Abstract
Pharmacogenomics (PGx) relates to the study of genetic factors determining variability in drug response. Implementing PGx testing in paediatric patients can enhance drug safety, helping to improve drug efficacy or reduce the risk of toxicity. Despite its clinical relevance, the implementation of PGx testing in paediatric practice to date has been variable and limited.
As with most paediatric pharmacological studies, there are well‐recognised barriers to obtaining high‐quality PGx evidence, particularly when patient numbers may be small, and off‐label or unlicensed prescribing remains widespread. Furthermore, trials enrolling small numbers of children can rarely, in isolation, provide sufficient PGx evidence to change clinical practice, so extrapolation from larger PGx studies in adult patients, where scientifically sound, is essential.
This review paper discusses the relevance of PGx to paediatrics and considers implementation strategies from a child health perspective. Examples are provided from Canada, the Netherlands and the UK, with consideration of the different healthcare systems and their distinct approaches to implementation, followed by future recommendations based on these cumulative experiences.
Improving the evidence base demonstrating the clinical utility and cost‐effectiveness of paediatric PGx testing will be critical to drive implementation forwards. International, interdisciplinary collaborations will enhance paediatric data collation, interpretation and evidence curation, while also supporting dedicated paediatric PGx educational initiatives. PGx consortia and paediatric clinical research networks will continue to play a central role in the streamlined development of effective PGx implementation strategies to help optimise paediatric pharmacotherapy.
Keywords: children, personalised medicine, pharmacogenetics, precision medicine
1. PHARMACOGENOMICS: AN INTRODUCTION
Pharmacogenomics (PGx) relates to the study of genetic factors determining variability in drug response, in terms of both efficacy and toxicity. 1 It has long been known that individuals respond differently to medicines and, over the last two decades, numerous peer‐reviewed publications have promised imminent benefits of precision medicine, with many highlighting pharmacogenomic strategies as a core component of this long awaited revolution in healthcare. 2 There is extensive literature dedicated to the clinical utility of PGx, which is largely focused on adult patients, 3 , 4 , 5 on whom the majority of PGx research is based. The challenges to implementation of pharmacogenomic testing within routine healthcare are well described. 6 , 7 , 8 , 9 The importance of PGx has been recognised by the drug regulatory agencies, with the European Medicines Agency (EMA) describing PGx as an “integral part of the development and post‐authorization (marketing) phase for a number of medicines, with significant impact on the management of their benefits and risks in clinical use”. 10 , 11 , 12 However, the perceived relevance and familiarity of PGx to healthcare professionals working in paediatrics remain limited in most settings, 13 and the implementation of pharmacogenomic testing in paediatrics is also limited. 14
This review paper discusses the relevance of PGx to paediatrics and considers the pros and cons of different implementation strategies from a child health perspective. Examples are provided from Canada, the Netherlands, and the UK, with consideration of the distinct healthcare systems, different approaches to implementation and its coordination at a national level, followed by future recommendations based on these experiences to date.
1.1. Pharmacogenomic testing: An overview
The aim of a pharmacogenomic test is to improve either the safety or effectiveness of a pharmacological therapy, or both. This is achieved by using a patient's pharmacogenetic data (i.e., the elements of their genetic information that are of relevance to drug therapy) to inform prescription decision making. 15 To be deemed relevant to clinical practice, the genetic information obtained from a PGx test must be a robust predictor of drug response; 1 however, the degree of the genetically determined variability in drug response can vary considerably (ranging from 20–95% depending on the drug concerned 16 ). In complex disease genetics (e.g., hypertension, type 2 diabetes, schizophrenia), there are multiple genes of interest, each producing small individual effects. However, exposure of the body to medication is a new event, in evolutionary terms, and there is evidence that genetic variants affecting medicines have, overall, larger effect sizes aiding their potential clinical utility. 17
A PGx test result may detect a genetic predisposition to an adverse drug reaction (ADR), or differentiate between drug responders and drug non‐responders, or it may indicate that a different dose of the drug—or in some cases a different class of drug altogether—is required (Table 1). 1 Actionable PGx results are those that would alter the choices made by the prescriber, and that would significantly alter the balance of benefit to harm for the individual patient. 22 In many adult PGx panel‐based studies, actionable PGx variants have been identified in more than 90% of patients. 23 , 24 Evidence‐based classifications of actionability of drug/gene pairs are available from PGx practice guidelines consortia, which are discussed further below.
TABLE 1.
Examples of PGx stratification of patient groups that are clinically relevant to paediatrics 18
| PGx patient group | Clinically relevant examples |
|---|---|
| Responders | Ivacaftor therapy for cystic fibrosis patients with specified CFTR mutations: G551D, G1244E, G1349D, G178R, G551S, S1251N, S1255P, S549N or S549R |
| Non‐responders | CYP2D6 poor metabolisers: Limited conversion of codeine to morphine: Recommend prescription of alternative analgesic instead |
| Differential responders | CYP2C19 polymorphisms affect voriconazole pharmacokinetics: Genotype‐guided dosing can help to optimise paediatric voriconazole therapy 19 |
| CYP2D6 ultra‐rapid metabolisers have increased conversion of codeine to morphine, and this PGx variability is an important factor contributing to the EMA decision to make codeine use contraindicated in children under 12 years old 20 | |
| At risk of severe ADR | TPMT genetic polymorphisms resulting in TPMT deficiency can predispose to potentially fatal myelotoxicity with thiopurine therapy (e.g., 6‐MP; discussed further in text below) |
| HLA‐B*5701 allele predisposes to abacavir hypersensitivity: As the reaction is severe, the drug is contraindicated. | |
| HLA‐B*15:02 allele predisposes to carbamazepine induced Stevens–Johnson syndrome (SJS) and toxic epidermal necrolysis (TEN), in patients from Southeast Asian countries 21 |
Abbreviations: CFTR, cystic fibrosis transmembrane conductance regulator; EMA, European Medicines Agency; TPMT, thiopurine S‐methyltransferase; 6‐MP, 6‐mercaptopurine.
PGx testing can be either reactive or pre‐emptive. Reactive testing occurs at (or close to) the point of prescription of a drug paired with a known pharmacogene (i.e., the gene of relevance to the clinical pharmacology of that particular compound). This process requires a prompt turnaround time to ensure the result is available in time to inform the relevant prescription. 19 In contrast, pre‐emptive testing involves prospective PGx testing, before prescription of the relevant drug(s) is required. Here, results must be accessible within the electronic health record (EHR), and future prescribers need to be aware of the existence of actionable PGx results, and know what action to take. 19 In addition, when utilising an EHR, clinical decision support (CDS) systems are often also used. At least 20 different pharmacogenomic CDS systems have been, or are being, developed, with the majority embedded within EHR systems. 20 An ideal CDS will inform the clinician of the clinically relevant PGx finding before they prescribe, including drug–drug interactions, while at the same time being sophisticated enough to work with dose range checking processes (as PG may require dose alteration to achieve clinical efficacy, while avoiding alert fatigue in the users). Some evaluation of CDS for pharmacogenomics has been presented, but more will be required. 20
The approaches for PGx testing can include targeted genotyping technologies, which may focus on a single pharmacogene (if reactive testing is chosen) or a number of pharmacogenes within a PGx panel, 21 or may employ next generation sequencing techniques (e.g., whole exome or whole genome sequencing), which can be used with virtual (bioinformatics‐based) PGx panels, 18 , 25 , 26 , 27 , 28 , 29 although the latter is largely restricted to research contexts at present.
1.2. PGx practice guidelines consortia
The gap in the use of research discoveries to guide clinical practice—the so‐called “Valley of Death”—is well known. To address this, and aid the dissemination of PGx data and implementation into clinical practice, a number of PGx consortia have evolved. These include CPIC, The Pharmacogenomics Knowledge Base (PharmGKB), the Canadian Pharmacogenomics Network for Drug Safety (CPNDS) and the Dutch Pharmacogenetics Working Group. 30 , 31 , 32 , 33 Consortia members include representatives from many different disciplines, including physicians, pharmacists and clinical scientists, with relevant expertise in clinical pharmacology and therapeutics (CPT), genetics and numerous relevant subspecialty areas. These consortia disseminate evidence‐based, peer‐reviewed, updated and curated PGx practice guidelines, which consider all aspects of PGx including variation in risk according to ethnicity. Clear, well‐referenced knowledge summaries are also published, for example those dedicated to so‐called Very Important Pharmacogenes (VIPgenes: defined as genes with well‐documented information regarding the relationship with a drug's pharmacokinetics/pharmacodynamics [PKPD]). 33 Notably, these consortia also provide information about when a PGx test should not be used to guide prescribing (denoted, for example, by CPIC assigning actionability levels C and D, which signifies that a specified gene/drug pair is not actionable). 31 There are methodological differences in the evidence appraisal approaches used by different consortia, which warrant more detailed consideration when they lead to differing recommendations regarding the actionability of PGx variants encountered in clinical practice. 34
2. THE RATIONALE FOR IMPLEMENTING PGX TESTING IN CHILDREN
While the rationale underlying PGx testing is largely equivalent for adults and children, the relevant principles are summarised here from the paediatric perspective. An actionable PGx result reported in a child should change the choices made by a paediatrician (or a physician looking after the same patient in future during adulthood) if prescribing the relevant drug(s). Therefore, PGx information needs to be available and interpretable according to updated evidence‐based paediatric PGx recommendations. While it is true that PGx will not be directly relevant for every child, nor will it be relevant for every prescription. This does not mean, however, that appropriate PGx testing should not be available as part of routine care for those children for whom it is relevant. It also should not matter if the prescribing clinician is the one who ordered the test, or is even located in the same institution, as these data should be shared to all prescribers to avoid unnecessary repetition of tests (which entails potential harms for the child and additional financial costs). Currently, the limited availability of PGx testing and clear paediatric PGx guidelines presents a major obstacle, in addition to the need for enhanced PGx evidence in children.
