Abstract
A protein serine/threonine kinase, AfsK, and its target protein AfsR globally control physiological and morphological differentiation in the bacterial genus Streptomyces. A protein (KbpA) of 252 amino acids encoded by an open reading frame in a region upstream of afsK in Streptomyces coelicolor A3(2) was identified as an AfsK-interacting protein. The interaction site of AfsK was in the N-terminal portion containing the kinase catalytic domain. KbpA bound a nonphosphorylated form of AfsK and inhibited its autophosphorylation at serine and threonine residues. KbpA in the reaction mixture containing AfsK and AfsR also inhibited the phosphorylation of AfsR by AfsK, presumably because KbpA inhibited the conversion from the inactive, nonphosphorylated form of AfsK to the active, phosphorylated form. kbpA was transcribed throughout growth, and the transcription was enhanced when production of actinorhodin had already started. KbpA thus appeared to play an inhibitory role in a negative feedback system in the AfsK-AfsR regulatory pathway. Consistent with these in vitro observations, kbpA served as a repressor for actinorhodin production in S. coelicolor A3(2); disruption of kbpA greatly enhanced actinorhodin production, and overexpression of kbpA reduced the production.
In the gram-positive, soil-living, filamentous bacteria Streptomyces coelicolor A3(2) and Streptomyces griseus, a serine/threonine kinase, AfsK, and its target protein AfsR control secondary metabolism and morphological development, respectively (17, 25). Disruption of either one of the two genes reduced actinorhodin production in S. coelicolor A3(2) (9, 17). In S. griseus, disruption of either afsK or afsR resulted in the failure of aerial mycelium formation on medium containing glucose at concentrations higher than 1% (25). Even under the usual culture conditions in the laboratory, the AfsK-AfsR system in S. coelicolor A3(2) contributes considerably to pigment production because of reduced production of the pigment by afsK and afsR mutants. The wide distribution of afsK and afsR in various Streptomyces spp. (9, 17) shows their general and important roles in the regulation of secondary metabolism and morphogenesis in this genus. Biochemical and genetic studies of AfsK-AfsR have led us to assume that AfsK on the inner side of the membrane activates its own kinase activity by autophosphorylating its serine and threonine residues on sensing some external stimuli and then phosphorylates serine and threonine residues of AfsR; the phosphorylated AfsR serves as a transcriptional factor for many genes required for antibiotic production and morphogenesis (2, 6). Wietzorrek and Bibb (28) pointed out the possibility that AfsR is a DNA-binding protein. The genome project for S. coelicolor A3(2) has predicted the presence of more than 20 AfsK homologues with a serine/threonine kinase catalytic domain (www.sanger.ac.uk/Projects/S coelicolor/), and some AfsK homologues have been cloned from this species (21, 26) and other Streptomyces spp. (20, 27). Although some of these kinases are perhaps capable of autophosphorylation, it is not known what their targets are and how their autophosphorylation is controlled.
In S. griseus, the AfsK-AfsR system is involved in aerial mycelium and spore formation in response to glucose in the medium (25). However, transcription of neither afsK nor afsR depended on the concentration of glucose. These observations led us to assume that autophosphorylation of AfsK and phosphorylation of AfsR by AfsK may be controlled by some mechanism other than one at the transcriptional level. We focused on an open reading frame (ORF) that is located upstream of afsK and conserved in S. coelicolor A3(2) and S. griseus. Because the AfsK-AfsR system in S. coelicolor A3(2) could be assessed by monitoring actinorhodin (blue pigment) production, we studied the possible relationship between the conserved ORF and the AfsK-AfsR system in S. coelicolor A3(2). The ORF product was found to bind specifically to the nonphosphorylated form of AfsK and inhibit its autophosphorylation. Consistent with this in vitro observation, gene disruption and overexpression experiments suggested a role of this ORF as a repressor for actinorhodin production. We here describe control by this ORF product, named KbpA (for AfsK-binding protein A), of AfsK autophosphorylation by means of protein-protein interaction.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Escherichia coli JM109 and pUC19 (29) were used for DNA manipulation. E. coli AD494(DE3) and pET32a(+), purchased from Novagen, were used for expression of proteins fused to thioredoxin (TRX). E. coli BL21 and pGEX5X-1, purchased from Amersham Pharmacia Biotech, were used for expression of proteins fused to glutathione S-transferase (GST). E. coli BL21 trxB(DE3), purchased from Novagen, was used for preparation of 32P-labeled AfsK fusion proteins. E. coli JM110 dam dcm, deficient in the methylases, was purchased from Takara Shuzo. A high-copy-number plasmid, pIJ6021, containing a thiostrepton-inducible promoter was obtained from M. J. Bibb (24). S. coelicolor A3(2) was routinely cultured at 30°C on Trypto-Soya broth (TSB) (Nissui), which was supplemented with 10 μg of thiostrepton or kanamycin per ml when necessary.
Construction of plasmids.
DNA manipulations in E. coli were as described by Maniatis et al. (16), and those in Streptomyces were as described by Hopwood et al. (8). The kbpA coding sequence was amplified with the two primers 5′-GCCGAATTCCATATGGCCGAAAACGGGGCATTC-3′ (the underlined and italic letters indicate EcoRI and NdeI sites, respectively; the boldface letters indicate the start codon of kbpA) and 5′-GGCAAGCTTCTCGAGTCAGCGTCGCAGCAGCGCGAAC-3′ (the underlined and italic letters indicate HindIII and XhoI sites, respectively; the boldface letters indicate the stop codon of kbpA) and cloned between the EcoRI and HindIII sites of pUC19, resulting in pUC19-kbpA. After the kbpA coding sequence had been checked for errors in amplification, it was inserted between the EcoRI and HindIII sites of pET32a(+) to construct pTRX-KbpA. Similarly, kbpA as an EcoRI-XhoI fragment was inserted in pGEX5X-1 to generate pGST-KbpA. The kbpA sequence as an NdeI-HindIII fragment was also inserted in pIJ6021 to construct pIJ6021-kbpA.
