Abstract
Background
Inflammatory bowel disease (IBD) is a chronic inflammatory disease of the gastrointestinal tract. We aimed to investigate, for the first time, the expression profile of serum level of LncRNA THRIL and MiR-125b in IBD patients and their relations with patient’s clinical and biochemical investigations.
Methods
Our study included 210 subjects divided into 70 healthy subjects considered as control group (male and female), 70 patients with ulcerative colitis (UC), and 70 patients with Crohn’s disease (CD). Blood samples were obtained from all subjects. Expression of LncRNA THRIL and MiR-125b in serum was detected by Quantitative real time PCR (qRT-PCR).
Results
Our results showed a significant increase in the fold change of LncRNA THRIL in UC patients (Median = 11.11, IQR; 10.21–12.45, P<0.001) and CD patients (Median = 5.87, IQR; 4.57–7.88, P<0.001) compared to controls. Meanwhile there was a significant decrease in the fold change of MiR-125b in UC patients (Median = 0.36, IQR; 0.19–0.61, P<0.001) and CD patients (Median = 0.69, IQR; 0.3–0.83, P<0.001) compared to controls. Furthermore, there was a negative significant correlation between LncRNA THRIL and MiR-125b in UC patients (r = -0.28, P = 0.016) and in CD patients (r = -0.772, P<0.001). ROC curve analysis was done showing the diagnostic value of these markers as predictors in differentiating between cases of UC, CD, and control.
Conclusion
Serum LncRNA THRIL and MiR-125b could be used as potential biomarkers for diagnosis and prognosis of ulcerative colitis and Crohn’s disease.
Introduction
Inflammatory bowel disease is a chronic inflammatory disease of the gastrointestinal tract that is classified into two types: ulcerative colitis (UC) and Crohn’s disease (CD), CD is characterized by patchy transmural inflammatory patterns of any portion along the intestinal wall affecting the whole thickness of the intestinal layers, While in case of UC the inflammatory process limited to large intestine affecting the innermost layers of the mucosa. The most frequent symptoms of these disorders are bloody diarrhea, abdominal pain, malabsorption, fatigue [1]. The condition could be complicated by intestinal fistula, intestinal obstruction, abdominal abscesses, and increased incidence of malignancy [2, 3]. Pathogenesis of IBD is still unclear but many factors interaction may play a role as genetic predisposition, microbial infection, and environmental factors [1].
Long non-coding RNAs (lncRNA) are transcripts) that contain more than 200 nucleotides and not translated into protein; they are processed by RNA polymerase II [4]. It was reported that lncRNA regulates cell function and biological processes in intestinal diseases such as irritable bowel syndrome [5], Hirschsprung’s disease [6], and IBD [7]. Many studies demonstrate the role of LncRNAs in IBD. For example, LncRNA CCAT1 promote IBD malignancy by downregulating miR-185-3p [8], LncRNA KIF9-AS1 promotes cell apoptosis by targeting the microRNA-148a-3p [9], LncRNA H19 act as a Competing Endogenous RNA to Regulate AQP Expression in the Intestinal Barrier of IBS-D Patients [10]. Haberman et al. reported 15 expressed LncRNAs differentially expressed in the tissue samples obtained from the patients with CD when compared to healthy control [11], another study by Wang et al. found that LINC01272 was highly expressed in peripheral blood and tissues of IBD [12]. The exact role of LncRNA is still unclear; it may act through chromatin remodeling, regulation of protein activity and its stability [13, 14].
LncRNA THRIL (TNFα and heterogeneous nuclear ribonucleoprotein L (hnRNPL) related immunoregulatory lincRNA) binds to the promoter region of the TNF-a forming RNA-protein complex inducing the expression of TNF-a, which contribute to the inflammatory process [15], and its dysregulation characterizes the autoimmune and inflammatory diseases. Chen et al also showed that knockdown of LncRNA THRIL decreases the levels of TNFa, IL1b, IL6, macrophages and neutrophils counts [16].
MicroRNA is a small single-stranded non-coding RNA molecule (containing about 22 nucleotides) that regulates gene expression through base pairing with complementary sequences within mRNA molecules [17]. Their abnormal activity has been demonstrated in many diseases including inflammatory and immunological disorders; Many studies reported that miRNA is associated with other pathophysiological factors of IBD, they may act by increasing or decreasing the intensity of the inflammatory process [18, 19], or by strengthening or weakening the intestinal barrier [20–23].
MiR-125b is transcribed from two loci located on chromosomes 11q23 (hsa-miR-125b-1) and 21q21 (hsa-miR-125b-2) [24]. It regulates the proliferation and differentiation of tumor cells and could be used as a diagnostic biomarker for early-stage cervical cancer and rheumatoid arthritis [25, 26].
The aim of this work was to evaluate the relative expression levels of serum LncRNA THRIL and MiR-125b in IBD patients and their relations with patients’ clinical and biochemical investigations.
Materials and methods
Subjects
Our study included 210 subjects divided into 70 healthy subjects considered as controls, 70 patients with ulcerative colitis, and 70 patients with Crohn’s disease. Patients were selected from outpatient clinics and inpatients of Tropical and Internal Medicine departments, Fayoum University Hospital, Fayoum University, Egypt. The study was revised and approved by the Ethical Committee of Faculty of Medicine, Fayoum University. Informed consent was obtained from all participants before sample collection.
The diagnosis of inflammatory bowel disease (IBD) with its 2 main subtypes, Crohn’s disease, and ulcerative colitis is based on patient history, clinical symptoms, radiological, and endoscopic criteria followed by histopathological examination of biopsies collected from each anatomic segment (rectum; sigmoid, left, transverse, and right colon; and ileum) according to European Crohn’s and Colitis Organization (ECCO) guidelines [27].
The Crohn’s Disease Activity Index (CDAI)
CDAI was used for evaluating the disease severity of CD [28]. This index is scored on a scale from 0 to 1100 and includes abdominal pain, general wellbeing, complications, abdominal mass, anemia, and weight change. The patients with CD can be divided into
Asymptomatic remission (CDAI < 150)
Mild-to-moderate CD (150–220)
Moderate-to-severe CD (220–450)
Severe-fulminant disease (>450).
The Mayo score
It was used for grading the severity of UC. The Score is based on four parameters: stool frequency, rectal bleeding, endoscopic findings, and Physician rating of disease activity [29]. Total criteria point count: Scores range from 0 to 12, with higher scores indicating more severe disease.
Stool pattern: The patient reports a normal number of daily stools (0 points), 1 to 2 more stools than normal (1 point), 3 to 4 more stools (2 points), 5 or more stools (3 points).
Rectal bleeding: None (0 points), Blood streaks seen in the stool less than half the time (1 point), all stools contain blood (2 points), Presence of pure blood (3 points).