To overcome this, scaling up of new, validated testing strategies for routine implementation within healthcare systems is required, but can present numerous challenges. Traditionally, paediatric pharmacology research studies and implementation strategies to update drug labelling and evidence‐based guidelines have often faced long delays in comparison to the equivalent timelines in adult medicine. 35 Typically, this delay arises due to various factors, including practical, ethical and financial barriers to research in children, many of which are now largely historical, thanks to coordinated global efforts to improve the pace of paediatric medicines research. 36
The majority of prescriptions for children are prescribed in the community, 37 with medicines most commonly prescribed by a GP or Community Paediatrician. In contrast, the focus of interest for PGx, at least at first, is likely to be in prescriptions issued for medicines with narrow therapeutic ranges, variable efficacy, dose limiting toxicity, relatively frequent PGx‐related ADRs, or intensive monitoring requirements. Therefore, the majority of paediatric prescriptions for which PGx is relevant will, at least initially, be in hospitals. In keeping with this, selected PGx testing is already routinely available in certain specialist settings, such as in paediatric haemato‐oncology where thiopurine S‐methyltransferase (TPMT) testing is routine (or mandatory) for patients due to receive 6‐mercaptopurine therapy. 38 Relevant stakeholders from both primary and secondary care settings need to be prospectively involved in implementation planning, as discussed further below.
With the mainstreaming of genomic medicine rapidly changing the healthcare landscape, 39 increasing numbers of children and newborns are undergoing whole genome sequencing (WGS). 40 In particular, the role of diagnostic WGS (or WES [whole exome sequencing]) in paediatric and neonatal intensive care settings is growing, 41 , 42 and in addition, pre‐existing WES/WGS data can be re‐analysed to investigate PGx variants. 43 There have also been recent discussions about whether WGS may be introduced as part of routine newborn screening. 44 , 45 Since PGx testing can either be pre‐emptively integrated into WGS for diagnostic reasons—or introduced as a separate “routine” (or reactive) test in its own right (e.g., when WGS is not available/required)—the concepts behind the development of a PGx “passport” 46 with a lifelong PGx record 47 have become a reality that urgently needs further exploration within paediatric healthcare policy and practice. With appropriate data retention policies, WGS could represent the once in a lifetime test after which it would be possible to routinely access the PGx data at any point in a child's lifetime when they need a relevant drug prescribing. Such an approach would require the development of robust standard operating procedures (SOPs) that incorporate prospectively planned periodic data re‐analysis to identify PGx variants which have since become clinically actionable (following new research findings), as this information would need to be updated in individuals' PGx records in a timely fashion.
Beyond the adoption of widespread genomic testing into healthcare services, there are other ways in which PGx testing for children is likely to emerge. Home PGx testing is already available to (adult) patients themselves via direct‐to‐consumer (DTC) routes 48 , 49 (although not all DTC results are consistent with regulatory guidelines). The utility of these services in children is not clear and it is likely that their availability (and related legislation) will vary in different jurisdictions, but it will be important to recognise that parents may seek to use them for their children.
2.1. Assessing PGx evidence
Expert PGx consortia have researched and summarised much of the PGx evidence that is available to date, 50 , 51 considering data from the numerous previous randomised controlled trials (RCTs) in (mainly adult) patients that have demonstrated the clinical utility of PGx testing. As implementation initiatives progress, it will be important to synthesise and analyse the cumulative evidence from PGx studies involving children, which will require consideration of both the quantity and quality of evidence available. Requiring separate paediatric (or adult) RCTs to establish the clinical relevance of every single PGx gene/drug pair would prove impractical and unethical, as well as being unnecessary. 52 It also ignores the potential benefits of using either PGx panel or approaches based on next generation sequencing (NGS). It would be logical to approach PGx information (using data from either drug development or post‐marketing studies) on the basis of sound pharmacological principles, to include appropriate prescribing actions according to specified PGx results, in the same way that drug dose recommendations are often adjusted for patients with renal or hepatic dysfunction, where necessary, without separate trials in these special populations. 52 Aronson et al. recently highlighted the inadequate appreciation and utilisation of mechanistic evidence in drug approval processes (where mechanistic evidence refers to evidence of different types, e.g. in vivo, ex vivo, in vitro, clinical, observational or simulation studies, that supports the existence or details of a particular pharmacological mechanism), in association with an overemphatic focus on the results of clinical studies/trials 53 ; within a pharmacogenomic context, there is a risk that this same phenomenon exacerbates the ongoing delays in PGx implementation. 54
2.2. The need for separate paediatric PGx evidence
When reflecting on the need for more paediatric data in order to implement evidence‐based PGx testing in children, it is important to consider two fundamental questions: firstly, is separate paediatric PGx evidence always needed? Secondly, if separate paediatric PGx studies are not always essential, when is it appropriate to extrapolate adult pharmacogenomic data to adolescents, children or even infants? In the field of paediatric clinical pharmacology, the traditional mantra had always been that ‘children are not small adults’, 55 although it has been increasingly recognised that, in terms of pharmacokinetics, it can be argued that “children are small adults, neonates are immature children”. 56 For those drugs where there is an actionable pharmacogene, this will typically be because of the pharmacogenetic effect on either the PK or PD of the drug in question.
Given that most PGx data have been generated in adult populations, 14 , 57 it therefore needs to be established, to what extent (if any) adult PGx data can safely be extrapolated to the paediatric population for each relevant drug/gene pair and the related prescribing indication(s). Several factors must be considered to determine the appropriateness of extrapolation, including the natural history of the disease progression and response to the proposed therapeutic intervention, the likely (or known) exposure–response profile, and the applicability of adult PD measures to children. 58 , 59 The EMA has acknowledged in its reflection paper on paediatric extrapolation that “gaps in knowledge of intrinsic factors related to organ maturation and ontogeny of enzymatic and transport functions or pharmacogenetics […], particularly in the youngest age groups of the paediatric population are sources of uncertainties and can affect the reliability in the predictions”. 60 It will be essential for researchers to further characterise the role of ontogeny in relation to developmental PGx, particularly with respect to the age‐related changes in expression of drug‐metabolising enzymes (DME) and transporters, exploring how these dynamic processes influence the clinical relevance of known pharmacogenes from birth to adulthood. 61 , 62 This research will be especially important in the youngest age groups (i.e., patients under 2 years of age) where pharmacological variability—particularly that derived from DME ontogeny—is most pronounced, and also in those settings where the indication for a particular drug is unique to paediatrics.
Evidence should also be sought to validate known PGx associations in children 57 and, where possible, to demonstrate the conditions determining suitability of extrapolation using different PGx datasets including data from both adults and children. Given the paediatric PGx data that exist and the implementation programmes that have already evolved, it would be valuable to reach consensus regarding the acceptability of adopting (or adapting) paediatric PGx guidelines from other countries or institutions, when there are examples already demonstrating the effectiveness and utility of PGx testing in children. 63 Progress will improve when there is consensus regarding the definition of the threshold of PGx evidence needed for implementation. 64 This process would benefit from involvement of international specialist paediatric, clinical genetics and pharmacological societies, developing collaborative guidelines together with associated educational materials and mutual endorsement, in order to help accelerate implementation processes, as has previously been achieved in other clinical areas. 65 , 66 , 67 , 68 , 69 Ethically, it is important to avoid unnecessary duplication of paediatric PGx research whenever possible 70 ; it is therefore essential to synthesise existing evidence from PGx research and implementation, so that the lessons learned can enable adoption elsewhere to be streamlined and accelerated.
3. IMPLEMENTING PGX TESTING IN PAEDIATRICS: STRATEGIES AND CHALLENGES
The translation of scientific knowledge into healthcare policy and practice is a complex process, 71 but strategic change is now supported by the ever‐growing field of implementation science. 72 When planning paediatric PGx policy, healthcare leaders, managers and PGx experts must consider the issues described above in addition to viewpoints of different stakeholders, expected benefits, unintended consequences and cost. Examples of key issues that need addressing are briefly summarised in Table 2. Rather than describing different implementation strategies and challenges conceptually, 73 specific examples are given below based on the paediatric PGx implementation experiences and future plans from three countries.
TABLE 2.