For placing the afsR coding sequence in pGEX5X-1, a procedure similar to that used for constructing pGST-AfsR-g (25) was employed. The pGST-AfsR thus constructed directed the synthesis of an AfsR protein fused to GST. For construction of pTRX-AfsK and pGST-AfsK, directing the synthesis of TRX-AfsK and GST-AfsK, respectively, the afsK sequence was divided into two. The sequence for the N-terminal portion was amplified by PCR with the primers 5′-GCCGAATTCATGGTGGATCAGCTGACGCAG-3′ (the underlined and boldface letters indicate an EcoRI site and the initiation codon of afsK [originally GTG], respectively) and 5′-GGCAAGCTTTCAGCGGCCGCCGGCCGTGGTGGCGGGC-3′ (the underlined and italic letters indicate HindIII and NotI sites, respectively; the boldface letters indicate an artificial stop codon), and the EcoRI-HindIII fragment was inserted in pUC19 to construct pUC19-AfsKΔC, containing the region from Met-1 to Arg-311. The sequence for the C-terminal portion was amplified with 5′-GCCGAATTCGGCGGCCGCGGCCACGGCCACGGCC-3′ and 5′-GGCAAGCTTCTCGAGTCACGTCGTACGGGCGGTCCCCGTG-3′ (the italic and boldface letters indicate an XhoI site and the stop codon of afsK, respectively; the underlined letters indicate restriction sites used for cloning) and inserted between the EcoRI and HindIII sites of pUC19, resulting in pUC19-AfsKΔN, containing the region from Gly-309 to the stop codon of afsK. The EcoRI-NotI fragment from pUC19-AfsKΔC, the NotI-HindIII fragment from pUC19-AfsKΔN, and the EcoRI-HindIII fragment from pET32a(+) were connected by three-fragment ligation to construct pTRX-AfsK, which would direct the synthesis of TRX-His6-S tag-AfsK. The EcoRI-NotI fragment from pUC19-AfsKΔC, the NotI-XhoI fragment from pUC19-AfsKΔN, and the EcoRI-XhoI fragment from pGEX5X-1 were similarly connected to construct pGST-AfsK. pTRX-KΔCwt, which would direct the synthesis of the kinase domain (Met-1 to Arg-311) of AfsK, was constructed by inserting the EcoRI-HindIII fragment from pUC19AfsKΔC in pET32a(+). For site-directed mutagenesis to replace Lys-44 with Ala, 5′-CGGCGCGTGGCGATCGCGACGGTGCGC-3′ (the nucleotides in boldface were originally AA) as a mutant primer and the Mutan-Super Express Km kit (Takara Shuzo) were used according to supplier's manual. The EcoRI-HindIII fragment containing the mutation was inserted in pET32a(+) to construct pTRX-KΔCK44A with essentially the same construction as pTRX-KΔCwt.
Production and preparation of GST- and TRX-fused proteins.
An overnight culture (1 ml) of E. coli harboring each of the expression plasmids was inoculated in 9 ml of L broth. After cultivation at 30°C for 2 h, isopropyl-β-d-thiogalactopyranoside (IPTG) was added at a final concentration of 1 mM and the culture was continued at 30°C for 3 h, except for E. coli harboring pGST-AfsK. GST-AfsK appeared to readily form inclusion bodies and was cultured at 20°C for 8 h. The E. coli cells were harvested by centrifugation, suspended in 0.5 ml of buffer A (10 mM Tris-HCl [pH 7.2] and 10% glycerol), and sonicated to prepare soluble fractions by centrifugation at 24,000 × g for 30 min. TRX and TRX-KbpA were purified with an Ni-nitrilotriacetic acid Spin kit (Qiagen) for GST pull-down assays. GST, GST-KbpA, and GST-AfsR were purified with a MicroSpin GST purification module (Amersham Pharmacia Biotech) for in vitro phosphorylation assay. TRX-AfsK, TRX-KΔCwt, and TRX-KΔCK44A were mainly produced as inclusion bodies, and these were solubilized with 6 M urea, purified with the Ni-bound resin, and finally refolded into active forms by gradual dialysis against buffer A, as described previously (25). Protein concentrations were measured with the Bio-Rad protein assay kit using bovine serum albumin as the standard.
AfsK-KbpA interaction assay.
The soluble fraction (10 μg of protein) containing GST-AfsK was incubated with 10 μl of glutathione-Sepharose 4B (Amersham Pharmacia Biotech) at 4°C for 1 h in 0.5 ml of pull-down assay buffer (PDA buffer) (10 mM Tris-HCl [pH 7.2], 100 mM NaCl, 1 mM MgCl2, 0.025% β-mercaptoethanol, 1% Triton X-100, and 10% glycerol). Glutathione-Sepharose was collected by centrifugation, and the pellet was washed twice with 1 ml of PDA buffer. The pellet was suspended in 0.5 ml of PDA buffer, and 5 μg of purified TRX-KbpA was added. After incubation at 4°C for 1 h, the Sepharose was collected by centrifugation and washed three times with 1 ml of PDA buffer. The pellet was suspended in 15 μl of sodium dodecyl sulfate (SDS) loading buffer (58 mM Tris-HCl [pH 6.8], 1.7% SDS, 6% glycerol, 100 mM dithiothreitol, and 0.002% bromophenol blue) and boiled for 5 min to release GST complexes from the Sepharose. A portion (10 μl) of the supernatant obtained by centrifugation of the boiled sample was subjected to SDS–12.5% polyacrylamide gel electrophoresis (PAGE). After the proteins had been transferred to a nitrocellulose membrane, TRX-KbpA was detected by Western blotting with S protein horseradish peroxidase (HRP) conjugate (Novagen) and the ECL Western blotting reagents (Amersham Pharmacia Biotech). The membrane was reprobed with anti-GST antibody HRP conjugate (Santa Cruz Biotechnology) to detect the GST-fused proteins. As negative controls, the soluble fractions containing GST and GST-AfsR, instead of GST-AfsK, were used. TRX itself, instead of TRX-KbpA, was also used as a negative control. The interactions of TRX-KΔCwt and TRX-KΔCK44A with GST-KbpA were similarly assayed, but with the proteins purified with the Ni or GST resins, as described above.
