Abstract
The World Health Organization (WHO) estimates that the prevalence of human papillomaviruses (HPV) infection is between 9% and 13% of the world population and only in the United States, more than 6.2 million are positive every year. There are more than 100 types of HPV, among them, two serotypes (16 and 18) are related to 70% of cervical cancers and precancerous cervical lesions. The vaginal microbiota could play a considerable role in HPV infection and the genesis of cervical tumors caused by HPV. Moreover, bacteria are strongly associated with vaginal inflammation and oncogenic mutations in human cells. We aim to investigate whether HPV infection could influence the bacterial microbiota composition in the uterine cervix. A total of 31 women were enrolled in this study. The vaginal swabs were collected; the HPV‐DNA was extracted with QIAamp DNA Microbiome. The V3–V4–V6 region of the 16S rDNA gene was amplified by polymerase chain reaction (PCR) followed by sequencing with MiSeq Illumina. The main phylum identified in the vaginal microbiota were Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria. The phylum of Actinobacteria, Proteobacteria, and Bacteroides was more represented in HPV‐positive patients. Lactobacilli represented the dominant genus, with a high percentage of Lactobacilli iners, Lactobacilli jensenii, and Lactobacilli crispatus as species. Gardnerella vaginalis, Enterococcus spp., Staphylococcus spp., Proteus spp., and Atopobium were the most represented in HPV‐positive patients. An altered vaginal microbiota might play a functional role in HPV cervical infection, progression, and clearance. The relationship between infection and microbiota could spur the development of new probiotics. However, further studies are needed to clarify the role of the vaginal microbiota in HPV infection.
Keywords: bacteria, microbiota, papillomavirus infections, uterine cervical neoplasms
1. INTRODUCTION
Human papillomaviruses (HPV) are double‐stranded DNA (dsDNA) viruses belonging to the Papillomaviridae family. HPV infections are mainly sexually transmitted and are particularly frequent in the world population with a prevalence in women aged under 25 years (17%). 1 , 2 In the greatest cases, the infection generates by these viruses results asymptomatic, and the infection regresses autonomously in a short time. 3 Otherwise, the persistence of the infection increases the probability of the appearance of dysplastic lesions from low to high grade (cervical intra‐epithelial neoplasia [CIN] 1‐2‐3), up to the neoplastic transformation. However, the main genotypes of Papilloma Virus responsible for tumor forms are 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59. 4 , 5 The genotypes 16 and 18 are the most frequent and alone account for 70% of cases of cancers of the uterine cervix. 6 However, cases induced by phylogenetically related genotypes 16 and 18, such as 45, 33, and 35, have been increasing, probably also because of previous vaccinations against only high‐risk genotypes 16 and 18. 7 , 8 , 9
The absence of a therapy and the molecular characteristics of the infection maintain a critical interest in the related HPV pathology. Several studies have been focusing on the new risk factors and, also on analyzing the viral persistence with the consequent genomic integration in the cells of the uterine cervix and the triggering of neo‐oncogenic processes. 10 , 11 , 12 In this regard, today the scientific community pays much attention to the role that the microbiota can play in countering the infection. 13 , 14
It has been shown that many factors may increase the risk of genital HPV infection, such as the number of sexual partners, sexual behavior, and coinfection with other viruses sexually transmitted. 15 In the last few decades, thanks to the introduction of the next‐generation sequencing (NGS) it has been possible to perform the phylogenetic and taxonomic characterization of the microbial communities present in living organisms, with insights on the vaginal microbiota and its possible designation as a potential risk factor in HPV infection. 16