Logistics of implementing PGx testing in paediatrics
| Issue | Challenge | Potential solution |
|---|---|---|
| Indication for PGx testing | Clinicians and pharmacists may feel uncertain when testing is required | Clear paediatric PGx guidelines with integration into electronic prescribing systems and protocols, with PGx champions in each clinical area |
| Reporting | Insufficient standardisation of PGx reports will impede interpretation and use of results | Standardised PGx report format, with educational modules to support prescribers, and local PGx web portal and helpline for queries |
| PGx result transfer | Inadequate mechanisms for data transfer/retention between different healthcare IT systems | Unified or interoperable EHRs between primary and secondary care and pharmacists in which PGx data is stored life long |
| Data retention | PGx results may get lost and the information will not be retained in the patient's lifelong EHR | Use of PGx cards or PGx QR codes linked to smartphone app (compatible with national health systems) and IT to enable linkage to local/centralised lifelong EHR |
| Data curation | Research updating PGx knowledge will not be checked against historical PGx results | PGx data repositories will allow original data to be revisited and reports updated periodically |
| Accountability | Prescribers including physicians, pharmacists, nurse prescribers, may not know how to use PGx information and it will be wasted | Proactive multidisciplinary education with CDS tools embedded in e‐prescribing software alerting prescribers to actionable PGx variants |
| Coding | PGx testing and results are not linked to appropriate standardised clinical coding terms | Coding dictionaries need to be updated in discussion with PGx experts |
| Cost | It is unclear who should pay for PGx testing | Cost allocations need predefining during implementation planning |
| Cascade testing | There are ethical issues surrounding the implications for family members once actionable PGx results are known | Guidelines and SOPs should clarify when testing of a patient's relatives is recommended and how this will be communicated to relevant parties |
Abbreviations: PGx, pharmacogenomics; EHR, electronic health record; QR, quick response; CDS, clinical decision support; SOP, standard operating procedure.
3.1. Implementation experience to date and lessons learned
The next section summarises relevant examples of PGx implementation from different countries, with reference to the structure of the healthcare system where relevant, and the key lessons learned.
3.1.1. Canada
Direct provision of healthcare in Canada is a provincial responsibility, with federal oversight of national issues such as drug and device approval. Thus, it could be said that Canada has 14 healthcare systems, and consequently implementation of PGx strategies varies between provinces, with some provinces being much more active in this space than others, with the partial exception of diseases treated by national networks, for example children with cancer who are treated pursuant to national guidelines. It should be noted that, even with these guidelines in place and despite TPMT genotyping being considered a standard of care, availability still varies between provinces. As in many places, the initial push for pharmacogenetic testing came from academic researchers, who in the case of Canada have significant federal grant support.
Pharmacogenetic testing in broader clinical practice began based on discovery, replication and validation work completed by the Canadian Pharmacogenomics Network for Drug Safety (CPNDS) and with other research groups. Clinicians wanted to know why specific genes were being tested from the range of genetic variants reported in the literature. Development of peer‐reviewed, multidisciplinary pharmacogenetic clinical practice guidelines based on systematic reviews and AGREE II 74 were developed for cisplatin 75 and anthracyclines, 76 and published thiopurine guidelines were used. 77 , 78 For the cisplatin and anthracycline guidelines, key questions to be answered were: (1) Who should be tested? (2) Which variants should be tested for? and (3) What therapeutic recommendations should be made if variants are found?
The CPNDS began the implementation of pharmacogenetic testing into clinical practice in 2012, focusing specifically on the implementation of tests in paediatric oncology at ten paediatric oncology centres in seven of the ten provinces in Canada. TPMT variants strongly associated with hearing loss secondary to cisplatin therapy (rs1142345, rs1800460, rs1800462, rs56161402, rs6921269) 79 , 80 UGT1A6, SLC28A3 and RARG variants strongly associated with anthracycline‐induced cardiotoxicity (rs17863783, rs7853758 and rs2229774) 81 , 82 , 83 and TPMT and NUDT15 variants for thiopurine‐induced myelosuppression (rs1142345, rs1800460, rs1800462, rs56161402, rs6921269, rs116855232, rs147390019, rs186364861). 77 , 78 Currently, 827 pharmacogenetic tests have been ordered and results returned in paediatric oncology across Canada.
CPNDS also established a research programme to understand how best to return pharmacogenetic results for robust markers associated with drug outcomes within the three most commonly used classes of drugs: analgesics, antibiotics and psychotropic drugs. Robustness was defined as: information on pharmacogenetic variants included in the drug label by a regulator (e.g., Health Canada, the US Food and Drug Administration, European Medicines Agency); or published, peer‐reviewed pharmacogenetic clinical practice guidelines from an established expert group (CPIC, DPWG or CPNDS); or variants with drug outcome associations with odds ratio ≥ 3 in at least three independent populations (see Table 3). To date, 222 pharmacogenetic tests have been ordered and results returned for these three classes of drugs in three provinces (British Columbia, Ontario, Quebec). Like in other countries such as the US, commercial pharmacogenetic panels are available and accessed throughout Canada by some patients. Some private insurers are paying for pharmacogenetic testing in specific instances.
TABLE 3.
CPNDS PGx testing panel for antibiotics, analgesics and mental health drugs
| Drugs | Adverse drug reactions | Genes | Rationale | ||
|---|---|---|---|---|---|
| ANTIBIOTICS PGx PANEL | |||||
| Aminoglycosides | Hearing loss, deafness | MT‐RNR1 | |||
| Dapsone/sulfonamides | Haemolytic anaemia | G6PD | |||
| Rifampin/isoniazid/pyrazinamide | Serious liver injury | NAT2 | |||
| ANALGESICS PGx PANEL | |||||
| Codeine | CNS depression, therapeutic failure, death | CYP2D6 | |||
| Hydrocodone | CYP2D6 | ||||
| Oxycodone | CNS depression, death | CYP2D6 | |||
| Tramadol | CYP2D6 | ||||
| MENTAL HEALTH PGx PANEL | |||||
| Mood disorders | |||||
| Carbamazepine | Severe cutaneous reactions | HLA‐B, HLA‐A | |||
| Phenytoin | HLA‐B, CYP2C9 | ||||
| ADHD | |||||
| Atomoxetine | Therapeutic failure | CYP2D6 | |||
| Antidepressants | |||||
| SSRIs (e.g., paroxetine) | Therapeutic failure | CYP2D6, CYP2C19 | |||
| SNRIs (e.g., venlafaxine) | CYP2D6 | ||||
Abbreviations: PGx, pharmacogenomics; SNRI, serotonin‐norepinephrine reuptake inhibitor; SSRI, selective serotonin reuptake inhibitors.
3.1.2. The Netherlands
A landmark event in the clinical implementation of pharmacogenetics in the Netherlands was made in 2005 when the Royal Dutch Pharmacist Association (KNMP) founded the Dutch Pharmacogenetics Working Group. 32 This multidisciplinary working group consisting of physicians and pharmacists writes pharmacogenetic guidelines based on systematic literature reviews of drug/gene pairs. To date, more than 100 different drug/gene pairs have been assessed and guidelines are available for 47 drugs. Recently, the DPWG guidelines were endorsed by the European Association of Clinical Pharmacology and Therapeutics 84 and the European Association of Hospital Pharmacists. 85
Originally DPWG recommendations focused on patients with a known genotype. However, DPWG has also started to recommend testing, since the creation of the Clinical Implication Score, a system that identifies the drugs for which specific PGx testing is needed prior to prescribing. 86 The score is assigned to all actionable drug/gene pairs and has three categories for testing: “potentially beneficial,” “beneficial,” and “essential”. Currently the score “essential” is assigned to 14 drug/gene combinations comprising 11 drugs including clopidogrel (CYP2C19), azathioprine/6‐MP (TPMT) and capecitabine/5‐FU (DPYD). The utility of DPYD in paediatrics is limited as fluoropyrimidines are only used to treat rarer solid tumours in this population (e.g., naso‐pharyngeal carcinoma). The clinical recommendations of the DPWG are available at point of care through incorporation in more than 90% of electronic prescribing systems and pharmacy order entry systems.
The most widely adopted PGx test in the Netherlands is testing for variants in DPYD to prevent fluoropyrimidine toxicity. A recent evaluation reported that over 85% of patients were tested prior to start of treatment. 87 Other PGx tests that are generally well accepted include TPMT (mercaptopurine and azathioprine in paediatrics), CYP2C19 (clopidogrel) and UGT1A1 (irinotecan), but the level of implementation is highly variable with specialised and academic centres having the highest adoption rates. PGx testing is mostly performed by 10–15 laboratories that are part of clinical chemistry, hospital pharmacy or clinical genetics departments. In recent years, the number of tests performed in primary care has expanded and in response the Dutch College of General Practitioners has issued a point of view on pharmacogenetics. 88 More recently also the Dutch Society for Psychiatry has released a PGx guidance document. 89
Most PGx testing in the Netherlands is reactive, i.e. in response to the prescription of a drug with a potential PGx recommendation. However, in paediatric clinical genetics WES has become standard practice and, as mentioned above, this offers the opportunity to repurpose existing diagnostic WES data for pharmacogenomics to enable pre‐emptive testing. Recently it was shown that meaningful pharmacogenetic profiles for seven of 11 important pharmacogenes can be successfully extracted, 43 although there were limitations with this technique, with some variations in established pharmacogenes (e.g., CYP2C19 and CYP2D6) not identified.
3.1.3. United Kingdom
Currently paediatric PGx testing in the UK mainly takes place in specialist contexts or research studies, examples of which are given below.
