Abstract
Crops with reduced nutrient and water requirements are urgently needed in global agriculture. Root growth angle plays an important role in nutrient and water acquisition. A maize diversity panel of 481 genotypes was screened for variation in root angle employing a high‐throughput field phenotyping platform. Genome‐wide association mapping identified several single nucleotide polymorphisms (SNPs) associated with root angle, including one located in the root expressed CBL‐interacting serine/threonine‐protein kinase 15 (ZmCIPK15) gene (LOC100285495). Reverse genetic studies validated the functional importance of ZmCIPK15, causing a approximately 10° change in root angle in specific nodal positions. A steeper root growth angle improved nitrogen capture in silico and in the field. OpenSimRoot simulations predicted at 40 days of growth that this change in angle would improve nitrogen uptake by 11% and plant biomass by 4% in low nitrogen conditions. In field studies under suboptimal N availability, the cipk15 mutant with steeper growth angles had 18% greater shoot biomass and 29% greater shoot nitrogen accumulation compared to the wild type after 70 days of growth. We propose that a steeper root growth angle modulated by ZmCIPK15 will facilitate efforts to develop new crop varieties with optimal root architecture for improved performance under edaphic stress.
Keywords: crown root, GWAS, phenotyping, root architecture
Short abstract
Genome‐wide association mapping identified CBL‐interacting serine/threonine‐protein kinase 15 (ZmCIPK15) gene that controls root angle and is associated with increased deep nitrogen capture and plant performance in low nitrogen environments.
1. INTRODUCTION
Plant productivity is often limited by suboptimal nutrient and water availability in natural and managed ecosystems. Root system architecture has important effects on plant fitness in environments with suboptimal water and nutrient availability (Lynch, 2018, 2019; Lynch & Brown, 2012; York et al., 2013). Root architecture determines the spatiotemporal distribution of root exploration in the heterogeneous matrix of the soil and the ability of plant roots to obtain mobile and immobile resources (Hirel et al., 2007; Lynch, 1995, 2013; Lynch & Brown, 2012). Root architectural phenes, including root angle (Bonser et al., 1996; Dathe et al., 2016; Trachsel et al., 2013; York et al., 2013) influence root distribution, plant performance and soil resource acquisition under nutrient and water stress. A phene is elementary and unique at its level of biological organization (Lynch & Brown, 2012); ‘phene’ is to ‘phenotype’ as ‘gene’ is to ‘genotype’ (Serebrovsky, 1925; York et al., 2013). Understanding the role of root phenes, particularly the gravitropic response of roots will facilitate the development of crop varieties with more efficient resource capture by roots under conditions of suboptimal water and nutrient availability (Kell, 2011; Lynch, 2019; Lynch & Brown, 2012).
The growth angle of roots plays an important role in the placement of roots in specific soil domains (Forde, 2009; Lynch, 1995, 2019; Lynch et al., 2011; Lynch & Wojciechowski, 2015; Manschadi et al., 2008; Singh et al., 2010). Root growth angle affects rooting depth in maize (Trachsel et al., 2013), rice (Kato et al., 2006), sorghum (Mace et al., 2012), common bean (Bonser et al., 1996) and wheat (Oyanagi, 1994). Root growth angle and depth have direct effects on biomass accumulation, nutrient acquisition and water capture in drought and other edaphic stress conditions (Campos et al., 2004; Hammer et al., 2009; Lynch & Brown, 2001; Manschadi et al., 2008). A shallow root system enhances root exploration in the topsoil, where the availability of immobile nutrients such as phosphorus is greatest. Plants with shallow root angles are better suited to capture immobile nutrients and have been shown to improve phosphorus acquisition in maize, soybean and common bean (Bonser et al., 1996; Jing et al., 2004; Liao et al., 2001; Lynch & Brown, 2001; Zhu et al., 2005). In contrast, plants with steep root angles are better suited to capture mobile nutrients and water available in deep soil domains over the growing season due to leaching and soil drying from the surface (Lynch, 2013, 2018, 2019; Trachsel et al., 2013). In addition, environmental factors, including soil water availability (Oyanagi et al., 1993) and soil nitrogen (Trachsel et al., 2013) can alter root angle. However, evidence for enhanced nutrient acquisition by plants with steeper root growth angles is based on contrasting natural variation in root phenes. Here, we use near‐isogenic lines contrasting in root angle, modulated by expression of a single gene, to demonstrate how root angle affects nitrogen capture.
Genetic variation for root growth angle has been observed in nodal roots in maize (Bayuelo‐Jiménez et al., 2011; Giuliani et al., 2005; Nakamoto et al., 1991; Peñagaricano et al., 2012), sorghum (Tsuji Wataru et al., 2005), foxtail millet (Nakamoto et al., 1991) and rice (Kato et al., 2006) and seminal roots in barley (Hargreaves et al., 2008) and wheat (Manschadi et al., 2008; Oyanagi, 1994). Detection of genetic loci associated with root angle for plants grown in the greenhouse and field (Liao et al., 2004; Norton & Price, 2009; Omori & Mano, 2007; Uga et al., 2013) and identification of gene expression networks (Vidal et al., 2010) indicate a strong genetic component controlling root growth angle. DRO1, a gene associated with deeper rooting in rice, enabled greater nitrogen uptake (Arai‐sanoh et al., 2014), greater water uptake (Uga et al., 2013) and subsequently greater yield. In addition to DRO1, a few genetic loci have been detected for root angle, including bending angle in rice (Giri et al., 2018; Norton & Price, 2009), basal root growth angle in common bean (Liao et al., 2004), nodal root angle in maize (Guingo et al., 1998; Omori & Mano, 2007; Schneider et al., 2020b), the plastic response of nodal root angle in maize (Schneider et al., 2020b) and nodal root angle in sorghum (Mace et al., 2012).
The maize root system consists of three major root classes: primary, seminal (or seed‐borne) and nodal (shoot‐borne) roots, which all produce lateral (root‐borne) roots of the first and second order. Typically, maize plants develop up to six root‐bearing nodes emerging belowground (crown roots) and up to three additional root‐bearing nodes emerging aboveground (brace roots) (Hochholdinger et al., 2004; Hoppe et al., 1986) and collectively, brace and crown roots are referred to as nodal roots. In field‐grown maize, critical nitrogen uptake occurs during the development of these later, younger nodes, which contribute to the majority of nitrogen uptake beginning around 4 weeks after planting at the 10‐ to 14‐leaf stage (DeBruin et al., 2017).
Here, we used genome‐wide association mapping (GWAS) in mature, field‐grown maize to identify a candidate gene controlling root angle in maize. The Wisconsin Diversity (WiDiv) association panel has been genotyped with a high marker density and has very small linkage blocks, allowing for precise detection of candidate genes and the identification of rare alleles (Hansey et al., 2011). We confirmed the function of the candidate gene through knock‐out mutants. We utilized functional‐structural modelling as well as empirical observations of plants grown in controlled environment mesocosms and in the field to confirm the functional utility of root growth angle for enhanced nitrogen capture and the functional relevance of this gene for improved nitrogen uptake and increased plant biomass in low nitrogen environments.
2. MATERIALS AND METHODS
Root growth angle was phenotyped in 481 lines from the WiDiv Panel, comprised of lines displaying uniformity and vigour that reach grain physiological maturity in the upper Midwest region of the United States. Experiments were conducted at the Ukulima Root Biology Centre (URBC) in Alma, Limpopo, South Africa (24° 330,012 S, 28° 072584 E) under standard agronomic practices. The experiments were conducted on a Clovelly loamy sand (Typic Ustipsamment). Experiments were conducted during January to April of 2010, 2011 and 2012 and during November to February of 2013. Row width was 75 cm and distance within a row was 23 cm. The WiDiv panel (Table S1) was grown with two replications in a randomized complete block design with single row plots (20 plants per plot). In all trials, levels of macro and micronutrients were adjusted to meet the requirements for maize production as determined by soil tests at the beginning of the growing seasons. The trials were irrigated using a centre pivot system. Pest control was carried out as needed.
Evaluations of maize root crowns were performed based on the shovelomics method (Trachsel et al., 2011). At anthesis, three representative plants were selected and excavated from each plot. Root crowns were washed with low pressure water and root growth angle was measured at the youngest emerged node as degrees from the soil line and automatically computed using algorithms in the Digital Imaging of Root Traits (DIRT) software (Bucksch et al., 2014; Das et al., 2015). Spearman and Pearson correlations between years and replications suggested that data could be combined using best linear unbiased predictors (BLUPs). BLUPs across all years were calculated and used for subsequent analysis. Residuals were transformed according to box cox analysis. Statistical analyses were performed in R version 3.1.2 (R Core Team, 2018).
Angle phenotypes from four field seasons were used in a Multiple Loci Linear Mixed Model for GWAS analysis (Zhang et al., 2010) implemented in the FarmCPU R package (Liu et al., 2016). The model used 523,602 single nucleotide polymorphism (SNP) markers (Mazaheri et al., 2019). Allelic effects are estimated relative to the minor allele and a minimum minor allele frequency of 0.05 was used.
The following linear model was used for GWAS analysis of the WiDiv Panel:
where Y i is the observation of the ith individual; Mi1, Mi2,…Mit are the genotypes of the t pseudo QTNs, initiated as an empty set; b1, b2,…, bj are the corresponding effects of the pseudo QTNs; S ij is the genotype of the ith individual and jth genetic marker; d j is the corresponding effect of the jth genetic marker and e i is the residuals having a distribution with zero mean and variance of σ 2 e . All effects were considered random. Significant SNPs were identified based on a genome‐wide corrected Bonferroni threshold of –log(p) = 7.02.
R Software (version 3.2.4)(Core Team, 2018), Bioconductor (Bates et al., 2002) and MaizeGDB (Lawrence, 2005) were used to annotate genes. Candidate genes identified through significant GWAS hits were detected based on the physical position of genes in the version 4 B73 (AGPv4) reference sequence assembly (Jiao et al., 2017).
A linkage disequilibrium (LD) analysis was performed to confirm that ZmCIPK15 (Zm00001d033316) is not linked to neighbouring genes. Genotypic data in HapMap format was used (Mazaheri et al., 2019), converted to vcf format using TASSEL version 5.0 (Bradbury et al., 2007) and numericalized using the rMVP package in R (Yin et al., 2021). During numericalization, the missing genotypes (<0.22%) were imputed to the major allele. LD was calculated as the squared correlation coefficient (r 2) between two loci. The LD analysis was performed 100 Kb upstream and downstream of Zm00001d033316 and was implemented using the gpart R package (Kim et al., 2019).
The zmcipk mutant allele stock (mu1046464::Mu, stock ID: UFMu‐06162) was obtained from the Maize Genetics Stock Centre‐Uniform Mu collection (McCarty et al., 2005). Seeds from the stock centre were grown at West Madison Agricultural Research Station during the summer of 2014 and genotyped for the transposon insertion. DNA was isolated by CTAB method and all primers were designed by Primer 3 based on B73 reference sequence. Plants carrying the mutant allele were identified by genotyping using an outward‐facing primer in the TIR of the Mutator transposon, TIR6 (5′‐AGAGAAGCCAACGCCAWCGCCTCYATTTCGTC ‐3′) (Settles et al., 2007) and the ZmCIPK gene‐specific primer ZmCIPK F1 (5′‐TTGGCACCACCAAGGCGCACCCTGTA −3′). The wild‐type allele was identified by using the gene‐specific primer set ZmCIPK F1 and ZmCIPK R1 (5′‐ CGTCCGCCTTGGCGCCGTCGT −3′). All PCR conditions were 95°C for 30 s, 63°C for 30 s, 72°C for 45 s, repeated for 30 cycles. A homozygous population positive for the transposon insertion was generated after three generations of self‐pollination and was used for phenotype analysis.
