Abstract
The LasR-dependent and RhlR-dependent quorum-sensing systems are global regulators of gene expression in Pseudomonas aeruginosa. Previous studies have demonstrated that promoter elements of the quorum-sensing-controlled genes lasB and hcnABC are important in density-dependent regulation. We have identified LasR- and RhlR-dependent determinants in promoters of quorum-sensing-controlled genes qsc102, qsc117 (acpP), and qsc131 (phzA to -G) by in silico, deletion, point-mutational, and primer extension analyses. Each of these genes (in addition to lasI and rsaL) is activated by LasR, and qsc117 and qsc131 also respond to RhlR. Point mutations in the promoters of the LasR-specific gene, qsc102, relax specificity so that this promoter can respond to RhlR in addition to LasR. Our findings indicate that quorum-sensing-controlled promoters in P. aeruginosa are either specific for LasR or respond to both LasR and RhlR and that critical bases in the promoter elements determine specificity.
The opportunistic human pathogen Pseudomonas aeruginosa possesses two quorum-sensing systems: the LasR-LasI system and the RhlR-RhlI system. These systems are global regulatory elements that control the expression of approximately 50 to 200 genes (26). LasI catalyzes the synthesis of N-(3-oxododecanoyl)-l-homoserine lactone (3OC12-HSL) (16), and LasR is a 3OC12-HSL-dependent transcriptional activator (11). RhlI catalyzes the synthesis of the signal N-butyryl-l-homoserine lactone (C4-HSL) (17), and RhlR is a C4-HSL-dependent transcriptional activator (3). These two quorum-sensing systems are interrelated in that LasR activates the expression of the rhlR and rhlI genes (13, 19, 26).
RhlR and LasR are members of a family of transcription factors that influence expression of genes with specific palindromic sequences in their promoter regions. These sequences have been called lux-box-like sequences. The lux box is a 20-bp inverted repeat centered at −41.5 bp from the start of the Vibrio fischeri lux operon (6). Inverted repeats similar to the lux box have been identified in the promoter regions or putative promoter regions of a number of quorum-sensing-controlled (qsc) genes of P. aeruginosa. lux-box-like elements have been shown to be involved in P. aeruginosa quorum sensing in only two cases. One case is lasB, which is controlled primarily by RhlR but which is also influenced by LasR (15, 26). Mutational analyses have demonstrated that there are two lux-box-like elements involved in LasR control of lasB (1, 22). The other case is the promoter of the hydrogen cyanide (hcn) operon (20). Both LasR and RhlR can exert control over this operon (20, 26). There are genes that appear to be controlled solely by 3OC12-HSL–LasR but not C4-HSL–RhlR, and there are genes that appear to be controlled primarily by C4-HSL, but these genes also respond to 3OC12-HSL (5, 24, 26). We do not understand the promoter specificity determinants for 3OC12-HSL–LasR versus those for C4-HSL–RhlR.
Some P. aeruginosa qsc genes are regulated by additional factors. Although they require acyl-HSL signals for activation, they are repressed until stationary phase even in the presence of acyl-HSLs (26). These are termed late qsc genes. Other genes, early qsc genes, show activation upon addition of acyl-HSL signals even in early logarithmic phase. We have analyzed the promoters of several P. aeruginosa early qsc genes. These promoters all possess lux-box-like elements that we show are involved in qsc transcription. Furthermore, we show that two bases in lux-box-like elements define LasR-specific promoters.