For preparation of 32P-TRX-KΔCwt and TRX-KΔCK44A, E. coli BL21 trxB(DE3) harboring pTRX-KΔCwt or pTRX-KΔCK44A was cultured in the presence of 100 μCi of phosphorus-32 (Amersham Pharmacia Biotech). The E. coli cells were collected and disrupted with the BugBuster protein extraction reagent containing Benzonase nuclease (Novagen). The TRX proteins were solubilized and purified as described above.
In vitro phosphorylation assay.
The standard reaction mixture, containing 15 pmol of TRX-AfsK in 10 mM Tris-HCl (pH 7.2)–5 mM MnCl2–10 mM MgCl2–0.1 mM ATP–10 μCi of [γ-32P]ATP–1 mM dithiothreitol, was incubated at 30°C for 5 min. For phosphorylation of GST-AfsR by TRX-AfsK, 30 pmol of GST-AfsR was added. Autophosphorylation of TRX-AfsK and GST-AfsR phosphorylation by TRX-AfsK were examined in the presence of 300 pmol of GST-KbpA. In this case, the reaction mixture without ATP and MgCl2 was placed on ice for 1 h to allow formation of the complex between TRX-AfsK and GST-KbpA. The reaction was started by adding ATP and MgCl2 and continued at 30°C for 5 min. After separation of the proteins by SDS-PAGE, the gel was placed on a Fuji BAS2000 image analyzer. Phosphoamino acid analysis by one-dimensional electrophoresis on a cellulose thin-layer plate was carried out with hydrolyzed samples, as described previously (4, 12).
Disruption of chromosomal kbpA.
A 1,014-bp region upstream of kbpA was amplified by PCR with primers 5′-GGCGAATTCGGGGTCGTCACGGTGCTGAACTTC-3′ (the underlining indicates an EcoRI site) and 5′-GCCAAGCTTTCTAGACATGCCGTCAAAGTAACCGC-3′ (the underlined and italic letters indicate HindIII and XbaI sites, respectively; the boldface letters indicate the start codon of kbpA). A 1,154-bp region downstream of kbpA was amplified with 5′-GGCGAATTCTCTAGATGACCCGGCGGCCACGGCG-3′ (the underlined and italic letters indicate EcoRI and XbaI sites, respectively; the boldface letters indicate the stop codon of kbpA) and 5′-GCCAAGCTTGGATCCGCGGCCGCCGGCCGTGGTGGCGGGCTTG-3′ (the underlined and italic letters indicate HindIII and BamHI sites, respectively). The regions upstream and downstream of kbpA were each cloned between the EcoRI and HindIII sites of pUC19. The two regions in the pUC19 plasmids were then connected by use of the XbaI sites, generating pDisKbpA, which contained a 2,192-bp insertion with complete deletion of the kbpA sequence. The thiostrepton resistance (tsr) gene, obtained as a BamHI-HindIII fragment from pKU209 (11), was inserted between the BamHI and HindIII sites of pDisKbpA. The circular form of the resultant plasmid prepared from E. coli JM110 was alkali denatured (22) and introduced by protoplast transformation into S. coelicolor A3(2) M130 to isolate mutants in which the whole plasmid was integrated in the chromosome by homologous recombination. Thiostrepton-resistant colonies were incubated on TSB for 7 days in the absence of thiostrepton, and spores were recovered. Of the isolated spores, thiostrepton-sensitive colonies were selected to obtain mutants containing the deletion of kbpA due to double crossover. Correct deletion of the kbpA sequence was checked by Southern hybridization with the 0.7-kb EcoRI-HindIII fragment containing kbpA on pUC19-kbpA and the 2.3-kb EcoRI-SphI fragment containing kbpA and part of afsK (Fig. 1B).
FIG. 1.
Effects of overexpression and disruption of kbpA in S. coelicolor A3(2) M130. (A) Strain M130 harboring pIJ6021 produces the blue pigment actinorhodin, whereas that harboring kbpA on pIJ6021 produces a much smaller amount of the pigment. (B) Schematic representation of the chromosomal kbpA disruption. The kbpA coding region is completely deleted so that this deletion does not affect the promoter in front of kbpA or afsK. Abbreviations: Ec, EcoRI; Not, NotI; Sp, SphI; Xb, XbaI. (C) The kbpA disruptant produces a larger amount of actinorhodin than strain M130.
Transcriptional analysis of kbpA.
RNA was purified from mycelium grown on TSB agar medium (13). For S1 nuclease mapping of kbpA, a 342-bp fragment was prepared by PCR with 5′-AGCATTTCGCTGAGGCAGTCGAGCAGTTTC-3′ and 5′-TCCTGGAAGGTCCAGCCGAACAGCTCGCCG-3′ (corresponding to positions −217 to +125, taking the transcriptional start point of kbpA as +1, which was later determined) and used as the 32P-labeled probe, as described previously (13). Transcription of afsK and hrdB was determined as described previously (25). For reverse transcription-PCR, two primers containing the start codons of kbpA and afsK were used: 5′-ATGGCCGAAAACGGGGCATTCGAGAAG-3′ (the boldface letters indicate the start codon of kbpA) and 5′-ATCGTGCTGCGTCAGCTGATCCACCAC-3′ (the boldface letters indicate the start codon of afsK).
