Abstract
De novo point mutations in GNAO1, gene encoding the major neuronal G protein Gαo, have recently emerged in patients with pediatric encephalopathy having motor, developmental, and epileptic dysfunctions. Half of clinical cases affect codons Gly203, Arg209, or Glu246; we show that these mutations accelerate GTP uptake and inactivate GTP hydrolysis through displacement Gln205 critical for GTP hydrolysis, resulting in constitutive GTP binding by Gαo. However, the mutants fail to adopt the activated conformation and display aberrant interactions with signaling partners. Through high-throughput screening of approved drugs, we identify zinc pyrithione and Zn2+ as agents restoring active conformation, GTPase activity, and cellular interactions of the encephalopathy mutants, with negligible effects on wild-type Gαo. We describe a Drosophila model of GNAO1 encephalopathy where dietary zinc restores the motor function and longevity of the mutant flies. Zinc supplements are approved for diverse human neurological conditions. Our work provides insights into the molecular etiology of GNAO1 encephalopathy and defines a potential therapy for the patients.
Encephalopathy mutations destabilize Gαo nucleotide binding, structure, and signaling, features corrected by dietary zinc.
INTRODUCTION
Heterotrimeric G proteins are the immediate cytoplasmic signaling transducers of G protein–coupled receptors (GPCRs), the largest receptor class in animals and the major target of modern drugs (1). Composed of the Gα, β, and γ subunits, they interact with receptors when the Gα is loaded with GDP (guanosine diphosphate) to undergo the activated GPCR-induced GDP-to-GTP (guanosine triphosphate) exchange and dissociation into the Gα-GTP and Gβγ components, both competent to transmit the signal further downstream (2). With time, intrinsic guanosine triphosphatase (GTPase) activity of the Gα subunits leads to GTP hydrolysis; the activity further sped up by the dedicated RGS (regulator of G protein signaling) proteins (3). The resultant Gα-GDP can reload with GTP (4) or complex back with Gβγ, thus closing the G protein activation-deactivation loop (2).
Of the 16 human Gα subunits, Gαo is the major neuronal representative, transmitting the signals from numerous GPCRs in developing and adult brain. In 2013, the first cases were reported on patients with pediatric encephalopathy harboring de novo mutations in GNAO1, the gene encoding Gαo (5). This discovery was followed by an avalanche of clinical analyses that cumulatively led to the recognition of GNAO1 encephalopathy as a spectrum of neurodevelopmental disorders manifesting as motor dysfunction, epileptic seizures, and developmental delay first appearing mostly in infancy (6–8). Although Gαo, in addition to neurons, is also strongly expressed in glial cells (www.proteinatlas.org), disease modeling in mice (9) and brain organoids (10) demonstrated strong effects of GNAO1 mutations on neuronal rather than glial differentiation, highlighting neurons as the main target of Gαo malfunctioning in the disease. Gαo couples to many neuronal GPCRs, such as D2 dopamine, μ-opioid, M2 muscarinic, α2-adrenergic, and more. As these GPCRs belong to the inhibitory receptors, Gαo misfunctioning induced by mutations may be expected to disbalance the equilibrium formed by the stimulatory and inhibitory GPCR signaling and thus contribute to the disease manifestations (11).
As of today, about 200 patients worldwide have been identified to harbor a mutation in GNAO1 (https://gnao1.org/). With the advances in genetic analysis application in clinical practice, the reported GNAO1 encephalopathy cases will continue to multiply. While most of the mutations being single–amino acid substitutions spread over the coding sequence of the gene, the codons Gly203, Arg209, and Glu246 emerge as the disease mutation hotspots, together taking the share of 45 to 68% in recent surveys (6–8). Of these amino acid residues, Gly203 is located near the GTP-binding pocket of Gαo, while Arg209 and Glu246 form a salt bridge, playing an important role in the adoption of the activated G protein conformation (Fig. 1A) (12).
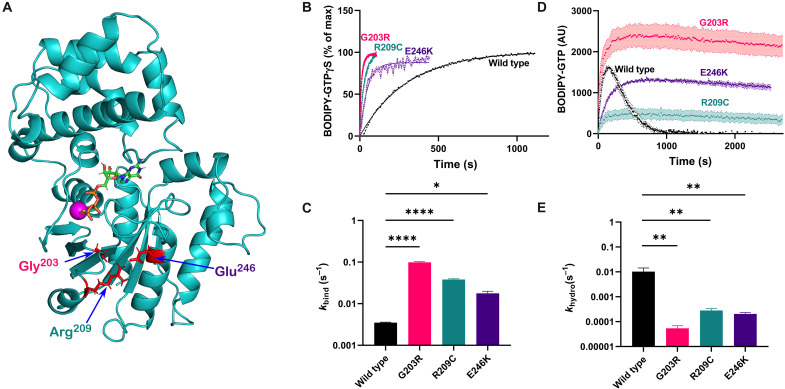
Fig. 1. Mutations in positions 203, 209, and 246 result in constitutive GTP loading of Gαo.