MATERIALS AND METHODS
Bacterial strains, plasmids, growth conditions, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. P. aeruginosa and Escherichia coli were grown at 37°C in Luria-Bertani (LB) broth or LB agar (23) unless otherwise indicated. We used E. coli DH5α for cloning and plasmid propagation, and E. coli JM109 was used as a heterologous host for P. aeruginosa gene expression studies. For plasmid selection and maintenance, antibiotics were added to growth media at the following concentrations: ampicillin, 100 μg/ml; carbenicillin, 300 μg/ml; chloramphenicol, 30 μg/ml; HgCl2, 15 μg/ml; tetracycline, 20 μg/ml for E. coli and 50 μg/ml for P. aeruginosa. Acyl-HSLs were added at concentrations of 10 μM for C4-HSL and 2 μM for 3OC12-HSL for P. aeruginosa and 100 nM 3OC12-HSL for E. coli.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| P. aeruginosa | ||
| PAO1 | Wild type | 12 |
| PAO-R1 | ΔlasR derivative of PAO1; Tcr | 10 |
| PDO111 | ΔrhlR derivative of PAO1; Hgr | 3 |
| PAO-JP3 | ΔlasR ΔrhlR derivative of PAO1; Hgr Tcr | 18 |
| PAO-MW1 | ΔlasI ΔrhII derivative of PAO1; Hgr Tcr | 26 |
| E. coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17 deoR gyrA96 thi-1 relA1 supE44 | 23 |
| JM109 | F− (traD36 proAB+ laclqlacZΔM15) endA1 recA1 hsdR17 (rK− mK+) supE44 thi-1 gyrA96 relA1 Δ(lac-proAB) | 28 |
| Plasmids | ||
| pQF50 | lacZ transcriptional fusion vector; Apr | 7 |
| pACYC184 | General cloning vector; Cmr Tcr | 4 |
| pECP11 | pACYC184 carrying ptac-rhlR; Cmr | E. C. Pesci |
| pPCS11 | pACYC184 carrying ptac-lasR; Cmr | 22 |
| pMW302D | pQF50 carrying qsc117 with partial deletion of the NNCT-(N)12-AGNN; Apr | This study |
| pMW302E | pQF50 carrying wild-type qsc117 promoter from −54 to +88 relative to the primary transcriptional start site; Apr | This study |
| pMW302F | pMW302E with A-to-C change at NNCT-(N)12-AGNN position 8; Apr | This study |
| pMW302G | pMW302E with T-to-G change at NNCT-(N)12-AGNN position 13; Apr | This study |
| pMW302H | pMW302E with A-to-C change at NNCT-(N)12-AGNN position 8 and T-to-G change at position 13; Apr | This study |
| pMW302I | pMW302E with C-to-T change at NNCT-(N)12-AGNN position 3; Apr | This study |
| pMW308C | pQF50 carrying wild-type qsc102 promoter from −58 to +196 relative to the transcriptional start; Apr | This study |
| pMW308D | pQF50 containing wild-type qsc102 promoter with partially deleted NNCT-(N)12-AGNN; Apr | This study |
| pMW308E | pMW308C with C-to-A mutation at position 8; Apr | This study |
| pMW308F | pMW308C with G-to-T mutation NNCT-(N)12-AGNN at position 13; Apr | This study |
| pMW308G | pMW308C with C-to-A mutation at NNCT-(N)12-AGNN position 8 and G-to-T mutation at position 13; Apr | This study |
| pMW308I | pMW308C with C-to-T mutation at NNCT-(N)12-AGNN position 3; Apr | This study |
| pMW303C | pQF50 carrying qsc131 promoter from −59 to +326 relative to the transcriptional start site; Apr | This study |
| pMW303D | pQF50 carrying qsc131 promoter with partially deleted NNCT-(N)12-AGNN; Apr | This study |
| pMW303F | pMW303C with C-to-T mutation at NNCT-(N)12-AGNN position 3; Apr | This study |
| pMW312 | pQF50 carrying rsaL promoter from −82 to +29 relative to the translational start of rsaL; Apr | This study |
| pMW312B | pMW312 with a C-to-T mutation at NNCT-(N)12-AGNN position 3; Apr | This study |
| pMW313 | pQF50 carrying the lasI promoter from −57 to +55 relative to the transcriptional start site; Apr | This study |
| pMW313B | pMW313 with a C-to-T mutation at NNCT-(N)12-AGNN position 3; Apr | This study |
DNA manipulations.
Standard methods were used to manipulate plasmids and DNA fragments (2). Restriction endonucleases and DNA modification enzymes were purchased from New England Biolabs (Beverly, Mass.). Plasmids were isolated using spin miniprep kits (Qiagen, Chatsworth, Calif.), and DNA fragments were purified using Qiaquick PCR purification kits (Qiagen). Chromosomal DNA from P. aeruginosa was prepared using a DNeasy tissue kit (Qiagen). DNA fragments were excised and purified from agarose gels using GeneClean spin kits (Bio 101, La Jolla, Calif.), and PCR was performed with an Expand Long Template PCR system (Roche, Indianapolis, Ind.). DNA was sequenced at the University of Iowa DNA core facility by standard automated-sequencing technology.
Primer extension analysis.
