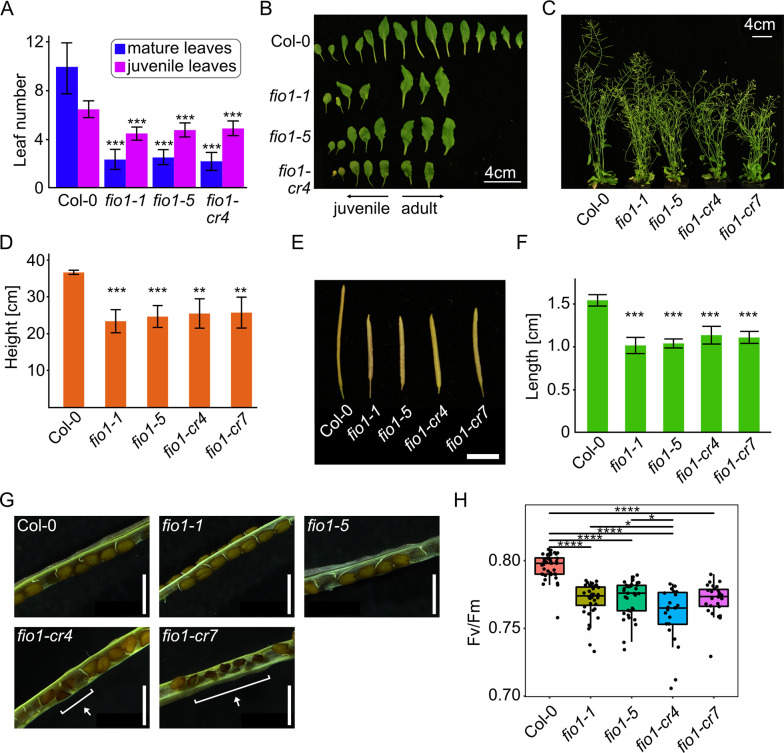
Precocious flowering upon loss of FIO1 function is a striking phenotype that appears early in development. To characterize fio1 mutants that lack large fractions of the FIO1 protein and compare them to the previously characterized mutants, we recorded the flowering of the panel of novel CRISPR alleles that we generated in this study (Fig 2). This analysis revealed that all fio1 mutants, regardless of the genetic background (here Col-0 or Ler), flowered unanimously early (S3 Fig). A more detailed analysis of the fio1-cr4 mutant that is lacking most of the FIO1 protein, showed that it flowered as early as fio1-1 and fio1-5 compared to Col-0 wild type plants. In addition to recording the number of leaves produced at the bolting stage, we also collected all leaves and found that fio1 mutant plants produce fewer juvenile and mature leaves, indicating that both growth phases (juvenile and adult phase) are accelerated in fio1 mutants compared to wild type (Fig 3A and 3B). When grown to full maturity, we also detected that fio1 mutant plants were significantly shorter than wild type plants and also appeared bushier (Fig 3C and 3D). Siliques produced by the fio1 mutant plants were also notably shorter (Fig 3E and 3F) and we were wondering if this could be due to a higher rate of early seed abortion. A closer inspection of the siliques revealed normal seeds in Col-0 wild type and fio1-1 and fio1-5 mutant plants, whereas fio1-cr4 and fio1-cr7 plants showed fractions of improperly developed shriveled seeds. The fio1-cr4 and fio1-cr7 mutants both carry large structural deletions in the FIO1 gene and this latter finding suggests that they are both complete loss-of-function alleles. The fio1-1 and fio1-5 alleles, on the other hand, might have residual FIO1 enzyme activity or participate in FIO1 protein complexes that do not involve its methyltransferase activity. Besides the smaller stature and earlier flowering, fio1 mutant plants also appeared paler in color compared to wild type plants. To determine whether fio1 mutants are additionally impaired in photosynthetic performance, we measured the Fv/Fm ratio using pulse amplitude modulated (PAM) fluorometry. PAM measurements confirmed defects in photosynthetic performance of fio1 mutant plants and all tested mutants showed consistently lower Fv/Fm ratios (Fig 3H).
Fig 3. fio1 mutants display a pleiotropic phenotype.
(A) Number of mature and juvenile leaves of plants grown under LD at the bolting stage. Plotted are the means with bars denoting +/- SD with N = 11–17. Asterisks represent significance level between values determined by two-sample T-Test. (B) Rosette leaf morphology of representative WT and fio1 mutants at the WT bolting stage. (C) Branching phenotype of 44-day old WT and fio1 mutant plants grown in LD. (D) Height of tallest inflorescence of WT and fio1 mutant plants at senescence. Plotted are the means with bars denoting +/- SD with N = 3–6. Asterisks represent significance level between values determined by two-sample T-Test. (E) Silique morphology at maturation. One representative silique from each line. Scale bar is 5 mm. (F) Silique length at maturation. Plotted are the means with bars denoting +/- SD with N = 10–12 and 3 biological replicates of the WT and 4 biological replicates per fio1 mutant line. Asterisks represent significance level between values determined by two-sample T-Test. (G) Opened siliques from WT and fio1 mutants. Highlighted are aborted seeds of fio1-cr4 and fio1-cr7. Scale bar is 1 mm. (H) Photosynthetic efficiency of WT and fio1 mutant seedlings expressed as variable fluorescence/maximum fluorescence (Fv/Fm), N = 23–41. Asterisks represent significance level between values determined by two-sample T-Test. For all plots, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
The loss of FIO1 function affects multiple flowering pathways
A previous genetic screen for regulators of flowering resulted in the identification of the fio1-1 mutant that exhibited early flowering in both long- and short-day conditions [13]. A knock-down mutation caused by a T-DNA insertion in the 5’-region of the FIONA1 gene [9, 16] showed a similar phenotype. The fio1-1 mutant was shown to have elevated levels of both CONSTANS (CO) and FLOWERING LOCUS T (FT) mRNA. CO is a photoperiod-sensitive transcription factor that accumulates in response to long days to activate FT [17], which in turn acts as a florigen to induce flowering [18, 19]. The flowering phenotype of fio1-1 was ascribed to changes in period length of the central oscillator. Consistent with previous findings, we found that levels of both CO and FT were elevated in fio1-1 and fio1-5 (S4A and S4B Fig). A genetic interaction study revealed that miP1a miP1b fio1-5 triple mutant plants also flowered early like fio1-5 mutant plants. The combination of fio1 mutants with either co and ft mutants as in fio1 co and fio1 ft, revealed a promotion of flowering (S4C and S4D Fig) in both short days and long days. These results unequivocally show that the function of FIO1 is independent of the function of miP1a and partially independent of the photoperiod flowering time pathway.
Transcriptome analysis of fio1-1 and fio1-5 mutant plants reveal substantial gene expression changes
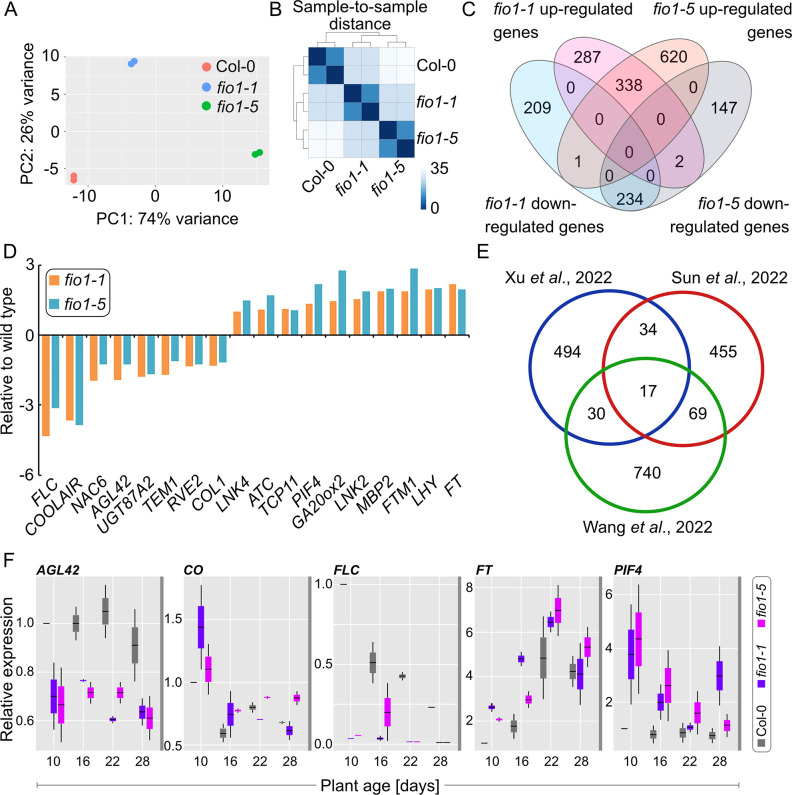
To obtain a better understanding of how FIO1 affects flowering, we performed an RNA-seq experiment with Col-0, fio1-1 and fio1-5 mutant plants to identify differentially expressed genes. RNA of two biological replicates of 14 day-old seedlings was isolated and sequenced on an Illumina HiSeq instrument. After removing low-quality reads, an average of 91.47% of the filtered reads was mapped to the Arabidopsis thaliana reference genome. Principal component analysis (PCA) and hierarchical cluster analysis (HCA) revealed that the individual biological replicates clustered closely together (Fig 4A and 4B), indicating a high degree of experimental reproducibility. Interestingly, fio1-1 and fio1-5 were also distinct from each other and wild type, indicating that although they show a similar flowering phenotype they might differ at the molecular level.