Endoscopic findings: Normal or inactive colitis seen (0 points), Mild colitis: mild erythema, decrease in vascularity (1 point), Moderate colitis: marked erythema, erosions seen (2 points), Severe colitis: spontaneous bleeding (3 points).
Physician rating of disease activity: Normal (0 points), Mild colitis (1 point), Moderate colitis (2 points), severe colitis (3 points).
Samples collection
Six-milliliter blood samples were drawn from all participants and collected in two tubes; one of them containing Ethylene Diamine TetraAcetic Acid (EDTA) for complete blood count (CBC) and Erythrocyte sedimentation rate (ESR) assay, the second tube left to clot for 15 min, centrifuged at 4000_g for 10 min, collect the serum, and stored at -80°C for all serological tests, including molecular biology techniques.
RNA extraction
RNAs were extracted from serum using the miRNeasy Serum/Plasma Kit (Qiagen, Valenica, CA, USA) extraction kits following the manufacturer protocol, RNA concentration and purity were determined by Nano Drop2000 (Thermo Scientific, USA).
Reverse transcription reactions
Reverse transcription was carried out on total RNA in a final volume of 20 μL(11 μl RNA + 2 μl genomic DNA elimination (GE)+ 7μl reverse-transcription mix). RNAs were reverse transcribed by RT-PCR kit into cDNAs using RvertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions.
Quantitative real-time polymerase chain reaction
The cDNAs templates were amplified by Quantitative RT-PCR using the miScript SYBR Green PCR Kit (Qiagen, Germany). Regarding MiR-125b, the total volume was 25 μL per reaction using the specific MiR-125b primer (catalog numbers MS00006629 Lot. Number 20151214121). SNORD 68 was used to normalize the expression and for relative quantification of MiR-125b. While the total volume for LncRNA THRIL was 20 μL per reaction using the specific LncRNA THRIL primer (catalog numbers 330701LPH42418A) and GADPH was used to normalize the expression and for relative quantification of LncRNA THRIL. The qRT-PCR for both was programmed for the following cycling conditions: Initial activation step for 15 min at 95 °C then cycling denaturation for 15 sec at 94 °C, annealing for 30 sec at 55 °C, and extension for 30 sec at 70 °C, and these steps were repeated for 40 cycles in Rotor-gene qRT-PCR system thermocycler (Qiagen, USA). The relative expression of RNAs was calculated by the 2-ΔΔCt method [30].
Statistical analysis
The collected data were arranged and statistically analyzed using SPSS software with statistical computer package version 25 (SPSS). For quantitative data, the mean, median, SD, and interquartile range were calculated. If the variable was not normally distributed, the Mann–Whitney U test or the Kruskal–Wallis test was used for comparison between any two groups or three groups, respectively. Otherwise, one-way ANOVA was used. Qualitative data were presented as number and percentages. Chi-square (χ2) was used as a test of significance. Spearman’s correlation was run to identify the relation of LNC THRIL and MiR-125b with study parameters. (ROC) curves were used to determine the cutoff point, which shows the highest sensitivity and specificity of LNC THRIL and MiR-125b in differentiating between different study groups. P values <0.05 were considered as statistically significant.
Results
Demographic, clinical and laboratory characteristics of the study groups
There was no significant difference between patients and controls as regards age (p = 0.106) and sex (P = 0.868). Results showed a highly statistically significant difference between UC, CD, and control groups as regards Hb (P<0.001), Total leucocytic count (P<0.001), Platelets number (P<0.001) with high level in CD and Albumin level (P<0.001) with a low level in CD. There was also a highly statistically significant difference between UC, CD groups as regards incidence of diabetes (P = 0.004), musculoskeletal complications (P = 0.006), Hematocrit (P = 0.041), CRP (P <0.001) and ESR (P = 0.023) with high level in CD group and as regards patients on salicalyates (P = 0.002) with a high level in UC (Table 1).
Table 1. Demographic, biochemical and clinical characteristics of the study groups.
| Ulcerative colitis | Crohn’s Disease | Control | P-value | |||||
|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | |||
| Age | 32.1 | 3.7 | 33.2 | 1.7 | 30 | 2.3 | 0.106 | |
| Duration of illness | 3.8 | 0.3 | 4.6 | 0.1 | . | . | 0.436 | |
| No | % | No | % | No | % | |||
| sex | Female | 28 | 40.0% | 26 | 37.1% | 25 | 35.7% | 0.868 |
| male | 42 | 60.0% | 44 | 62.9% | 45 | 64.3% | ||
| smoker | no | 54 | 77.1% | 48 | 68.6% | 59 | 84.3% | 0.089 |
| yes | 16 | 22.9% | 22 | 31.4% | 11 | 15.7% | ||
| coffee consumption | no | 58 | 82.9% | 48 | 68.6% | 54 | 77.1% | 0.136 |
| yes | 12 | 17.1% | 22 | 31.4% | 16 | 22.9% | ||
| diabetes | no | 62 | 88.6% | 68 | 97.1% | 0.004* | ||
| yes | 8 | 11.4% | 2 | 2.9% | ||||
| hypertension | no | 68 | 97.1% | 68 | 97.1% | 0.361 | ||
| yes | 2 | 2.9% | 2 | 2.9% | ||||
| Other comorbidities | congenital heart disease (TS) | 0 | 0.0% | 2 | 2.9% | |||
| Ischemic heart disease | 0 | 0.0% | 2 | 2.9% | ||||
| stroke | 0 | 0.0% | 2 | 2.9% | ||||
| thyrotoxic | 2 | 2.9% | 0 | 0.0% | ||||
| no | 68 | 97.1% | 64 | 91.4% | ||||
| Extraintestinal | ||||||||
| Hepatobiliary | 6 | 8.6% | ||||||
| Endocrine | 2 | 2.9% | ||||||
| Arthralgia | 4 | 5.7% | ||||||
| Thromboembolic | 10 | 14.3% | ||||||
| MSK | 22 | 31.4% | 38 | 54.3% | 0.006* | |||
| Eye | 10 | 14.3% | 8 | 11.4% | 0.614 | |||
| History (Crohn’s) | ||||||||
| ileal strictures | 10 | 4.8% | ||||||
| rectal fistula | 20 | 9.5% | ||||||
| Perforation | 8 | 3.8% | ||||||
| Intestinal obstruction | 12 | 5.7% | ||||||
| surgical resection | 8 | 3.8% | ||||||
| colocutaneous fistula | 8 | 3.8% | ||||||
| perianal abcess or fistula | 4 | 1.9% | ||||||
| colonic stricture | 4 | 1.9% | ||||||
| psoas abcess | 6 | 2.9% | ||||||
| Treatment | ||||||||
| Salicalyates | 42 | 60.0% | 24 | 34.3% | 0.002* | |||
| Steroids | 38 | 54.3% | 30 | 42.9% | 0.176 | |||
| Azathioprine | 18 | 25.7% | 20 | 28.6% | 0.704 | |||
| Infliximab | 10 | 14.3% | 16 | 22.9% | 0.192 | |||
| Mayo score | 7.6 | 3.6 | . | . | . | . | ||
| CDAI | . | . | 267.3 | 130.9 | . | . | ||
| Hb gl/dl | 11.2 | 2 | 12.9 | 2 | 12 | 1.2 | <0.001* | |
| HCT | 34.4 | 6.1 | 36.5 | 5.9 | . | . | 0.041* | |
| TLC | 8.3 | 4.8 | 8.5 | 3.9 | 5.5 | 1.9 | <0.001* | |
| Neutrophil count | 60.7 | 16.6 | 60.5 | 12.1 | . | . | 0.936 | |
| Platelets | 317.3 | 107.4 | 348.2 | 126 | 210.1 | 34.8 | <0.001* | |
| CRP | 14.2 | 12.2 | 26.8 | 26.1 | . | . | <0.001* | |
| ESR | 26.5 | 18.5 | 35.5 | 27.3 | . | . | 0.023* | |
| Albumin | 3.7 | 0.8 | 3.6 | 0.7 | 4.6 | 0.3 | <0.001* | |
Description of fold change of MiR-125b and Lnc THRIL among study groups
Results showed significant differences between the patients’ groups and control group regarding Lnc THRIL and MiR-125b with the fold change of Lnc THRIL was significantly up-regulated in UC patients (Median = 11.11, IQR; 10.21–12.45, P<0.001) and CD patients (Median = 5.87, IQR; 4.57–7.88, P<0.001) compared to controls. Meanwhile, the fold change of MiR-125b was significantly down-regulated in UC patients (Median = 0.36, IQR; 0.19–0.61, P<0.001) and CD patients (Median = 0.69, IQR; 0.3–0.83, P<0.001) compared to controls (Table 2).