Primer extension analysis of the genes qsc102, qsc117, and qsc131 was according to standard methods (2). RNA was prepared from P. aeruginosa PAO1 and PAO-MW1 using the Trizol reagent (Life Technologies, Grand Island, N.Y.). RNA was extracted from cultures at an optical density at 600 nm (OD600) of 2.0. The initial culture OD600 was 0.1. The extension primers for qsc102, qsc117, and qsc131 were 5′-GTCAGGCGTGGATAGCTTGTC-3′, 5′-TCGCATTCCTCCACGCCGAAC-3′, and 5′-GTTAAGGTGCGACAGACGAGG-3′, respectively. Each primer was 5′ end labeled using [γ32-P]dATP and a KinaseMax kit (Ambion, Austin, Tex.). 32P-labeled primers were annealed to 5 to 10 μg of P. aeruginosa RNA and extended using a First-Strand cDNA synthesis kit (Amersham, Piscataway, N.J.). DNA sequences were obtained using plasmid templates, and the oligonucleotides were used for the primer extension. Sequencing was with α-35S-dATP and a Sequenase, version 2.0, DNA sequencing kit (U.S. Biochemicals, Cleveland, Ohio). DNAs were resolved on 8 M urea–8% polyacrylamide gels.
Plasmid constructions.
The parent plasmid for all lacZ fusions was pQF50. Promoter fragments were generated from P. aeruginosa genomic DNA or plasmid templates by using PCR. Specific nucleotide changes within promoter fragments were incorporated into the oligonucleotide primers used in PCR amplification. For cloning, promoter fragments were end polished with T4 polymerase and 5′ phosphorylated with T4 polynucleotide kinase. The resulting fragments were blunt end cloned into SmaI-digested, phosphatase-treated pQF50, and correct orientations were identified by PCR analysis. After cloning, PCR-generated promoter fragments were verified by DNA sequencing.
Monitoring promoter activity in P. aeruginosa and E. coli.
Transformation of P. aeruginosa was as described previously (27). For lacZ expression studies, mid-logarithmic-phase cultures (OD600 of 0.2 to 0.5) were diluted 1:1,000 in LB broth and allowed to grow for 17 h at 37°C, at which point β-galactosidase activity was measured as described by Miller (14). With the acyl-HSL signal generation mutant P. aeruginosa PAO-MW1, cultures were grown in the presence and absence of 3OC12-HSL and C4-HSL. The starting OD600 was 0.1, and β-galactosidase activity was measured after 7 h at 37°C.
To monitor expression of qsc promoters in a heterologous host, we used a two-plasmid system in E. coli JM109. Overnight cultures grown in supplemented A medium (23) were diluted to an OD600 of 0.1 in fresh A medium. Cultures were grown to an OD600 of 0.2. At this culture density, 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added, and bacteria were grown in the presence and absence of 3OC12-HSL or C4-HSL. β-Galactosidase activities were assayed using a luminescent microtiter dish assay (26).
RESULTS
Primer extension analysis of three qsc genes.
To begin our study of qsc promoter elements, we determined the transcript start sites for three early genes by primer extension. These genes were qsc102, which responds only to 3OC12-HSL, and qsc117 and phzA, which respond to either 3OC12-HSL or C4-HSL but which require both for full activation in P. aeruginosa (26). Primer extension products were obtained with RNA isolated from the parent strain, PAO1 (Fig. 1), and with acyl-HSL signal generation mutant strain PAO-MW1 grown in the presence of 3OC12-HSL and C4-HSL, but not with PAO-MW1 grown without autoinducers (data not shown).
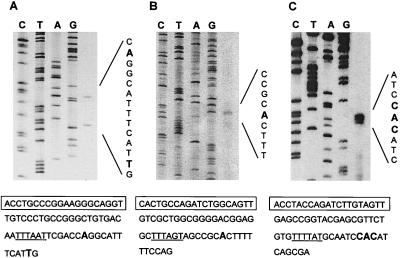
FIG. 1.
Primer extension analysis of three qsc transcripts. (Top) Polyacrylamide gels of qsc102 (A), qsc117 (B), and phzA (C). Sequencing ladders are shown on the left, and primer extension products from P. aeruginosa PAO1 RNA are on the right. (Bottom) DNA sequences of the promoter regions. Transcription start sites are in boldface, putative −10 regions are underlined, and lux-box-like elements are boxed.
For qsc102, two primer extension products were evident (Fig. 1A). The start of the longest product mapped to a position 211 bp from the predicted translation start site of qsc102. The other product was 11 bp shorter. The sequence at −10 from the start of the longer of the two transcripts was characteristic of a −10 hexamer in a promoter (TTTAAT). A lux-box-like element is centered at −44.5 from the start of the long transcript.
For qsc117 there was a single primer extension product (Fig. 1B). The start of this transcript mapped to a position 39 bp from the predicted translational start site. A −10-like hexamer (TTTAGT) is upstream of the start of the qsc117 transcript, and a lux-box-like sequence is centered at −40.5.