In several studies, a close correlation between health and microbiota has been demonstrated. By characterizing the vaginal microbiota, it is possible to define a state of eubiosis, indicative of good microbial health, and a state of dysbiosis, indicative of microbial imbalance. 17 The state of eubiosis is favored by the predominate of species belonging to the Lactobacillus spp., which reduced the richness and bacterial diversity. 18 The lactobacillus spp. through the production of lactic acid and hydrogen peroxide, they maintain a low pH, 19 which disadvantages the colonization of pathogenic species responsible for bacterial vaginosis, such as Chlamydia trachomatis, Neisseria gonorrhoeae, and Gardnerella vaginalis. 20 A healthy microbiota can prevent many urogenital diseases, such as urinary tract infections, 21 human immunodeficiency virus infection (HIV), 22 infections by Candida, 23 sexually transmitted diseases. 24 However, the lactate produced by lactobacilli increases the viscosity of cervical mucus that traps the viral particles and inhibits the access of papillomavirus to basal keratinocytes, assuming an essential role in the maintenance of the cervical epithelial barrier. In this respect, most studies have evaluated the link between the vaginal microbiota, HPV infection, and cervical disease, demonstrating that changes in the composition of the vaginal microbiota, with reduced mucus production and consequent reduction of viral capture, can be associated with the acquisition, reactivation or late clearance of cervical HPV infection 25 , 26 and progression of CIN. 27 , 28 Some studies have shown the association between greater vaginal microbiota diversity and the development of squamous intraepithelial lesions (SIL). 29 Dols et al. affirm that in women with HPV infection, the prevalence of Lactobacillus crispatus has been significantly reduced. 30 Oh et al. indicated that a cervical microbiota predominated by Atopobium vaginae, Gardnerella vaginalis, and Lactobacillus iners was associated with an almost sixfold increase in SIL risk. 25 These studies suggest a potential role of the vaginal microbiota in cervical infection and carcinogenesis. Since the vaginal microbiome plays an essential role in urogenital pathologies, we speculated that a different vaginal microbial environment could facilitate HPV infection. Therefore, the purpose of this study is to distinguish the microbial profile of women with HPV infection from that of HPV‐negative women and to evaluate the possible association of species present only in the case of an ongoing infection HPV (Figure 1).
Figure 1.
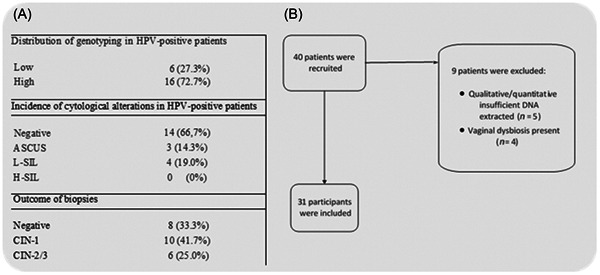
(A) Distribution table of genotyping, the incidence of cytological changes, and biopsy results in HPV‐positive patients. (B) Flow chart of the study. ASCUS, atypical squamous cells of undetermined significance; CIN, cervical intraepithelial neoplasia; HPV, human papillomaviruses; H‐SIL, high‐grade squamous intraepithelial lesions; L‐SIL, low‐grade squamous intraepithelial lesions.
2. MATERIALS AND METHODS
2.1. Study population
Between October 2019 and April 2020, we recruited women with a cytological diagnosis of HPV cervical lesions from the Department of Obstetrics and Gynecology of the University of Campania “Luigi Vanvitelli” in Naples (Italy). Institutional Review Board approved the study. All patients gave informed consent. All experiments were performed following the approved guidelines. This study was conducted following the principles of the Helsinki Declaration. We included premenopausal nonpregnant women between 25 and 50 years. The inclusion criteria were: (i) cytological diagnosis of LSIL or HSIL, (ii) no use of vaginal probiotics for 30 days before the start of this study, (iii) absence of genital tract infection, and (iv) no sexual intercourse 72 h before the qualification test. The control group was composed of women attending our Department and without HPV cervical infection. The exclusion criteria were: (i) vaginal bleeding, (ii) pregnancy, (iii) history of other types of cancer, (iv) diseases of the immune system, and (v) bacterial vaginosis.
2.2. Sample collection
Cervical cytology samples were collected by three gynecologists using the Abbott cervix‐collect specimen collection kit (Abbott) and transported in Prep® PreservCyt® Solution (HOLOGIC™). These specimens were stored at 15°C to 20°C and transported to the laboratory within 24 h of collection. Samples for vaginal microbiota analysis were collected from the lateral and posterior fornix using a nonlubricating disposable swab containing BBLTM CultureSwab liquid Amies (Swab code 490CE. AM‐COPAN, Becton Dickinson), immediately stored at 4°C and processed within 48 h of collection.