Table 2. Description of fold change of MiR-125b and Lnc THRIL among study groups.
| Ulcerative colitis | Crohn’s | Control | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Median | IQR | Median | IQR | Median | IQR | ||||
| MiR-125b | 0.36 | 0.19 | 0.61 | 0.69 | 0.31 | 0.83 | 1 | 1 | 1 |
| P-values | 0.004* | ||||||||
| <0.001* | |||||||||
| <0.001* | |||||||||
| Lnc THRIL | 11.11 | 10.21 | 12.45 | 5.87 | 4.57 | 7.88 | 1 | 1 | 1 |
| P-values | <0.001* | ||||||||
| <0.001* | |||||||||
| <0.001* | |||||||||
Relations between Lnc THRIL and MiR-125b and clinical data in UC and CD patients
We found that the level of LncRNA THRIL was significantly higher in nonsmoker UC patients than in smokers (P = 0.012) and the level of MiR-125b was significantly higher in patients with endocrinal complication (P = 0.041) (Table 3). Meanwhile in CD patients, results showed that the level of LncRNA THRIL was significantly higher in females (P <0.001), nondiabetic patients (P <0.001), patients with intestinal perforation (P = 0.002), and patients with colonic stricture (P = 0.026). As regards MiR-125b, it was significantly low in females (P = 0.002), patients on salicylates (P = 0.040), patients with colonic stricture (P = 0.002) and significantly high in patients with thromboembolic complications (P = 0.007), musculoskeletal complications (P = 0.021), rectal fistula (P = 0.002) and perforation (P = 0.001) (Table 4).
Table 3. Relations between Lnc THRIL and MiR-125b and clinical data in UC patients.
| miR-125b | Lnc THRIL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Median | IQR | P-value | Median | IQR | P-value | ||||
| Sex | Female | 0.27 | 0.18 | 0.6 | 0.208 | 10.89 | 10.35 | 11.4 | 0.414 |
| male | 0.49 | 0.23 | 0.63 | 11.12 | 10.21 | 12.45 | |||
| Smoker | yes | 0.33 | 0.22 | 0.57 | 0.911 | 10.21 | 10.18 | 10.88 | 0.012* |
| no | 0.41 | 0.19 | 0.61 | 11.19 | 10.76 | 12.45 | |||
| Coffee consumption | yes | 0.34 | 0.12 | 0.51 | 0.417 | 11.78 | 11.05 | 12.53 | 0.109 |
| no | 0.41 | 0.21 | 0.63 | 10.94 | 10.18 | 12.29 | |||
| Diabetes | yes | 0.57 | 0.31 | 0.67 | 0.284 | 10.18 | 9.46 | 11.75 | 0.159 |
| no | 0.34 | 0.19 | 0.6 | 11.12 | 10.21 | 12.45 | |||
| Hypertension | yes | 0.3 | 0.28 | 0.31 | 0.720 | 11.05 | 11 | 11.11 | 0.902 |
| no | 0.39 | 0.19 | 0.62 | 11.12 | 10.19 | 12.45 | |||
| Hepatobiliary | yes | 0.29 | 0.11 | 0.36 | 0.101 | 12.29 | 8.25 | 12.45 | 0.976 |
| no | 0.45 | 0.2 | 0.63 | 11.06 | 10.21 | 12.37 | |||
| Endocrine | yes | 0.73 | 0.7 | 0.76 | 0.041* | 10.38 | 10 | 10.76 | 0.239 |
| no | 0.35 | 0.19 | 0.61 | 11.12 | 10.21 | 12.45 | |||
| MC | yes | 0.62 | 0.59 | 0.64 | 0.225 | 11.02 | 10.7 | 11.35 | 0.901 |
| no | 0.35 | 0.19 | 0.61 | 11.11 | 10.19 | 12.45 | |||
| MSK | yes | 0.34 | 0.23 | 0.55 | 0.586 | 10.79 | 10.18 | 11.4 | 0.054 |
| no | 0.43 | 0.18 | 0.64 | 11.12 | 10.76 | 12.49 | |||
| Eye | yes | 0.23 | 0.23 | 0.41 | 0.202 | 11.12 | 10.89 | 11.4 | 0.788 |
| no | 0.43 | 0.19 | 0.64 | 11.06 | 10.19 | 12.45 | |||
| Salicalyates | yes | 0.49 | 0.23 | 0.64 | 0.179 | 10.94 | 10.18 | 11.4 | 0.358 |
| no | 0.32 | 0.18 | 0.57 | 11.19 | 10.21 | 12.45 | |||
| Steroids | yes | 0.3 | 0.18 | 0.63 | 0.662 | 11.11 | 10.18 | 12.45 | 0.482 |
| no | 0.45 | 0.24 | 0.59 | 11.07 | 10.76 | 12.37 | |||
| Azathioprine | yes | 0.5 | 0.23 | 0.63 | 0.404 | 11.01 | 10.18 | 11.12 | 0.194 |
| no | 0.33 | 0.18 | 0.61 | 11.12 | 10.21 | 12.45 | |||
| Infliximab | yes | 0.31 | 0.19 | 0.64 | 0.626 | 11.12 | 10.18 | 11.35 | 0.699 |
| no | 0.39 | 0.21 | 0.61 | 11.06 | 10.21 | 12.45 | |||
Table 4. Relations between Lnc THRIL and MiR-125b and clinical data in CD patients.