There were three transcripts evident in the analysis of phzA (Fig. 1C). These started at 334, 335, and 336 bp from the predicted translation start site. There is a −10-like hexamer (TTTTAT) positioned appropriately upstream of the transcription start site suggested by the primer extension analysis, and there is a lux-box-like sequence centered at −44 to −46 with respect to the transcript start sites.
The lux-box-like sequences positioned 40 to 60 bp upstream of each transcript start showed considerable dyad symmetry (16 to 18 bp of 20 bp) and considerable sequence variation. The three 20-bp sequences matched the minimal consensus sequence (NNCT-[N]12-AGNN) for a P. aeruginosa qsc regulatory element (26), with the proximal and distal nucleotide pairs possessing dyad symmetry. To define a minimal promoter sequence and to test the hypothesis that the lux-box-like sequences were cis-acting qsc gene regulatory elements, we carried out deletion and point mutation analyses.
Definition of promoter elements by analysis of deletion and point mutations.
We constructed plasmids containing P. aeruginosa DNA extending from 4 bp upstream of the lux-box-like elements of qsc102, qsc117, and phzA (pMW308C, pMW302E, and pMW303C). These plasmids contain lacZ transcription fusions in the qsc gene coding regions. We assessed promoter strength by measuring β-galactosidase activity in P. aeruginosa with different quorum-sensing mutations. For all of these reporters, promoter strength in a P. aeruginosa lasR rhlR double mutant (JP3) was <10% of the promoter strength in the parental strain, PAO1 (Table 2). This established that the plasmids contain the P. aeruginosa promoter DNA required for quorum-sensing-dependent transcription activation.
TABLE 2.
Activities of the qsc102, qsc117, phzA, and qsc102 8A-13T promoters in P. aeruginosa quorum-sensing mutants
| Strain (relevant genotype or acyl-HSL[s] added) | β-Galactosidase activitya ± SEM (% parent) of:
|
|||
|---|---|---|---|---|
| qsc102 | qsc117 | phzA | qsc102 8A-13T | |
| PAO1 (wild type) | 94 ± 8 | 1,479 ± 59 | 1,210 ± 50 | 862 ± 82 |
| PAO-R1 (ΔlasR) | 9 ± 3 (10) | 54 ± 10 (4) | 12 ± 3 (1) | 19 ± 4 (2) |
| PDO111 (ΔrhlR) | 117 ± 3 (124) | 163 ± 16 (11) | 61 ± 6 (5) | 164 ± 15 (19) |
| PAO-JP3 (ΔlasR ΔrhlR) | 7 ± 1 (7) | 63 ± 9 (4) | <1 (<1) | 19 ± 8 (2) |
| PAO-MW1 (Δlas1 Δrhl1) | 4 ± 1 | 63 ± 14 | 15 ± 2 | 5 ± 1 |
| PAO-MW1 (3OC12-HSL) | 81 ± 3 | 329 ± 15 | 278 ± 13 | 146 ± 17 |
| PAO-MW1 (3OC12-HSL + C4-HSL) | 68 ± 2 | 1,360 ± 23 | 632 ± 14 | 592 ± 37 |
Expressed as Miller units.
Transcription from the qsc102 promoter was low in a lasR mutant (Table 2) but high in an rhlR mutant. Addition of 3OC12-HSL to strain PAO-MW1, a signal generation mutant, resulted in the induction of the qsc102-lacZ fusion. Addition of both 3OC12-HSL and C4-HSL did not result in further induction (Table 2). This is consistent with our previous finding that qsc102 transcription is regulated by 3OC12-HSL and not by C4-HSL (26).
For qsc117 and phzA, promoter activity was reduced by mutations in lasR or rhlR or both (Table 2). Although reduced compared to the activity in the parent, the activity in the rhlR mutant was greater than that in the lasR mutant. One explanation for this is that rhlR transcription depends on LasR but that lasR transcription does not depend on RhlR (19). Addition of 3OC12-HSL to signal generation mutant strain PAO-MW1 resulted in a small induction of the qsc117-lacZ fusion. Addition of both 3OC12-HSL and C4-HSL resulted in a much larger induction (Table 2). The signal addition results are consistent with previous studies of qsc-lacZ chromosomal fusions (26). One explanation for the results with the lasR and rhlR mutants is that the qsc117 and phzA promoters can interact with either LasR or RhlR.