In paediatric haemato‐oncology, TPMT PGx testing is now routine for patients due to receive mercaptopurine as part of the chemotherapy regime for childhood acute lymphoblastic leukaemia (ALL). 38 The implementation of routine testing followed many years of research, 90 which had investigated TPMT genetic polymorphisms in relation to clinically relevant endpoints, including the prevention of potentially fatal myelotoxicity associated with TPMT deficiency, optimal dosing of mercaptopurine, 91 ALL clinical outcomes 92 , 93 and the importance of genotype–phenotype correlation. 94 The details of TPMT pharmacogenomics are reviewed elsewhere 77 and the significance of standardising nomenclature has been emphasised. 95 , 96 Notably, the infrastructure to conduct this TPMT research efficiently in the UK was in place because of the pre‐existing clinical trials networks for childhood cancers; paediatric oncology has been recognised internationally as the “subspecialty in which research defines the standard of care”. 97 Testing of NUDT15 is not currently standard of care in the UK, but testing can be accessed if required. A pre‐existing paediatric clinical research network which fully integrates medical research into routine patient care, with ongoing trials into which PGx studies can be incorporated, is invaluable. 98 Similar trials networks and collaborative clinical groups will remain pivotal in facilitating PGx research and also provide a robust infrastructure within which to disseminate and implement new PGx recommendations. 99 , 100 , 101
Some research studies aim specifically to implement PGx within defined routine care settings, such as the Pharmacogenetics to Avoid Loss Of Hearing (PALOH) study. 102 , 103 PALOH is a clinical implementation study aiming to investigate a new point‐of‐care (POC) PGx testing device to identify neonates at risk of aminoglycoside‐induced hearing loss (secondary to the genetic variant m.1555A>G). 102 This study is recruiting in Liverpool and Manchester, but the findings will be of relevance to neonatal units across the country, particularly if they support the introduction of the new POC PGx test as part of standard neonatal care. PALOH is funded by the NIHR Invention for Innovation (i4i) programme, which is focused on early‐stage collaborative studies, involving academic, NHS and industry partners, and deemed to have commercial potential with scope for future implementation within the NHS. 104 , 105 The study is also supported by the NIHR Manchester Biomedical Research Centre (BRC) 103 , 106 in partnership with a charity 107 and a local small–medium enterprise (SME). The NIHR BRCs all involve collaborative partnerships between universities and NHS teaching hospitals, with the aim of translating scientific research into patient benefit. 108 , 109 This example again demonstrates the importance of appropriate research networks with an adequately resourced infrastructure to support delivery of PGx studies.
Historically, PGx testing approaches and availability within the UK were somewhat heterogeneous and non‐standardised. 110 However, there have recently been large changes to the Genomic Medicine infrastructure in the NHS, which will impact upon future PGx testing implementation. NHS England delivered a report in 2015 describing its focus on “improving outcomes through personalised medicine”, which included the aim of delivering whole genome sequencing for specific conditions by 2020. 111 To support this aim, a series of multidisciplinary genomic education initiatives have been implemented, which are discussed further below. A dedicated PGx working group has been created composed of national experts aiming to prioritise actionable gene/drug pairs. 112 At the time of writing, the formal recommendations of this working group are expected to be published in the near future and, while initial phases are understood to have focused on implementation of PGx testing in adults, it is anticipated that consideration of paediatrics will follow soon afterwards. The infrastructure provided by the NHS Genomic Medicine Service, underpinned by a coordinated national network of Genomic Laboratory Hubs and a National Genomic Test Directory, into which PGx testing can be integrated, has the potential to support delivery of an efficient, cost‐effective national paediatric PGx testing programme.
4. RECOMMENDATIONS FOR PGX IMPLEMENTATION APPROACHES IN PAEDIATRICS
The adoption of PGx guidelines into normal clinical practice will require careful orchestration of national, regional and institutional implementation strategies; these should ideally include CDS tools fully integrated into the electronic health record (EHR), 113 delivered in parallel with a specialist PGx consulting service. Recommendations regarding paediatric PGx needs and implementation strategies do not necessarily need to be segregated from the activities of PGx consortia dedicated to developing PGx practice guidelines and the approaches used for adult patients; for example, in the United States, three children's hospitals were involved from August 2012 in phase II of the Electronic Medical Records and Genomics (eMERGE) Network, 114 , 115 which includes extensive PGx implementation research. 116 A review of the integration of PGx into the US system has also recently been published. 117 There are clear advantages of integrating paediatric PGx implementation into nationwide approaches, rather than postponing paediatric initiatives until an unknown future point, in order to avoid children experiencing unnecessary delays in receiving the benefits of pharmacogenomics in practice.
It is recommended that paediatric PGx implementation planning in different nations is informed by the available evidence, experience and implementation science. Programme delivery should be continually monitored for clinically relevant outcomes, evaluating predefined metrics of success and cost‐effectiveness. As the costs of genomic testing have diminished, 118 PGx testing has become more affordable and therefore more accessible. This apparent affordability does not alone guarantee cost‐effectiveness or clinical utility, and the pharmacoeconomics of proposed paediatric PGx implementation strategies will need detailed evaluation. 119 , 120 Prompt information sharing, either through peer‐reviewed publications or paediatric PGx networks, will enable the collaborative PGx community to learn together to build adaptable, responsive implementation models that can be applied to different healthcare systems. 121 , 122 Implementation initiatives should consider paediatric drug utilisation patterns in different countries and how these will impact upon the practical relevance of PGx recommendations. It will also be important to continue to share lessons learned about implementation across borders so that paediatric PGx benefits can become available globally when resources permit, as previously advocated by the PharmacoGenetics for Every Nation Initiative. 123 , 124
Effective implementation will require early input from key stakeholders, some of whom will vary depending on local and institutional contexts. Our recommendations for stakeholders to invite to PGx planning teams are summarised in Table 4. The importance of advocacy for implementation from experts within the relevant specialty fields needs to be recognised and incorporated into planned educational strategies, together with identification of PGx champions in each discipline who can help to support implementation on the front line. Suggested members for the paediatric PGx multidisciplinary team are shown in Figure 1.
TABLE 4.
Stakeholders to invite in paediatric PGx implementation planning
| Group | Examples of inclusion or recommended representatives |
|---|---|
| Physicians | |
| Paediatricians | Both subspecialist consultants and general paediatricians should be involved in implementation planning |
| Clinical geneticists | Geneticists and genetic counsellors with PGx expertise 125 |
| Clinical pharmacologists | Physicians and pharmacists with expertise in paediatric clinical pharmacology and/or PGx |
| GPs and community doctors | General GPs and those with a special interest in child health |
| Pharmacists | |
| Hospital | Including representation from specialist hospitals and local hospitals |
| Community | Representatives from general community pharmacies |
| Academic and laboratory experts | |
| Scientists | PGx experts from genomic laboratories and clinical academia |
| Nurses | |
| Nurse prescribers | Advanced nurse practitioners and clinical nurse specialists who prescribe for children in relevant contexts |
| Other groups | |
| Patients | Lay representation on PGx working groups and committees |
| Funders | Include management representation and engage early with commissioners |
| Trial coordinators | Research network leads and trial coordinators can advise on integrating planning PGx studies into existing paediatric research networks |
FIGURE 1.
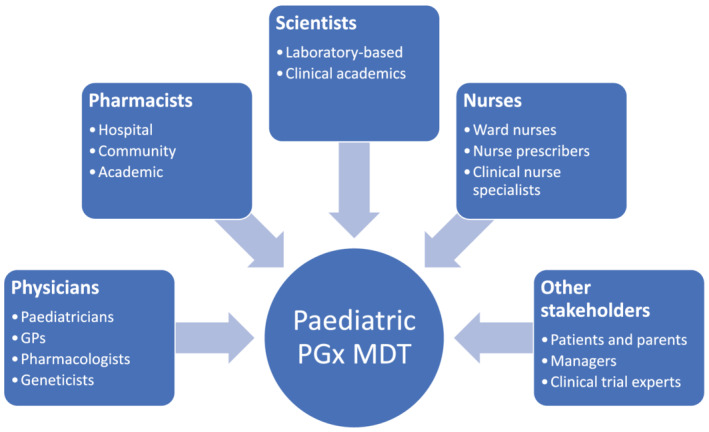
Healthcare professions and other stakeholders who could contribute to a paediatric pharmacogenomics multidisciplinary team (MDT) meeting
4.1. Embedding education in PGx programmes
A critical aspect of implementing and “mainstreaming” paediatric PGx testing will be ensuring that concise educational material is developed for all stakeholders in parallel with the implementation plan. Relevant stakeholders will include practising paediatricians (including hospital consultants, registrars/residents and community paediatricians), general practitioners, hospital/community pharmacists, nurses and PGx analytical laboratory scientists. Educational programmes need to provide adequate information about the principles behind the new testing recommendations, indications, cost and, most importantly, the clinical relevance to patients. Inadequate or ill‐timed multiprofessional education could jeopardise the success of implementation, for example through contributing to inappropriate use of PGx testing, lack of uptake or misinterpretation of results, and therefore a well‐planned education strategy must be embedded within the implementation plan. The importance of this extends beyond pharmacogenomics alone, and to improve “genomic medicine preparedness” (in its broadest sense) among healthcare professionals, interprofessional, interdisciplinary educational programmes should be developed 126 and the material should concomitantly be adapted for undergraduate students in each discipline.