Seeds were germinated and RNA was extracted for semi‐quantitative RT‐PCR analysis. CIPK::Mu and W22 wild‐type seeds were surface sterilized in 50 ml of 20% v/v NaOCl with 20 μl of Tween20 for 20 min and rinsed with sterile deionized water. Sterilized seeds were germinated in rolled germination paper (Number 78, Anchor Paper Company, St. Paul, MN). Six seeds were placed on a germination paper 4 cm from the top edge of the paper and spaced 4 cm apart. The rolled germination paper was placed vertically in a 20 cm × 10 cm × 30 cm stainless steel tank. Water was added to a depth of 2.5 cm. Seedlings were germinated in a Percival at 21°C with 18:6 h light–dark cycle for 7 days. Whole primary roots from three 7‐day seedlings were pooled into a 1.6 ml microfuge tube, flash frozen in liquid nitrogen and stored at −80°C for RNA extraction.
For RNA extraction, root samples were ground to a fine powder in the 1.6 ml microfuge tubes using a 7 cm polypropylene pellet pestle (Grainger). RNA was extracted from samples using IBI Total RNA Mini Kit (Plant) with β‐mercaptoethanol according to the manufacturer instructions. DNA contamination was removed from RNA samples using TURBO DNA‐free Kit (Invitrogen) according to manufacturer instructions.
Complementary DNA (cDNA) was synthesized from DNAse‐treated RNA using SuperScript IV First‐Strand Synthesis System with Oligo‐dT primers according to manufacturer instructions. cDNA samples were tested for genomic DNA contamination by PCR using 2× Platinum II hot‐Start PCR Master Mix (Thermo Fisher Scientific). The primer pairs (ZmAct1_F: AATGGCACTGGAATGGTCAA and ZmAct1_R: CTCTTGGCCTGAGCCTCATC) were used to detect β‐Actin1 (Zm00001d010159) spanning an intron. Amplification from gDNA templates yielded a 237 bp PCR amplicon while amplification from cDNA yielded a 152 bp PCR amplicon. cDNA samples were verified to be free of gDNA contamination prior to use for analysis of ZmCIPK15 expression by semiquantitative reverse transcription PCR (sqRT‐PCR). The primer pairs (ZmCIPK15_F: TCATCCTCTTCGTCCTCCTC and ZmCIPK15_R: ACATGTCGGCGATGTTGG) were used to detect ZmCIPK15 and yielded a 63 bp PCR amplicon. One‐hundred ng of cDNA was used to make a 100 μl PCR master mix using 2X Platinum II hot‐Start PCR Master Mix (Thermo Fisher Scientific). Fifty ng of gDNA was used to make a 100 μl PCR mater mix for the W22 wild‐type control reactions using 2× Platinum II hot‐Start PCR Master Mix (Thermo Fisher Scientific). The master mix was separated into two 50 μl aliquots to make replicate sets. Conditions were set according to manufacturer instructions with the annealing temperature set to 60°C. One PCR set cycled for 35 cycles while the second PCR set cycled for 45 cycles and then separated by 1.5% agarose gel electrophoresis.
Greenhouse and field experiments were used to confirm the phenotype of the mutant and wild‐type plants and to test the functional utility of changes in root angle for nitrogen capture. Greenhouse mesocosm experiments were conducted in a greenhouse at University Park, PA (40° 45′36.0″ N, 73° 59′ 2.4″ W). Plants were grown with a 14 h photoperiod, the temperature was maintained at approximately 28°C/26°C day/night, 40% RH and light (LED) photosynthetic photon flux density of 500 μmol m−2 s−1 at the sixth leaf. For each experiment, four replications of cipk15 mutant and wild‐type genotypes were planted in a split plot design in two treatments (high and low nitrogen or well‐watered and water‐stressed). Plants were grown in individual mesocosms constructed out of polyvinyl chloride cylinders with an inner diameter of 15.5 cm and height of 1.54 m and lined with transparent 6 mm high‐density polyethylene film to facilitate root sampling. Each mesocosm was filled with a 30 L growth medium consisting of 50% commercial grade medium sand (US Silica), 27% horticultural grade fine vermiculite (D3, Whittemore Companies Inc., Lawrence), 18% field soil and 5% horticultural grade super coarse perlite (Whittemore Companies Inc.), by volume.
Nitrogen stress experiments were conducted from April to May 2017. Plants were fertigated with nutrient solutions providing sufficient or insufficient N (Table S2) using drip rings Nutrient solutions were adjusted to pH 6.0 using KOH pellets, and maintained at this pH with KOH or HCl as needed. Each mesocosm was fertigated with 100 ml per mesocosm daily.
Water stress experiments were conducted from June to July 2017. Mineral nutrients were provided by mixing the medium with 70 g per mesocosm of Osmocote Plus fertilizer (5–6 months release, Scotts‐Sierra Horticultural Products Company, Marysville) consisting of (%): NO3 (8) NH+ 4(7), P (9), K (12), S (2.3), B (0.02) Cu (0.05), Fe (0.68), Mn (0.06), Mo (0.02) and Zn (0.05). Each mesocosm was irrigated with approximately 100 ml water daily. For the water stress treatment, water was withheld starting 14 days after planting until the end of the experiment.
For all experiments, two seeds were directly sown into each mesocosm and plants were thinned 1 week after planting. Each mesocosm was saturated with 2.5 L of water 1 day prior to planting. At harvest (i.e., 43–45 days after planting), the shoot was removed, the plastic liner was extracted from the mesocosm, cut open and the roots were washed by rinsing the medium away with low pressure water. Root crowns were imaged node by node according to (York & Lynch, 2015).
Six field experiments were used to study the cipk15 mutant and wild‐type (W22) genotype. Genotypes were planted in a randomized split plot design in two treatments per experiment (high and low nitrogen or well‐watered and water‐stressed) in four replications. Experiments at the Russell E. Larson Agricultural Research Centre in Rock Springs, Pennsylvania, United States (PSU) (40°42′N, 77°57′W) were conducted from May to August 2016, 2017, 2018 and 2019. Each genotype was planted in a 3‐row plot consisting of 60 plants per plot. Row width was 75 cm and distance between plants within a row was 23 cm. Two experiments (2018 and 2019) were performed to study angle phenotypes in well‐watered and water‐stress in the rainout shelter structures. Four experiments (2016, 2017, 2018 and 2019) were performed to study angle phenotypes in low and high nitrogen environments. At an thesis, two plants per plot were evaluated using the shovel omics method (Trachsel et al., 2011). Shoot biomass was collected, and shoot material was dried at 60°C. The root angle at each node was imaged according to (York & Lynch, 2015). Additional root phenes, including root diameter and lateral branching density and length were quantified using DIRT software (Das et al., 2015). Crown root number was phenotyped manually.
For nitrogen experiments, the experiments included four blocks with each block being a separate 0.4 ha area split across two fields. On one half of the field, no nitrogen fertilizer had been applied since 2010 and this was designated as the low N treatment (10 mg kg−1). The high nitrogen side of the field had 146 kg ha−1 nitrogen applied annually before planting. Within the high and low nitrogen treatment split‐plots, the genotypes were randomized.
The ability of roots to acquire deep soil nitrogen was studied by deep injection of 15NO3 − in the field in 2015 and 2016 in nitrogen experiments. At 60 days after planting, three representative plants were selected and a polyvinyl chloride (PVC) pipes (length of 75 cm and a diameter of 5 cm) were used to inject 15NO3 − between adjacent plants within a planting row. Prior to injections, a soil sampling probe was used to excavate a cylinder of soil to a depth of 45 cm. A PVC pipe was inserted into the hole and the 15NO3 − solution was injected into the hole. Each sampling point received 10 ml of K15NO3 − solution (0.46 mg 15N ml−1, 98% 15N enriched). Following injection, the hole was filled with field soil to prevent roots from growing down into the hole. Ten days after 15NO3 − injection, the shoot biomass of an adjacent plant was harvested, and dried for 15N and total N analysis. Leaf and stem tissues were dried at 120°C for 5 days, ground and 3 mg of ground tissue were analysed for tissue nitrogen content using an elemental analyser (SeriesII CHNS/O 440 Analyser 2400, PerkinElmer). 15N in plant tissue was analysed using a PDZ Europa 441 ANCA‐GSL elemental analyser interfaced to a PDZ Europa 20–20 isotope ratio mass 442 spectrometer (Sercon Ltd.) at the Stable Isotope Facility, University of 443 California at Davis, United States (http://stableisotopefacility.ucdavis.edu/).
For the water‐stress field experiment, the shelters (10 × 30 m) were covered with a clear polyethylene film and were automatically triggered by rainfall to cover the plots, excluding natural precipitation from 3 weeks after planting until grain harvest. Adjacent non‐sheltered plots were drip‐irrigated as needed to provide unstressed comparisons.
Models representing the cipk15 and W22 angle phenotypes in low nitrogen environments were constructed using the functional‐structural plant model OpenSimRoot, a heuristic model (i.e., a model, which seeks to test the adequacy of a logic model rather than a model, which seeks to precisely predict empirical observations), which simulates root system growth, nitrate uptake, nitrate depletion and nitrate leaching in three dimensions over time. OpenSimRoot is an open‐source model and mathematical description of root function and growth (Postma et al., 2017). Soil nitrate and water movement are modelled using SWMS_3D (Šimůnek et al., 1995). Models representing a single maize plant grown in a monoculture were created by simulating a rectangular prism of soil that was 76 cm wide, 15 cm long and 150 cm deep, and roots that reached the vertical barriers were reflected to represent the roots of neighbouring plants. Existing maize parameters based on empirical data were modified to incorporate crown root angle measurements taken on both genotypes in field and greenhouse environments. Previous parameters for nitrate concentration by depth, organic matter by depth and precipitation for the Rock Springs environment were used to simulate a high precipitation growing season with approximately 165 kg ha−1 available nitrate as described by (Dathe et al., 2016). Models simulated a period of 40 days after germination.
2.1. SNP mapping and analysis
MaizeGDB was used to identify chromosome number, gene start and gene end positions for GRMZM2G472643 (associated with Zm00001d033316 gene model for assembly version 4.0). These coordinates were used to parse SNP information of 481 genotypes (Table S3; part of WIDIV collection) overlapping with those present in the phenotypic data used for GWAS. Exon sequence information of Zm00001d033316_T001 from EnsemblePlants was next used to retain the SNPs that are present only within the coding sequence (CDS). Next, for each genotype, the SNP nucleotide at given SNP position was substituted in the native CDS and translated to the variant amino acid sequence. Further, the native amino acid sequence (translated from CDS) was compared with the variant amino acid sequence to determine sense or mis‐sense amino acid substitutions within each genotype. Only significant SNPs (using Welch's t‐test, p‐value <0.05) resulting into mis‐sense amino acid substitutions in more than one genotype were further studied for their observed effects on root growth angle selected SNPs were mapped on the predicted structure.