Fig 4. Transcriptome changes observed in fio1 mutants.
(A) Principal component analysis (PCA) plot displaying the different RNA-seq performed using DESeq2 rlog-normalized RNA-seq data. Plotted is the percentage of variance for each component. (B) Hierarchical clustering analysis (HCA) of the different RNA-seq libraries. The heatmap was built using the DEseq2 package. Samples were clustered using HCA performed with DESeq2 rlog-normalized RNA-seq data, and the dendrogram represents the clustering results. The heatmap illustrates the pairwise distances between the different samples, with higher similarity indicated by higher intensity of color. (C) Venn diagram showing the overlap of differentially expressed genes in fio1-1 and fio1-5 compared to the wild type. The absolute value of log2 FC (fold change; fio1 mutant / WT) ≥ 1.0 and adjusted P-value (false discovery rate; FDR) ≤ 0.05. (D) RNA-seq showing the expression levels of flowering related genes in fio1-1 and fio1-5 compared to the wild type. The absolute value of log2 FC (fold change; fio1 mutant / WT) ≥ 1.0 and adjusted P-value (false discovery rate; FDR) ≤ 0.05. (E) Comparative analysis of differentially expressed genes identified by RNA-seq and nanopore-sequencing in three different studies. P-Value < 0.01, FC > 2. The Venn diagram depicts the overlap of the three datasets. (F) Gene expression analysis of candidate genes over a developmental time course. Plants were grown in LD conditions and samples were harvested before the end of the long day, 10, 16, 22 and 28 days after germination. Box plots depict relative expression levels of respective genes to the UBQ10 and TIP41 gene of two biological replicates with three technical replicates each.
To identify differentially expressed genes (DEGs) in fio1-1 and fio1-5 we used limma-voom [20] with a fold change cutoff of 2.0 or more. In total, we identified 627 and 959 up-regulated genes in fio1-1 plants and fio1-5 plants respectively (P value < 0.05 and adjusted P value < 0.05; S1 Table). We found 1071 DEGs in fio1-1 and 1342 DEGs in fio1-5 with an overlap of 338 up-regulated genes and 234 down-regulated genes (Fig 4C). Of these deregulated transcripts, 18 were associated with regulation of flowering (Fig 4D), including flowering repressors FLOWERING LOCUS C (FLC) and TEMPRANILLO1 (TEM1) whose mRNA levels were significantly reduced in fio1 mutant plants and flowering activators such as PHYTOCHROME INTERACTING FACTOR4 (PIF4), FT and LATE ELONGATED HYPOCOTYL (LHY) whose mRNA levels were significantly increased in fio1 mutant plants (Fig 4D). These findings are in agreement with the early flowering phenotype of fio1 mutant plants. Comparative analysis of RNAseq data of two recent studies [9, 21] produced only a limited overlap (Fig 4E) which is likely due to differences in growth conditions and stages of development that were used for the analysis. To validate some of the differentially expressed genes that we had identified in our analysis, we tested the expression of AGL42, CO, FLC, FT and PIF4 by qRT-PCR at four timepoints during plant development (Fig 4F). The expression of AGL42 and FLC was consistently low in fio1 mutants at all growth stages analyzed, while CO and FT levels were higher in fio1 mutants only during the early stages of development and not after the floral transition had occurred (Fig 4F). Expression of the floral thermoregulator PIF4 was increased in fio1 mutants at all stages that were analyzed. The findings that PIF4 is deregulated in fio1 mutants is in line with fio1 mutants showing defects in the shade avoidance response (S5 Fig). When germinated in white light conditions, fio1 seedlings develop elongated hypocotyls and in shade elongate even more than wild type seedlings which is indicative of shade hypersensitivity.
FIO1 acts as m6A -methyltransferase and methylates predominantly the 3’UTR of mRNAs
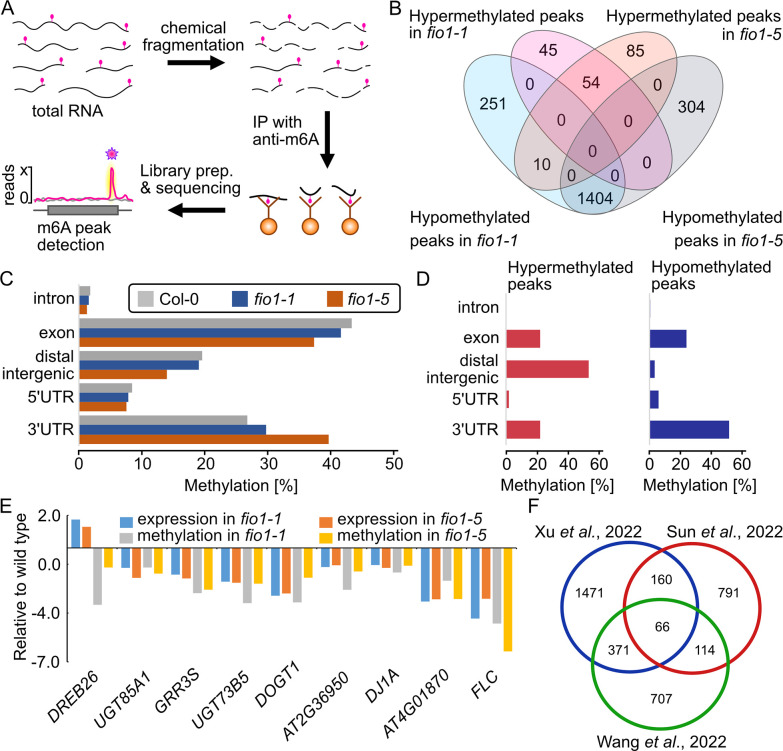
The presence of the DUF890 domain suggests that FIO1 acts as a genuine m6A methyltransferase. To identify the FIO1 RNA substrates, we employed a modified version of methylated RNA-immunoprecipitation (meRIP) followed by deep sequencing that was described earlier (Fig 5A) [22]. To determine methylation positions (m6A peaks) we used MACS [23] with a false discovery rate (FDR) ≤ 0.05 and enrichment of ≥ 2-fold of sequence reads. In summary, we identified 3,025 m6A-methylation peaks in wild type, 2,088 in fio1-1 and 2,109 in fio1-5 (S2 Table). In fio1-1 plants and fio1-5 plants we identified 99 and 149 peaks, respectively, with increased m6A level compared to wild type. In contrast, a total of 1,665 m6A methylation peaks in fio1-1 and 1,708 peaks in fio1-5 were decreased or absent compared to the wild type (Fig 5B). These findings suggest that FIO1 methylates mRNAs. When assessing the localization of the m6A -peaks globally in wild type, fio1-1 and fio1-5, we observed more peaks in exons of fio1 mutants and a reduced number of peaks in the 3’UTR of fio1 mutants compared to wild type (Fig 5C). The differential m6A peak distribution analysis (wild type versus fio1 mutants) revealed a massive over-representation of hypomethylated peaks in 3’UTRs in fio1 mutants compared to wild type (Fig 5D). These findings indicate that FIO1 acts as m6A methyltransferase and methylates the 3’UTRs of its target substrates. To explore a potential connection between m6A -methylation and RNA stability we compared our mRNA-seq and MeRIP datasets. In total we found nine genes containing hypomethylated peaks, eight of which were expressed at lower levels while one was expressed at higher level in fio1 mutants compared to the wild type (Fig 5E). Additional comparative analysis of meRIPseq and nanopore-sequencing data of two recent studies [9, 21] again produced only a limited overlap (Fig 5F) which could indicate differences in growth conditions and stages of development that were used for the analysis.