Inspiration can be taken from various pre‐existing educational initiatives, including, for example, the Health Education England (HEE) Genomics Education Programme, which has developed high‐quality educational modules targeted towards professionals and students from different disciplines, and these educational programmes have been formally recognised as a core component of implementing Genomic Medicine within the NHS. 127 The Ubiquitous Pharmacogenomic (U‐PGx) e‐learning platform also provides open access online learning materials. 128 Furthermore, there is a wealth of educational literature relating to pharmacogenomics programmes in many different settings, 129 , 130 , 131 , 132 , 133 , 134 including undergraduate curricula, 135 and it will be important to tailor pre‐existing material specifically towards paediatrics in order to keep it relevant and to make it suitable for CPD (continuing professional development) at a postgraduate level.
4.2. Integrating research with implementation
In addition to education, PGx research needs to be similarly embedded in any paediatric implementation strategy. The evidence based supporting PGx recommendations will constantly grow and protocols will, as always, need to be regularly updated. Demands for ever more research prior to implementation, in order to increase the quantity of evidence available to inform the implementation process, need to be weighed against the risks of further delays to implementation and the right balance must be struck. 54 Integrating research into implementation, for example through prospective ethical approvals for gathering PGx data into curated repositories, can help to overcome this barrier and develop a systematic infrastructure for improving the population PGx evidence base.
5. THE FUTURE
Continued international collaboration and cooperation will enable the PGx community to realise the potential of precision medicine to contribute meaningfully to optimal pharmacotherapy for children. 136 As the omics scientific technologies continue to grow, the remit of PGx will expand to incorporate understanding of new domains such as pharmacotranscriptomics and metabolomics. 137 Updated, curated data‐sharing initiatives underpinned by robust information governance will prove invaluable in advancing paediatric PGx science. Ultimately, evidence‐based PGx in practice will aim to provide clear and timely PGx results to paediatric prescribers, supported by high quality genomic education resources, and continually informed by a cycle of improvement incorporating the latest results of research, audit and stakeholder feedback. It will also be important to work towards harmonisation of paediatric recommendations across the PGx consortia and regulatory agencies to help build internationally recognised PGx standards. 34 , 138
6. CONCLUSIONS
Children deserve the benefits of genomic medicine including PGx‐informed therapies based on strong evidence coupled with affordable implementation. It is inevitable that there are many challenges when introducing new paediatric pharmacogenomic testing strategies into any complex healthcare system. However, with international collaboration and evidence curation, to synthesise implementation success stories from different countries, we can avoid unnecessary duplication of research and develop streamlined approaches to pharmacogenomic implementation strategies. As the genomic medicine revolution is well underway, collaborative efforts of the pharmacogenomic community will continue to bring PGx benefits to the bedside, supported by the infrastructure of well‐funded paediatric research networks. It is essential that a pro‐active dialogue with the paediatric workforce continues to be nurtured, to ensure children benefit from improved access to pharmacogenomic testing delivered with a cost‐effective, evidence‐based and sustainable strategy.
COMPETING INTERESTS
G.G. holds a Genomic Applications Partnership Program grant from Genome Canada and Genome British Columbia with required matching funding from a commercial partner provided by Dynacare.
A.H.M. has received research grants outside the submitted work from GSK, Boehringer Íngelheim and Vertex, is the PI of a P4O2 (Precision Medicine for more Oxygen) public private partnership sponsored by Health Holland involving many private partners that contribute in cash and/or in kind (Boehringer Ingelheim, Breathomix, Fluidda, Ortec Logiqcare, Philips, Quantib‐U, Roche, Smartfish, SODAQ, Thirona, TopMD and Novartis), and she has served on advisory boards for AstraZeneca, GSK and Boehringer Ingelheim with money paid to her institution.
M.R. holds the CIHR‐GSK Chair in Paediatric Clinical Pharmacology at the University of Western Ontario and is a co‐investigator on a Genomic Applications Partnership Program grant from Genome Canada and Genome British Columbia with required matching funding from a commercial partner provided by Dynacare. M.R. holds unrelated grants from the National Science and Engineering Council, the Canadian Institutes of Health Research and the Academic Medical Association of Southwestern Ontario.
B.C. holds a Genomic Applications Partnership Program grant from Genome Canada and Genome British Columbia with required matching funding from a commercial partner provided by Dynacare. B.C. is also an advisor for pharmacogenetic testing at the UnitedHealth Group.
All the other authors declare they have no conflicts of interest.
CONTRIBUTORS
This study was conceived by C.I.S.B. All authors contributed to the writing, editing and overall content of the manuscript.
ACKNOWLEDGEMENTS
C.I.S.B. and T.H. have been supported by the National Institute for Health Research (NIHR) Biomedical Research Centre at King's College London (KCL). D.H. has been supported by the NIHR Alder Hey Clinical Research Facility (CRF). This is a summary of independent research carried out at the National Institute for Health Research (NIHR) Alder Hey Clinical Research Facility. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. C.I.S.B. is funded by the National Institute for Health Research as an Academic Clinical Fellow. The research conducted by B.C. and G.G. at the Canadian Pharmacogenomics Network for Drug Safety (CPNDS) at the University of British Columbia is funded by Genome Canada, Genome BC, the Canadian Institutes of Health Research, with additional support from British Columbia's Provincial Health Services Authority, BC Children's Hospital Foundation and Health Canada.
Barker CIS, Groeneweg G, Maitland‐van der Zee AH, et al. Pharmacogenomic testing in paediatrics: Clinical implementation strategies. Br J Clin Pharmacol. 2022;88(10):4297-4310. doi: 10.1111/bcp.15181
[Correction added on 31 March 2022, after first online publication: The email address of the corresponding author has been updated.]
REFERENCES
- 1. Alfirevic A, Pirmohamed M. Pharmacogenetics and pharmacogenomics. In: Kumar D, Antonarakis S, eds. Medical and Health Genomics. Oxford: Academic Press; 2016:121‐137. [Google Scholar]
- 2. Wolf CR, Smith G, Smith RL. Science, medicine, and the future: pharmacogenetics. BMJ. 2000;320(7240):987‐990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chenoweth MJ, Giacomini KM, Pirmohamed M, et al. Global pharmacogenomics within precision medicine: challenges and opportunities. Clin Pharmacol Ther. 2020;107(1):57‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Klein ME, Parvez MM, Shin JG. Clinical implementation of pharmacogenomics for personalized precision medicine: barriers and solutions. J Pharm Sci. 2017;106(9):2368‐2379. [DOI] [PubMed] [Google Scholar]
- 5. Caraballo PJ, Hodge LS, Bielinski SJ, et al. Multidisciplinary model to implement pharmacogenomics at the point of care. Genet Med. 2017;19(4):421‐429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shuldiner AR, Relling MV, Peterson JF, et al. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: overcoming challenges of real‐world implementation. Clin Pharmacol Ther. 2013;94(2):207‐210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Health Education England Genomics Education Programme . Pharmacogenomics: three challenges to the NHS. Available at: https://www.genomicseducation.hee.nhs.uk/blog/pharmacogenomics-three-challenges-to-the-nhs/. Accessed June 4, 2020.
- 8. Karnes JH, van Driest S, Bowton EA, et al. Using systems approaches to address challenges for clinical implementation of pharmacogenomics. WIREs Syst Biol Med. 2014;6(2):125‐135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Owusu Obeng A, Fei K, Levy K, et al. Physician‐reported benefits and barriers to clinical implementation of genomic medicine: a multi‐site IGNITE‐network survey. J Pers Med. 2018;8(3):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ehmann F, Caneva L, Papaluca M. European Medicines Agency initiatives and perspectives on pharmacogenomics. Br J Clin Pharmacol. 2014;77(4):612‐617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. European Medicines Agency (EMA) . Guideline on the use of pharmacogenetic methodologies in the pharmacokinetic evaluation of medicinal products. 2011. EMA/CHMP/37646/2009. https://www.ema.europa.eu/documents/scientific-guideline/guideline-use-pharmacogenetic-methodologies-pharmacokinetic-evaluation-medicinal-products_en.pdf. Accessed June 2020.
- 12. US Department of Health and Human Services Food and Drug Administration (FDA) . Guidance for Industry. Clinical Pharmacogenomics: Premarket Evaluation in Early‐Phase Clinical Studies and Recommendations for Labeling. 2013. https://www.fda.gov/files/drugs/published/Clinical-Pharmacogenomics--Premarket-Evaluation-in-Early-Phase-Clinical-Studies-and-Recommendations-for-Labeling.pdf. Accessed June 2020.