2.3. HPV genotyping
All enrolled women have been examined both with an HPV genotyping test and with the Thin Prep cytology test (TCT). The HPV detection was performed using the linear array (Roche Molecular Diagnostics), a qualitative in vitro test for the detection of HPV in clinical specimens. These tests utilize amplification of target DNA by polymerase chain reaction (PCR) and nucleic acid hybridization and detects 37 anogenital HPV‐DNA genotypes [6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 42, 45, 51–56, 58, 59, 61, 62, 64, 66–73 (MM9), 81, 82 (MM4), 83 (MM7), 84 (MM8), IS39, and CP6108] in cervical cells collected in PreservCyt solution. Three experienced cytology experts performed cervical liquid‐based cytology tests and formulated cytological reports according to the Bethesda System.
2.4. Total bacterial genomic DNA extraction
The vaginal swabs obtained were suspended in transport buffer by vertexing and transferred to a sterile DNase/RNase‐free 2.0 ml tube for enzymatic lysis. DNA isolation from the swabs. Total DNA was extracted from cells using a DNA isolation kit (QIAamp DNA microbiome kit, cat. no. 51704; Qiagen®), according to the manufacturer's protocols. The total genomic DNA quality was assessed by 1.8% agarose gel electrophoresis, and the DNA concentration was measured using a Qubit 2.0 Fluorometer (Life Technology) and normalized to 1 ng/µl.
2.5. Illumina MiSeq sequencing of 16S rRNA gene amplicons
The V3–V4–V6 hypervariable regions of the 16S rRNA gene were amplified using the Arrow Microbiota Solution B kit (cod. AD‐002.024). Amplification was performed in 20‐μl reactions with amp mix for “PCR target” and 5 µl of DNA extract. The reactions were performed using a SimpliAmp Thermal Cycler (Applied Biosystems®, #A24812) under the following thermal profile: 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, and one cycle of 72°C for 5 min and a 4°C hold. PCR products were examined by 1.6% agarose gel electrophoresis and then purified using AMPure Beads XP (cat. n. A63880, Beckman Coulter, Inc.). The amplicons' quality was assessed by 1.6% agarose gel electrophoresis, a product visible as a band with a molecular weight of approximately 840 bp. A 6‐bp barcode sequence was added to the ends of both the forward and reverse primers in the PCR “index.” PCR “index” products were examined and then purified as described above. Amplicons targets were normalized to 10 nM and used to prepare the library for sequencing. The library was processed using kit MiSeq Reagent Nano Kit v2 (500‐cycles) (cat. no: MS‐103‐1003, Illumina Inc.), following the user guidelines for the system MiSeq Illumina (“Denature ana Dilute Libraries Guide”‐MiSeq System). The library was corrected by adding 5% of Phix Control v3 (cat. n. FC‐110‐3001, Illumina Inc.) and sequencing using an Illumina MiSeq platform. The amplicons were pooled, and sequencing was conducted at Ames Center (Naples, Italy), using an Illumina MiSeq sequencing system (Illumina). The data obtained were processed with the MicrobAT (Microbiota Analysis Tool) system of the SmartSeq S.r.l. 16S rRNA gene sequence analysis. Sequence analysis sequences of low quality with a lower quality score of 20, a length less than 50 bp, or any misalignments to the primer or barcode containing chimeras have been removed.
2.6. Statistical analysis
Using the 16S rRNA gene sequencing data, the frequency of bacteria occurrences was calculated by multiplying the total number of present bacteria with the percentage of bacterial concentration in the cervical swabs. The statistical analysis was performed using k‑means cluster analysis (k‑means clustering algorithm) and Statistical 12.0 software (Stat Soft Inc.). Continuous variables were analyzed with the t test and categorical variables were analyzed with the χ 2 test. p < 0.05 were considered statistically significant.