2.2. Structure modelling
The amino acid sequence of GRMZM2G472643 was used for structure prediction using homology modelling in Phyre2 and SWISS‐Model servers. A homology modelling approach was chosen over de novo structure prediction from first principles as the gene of interest was inferred to have kinase activity and function in signal transduction. Protein domain analysis also predicted the presence of a protein kinase (catalytic) domain and a CBL interacting (regulatory) domain, both of which individually were used in the structure prediction algorithms.
3. RESULTS
3.1. Field phenotyping and GWAS reveals key loci controlling crown root angle in maize
Large variation for root angle was observed in the WiDiv Panel and crown root growth angle ranged from 10 to 90° (angle measured from horizontal) (Figure 1, Table S4). Root angle was relatively heritable and had a broad‐sense heritability of 0.64 (Schneider, Klein, Hanlon, Nord, et al., 2020b). GWAS results revealed that gene model Zm00001d033316 on chromosome 1 included the most significant SNP and was, therefore, selected for additional analysis (Figure 2a). This gene is highly expressed in root tissues, including the basal section of the primary root at 7 DAS and crown roots at nodes 1–5 at the V7 and V13 growth stage (Stelpflug et al., 2015) (Figure 2b). rs1_258847118 is located on chromosome 1 at position 258,847,118 (Zm00001d033316) and encodes a CBL‐interacting serine/threonine‐protein kinase 15 (LOC100285495). This genomic region exhibits rapid LD decay and is not in strong linkage with upstream or downstream gene models (Figure S1). GWAS results demonstrated that this SNP has an effect size of 2.8° (Schneider, Klein, Hanlon, Nord, et al., 2020b).
Figure 1.

Variation is observed in crown root angle in maize. (a) Image of demonstrating the measurement of maize crown root angle. (b) Variation in crown root angle in the Wisconsin Diversity Panel in four field environments under no stress. (c) Variation in crown root angle in maize ranging from shallow angle to steep angle. Scale bar represents 2.5 cm [Colour figure can be viewed at wileyonlinelibrary.com]
Figure 2.

Genome‐wide association study (GWAS) was performed on crown root angle data. (a) Manhattan plot describes FARMCPU GWAS results using ~500,000 SNP markers from crown root angle analysis. Horizontal line represents a Bonferroni‐corrected genome‐wide threshold. We detected a significant SNP on chromosome one. (b) CIPK is root expressed with higher expression in crown root nodes 1–3 (data replotted from Stelpflug et al., 2015). DAS, days after sowing [Colour figure can be viewed at wileyonlinelibrary.com]
3.2. Identification of missense alleles in ZmCIPK associated with steeper crown root angle
We aimed to understand the effects of the polymorphism on the protein structure and function of ZmCIPK15 in relation to the differences in root angles between ecotypes from two allelic groups. Here we focused on all of the SNPs within the ZmCIPK15 gene, including the most significant SNP identified from the GWAS. Using chromosome number (chromosome 1), gene start (258845263) and gene end (258848552) positions, we identified in total 72 SNPs within the genomic region encoding GRMZM2G472643, distributed within either the 5′ untranslated region (5′ UTR; 10), coding sequence (CDS; 43) and 3′ UTR (19). Among the 43 SNPs located in the CDS, 28 resulted in sense mutations (Table S5, green filled), 11 mis‐sense mutations (Table S5 red filled) and 4 were unresolved (Table S5, grey filled) due to non‐resolved nucleotides in the SNP information. Out of the mis‐sense substitution SNPs, only seven showed significant deviation in the mean crown root angle phenotype between the two populations of allelic variants (Figure S2, Table 1). Out of these, three SNPs (rs1_258846296, rs1_258846449 and rs1_258847118; Figure S2, red outlined; Table S6) occurred together in the same set of 16 genotypes (i.e., none occurred independently of the others) and exhibited steeper angle compared to the remaining 465 genotypes. These three SNPs are termed ‘co‐occurring SNPs’ in further analysis. SNP rs1_258847118 was among the most significant SNPs identified in the GWAS analysis. The remaining four SNPs (rs1_258846294, rs1_258846443, rs1_258846899 and rs1_258847073; termed ‘individually occurring SNPs’; Table 1) showed significant difference in the crown root angle distribution compared to the respective major allele's angle distribution.
Table 1.
SNPs resulting in mis‐sense substitutions with their amino acid variants, and mean of crown root angles of major and minor alleles among 481 genotypes
| SNPs | Amino acid | Mean of crown angle distribution | P‐value | ||
|---|---|---|---|---|---|
| Major allele | Minor allele | Major allele | Minor allele | ||
| rs1_258846294 | G | A | 60.5630308 | 62.9058645 | 0.034979234 |
| rs1_258846296 | P | S | 60.7181986 | 57.9570191 | 0.028526093 |
| rs1_258846443 | S | A | 60.7106075 | 59.6974184 | 0.029115474 |
| rs1_258846449 | L | V | 60.7181986 | 57.9570191 | 0.028526093 |
| rs1_258846575 | S | T | 60.4793597 | 61.4387751 | 0.14816344 |
| rs1_258846899 | V | I | 60.6584977 | 56.792802 | 0.017326689 |
| rs1_258847073 | G | S | 60.7181986 | 57.9570191 | 0.01071663 |
| rs1_258847118 | G | S | 60.6196284 | 60.5976835 | 0.028526093 |
| rs1_258847336 | M | I | 60.5689523 | 61.4595514 | 0.474282159 |
| rs1_258847421 | P | T | 60.5630308 | 62.9058645 | 0.092446822 |
Note: p‐values show significance based on Welch's test between crown root angle distributions of minor and major alleles.
3.3. Mapping missense alleles onto ZmCIPK protein domains
To uncover how the 3 ‘co‐occurring SNPs’ detailed above impact CIPK protein function, we mapped the resulting missense alleles onto the kinase's individual domains. Tables 2 and 3 list the top templates identified in the Phyre2 and SWISS‐Model algorithms for structural modelling of maize CIPK15. Both these servers rank the same template structures as top candidates, namely CBL‐Interacting Serine/Threonine protein kinase 23 (CIPK23, PDB ID 4CZT) from Arabidopsis thaliana and an uncharacterized protein T20L15_90 (PDB ID 2ZFD) for building the structural models (Figure 3) for the protein kinase and CBL‐interacting domains of maize CIPK, respectively. Next, the three co‐occurring and four individually present SNPs identified in the SNP mapping analysis were mapped on to these models (Figure 3a,c,d). The mapped SNP positions highlight the putative effect of the SNP on the function of the CIPK protein in maize. The mu insertion is located between amino acids 63 and 66 (Figure 3). Interestingly, at the 61st and 63rd amino acid positions, the SNPs are predicted to destabilize the kinase domain likely affecting the binding of ATP to the kinase domain (see Section 4).
Table 2.
Structural modelling results and statistics from SWISS‐Model and Phyre2 servers for the protein kinase (catalytic) domain of maize CIPK. Here, GMQE score (values ranging between 0 and 1) is a quality estimation of the accuracy of a model built with target‐template alignment and the coverage of the target. Higher numbers indicate higher reliability. QMEAN score is an estimate of the ‘degree of nativeness’ of the structural features. Score of zero indicates good agreement between the model structure and experimental structures of similar size
| SWISS‐model | ||||
|---|---|---|---|---|
| Templates | Sequence identity | Sequence coverage | GMQE | QMEAN |
| CBL‐interacting serine/threonine‐protein kinase 23 (CIPK23) (PDB 4czt.2) | 52.87 | 95% | 0.79 | −0.92 |
| CIPK24 (PDB 4d28.2) | 47.27 | 93% | 0.75 | −2.70 |
| Serine/threonine‐protein kinase MARK2 (PDB 2wzj.1) | 38.93 | 96% | 0.71 | −3.15 |
| Phyre2 | ||||
| Templates | Sequence identity | Sequence coverage | Raw score | |
| CBL‐interacting serine/threonine‐protein kinase 23 (CIPK23) (PDB 4czu.C) | 53 | 94% | 397 | |
| Serine/threonine‐protein kinase MARK1 (PDB c6c9) | 39 | 95% | 390 | |
Table 3.
Structural modelling results and statistics from SWISS‐Model and Phyre2 servers for the CBL‐interacting (regulatory) domain of maize CIPK
| SWISS‐model | ||||
|---|---|---|---|---|
| Templates | Sequence identity | Sequence coverage | GMQE | QMEAN |
| Putative uncharacterized protein T20L15_90 (PDB 2zfd.1) | 33.33 | 75% | 0.52 | −3.51 |
| CIPK24 (PDB 4ehb.1) | 23.02 | 81% | 0.54 | −2.00 |
| Serine/threonine‐protein kinase MARK1 (PDB 6c9d.1) | 14.78 | 74% | 0.41 | −3.74 |
| Phyre2 | ||||
| Templates | Sequence identity | Sequence coverage | Raw score | |
| Putative uncharacterized protein T20L15_90 (PDB 2zfd.1) | 33.33 | 76% | 161 | |
| CIPK24 (PDB 4ehb.1) | 27 | 78% | 157 | |
Note: Here, GMQE score (values ranging between 0 and 1) is a quality estimation of the accuracy of a model built with target‐template alignment and the coverage of the target. Higher numbers indicate higher reliability. QMEAN score is an estimate of the ‘degree of nativeness’ of the structural features. Score of zero indicates good agreement between the model structure and experimental structures of similar size.
Figure 3.