| miR-125b | Lnc THRIL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Median | IQR | P-value | Median | IQR | P-value | ||||
| Sex | Female | 0.31 | 0.27 | 0.78 | 0.002* | 7.88 | 5.77 | 8.23 | <0.001* |
| male | 0.78 | 0.4 | 0.92 | 5.53 | 4.43 | 6.47 | |||
| Smoker | yes | 0.69 | 0.22 | 0.96 | 0.693 | 5.87 | 4.43 | 7.21 | 0.510 |
| no | 0.67 | 0.31 | 0.78 | 5.9 | 4.67 | 7.97 | |||
| Coffee consumption | no | 0.62 | 0.25 | 0.8 | 0.394 | 5.55 | 4.43 | 6.89 | 0.100 |
| yes | 0.74 | 0.4 | 0.83 | 6.02 | 4.81 | 7.97 | |||
| Diabetes | yes | 0.74 | 0.7 | 0.8 | 0.720 | 2.07 | 2 | 2.13 | <0.001* |
| no | 0.67 | 0.3 | 0.83 | 5.95 | 4.76 | 7.88 | |||
| Hypertension | yes | 0.37 | 0.35 | 0.4 | 0.518 | 5.21 | 5 | 5.42 | 0.438 |
| no | 0.7 | 0.3 | 0.83 | 5.95 | 4.57 | 7.88 | |||
| Arthralgia | yes | 0.59 | 0.4 | 0.78 | 1.000 | 5.76 | 5.5 | 6.02 | 0.817 |
| no | 0.69 | 0.3 | 0.83 | 5.87 | 4.57 | 7.88 | |||
| MSK | yes | 0.69 | 0.3 | 0.83 | 0.021* | 8.12 | 5.42 | 9.01 | 0.066 |
| no | 0.30 | 0.19 | 0.35 | 5.82 | 4.56 | 7.54 | |||
| Thromboembolic | yes | 0.78 | 0.31 | 0.83 | 0.007* | 7.21 | 5.66 | 8.23 | 0.070 |
| no | 0.31 | 0.23 | 0.4 | 5.82 | 4.54 | 7.65 | |||
| MC | yes | 0.78 | 0.31 | 0.83 | 0.585 | 5.55 | 4.54 | 7.65 | 0.383 |
| no | 0.78 | 0.4 | 0.92 | 5.95 | 5.46 | 7.9 | |||
| Eye | yes | 0.4 | 0.25 | 0.78 | 0.363 | 5.82 | 4.65 | 7.5 | 0.912 |
| no | 0.71 | 0.47 | 1.26 | 5.87 | 4.57 | 7.88 | |||
| Salicalyates | yes | 0.69 | 0.27 | 0.83 | 0.040* | 5.23 | 4.5 | 7.06 | 0.166 |
| no | 0.81 | 0.4 | 0.94 | 6.02 | 4.76 | 8 | |||
| Steroids | yes | 0.64 | 0.23 | 0.78 | 0.311 | 5.5 | 4.57 | 7.88 | 0.962 |
| no | 0.54 | 0.3 | 0.96 | 5.95 | 4.55 | 7.68 | |||
| Azathioprine | yes | 0.7 | 0.27 | 0.78 | 0.214 | 5.97 | 4.57 | 7.65 | 0.959 |
| no | 0.66 | 0.4 | 0.96 | 5.87 | 4.76 | 7.88 | |||
| Infliximab | yes | 0.69 | 0.3 | 0.78 | 0.456 | 5.79 | 4.67 | 7.39 | 0.978 |
| no | 0.78 | 0.43 | 0.85 | 5.87 | 4.43 | 7.93 | |||
| Ileal strictures | yes | 0.78 | 0.78 | 0.96 | 0.098 | 5.87 | 4.76 | 6.47 | 0.946 |
| no | 0.59 | 0.3 | 0.78 | 5.9 | 4.57 | 7.88 | |||
| Rectal fistula | yes | 0.81 | 0.64 | 0.96 | 0.002* | 5.39 | 4.43 | 6.47 | 0.145 |
| no | 0.40 | 0.23 | 0.78 | 6.02 | 4.76 | 7.93 | |||
| Perforation | yes | 1.08 | 0.76 | 1.53 | 0.001* | 6.02 | 5.05 | 7.93 | 0.002* |
| no | 0.54 | 0.27 | 0.78 | 4.27 | 3.28 | 5.1 | |||
| Intestinal obstruction | yes | 0.78 | 0.31 | 0.83 | 0.562 | 4.55 | 4.11 | 7.93 | 0.152 |
| no | 0.64 | 0.3 | 0.78 | 6.02 | 5.05 | 7.65 | |||
| Surgical resection | yes | 0.8 | 0.5 | 0.83 | 0.383 | 4.44 | 4.05 | 6.6 | 0.121 |
| no | 0.64 | 0.3 | 0.78 | 6.02 | 5.05 | 7.88 | |||
| Colocutaneous fistula | yes | 0.57 | 0.33 | 0.78 | 0.578 | 5.9 | 3.95 | 7.23 | 0.853 |
| no | 0.69 | 0.3 | 0.83 | 5.87 | 4.57 | 7.88 | |||
| Perianal abcess or fistula | yes | 0.45 | 0.12 | 0.78 | 0.229 | 6.11 | 4.33 | 7.88 | 0.779 |
| no | 0.69 | 0.31 | 0.83 | 5.87 | 4.76 | 7.65 | |||
| Colonic stricture | yes | 0.17 | 0.12 | 0.23 | 0.002* | 8.06 | 7.88 | 8.23 | 0.026* |
| no | 0.74 | 0.31 | 0.83 | 5.77 | 4.57 | 7.43 | |||
| Psoas abcess | yes | 0.74 | 0.22 | 0.83 | 0.959 | 4.11 | 2.13 | 8.44 | 0.188 |
| no | 0.67 | 0.31 | 0.8 | 5.95 | 4.88 | 7.77 | |||
Correlations of Lnc THRIL and MiR-125b with study parameters among the patients
In UC patients, our results showed that there were negative significant correlations between LncRNA THRIL and each of MiR-125b (r = -0.28, P = 0.016), duration of illness (r = -0.35, P = 0.020), and ESR (r = -0.24, P = 0.042) and between MiR-125b and both ESR(r = -0.38, P = 0.001) and neutrophil count (r = -0.24, P = 0.039) (Table 5). In CD patients, our results showed that there were negative significant correlations between LncRNA THRIL and each of MiR-125b (r = -0.77, P<0.001), age (r = -0.23, P = 0.048), TLC (r = -0.41, P<0.001), neutrophil count (r = -0.42, P<0.001), platelets (r = -0.29, P = 0.014), and CRP (r = -0.35, P = 0.002), and positive significant correlations between MiR-125b and both TLC (r = 0.37, P = 0.001) and platelets (r = -0.29, P = 0.012) (Table 6).