3. RESULTS
We recruited 25 HPV‐infected women [HPV(+), n = 25] and 6 women without HPV infection [HPV(–), n = 6]. No difference in the average age of the population was determined (p = 0.807). The analysis of the data following the identification of the vaginal microbiota showed the presence of four dominants Phyla: (i) Firmicutes, (ii) Actinobacteria, (iii) Bacteroidetes, and (iv) Proteobacteria (Figure 2). The Firmicutes were the most abundant phylum, equal to 90.7% and 87.4% in the control group (HPV‐negative) and in the HPV‐infected group (HPV‐positive), respectively. The percentage of Bacteroidetes (0.2% vs. 0.6%), Proteobacteria (0.6% vs. 1.8%) (p = 0.03549), and Actinobacteria (0.3% vs. 0.8%) was increased in patients with HPV infection compared with the group control. The analysis of the classes of bacteria revealed the abundant presence of Gammaproteobacteria in HPV‐positive patients (0.071% vs. 0.951%). At the genus level, the Lactobacillus represented the dominant population in the two groups (99.2% vs. 84.5%) (Figure 3). In contrast, HPV infection had a positive effect on the abundance of Enterococcus, and Staphylococcus, with a relative percentage of 6.9% and 4.2%, respectively, and relative absence in controls. Furthermore, a significant gender difference was Gardnerella (1.7%) and Morganella (1%), significantly more abundant in the HPV‐positive group than in the HPV‐negative group with much lower percentages, 0.1%, and 0.01%, respectively. Further analysis carried out at the species level revealed, in the controls, the presence of a subpopulation of Lactobacillus crispatus, Lactobacillus iners, and Lactobacillus taiwanensis were more abundant, while the Gardnerella vaginalis, Morganella morganii, Lactobacillus acidophilus species had not been identified. However, in the HPV‐positive group, the predominant bacterial species were Lactobacillus acidophilus and Lactobacillus iners, with a greater abundance of Gardnerella vaginalis and Morganella morganii Finally, a relative abundance of Enterococcus faecalis and Staphylococcus haemolyticus was found in the HPV‐positive group, almost absent in the control group.
Figure 2.
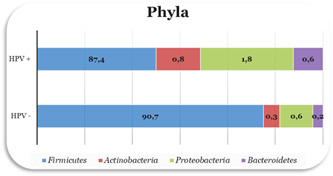
Analysis for bacterial phyla (%) of the vaginal microbiota in according to HPV status. HPV, human papillomaviruses.
Figure 3.
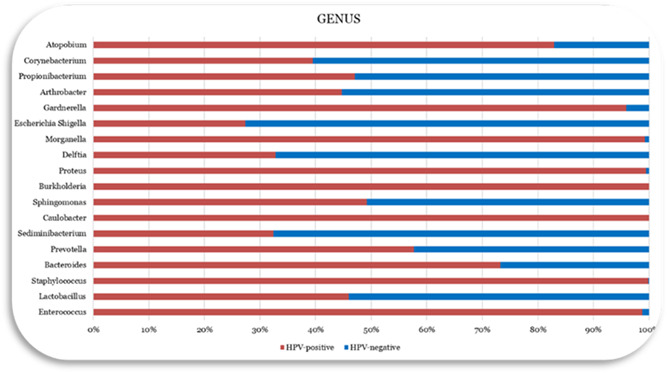
Bacterial genus (%) analysis of the vaginal microbiota.