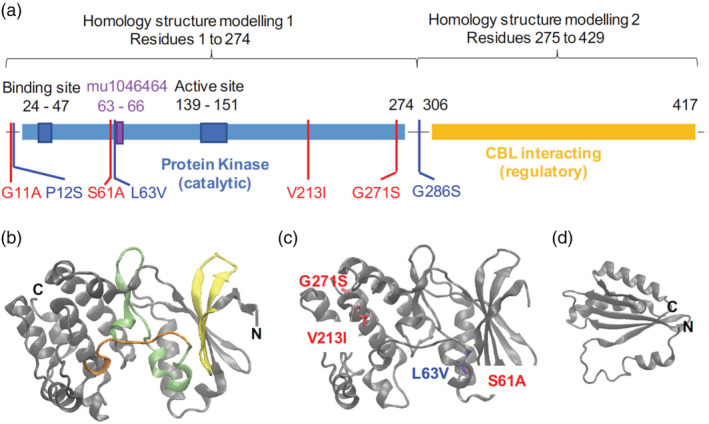
(a) Predicted domain organization and mapping of active sites of Maize CIPK. The CIPK protein consists of two domains, which were used separately in homology modelling to predict the structure of the two domains. The N‐terminal kinase domain catalyses the conversion of ATP to ADP and consists of an ATP binding site between residues 24 and 47 and the active site in the region around residues 139–151. The C‐terminal domain is involved in protein–protein interaction with CBL proteins and thus acts as a regulator of CIPK activity. SNPs represented in Figure S2 are mapped over the domain structure and indicate positions of individually occurring (in red, rs1_258846294 ‐ > G11A, rs1_258846443 ‐ > S61A, rs1_258846899 ‐ > V213 I and rs1_258847073 ‐ > G271S) and co‐occurring (in blue, rs1_258846296 ‐ > P12S, rs1_258846449 ‐ > L63V and rs1_258847118 ‐ > G286S) SNPs. Magenta box indicates the position of mutant allele stock (mu1046464::Mu, stock ID: UFMu‐06162) between residues 63–66. (b–d) Homology models for the Protein kinase (b–c) and CBL‐interacting (d) domains of maize CIPK. ‘C’ and ‘N’ notations on the models represent the C‐ and N‐termini of the respective domains. The Protein kinase domain in (b) is colour‐coded according to residues involved in kinase activity. Green residues indicate the active site, orange residues are part of the activation loop and yellow residues are involved in nucleotide binding. Locations of amino acid substitutions caused due to individual and co‐occurring SNPs are indicated on the structural models in (c). G11A, P12S and G286S are not indicated as they do not occur within the boundaries of the modelling outputs [Colour figure can be viewed at wileyonlinelibrary.com]
3.4. Greenhouse and field studies confirm that ZmCIPK controls root angle
To validate the functional importance of the ZmCIPK15 gene, a Mu transposon insertional mutant, mu1046464::Mu (stock: UFMu‐06162) was identified. A homozygous cipk15 mutant population was generated and the transposon was verified to be inserted 189 bp downstream of the transcriptional start site in exon 1 of the Zm00001d033316 gene (Figure S3). Homozygous cipk15 mutants were phenotyped in greenhouse mesocosms and in six field environments (high and low nitrogen environments over 4 years and well‐watered and water deficit environments over 2 years). Homozygous cipk15 mutants had a significantly steeper root growth angle compared to W22 wild‐type plants. In the greenhouse, the growth angles of second node roots of mutant plants were on average 9° steeper under high nitrogen and 15° steeper under low nitrogen compared to the wild‐type plants. The growth angles of fourth node roots were on average 7° steeper in high nitrogen and 4° steeper under low nitrogen (Figure 4c,d; Table S7). On average, nitrogen stress reduced shoot biomass in cipk15 mutant and wild‐type genotypes by 43% compared to high nitrogen conditions (Table S8).
Figure 4.
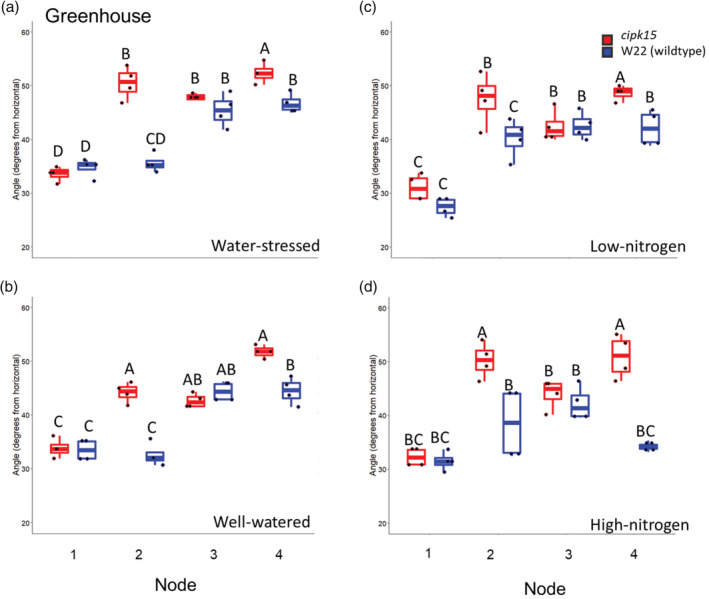
Greenhouse results comparing the root angle of the zmcipk15 mutant and wild type. zmcipk15 mutant demonstrated significantly steeper root angles at nodes two and four in (a) Water‐stressed, (b) Well‐watered, (c) Low‐nitrogen and (d) High‐nitrogen regimes. Letters above the columns show significant differences between genotypes and treatments within a node according to a Tukey's HSD (honest significant difference) test. In the boxplot, the box represents the interquartile range with the median, lower (Q1) and upper (Q3) quartile. The ‘whiskers’ represent the maximum and minimum value [Colour figure can be viewed at wileyonlinelibrary.com]
The growth angles of second node roots of mutant plants were 12° steeper in well‐watered conditions and 10° steeper in water‐stressed conditions compared to the wild‐type plants in the greenhouse. In the greenhouse, the growth angles of fourth node roots of homozygous mutant plants were on average 2° steeper in well‐watered conditions and 6° steeper in water‐stressed conditions compared to the wild‐type plants (Figure 4a,b; Table S7). On average, the water‐stress treatment reduced shoot biomass of both the cipk15 mutant and wild‐type genotypes by 51% compared to plants grown in well‐watered environments. No biomass differences were observed between the cipk15 mutant and wild‐type genotype in the water‐stress or well‐watered treatment (Table S8).
In the field in 2016, 2017 and 2019, the growth angle of mutant plants in high and low nitrogen conditions was 8° steeper in node two and 11° steeper at node three compared to wild‐type plants (Figure 5). In 2018, the effect of nitrogen stress was weak and no significant differences were observed in angle between genotypes and this data was excluded from further analysis (Figure 6c,d). Over 3 years in the field, cipk15 mutant plants had 18% greater shoot biomass in low nitrogen compared to wild‐type genotypes (Figure 7a). No difference in shoot biomass was observed between cipk15 mutant and wild‐type plants in high nitrogen (Table S8).
Figure 5.

Images of root architecture in the field. The cipk15 mutant genotype had significantly steeper angles at nodes 2 and 3 compared to the wild‐type genotype. Plants were grown in low nitrogen conditions. Scale bar represents 2.5 cm [Colour figure can be viewed at wileyonlinelibrary.com]
Figure 6.

Field study results comparing the root angle of the zmcipk15 mutant and wild type. (a) Two field studies confirm that the zmcipk15 mutant had significantly steeper angles at nodes two and three in water‐stressed conditions. (b) No significant differences in root angle were observed between the zmcipk15 and the wild type in well‐watered conditions within a node. (c) Four field studies confirm that the zmcipk15 mutant had significantly steeper angles at node two and three in low‐nitrogen conditions. (d) No significant differences in root angle were observed between the zmcipk15 and the wild type in high‐nitrogen regimes within a node. Letters above the columns show significant differences between genotypes and treatments within a node according to a Tukey's HSD (honest significant difference) test. In the boxplot, the box represents the interquartile range with the median, lower (Q1) and upper (Q3) quartile. The ‘whiskers’ represent the maximum and minimum value [Colour figure can be viewed at wileyonlinelibrary.com]
Figure 7.
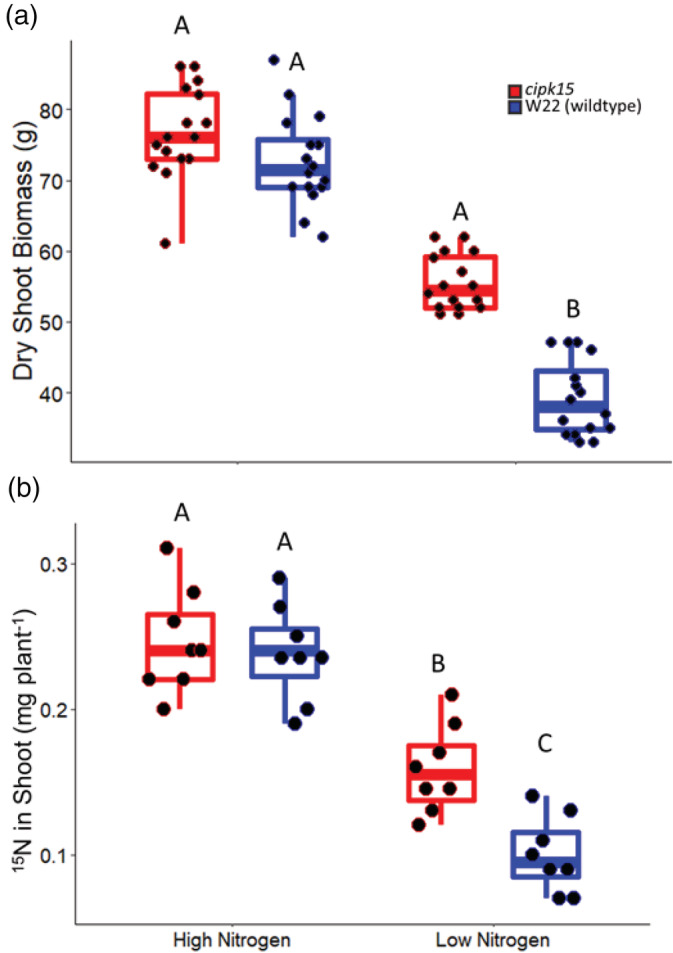
(a) In high nitrogen conditions, the cipk15 mutant and W22 wild‐type plants had no significant differences in dry shoot biomass. In low nitrogen conditions, cipk15 mutants had significantly greater dry shoot biomass when compared to W22 wild type. (b) In high nitrogen conditions, the cipk15 mutant and W22 wild‐type plants had no significant differences in deep nitrogen uptake. In low nitrogen conditions, cipk15 mutants had significantly greater deep nitrogen uptake when compared to W22 wild type. Letters above the columns show significant differences between genotypes and treatments according to a Tukey's HSD (honest significant difference) test. In the boxplot, the box represents the interquartile range with the median, lower (Q1) and upper (Q3) quartile. The ‘whiskers’ represent the maximum and minimum value [Colour figure can be viewed at wileyonlinelibrary.com]
In the field in 2017 and 2018, the growth angle of mutant plants in water‐stress conditions was 4° steeper at node two and 14° steeper at node three compared to wild‐type plants. There were no significant differences in root angle between cipk15 mutant and wild‐type plants in well‐watered conditions at any node (Figure 6a,b; Table S7). There were no significant differences in shoot biomass between cipk15 mutant and wild‐type plants in well‐watered or water‐stress conditions (Table S8). In all experiments and under all growth conditions, no differences in stem diameter, root diameter, crown root number and lateral branching density and length were observed between cipk15 mutant and wild‐type plants (Table S9). Interestingly, we did not observe any differences in ZMCIPK15 expression by semi‐quantitative RT‐PCR between the cipk15 mutant and the wild type (Figure S4).
3.5. Changes in root growth angle attributed to ZmCIPK enhances deep nitrogen capture and improves plant performance in low nitrogen in silico
OpenSimRoot simulated nitrogen uptake and plant biomass accumulation in high and low nitrogen conditions. Since the cipk15 mutants displayed different angle phenotypes in the field and greenhouse, these angle phenotypes were simulated separately and named ‘cipk15 field’ and ‘cipk15 greenhouse’, respectively. At 40 days after planting in low nitrogen, simulated plants with root growth angles representing cipk15 greenhouse plants had 8% greater nitrogen uptake and 3% greater shoot biomass compared to simulated wild‐type plants. Simulated cipk15 field plants with root growth angles reflective of field‐grown cipk15 plants had 4% greater nitrogen uptake and 11% greater biomass compared to simulated wild‐type plants. In environments with greater nitrogen availability, there was no predicted difference in nitrogen capture or plant performance between simulated plants representing cipk15 growth angles in the field and greenhouse and wild‐type plants (Figure 8). Simulation results were supported by field studies. In 2017, cipk15 mutant plants had 29% greater 15N nitrogen uptake in low nitrogen conditions compared to wild‐type genotypes (Figure 7b; Table S10).