Nucleotide sequence accession number.
The nucleotide sequence of kbpA has been deposited in the DDBJ, EMBL, and GenBank databases under accession number D45382.
RESULTS
Repression of actinorhodin production by kbpA.
The ORF products (KbpAs) that are encoded by a region upstream of afsK in S. coelicolor A3(2) and S. griseus showed 47% identity to each other. The KbpA of 252 amino acids in S. coelicolor A3(2) also showed 36 and 33% identity to SgaA, which is involved in suppression of the growth disturbance caused by high osmolality in S. griseus (1), and to DnrV, which is located in the doxorubicin biosynthetic gene cluster in Streptomyces peucetius (15), respectively. The functions of these products, however, are not understood. In the database of the S. coelicolor A3(2) genome, there are six KbpA homologues (5H1.9.C, 9B10.0C, 2G4.09, F1.07, 4C6.24C, and 4G10.18C). Since in prokaryotes, genes for the same biological function comprise a cluster in most cases, we examined a possible role of kbpA in actinorhodin production in S. coelicolor A3(2). Overexpression of kbpA in strain M130 by means of placing kbpA under the control of the thiostrepton-inducible promoter tip in a high-copy-number plasmid, pIJ6021, severely reduced actinorhodin production on TSB agar (Fig. 1A). The reduction was independent of the nutritional conditions, because strain M130 harboring pIJ6021-kbpA produced a much smaller amount of actinorhodin on media containing various carbon and nitrogen sources. The degree of reduction was almost the same as that in afsK disruptants (data not shown). On the other hand, complete deletion of the kbpA coding region (Fig. 1B) caused overproduction of actinorhodin on various media (Fig. 1C). This mutant also produced undecylprodigiosin at an earlier stage and accumulated it in a larger amount than strain M130 (data not shown). The ΔkbpA mutant produced spores normally, which showed that the mutation did not affect morphological differentiation. The AfsK-AfsR system in S. coelicolor A3(2) caused no detectable effect on morphogenesis (7, 17). We thus concluded that kbpA acted as a repressor for actinorhodin production, irrespective of the culture conditions.
Transcription of kbpA.
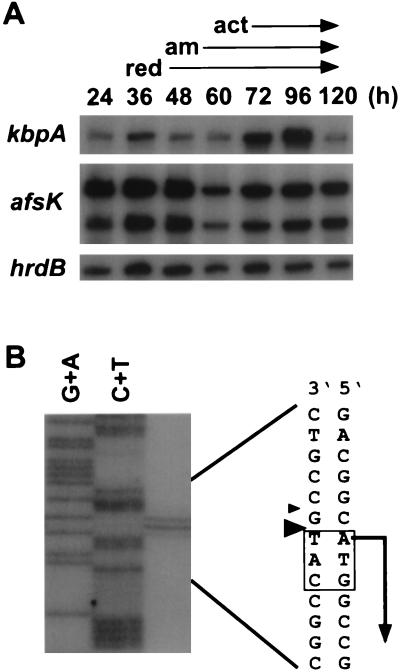
Because of the involvement of kbpA in actinorhodin production, we determined the course of its transcription, in relation to actinorhodin production, using RNA from mycelium grown on agar medium. afsK was constantly transcribed from two promoters throughout growth. Constant transcription from two promoters was also observed for the afsK gene of S. griseus (2). hrdB mRNA, encoding the major sigma factor of RNA polymerase, is known to be transcribed constantly. kbpA was transcribed from a single promoter throughout growth, but transcription was enhanced when production of both undecylprodigiosin and actinorhodin and aerial mycelium formation had already started (Fig. 2A). It thus appeared that under usual culture conditions, KbpA repressed secondary metabolism when these pigments had been produced and accumulated. High-resolution S1 mapping revealed the transcriptional start point to be the first A of the start codon (Fig. 2B). A sequence, TACTTT, similar to the −10 consensus sequence was present at an appropriate position from the transcriptional start point, but no sequence similar to the −35 consensus sequence was present. Transcription of leaderless mRNA is not uncommon in Streptomyces. For example, the 23S rRNA methylase gene mediating erythromycin resistance in Streptomyces erythraeus (3) and afsA, which probably encodes an A-factor biosynthetic enzyme in S. griseus (10), are transcribed from leaderless transcripts.
FIG. 2.
Transcription of kbpA. (A) Low-resolution S1 mapping of kbpA, afsK, and hrdB. kbpA and hrdB contain a single promoter, and afsK contains two promoters. On agar medium, production of the red pigment undecylprodigiosin (red) and the blue pigment actinorhodin (act) started at 48 and 72 h, respectively. Aerial mycelium (am) was formed at 60 h. (B) High-resolution S1 mapping of kbpA. In this particular experiment, the strongest signal of the S1-protected fragments corresponds to the second residue of the translational start codon ATG, since S1-protected fragments run slower than the chemically cleaved fragments (23). It is reasonable, however, to assume that transcription of kbpA starts at the first nucleotide of the ATG.
kbpA and afsK are separated by 219 nucleotides, which suggested that the genes were transcribed independently. Reverse transcription-PCR with primers containing the start codons of kbpA and afsK yielded only a weak signal representing a 1,004-bp DNA fragment after 30 cycles of amplification (data not shown). We thus concluded that kbpA and afsK were transcribed mainly from their own promoter and that very little kbpA transcript leaked into afsK.
Direct interaction of AfsK with KbpA.