4. DISCUSSION
Our study addressed an indeterminate topic about the association between vaginal microbiota and HPV cervical infection in women. We supposed that a vaginal dysbiosis might play a key role in HPV infection, progression, and clearance. Microbiome diversity in most sites of the human body act as a health condition. 31 However, the status of health for the vaginal microbiome is more related to low microbial heterogeneity and dominance by Lactobacillus spp. 32 , 33 Genetic factors could also influence mucosal immunity and the impact on microbiome diversity. Given the above, Caucasian, and Asian women exhibit a significantly greater prevalence of Lactobacillus spp., compared with Hispanic and Black women. 34 , 35 Female hormones have a critical impact on vaginal microbial diversity. Reduced estrogen in menopause alters the vaginal microbiome through mucosal atrophy and consequent Lactobacillus spp. depletion and increased diversity. 36 This genus produces enzymes capable of glycogen fermentation, thus producing large amounts of lactic acid. The consequent low pH exerts protection for the cellular metabolic function of the cervix and the vagina and negatively impacts the growth of potentially pathogenic species, such as Chlamydia trachomatis, Neisseria gonorrhoeae, and Gardnerella vaginalis. The persistence of the virus is essential for the development of cervical cancer and factors such as age, immunodeficiency, and smoking is associated with a reduced virus clearance. 37 Recent studies have underlined the emerging role of HPV acquisition and clearance and the development of cervical precancer lesions through increased diversity of vaginal microbiome combined with a reduced relative abundance of Lactobacillus spp. 16 Several bacteria have been related to vaginal health diseases, such as bacterial vaginosis, pelvic inflammatory disease, and sexually transmitted infections. 38 , 39 The vaginal health assay of our study presents the relative abundance of bacteria positively associated with bacterial vaginosis, such as Morganella or Gardnerella species, as well as those negatively associated with that condition such as Lactobacillus spp. Thanks to the introduction of NGS in research, the possibility has opened up to identify specific ecological niches for microorganisms within living organisms. It has been reported that highly conserved regions of the 16S rRNA gene (V1–V9) allow for the phylogenetic and taxonomic characterization of the microbial communities analyzed. 40 In this study, the hypervariable V4–V5–V6 regions of 16S rRNA were used, which allowed for the identification of microbial species present in the vaginal microbiota of women with HPV infection and healthy women without any vaginal infection. We observed that HPV infection presented an increased vaginal bacterial richness and diversity. Lactobacillus represented the most abundant genus in both groups. However, HPV infection influenced the abundance of some species. Overall, more anaerobic bacteria were associated with HPV infection, and the lower the percentage of Lactobacillus present. HPV infection has enriched some specific genera, including Bifidobacterium, Bacillus, Megasphaera, Prevotella, Gardnerella, and Dialister, identified as more abundant taxa in HPV‐infected populations than in the healthy population. Other studies confirm this finding with the association of these species with HPV infection. 41 , 42 , 43 However, other studies have indicated that HPV is not necessarily sufficient to induce changes in the cervicovaginal microbiota. 44 , 45 In general, a microenvironment with a high percentage of anaerobic bacteria and a lower percentage of Lactobacillus spp. is more susceptible to HPV infections. However, the roles of these microorganisms are unclear and should be studied further. The strength of the study was the recruitment in one single center, and the quality of the data collected through the latest generation sequencing (NGS). The limitation of the study was the small sample size and vaginal intercourse without the use of barrier contraception.
5. CONCLUSIONS
The vaginal microbiota seems to act as a key role in the acquisition, persistence, and clearance of HPV in the human vagina. Our results support the hypothesis that HPV infection increases vaginal bacterial diversity leading to a differentially altered vaginal microbiota in women with HPV infection compared with healthy women. These data may represent the opportunity for the development of novel therapeutic agents 46 in the form of probiotics, to prevent HPV infection and increase its clearance. However, further investigations are required to deepen the role of the vaginal microbiome in HPV infection.
AUTHOR CONTRIBUTIONS
Conceptualization, writing—review, data curation, and editing: Biagio Santella. Writing—review: Antonio Schiattarella. Supervision: Maria Teresa Schettino. Supervision and funding acquisition: Gianluigi Franci, Pasquale De Franciscis, Nicola Colacurci, and Massimiliano Galdiero. All authors have read and agreed to the published version of the manuscript.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
ETHICS STATEMENT
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the local ethical (no. 815–30/12/2019).
ACKNOWLEDGMENTS
The authors would like to thank the staff of the U.O.C University Hospital of Campania “Luigi Vanvitelli” in Naples, Italy and the staff of University Hospital “San Giovanni di Dio e Ruggi d'Aragona”, for their contributions. Open Access Funding provided by Universita degli Studi della Campania Luigi Vanvitelli within the CRUI‐CARE Agreement.
Santella B, Schettino MT, Franci G, et al. Microbiota and HPV: the role of viral infection on vaginal microbiota. J Med Virol. 2022;94:4478‐4484. 10.1002/jmv.27837
Biagio Santella and Maria Teresa Schettino contributed equally to this paper as first authors.