Figure 8.
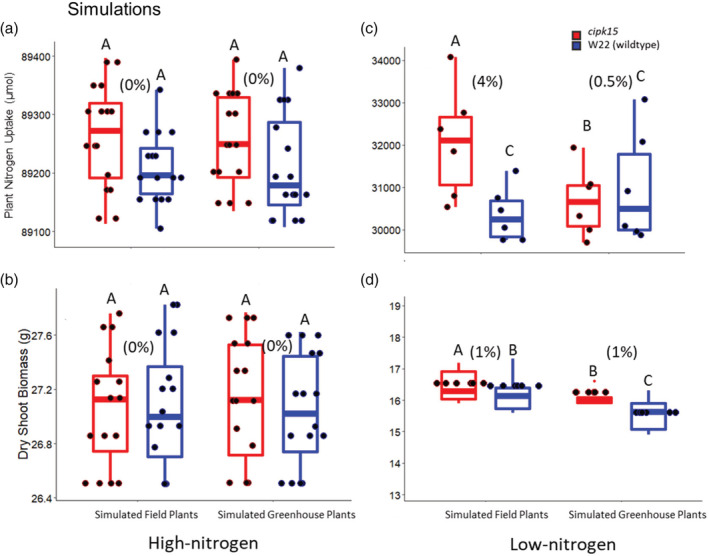
OpenSimRoot simulated nitrogen uptake and shoot biomass of the cipk15 mutant and wild‐type (WT) plant in high (a, b) and low (c, d) nitrogen conditions. Simulated field plants had root growth angles representing cipk15 and WT field‐grown plants. Simulated greenhouse plants had root growth angles representing cipk15 and WT greenhouse‐grown plants. cipk15 has greater plant nitrate uptake and greater shoot biomass in low nitrogen environments compared to the wild type (WT). Numbers in parenthesis represent the percent difference between the cipk15 and WT for each treatment. No differences in plant nitrate uptake or shoot biomass was predicted between cipk15 and WT in high nitrogen environments. Letters above the columns show significant differences between genotypes within an environment according to a Tukey's HSD (honest significant difference) test. In the boxplot, the box represents the interquartile range with the median, lower (Q1) and upper (Q3) quartile. The ‘whiskers’ represent the maximum and minimum value [Colour figure can be viewed at wileyonlinelibrary.com]
4. DISCUSSION
Our objective was to determine the role of root angle for nitrogen capture and identify key regulatory loci controlling crown root angle in maize employing a high‐throughput field phenotyping and genome wide association mapping approach. We observed large variation in crown root growth angle and identified novel genomic regions and significant SNPs controlling crown root angle in maize. A significant SNP detected in GWAS was located in the CBL‐interacting serine/threonine‐protein kinase 15 gene. Functional evidence for a CBL‐interacting serine/threonine‐protein kinase 15 as a regulator controlling root angle in maize was provided by phenotypic changes in an insertional mutant phenotype (Figure 5; Figure S1). Changes in angle attributed to this gene resulted in functional differences in nitrogen capture and plant performance in silico and in the field (Figures 4, 5, 6, 7, 8).
Root angle has important implications for soil resource capture. In nutrient and water stress conditions, root angle influenced root depth, and therefore, plant performance (Bonser et al., 1996; Dathe et al., 2016; Trachsel et al., 2013; Uga et al., 2011; York et al., 2013). Steep growth angles enable deeper rooting and the capture of mobile soil resources, including nitrogen and water, in deep soil domains (Dathe et al., 2016; Trachsel et al., 2013). Root phenotypes that explore deep soil domains enhance the capture of deep resources like water and nitrogen in most agricultural systems, since these resources are often more available at depth later in the season, when plant demand is greatest (Gowda et al., 2011; Henry et al., 2011; Manschadi et al., 2006). Identifying the genetic control and functional significance of genes controlling root angle will aid in developing crop varieties with increased root depth and deep soil resource capture.
In many maize genotypes, nodal root growth angles are steeper in younger nodes and shallow in older nodes (Feldman, 1994; York & Lynch, 2015). The production of crown roots with progressively steeper growth angles results in a root architecture that it is initially shallow, which coincides with the availability of water, nitrogen and phosphorus in the topsoil during seedling establishment. The emergence of roots with progressively steeper growth angles over time coincides with the availability of water and nitrogen in deeper soil strata as the season progresses (Lynch, 2018). Root phenotypes that explore deep soil domains enhance the capture of mobile soil resources located in deep soil domains later in the growth season (Gowda et al., 2011; Henry et al., 2011; Manschadi et al., 2006). In addition, throughout the growth season, the continual development of crown and brace roots from stem nodes and tillers in maize enable the exploration of shallow soil and capture of shallow soil resources, like phosphorus, an immobile soil nutrient and water from intermittent rainfall, despite a steep growth angle.
Crown root angle is important for plant stress tolerance, however, root angle and other root phenes do not function in isolation (Miguel et al., 2015; York et al., 2013). Phene synergisms exist between those that reduce the metabolic cost of the root and those that affect the placement of roots in the soil domain. In bean, basal root growth angle interacts with root hair density and length to determine the placement of root hairs in the soil profile and increase plant growth up to twice the expected additive effects (Miguel et al., 2015; York et al., 2013). In common bean, plants with a shallow growth angle and few nodes of basal roots had significantly greater nitrogen uptake than plants with a shallow growth angle and many nodes of basal roots (Rangarajan et al., 2018). Maize crown root angle would be expected to interact with other phenes such as crown root number and anatomy to regulate acquisition of limiting resources. Maize root nodes are under distinct genetic control. Recent studies have reported a weak relationship among phenes of roots on different nodal positions (Colombi et al., 2015; Yang et al., 2019; York & Lynch, 2015). Phenotypes of young crown roots emerging continually throughout the growth season on older plants may not reflect root phenotypes of older crown roots or embryonic roots. For example, root angle varies by node, and in some studies nodal root growth angles were steeper in younger nodes (York & Lynch, 2015). Our study suggests that roots originating from different nodes may be under distinct genetic control. Phenotyping root features on individual nodes are essential for understanding the genetic control and functional utility of root phenes. In maize, the variation in root phenes within the root system is difficult to study because the outer, younger nodes occlude the older roots in the interior of the crown. Imaging of intact, mature root crowns prevents measurements of the older, obstructed part of the root system (Trachsel et al., 2011, 2013; York & Lynch, 2015). Measuring nodal root growth angle on only the visible younger nodes is not a reliable way to predict the overall pattern of growth angles among nodes (York & Lynch, 2015). Several methods have been used to measure root phenotypes on individual nodes, including destructive node‐by‐node phenotyping (York & Lynch, 2015) and imaging root crowns split lengthwise in half (Colombi et al., 2015). Root originating from old nodes would be expected to contribute more to soil resource acquisition later in development, as these roots are located deeper in the soil profile where nitrogen and water availability are greatest.
The growth system and environment have implications for the interpretation of the utility of root phenotypes and their application in plant breeding. With a few exceptions (e.g., (Schneider et al. 2020a, 2020b; Zheng et al., 2019), the majority of QTL and GWAS studies on roots use artificial growth systems that do not represent field conditions. Studies performed in artificial systems cannot adequately predict root phenotypes and their relationship to nutrient uptake in the field (e.g., [Nestler et al., 2016]). In the current study, cipk15 mutants grown in the greenhouse and field had differences in root angles at individual nodal positions. In the greenhouse, cipk15 mutants had significantly steeper angles at nodes two and four, with the strongest angle differences in node two in high nitrogen, low nitrogen, drought and well‐watered treatments (Figure 4). In the field, the cipk15 mutants had significantly steeper roots at nodes two and three drought and low nitrogen environments (Figure 6). These small differences in angle expression could be attributed to differences in the growth environment, the spatiotemporal location of nitrogen in the soil and differences in soil bulk density between the greenhouse and field.
In the current study, differences in the growth angle of cipk15 mutants in nitrogen stress in the field resulted in greater deep nitrogen acquisition and greater plant biomass in low nitrogen conditions compared to the wild type. In contrast, differences in the growth angle of cipk15 mutants in drought did not translate to improved plant water status or plant biomass compared to wild‐type plants. No growth differences were observed between cipk15 mutant and wild‐type plants in the greenhouse under any treatment. Steeper nodal root growth angles are beneficial in many agricultural environments for the capture of mobile soil resources located deep in the soil profile, like water and nitrogen. Greenhouse mesocosm studies do not replicate the distribution of nitrogen in the soil profile as nitrogen is not leached into deep soil throughout growth, rather the plants receive low levels of nitrogen through regular fertigation. In addition, drip irrigation regimes in the greenhouse and field may not represent the natural spatiotemporal distribution of water in the soil profile, and therefore, may not provide a resource advantage for deep roots. We propose that a steeper root growth angle in drought enables enhanced capture of deep water in drought environments, especially in terminal drought scenarios.
No differences in root angle were observed between the cipk15 mutant and the wild‐type plant in the field under non‐stress conditions. Edaphic stress, including drought and low nitrogen (Oyanagi et al., 1993; Trachsel et al., 2013) have been shown to alter root angle. Root angle is a plastic phene that is responsive to the environment (Huang et al., 2018; Schneider, Klein, Hanlon, Nord, et al., 2020b). The plastic response of a steeper angle in nitrogen stress may be an adaptive strategy for increased deep nitrogen capture (Trachsel et al., 2013).
Here, we identify CIPK as an important gene regulating crown root growth angle in maize. Lines overexpressing AtCIPK23 demonstrated enhanced drought stress tolerance in tobacco (Lu et al., 2018). Calcium signals have known roles in mediating a variety of plant responses to external stimuli and physiological processes (Harper, 2001; Knight & Knight, 2001). Calcium‐binding proteins, including calcineurin B‐like (CBL) proteins, are important relays in calcium signalling in the plants (Kolukisaoglu et al., 2014). CIPKs are targets of calcium signals sensed and transduced by CBL proteins and have important implications in abiotic stress tolerance (Kolukisaoglu et al., 2014). AtCIPK4 a homolog for ZmCIPK15, has been shown to be down‐regulated in drought and cold stress (Kanwar et al., 2014). In addition, the CBL‐CIPK network has been demonstrated to be involved in nutrient signalling responses, including differential expression in response to low potassium availability (Luan, 2009), low nitrogen availability (Hu et al., 2009), drought, heat stress and cold stress (Chen et al., 2011). Cytosolic calcium levels have been shown to be an important regulator in root bending and angle establishment (Monshausen et al., 2009). In addition, CIPK23 in Arabidopsis has implications in ammonium transport (Straub et al., 2017) and its expression is transiently induced by nitrate availability (Ho et al., 2009). In maize, there are 43 putative ZmCIPK genes in the B73 inbred line (Chen et al., 2011). Therefore, ZmCIPK15 was selected as a target gene controlling maize root angle.