Fig 5. FIONA1 acts as m6A-methyltransferase in Arabidopsis.
(A) Depiction of the meRIP-seq method. In brief, total RNA was isolated from seedlings and subsequently fragmented into small (100bp) fragments. After immunoprecipitation with an m6A -specific antibody, Illumina short-read sequencing libraries were generated and sequenced. After mapping all reads to the Arabidopsis genome, m6A peak regions (pink star) could be identified. (B) Venn diagram showing the overlap of the hypermethylated and hypomethylated m6A peaks identified in fio1-1, fio1-5 compared to Col-0 wild type plants. (C) Comparison of distribution of m6A peaks in different segments of wild-type (left panel), fio1-1 (middle panel) and fio1-5 (right panel) transcripts. The panels show pie charts presenting the percentages of m6A peaks in different transcript segments. (D) Comparison of distribution of m6A peaks in different segments of differently methylated peaks (left panel), hypermethylated peaks (middle panel) and hypomethylated peaks (right panel) in the overlap of fio1-1 and fio1-5 compared to wild type. The panels show pie charts presenting the percentages of m6A peaks in different transcript segments. (E) Expression levels and m6A methylation levels of the transcripts in the overlapping of RNAseq and MeRIPseq. Gene expression levels were derived from RNA-Seq data. m6A methylation levels were derived from MeRIPseq data. (F) Comparative analysis of hypomethylated m6A peaks in three different studies. MeRIP-seq cutoff RPM>5 and logFC<-0.5; Nanopore cutoffs FC>5 and FDR<0.05. The Venn diagram shows the overlap of the three datasets.
FLC is a prime target of FIO1
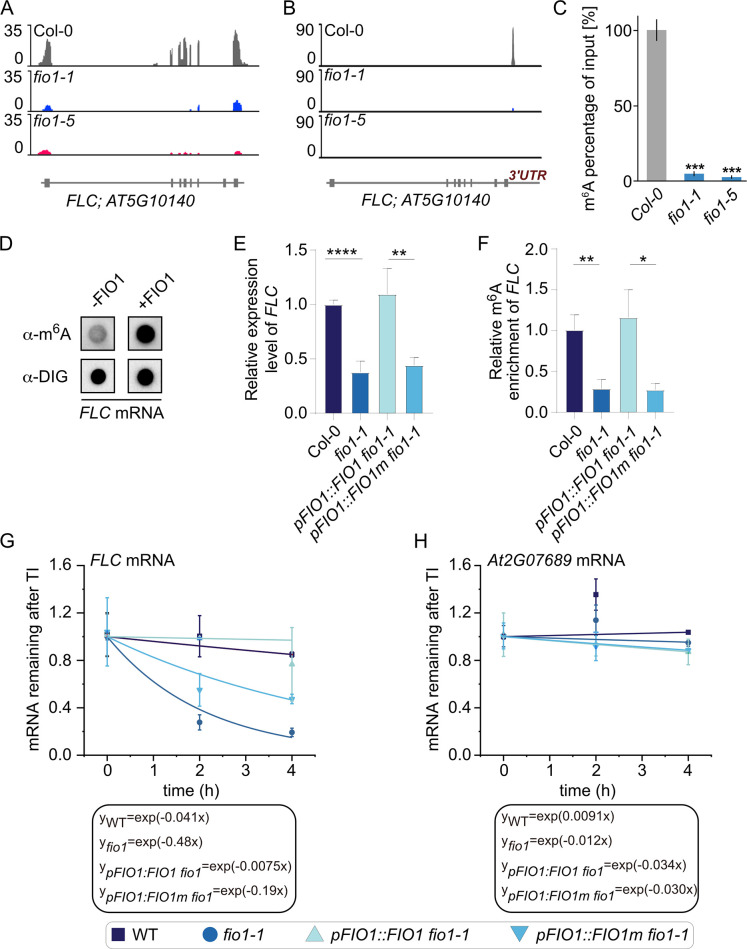
The mRNA of the flowering repressor FLOWERING LOCUS C (FLC) was identified as a prime methylation target of FIO1 (Fig 5E). We detected strongly decreased expression of FLC mRNA in fio1 mutants compared to wild type (Fig 6A) and the m6A peak that can be detected in wild type plants is absent in fio1-5 and strongly reduced in fio1-1 mutant plants (Fig 6B). To verify that FLC is indeed a bona fide methylation target of FIO1, we performed anti- m6A antibody immunoprecipitations (m6A -IP) of total RNA from wild type (Col-0), fio1-1 and fio1-5 seedlings followed by qPCR (m6A -IP-qPCR). We found the relative amount of m6A methylated FLC mRNA was strongly decreased in both fio1 mutant plants (Fig 6D) confirming that FIO1 is the essential m6A methyltransferase that methylates the 3’UTR of FLC. To validate if FIO1 is capable of catalyzing the methylation reaction of FLC mRNA outside of a cellular context, we purified the Arabidopsis FIO1 enzyme from E. coli and carried out methylation reactions with in vitro synthesized digoxygenin-labelled FLC RNA (Fig 6D). In these assays, FIO1 can methylate FLC mRNA indicating that the effect is of a direct nature. We also tested the levels of FLC mRNA in seedlings and compared it to the enrichment of m6A methylation of FLC (Fig 6E and 6F). Wild type plants and fio1 mutants complemented with a transgene expressing FIO1 from its own promoter have higher levels of both FLC mRNA and FLC m6A methylation (Fig 6E and 6F). This contrasts the findings in fio1 mutants and fio1 mutants complemented with a transgene expressing mutant FIO1m from its own promoter that have lower levels of FLC mRNA and decreased levels of FLC m6A methylation (Fig 6E and 6F). Quantification of mRNA levels of FLC and of the control gene AT2G07689 after transcriptional inhibition in wild type, fio1 mutants and previously described complementation lines were also carried out. We found decreased FLC mRNA stability in fio1 and in fio1 mutants complemented with the non-functional FIO1m (Fig 6G). In summary, our data reveal a role for FIO1 as FLC methylating enzyme.
Fig 6. FIONA1 acts as m6A-methyltransferase on FLC.
(A) RNA-seq coverage observed at the FLC locus. RNA-seq reads in Col-0 (grey), fio1-1 (blue) and fio1-3 (pink). Gene model depicts exons and introns. (B) MeRIP-seq coverage observed at the FLC locus. RNA-seq reads in Col-0 (grey), fio1-1 (blue) and fio1-3 (pink). Gene model depicts exons and introns. (C) Percentages of the m6A methylated FLC mRNA in input samples in the wild type, fio1-1 and fio1-3 measured by m6A-IP-qRT PCR. Values are the means ±SD. N = 4, ***P≤ 0.001. (D) In vitro methylation of FLC mRNA. FIO1 was purified as GST-tagged protein from E. coli and incubated with digoxygenin (DIG)-labeled in vitro produced FLC mRNA in the presence of SAM. Dot blot in the upper row show m6A methylated signal with the anti- m6A antibody. The dots in the lower row the signal of the anti-DIG antibody. Increased signal intensities were detected when FIO1 enzyme was present. (E) qPCR results showing the relative expression of FLC in 12-day-old Col-0, fio1, pFIO1:FIO1/fio1, and pFIO1:FIO1m/fio1 seedlings. Data are means ± SD for 3 biological replicates × 3 technical replicates. * p < 0.05, ** p < 0.01 by t test (two-tailed). (F) m6A-IP-qPCR results showing the relative m6A levels of FLC transcripts in 12-day-old Col-0, fio1, pFIO1:FIO1/fio1, and pFIO1:FIO1m/fio1 seedlings. Data are means ± SD for 3 biological replicates × 3 technical replicates. * p < 0.05, ** p < 0.01 by t test (two-tailed). (G) and (H) The mRNA lifetimes of FLC (G) in Col-0, fio1, pFIO1:FIO1/fio1, and pFIO1:FIO1m/fio1. The AT2G07689 (H) was used as the negative control. TI: transcription inhibition. Data are represented as means ± SD for 2 biological replicates × 3 technical replicates.