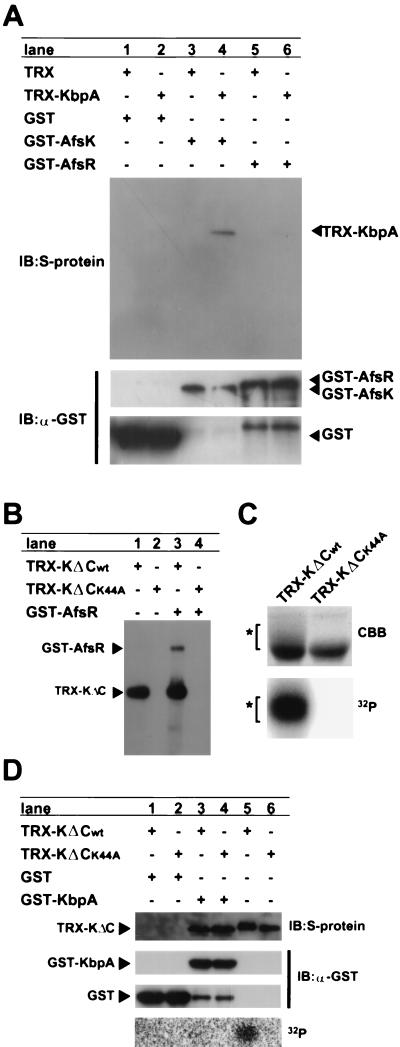
For examining possible KbpA-AfsK interaction, we produced TRX-KbpA with the structure of TRX-His6-S tag-KbpA (419 amino acids, 45 kDa), detectable with S protein HRP conjugate, and GST-AfsK (1,029 amino acids, 110 kDa), precipitable with glutathione-Sepharose. The S tag consisted of 15 amino acids derived from RNase S protein. TRX-KbpA was produced in the soluble fraction of E. coli and purified with His-bind resin. GST-AfsK was produced in a soluble fraction of E. coli when the cells were cultured at 20°C to avoid formation of inclusion bodies. After the soluble fraction (10 μg of protein) containing GST-AfsK had been incubated with 5 μg of purified TRX-KbpA, the GST complexes were pulled down with glutathione-Sepharose and separated by SDS-PAGE. TRX complexes and GST complexes were detected by Western blotting with the S protein HRP conjugate and the antibody for GST, respectively (Fig. 3A). This pull-down assay recovered TRX-KbpA (Fig. 3A, lane 4), indicating that GST-AfsK formed a complex with TRX-KbpA. The lack of recovery of proteins reactive with the antibody in the control experiments with TRX instead of TRX-KbpA or with GST instead of GST-AfsK showed that the complex was formed via parts of KbpA and AfsK. In addition, KbpA did not interact with AfsR, since the same experiment with GST-AfsR (1,223 amino acids, 132 kDa) instead of GST-AfsK did not recover TRX-KbpA.
FIG. 3.
Interaction of KbpA and AfsK. (A) The pull-down of GST-AfsK with glutathione-Sepharose coprecipitated TRX-KbpA, which was detected with S protein by Western blotting (immunoblotting [IB]) (lane 4). TRX-KbpA was not recovered when GST or GST-AfsR was used. The pull-down of GST-AfsR (132 kDa), GST-AfsK (110 kDa), and GST (28 kDa) with glutathione-Sepharose was apparent by Western blotting with the antibody for GST (α-GST). The small protein found in lanes 5 and 6 is a degradation product derived from GST-AfsR. (B) Autophosphorylation of TRX-KΔCwt (51 kDa) and phosphorylation of GST-AfsR by TRX-KΔCwt. TRX-KΔCwt (5 μg) was incubated at 30°C for 10 min in the presence of [γ-32P]ATP, subjected to SDS-PAGE, and analyzed by autoradiography. GST-AfsR (3 μg) in the reaction mixture was also phosphorylated. Neither autophosphorylation nor phosphorylation of GST-AfsR occurred for TRX-KΔCK44A. (C) SDS-PAGE of the TRX-KΔC proteins labeled in vivo. Coomassie brilliant blue (CBB) staining revealed smeared bands for TRX-KΔCwt, as indicated by an asterisk, which represent phosphorylated forms of TRX-KΔCwt, as found by autoradiography. (D) The pull-down of GST-KbpA (54 kDa) with glutathione-Sepharose coprecipitated TRX-KΔCwt (lane 3) and TRX-KΔCK44A (lane 4) without recovering smeared, phosphorylated forms of TRX-KΔCwt. The small protein recovered by anti-GST antibody in lanes 3 and 4 is a degradation product. GST itself did not pull down the TRX-KΔC proteins (lanes 1 and 2). TRX-KΔCwt gave smeared bands (lane 5), but TRX-KΔCK44A did not (lane 6).
During the pull-down assay, we noticed that the amount of TRX-KbpA recovered as a complex of TRX-KbpA and GST-AfsK appeared to depend on the degree of phosphorylation of GST-AfsK. The amount of TRX-KbpA recovered and detected by Western blotting was decreased as the amount of the phosphorylated form of GST-AfsK was increased. The degree of autophosphorylation of GST-AfsK in E. coli could be estimated by the intensity of smeared, slow-moving bands on SDS-PAGE. We examined this observation in detail by reducing the size of AfsK and by using a K44A mutant that lost the ability to autophosphorylate because of the mutation at one of the active-site residues, Lys-44. The TRX-KΔC proteins (478 amino acids, 51 kDa) contained the kinase domain (Met-1 to Arg-311). In vitro phosphorylation assay of TRX-KΔCwt revealed autophosphorylation and phosphorylation of AfsR (Fig. 3B). Phosphoamino acid analysis showed that both contained phosphorylated serine and threonine residues (data not shown), as does the native AfsK-AfsR phosphorylation system. No phosphorylation occurred for TRX-KΔCK44A.