The mapped SNP positions highlight the putative effect of the SNP on the function of the CIPK protein in maize. For example, both the S61A and L63V substitutions (SNPs rs1_258846443 and rs1_258846449, respectively) are located in a hydrophobic pocket near the nucleotide binding site of the CIPK kinase domain (Figure 3). Since S61 is located on the surface of the kinase domain, a polar to hydrophobic substitution here and the shorter valine side chain compared to that of leucine in the L63V substitution, may result in a slight destabilization of this hydrophobic pocket and affect the binding of ATP to the kinase domain. Similarly, V213 lies close to the active site of the kinase (Figure 3b,c) and even a conservative amino acid substitution from V to I (SNP rs1_258846899) at this site could modify the activity of the kinase due to the difference in the size of the side chains in V and I. Next, both the 271AA (SNP rs1_258847073) and 286AA (SNP rs1_258847118) positions lie at the interdomain boundary of the catalytic and regulatory domains of CIPK (Figure 3) and could possibly contribute to the interface interactions between these domains. G to S substitution at these positions due to the SNPs is a non‐conservative replacement, which often leads to more pronounced effects on protein function than conservative replacements such as L to V. Moreover, S could be a potential site of post‐translation phosphorylation, leading to an additional layer of regulatory control of the minor allele with the G to S substitution. Proline residues are generally present at sites where the protein backbone needs to turn, such as the turns between beta strands and thus the P12S substitution (SNP rs1_258846296) could thus disrupt the folding or stability of the N‐terminal half of CIPK. In addition, all of these SNPs have significant differences in crown root angle distributions between the minor and major alleles (Table 1). However, these conclusions require more extensive biochemical studies to validate the role of the SNPs and the missense substitutions on CIPK structure and function in vitro and in vivo.
Gene expression may not always be a good predictor of causative genetic loci. Here, we did not observe any differences in ZmCIPK15 expression by semi‐quantitative RT‐PCR between the cipk15 mutant and the wild type (Figure S4). Expression differences are not necessarily expected and an aberrant protein with a premature stop due to a transposon insertion can result in a clear phenotype even if expression is not compromised. Mu insertions have been shown to cause ectopic expression of genes with an insertion. The terminal inverted repeats (TIR) of the Mu transposon may have cryptic promoter activity, resulting in ectopic expression. This phenomenon was observed in the maize hcf106 Mu transposon allele where the mutant still produced Hcf106 transcripts (Barkan & Martienssen, 1991). In addition, observation of ectopic expression of Lg3 in an lg3 Mu insertion allele has also been reported (Girard & Freeling, 2000).
The ability of plants with a steeper root growth angle to acquire nitrogen from deep soil domains and improve plant growth in low nitrogen environments has important implications for the development of more productive crop cultivars. A relatively small change in root growth angle of specific nodes, regulated by one gene, has large implications in nitrogen uptake and plant performance. ZmCIPK15 regulates a approximately 10° change in two root‐bearing nodes, which resulted in 18% greater shoot biomass and 29% nitrogen uptake in the field. Recent studies have demonstrated that improved crop productivity is associated with root phenotypes. Over the past century, U.S. commercial maize lines have developed shallower axial root growth angles, and other architectural and anatomical changes that are associated with increased N efficiency (York et al., 2015).
Breeding programmes focused on superior root phenotypes have the potential to improve plant performance in edaphic stresses for a variety of crops (Jung & McCouch, 2013; Lynch, 2013, 2018, 2019; Wasson et al., 2012; York et al., 2013), but conventional breeding programmes typically do not focus on root phenotypes due to the limitations and challenges of root phenotyping and the complex influence of the environment on plant growth (Lobet et al., 2019; Tuberosa et al., 2003). However, detection and identification of genetic loci associated with root architectural phenotypes may assist the breeding of crops more tolerant to a variety of edaphic stresses (de Dorlodot et al., 2007). During vegetative and reproductive phases of maize growth, crown roots are the most important part of the root system for soil resource acquisition (DeBruin et al., 2017; Lynch, 2013). However, until recently, few methods have existed to identify and characterize root architectural phenotypes quickly and efficiently, which has been a limitation in the identification genetic loci associated with root phenotypes. Advancements in high‐throughput root phenotyping enable focused efforts to phenotype the distribution and magnitude of root phenes in many different plant species. The development of the ‘shovelomics’ method has improved our ability to visualize and quantify root architecture and its relationship to plant productivity. In addition, technologies such as imaging‐based, high‐throughput protocols for phenotyping field‐grown plants can increase both the speed and reproducibility of the trait estimation pipeline (Bucksch et al., 2014; Das et al., 2015). A major bottleneck to GWAS studies is identifying relevant candidate genes associated with the trait of interest. Many genes are not annotated, which poses a challenge to the relevance and selection of candidate genes for follow‐up studies. There is a clear need for the verification and confirmation of candidate gene functions as they have a greater utility in additional functional studies compared to SNPs or other markers.
In this project, we report a gene regulating maize crown root angle in the field. This genomic region of interest includes CIPK15, which we demonstrate to regulate crown root angles in the greenhouse and field. Steeper crown roots improve water and nitrogen capture; therefore, crown root angle is a promising target for selection in breeding programmes aiming to improve crop resilience to edaphic stresses.
CONFLICT OF INTEREST
The authors have no conflict of interest to report.
Supporting information
Table S1. List of inbred maize genotypes in the Wisconsin Diversity (WiDiv) Panel
Table S2. Nutrient solutions used in mesocosm experiments
Table S3. Overlapping genotypes between the Wisconsin Diversity Panel and SNP set
Table S4. Root angle phenotypes of the Wisconsin Diversity Panel
Table S5. Mis‐sense mutants of SNPs located in the coding region
Table S6. Co‐occurring SNPs and individually occurring SNPs in genotypes with steep angles
Table S7. cipk15 mutant and wild‐type angles in the field and greenhouse. Data presented are raw data per plot in the field or per individual greenhouse mesocosm
Table S8. Dry shoot biomass of cipk15 mutant and wild type in field and greenhouse experiments. Data presented are raw data per plot in the field or per individual greenhouse mesocosm
Table S9. cipk15 mutant and wild‐type architectural phenotypes in the field and greenhouse. Data presented are raw data per plot in the field or per individual greenhouse mesocosm
Table S10. 15N uptake data from the field. Data presented are raw data per plot in the field
Figure S1. Linkage disequilibrium heatmaps displaying Zm00001d033316 in a 100 Kb window upstream and downstream
Figure S2. SNPs resulting in mis‐sense amino acid substitutions in ZmCIPK and their associated crown root angle phenotypes
Figure S3. Identification of mu1046464::Mu insertion position
Figure S4. ZmCIPK15 expression by semi‐quantitative RT‐PCR between the cipk15 mutant and the wild type
ACKNOWLEDGEMENTS
We thank Bob Snyder, Curtis Frederick and Johan Prinsloo for technical support. This research was supported by the Howard G. Buffet Foundation, USDOE ARPA‐E ROOTS Award Number DE‐AR0000821 and the USDA National Institute of Food and Agriculture Federal Appropriations under Project PEN04732. ANB is supported by Anne McLaren Fellowship from the University of Nottingham. RB is supported by BBSRC Discovery and Future Food Beacon Nottingham Research Fellowships.
Schneider, H. M. , Lor, V. S. N. , Hanlon, M. T. , Perkins, A. , Kaeppler, S. M. , Borkar, A. N. , Bhosale, R. , Zhang, X. , Rodriguez, J. , Bucksch, A. , Bennett, M. J. , Brown, K. M. , & Lynch, J. P. (2022). Root angle in maize influences nitrogen capture and is regulated by calcineurin B‐like protein (CBL)‐interacting serine/threonine‐protein kinase 15 ( ZmCIPK15 ). Plant, Cell & Environment, 45, 837–853. 10.1111/pce.14135
Funding information Anne McLaren Fellowship; BBSRC Discovery and Future Food Beacon Nottingham Research Fellowship; Howard G Buffett Foundation; US Department of Energy ARPA‐e ROOTS, Grant/Award Number: DE‐AR0000821; USDA National Institute of Food and Agriculture Federal Appropriations, Grant/Award Number: PEN04732
DATA AVAILABILITY STATEMENT
All data are available upon request to the corresponding author.
REFERENCES
- Arai‐sanoh, Y. , Takai, T. , Yoshinaga, S. , Nakano, H. , Kojima, M. , Sakakibara, H. , … Uga, Y. (2014). Deep rooting conferred by DEEPER ROOTING 1 enhances rice yield in paddy fields. Scientific Reports, 4, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan, A. , & Martienssen, R. (1991). Inactivation of maize transposon mu suppresses a mutant phenotype by activating an outward‐reading promoter near the end of Mu1. Proceedings of the National Academy of Sciences, 88, 3502–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates, D., Carey, V., Dettling, M., Dudoit, S., Ellis, B., Gautier, L., … Huber, W. (2002) Bioconductor. Retrieved from http://www.bioconductor.org.