Direct RNA sequencing of wild type and fio1 mutant plants confirms altered FLC mRNA levels in fio1 mutants
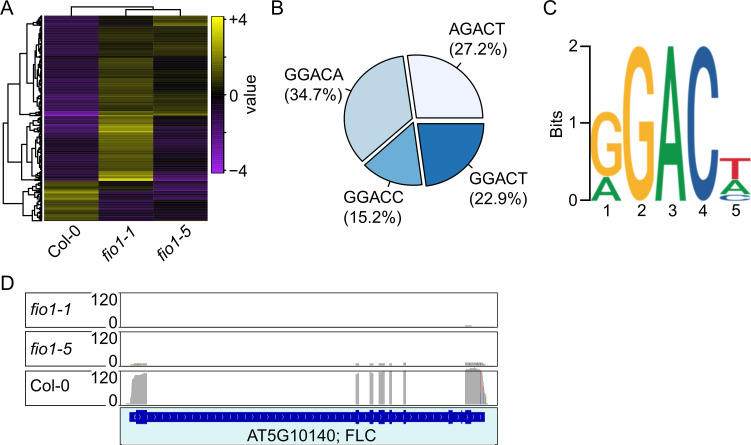
To determine the genome-wide m6A methylation changes in fio1 loss of function mutants compared to wild type and to validate FLC methylation and stability in an unbiased fashion, we employed Nanopore direct RNA sequencing. This analysis showed that fio1 mutants differ in the levels of transcriptome changes (Fig 7A). Furthermore, in Col-0 wild type plants, the majority (34.7%) of m6A methylations occurred in the GGACA element, followed by AGACT (27.2%), GGACT (22.9%) and GGACC (15.25) (Fig 7B). In summary, our work defined the Arabidopsis consensus m6A methylation site as RGACH, in which R represents A or G and H all nucleotides except G, which corresponds with the RRACH element that had previously been identified [2]. FIONA1 is a methyltransferase that adds methyl-groups to adenine bases of RNAs. Messenger-RNAs that are targets of FIO1 are therefore expected to be hypomethylated in a situation of lost or reduced FIO1 activity. Our direct RNA-sequencing approach yielded 74 genes that were hypomethylated in fio1-1 mutants compared to wild type and 63 genes in fio1-5 (S4 Table). Another recent direct RNA-sequencing study of the fio1-2 knock-down mutant revealed over 2000 hypomethylated transcripts in Arabidopsis [9]. The comparison with our datasets identified in total 28 hypomethylated transcripts that are detected in at least two mutants (Table 1). FLC expression was shown to be significantly reduced in both fio1-1 and fio1-5 mutants and meRIP-seq detected m6A methylation in the 3’UTR of FLC (Figs 4D, 5E and 6A–6C). In agreement with these latter results, direct RNA-sequencing confirmed that FLC mRNA is depleted in both fio1-1 and fio1-5 mutants (Fig 7D).
Fig 7. Direct RNA-sequencing analysis.
(A) Cluster analysis of differentially expressed transcripts in the three different genotypes. (B) Distribution of m6A methylations detected by direct RNA-sequencing. (C) Logo of the conserved m6A sequence motif detected by direct RNA-sequencing. (D) Sequence coverage observed at the FLC locus. Direct RNA-seq reads in Col-0, fio1-1 and fio1-5. Gene model depicts exons and introns.
Table 1. Comparative analysis of hypomethylated transcripts in fio1-1, fio1-2 and fio1-5 relative to wild type Col-0.
| Arabidopsis Gene Identifier (AGI) |
Hypomethylated | Annotation | ||
|---|---|---|---|---|
| AT1G12840 | fio1-1 | fio1-5 | DET3, ATVHA-C, ARABIDOPSIS THALIANA VACUOLAR ATP SYNTHASE SUBUNIT C, DE-ETIOLATED 3 | |
| AT1G19980 | fio1-1 | fio1-5 | no symbol available | |
| AT1G52040 | fio1-1 | fio1-5 | MBP1, ATMBP, myrosinase-binding protein 1 | |
| AT1G52710 | fio1-1 | fio1-5 | no symbol available | |
| AT1G76730 | fio1-1 | fio1-5 | COG0212, Clusters of Orthologous group 212 | |
| AT2G18050 | fio1-1 | fio1-5 | HIS1-3, histone H1-3 | |
| AT2G40480 | fio1-1 | fio1-5 | no symbol available | |
| AT5G18790 | fio1-1 | fio1-5 | no symbol available | |
| AT5G56860 | fio1-1 | fio1-5 | GNC, GATA21, GATA TRANSCRIPTION FACTOR 21 | |
| AT5G64860 | fio1-1 | fio1-5 | AtDPE1, DPE1, disproportionating enzyme | |
| AT1G50250 | fio1-1 | fio1-2 | FTSH1, FTSH protease 1 | |
| AT1G52400 | fio1-1 | fio1-2 | BGL1, ATBG1, BGLU18, A. THALIANA BETA-GLUCOSIDASE 1 | |
| AT1G63770 | fio1-1 | fio1-2 | no symbol available | |
| AT2G30520 | fio1-1 | fio1-2 | RPT2, ROOT PHOTOTROPISM 2 | |
| AT2G47940 | fio1-1 | fio1-2 | DEG2, DEGP2, EMB3117 | DEGP protease 2, | |
| AT3G10060 | fio1-1 | fio1-2 | no symbol available | |
| AT3G51950 | fio1-1 | fio1-2 | no symbol available | |
| AT5G42650 | fio1-1 | fio1-2 | CYP74A, AOS, DDE2, allene oxide synthase, DELAYED DEHISCENCE 2, CYTOCHROME P450 74A | |
| AT5G66190 | fio1-1 | fio1-2 | LFNR1, ATLFNR1, FNR1, leaf-type chloroplast-targeted FNR 1, LEAF FNR 1 | |
| AT1G67480 | fio1-2 | fio1-5 | no symbol available | |
| AT2G22990 | fio1-2 | fio1-5 | SNG1, SCPL8, sinapoylglucose 1 | |
| AT4G19110 | fio1-2 | fio1-5 | no symbol available | |
| AT4G19160 | fio1-2 | fio1-5 | no symbol available | |
| AT5G25265 | fio1-2 | fio1-5 | HPAT1, hydroxyproline O-arabinosylatransferase 1 | |
| AT5G57560 | fio1-2 | fio1-5 | XTH22, TCH4, Touch 4, xyloglucan endotransglucosylase/hydrolase 22 | |
| AT2G01490 | fio1-1 | fio1-2 | fio1-5 | PAHX | phytanoyl-CoA 2-hydroxylase |
| AT2G28900 | fio1-1 | fio1-2 | fio1-5 | OEP16, OEP16-1, ATOEP16-L, ATOEP16-1, outer plastid envelope protein 16–1 |
| AT4G08950 | fio1-1 | fio1-2 | fio1-5 | EXO, EXORDIUM |
Discussion
The precise timing of the floral transition is crucial for reproductive success. Premature as well as delayed flowering can result in seed dispersal at times where the offspring will be facing suboptimal conditions for survival and reproduction. This could either be due to the absence of pollinators or adverse environmental conditions. Therefore, a highly integrative network of transcription factors, but also epigenetic regulators, operate to ensure that flowering occurs in the most optimal conditions.
Methylation of mRNA is crucial for various functions within the cell. The m6A methylation of mRNA is an ancient molecular process and its disruption strongly compromises cellular functions. Strong reduction of the global m6A methylome early in plant development, as seen in mutants lacking the METTL3-homolog MTA, causes embryonic arrest [4]. Partial complementation of the mta mutant resulted in plants with compromised m6A levels that showed pleiotropic phenotypes such as reduced apical dominance and missing floral organs [24]. These latter results suggest that more subtle reductions of the global m6A levels are not detrimental to plant development. We provide further support of this by showing that the loss-of-function mutants of FIO1, a protein that is not essential for plant development, have only a subtle effect on the global m6A-methylome. However, CRISPR-induced mutants that caused larger genomic deletions in the FIO1 gene (here fio1-cr4 and fio1-cr7) showed a low-frequency seed abortion phenotype that resembles mta mutants (Fig 3G). Furthermore, in contrast to the effect that the loss of its homolog has on animal development, FIO1 is not essential and causes hypomethylation of specific transcripts. These hypomethylated mRNAs can then be stabilized, or destabilized, or mis-spliced. Affected transcripts that encode transcription factors or other regulators that are either mis-spliced of mis-methylated can subsequently induce alterations of circadian rhythms, cause changes in the production of hormones, or mis-regulation of other biological processes. It might be important to note that the pleiotropic phenotype of fio1 mutant plants includes short stature, higher degree of shoot branching (Fig 3C) and a constitutive shade avoidance response (S5 Fig). A commonality of these phenotypes is that they relate to alterations in the levels of plant hormones, especially cytokinin and auxin. Wang et al. [21] found that fio1 loss-of-function leads to an enrichment of hypomethylated genes associated with cytokinin signaling and ethylene response, potentially linking the phenotype we observe to the effect of FIO1 at the molecular level.