- 13. Liko I, Lee YM, Stutzman DL, et al. Providers' perspectives on the clinical utility of pharmacogenomic testing in pediatric patients. Pharmacogenomics. 2021;22(5):263‐274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maagdenberg H, Vijverberg SJH, Bierings MB, et al. Pharmacogenomics in pediatric patients: towards personalized medicine. Paediatr Drugs. 2016;18(4):251‐260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rollinson V, Turner RM, Pirmohamed M. Pharmacogenomics: an overview. Clin Pharm. 2017;9(10). [Google Scholar]
- 16. Arwood MJ, Chumnumwat S, Cavallari LH, Nutescu EA, Duarte JD. Implementing pharmacogenomics at your institution: establishment and overcoming implementation challenges. Clin Transl Sci. 2016;9(5):233‐245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maranville JC, Cox NJ. Pharmacogenomic variants have larger effect sizes than genetic variants associated with other dichotomous complex traits. Pharmacogenomics J. 2016;16(4):388‐392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mizzi C, Peters B, Mitropoulou C, et al. Personalized pharmacogenomics profiling using whole‐genome sequencing. Pharmacogenomics. 2014;15(9):1223‐1234. [DOI] [PubMed] [Google Scholar]
- 19. Roden DM, van Driest SL, Mosley JD, et al. Benefit of preemptive pharmacogenetic information on clinical outcome. Clin Pharmacol Ther. 2018;103(5):787‐794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hinderer M, Boeker M, Wagner SA, et al. Integrating clinical decision support systems for pharmacogenomic testing into clinical routine—a scoping review of designs of user‐system interactions in recent system development. BMC Med Inform Decis Mak. 2017;17(1):81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shahandeh A, Johnstone DM, Atkins JR, et al. Advantages of array‐based technologies for pre‐emptive pharmacogenomics testing. Microarrays (Basel). 2016;5(2):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aronson JK. Adjusting therapeutic dosage regimens to optimise the balance of benefit to harm. Clin Med. 2005;5(1):16‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van Driest S, Shi Y, Bowton EA, et al. Clinically actionable genotypes among 10,000 patients with preemptive pharmacogenomic testing. Clin Pharmacol Ther. 2014;95(4):423‐431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bush WS, Crosslin DR, Owusu‐Obeng A, et al. Genetic variation among 82 pharmacogenes: the PGRNseq data from the eMERGE network. Clin Pharmacol Ther. 2016;100(2):160‐169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gordon AS, Fulton RS, Qin X, Mardis ER, Nickerson DA, Scherer S. PGRNseq: a targeted capture sequencing panel for pharmacogenetic research and implementation. Pharmacogenet Genomics. 2016;26(4):161‐168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cohn I, Paton TA, Marshall CR, et al. Genome sequencing as a platform for pharmacogenetic genotyping: a pediatric cohort study. NPJ Genom Med. 2017;2(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ahn E, Park T. Analysis of population‐specific pharmacogenomic variants using next‐generation sequencing data. Sci Rep. 2017;7(1):8416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cousin MA, Matey ET, Blackburn PR, et al. Pharmacogenomic findings from clinical whole exome sequencing of diagnostic odyssey patients. Mol Genet Genomic Med. 2017;5(3):269‐279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee SB, Wheeler MM, Thummel KE, Nickerson DA. Calling star alleles with Stargazer in 28 pharmacogenes with whole genome sequences. Clin Pharmacol Ther. 2019;106(6):1328‐1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89(3):464‐467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Relling MV, Klein TE, Gammal RS, Whirl‐Carrillo M, Hoffman JM, Caudle KE. The Clinical Pharmacogenetics Implementation Consortium: 10 years later. Clin Pharmacol Ther. 2020;107(1):171‐175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Swen JJ, Wilting I, Goede AL, et al. Pharmacogenetics: from bench to byte. Clin Pharmacol Ther. 2008;83(5):781‐787. [DOI] [PubMed] [Google Scholar]
- 33. Thorn CF, Klein TE, Altman RB. PharmGKB: the pharmacogenomics knowledge base. Methods Mol Biol. 2013;1015:311‐320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bank PCD, Caudle KE, Swen JJ, et al. Comparison of the guidelines of the Clinical Pharmacogenetics Implementation Consortium and the Dutch Pharmacogenetics Working Group. Clin Pharmacol Ther. 2018;103(4):599‐618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Samiee‐Zafarghandy S, Mazer‐Amirshahi M, van den Anker JN. Trends in paediatric clinical pharmacology data in US pharmaceutical labelling. Arch Dis Child. 2014;99(9):862‐865. [DOI] [PubMed] [Google Scholar]
- 36. Turner MA, Catapano M, Hirschfeld S, Giaquinto C, Global Research in Paediatrics . Paediatric drug development: the impact of evolving regulations. Adv Drug Deliv Rev. 2014;73:2‐13. [DOI] [PubMed] [Google Scholar]
- 37. Helms PJ, Ekins Daukes S, Taylor MW, Simpson CR, McLay JS. Utility of routinely acquired primary care data for paediatric disease epidemiology and pharmacoepidemiology. Br J Clin Pharmacol. 2005;59(6):684‐690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lennard L. Implementation of TPMT testing. Br J Clin Pharmacol. 2014;77(4):704‐714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Scherer SW. Genomic medicine goes mainstream. NPJ Genom Med. 2016;1(1):15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Peplow M. The 100 000 Genomes Project. BMJ. 2016;353:i1757. [DOI] [PubMed] [Google Scholar]
- 41. French CE, Delon I, Dolling H, et al. Whole genome sequencing reveals that genetic conditions are frequent in intensively ill children. Intensive Care Med. 2019;45(5):627‐636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kingsmore SF, Cakici JA, Clark MM, et al. A randomized, controlled trial of the analytic and diagnostic performance of singleton and trio, rapid genome and exome sequencing in ill infants. Am J Hum Genet. 2019;105(4):719‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. van der Lee M, Allard WG, Bollen S, et al. Repurposing of diagnostic whole exome sequencing data of 1,583 individuals for clinical pharmacogenetics. Clin Pharmacol Ther. 2020;107(3):617‐627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kingsmore SF. Newborn testing and screening by whole‐genome sequencing. Genet Med. 2016;18(3):214‐216. [DOI] [PubMed] [Google Scholar]
- 45. King JS, Smith ME. Whole‐genome screening of newborns? The constitutional boundaries of state newborn screening programs. Pediatrics. 2016;137(Suppl 1):S8‐S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van der Wouden CH, van Rhenen MH, Jama WOM, et al. Development of the PGx‐passport: a panel of actionable germline genetic variants for pre‐emptive pharmacogenetic testing. Clin Pharmacol Ther. 2019;106(4):866‐873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Samwald M, Adlassnig KP. Pharmacogenomics in the pocket of every patient? A prototype based on quick response codes. J Am Med Inform Assoc. 2013;20(3):409‐412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bloss CS, Schork NJ, Topol EJ. Direct‐to‐consumer pharmacogenomic testing is associated with increased physician utilisation. J Med Genet. 2014;51(2):83‐89. [DOI] [PubMed] [Google Scholar]
- 49. Lu M, Lewis CM, Traylor M. Pharmacogenetic testing through the direct‐to‐consumer genetic testing company 23andMe. BMC Med Genomics. 2017;10(1):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Caudle KE, Gammal RS, Whirl‐Carrillo M, Hoffman JM, Relling MV, Klein TE. Evidence and resources to implement pharmacogenetic knowledge for precision medicine. Am J Health Syst Pharm. 2016;73(23):1977‐1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hippman C, Nislow C. Pharmacogenomic testing: clinical evidence and implementation challenges. J Pers Med. 2019;9(3):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gammal RS, Caudle KE, Klein TE, Relling MV. Considerations for pharmacogenomic testing in a health system. Genet Med. 2019;21(8):1886‐1887. [DOI] [PubMed] [Google Scholar]
- 53. Aronson JK, la Caze A, Kelly MP, Parkkinen VP, Williamson J. The use of mechanistic evidence in drug approval. J Eval Clin Pract. 2018;24(5):1166‐1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Krebs K, Milani L. Translating pharmacogenomics into clinical decisions: do not let the perfect be the enemy of the good. Hum Genomics. 2019;13(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stephenson T. How children's responses to drugs differ from adults. Br J Clin Pharmacol. 2005;59(6):670‐673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Anderson BJ, Holford NH. Understanding dosing: children are small adults, neonates are immature children. Arch Dis Child. 2013;98(9):737‐744. [DOI] [PubMed] [Google Scholar]
- 57. Ramsey LB, Brown JT, Vear SI, Bishop JR, van Driest SL. Gene‐based dose optimization in children. Annu Rev Pharmacol Toxicol. 2020;60(1):311‐331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Germovsek E, Barker CIS, Sharland M, Standing JF. Pharmacokinetic‐pharmacodynamic modeling in pediatric drug development, and the importance of standardized scaling of clearance. Clin Pharmacokinet. 2019;58(1):39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dunne J, Rodriguez WJ, Murphy MD, et al. Extrapolation of adult data and other data in pediatric drug‐development programs. Pediatrics. 2011;128(5):e1242‐e1249. [DOI] [PubMed] [Google Scholar]
- 60. European Medicines Agency (EMA) . Reflection paper on the use of extrapolation in the development of medicines for paediatrics. EMA/189724/2018. 2018. https://www.ema.europa.eu/documents/scientific-guideline/adopted-reflection-paper-use-extrapolation-development-medicines-paediatrics-revision-1_en.pdf. Accessed June 2020.