Table 5. Correlations of Lnc THRIL and MiR 125b with study parameters among UC patients.
| MiR-125b | Lnc THRIL | ||
|---|---|---|---|
| Lnc THRIL | r | - 0.288 | |
| P-value | 0.016* | ||
| Age | r | 0.013 | -0.054 |
| P-value | 0.913 | 0.659 | |
| Duration of illness | r | 0.148 | -0.357 |
| P-value | 0.351 | 0.020* | |
| Hb gl/dl | r | -0.033 | 0.023 |
| P-value | 0.788 | 0.847 | |
| HCT | r | -0.020 | 0.055 |
| P-value | 0.869 | 0.651 | |
| TLC | r | -0.183 | -0.225 |
| P-value | 0.129 | 0.061 | |
| Neutrophil count | r | -0.248 | 0.076 |
| P-value | 0.039* | 0.531 | |
| Platelets | r | 0.020 | -0.185 |
| P-value | 0.869 | 0.125 | |
| CRP | r | -0.201 | -0.221 |
| P-value | 0.095 | 0.066 | |
| ESR | r | -0.380 | -0.243 |
| P-value | 0.001* | 0.042* | |
| Albumin(mg/dl) | r | 0.076 | 0.049 |
| P-value | 0.533 | 0.688 | |
| Mayo score | r | -0.170 | -0.210 |
| P-value | 0.159 | 0.081 |
Table 6. Correlations of Lnc THRIL and MiR 125b with study parameters among CD patients.
| MiR-125b | Lnc THRIL | ||
|---|---|---|---|
| Lnc THRIL | r | -0.772 | |
| P-value | <0.001* | ||
| Age | r | 0.016 | -0.237 |
| P-value | 0.893 | 0.048* | |
| Duration of illness | r | -0.134 | 0.024 |
| P-value | 0.387 | 0.876 | |
| Hb gl/dl | r | 0.139 | -0.048 |
| P-value | 0.252 | 0.695 | |
| HCT | r | 0.019 | 0.052 |
| P-value | 0.874 | 0.667 | |
| TLC | r | 0.378 | -0.416 |
| P-value | 0.001* | <0.001* | |
| Neutrophil count | r | 0.165 | -0.428 |
| P-value | 0.171 | <0.001* | |
| Platelets | r | 0.299 | -0.293 |
| P-value | 0.012* | 0.014* | |
| CRP | r | 0.109 | -0.359 |
| P-value | 0.371 | 0.002* | |
| ESR | r | -0.024 | -0.138 |
| P-value | 0.844 | 0.256 | |
| Albumin | r | -0.129 | 0.189 |
| P-value | 0.289 | 0.118 | |
| CDAI | r | 0.195 | -0.130 |
| P-value | 0.105 | 0.285 |
Receiver operating characteristic of sensitivity and specificity of Lnc THRIL and MiR-125b in the study groups
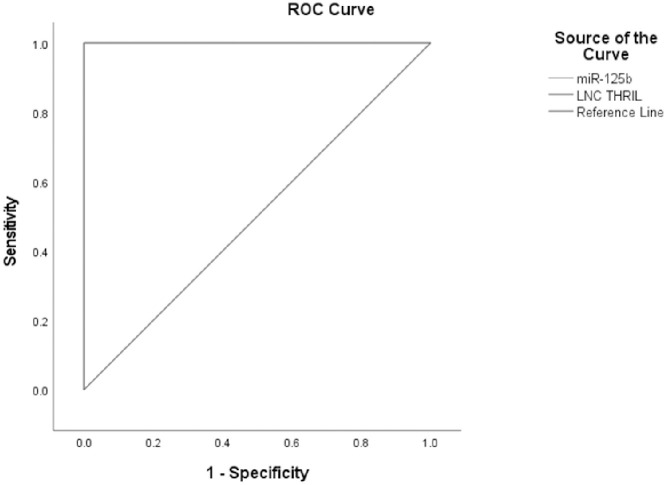
Fig 1 illustrates the ROC curve of Lnc THRIL and MiR-125b in the UC group, showing the diagnostic value of these markers as predictors in differentiating between cases of UC and control. Lnc THRIL; AUC = 1.000, P<0.0001, cut off point 4.62, sensitivity 100%, specificity 100.0%. MiR-125b; AUC = 1.000, P<0.0001, cut off point 0.94, sensitivity 100%, specificity 100.0%.
Fig 1. Receiver operating characteristic of sensitivity and specificity of Lnc THRIL and MiR-125b in UC patients vs. controls.
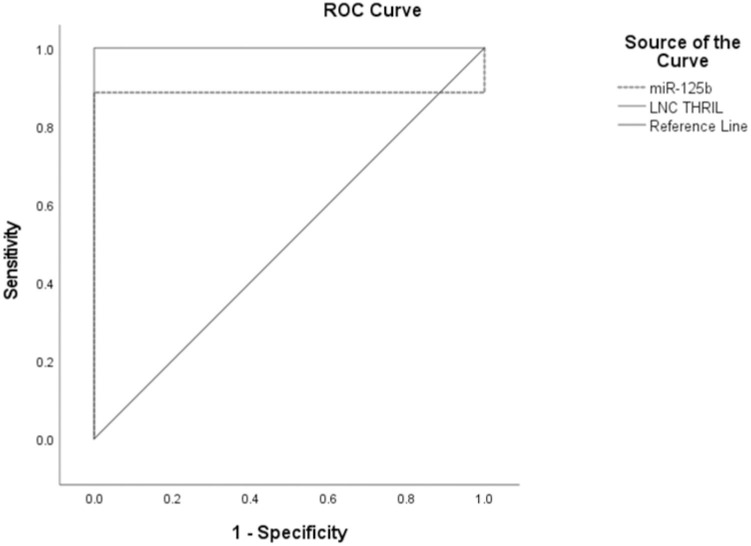
Fig 2 illustrates the ROC curve of Lnc THRIL and MiR-125b in the CD group, showing the diagnostic value of these markers as predictors in differentiating between cases of CD and control. Lnc THRIL; AUC = 1.000, P<0.0001, cut off point 1.57, sensitivity 100%, specificity 100.0%. MiR-125b; AUC = 0.886, P<0.0001, cut off point 0.98, sensitivity 88.6%, specificity 100.0%.
Fig 2. Receiver operating characteristic of sensitivity and specificity of Lnc THRIL and MiR-125b in CD patients vs. controls.