Contributor Information
Antonio Schiattarella, Email: antonio.schiattarella@unicampania.it.
Massimiliano Galdiero, Email: massimilianogaldiero@unicampania.it.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Satterwhite CL, Torrone E, Meites E, et al. Sexually transmitted infections among US women and men: prevalence and incidence estimates, 2008. Sex Transm Dis. 2013;40:187‐193. 10.1097/OLQ.0b013e318286bb53 [DOI] [PubMed] [Google Scholar]
- 2. Insinga RP, Perez G, Wheeler CM, et al. Incident cervical HPV infections in young women: transition probabilities for CIN and infection clearance. Cancer Epidemiol Biomarkers Prev. 2011;20:287‐296. 10.1158/1055-9965.EPI-10-0791 [DOI] [PubMed] [Google Scholar]
- 3. Castellsague X. Natural history and epidemiology of HPV infection and cervical cancer. Gynecol Oncol. 2008;110:S4‐S7. 10.1016/j.ygyno.2008.07.045 [DOI] [PubMed] [Google Scholar]
- 4. Munoz N, Castellsague X, de Gonzalez AB, Gissmann L. Chapter 1: HPV in the etiology of human cancer. Vaccine. 2006;24(Suppl 3):S3/1‐S3/10. 10.1016/j.vaccine.2006.05.115 [DOI] [PubMed] [Google Scholar]
- 5. Munoz N, Bosch FX, de Sanjose S, et al. International Agency for Research on Cancer Multicenter Cervical Cancer Study, G. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518‐527. 10.1056/NEJMoa021641 [DOI] [PubMed] [Google Scholar]
- 6. Burd EM. Human papillomavirus and cervical cancer. Clin Microbiol Rev. 2003;16:1‐17. 10.1128/cmr.16.1.1-17.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thompson EL, Wheldon CW, Rosen BL, Maness SB, Kasting ML, Massey PM. Awareness and knowledge of HPV and HPV vaccination among adults ages 27‐45 years. Vaccine. 2020;38:3143‐3148. 10.1016/j.vaccine.2020.01.053 [DOI] [PubMed] [Google Scholar]
- 8. Schwarz TF, Huang LM, Valencia A, et al. A ten‐year study of immunogenicity and safety of the AS04‐HPV‐16/18 vaccine in adolescent girls aged 10‐14 years. Hum Vaccin Immunother. 2019;15:1970‐1979. 10.1080/21645515.2019.1625644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Martel C, Plummer M, Vignat J, Franceschi S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int J Cancer. 2017;141:664‐670. 10.1002/ijc.30716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu Z, Ma D. The precision prevention and therapy of HPV‐related cervical cancer: new concepts and clinical implications. Cancer Med. 2018;7:5217‐5236. 10.1002/cam4.1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brant AC, Menezes AN, Felix SP, de Almeida LM, Sammeth M, Moreira MAM. Characterization of HPV integration, viral gene expression and E6E7 alternative transcripts by RNA‐Seq: a descriptive study in invasive cervical cancer. Genomics. 2019;111:1853‐1861. 10.1016/j.ygeno.2018.12.008 [DOI] [PubMed] [Google Scholar]
- 12. Oyervides‐Munoz MA, Perez‐Maya AA, Rodriguez‐Gutierrez HF, et al. Understanding the HPV integration and its progression to cervical cancer. Infect Genet Evol. 2018;61:134‐144. 10.1016/j.meegid.2018.03.003 [DOI] [PubMed] [Google Scholar]
- 13. Li Y, Yu T, Yan H, et al. Vaginal microbiota and HPV infection: novel mechanistic insights and therapeutic strategies. Infect Drug Resist. 2020;13:1213‐1220. 10.2147/IDR.S210615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Borgogna JC, Shardell MD, Santori EK, et al. The vaginal metabolome and microbiota of cervical HPV‐positive and HPV‐negative women: a cross‐sectional analysis. BJOG. 2020;127:182‐192. 10.1111/1471-0528.15981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Panatto D, Amicizia D, Trucchi C, et al. Sexual behaviour and risk factors for the acquisition of human papillomavirus infections in young people in Italy: suggestions for future vaccination policies. BMC Public Health. 2012;12:623. 10.1186/1471-2458-12-623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mitra A, MacIntyre DA, Marchesi JR, Lee YS, Bennett PR, Kyrgiou M. The vaginal microbiota, human papillomavirus infection and cervical intraepithelial neoplasia: what do we know and where are we going next? Microbiome. 