Are the lux-box-like elements in the vicinity of −40 to −60 from the starts of the qsc102, qsc117, and phzA promoters required for quorum-sensing-dependent gene activation? To address this question, we generated site-specific point mutations in the lux-box-like elements. Mutations in conserved nucleotides of minimal sequence NNCT-(N)12-AGNN have been shown to be critical for quorum-sensing control of lasB (1, 22). Thus we changed the conserved C to T in these promoters and measured quorum-sensing-activated transcription. Replacement of the C with a T resulted in a significant decrease in signal activation for all three promoters, and deletion of portions of the lux-box-like elements resulted in a severe decrease in signal-activated transcription (Fig. 2). These data support the hypothesis that the lux-box-like sequences of qsc102, qsc117, and phzA are cis-acting elements required for quorum-sensing control of transcription.
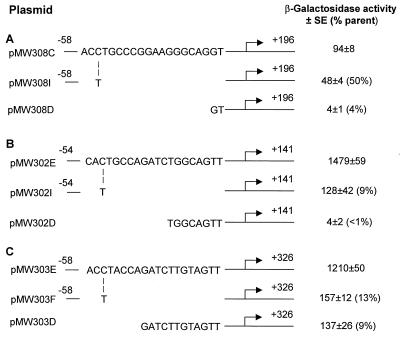
FIG. 2.
Effects of deletions and point mutations on the expression of qsc102 (A), qsc117 (B), and phzA (C) in P. aeruginosa PAO1. The starting bases with respect to the transcript start site are shown at the left (for phzA the number is for the longest transcript). The sites of lacZ insertion are given on the right, also with respect to the transcription start sites. β-Galactosidase activities are in Miller units (means ± standard errors of the means [SE]). Numbers in parentheses are percentages of wild-type promoter activity (pMW308C, pMW302E, and pMW303E in PAO1). In lasI rhlI signal synthesis mutant P. aeruginosa PAO-MW1, the activities of the wild-type qsc promoters (Table 2) were indistinguishable from the activities of the promoters with point mutations or deletions and were <10% of the activities in wild-type P. aeruginosa PAO1. Thus the differences in promoter strengths shown are not a reflection of changes in the basal transcription levels as monitored in the signal synthesis mutant.
LasR specificity determinants in P. aeruginosa qsc promoters.
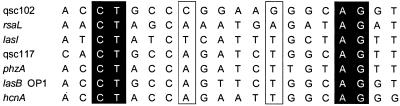
Palindromic lux-box-like sequences can be found in the putative promoter regions of a number of early qsc genes (26). The available evidence is consistent with the hypothesis that some of these genes, the class I genes (qsc102, for example), respond to LasR specifically and that others, the class III genes (qsc117 for example), require a functional LasR and a functional RhlR for full induction. An alignment shows that the class III genes qsc117 and phzA have an A at position 8 and a complementary T at position 13 of the lux-box-like sequence and that the class I gene qsc102 does not have the 8A-13T lux-box-like sequence motif (Fig. 3). The palindromic sequences of other class III promoters for which transcript start site information exists also show the 8A-13T motif (1, 20, 22).
FIG. 3.
An alignment of the NNCT-(N)12-AGNN elements of qsc102, rsaL, lasI, qsc117, phzA, lasB, and hcnA. Nucleotides in black background represent bases present in all qsc elements, and those boxed are putative specificity determinants.
To test the hypothesis that the bases at positions 8 and 13 are specificity determinants for class I promoters, we used site-directed mutagenesis to construct reporter plasmids. We constructed a plasmid with base 8 of the qsc102 lux-box-like element converted from a C to an A, a plasmid with the base at position 13 converted from a G to a T, and a plasmid with an A at position 8 and a T at position 13. Neither of the promoters with single base substitutions showed an altered specificity. Induction depended solely on LasR (data not shown). The promoter with both the 8A and 13T mutations (called qsc102 8A-13T) responded in a fashion similar to class III promoters such as the qsc117 promoter (Table 2). The promoter was stronger than the parent qsc102 promoter, and activity was reduced not only in a lasR mutant but also in an rhlR mutant.