To confirm that the smeared bands observed for TRX-KΔCwt represented the phosphorylated forms, we purified the TRX-KΔC proteins from recombinant E. coli cells grown in the presence of 32P-labeled inorganic phosphate. TRX-KΔCwt showed a smeared pattern on SDS-PAGE, but TRX-KΔCK44A did not (Fig. 3C). Autoradiography of the gel indicated the presence of 32P in the smeared bands of TRX-KΔCwt and the absence of 32P in TRX-KΔCK44A.
We did pull-down assays to examine the interaction between GST-KbpA and TRX-KΔCwt or TRX-KΔCK44A (Fig. 3D). TRX-KΔC proteins labeled in vivo were used. After incubation, the GST complexes were precipitated with glutathione-Sepharose and separated by SDS-PAGE. TRX complexes and GST complexes were detected by Western blotting with S protein and the antibody for GST, respectively. Both TRX-KΔCwt and TRX-KΔCK44A were coprecipitated with GST-KbpA (Fig. 3D, lanes 3 and 4). Only a sharp band, with no smeared bands, was detected for TRX-KΔCwt, although the TRX-KΔCwt preparation contained smeared bands (lane 5). These observations suggested that GST-KbpA interacted only with the nonphosphorylated form of AfsK. In fact, the TRX-KΔCwt that was coprecipitated with GST-KbpA contained no radioactivity.
Inhibition of AfsK autophosphorylation by KbpA.
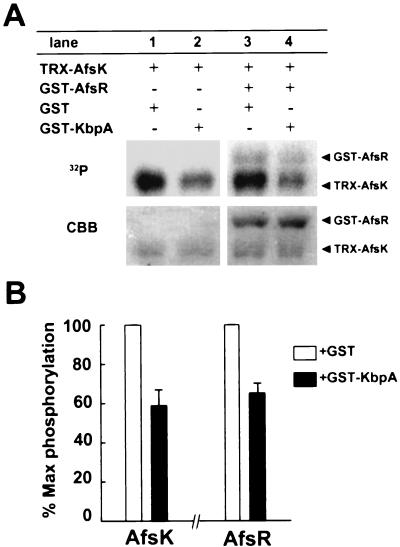
Because KbpA was found to bind the kinase domain in the nonphosphorylated form of AfsK, we examined the effect of KbpA binding to AfsK on autophosphorylation. Incubation of TRX-AfsK (966 amino acids, 102 kDa) in the presence of [γ-32P]ATP yielded the autophosphorylated form (Fig. 4A), because the TRX-AfsK preparation contained a large population of the nonphosphorylated form. GST-KbpA inhibited this phosphorylation by about 40% when analyzed with an image analyzer (Fig. 4B). The inhibition of autophosphorylation to a small extent may be attributed to the equilibrium of association and dissociation between AfsK and KbpA and rapid autophosphorylation of dissociated AfsK. Coincubation of TRX-AfsK and GST-AfsR yielded a phosphorylated form of GST-AfsR, in addition to the autophosphorylated form of TRX-AfsK. The presence of GST-KbpA inhibited the phosphorylation of AfsR by AfsK, because GST-KbpA reduced the amount of the phosphorylated, active form of TRX-AfsK by inhibiting the autophosphorylation. The small extent of inhibition can be attributed to the autophosphorylated, active AfsA that was inevitably present in a small amount in the AfsK preparation. GST-KbpA also inhibited autophosphorylation of TRX-KΔCwt to a similar extent (data not shown). It is thus apparent that KbpA binds the kinase domain of AfsK and inhibits its autophosphorylation.
FIG. 4.
Inhibition of AfsK autophosphorylation by KbpA. (A) Incubation of TRX-AfsK (15 pmol) in the presence of [γ-32P]ATP and GST (300 pmol) yielded the autophosphorylated form (lane 1). The presence of GST-KbpA (300 pmol) instead of GST inhibited the autophosphorylation. Phosphorylation of GST-AfsR (30 pmol) was also inhibited by GST-KbpA but not by GST (lane 4). The amounts of the proteins contained in the reaction mixture were monitored by staining the gel with Coomassie brilliant blue (CBB). (B) The gels were analyzed on an image analyzer, and the inhibition of autophosphorylation of TRX-AfsK and phosphorylation of GST-AfsR by TRX-AfsK was plotted. The degrees of inhibition are the means of values obtained from three independent experiments. Error bars indicate standard deviations.