The precocious flowering phenotype is the most striking but fio1 mutants additionally display a constitutive shade-avoidance phenotype, earlier senescence, and paler leaves [13]. In accordance with these phenotypes, our RNA-seq study revealed that several genes encoding circadian clock regulators and positive regulators of flowering time were upregulated in the fio1 mutant background (e.g. LHY, PIF4). In contrast, several of the downregulated transcripts encoded transcription factors that repress flowering (Fig 4D and S1 Table).
Genetically, flowering is controlled by distinct pathways that interact at multiple levels to integrate inputs from all pathways. This integration ensures flowering occurs at the optimal time. The photoperiod pathway controls flowering in response to daylength and involves the B-Box zinc finger transcription factor CONSTANS (CO) which, in Arabidopsis, is stabilized at the end of long days [17]. CO positively regulates the expression of FLOWERING LOCUS T (FT) [25], encoding a mobile protein that travels to the shoot meristem to induce flowering [18]. FIO1 acts partially through the photoperiod pathway and the early flowering phenotype of fio1 mutants correlates with increased levels of both CO and FT mRNAs (S4 Fig) as well as increased levels of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) [9]. Our genetic interaction studies have shown that mutations in both CO and FT can partially suppress the early flowering effect of fio1 mutants. Consistent with our findings, the soc1 mutant has also been shown to partially suppress the early flowering phenotype of fio1-2 mutant plants [9]. Taken together, these data support a model that assumes an indirect effect of the photoperiod pathway in the control of flowering by FIO1.
The spatial and temporal aspects of FIO1 function are currently unknown. It seems possible that depending on the developmental stage, the organ, or the age of the plant, that FIO1-executed m6A could differ and have different consequences downstream (directed mRNA modification versus changes in splicing patterns). Our analysis of the temporal changes in gene expression (Fig 4F) supports this notion. This could also explain contrasting results between papers, for example Xu et al. reported a consensus target sequence of YHm6AGA, which is significantly different from what has been reported here and by others, but their analysis was of 6-day old plants compared to 14- or 12-day old plants that were used here and in Wang et al., respectively.
Our RNA-sequencing data identified both up- and downregulated transcripts in fio1 mutants compared to wild type. However, the overlap between the set of de-regulated transcripts identified in fio1-2 mutants [9] is very limited. The latter fact can be attributed to the different types of mutations that were analyzed. While our study capitalized on mutant variants that are likely enzyme-dead and loss-of-function alleles, fio1-2 is a T-DNA insertion line that still expresses FIO1 mRNA, although at a lower level. Alternatively, the observed differences could be technical in nature, the result of either of the different sequencing approaches that were chosen or the growth conditions in which plants were cultivated. As described before, we have currently no knowledge on the spatial and temporal aspects of FIO1 function and differences in the circadian activity of FIO1 might also exist.
Our meRIP-sequencing approach further confirmed that FIO1 is likely not the main factor in the m6A modification of mRNAs but a more selective methyltransferase that modifies specific mRNAs. This assumption is supported by the finding that loss-of-function mutants are viable and able to produce mostly fertile offspring. Interestingly, despite the much higher number of differentially methylated transcripts in the fio1-2 mutant [9], the comparison of the differentially hypomethylated transcripts compared to those in fio1-1 and fio1-5 (this study) produced only a very moderate overlap (Fig 7C). Again, this might be due to the application of different methods or an indication that the reduction of FIO1 activity affects the m6A methylome more strongly than does the complete loss. A recent paper characterized the effect of the loss of FIO1 function on global splicing patterns and profound changes were identified [16]. The authors relate these changes to defects in U6 snRNA m6A modification, rather than being a direct consequence of loss of FIO1 activity.
The analysis of the m6A consensus in fio1-2 identified the YHAGA motif, which is significantly different to the RRACH motif that has been described in both plants and animals [2, 26], and to the RGACH consensus sequence that we identify in this work (Fig 7B) and the motifs identified by Parker et al [16].
Detailed analysis of specific transcripts that are differentially methylated and differentially expressed led us to the flowering regulator FLC. Regardless of whether the contribution of FLC methylation contributes only marginally to the early flowering response of fio1 mutants, our work unequivocally demonstrates that FIO1 is the m6A-methyltransferase that methylates the 3’UTR of FLC mRNA. We show that the failure to methylate FLC mRNA targets it for rapid degradation, hence the absence of FLC mRNA in fio1 mutants (Fig 6). Further characterization of the relationship between FIO1 and the biology of FLC will lead to insights into the function of its 3’-end methylation.
Our analyses focused on the role of methylation of mRNAs and the impact on the regulation of flowering. We cannot rule out confounding effects that the loss of FIO1 may have on the methylation and regulation of the non-coding transcriptome. Such effects and changes in splicing patterns might also greatly contribute to the pleiotropic phenotype of fio1 mutant plants and further characterization is needed to shed light on these processes.
Methods
Plant materials and growth conditions
Arabidopsis thaliana genotypes used in the study were, if not otherwise stated, in the Columbia Col-0 background. Double and triple mutant plants, such as fio1 co-sail, fio1 ft10 and fio1 miP1a miP1b were generated by genetic crossing. For flowering experiments, seeds were stratified 48 h at 4°C, and grown on soil in a plant growth chamber under long daylight conditions (16 h light / 8 h dark), or short daylight conditions (8 h light / 16 h dark) at 22°C day / 20°C night. Flowering time was measured by counting the number of rosette leaves at the bolting stage.
For RNA-seq, MeRIP-seq, Nanopore direct sequencing, and qPCR, 14-day old seedlings were collected. Seeds were sterilized in 70% ethanol and sown on 1/2 Murashige and Skoog (MS) medium plates with 0.8% agar and kept at 4°C for 48 hours in darkness for stratification and then grown at (22°C day / 20°C night) and 70% humidity under long daylight conditions (16 h light / 8 h dark).
Loss-of-function mutants of fio1 were generated using the CRISPR/Cas9 vector pKI1.1R, containing the Cas9 expression cassette (RPS5Ap::Cas9:HspT), a sgRNA expression cassette (U6.26p::AarI_site:sgRNA) and, for selection the RFP expression cassette (OLE1p::OLE1:TagRFP). Single-guide RNAs (sgRNAs) were designed using the web tool CRISPR-P v 2.0 [27]. Vectors with sgRNAs were generated according to the published description [28]. To create mutants with deletions, two to three Agrobacterium strains GV3101 pMD90 with different sgRNAs (S3 Table) were pooled and transformed into wild type plants via floral dip. RFP-positive seeds were selected using a Leica MZFLIII stereomicroscope equipped with RFP filters. Deletions were detected by PCR based sequencing.
Mapping-by-sequencing
91.99% sequenced reads were mapped by Bowtie2 (v2.1.0)[29] using the TAIR9 genome assembly and TAIR10 annotation from Phytozome v10.3 (phytozome.org). SNP calling was performed using samtools and BCFtools (v0.1.19)[30, 31]. 1118 (Chr1: 203, Chr2: 194, Chr3: 247, Chr4: 189, Chr5: 285) background corrected EMS-induced SNP markers were identified by SHOREmap[32] (v3.2) using standard settings. Finally, the mutations indicated a mapping interval of 7 Mb Kb on chromosome 2, containing 84 mutations. The trend line is the average of all SNP allele frequencies in a sliding window (size: 2,500 Kb; step: 100 Kb). Mapping-by-sequencing data has been deposited in NCBI’s Gene Expression Omnibus under GEO Series accession no. GSE171924.