We monitored qsc102, qsc117, and qsc102 8A-13T promoter activity in E. coli to test two hypotheses. The first hypothesis is that quorum sensing controls qsc102, qsc117, and qsc102 8A-13T directly, and the second hypothesis is that qsc102 responds to LasR but not RhlR, whereas qsc117 and qsc102 8A-13T are less specific and can respond to either LasR or RhlR. We monitored promoter activity in E. coli containing a ptac-lasR plasmid (pPCS11) or a ptac-rhlR plasmid (pECP11) plus the qsc102-lacZ reporter plasmid. The qsc102 promoter was activated by LasR and 3OC12-HSL but was not activated by RhlR and C4-HSL (Table 3). The qsc117 and qsc102 8A-13T promoters were activated by either LasR and 3OC12-HSL or RhlR and C4-HSL (Table 3). The LasR- or RhlR-dependent activation in recombinant E. coli supports the conclusion that activation is direct. The lack of qsc102 activation by RhlR supports the conclusion that the qsc102 promoter responds to LasR but not RhlR. The activation of qsc117 and qsc102 8A-13T by both LasR and RhlR further substantiates that these promoters are less specific than qsc102 and that the 8C→A and 13G→T mutations in the qsc102 promoter converted it from a class I (LasR-controlled) to a class III (LasR- and RhlR-controlled) qsc gene.
TABLE 3.
Activation of qsc102, qsc117, and qsc102 8A-13T promoter-lacZ fusions by the las and rhl systems in E. coli
| Activatorb | β-Galactosidase activitya ± SEM of:
|
||
|---|---|---|---|
| qsc102 | qsc117 | qsc102 8A-13T | |
| LasR | 4 ± 1 | 35 ± 5 | 12 ± 3 |
| LasR + 3OC12-HSL | 140 ± 5 | 107 ± 7 | 128 ± 2 |
| Vector + 3OC12-HSL | 2 ± 1 | 15 ± 2 | 20 ± 2 |
| RhlR | 3 ± 1 | 30 ± 4 | 9 ± 1 |
| RhlR + C4-HSL | 4 ± 1 | 132 ± 5 | 97 ± 2 |
| Vector + C4-HSL | 2 ± 2 | 16 ± 5 | 21 ± 2 |
Expressed as relative light units/OD660.
The control vector was pACYC184, and lasR and rhlR were supplied on pPCS11 and pECP11, respectively.
We were unable to convert the class III qsc117 promoter into a class I promoter. Changes in the lux-box-like element base 8 from an A to a C or base 13 from a T to a G resulted in very low promoter activity. In fact, when we modified the qsc117 promoter in pMW302E by site-specific mutagenesis so that the 20-bp inverted repeat matched that of qsc102, the promoter was inactive (data not shown).
Predictive value of the minimal consensus sequences for lux-box-like elements in P. aeruginosa early qsc genes.
Previous studies have established that lasI, which codes for the 3OC12-HSL synthase, is positively autoregulated by 3OC12-HSL and LasR and is a class I gene (24). The lasI gene is adjacent to and divergently transcribed from rsaL, another qsc gene (5). Prior to our ability to define a minimal consensus for the lux-box-like element of P. aeruginosa qsc promoters, NNCT-(N)12-AGNN, with 16 or more bases showing dyad symmetry, LasR-binding elements were predicted for lasI and rsaL, but they did not match the consensus. We hypothesized that there should be at least one NNCT-(N)12-AGNN motif in the lasI-rsaL intergenic region, and a search of the region revealed such a sequence (Fig. 3). This element is centered at −40.5 from the lasI transcription start site (24). The sequence does not show the 8A-13T motif characteristic of promoters that respond to both LasR and RhlR. Thus we predict that the promoter is LasR specific.
The transcription start site of rsaL has not been mapped, but the lux-box-like element is nearly equidistant from the rsaL and lasI open reading frames (56 and 55 bp, respectively). Thus it is quite conceivable that the lux-box-like element that we have identified serves in the bidirectional quorum activation of both lasI and rsaL. To test our hypothesis that the predicted lux-box-like sequence serves in quorum activation of both lasI and rsaL, we constructed lacZ reporter plasmids. One plasmid, pMW312, contains a rsaL-lacZ fusion and begins 63 bp upstream of the rsaL coding region. The other plasmid, pMW313, contains a lasI-lacZ fusion and begins 62 bp upstream of the lasI coding region. The lacZ gene was induced by addition of 3OC12-HSL, but not C4-HSL, to the lasI rhlI signal generation mutant PAO-MW1, containing either the lasI reporter (4-fold) or the rsaL reporter (25-fold) (Table 4). Thus the minimal promoter elements present on pMW312 and pMW313 are sufficient for activation by quorum-sensing signal 3OC12-HSL. The experiment is also a confirmation that rsaL and lasI do not respond to C4-HSL–RhlR.
TABLE 4.