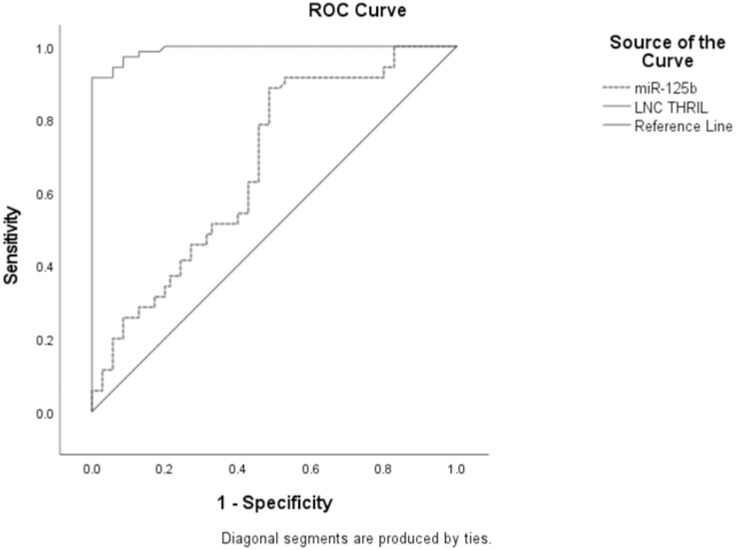
Fig 3 illustrates the ROC curve of Lnc THRIL and MiR-125b showing the diagnostic value of these markers as predictors in differentiating between CD and UC cases. Lnc THRIL; AUC = 0.992, P<0.0001, cut off point 9.49, sensitivity 91.4%, specificity 100.0%. MiR-125b; AUC = 0.677, P<0.0001, cut off point 0.67, sensitivity 88.6%, specificity 51.4%.
Fig 3. Receiver operating characteristic of sensitivity and specificity of Lnc THRIL and MiR-125b in UC vs. CD patients.
Discussion
Inflammatory bowel disease (IBD) is characterized by repetitive episodes of inflammation of the gastrointestinal tract, its exact cause remains unclear, knowing the pathogenesis of IBD will help to develop new strategies for therapies and reduce the incidence of complications. Interaction between the gastrointestinal microbiome and host immune system has been evidenced as a cause [31].
The role of non-coding RNAs (ncRNAs) has been reported in IBD. NcRNAs include microRNAs (miRNAs), long noncoding RNAs (lncRNAs), and circular RNAs (circRNAs) [32, 33]. Our study focused on the expression profile of LncRNA THRIL and MiR-125b in IBD and their relation with patient’s clinical and biochemical investigations.
THRIL (TNF- α and hnRNPL immunoregulatory lncRNA) is involved in innate immunity through the regulation of TNF -α expression level by forming a complex that bind TNF- α gene promotor region resulting in its induction, TNF- α is one of the cytokines that collaborate in the inflammatory process and its dysregulation characterizes autoimmune diseases [34]. It is implicated in a wide range of cellular processes including cell proliferation, survival, and death. In addition, TNF-α signaling is associated with the regulation of several inflammatory pathways including the cyclooxygenase-2 (COX-2) and induce nitric oxide synthase (iNOS) pathways [35]. Several studies investigating the role of anti-TNF-α therapy in modulating the consequences of IBD by different mechanisms as inhibiting activation of immune cells [36], downregulates the expression of cell adhesion molecules and proinflammatory cytokines [37] and has a favorable effect on the gut microbiome [38].
Our study showed a significant difference between the patients’ groups and control group regarding Lnc THRIL with the fold change of Lnc THRIL was significantly up-regulated in UC patients (Median = 11.11, IQR; 10.21–12.45, P<0.001) and CD patients (Median = 5.87, IQR; 4.57–7.88, P<0.001) compared to controls. As regards MiR-125b. Our study showed that the fold change of MiR-125b was significantly down-regulated in UC patients (Median = 0.36, IQR; 0.19–0.61, P<0.001) and CD patients (Median = 0.69, IQR; 0.3–0.83, P<0.001) compared to controls, This dysregulation could be explained by that TRAF6(TNF receptor associated factor 6) and TNFAIP3 (TNF alpha induced protein 3) also referred to as A20 are the two key signaling molecules involved in the NFκB pathway, and these genes carry complementary binding sites for miR-125b in their 3′UTRs and the role of NFκB pathway activation in several autoimmune diseases including IBD and various cancers was previously confirmed [39].
Increased NF-κB expression in mucosal macrophages increases the levels of pro-inflammatory cytokines such as TNF-α, IL-1 and IL-6 resulting in the mucosal cells damage, and it increases expression of intercellular adhesion molecule-1 in colonic epithelial cells that contributes to the recruitment of neutrophil granulocytes to the site of inflammation [40]. This imbalance between excessive secretion of pro-inflammatory cytokines and relative insufficient secretion of anti-inflammatory cytokines is linked to the development of non-specific inflammatory responses in the intestine [41]. Our data disagreed with a report done by Valmiki S et al who showed that MiR125b was significantly up-regulated in UC patients as compared to controls [42].
Moreover, The study showed that there was a negative significant correlation between LncRNA THRIL and MiR-125b in both UC(r = -0.28, P = 0.016) and CD patients(r = -0.77, P<0.001), This finding was previously confirmed by Liu et al who showed that Long non-coding RNA THRIL promotes lipopolysaccharide (LPS) induced inflammatory injury by down-regulating microRNA-125b in ATDC5 cells which act as cell line model of cartilage extracellular matrix neosynthesis and maturation [43]. Song et al., also showed that LncRNA THRIL expression was negatively correlated with miR-125b expression in allergic rhinitis patients [44]. Several studies demonstrated that lncRNAs can exert many cellular functions by interacting with miRNAs as inhibitors or RNA decoys to reduce miRNA production and availability. These interactions play an important role in intestinal epithelial homeostasis [45]. We detected negative significant correlations between MiR-125b and both ESR and neutrophil count in UC patients which agreed with Hruskova et al., who observed negative correlation between expression of miR-125b and the parameters of disease activity and detected inverse correlation between miR-125b and ESR (r = -0.268, P = 0.042) [46]. This finding supports miR-125b’s inhibitory effect on the expression of pro-inflammatory cytokines, cell proliferation, and apoptosis [47].
We also detected positive significant correlations between MiR-125b and both TLC and platelets in CD patients, which agreed with Marina et al., who showed that Hematopoietic cells benefit from miR-125b overexpression in terms of proliferation and CBC results obtained 16 weeks posttransplant revealed an increase in WBC in mice expressing miR-125b compared to control mice. There were statistically significant increases in neutrophils in particular [48].