2016;4:58. 10.1186/s40168-016-0203-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barrientos‐Duran A, Fuentes‐Lopez A, de Salazar A, Plaza‐Diaz J, Garcia F. Reviewing the composition of vaginal microbiota: inclusion of nutrition and probiotic factors in the maintenance of eubiosis. Nutrients. 2020;12:12. 10.3390/nu12020419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rinninella E, Raoul P, Cintoni M, et al. What is the healthy gut microbiota composition? a changing ecosystem across age, environment, diet, and diseases. Microorganisms. 2019;7:7. 10.3390/microorganisms7010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Amabebe E, Anumba DOC. The vaginal microenvironment: the physiologic role of Lactobacilli. Front Med. 2018;5:181. 10.3389/fmed.2018.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Skarin A, Sylwan J. Vaginal lactobacilli inhibiting growth of Gardnerella vaginalis, Mobiluncus and other bacterial species cultured from vaginal content of women with bacterial vaginosis. Acta Pathol Microbiol Immunol Scand B. 1986;94:399‐403. 10.1111/j.1699-0463.1986.tb03074.x [DOI] [PubMed] [Google Scholar]
- 21. Messano GA. Inadequate antibiotic therapy of genitourinary tract infections could be responsible for viral sexually transmitted diseases. Ann Ig. 2013;25:553‐554. [PubMed] [Google Scholar]
- 22. Chehoud C, Stieh DJ, Bailey AG, et al. Associations of the vaginal microbiota with HIV infection, bacterial vaginosis, and demographic factors. AIDS. 2017;31:895‐904. 10.1097/QAD.0000000000001421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu MB, Xu SR, He Y, et al. Diverse vaginal microbiomes in reproductive‐age women with vulvovaginal candidiasis. PLoS One. 2013;8:e79812. 10.1371/journal.pone.0079812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Borgdorff H, Tsivtsivadze E, Verhelst R, et al. Lactobacillus‐dominated cervicovaginal microbiota associated with reduced HIV/STI prevalence and genital HIV viral load in African women. ISME J. 2014;8:1781‐1793. 10.1038/ismej.2014.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Oh HY, Kim BS, Seo SS, et al. The association of uterine cervical microbiota with an increased risk for cervical intraepithelial neoplasia in Korea. Clin Microbiol Infect. 2015;21:674.e671‐674.e679. 10.1016/j.cmi.2015.02.026 [DOI] [PubMed] [Google Scholar]
- 26. Gillet E, Meys JF, Verstraelen H, et al. Bacterial vaginosis is associated with uterine cervical human papillomavirus infection: a meta‐analysis. BMC Infect Dis. 2011;11:10. 10.1186/1471-2334-11-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Piyathilake CJ, Ollberding NJ, Kumar R, Macaluso M, Alvarez RD, Morrow CD. Cervical microbiota associated with higher grade cervical intraepithelial neoplasia in women infected with high‐risk human papillomaviruses. Cancer Prev Res. 2016;9:357‐366. 10.1158/1940-6207.CAPR-15-0350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Watts DH, Fazarri M, Minkoff H, et al. Effects of bacterial vaginosis and other genital infections on the natural history of human papillomavirus infection in HIV‐1‐infected and high‐risk HIV‐1‐uninfected women. J Infect Dis. 2005;191:1129‐1139. 10.1086/427777 [DOI] [PubMed] [Google Scholar]
- 29. Curty G, de Carvalho PS, Soares MA. The role of the cervicovaginal microbiome on the genesis and as a biomarker of premalignant cervical intraepithelial neoplasia and invasive cervical cancer. Int J Mol Sci. 2019;21:21. 10.3390/ijms21010222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dols JA, Reid G, Kort R, et al. PCR‐based identification of eight Lactobacillus species and 18 hr‐HPV genotypes in fixed cervical samples of South African women at risk of HIV and BV. Diagn Cytopathol. 2012;40:472‐477. 10.1002/dc.21786 [DOI] [PubMed] [Google Scholar]