Activation of lasI and rsaL in a P. aeruginosa lasI rhlI double mutant by addition of acyl-HSL
| Fusion (plasmid) | β-Galactosidase activity (Miller units)a ± SEM with:
|
||
|---|---|---|---|
| No addition | 3OC12-HSL | 3OC12-HSL + C4-HSL | |
| lasI-lacZ (pMW313) | 5 ± 1 | 19 ± 1 (4-fold) | 18 ± 1 (4-fold) |
| rsaL-lacZ (pMW312) | 231 ± 31 | 5,668 ± 165 (25-fold) | 5,755 ± 546 (25-fold) |
β-Galactosidase activities of the lasI-lacZ and rsaL-lacZ fusions were 17 and 5,272 miller units, respectively, in wild-type P. aeruginosa PAO1. Values in parentheses are fold increases over activity with no addition.
To test the hypothesis that the lux-box-like sequence in the lasI-rsaL intergenic region is involved in the activation of lasI and rsaL, we generated a point mutation in the third base of the element with respect to lasI (pMW313B) and a point mutation in the third base with respect to rsaL (pMW312B). The point mutations severely decreased 3OC12-HSL–LasR-dependent expression of the rsaL-lacZ and lasI-lacZ fusions to 4 and 30% of parental levels, respectively. Thus we believe that there is now sufficient information to make predictions about promoter elements of early qsc genes and whether these genes will respond to LasR specifically or to either RhlR or LasR.
DISCUSSION
qsc genes have been grouped depending on whether they are fully activated by 3OC12-HSL, whether they require both 3OC12-HSL and C4-HSL for full activation, and whether the acyl-HSL signals can activate them in logarithmic phase (early genes) or stationary phase (late genes) (26). To begin to understand promoter elements of qsc genes, we have examined several early genes. We studied highly activated 3OC12-HSL-dependent early gene qsc102, and we studied two highly activated genes that require both signals for full induction, qsc117 and phzA. The transcript start sites were mapped by primer extension (Fig. 1), and we identified palindromic lux-box-like sequences in the −40 to −50 regions with respect to the start sites of the promoters (Fig. 3). Our genetic dissections of the promoter regions of these genes indicate that DNA including the palindromic region and a few base pairs upstream is sufficient for quorum signal activation. They also indicate that the specific sequences of the palindromic regions are important for quorum control (Fig. 2; Tables 1 and 2). We refer to the quorum control element as the NNCT-(N)12-AGNN element to signify the minimal conserved unit.
The positions of the NNCT-(N)12-AGNN elements, centered in the −40 regions, suggest that they serve as activator binding sites and that the activators are the ambidextrous type, functioning at these promoters by making contact with the RNA polymerase α C-terminal domain and with some other part of RNA polymerase (6, 8, 21, 25). We cannot conclude that all P. aeruginosa qsc genes contain NNCT-(N)12-AGNN elements in the −40 region. We suspect that some of the qsc genes showing small signal responses (for example, qsc104, which shows about 8-fold induction compared to qsc102, which shows about 400-fold induction) (26) may have different promoter arrangements.
Our promoter analysis revealed that the qsc117 and phzA NNCT-(N)12-AGNN elements showed an additional similarity, an A at position 8 and a T at position 13. These genes require both acyl-HSL signals for full induction, and we have shown that they can respond to either of signal receptors LasR and RhlR (Tables 2 and 3). The qsc102 gene responds only to LasR and does not have the 8A-13T motif (Tables 2 and 3; Fig. 3). When we placed an 8A-13T motif in the qsc102 promoter, its behavior was altered such that it responded to either LasR or RhlR. Activity was similar to the activity of the phzA and qsc117 promoters (Tables 2 and 3). The 8A-13T element appears to relax the specificity of the qsc102 promoter so that it will respond to either LasR or RhlR. Our attempt to convert qsc117 into a LasR-specific promoter failed. Even replacement of the NNCT-(N)12-AGNN element of qsc117 with that of qsc102 did not yield a LasR-specific promoter. Rather it resulted in the inactivation of the promoter. Thus it is clear that there are elements that remain to be defined in P. aeruginosa qsc promoters.
The analysis of qsc102, qsc117, and phzA afforded us an ability to learn about other promoters. We tested the predictive power of our analysis by examining the rsaL-lasI intergenic region. These two genes are early qsc genes activated solely by 3OC12-HSL (5, 24). Previous examinations of the intergenic region between rsaL and lasI were prior to the definition of the NNCT-(N)12-AGNN minimal consensus. The previous studies suggested that another region might represent the LasR-binding region (5). There is a sequence matching the NNCT-(N)12-AGNN consensus, and, as expected of a LasR-specific promoter, it does not possess an 8A-13T element (Fig. 3). Our genetic analysis of the promoter region suggests that the NNCT-(N)12-AGNN motif is in fact an element required for bidirectional qsc control of rsaL and lasI by LasR. This is very similar to the bidirectional quorum-sensing control of the divergently transcribed traA and traC genes in Agrobacterium tumefaciens (9).