Data Availability
All relevant data are within the paper.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Guan Q. (2019). A Comprehensive Review and Update on the Pathogenesis of Inflammatory Bowel Disease. J Immunol Res. doi: 10.1155/2019/7247238 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stidham RW, Higgins PDR. (2018) Colorectal Cancer in Inflammatory Bowel Disease. Clin Colon Rectal Surg. 31: 168–783. doi: 10.1055/s-0037-1602237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Habibi F, Habibi ME, Gharavinia A, et al. (2017) Quality of life in inflammatory bowel disease patients: a cross-sectional study. J Res Med Sci. 22:104. doi: 10.4103/jrms.JRMS_975_16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen L. L. (2016). Linking long noncoding RNA localization and function. Trends Biochem. Sci. 41, 761–772. doi: 10.1016/j.tibs.2016.07.003 [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Zhang H, Zhang W, Zhang Y, Wang W, Nie L. (2020). LncRNA XIST modulates 5-hydroxytrytophan-induced visceral hypersensitivity by epigenetic silencing of the SERT gene in mice with diarrhea-predominant IBS. Cell Signal. 73:109674. doi: 10.1016/j.cellsig.2020.109674 [DOI] [PubMed] [Google Scholar]
- 6.Cai P, Li H, Huo W, Zhu H, Xu C, Zang R, et al. (2018). Aberrant expression of LncRNA-MIR31HG regulates cell migration and proliferation by affecting miR-31 and miR-31\ in Hirschsprung’s disease. J Cell Biochem; 119:8195–8203. doi: 10.1002/jcb.26830 [DOI] [PubMed] [Google Scholar]
- 7.Lucafò M, Di Silvestre A, Romano M, Avian A, Antonelli R, Martelossi S, et al. (2018). Role of the long non-coding RNA growth arrest-specific 5 in glucocorticoid response in children with inflammatory bowel disease. Basic Clin Pharmacol Toxicol. 122:87–93. doi: 10.1111/bcpt.12851 [DOI] [PubMed] [Google Scholar]
- 8.Ma D, Cao Y, Wang Z, He J, Chen H, Xiong H et al.(2019).CCAT1 lncRNA Promotes Inflammatory Bowel Disease Malignancy by Destroying Intestinal Barrier via Downregulating miR-185-3p. Inflamm Bowel Dis. Apr 11; 25(5):862–874. doi: 10.1093/ibd/izy381 [DOI] [PubMed] [Google Scholar]
- 9.Yao J, Gao R, Luo M, Li D, Guo L, Yu Z et al.(2021). Long noncoding RNA KIF9-AS1 promotes cell apoptosis by targeting the microRNA-148a-3p/suppressor of cytokine signaling axis in inflammatory bowel disease. Eur J Gastroenterol Hepatol. Epub ahead of print. doi: 10.1097/MEG.0000000000002309 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chao G, Wang Z, Yang Y and Zhang S (2021) LncRNA H19 as a Competing Endogenous RNA to Regulate AQP Expression in the Intestinal Barrier of IBS-D Patients. Front. Physiol. 11:602076. doi: 10.3389/fphys.2020.602076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haberman Y., BenShoshan M., di Segni A. et al., (2018) Long ncRNA landscape in the ileum of treatment-naive early-onset Crohn disease. Inflammatory Bowel Diseases, vol. 24, no. 2, pp. 346–360,. doi: 10.1093/ibd/izx013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S., Hou Y., Chen W. et al. (2018) “KIF9-AS1, LINC01272 and DIO3OS lncRNAs as novel biomarkers for inflammatory bowel disease,” Molecular Medicine Reports, vol. 17(2): 2195–2202,. doi: 10.3892/mmr.2017.8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu F., Huang Y., Dong F. & Kwon J. H. (2016). Ulcerative colitis-associated long noncoding RNA, BC012900, regulates intestinal epithelial cell apoptosis. Inflamm. Bowel Dis. 22, 782–795. [DOI] [PubMed] [Google Scholar]
- 14.Quinn J. J. & Chang H. Y. (2016). Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62. doi: 10.1038/nrg.2015.10 [DOI] [PubMed] [Google Scholar]
- 15.Li Z, Chao TC, Chang KY, et al. (2014). The long noncoding RNA THRIL regulates TNFa expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A. 111 (3):1002–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H, Hu X, Li R, et al. (2020).LncRNA THRIL aggravates sepsis induced acute lung injury by regulating miR-424/ROCK2 axis. Mol Immunol.;126:111–119. doi: 10.1016/j.molimm.2020.07.021 [DOI] [PubMed] [Google Scholar]
- 17.Chen PY, Meister G. (2005): MicroRNA-guided posttranscriptional gene regulation. Biol Chem. 386:1205–1218. doi: 10.1515/BC.2005.139 [DOI] [PubMed] [Google Scholar]
- 18.Zhao Y, Ma T, Chen W, Chen Y, Li M, Ren L, et al. (2016). MicroRNA-124 Promotes Intestinal Inflammation by Targeting Aryl Hydrocarbon Receptor in Crohn’s Disease. J Crohns Colitis. 10: 703–12. doi: 10.1093/ecco-jcc/jjw010 [DOI] [PubMed] [Google Scholar]
- 19.Pierdomenico M, Cesi V, Cucchiara S, Vitali R, Prete E, Costanzo M, et al. (2016).NOD2 Is Regulated By Mir-320 in Physiological Conditions but this Control Is Altered in Inflamed Tissues of Patients with Inflammatory Bowel Disease. Inflamm Bowel Dis. 22: 315–26. doi: 10.1097/MIB.0000000000000659 [DOI] [PubMed] [Google Scholar]
- 20.Shen Y, Zhou M, Yan J, Gong Z, Xiao Y, Zhang C, et al. (2017). miR-200b inhibits TNF-alpha-induced IL-8 secretion and tight junction disruption of intestinal epithelial cells in vitro. Am J Physiol Gastrointest Liver Physiol. 312:G123–G32. [DOI] [PubMed] [Google Scholar]
- 21.Haines RJ, Beard RS Jr., Eitner RA, Chen L, Wu MH. (2016). TNFalpha/IFNgamma Mediated Intestinal Epithelial Barrier Dysfunction Is Attenuated by MicroRNA-93 Downregulation of PTK6 in Mouse Colonic Epithelial Cells. PLoS One.; 11: e0154351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zou T, Jaladanki SK, Liu L, Xiao L, Chung HK, Wang JY, et al. (2016). H19 Long Noncoding RNA Regulates Intestinal Epithelial Barrier Function via MicroRNA 675 by Interacting with RNA-Binding Protein HuR. Mol Cell Biol. 36: 1332–1341. doi: 10.1128/MCB.01030-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang B, Tian Y, Jiang P, Jiang Y, Li C, Liu T, et al. (2017).MicroRNA-122a Regulates Zonulin by Targeting EGFR in Intestinal Epithelial Dysfunction. Cell Physiol Biochem.; 42: 848–58. doi: 10.1159/000478629 [DOI] [PubMed] [Google Scholar]
- 24.Long YS, Deng GF, Sun XS, et al. (2011). Identification of the transcriptional promoters in the proximal regions of human microRNA genes. Mol Biol Rep. 38:4153–4157. doi: 10.1007/s11033-010-0535-y [DOI] [PubMed] [Google Scholar]