- Bayuelo‐Jiménez, J. S. , Gallardo‐Valdéz, M. , Pérez‐Decelis, V. A. , Magdaleno‐Armas, L. , Ochoa, I. , & Lynch, J. P. (2011). Genotypic variation for root traits of maize (Zea mays L.) from the Purhepecha plateau under contrasting phosphorus availability. Field Crops Research, 121, 350–362. [Google Scholar]
- Bonser, A. , Lynch, J. , & Snapp, S. (1996). Effect of phosphorus deficiency on growth angle of basal roots in Phaseolus vulgaris . New Phytologist, 132, 281–288. [DOI] [PubMed] [Google Scholar]
- Bradbury, P. J. , Zhang, Z. , Kroon, D. E. , Casstevens, T. M. , Ramdoss, Y. , & Buckler, E. S. (2007). TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics, 23, 2633–2635. [DOI] [PubMed] [Google Scholar]
- Bucksch, A. , Burridge, J. , York, L. M. , Das, A. , Nord, E. , Weitz, J. S. , & Lynch, J. P. (2014). Image‐based high‐throughput field phenotyping of crop roots. Plant Physiology, 166, 470–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos, H. , Cooper, M. , Habben, J. E. , Edmeades, G. O. , & Schussler, J. R. (2004). Improving drought tolerance in maize: A view from industry. Field Crops Research, 90, 19–34. [Google Scholar]
- Chen, X. , Gu, Z. , Xin, D. , Hao, L. , Liu, C. , Huang, J. , … Zhang, H. (2011). Identification and characterization of putative CIPK genes in maize. Journal of Genetics and Genomics, 38, 77–87. [DOI] [PubMed] [Google Scholar]
- Colombi, T. , Kirchgessner, N. , Andrée, C. , Marié, L. , York, L. M. , Lynch, J. P. , & Hund, A. (2015). Next generation shovelomics: Set up a tent and REST. Plant and Soil, 388, 1–20. [Google Scholar]
- Das, A. , Schneider, H. , Burridge, J. , Ascanio, A. K. M. , Wojciechowski, T. , Topp, C. N. , … Bucksch, A. (2015). DIRT: A high‐throughput computing and collaboration platform for field‐based plant phenomics. Plant Methods, 11, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dathe, A. , Postma, J. , Postma‐Blaauw, M. , & Lynch, J. (2016). Impact of axial root growth angles on nitrogen acquisition in maize depends on environmental conditions. Annals of Botany, 118, 401–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Dorlodot, S. , Forster, B. , Pagès, L. , Price, A. , Tuberosa, R. , & Draye, X. (2007). Root system architecture: Opportunities and constraints for genetic improvement of crops. Trends in Plant Science, 12, 474–481. [DOI] [PubMed] [Google Scholar]
- DeBruin, J. L. , Schussler, J. R. , Mo, H. , & Cooper, M. (2017). Grain yield and nitrogen accumulation in maize hybrids released during 1934 to 2013 in the US Midwest. Crop Science, 57, 1431–1446. [Google Scholar]
- Feldman L. (1994) The maize root. In: Freeling M, Walbot V, eds. In The maize handbook. (eds Freeling M. & Walbot V.), pp. 29–37. New York, NY: Springer; [Google Scholar]
- Forde, B. G. (2009). Is it good noise? The role of developmental instability in the shaping of a root system. Journal of Experimental Botany, 60, 3989–4002. [DOI] [PubMed] [Google Scholar]
- Girard, L. , & Freeling, M. (2000). Mutator‐suppressible alleles of rough sheath1 and liguleless2 in maize reveal multiple mechanisms for suppression. Genetics, 154, 437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri, J. , Bhosale, R. , Huang, G. , Pandey, B. K. , Parker, H. , Zappala, S. , … Bennett, M. J. (2018). Rice auxin influx carrier OsAUX1 facilitates root hair elongation in response to low external phosphate. Nature Communications, 9, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliani, S. , Sanguineti, M. C. , Tuberosa, R. , Bellotti, M. , Salvi, S. , & Landi, P. (2005). Root‐ABA1, a major constitutive QTL, affects maize root architecture and leaf ABA concentration at different water regimes. Journal of Experimental Botany, 56, 3061–3070. [DOI] [PubMed] [Google Scholar]
- Gowda, V. R. P. , Henry, A. , Yamauchi, A. , Shashidhar, H. E. , & Serraj, R. (2011). Root biology and genetic improvement for drought avoidance in rice. Field Crops Research, 122, 1–13. [Google Scholar]
- Guingo, E. , Yannick, H. , & Charcosset, A. (1998). Genetic analysis of root traits in maize. Agronomie, 18, 225–235. [Google Scholar]
- Hammer, G. L. , Dong, Z. , McLean, G. , Doherty, A. , Messina, C. , Schussler, J. , … Cooper, M. (2009). Can changes in canopy and/or root system architecture explain historical maize yield trends in the U.S. corn belt. Crop Science, 49, 299. [Google Scholar]
- Hansey, C. N. , Johnson, J. M. , Sekhon, R. S. , Kaeppler, S. M. , & de Leon, N. (2011). Genetic diversity of a maize association population with restricted phenology. Crop Science, 51, 704–715. [Google Scholar]
- Hargreaves, C. E. , Gregory, P. J. , & Bengough, A. G. (2008). Measuring root traits in barley (Hordeum vulgare ssp. vulgare and ssp. spontaneum) seedlings using gel chambers, soil sacs and X‐ray microtomography. Plant and Soil, 316, 285–297. [Google Scholar]
- Harper, J. F. (2001). Dissecting calcium oscillators in plant cells. Trends in Plant Science, 6, 395–397. [DOI] [PubMed] [Google Scholar]
- Henry, A. , Gowda, V. R. P. , Torres, R. O. , Mcnally, K. L. , & Serraj, R. (2011). Variation in root system architecture and drought response in rice (Oryza sativa): Phenotyping of the OryzaSNP panel in rainfed lowland fields. Field Crops Research, 120, 205–214. [Google Scholar]
- Hirel, B. , Le Gouis, J. , Ney, B. , & Gallais, A. (2007). The challenge of improving nitrogen use efficiency in crop plants: Towards a more central role for genetic variability and quantitative genetics within integrated approaches. Journal of Experimental Botany, 58, 2369–2387. [DOI] [PubMed] [Google Scholar]
- Ho, C. , Lin, S. , Hu, H. , & Tsay, Y. (2009). CHL1 functions as a nitrate sensor in plants. Cell, 138, 1184–1194. [DOI] [PubMed] [Google Scholar]
- Hochholdinger, F. , Woll, K. , Sauer, M. , & Dembinsky, D. (2004). Genetic dissection of root formation in maize (Zea mays) reveals root‐type specific developmental programmes. Annals of Botany, 93, 359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe, D. C. , McCully, M. E. , & Wenzel, C. L. (1986). The nodal roots of Zea: Their development in relation to structural features of the stem. Canadian Journal of Botany, 64, 2524–2537. [Google Scholar]
- Hu, H. C. , Wang, Y. Y. , & Tsay, Y. F. (2009). AtCIPK8, a CBL‐interacting protein kinase, regulates the low‐affinity phase of the primary nitrate response. Plant Journal, 57, 264–278. [DOI] [PubMed] [Google Scholar]
- Huang, G. , Liang, W. , Sturrock, C. J. , Pandey, B. K. , Giri, J. , Mairhofer, S. , … Zhang, D. (2018). Rice Actin binding protein RMD controls crown root angle in response to external phosphate. Nature Communications, 9(1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, Y. , Peluso, P. , Shi, J. , Liang, T. , Stitzer, M. C. , Wang, B. , … Ware, D. (2017). Improved maize reference genome with single‐molecule technologies. Nature, 546, 524–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing, Z. , Jiabing, F. U. , Honq, A. O. , Yong, H. E. , Hai, N. , Yueming, H. U. , … Xiaolong, Y. A. N. (2004). Characterization of root architecture in an applied core collection for phosphorus efficiency of soybean germplasm, 49, 1611–1620. [Google Scholar]
- Jung, J. K. H. , & McCouch, S. (2013). Getting to the roots of it: Genetic and hormonal control of root architecture. Frontiers in Plant Science, 4, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwar, P. , Sanyal, S. K. , Tokas, I. , Yadav, A. K. , Pandey, A. , Kapoor, S. , & Pandey, G. K. (2014). Comprehensive structural, interaction and expression analysis of CBL and CIPK complement during abiotic stresses and development in rice. Cell Calcium, 56, 81–95. [DOI] [PubMed] [Google Scholar]
- Kato, Y. , Abe, J. , Kamoshita, A. , & Yamagishi, J. (2006). Genotypic variation in root growth angle in rice (Oryza sativa L.) and its association with deep root development in upland fields with different water regimes. Plant and Soil, 287, 117–129. [Google Scholar]
- Kell, D. B. (2011). Breeding crop plants with deep roots: Their role in sustainable carbon, nutrient and water sequestration. Annals of Botany, 108, 407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. A. , Brossard, M. , Roshandel, D. , Paterson, A. D. , Bull, S. B. , & Yoo, Y. J. (2019). Gpart: Human genome partitioning and visualization of high‐density SNP data by identifying haplotype blocks. Bioinformatics, 35, 4419–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, H. , & Knight, M. R. (2001). Abiotic stress signalling pathways: Specificity and cross‐talk. Trends in Plant Science, 6, 262–267. [DOI] [PubMed] [Google Scholar]
- Kolukisaoglu, U. , Weinl, S. , Blazevic, D. , Batistic, O. , Kulda, J. , Kudla, J. , & Kulda, J. (2014). Calcium sensors and their interacting protein kinases: Genomics of the arabidopsis and rice CBL‐CIPK signaling networks. Genome Analysis, 134, 43–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, C. J. (2005). The maize genetics and genomics database. The community resource for access to diverse maize data. Plant Physiology, 138, 55–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, H. , Rubio, G. , Yan, X. , Cao, A. , Brown, K. M. , & Lynch, J. P. (2001). Effect of phosphorus availability on basal root shallowness in common bean. Plant and Soil, 232, 69–79. [PubMed] [Google Scholar]
- Liao, H. , Yan, X. , Rubio, G. , Beebe, S. E. , Blair, M. W. , & Lynch, J. P. (2004). Genetic mapping of basal root gravitropism and phosphorus acquisition efficiency in common bean. Functional Plant Biology, 31, 959–970. [DOI] [PubMed] [Google Scholar]
- Liu, X. , Huang, M. , Fan, B. , Buckler, E. , & Zhang, Z. (2016). Iterative usage of fixed and random effect models for powerful and efficient genome‐wide association studies. PLoS Genetics, 12, e1005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobet, G. , Paez‐Garcia, A. , Schneider, H. , Junker, A. , Atkinson, J. A. , & Tracy, S. (2019). Demystifying roots: A need for clarification and extended concepts in root phenotyping. Plant Science, 282, 11–13. [DOI] [PubMed] [Google Scholar]
- Lu, L. , Yang, L. , Li, K. , Lu, Y. , & Li, L. (2018). Ectopic expression of AtCIPK23 enhances drought tolerance via accumulating less H2O2 in transgenic tobacco plants. Genetics and Molecular Research, 17, 1. [Google Scholar]
- Luan, S. (2009). The CBL‐CIPK network in plant calcium signaling. Trends in Plant Science, 14, 37–42. [DOI] [PubMed] [Google Scholar]
- Lynch, J. (2019). Root phenotypes for improved nutrient capture: An underexploited opportunity for global agriculture. New Phytologist, 223, 548–564. [DOI] [PubMed] [Google Scholar]
- Lynch, J. , & Brown, K. (2001). Topsoil foraging—An architectural adaptation of plants to low phosphorus availability. Plant and Soil, 237, 225–237. [Google Scholar]
- Lynch, J. P. (1995). Root architecture and plant productivity. Plant Physiology, 109, 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, J. P. (2013). Steep, cheap and deep: An ideotype to optimize water and N acquisition by maize root systems. Annals of Botany, 112, 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, J. P. (2018). Rightsizing root phenotypes for drought resistance. Journal of Experimental Botany, 69, 3279–3292. [DOI] [PubMed] [Google Scholar]