Our current view is that early qsc genes can respond to LasR. Some of these genes, for example, qsc102, are LasR specific, and, if there is a response to RhlR, we cannot detect it. Other genes, for example, qsc117 and phzA, respond to either RhlR or LasR. We believe these less-specific genes respond primarily to RhlR in P. aeruginosa. LasR is required for induction of the RhlR system (19, 26). Thus both systems are required for full induction of genes such as qsc117 and phzA, but, because of the loose specificity, there is some response to either LasR or RhlR alone. We believe that there may be RhlR-specific genes, for example, qsc132, which unlike qsc117 and phzA show no detectable response to 3OC12-HSL alone (26). The qsc132 gene is a late gene, and a detailed analysis of its promoter has not been carried out.
ACKNOWLEDGMENTS
This research was supported by a grant from the National Institutes of Health (GM59026). M.W. was supported by a National Science Foundation Research Training grant (DBI9602247) and by a United States Public Health Service Training grant (732 GM8365).
REFERENCES
- 1.Anderson R M, Zimprich C A, Rust L. A second operator is involved in Pseudomonas aeruginosa elastase (lasB) activation. J Bacteriol. 1999;181:6264–6270. doi: 10.1128/jb.181.20.6264-6270.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley & Sons, Inc.; 1997. [Google Scholar]
- 3.Brint J M, Ohman D E. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang A C, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Kievit T, Seed P C, Nezezon J, Passador L, Iglewski B H. RsaL, a novel repressor of virulence gene expression in Pseudomonas aeruginosa. J Bacteriol. 1999;181:2175–2184. doi: 10.1128/jb.181.7.2175-2184.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egland K A, Greenberg E P. Quorum sensing in Vibrio fischeri: elements of the luxI promoter. Mol Microbiol. 1999;31:1197–1204. doi: 10.1046/j.1365-2958.1999.01261.x. [DOI] [PubMed] [Google Scholar]
- 7.Farinha M A, Kropinski A M. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J Bacteriol. 1990;172:3496–3499. doi: 10.1128/jb.172.6.3496-3499.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuqua W C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 9.Fuqua W C, Winans S C. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J Bacteriol. 1996;178:435–440. doi: 10.1128/jb.178.2.435-440.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gambello M J, Iglewski B H. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gambello M J, Kaye S, Iglewski B H. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect Immun. 1993;61:1180–1184. doi: 10.1128/iai.61.4.1180-1184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holloway B W, Krishnapillai V, Morgan A F. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhlR to expression of the stationary-phase sigma factor RpoS. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 14.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 15.Ochsner U A, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearson J P, Gray K M, Passador L, Tucker K D, Eberhard A, Iglewski B H, Greenberg E P. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearson J P, Passador L, Iglewski B H, Greenberg E P. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pearson J P, Pesci E C, Iglewski B H. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pesci E C, Pearson J P, Seed P C, Iglewski B H. Regulation of las and rhl quorum-sensing systems in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pessi G, Haas D. Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J Bacteriol. 2000;182:6940–6949. doi: 10.1128/jb.182.24.6940-6949.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhodius V A, Busby S J. Positive activation of gene expression. Curr Opin Microbiol. 1998;1:152–159. doi: 10.1016/s1369-5274(98)80005-2. [DOI] [PubMed] [Google Scholar]
- 22.Rust L, Pesci E C, Iglewski B H. Analysis of the Pseudomonas aeruginosa elastase (lasB) regulatory region. J Bacteriol. 1996;178:1134–1140. doi: 10.1128/jb.178.4.1134-1140.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Seed P C, Passador L, Iglewski B H. Activation of the Pseudomonas aeruginosa lasI gene by LasR and the Pseudomonas autoinducer PAI: an autoinduction regulatory hierarchy. J Bacteriol. 1995;177:654–659. doi: 10.1128/jb.177.3.654-659.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevens A M, Fujita N, Ishihama A, Greenberg E P. Involvement of the RNA polymerase alpha-subunit C-terminal domain in LuxR-dependent activation of the Vibrio fischeri luminescence genes. J Bacteriol. 1999;181:4704–4707. doi: 10.1128/jb.181.15.4704-4707.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whiteley M, Lee K M, Greenberg E P. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whiteley M, Parsek M R, Greenberg E P. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J Bacteriol. 2000;182:4356–4360. doi: 10.1128/jb.182.15.4356-4360.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]