- 61. Neville KA, Becker ML, Goldman JL, Kearns GL. Developmental pharmacogenomics. Paediatr Anaesth. 2011;21(3):255‐265. [DOI] [PubMed] [Google Scholar]
- 62. Brouwer K, Aleksunes LM, Brandys B, et al. Human ontogeny of drug transporters: review and recommendations of the Pediatric Transporter Working Group. Clin Pharmacol Ther. 2015;98(3):266‐287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ramsey LB, Prows CA, Zhang K, et al. Implementation of pharmacogenetics at Cincinnati Children's Hospital Medical Center: lessons learned over 14 years of personalizing medicine. Clin Pharmacol Ther. 2019;105(1):49‐52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Van Driest SL, McGregor TL. Pharmacogenetics in clinical pediatrics: challenges and strategies. Pers Med. 2013;10(7):661‐671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. International Pediatric Nephrology Association (IPNA) . Endorsed Guidelines. https://ipna-online.org/resources/guidelines/endorsed-guidelines/. Accessed June 2020.
- 66. Galiè N, Humbert M, Vachiery JL, et al. 2015 ESC/ERS Guidelines for the diagnosis and treatment of pulmonary hypertension: The Joint Task Force for the Diagnosis and Treatment of Pulmonary Hypertension of the European Society of Cardiology (ESC) and the European Respiratory Society (ERS): endorsed by: Association for European Paediatric and Congenital Cardiology (AEPC), International Society for Heart and Lung Transplantation (ISHLT). Eur Respir J. 2015;46(4):903‐975. [DOI] [PubMed] [Google Scholar]
- 67. Pfaar O, Bachert C, Bufe A, et al. Guideline on allergen‐specific immunotherapy in IgE‐mediated allergic diseases: S2k Guideline of the German Society for Allergology and Clinical Immunology (DGAKI), the Society for Pediatric Allergy and Environmental Medicine (GPA), the Medical Association of German Allergologists (AeDA), the Austrian Society for Allergy and Immunology (ÖGAI), the Swiss Society for Allergy and Immunology (SGAI), the German Society of Dermatology (DDG), the German Society of Oto‐ Rhino‐Laryngology, Head and Neck Surgery (DGHNO‐KHC), the German Society of Pediatrics and Adolescent Medicine (DGKJ), the Society for Pediatric Pneumology (GPP), the German Respiratory Society (DGP), the German Association of ENT Surgeons (BV‐HNO), the Professional Federation of Paediatricians and Youth Doctors (BVKJ), the Federal Association of Pulmonologists (BDP) and the German Dermatologists Association (BVDD). Allergo J Int. 2014;23(8):282‐319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Turner D, Levine A, Escher JC, et al. Management of pediatric ulcerative colitis: joint ECCO and ESPGHAN evidence‐based consensus guidelines. J Pediatr Gastroenterol Nutr. 2012;55(3):340‐361. [DOI] [PubMed] [Google Scholar]
- 69. Vaughan JI, Jeffery HE, Raynes‐Greenow C, et al. A method for developing standardised interactive education for complex clinical guidelines. BMC Med Educ. 2012;12(1):108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Regulation (EC) No. 1901/2006 of The European Parliament and of the Council of 12 December 2006 on medicinal products for paediatric use. http://ec.europa.eu/health/files/eudralex/vol-1/reg_2006_1901/reg_2006_1901_en.pdf. Accessed June 2020.
- 71. Grol R, Grimshaw J. From best evidence to best practice: effective implementation of change in patients' care. Lancet. 2003;362(9391):1225‐1230. [DOI] [PubMed] [Google Scholar]
- 72. Wensing M, Grol R. Knowledge translation in health: how implementation science could contribute more. BMC Med. 2019;17(1):88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Giri J, Moyer AM, Bielinski SJ, Caraballo PJ. Concepts driving pharmacogenomics implementation into everyday healthcare. Pharmgenomics Pers Med. 2019;12:305‐318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Brouwers MC, Kho ME, Browman GP, et al. AGREE II: advancing guideline development, reporting and evaluation in health care. CMAJ. 2010;182(18):E839‐E842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lee JW, Pussegoda K, Rassekh SR, et al. Clinical practice recommendations for the management and prevention of cisplatin‐induced hearing loss using pharmacogenetic markers. Ther Drug Monit. 2016;38(4):423‐431. [DOI] [PubMed] [Google Scholar]
- 76. Aminkeng F, Ross CJD, Rassekh SR, et al. Recommendations for genetic testing to reduce the incidence of anthracycline‐induced cardiotoxicity. Br J Clin Pharmacol. 2016;82(3):683‐695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Relling MV, Schwab M, Whirl‐Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium guideline for thiopurine dosing based on TPMT and NUDT15 genotypes: 2018 update. Clin Pharmacol Ther. 2019;105(5):1095‐1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Children's Oncology Group (COG) . AALL0232: High Risk B‐precursor Acute Lymphoblastic Leukemia (ALL). A Phase III Group‐Wide Study. Children's Oncology Group (COG), Arcadia, CA, USA, 2008.
- 79. Ross CJ, Katzov‐Eckert H, Dubé MP, et al. Genetic variants in TPMT and COMT are associated with hearing loss in children receiving cisplatin chemotherapy. Nat Genet. 2009;41(12):1345‐1349. [DOI] [PubMed] [Google Scholar]
- 80. Pussegoda K, Ross CJ, Visscher H, et al. Replication of TPMT and ABCC3 genetic variants highly associated with cisplatin‐induced hearing loss in children. Clin Pharmacol Ther. 2013;94(2):243‐251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Visscher H, Ross CJD, Rassekh SR, et al. Pharmacogenomic prediction of anthracycline‐induced cardiotoxicity in children. J Clin Oncol. 2012;30(13):1422‐1428. [DOI] [PubMed] [Google Scholar]
- 82. Visscher H, Ross CJD, Rassekh SR, et al. Validation of variants in SLC28A3 and UGT1A6 as genetic markers predictive of anthracycline‐induced cardiotoxicity in children. Pediatr Blood Cancer. 2013;60(8):1375‐1381. [DOI] [PubMed] [Google Scholar]
- 83. Aminkeng F, Bhavsar AP, Visscher H, et al. A coding variant in RARG confers susceptibility to anthracycline‐induced cardiotoxicity in childhood cancer. Nat Genet. 2015;47(9):1079‐1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. European Association for Clinical Pharmacology and Therapeutics (EACPT) . https://www.eacpt.eu/. Accessed April 2021.
- 85. European Association of Hospital Pharmacists (EAHP) . https://www.eahp.eu/. Accessed April 2021.
- 86. Swen JJ, Nijenhuis M, van Rhenen M, et al. Pharmacogenetic information in clinical guidelines: the European perspective. Clin Pharmacol Ther. 2018;103(5):795‐801. [DOI] [PubMed] [Google Scholar]
- 87. Lunenburg CA, van Staveren MC, Gelderblom H, Guchelaar HJ, Swen JJ. Evaluation of clinical implementation of prospective DPYD genotyping in 5‐fluorouracil‐ or capecitabine‐treated patients. Pharmacogenomics. 2016;17(7):721‐729. [DOI] [PubMed] [Google Scholar]
- 88. Nederlands Huisartsen Genootschap . Pharmacogenetic research in general practice. 2020. https://richtlijnen.nhg.org/medisch-inhoudelijke-nhg-standpunten/farmacogenetisch-onderzoek-de-huisartspraktijk. Accessed June 23, 2021.
- 89. Dutch Association for Psychiatry . Leidraad farmacogenetica voor de dagelijkse psychiatrische praktijk. 2020. https://www.nvvp.net/stream/leidraad-farmacogenetica-voor-de-dagelijkse-psychiatrische-praktijk.pdf. Accessed June 23, 2021.
- 90. Research Excellence Framework . Impact Case Studies. Safer treatment of childhood leukaemia through improved delivery of thiopurine drugs. 2014. https://impact.ref.ac.uk/casestudies/CaseStudy.aspx?Id=12333. Accessed June 2020.
- 91. Lennard L, Cartwright CS, Wade R, Vora A. Thiopurine dose intensity and treatment outcome in childhood lymphoblastic leukaemia: the influence of thiopurine methyltransferase pharmacogenetics. Br J Haematol. 2015;169(2):228‐240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lennard L, Cartwright CS, Wade R, Vora A. Thiopurine methyltransferase and treatment outcome in the UK acute lymphoblastic leukaemia trial ALL2003. Br J Haematol. 2015;170(4):550‐558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lilleyman JS, Lennard L. Mercaptopurine metabolism and risk of relapse in childhood lymphoblastic leukaemia. Lancet. 1994;343(8907):1188‐1190. [DOI] [PubMed] [Google Scholar]
- 94. Lennard L, Cartwright CS, Wade R, Richards SM, Vora A. Thiopurine methyltransferase genotype‐phenotype discordance and thiopurine active metabolite formation in childhood acute lymphoblastic leukaemia. Br J Clin Pharmacol. 2013;76(1):125‐136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Appell ML, Berg J, Duley J, et al. Nomenclature for alleles of the thiopurine methyltransferase gene. Pharmacogenet Genomics. 2013;23(4):242‐248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. TPMT Nomenclature Committee . Thiopurine methyltransferase (TPMT) nomenclature website. http://www.imh.liu.se/tpmtalleles. Accessed June 2020.