- Lynch, J. P. , & Brown, K. M. (2012). New roots for agriculture: Exploiting the root phenome. Philosophical Transactions of the Royal Society Series B, 367, 1598–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, J. P. , Burton, A. L. , Brown, K. M. , & Lynch, J. P. (2011). Root phenes for enhanced soil exploration and phosphorus acquisition: Tools for future crops. Plant Physiology, 156, 1041–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, J. P. , & Wojciechowski, T. (2015). Opportunities and challenges in the subsoil: Pathways to deeper rooted crops. Journal of Experimental Botany, 66, 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace, E. S. , Singh, V. , Van Oosterom, E. J. , Hammer, G. L. , Hunt, C. H. , & Jordan, D. R. (2012). QTL for nodal root angle in sorghum (Sorghum bicolor L. Moench) co‐locate with QTL for traits associated with drought adaptation. Theoretical and Applied Genetics, 124, 97–109. [DOI] [PubMed] [Google Scholar]
- Manschadi, A. M. , Christopher, J. , deVoil, P. , & Hammer, G. L. (2006). The role of root architectural traits in adaptation of wheat to water‐limited environments. Functional Plant Biology, 33, 823–837. [DOI] [PubMed] [Google Scholar]
- Manschadi, A. M. , Hammer, G. L. , Christopher, J. T. , & DeVoil, P. (2008). Genotypic variation in seedling root architectural traits and implications for drought adaptation in wheat (Triticum aestivum L.). Plant and Soil, 303, 115–129. [Google Scholar]
- Mazaheri, M. , Heckwolf, M. , Vaillancourt, B. , Gage, J. L. , Burdo, B. , Heckwolf, S. , … Kaeppler, S. M. (2019). Genome‐wide association analysis of stalk biomass and anatomical traits in maize. BMC Plant Biology, 19, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty, D. R. , Settles, A. M. , Suzuki, M. , Tan, B. C. , Latshaw, S. , Porch, T. , … Hannah, L. C. (2005). Steady‐state transposon mutagenesis in inbred maize. The Plant Journal, 44, 52–61. [DOI] [PubMed] [Google Scholar]
- Miguel, M. A. , Postma, J. A. , & Lynch, J. (2015). Phene synergism between root hair length and basal root growth angle for phosphorus acquisition. Plant Physiology, 167, 1430–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshausen, G. B. , Bibikova, T. N. , Weisenseel, M. H. , & Gilroy, S. (2009). Ca2+ regulates reactive oxygen species production and pH during mechanosensing in arabidopsis roots. Plant Cell, 21, 2341–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto, T. , Shimoda, K. , & Matsuzaki, A. (1991). Elongation angle of nodal roots and its possible relation to spatial root distribution in maize and foxtail millet. Japanese Journal of Crop Science, 60, 543–549. [Google Scholar]
- Nestler, J. , Keyes, S. D. , & Wissuwa, M. (2016). Root hair formation in rice (Oryza sativa L.) differs between root types and is altered in artificial growth conditions. Journal of Experimental Botany, 67, 3699–3708. [DOI] [PubMed] [Google Scholar]
- Norton, G. J. , & Price, A. H. (2009). Mapping of quantitative trait loci for seminal root morphology and gravitropic response in rice. Euphytica, 166, 229–237. [Google Scholar]
- Omori, F. , & Mano, Y. (2007). QTL mapping of root angle in F2 populations from maize B73 x teosinte Zea luxurians . Plant Root, 1, 57–65. [Google Scholar]
- Oyanagi, A. (1994). Gravitropic response growth angle and vertical distribution of roots of wheat (Triticum aestivum L.). Plant and Soil, 165, 323–326. [Google Scholar]
- Oyanagi, A. , Nakamoto, T. , & Morita, S. (1993). The gravitropic response of roots and the shaping of the root system in cereal plants. Environmental and Experimental Botany, 33, 141–158. [Google Scholar]
- Peñagaricano, F. , Weigel, K. A. , Rosa, G. J. M. , & Khatib, H. (2012). Inferring quantitative trait pathways associated with bull fertility from a genome‐wide association study. Frontiers in Genetics, 3, 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma, J. A. , Kuppe, C. , Owen, M. R. , Mellor, N. , Griffiths, M. , Bennett, M. J. , … Watt, M. (2017). OpenSimRoot: Widening the scope and application of root architectural models. New Phytologist, 215, 1274–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2018). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Version 3.3.1. URL https://www.R-project.org/. [Google Scholar]
- Rangarajan, H. , Postma, J. A. , & Lynch, J. J. P. (2018). Co‐optimisation of axial root phenotypes for nitrogen and phosphorus acquisition in common bean. Annals Botany, 122, 485–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, H. , Klein, S. , Hanlon, M. , Brown, K. , Kaeppler, S. , & Lynch, J. (2020a). Genetic control of root anatomical plasticity in maize. The Plant Genome, 13, e20003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, H. , Klein, S. , Hanlon, M. , Nord, E. , Kaeppler, S. , Brown, K. , … Lynch, J. (2020b). Genetic control of root architectural plasticity in maize. Journal of Experimental Botany, 71, 3185–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serebrovsky, A. S. (1925). “Somatic segregation” in domestic fowl. Journal of Genetics, 16, 33–42. [Google Scholar]
- Settles, A. M. , Holding, D. R. , Tan, B. C. , Latshaw, S. P. , Liu, J. , Suzuki, M. , … McCarty, D. R. (2007). Sequence‐indexed mutations in maize using the UniformMu transposon‐tagging population. BMC Genomics, 8, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šimůnek J., Huang K. & van Genuchten M.T. (1995) The SWMS_3D code for simulating water flow and solute transport in three‐dimensional variably saturated media. Research Report No. 139, US Salinity Laboratory, USDA, California.
- Singh, V. , Oosterom, E. J. , Jordan, D. R. , Messina, C. D. , Cooper, M. , & Hammer, G. L. (2010). Morphological and architectural development of root systems in sorghum and maize. Plant and Soil, 333, 287–299. [Google Scholar]
- Stelpflug, S. C. , Rajandeep, S. , Vaillancourt, B. , Hirsch, C. N. , Buell, C. R. , de Leon, N. , & Kaeppler, S. M. (2015). An expanded maize gene expression atlas based on RNA‐sequencing and its use to explore root development. The Plant Genome, 9, 314–362. [DOI] [PubMed] [Google Scholar]
- Straub, T. , Ludewig, U. , & Neuhäuser, B. (2017). The kinase CIPK23 inhibits ammonium transport in Arabidopsis thaliana . Plant Cell, 29, 409–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachsel, S. , Kaeppler, S. M. , Brown, K. M. , & Lynch, J. P. (2011). Shovelomics: High throughput phenotyping of maize (Zea mays L.) root architecture in the field. Plant and Soil, 341, 75–87. [Google Scholar]
- Trachsel, S. , Kaeppler, S. M. , Brown, K. M. , & Lynch, J. P. (2013). Maize root growth angles become steeper under low N conditions. Field Crops Research, 140, 18–31. [Google Scholar]
- Tuberosa, R. , Salvi, S. , Sanguineti, M. C. , Maccaferri, M. , Giuliani, S. , & Landi, P. (2003). Searching for quantitative trait loci controlling root traits in maize: A critical appraisal. Plant and Soil, 255, 35–54. [Google Scholar]
- Uga, Y. , Okuno, K. , & Yano, M. (2011). Dro1, a major QTL involved in deep rooting of rice under upland field conditions. Journal of Experimental Botany, 62, 2485–2494. [DOI] [PubMed] [Google Scholar]
- Uga, Y. , Sugimoto, K. , Ogawa, S. , Rane, J. , Ishitani, M. , Hara, N. , … Yano, M. (2013). Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nature Genetics, 45, 1097–1102. [DOI] [PubMed] [Google Scholar]
- Vidal, E. A. , Tamayo, K. P. , & Gutierrez, R. A. (2010). Gene networks for nitrogen sensing, signaling, and response in Arabidopsis thaliana. Wiley interdisciplinary reviews . Systems Biology and Medicine, 2, 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson, A. P. , Richards, R. A. , Chatrath, R. , Misra, S. C. , Prasad, S. V. S. S. , Rebetzke, G. J. , … Watt, M. (2012). Traits and selection strategies to improve root systems and water uptake in water‐limited wheat crops. Journal of Experimental Botany, 63, 3485–3498. [DOI] [PubMed] [Google Scholar]
- Wataru, T. , Inanaga, S. , Araki, H. , Morita, S. , An, P. , & Sonobe, K. (2005). Development and distribution of root systems in two grain sorghum cultivars originated from Sudan under drought stress. Plant Production Science, 8, 553–562. [Google Scholar]
- Yang, J. T. , Schneider, H. M. , Brown, K. M. , & Lynch, J. P. (2019). Genotypic variation and nitrogen stress effects on root anatomy in maize are node specific. Journal of Experimental Botany, 70, 5311–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, L. , Zhang, H. , Tang, Z. , Xu, J. , Yin, D. , Zhang, Z. , … Liu, X. (2021). rMVP: A memory‐efficient , visualization‐enhanced, and parallel‐accelerated tool for genome‐wide association study. Genomics, Proteomics & Bioinformatics In press. 10.1016/j.gpb.2020.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- York, L. M. , Galindo‐Castaneda, T. , Schussler, J. R. , & Lynch, J. P. (2015). Evolution of US maize (Zea mays L.) root architectural and anatomical phenes over the past 100 years corresponds to increased tolerance of nitrogen stress. Journal of Experimental Botany, 66, 2347–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York, L. M. , & Lynch, J. P. (2015). Intensive field phenotyping of maize (Zea mays L.) root crowns identifies phenes and phene integration associated with plant growth and nitrogen acquisition. Journal of Experimental Botany, 66, 5493–5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York, L. M. , Nord, E. A. , & Lynch, J. P. (2013). Integration of root phenes for soil resource acquisition. Frontiers in Plant Science, 4, 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Ersoz, E. , Lai, C.‐Q. , Todhunter, R. J. , Tiwari, H. K. , Gore, M. A. , … Buckler, E. S. (2010). Mixed linear model approach adapted for genome‐wide association studies. Nature Genetics, 42:355–360. 10.1038/ng.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Z. , Hey, S. , Jubery, T. , Liu, H. , Yang, Y. , Coffey, L. , … Schnable, P. S. (2019). Shared genetic control of root system architecture between Zea mays and Sorghum bicolor . Plant Physiology, 182, 00752.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J. , Kaeppler, S. M. , & Lynch, J. P. (2005). Topsoil foraging and phosphorus acquisition efficiency in maize (Zea mays). Functional Plant Biology, 32, 749–762. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of inbred maize genotypes in the Wisconsin Diversity (WiDiv) Panel
Table S2. Nutrient solutions used in mesocosm experiments
Table S3. Overlapping genotypes between the Wisconsin Diversity Panel and SNP set
Table S4. Root angle phenotypes of the Wisconsin Diversity Panel
Table S5. Mis‐sense mutants of SNPs located in the coding region
Table S6. Co‐occurring SNPs and individually occurring SNPs in genotypes with steep angles
Table S7. cipk15 mutant and wild‐type angles in the field and greenhouse. Data presented are raw data per plot in the field or per individual greenhouse mesocosm
Table S8. Dry shoot biomass of cipk15 mutant and wild type in field and greenhouse experiments. Data presented are raw data per plot in the field or per individual greenhouse mesocosm
Table S9. cipk15 mutant and wild‐type architectural phenotypes in the field and greenhouse. Data presented are raw data per plot in the field or per individual greenhouse mesocosm
Table S10. 15N uptake data from the field. Data presented are raw data per plot in the field
Figure S1. Linkage disequilibrium heatmaps displaying Zm00001d033316 in a 100 Kb window upstream and downstream
Figure S2. SNPs resulting in mis‐sense amino acid substitutions in ZmCIPK and their associated crown root angle phenotypes
Figure S3. Identification of mu1046464::Mu insertion position
Figure S4. ZmCIPK15 expression by semi‐quantitative RT‐PCR between the cipk15 mutant and the wild type
Data Availability Statement
All data are available upon request to the corresponding author.