FIO1 homology modeling
The methyltransferase domain of FIONA1 (UniProt accession code F4IGH3, residues 1–333) was modelled with Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2) using the Intensive modelling mode. The resulting homology model was aligned against the human crystal structure of the human FIONA1 homologue, METTL16 (PDB ID: 6DU4) for structural analysis.”
mRNA sequencing analysis
For RNAseq analysis, we collected two biological replicates of 14 day-old wild type (Col-0), fio1-1, fio1-5 seedlings. Total RNA was extracted from 100 A. thaliana seedlings for each line grown on a ½ MS agar plate using the Spectrum Plant Total RNA Kit (Sigma-Aldrich) following the manufacturer’s instructions. Total RNA was treated with DNAase I (RapidOut DNA Removal Kit, Thermo Scientific) according to the manufacturer’s instructions. Sequencing library preparation and sequencing on an Illumina HiSeq4000 instrument was performed by Novogene (Hongkong). About 3.7 Gb high-quality 150-bp paired-end reads were generated from each library. FastQC (Galaxy Version 0.72 + galaxy1) was initially run to assess the overall quality of all sample reads. Poor quality bases and adapters were filtered out using Trim Galore (Galaxy Version 0.6.3). The quality-filtered reads were aligned to the Arabidopsis thaliana reference genome (TAIR10) using HISAT282 (Version 2.1.0 + Galaxy4) with default parameters. HTseq (Galaxy Version 0.9.1) software was used to count the number of raw reads mapped to each of the genes. Differential expression analysis was performed with four analytical methods, DEseq 2 (Galaxy Version 2.11.40.6+galaxy1), edgeR (Galaxy Version 3.24.1+galaxy1), Limma-voom (Galaxy Version 3.38.3+galaxy3) and Limma-trend (Galaxy Version 3.38.3+galaxy3). All four statistical methods gave similar overall conclusions. We selected the most conservative results (Limma-voom; false discovery rate (FDR) = 0.05) for further investigation. Significance testing was performed using the Benjamini-Hochberg method[33]. Genes showing an absolute value of log2 FC (fold change; fio1 mutant / WT) ≥ 1.0 and adjusted P-value (false discovery rate; FDR) < 0.05 were considered as differentially expressed genes. RNAseq data generated in this study has been deposited in NCBI’s Gene Expression Omnibus under GEO Series accession no. GSE171926.
m6A RNA Immunoprecipitation sequencing (MeRIP-seq) and data analysis
MeRIP-seq was performed as described before[22] with modifications. Briefly, total RNA was extracted from 14 day-old Arabidopsis thaliana seedlings using the Spectrum Plant Total RNA Kit (Sigma-Aldrich) and treated with DNAase I (RapidOut DNA Removal Kit, Thermo Scientific). 300 μg of total RNA was mixed with 10×Fragmentation buffer (1 M Tris-HCl pH = 7.0, 1 M ZnCl2) and placed at 94°C for 5 min then snap cooled on ice for 5 minutes. The volume of fragmented RNA was then adjusted to 755 μl with RNase-free water. Next, 10 μL RNasin Plus RNase inhibitor (Promega, cat. no. N2611), 10 μL Ribonucleoside vanadyl complexes (RVC; 200 mM; Sigma-Aldrich, cat. no. R3380), 200 μL 5×IP buffer (50 mM Tris-HCl, 750 mM NaCl and 0.5% (vol/vol) Igepal CA-630), and 25 μL of m6A antibody (Synaptic Systems, cat. no. 202 003) were added to samples and samples were rotated at 4°C for 2 hours. After 2 hours, pre-blocked Protein A Dynabeads (Thermo Fisher, 1001D) was added to the RNA samples and rotated for an additional 2 hours at 4°C. After 2 hours, Dynabeads were pelleted using a magnetic stand and washed three times with 1 mL 1×IP buffer. RNA was eluted from Dynabeads by adding 98 μL elution buffer (20 mM Tris-HCl pH 7.5, 300 mM sodium acetate, 2 mM EDTA, 0.25% SDS), 2 μL of proteinase K (Thermo Fisher, AM2546) and then shaking for 1 hour at 37°C. All samples were precipitated using 3 M sodium acetate (pH 5.2) and 2.5 volumes of 100% ethanol and kept at -80°C overnight. Libraries were prepared using NEBNext Multiplex Small RNA Library Prep Set for Illumina (New England BioLabs, E7300S) according to the manufacturer’s instructions. Novogene (Beijing) performed sequencing on an Illumina HiSeq4000 instrument. About 3.0 Gb high-quality 150-bp paired-end reads were generated from each library. FastQC (Galaxy Version 0.72 + galaxy1) was initially run to assess the overall quality of all sample reads. Poor quality bases and adapters were filtered out using Trim Galore (Galaxy Version 0.6.3). The quality-filtered reads were aligned to the A. thaliana reference genome using HISAT2 (Version 2.1.0 + Galaxy4) with default parameters. To identify regions in which m6A modifications occurred, MACS [23] was used to call peaks on aligned files. The peaks showing an absolute value of log2 FC (fold change; fio1 mutant / WT) ≥ 0.5 and RPM ≥ 5 were considered as differentially modified peaks. MeRIPseq data generated in this study has been deposited in NCBI’s Gene Expression Omnibus under GEO Series accession no. GSE171928.
mRNA stability measurements
mRNA stability measurement assay was performed as previously described [34] with modification. Briefly, 12-day-old Arabidopsis seedlings grown on 1/2 MS medium were transferred to 10-cm Petri dishes containing 1/2 MS liquid medium at ZT13. After 30 min incubation, 0.2 mM actinomycin D was added to the buffer. The tissues were collected at 1 h after the transcription inhibitor was added; these samples are referred to as 0 h samples. The 2 h and 4 h samples were collected and immediately frozen in liquid nitrogen. The total RNA was isolated from these tissues, and the remaining mRNA levels were quantified by RT-qPCR with gene-specific qPCR primers. 18S RNA was used as the internal control, and AT2G07689 was used as a negative control [35]. Primers used for qPCR: AT2G07689-qF;CATTACGGCAAACCCGTGTC | AT2G07689-qR;GGCTAACGGGGGTATTCCTG | FLC-qF;GAGAACAAAAGTAGCCGACAAGTC | FLC-qR;GGATGCGTCACAGAGAACAGA | 18s-qF;GCGGCTTAATTTGACTCAACACG | 18s-qR;CCTGTTATTGCCTCAAACTTCC
Primers for FA-RIP-qPCR, m6A-IP-qPCR and mRNA stability assay: FLC-IP-qPCR-F;CTCCCACTACTTAATTAGCCACCTTA | FLC-IP-qPCR-R;CCCTTATCAGCGGAATAATTACATATC
Nanopore direct RNA sequencing
Total RNA was isolated as described above for mRNA-seq and direct RNA sequencing libraries were prepared by CD genomics using the Oxford Nanopore DRS protocol (SQK-RNA002, Oxford Nanopore Technologies). Samples were loaded into the Nanopore R9.4 sequencing micro-array and sequenced for 48–72 hrs using the PromethION sequencer (Oxford Nanopore Technologies). Read quality assessment, base calling and adapter trimming was carried out with the Guppy software (version 3.2.6). Nanofilt (version 2.7.1) was then used to remove low quality reads (Q-value < 7) and short-length reads (<50 bp). The clean reads were subsequently corrected using Fclmr2 (version 0.1.2). Minimap2 (version 2.17-r941) was used to map the clean reads to the A. thaliana genome and the alignment ratio of clean reads to the reference genes was calculated using Samtools (version 1.10). To identify m6A sites, the Tombo software de novo model together with MINES was used for calculation. Methylkit software was then used to analyze differential methylation sites (DML). Logistic regression test was used to detect differential methylation sites. Nanopore direct RNA-sequencing data has been deposited in NCBI’s Gene Expression Omnibus under GEO Series accession no. GSE212766.
RNA m6A immunoprecipitation RT-qPCR
Quantitative real-time PCR was performed to assess relative abundance of m6A RNA in the RIP samples. 300 μg total RNA was adjusted the volume to 1000 μl with 5×IP buffer (50 mM Tris-HCl, 750 mM NaCl and 0.5% (vol/vol) Igepal CA-630) and RNase-free water and incubated with 10 μg m6A antibody (Synaptic Systems, cat. no. 202 003, Goettingen, Germany). The mixture was rotated at 4°C for 2 h, then pre-blocked and washed Dynabead Protein A (Thermo Fisher, 1001D) were added and the mixture rotated for an additional 2 h at 4°C. After washing with IP buffer containing Ribonucleoside vanadyl complexes (RVC, Sigma, R3380-5ML) three times, the m6A IP RNA was eluted with 98 μL elution buffer (20 mM Tris-HCl pH 7.5, 300 mM sodium acetate, 2 mM EDTA, 0.25% SDS). 2 μL of proteinase K (Thermo Fisher, AM2546) was added and the RNA incubated for 1 hour at 37°C with gentle shaking. All samples were precipitated using 3 M sodium acetate (pH 5.2) and 2.5 volumes of 100% ethanol and kept at -80°C overnight. cDNA was synthesized by iScrip cDNA Synthesis Kit (Bio-Rad). qPCR analyses was done with Ultra SYBR Mixture with ROX (CWBIO) on a CFX384 Touch Real-Time PCR Detection System (Bio-Rad). qRT- PCR primers that were used to amplify FLC were: flc_qF: AGCCAAGAAGACCGAACTCA and flc_qR: TTTGTCCAGCAGGTGACATC.