- 97. Unguru Y. The successful integration of research and care: how pediatric oncology became the subspecialty in which research defines the standard of care. Pediatr Blood Cancer. 2011;56(7):1019‐1025. [DOI] [PubMed] [Google Scholar]
- 98. Turner MA, Attar S, de Wildt SN, Vassal G, Mangiarini L, Giaquinto C. Roles of clinical research networks in pediatric drug development. Clin Ther. 2017;39(10):1939‐1948. [DOI] [PubMed] [Google Scholar]
- 99. Rose AC, van't Hoff W, Beresford MW, Tansey SP. NIHR Medicines for Children Research Network: improving children's health through clinical research. Expert Rev Clin Pharmacol. 2013;6(5):581‐587. [DOI] [PubMed] [Google Scholar]
- 100. Lythgoe H, Price V, Poustie V, et al. NIHR Clinical Research Networks: what they do and how they help paediatric research. Arch Dis Child. 2017;102(8):755‐759. [DOI] [PubMed] [Google Scholar]
- 101. Moore TB, McCabe ERB. National collaborative study groups: structure, benefits gained and potential for rare genetic diseases. Genet Med. 2006;8(12):793‐796. [DOI] [PubMed] [Google Scholar]
- 102. ISRCTN Registry . Pharmacogenetics to avoid loss of hearing. ISRCTN13704894. http://www.isrctn.com/ISRCTN13704894. Accessed June 2020.
- 103. NIHR News . New rapid genetic test could prevent antibiotic‐related hearing loss in newborns. 2020. https://www.nihr.ac.uk/news/new-rapid-genetic-test-could-prevent-antibiotic-related-hearing-loss-in-newborns/23808. Accessed June 2020.
- 104. NIHR . Invention for Innovation. https://www.nihr.ac.uk/explore-nihr/funding-programmes/invention-for-innovation.htm. Accessed June 2020.
- 105. Marjanovic S, Krapels J, Sousa S, Castle‐Clarke S, Horvath V, Chataway J. The NIHR Invention for Innovation (i4i) Programme: a review of progress and contributions to innovation in healthcare technologies. Rand Health Q. 2015;5(2):4. [PMC free article] [PubMed] [Google Scholar]
- 106. NIHR . Manchester Biomedical Research Centre. https://www.manchesterbrc.nihr.ac.uk/. Accessed June 2020.
- 107. Action on Hearing Loss Charity. https://actiononhearingloss.org.uk/. Accessed June 2020.
- 108. National Institute for Health Research (NIHR) . Biomedical Research Centres (BRCs). https://www.nihr.ac.uk/explore-nihr/support/experimental-medicine.htm. Accessed June 2020.
- 109. Snape K, Trembath RC, Lord GM. Translational medicine and the NIHR Biomedical Research Centre concept. QJM. 2008;101(11):901‐906. [DOI] [PubMed] [Google Scholar]
- 110. Higgs J, Gambhir N, Ramsden SC, Poulton K, Newman WG. Pharmacogenetic testing in the United Kingdom genetics and immunogenetics laboratories. Genet Test Mol Biomarkers. 2010;14(1):121‐125. [DOI] [PubMed] [Google Scholar]
- 111. NHS England . Improving outcomes through personalised medicine. https://www.england.nhs.uk/wp-content/uploads/2016/09/improving-outcomes-personalised-medicine.pdf. Accessed June 2, 2020.
- 112. Health Education England Genomics Education Programme . Pharmacogenomics: a new normal for the NHS? https://www.genomicseducation.hee.nhs.uk/blog/pharmacogenomics-a-new-normal-for-the-nhs/. Accessed June 2, 2020.
- 113. Caraballo PJ, Sutton JA, Giri J, et al. Integrating pharmacogenomics into the electronic health record by implementing genomic indicators. J Am Med Inform Assoc. 2020;27(1):154‐158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Gottesman O, Kuivaniemi H, Tromp G, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med. 2013;15(10):761‐771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. National Human Genome Research Institute . eMERGE Phase II Pediatric Request For Applications (RFA). 2011. http://grants.nih.gov/grants/guide/rfa-files/RFA-HG-11-022.html. Accessed June 2020.
- 116. Herr TM, Peterson JF, Rasmussen LV, Caraballo PJ, Peissig PL, Starren JB. Pharmacogenomic clinical decision support design and multi‐site process outcomes analysis in the eMERGE Network. J Am Med Inform Assoc. 2019;26(2):143‐148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Gregornik D, Salyakina D, Brown M, Roiko S, Ramos K. Pediatric pharmacogenomics: challenges and opportunities: on behalf of the Sanford Children's Genomic Medicine Consortium. Pharmacogenomics J. 2021;21(1):8‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Dewey FE, Grove ME, Pan C, et al. Clinical interpretation and implications of whole‐genome sequencing. JAMA. 2014;311(10):1035‐1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Grosse SD. Economic analyses of genetic tests in personalized medicine: clinical utility first, then cost utility. Genet Med. 2014;16(3):225‐227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Phillips KA, Sakowski JA, Trosman J, Douglas MP, Liang SY, Neumann P. The economic value of personalized medicine tests: what we know and what we need to know. Genet Med. 2014;16(3):251‐257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Liu M, Vnencak‐Jones CL, Roland BP, et al. A tutorial for pharmacogenomics implementation through end‐to‐end clinical decision support based on ten years of experience from PREDICT. Clin Pharmacol Ther. 2021;109(1):101‐115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Brown JT, Ramsey LB, van Driest SL, Aka I, Colace SI. Characterizing pharmacogenetic testing among children's hospitals. Clin Transl Sci. 2021;14(2):692‐701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Roederer MW, McLeod HL. Applying the genome to national drug formulary policy in the developing world. Pharmacogenomics. 2010;11(5):633‐636. [DOI] [PubMed] [Google Scholar]
- 124. Roederer MW, Sanchez‐Giron F, Kalideen K, et al. Pharmacogenetics and rational drug use around the world. Pharmacogenomics. 2011;12(6):897‐905. [DOI] [PubMed] [Google Scholar]
- 125. Sturm AC, Sweet K, Manickam K. Implementation of a clinical research pharmacogenomics program at an academic medical center: role of the genetics healthcare professional. Pharmacogenomics. 2013;14(7):703‐706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Rubanovich CK, Cheung C, Mandel J, Bloss CS. Physician preparedness for big genomic data: a review of genomic medicine education initiatives in the United States. Hum Mol Genet. 2018;27(R2):R250‐R258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Health Education England Genomics Education Programme . Genomics education moves into the mainstream. https://www.genomicseducation.hee.nhs.uk/news/genomics-education-moving-into-the-mainstream/. Accessed June 4, 2020.
- 128. Just KS, Steffens M, Swen JJ, Patrinos GP, Guchelaar HJ, Stingl JC. Medical education in pharmacogenomics—results from a survey on pharmacogenetic knowledge in healthcare professionals within the European pharmacogenomics clinical implementation project Ubiquitous Pharmacogenomics (U‐PGx). Eur J Clin Pharmacol. 2017;73(10):1247‐1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Perry CG, Maloney KA, Beitelshees AL, et al. Educational innovations in clinical pharmacogenomics. Clin Pharmacol Ther. 2016;99(6):582‐584. [DOI] [PubMed] [Google Scholar]
- 130. Kuželički NK, Žitnik IP, Gurwitz D, et al. Pharmacogenomics education in medical and pharmacy schools: conclusions of a global survey. Pharmacogenomics. 2019;20(9):643‐657. [DOI] [PubMed] [Google Scholar]
- 131. Nutter SC, Galvez‐Peralta M. Pharmacogenomics: from classroom to practice. Mol Genet Genomic Med. 2018;6(3):307‐313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Weitzel KW, Aquilante CL, Johnson S, Kisor DF, Empey PE. Educational strategies to enable expansion of pharmacogenomics‐based care. Am J Health Syst Pharm. 2016;73(23):1986‐1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Adams SM, Anderson KB, Coons JC, et al. Advancing pharmacogenomics education in the core PharmD curriculum through student personal genomic testing. Am J Pharm Educ. 2016;80(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Brennan KS. Clinical implications of pharmacogenetic and microarray testing for advanced practice nurses. J Am Assoc Nurse Pract. 2015;27(5):246‐255. [DOI] [PubMed] [Google Scholar]
- 135. O'Brien TJ, Goodsaid F, Plack M, et al. Development of an undergraduate pharmacogenomics curriculum. Pharmacogenomics. 2009;10(12):1979‐1986. [DOI] [PubMed] [Google Scholar]
- 136. Manolio TA, Abramowicz M, Al‐Mulla F, et al. Global implementation of genomic medicine: we are not alone. Sci Transl Med. 2015;7(290):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Sa ACC, Sadee W, Johnson JA. Whole transcriptome profiling: an RNA‐Seq primer and implications for pharmacogenomics research. Clin Transl Sci. 2018;11(2):153‐161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Koutsilieri S, Tzioufa F, Sismanoglou DC, Patrinos GP. Unveiling the guidance heterogeneity for genome‐informed drug treatment interventions among regulatory bodies and research consortia. Pharmacol Res. 2020;153:104590. [DOI] [PubMed] [Google Scholar]
- 139. Parry CM, Hawcutt D. Fifteen‐minute consultation: pharmacogenomics: a guide for busy clinicians. Arch Dis Child Educ Pract Ed. 2020;105(2):107‐110. [DOI] [PubMed] [Google Scholar]