- 31. Sun Y, Li L, Xia Y, et al. The gut microbiota heterogeneity and assembly changes associated with the IBD. Sci Rep. 2019;9:440. 10.1038/s41598-018-37143-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Greenbaum S, Greenbaum G, Moran‐Gilad J, Weintraub AY. Ecological dynamics of the vaginal microbiome in relation to health and disease. Am J Obstet Gynecol. 2019;220:324‐335. 10.1016/j.ajog.2018.11.1089 [DOI] [PubMed] [Google Scholar]
- 33. Champer M, Wong AM, Champer J, et al. The role of the vaginal microbiome in gynaecological cancer. BJOG. 2018;125:309‐315. 10.1111/1471-0528.14631 [DOI] [PubMed] [Google Scholar]
- 34. Schettino MT, De Franciscis P, Schiattarella A, et al. Prevalence of HPV genotypes in South Europe: comparisons between an Italian and a Turkish unvaccinated population. J Environ Public Health. 2019;2019:8769735. 10.1155/2019/8769735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. MacIntyre DA, Chandiramani M, Lee YS, et al. The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci Rep. 2015;5:8988. 10.1038/srep08988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brotman RM, Shardell MD, Gajer P, et al. Association between the vaginal microbiota, menopause status, and signs of vulvovaginal atrophy. Menopause. 2014;21:450‐458. 10.1097/GME.0b013e3182a4690b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Munoz N, Bosch FX, Castellsague X, et al. Against which human papillomavirus types shall we vaccinate and screen? the international perspective. Int J Cancer. 2004;111:278‐285. 10.1002/ijc.20244 [DOI] [PubMed] [Google Scholar]
- 38. Donders GGG, Bellen G, Grinceviciene S, Ruban K, Vieira‐Baptista P. Aerobic vaginitis: no longer a stranger. Res Microbiol. 2017;168:845‐858. 10.1016/j.resmic.2017.04.004 [DOI] [PubMed] [Google Scholar]
- 39. Ravel J, Gajer P, Abdo Z, et al. Vaginal microbiome of reproductive‐age women. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4680‐4687. 10.1073/pnas.1002611107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu D, Wu M, Halpern A, et al. Stalking the fourth domain in metagenomic data: searching for, discovering, and interpreting novel, deep branches in marker gene phylogenetic trees. PLoS One. 2011;6:e18011. 10.1371/journal.pone.0018011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Di Paola M, Sani C, Clemente AM, et al. Characterization of cervico‐vaginal microbiota in women developing persistent high‐risk Human Papillomavirus infection. Sci Rep. 2017;7:10200. 10.1038/s41598-017-09842-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brotman RM, Shardell MD, Gajer P, et al. Interplay between the temporal dynamics of the vaginal microbiota and human papillomavirus detection. J Infect Dis. 2014;210:1723‐1733. 10.1093/infdis/jiu330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee JE, Lee S, Lee H, et al. Association of the vaginal microbiota with human papillomavirus infection in a Korean twin cohort. PLoS One. 2013;8:e63514. 10.1371/journal.pone.0063514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Godoy‐Vitorino F, Romaguera J, Zhao C, et al. Cervicovaginal fungi and bacteria associated with cervical intraepithelial neoplasia and high‐risk human papillomavirus infections in a hispanic population. Front Microbiol. 2018;9:2533. 10.3389/fmicb.2018.02533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bienkowska‐Haba M, Luszczek W, Myers JE, et al. A new cell culture model to genetically dissect the complete human papillomavirus life cycle. PLoS Pathog. 2018;14:e1006846. 10.1371/journal.ppat.1006846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zannella C, Shinde S, Vitiello M, et al. Antibacterial activity of indolicidin‐coated silver nanoparticles in oral disease. Appl Sci. 2020;10:1837. 10.3390/app10051837 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