Supporting information
(A) Phenotype of fio1-5 compared to the Col-0 wildtype when grown in LD conditions. (B) Determination of flowering by counting the number of rosette leaves (RLN = rosette leaf number) at the bolting stage in LD. Plotted are average leaf number +/- SD, ***p = <0.001, N = 10–14. (C) Nucleotide alignment showing the CRISPR-induced genomic deletion found in fio1-5. Gene model on top shows the relative positions of all three fio1 mutations.
(PDF)
The three mutants were mapped to the homology model of FIO1 (see Materials and Methods). The fio1-1 mutation involved the loss of five amino acids highlighted in pink, including the loss of a potential hydrogen bond between the threonine and asparagine. The sum8 mutation changes the serine (orange), which normally hydrogen bonds to a tryptophan, into an asparagine. The resulting larger side-chain of asparagine is unlikely to be accommodated in the constrained protein interior, leading to changes in the protein structure and loss of function. The fio1-5 mutation involves a large deletion (orange) and missense mutations (light cyan) in a partially buried alpha helix, which are very likely to disrupt protein folding and function.
(PDF)
Plants were grown in long day conditions (16h light, 8 hour dark) and the number of leaves were counted at the bolting stage. Depicted is the average +/- standard deviation. N = 10.
(PDF)
(A) and (B) Quantification of CO and FT in Col-0, fio1-1 and fio1-5 by qRT-PCR. Values are the means ±SD. N = 4. * P ≤ 0.01. (C) Phenotypes of miP1a miP1b, fio1-1, fio1-5, miP1a miP1b fio1-5, co-sail, co-sail fio1-1, co-sail fio1-5, ft10, ft10 fio1-1, ft10 fio1-5 and determination of flowering time by counting the number of rosette leaves at bolting compare to wild type, under long day conditions. RLN = number of rosette leaves at the bolting stage. Values are the means ±SD. N = 10 to 20. One-way ANOVA was carried out to test significance, **P ≤ 0.005, ***P≤ 0.001. (D) Phenotypes of miP1a miP1b, fio1-1, fio1-5, miP1a miP1b fio1-5, co-sail, co-sail fio1-1, co-sail fio1-5, ft10, ft10 fio1-1, ft10 fio1-5 and determination of flowering time by counting the number of rosette leaves at bolting compare to wild type, under short day conditions. RLN = number of rosette leaves at the bolting stage. Values are the means ±SD. N = 10 to 12. One-way ANOVA was carried out to test significance, ***P≤ 0.001.
(PDF)
Hypocotyl length of Col-0 wildtype and fio1-1 and fio1-5 mutants grown in either white light conditions or in far-red light enriched white light conditions (+FR). The fio1 mutants show a hypersensitivity response with increase hypocotyls in white light and even longer hypocotyls in shade conditions.
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank Peter Brodersen and Sebastian Marquardt for comments on the manuscript and Hong Gil Nam (DGIST, Korea) for providing fio1-1 mutant seeds.
Data Availability
RNAseq data generated in this study has been deposited in NCBI's Gene Expression Omnibus under GEO Series accession no. GSE171926. MeRIPseq data generated in this study has been deposited in NCBI's Gene Expression Omnibus under GEO Series accession no. GSE171928. Mapping-by-sequencing data has been deposited in NCBI's Gene Expression Omnibus under GEO Series accession no. GSE171924. Nanopore direct RNA-sequencing data has been deposited in NCBI's Gene Expression Omnibus under GEO Series accession no. GSE212766.
Funding Statement
We acknowledge funding through NovoCrops Centre (Novo Nordisk Foundation project number 2019OC53580 to S. W.), the Independent Research Fund Denmark (0136‐00015B and 0135‐00014B to S. W.), the Novo Nordisk Foundation (NNF18OC0034226 and NNF20OC0061440 to S. W.), and the funding of DataPLANT (NFDI 7/1 – 42077441) as part of the German National Research Data Infrastructure funded by the Deutsche Forschungsgemeinschaft (DFG – German Research Foundation). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20(10):608–24. Epub 2019/09/15. doi: 10.1038/s41580-019-0168-5 . [DOI] [PubMed] [Google Scholar]
- 2.Luo GZ, MacQueen A, Zheng G, Duan H, Dore LC, Lu Z, et al. Unique features of the m6A methylome in Arabidopsis thaliana. Nat Commun. 2014;5:5630. Epub 2014/11/29. doi: 10.1038/ncomms6630 ; PubMed Central PMCID: PMC4248235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10(2):93–5. Epub 2013/12/10. doi: 10.1038/nchembio.1432 ; PubMed Central PMCID: PMC3911877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008;20(5):1278–88. Epub 2008/05/29. doi: 10.1105/tpc.108.058883 ; PubMed Central PMCID: PMC2438467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Růžička K, Zhang M, Campilho A, Bodi Z, Kashif M, Saleh M, et al. Identification of factors required for m(6) A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 2017;215(1):157–72. Epub 2017/05/16. doi: 10.1111/nph.14586 ; PubMed Central PMCID: PMC5488176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, et al. The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell. 2017;169(5):824–35.e14. Epub 2017/05/20. doi: 10.1016/j.cell.2017.05.003 ; PubMed Central PMCID: PMC5502809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mendel M, Delaney K, Pandey RR, Chen KM, Wenda JM, Vågbø CB, et al. Splice site m(6)A methylation prevents binding of U2AF35 to inhibit RNA splicing. Cell. 2021. Epub 2021/05/01. doi: 10.1016/j.cell.2021.03.062 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arribas-Hernández L, Bressendorff S, Hansen MH, Poulsen C, Erdmann S, Brodersen P. An m(6)A-YTH Module Controls Developmental Timing and Morphogenesis in Arabidopsis. Plant Cell. 2018;30(5):952–67. Epub 2018/04/13. doi: 10.1105/tpc.17.00833 ; PubMed Central PMCID: PMC6002192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu T, Wu X, Wong CE, Fan S, Zhang Y, Zhang S, et al. FIONA1-Mediated m(6) A Modification Regulates the Floral Transition in Arabidopsis. Adv Sci (Weinh). 2022:e2103628. Epub 2022/01/07. doi: 10.1002/advs.202103628 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu C, Wu Z, Duan HC, Fang X, Jia G, Dean C. R-loop resolution promotes co-transcriptional chromatin silencing. Nat Commun. 2021;12(1):1790. Epub 2021/03/21. doi: 10.1038/s41467-021-22083-6 ; PubMed Central PMCID: PMC7979926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graeff M, Straub D, Eguen T, Dolde U, Rodrigues V, Brandt R, et al. MicroProtein-Mediated Recruitment of CONSTANS into a TOPLESS Trimeric Complex Represses Flowering in Arabidopsis. PLoS Genet. 2016;12(3):e1005959. Epub 2016/03/26. doi: 10.1371/journal.pgen.1005959 ; PubMed Central PMCID: PMC4807768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodrigues VL, Dolde U, Sun B, Blaakmeer A, Straub D, Eguen T, et al. A microProtein repressor complex in the shoot meristem controls the transition to flowering. Plant Physiol. 2021. Epub 2021/05/21. doi: 10.1093/plphys/kiab235 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J, Kim Y, Yeom M, Kim JH, Nam HG. FIONA1 is essential for regulating period length in the Arabidopsis circadian clock. Plant Cell. 2008;20(2):307–19. Epub 2008/02/19. doi: 10.1105/tpc.107.055715 ; PubMed Central PMCID: PMC2276451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorsett M, Westlund B, Schedl T. METT-10, a putative methyltransferase, inhibits germ cell proliferative fate in Caenorhabditis elegans. Genetics. 2009;183(1):233–47. Epub 2009/07/15. doi: 10.1534/genetics.109.105270 ; PubMed Central PMCID: PMC2746148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendel M, Chen KM, Homolka D, Gos P, Pandey RR, McCarthy AA, et al. Methylation of Structured RNA by the m(6)A Writer METTL16 Is Essential for Mouse Embryonic Development. Mol Cell. 2018;71(6):986–1000.e11. Epub 2018/09/11. doi: 10.1016/j.molcel.2018.08.004 ; PubMed Central PMCID: PMC6162343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker MT, Soanes BK, Kusakina J, Larrieu A, Knop K, Joy N, et al. m6A modification of U6 snRNA modulates usage of two major classes of pre-mRNA 5’ splice site. bioRxiv. 2022:2022.04.05.487178. doi: 10.1101/2022.04.05.487178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303(5660):1003–6. doi: 10.1126/science.1091761 WOS:000188918000043. [DOI] [PubMed] [Google Scholar]