- 25.Qiu H, Liang D, Liu L, Xiang Q, Yi Z, Ji Y. (2020). A novel circulating miRNA-based signature for the diagnosis and prognosis prediction of early-stage cervical cancer. Technol Cancer Rese Treat. 19:1–7. doi: 10.1177/1533033820970667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murata K, Furu M, Yoshitomi H, Ishikawa M, Shibuya H, Hashimoto M, et al. (2013). Comprehensive microRNA analysis identifies miR-24 and miR-125a-5p as plasma biomarkers for rheumatoid arthritis. PLoS One. 8:e69118. doi: 10.1371/journal.pone.0069118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magro F, Langner C, Driessen A, Ensari A, Geboes K, Mantzaris GJ, et al. (2013). European consensus on the histopathology of inflammatory bowel disease. J. Crohns Colitis. 7:827–851. doi: 10.1016/j.crohns.2013.06.001 [DOI] [PubMed] [Google Scholar]
- 28.Best W.R, Becktel J.M., Singleton J.W., et al. (1976). Development of a Crohn’s disease activity index. National Cooperative Crohn’s Disease Study Gastroenterology, 70 (3): 439–444. [PubMed] [Google Scholar]
- 29.Schroeder KW, Tremaine WJ, Ilstrup DM. (1987). Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. A randomized study. N Engl J Med. 317:1625–1629. doi: 10.1056/NEJM198712243172603 [DOI] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. (2001): Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) method. Methods. 25:402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 31.Larabi A, Barnich N, Nguyen HTT. (2020). New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy.16:38–51. doi: 10.1080/15548627.2019.1635384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wawrzyniak M. & Scharl M. (2018). Genetics and epigenetics of inflammatory bowel disease. Swiss Med. Wkly 148, e00015. doi: 10.4414/smw.2018.14671 [DOI] [PubMed] [Google Scholar]
- 33.Yarani R., Mirza A. H., Kaur S. & Pociot F. (2018). The emerging role of lncRNAs in inflammatory bowel disease. Exp. Mol. Med. 50, 1–14. doi: 10.1038/s12276-018-0188-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hur K, Kim SH, Kim JM. (2019). Potential Implications of Long Noncoding RNAs in Autoimmune Diseases. Immune Netw. Feb 25; 19(1):e4. doi: 10.4110/in.2019.19.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y., Soendergaard C., Bergenheim F.H., Aronoff D.M., Milne G., Riis L.B. et al. (2018).COX-2-PGE2 Signaling Impairs Intestinal Epithelial Regeneration and Associates with TNF Inhibitor Responsiveness in Ulcerative Colitis. EBioMedicine. 36: 497–507. doi: 10.1016/j.ebiom.2018.08.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang C., Shu W., Zhou G., Lin J., Chu F., Wu H., et al. (2018). Anti-TNF- α Therapy Suppresses Proinflammatory Activities of Mucosal Neutrophils in Inflammatory Bowel Disease. Mediators Inflamm.; 2018:1–12. doi: 10.1155/2018/3021863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olsen T., Rismo R., Gundersen M.D., Paulssen E.J., Johnsen K., et al. (2016). Normalization of mucosal tumor necrosis factor-α: A new criterion for discontinuing infliximab therapy in ulcerative colitis. Cytokine.; 79:90–95. doi: 10.1016/j.cyto.2015.12.021 [DOI] [PubMed] [Google Scholar]
- 38.Dovrolis N., Michalopoulos G., Theodoropoulos G.E., Arvanitidis K., Kolios G., Sechi L.A., et al. (2020). The Interplay between Mucosal Microbiota Composition and Host Gene-Expression is Linked with Infliximab Response in Inflammatory Bowel Diseases. Microorganisms.8:438. doi: 10.3390/microorganisms8030438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia L, Tan S, Zhou Y, Lin J, Wang H, Oyang L, et al. (2018). Role of the NFκB-signaling pathway in cancer. Onco Targets Ther. 11: 2063–2073. doi: 10.2147/OTT.S161109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang L, Walia B, Evans J, Gewirtz AT, Merlin D, Sitaraman SV. (2003).IL-6 induces NF-kappa B activation in the intestinal epithelia. J Immunol.;171:3194–201. doi: 10.4049/jimmunol.171.6.3194 [DOI] [PubMed] [Google Scholar]
- 41.Woodford-Richens K, Bevan S, Churchman M, Dowling B, Jones D, Norbury CG, et al.(2000) Analysis of genetic and phenotypic heterogeneity in juvenile polyposis. Gut.46:656–60. doi: 10.1136/gut.46.5.656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valmiki S., Ahuja V., Puri N., & Paul J. (2020). miR-125b and miR-223 Contribute to Inflammation by Targeting the Key Molecules of NFκB Pathway. Frontiers in medicine, 6, 313. doi: 10.3389/fmed.2019.00313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Guangyao & Wang Yongkun & Zhang Mingran et al. (2019). Long non-coding RNA THRIL promotes LPS-induced inflammatory injury by down-regulating microRNA-125b in ATDC5 cells. International Immunopharmacology. 66. 354–361. doi: 10.1016/j.intimp.2018.11.038 [DOI] [PubMed] [Google Scholar]
- 44.Song J, Liu D, Yin W (2022). lnc-THRIL and miR-125b relate to disease risk, severity, and imbalance of Th1 cells/Th2 cells in allergic rhinitis. Allergol Immunopathol (Madr). 1;50(3):15–23. [DOI] [PubMed] [Google Scholar]
- 45.Wang JY, Xiao L, Wang JY. (2017). Posttranscriptional regulation of intestinal epithelial integrity by noncoding RNAs. Wiley Interdiscip Rev RNA 8: e1399. doi: 10.1002/wrna.1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hruskova V, Jandova R, Vernerova L, et al. (2016). MicroRNA-125b: association with disease activity and the treatment response of patients with early rheumatoid arthritis. Arthritis Res Ther. 2;18(1):124. doi: 10.1186/s13075-016-1023-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng NL, Chen X, Kim J, Shi AH, Nguyen C, Wersto R, et al.(2015). MicroRNA-125b modulates inflammatory chemokine CCL4 expression in immune cells and its reduction causes CCL4 increase with age. Aging Cell.;14:200–8. doi: 10.1111/acel.12294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marina B, Marian H, Beiyan Z, Harvey F (2010). MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci U S A. 14; 107(50): 21558–21563. doi: 10.1073/pnas.1016611107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.