DISCUSSION
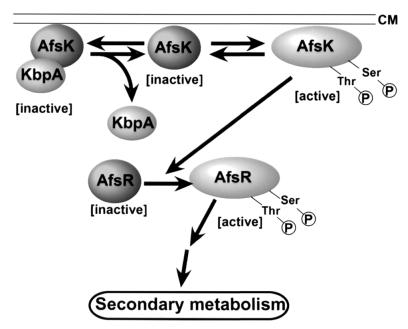
The in vitro experiments have shown that KbpA directly binds the nonphosphorylated form of AfsK and inhibits the autophosphorylation of AfsK. This is consistent with the in vivo observations that KbpA serves as a repressor for actinorhodin production. Also consistent with this are the observations that a mutant AfsK unable to autophosphorylate did not complement the reduced actinorhodin production by an afsK mutant (unpublished results). The kinase domain at the N-terminal portion of AfsK interacts with KbpA. Recovery of only nonphosphorylated forms of TRX-KΔCwt and TRX-KΔCK44A by the pull-down assay with GST-KbpA suggests that the presence of even a single phosphorylated site at either serine or threonine residue prevents KbpA from binding to AfsK, although the numbers and the exact positions of serine and threonine residues to be autophosphorylated are still unknown. We thus assume that the population of the phosphorylated, active form of AfsK is modulated by the amount of KbpA, as a result of which the degree of phosphorylation of AfsR is controlled (Fig. 5). At a later stage of growth under usual culture conditions on agar medium, KbpA binds the nonphosphorylated form of AfsK, which has not yet autophosphorylated or which has been dephosphorylated by protein phosphatases, and inhibits its autophosphorylation at serine and threonine residues. It is unclear whether the AfsK-KbpA complex is associated with the membrane. The nonphosphorylated AfsK is inactive and unable to activate the positive regulator AfsR by phosphorylating it at serine and threonine residues, as a result of which secondary metabolism, including actinorhodin production, is repressed. Thus, KbpA serves as an inhibitor in a negative feedback system in the AfsK-AfsR regulatory pathway. The ΔkbpA mutation, allowing AfsK activation by autophosphorylation, would result in accumulation of a larger amount of the phosphorylated form of AfsR and in overproduction of actinorhodin and undecylprodigiosin, as is observed in strains overexpressing afsR. A larger amount of KbpA would result in a decrease of the amount of the active form of AfsR and a reduction of the pigment production, as is found in afsR and afsK mutants. The hydropathy plot of KbpA excludes the possibility that it is a membrane protein. Therefore, KbpA seems not to control the localization of AfsK inside the hyphae, unlike the many kinase-anchoring proteins that determine the specificities and activities of kinase-mediated signaling pathways in eukaryotes (5, 14, 18). AfsK appears to bind loosely to the inner side of the membrane, because it is recovered from the membrane fraction by mild treatment with detergents during purification (17). A seven-repeat sequence in the C-terminal portion may be a motif to anchor a membrane protein, as pointed out by Nadvornik et al. (20).
FIG. 5.
Model of inhibition of AfsK autophosphorylation by KbpA.
kbpA is transcribed throughout growth, and its transcription is enhanced when production of actinorhodin and undecylprodigiosin has already started. Since afsK is transcribed constantly throughout growth, KbpA appears to put a brake on the unlimited production of the pigments that has been commenced by the AfsK-AfsR system. AfsK-AfsR seems to operate independently of the pathway-specific regulatory proteins to mainly control pigment production, such as ActII-ORF4 for actinorhodin and RedD for undecylprodigiosin (6), suggesting an inhibitory role of KbpA in a negative feedback system in the AfsK-AfsR regulatory pathway.
The genome project for S. coelicolor A3(2) revealed the presence of six KbpA homologues and more than 20 AfsK homologues containing a catalytic domain of protein serine/threonine kinases. AfsR is phosphorylated not only by AfsK but also by an additional kinase (17). We have recently found that two other kinases are capable of phosphorylation of AfsR at serine and threonine residues (unpublished results). By analogy with the eukaryotic systems, these AfsK homologues recognize their respective signals and transfer them to AfsR by means of phosphorylation. It is unclear whether KbpA and the six KbpA homologues discriminate and bind these kinases and modulate their activity. Functionally unknown KbpA homologues found in Streptomyces and various bacteria may be elucidated through an approach based on the assumption that KbpA homologues serve as a modulator by protein-protein interaction. Among KbpA homologues in various bacteria, PA1672, encoded by the region just downstream of sty1 encoding a serine/threonine kinase in Pseudomonas aeruginosa (19) (www.pseudomonas.com/), may serve as a StyI-binding protein.
ACKNOWLEDGMENTS
T. Umeyama was supported by the Japan Society for the Promotion of Science (JSPS). This work was supported by the Asahi Glass Foundation, by the “Research for the Future” Program of JSPS, and by the Bio Design Program of the Ministry of Agriculture, Forestry, and Fisheries of Japan (BDP-01-VI-2-1).
REFERENCES
- 1.Ando N, Ueda K, Horinouchi S. A Streptomyces griseus gene (sgaA) suppresses the growth disturbance caused by high osmolality and a high concentration of A-factor during early growth. Microbiology. 1997;143:2715–2723. doi: 10.1099/00221287-143-8-2715. [DOI] [PubMed] [Google Scholar]
- 2.Arias P, Fernandez-Moreno M A, Malpartida F. Characterization of the pathway-specific positive transcriptional regulator for actinorhodin biosynthesis in Streptomyces coelicolor A3(2) as a DNA-binding protein. J Bacteriol. 1999;181:6958–6968. doi: 10.1128/jb.181.22.6958-6968.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bibb M J, Janssen G R, Ward J M. Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene. 1986;41:E357–E368. doi: 10.1016/0378-1119(85)90220-3. [DOI] [PubMed] [Google Scholar]
- 4.Cooper J A, Sefton B M, Hunter T. Detection and quantification of phosphotyrosine in proteins. Methods Enzymol. 1983;99:387–402. doi: 10.1016/0076-6879(83)99075-4. [DOI] [PubMed] [Google Scholar]