- 18.Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, et al. FT Protein Movement Contributes to Long-Distance Signaling in Floral Induction of Arabidopsis. Science. 2007;316(5827):1030–3. doi: 10.1126/science.1141752 [DOI] [PubMed] [Google Scholar]
- 19.Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a Protein Is a Mobile Flowering Signal in Rice. Science. 2007;316(5827):1033–6. doi: 10.1126/science.1141753 [DOI] [PubMed] [Google Scholar]
- 20.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. Epub 2014/02/04. doi: 10.1186/gb-2014-15-2-r29 ; PubMed Central PMCID: PMC4053721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang C, Yang J, Song P, Zhang W, Lu Q, Yu Q, et al. FIONA1 is an RNA N(6)-methyladenosine methyltransferase affecting Arabidopsis photomorphogenesis and flowering. Genome Biol. 2022;23(1):40. Epub 2022/02/02. doi: 10.1186/s13059-022-02612-2 ; PubMed Central PMCID: PMC8802475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, Rechavi G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc. 2013;8(1):176–89. Epub 2013/01/05. doi: 10.1038/nprot.2012.148 . [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9(9):R137. Epub 2008/09/19. doi: 10.1186/gb-2008-9-9-r137 ; PubMed Central PMCID: PMC2592715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bodi Z, Zhong S, Mehra S, Song J, Graham N, Li H, et al. Adenosine Methylation in Arabidopsis mRNA is Associated with the 3’ End and Reduced Levels Cause Developmental Defects. Front Plant Sci. 2012;3:48. Epub 2012/05/29. doi: 10.3389/fpls.2012.00048 ; PubMed Central PMCID: PMC3355605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, et al. Distinct Roles of CONSTANS Target Genes in Reproductive Development of Arabidopsis. Science. 2000;288(5471):1613–6. doi: 10.1126/science.288.5471.1613 [DOI] [PubMed] [Google Scholar]
- 26.Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Höbartner C, et al. Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18(11):2004–14. Epub 2017/10/21. doi: 10.15252/embr.201744940 ; PubMed Central PMCID: PMC5666602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H, Ding Y, Zhou Y, Jin W, Xie K, Chen LL. CRISPR-P 2.0: An Improved CRISPR-Cas9 Tool for Genome Editing in Plants. Mol Plant. 2017;10(3):530–2. Epub 2017/01/17. doi: 10.1016/j.molp.2017.01.003 . [DOI] [PubMed] [Google Scholar]
- 28.Tsutsui H, Higashiyama T. pKAMA-ITACHI Vectors for Highly Efficient CRISPR/Cas9-Mediated Gene Knockout in Arabidopsis thaliana. Plant Cell Physiol. 2017;58(1):46–56. Epub 2016/11/20. doi: 10.1093/pcp/pcw191 ; PubMed Central PMCID: PMC5444565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. Epub 2012/03/06. doi: 10.1038/nmeth.1923 ; PubMed Central PMCID: PMC3322381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27(21):2987–93. Epub 2011/09/10. doi: 10.1093/bioinformatics/btr509 ; PubMed Central PMCID: PMC3198575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. Epub 2009/06/10. doi: 10.1093/bioinformatics/btp352 ; PubMed Central PMCID: PMC2723002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, et al. SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Meth. 2009;6(8):550–1. http://www.nature.com/nmeth/journal/v6/n8/suppinfo/nmeth0809-550_S1.html. doi: 10.1038/nmeth0809-550 [DOI] [PubMed] [Google Scholar]
- 33.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological). 1995;57(1):289–300. doi: 10.2307/2346101 [DOI] [Google Scholar]
- 34.Seo E, Yu J, Ryu KH, Lee MM, Lee I. WEREWOLF, a regulator of root hair pattern formation, controls flowering time through the regulation of FT mRNA stability. Plant Physiol. 2011;156(4):1867–77. Epub 2011/06/10. doi: 10.1104/pp.111.176685 ; PubMed Central PMCID: PMC3149934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei LH, Song P, Wang Y, Lu Z, Tang Q, Yu Q, et al. The m(6)A Reader ECT2 Controls Trichome Morphology by Affecting mRNA Stability in Arabidopsis. Plant Cell. 2018;30(5):968–85. Epub 2018/05/03. doi: 10.1105/tpc.17.00934 ; PubMed Central PMCID: PMC6002187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Phenotype of fio1-5 compared to the Col-0 wildtype when grown in LD conditions. (B) Determination of flowering by counting the number of rosette leaves (RLN = rosette leaf number) at the bolting stage in LD. Plotted are average leaf number +/- SD, ***p = <0.001, N = 10–14. (C) Nucleotide alignment showing the CRISPR-induced genomic deletion found in fio1-5. Gene model on top shows the relative positions of all three fio1 mutations.
(PDF)
The three mutants were mapped to the homology model of FIO1 (see Materials and Methods). The fio1-1 mutation involved the loss of five amino acids highlighted in pink, including the loss of a potential hydrogen bond between the threonine and asparagine. The sum8 mutation changes the serine (orange), which normally hydrogen bonds to a tryptophan, into an asparagine. The resulting larger side-chain of asparagine is unlikely to be accommodated in the constrained protein interior, leading to changes in the protein structure and loss of function. The fio1-5 mutation involves a large deletion (orange) and missense mutations (light cyan) in a partially buried alpha helix, which are very likely to disrupt protein folding and function.
(PDF)
Plants were grown in long day conditions (16h light, 8 hour dark) and the number of leaves were counted at the bolting stage. Depicted is the average +/- standard deviation. N = 10.
(PDF)
(A) and (B) Quantification of CO and FT in Col-0, fio1-1 and fio1-5 by qRT-PCR. Values are the means ±SD. N = 4. * P ≤ 0.01. (C) Phenotypes of miP1a miP1b, fio1-1, fio1-5, miP1a miP1b fio1-5, co-sail, co-sail fio1-1, co-sail fio1-5, ft10, ft10 fio1-1, ft10 fio1-5 and determination of flowering time by counting the number of rosette leaves at bolting compare to wild type, under long day conditions. RLN = number of rosette leaves at the bolting stage. Values are the means ±SD. N = 10 to 20. One-way ANOVA was carried out to test significance, **P ≤ 0.005, ***P≤ 0.001. (D) Phenotypes of miP1a miP1b, fio1-1, fio1-5, miP1a miP1b fio1-5, co-sail, co-sail fio1-1, co-sail fio1-5, ft10, ft10 fio1-1, ft10 fio1-5 and determination of flowering time by counting the number of rosette leaves at bolting compare to wild type, under short day conditions. RLN = number of rosette leaves at the bolting stage. Values are the means ±SD. N = 10 to 12. One-way ANOVA was carried out to test significance, ***P≤ 0.001.
(PDF)
Hypocotyl length of Col-0 wildtype and fio1-1 and fio1-5 mutants grown in either white light conditions or in far-red light enriched white light conditions (+FR). The fio1 mutants show a hypersensitivity response with increase hypocotyls in white light and even longer hypocotyls in shade conditions.
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
RNAseq data generated in this study has been deposited in NCBI's Gene Expression Omnibus under GEO Series accession no. GSE171926. MeRIPseq data generated in this study has been deposited in NCBI's Gene Expression Omnibus under GEO Series accession no. GSE171928. Mapping-by-sequencing data has been deposited in NCBI's Gene Expression Omnibus under GEO Series accession no. GSE171924. Nanopore direct RNA-sequencing data has been deposited in NCBI's Gene Expression Omnibus under GEO Series accession no. GSE212766.