- 5.Edwards A S, Scott J D. A-kinase anchoring proteins: protein kinase A and beyond. Curr Opin Cell Biol. 2000;12:217–221. doi: 10.1016/s0955-0674(99)00085-x. [DOI] [PubMed] [Google Scholar]
- 6.Floriano B, Bibb M. afsR is a pleiotropic but conditionally required regulatory gene for antibiotic production in Streptomyces coelicolor A3(2) Mol Microbiol. 1996;21:385–396. doi: 10.1046/j.1365-2958.1996.6491364.x. [DOI] [PubMed] [Google Scholar]
- 7.Hong S-K, Kito M, Beppu T, Horinouchi S. Phosphorylation of the afsR product, a global regulatory protein for secondary-metabolite formation in Streptomyces coelicolor A3(2) J Bacteriol. 1991;173:2311–2318. doi: 10.1128/jb.173.7.2311-2318.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: John Innes Foundation; 1985. [Google Scholar]
- 9.Horinouchi S, Kito M, Nishiyama M, Furuya K, Hong S-K, Miyake K, Beppu T. Primary structure of AfsR, a global regulatory protein for secondary metabolite formation in Streptomyces coelicolor A3(2) Gene. 1990;95:49–56. doi: 10.1016/0378-1119(90)90412-k. [DOI] [PubMed] [Google Scholar]
- 10.Horinouchi S, Suzuki H, Nishiyama M, Beppu T. Nucleotide sequence and transcriptional analysis of the Streptomyces griseus gene (afsA) responsible for A-factor biosynthesis. J Bacteriol. 1989;171:1206–1210. doi: 10.1128/jb.171.2.1206-1210.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kakinuma S, Takada Y, Ikeda H, Tanaka H, Omura S. Cloning of large DNA fragments, which hybridize with actinorhodin biosynthesis genes, from kalafungin and nanaomycin A methyl ester producers and identification of genes for kalafungin biosynthesis of the kalafungin producer. J Antibiot. 1991;44:995–1005. doi: 10.7164/antibiotics.44.995. [DOI] [PubMed] [Google Scholar]
- 12.Kamps M P, Sefton B M. Acid and base hydrolysis of phosphoproteins bound to Immobilon facilitates analysis of phosphoamino acids in gel-fractionated proteins. Anal Biochem. 1989;176:22–27. doi: 10.1016/0003-2697(89)90266-2. [DOI] [PubMed] [Google Scholar]
- 13.Kelemen G H, Brown G L, Kormanec J, Potuckova L, Chater K F, Buttner M J. The position of the sigma-factor genes, whiG and sigF, in the hierarchy controlling the development of spores chains in the aerial hyphae of Streptomyces coelicolor A3(2) Mol Microbiol. 1996;21:593–603. doi: 10.1111/j.1365-2958.1996.tb02567.x. [DOI] [PubMed] [Google Scholar]
- 14.Kolch W. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem J. 2000;351:289–305. [PMC free article] [PubMed] [Google Scholar]
- 15.Lomovskaya N, Otten S L, Doi-Katayama Y, Fonstein L, Liu X-C, Takatsu T, Inventi-Solari A, Filippini S, Torti F, Colombo A L, Hutchinson C R. Doxorubicin overproduction in Streptomyces peucetius: cloning and characterization of the dnrU ketoreductase and dnrV genes and the doxA cytochrome P-450 hydroxylase gene. J Bacteriol. 1999;181:305–318. doi: 10.1128/jb.181.1.305-318.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 17.Matsumoto A, Hong S-K, Ishizuka H, Horinouchi S, Beppu T. Phosphorylation of the AfsR protein involved in secondary metabolism in Streptomyces species by a eukaryotic-type protein kinase. Gene. 1994;146:47–56. doi: 10.1016/0378-1119(94)90832-x. [DOI] [PubMed] [Google Scholar]
- 18.Mochly-Rosen D, Gordon A S. Anchoring proteins for protein kinase C: a means for isozyme selectivity. FASEB J. 1998;12:35–42. [PubMed] [Google Scholar]
- 19.Mukhopadhyay S, Kapatral V, Xu W, Chakrabarty A M. Characterization of a Hank's type serine/threonine kinase and serine/threonine phosphoprotein phosphatase in Pseudomonas aeruginosa. J Bacteriol. 1999;181:6615–6622. doi: 10.1128/jb.181.21.6615-6622.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nadvornik R, Vomastek T, Janecek J, Technikova Z, Branny P. Pkg2: a novel transmembrane protein Ser/Thr kinase of Streptomyces granaticolor. J Bacteriol. 1999;181:15–23. doi: 10.1128/jb.181.1.15-23.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogawara H, Aoyagi N, Watanabe M, Urabe H. Sequences and evolutionary analyses of eukaryotic-type protein kinases from Streptomyces coelicolor A3(2) Microbiology. 1999;145:3343–3352. doi: 10.1099/00221287-145-12-3343. [DOI] [PubMed] [Google Scholar]
- 22.Oh S H, Chater K F. Denaturation of circular or linear DNA facilitates targeted integrative transformation of Streptomyces coelicolor A3(2): possible relevance to other organisms. J Bacteriol. 1997;179:122–127. doi: 10.1128/jb.179.1.122-127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sollner-Webb B, Reeder R H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. leavis. Cell. 1979;18:485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- 24.Takano E, White J, Thompson C J, Bibb M J. Construction of thiostrepton-inducible, high-copy-number expression vectors for use in Streptomyces spp. Gene. 1995;166:133–137. doi: 10.1016/0378-1119(95)00545-2. [DOI] [PubMed] [Google Scholar]
- 25.Umeyama T, Lee P-C, Ueda K, Horinouchi S. An AfsK/AfsR system involved in the response of aerial mycelium formation to glucose in Streptomyces griseus. Microbiology. 1999;145:2281–2292. doi: 10.1099/00221287-145-9-2281. [DOI] [PubMed] [Google Scholar]
- 26.Urabe H, Ogawara H. Cloning, sequencing and expression of serine/threonine kinase-encoding genes from Streptomyces coelicolor A3(2) Gene. 1995;153:99–104. doi: 10.1016/0378-1119(94)00789-u. [DOI] [PubMed] [Google Scholar]
- 27.Vomastek T, Nadvornik R, Janecek J, Technikova Z, Weiser J, Branny P. Characterization of two putative protein Ser/Thr kinases from actinomycete Streptomyces granaticolor both endowed with different properties. Eur J Biochem. 1998;257:55–61. doi: 10.1046/j.1432-1327.1998.2570055.x. [DOI] [PubMed] [Google Scholar]
- 28.Wietzorrek A, Bibb M. A novel family of proteins that regulate antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol Microbiol. 1997;25:1181–1184. doi: 10.1046/j.1365-2958.1997.5421903.x. [DOI] [PubMed] [Google Scholar]
- 29.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]