(A) Mutated amino acid residues (in red) in the overall structure of Gαo. The residues are either located in the switch II region of Gαo (Gly203) or represent the molecular latch fastening switch II and the α3 helix (Arg209, Glu246), performing key functions in uptake and hydrolysis of GTP (shown as a stick structure with standardly colored atoms, complexed with magnesium in magenta). (B and C) Representative curves (B) and quantification of the binding rate constant (kbind) (C) of BODIPY-labeled GTPγS binding to Gαo, wild type (WT) or mutated. All the mutants demonstrate strongly elevated rates, with G203R being the fastest. (D and E) Representative curves (D) and quantification of the hydrolysis rate constant (khydr) (E) characterizing the course of BODIPY-labeled GTP binding and hydrolysis by Gαo or its mutants by monitoring the formation and decay of the GTP-bound fraction of Gαo. Note the difference between (B) where the data are adjusted to the plateau to highlight the differences in the binding rates and (D) where the data are shown in raw fluorescence units, as needed for the proper khydr calculation. Note the log scale in the y axes in (C) and (E). Data in (B) to (E) are shown as means of at least three biological replicates ± SEM. *P < 0.05; **P < 0.01, and ****P < 0.0005 by one-way analysis of variance (ANOVA) followed by Dunnett’s multiple comparisons test.
No curative therapy exists for patients with GNAO1 encephalopathy. Symptomatic treatments for motor dysfunctions include, e.g., benzodiazepines and deep-brain stimulation, while, for epilepsy, treatments include antiepileptic drugs such as carbamazepine, levetiracetam, or valproic acid, but at best with partial effects (8, 13, 14). Development of eventual therapies, in turn, is delayed by the lack of understanding of the molecular etiology of the disease. Here, we probe the biochemical deficiency at the molecular core of GNAO1 encephalopathy, identify a drug correcting this deficiency, validate it in neuronal cells, and lastly show that the dietary supplementation of the drug rescues defects in a Drosophila model of the disease, identifying the potential therapeutic avenue to treat human patients.
RESULTS
Mutations in Gly203, Arg209, and Glu246 result in constitutive GTP binding by Gαo in vitro
To seek understanding of the molecular etiology of GNAO1 encephalopathy, we focused on the most frequent pathologic Gαo mutants and first probed their basic biochemical properties: GTP uptake and hydrolysis, in comparison to the wild-type protein. The GTP uptake analysis has so far been performed for three GNAO1 encephalopathy mutants: Q52P and Q52R displayed complete loss of the GTP uptake (14), while R209H was reported to display a faster GTP uptake (15). We applied the nonhydrolyzable fluorescent BODIPY-GTPγS (guanosine 5′-[γ-thio]triphosphate) (4, 16–18) to monitor the GTP uptake properties of the wild-type protein and three Gαo mutant variants: G203R, R209C, and E246K. This analysis reveals that the three mutants are much faster in uptaking GTP than the wild type (Fig. 1B). The calculated binding rate constant, kbind, increases 5-fold by the E246K mutation, 11-fold by the R209C mutation, and 28-fold by the G203R mutation over that of the wild-type Gαo (Fig. 1C).
The faster GTP uptake will lead to higher GTP residence of the G protein if its GTP hydrolysis rate is not proportionally increased; an accompanying decreased GTP hydrolysis will further aggravate the GTP residence of the G protein. We thus next applied a hydrolyzable fluorescent GTP analog, BODIPY-GTP, whose interaction with an active G protein is seen as a transient rise in fluorescence (indicative of the nucleotide binding) followed by a decay in fluorescence (indicative of GTP hydrolysis due to the lower quantum yield the resultant fluorophore-GDP on the protein) (4, 17, 18). We see that all three pathologic Gαo mutants reveal essentially abolished GTP hydrolysis as compared to the wild type (Fig. 1D). Calculation of the hydrolysis rate constant, khydr, confirms this assessment: khydr of R209C is reduced ca. 50-fold, of E246K ca. 100-fold, and of G203R ca. 300-fold as compared to the wild-type Gαo (Fig. 1E).
Thus, mutations in the three most frequently affected GNAO1 encephalopathy amino acid residues lead to a strong increase in the rate of GTP uptake accompanied by a gigantic drop in the rate of GTP hydrolysis. We thus must conclude that, biochemically, mutations in Gly203, Arg209, and Glu246 of Gαo lead to the constitutive GTP-binding state of the G protein, the molecular feature possibly at the basis of the etiology of the disease caused by these mutations.
Defective cellular interactions of the GNAO1 encephalopathy mutants
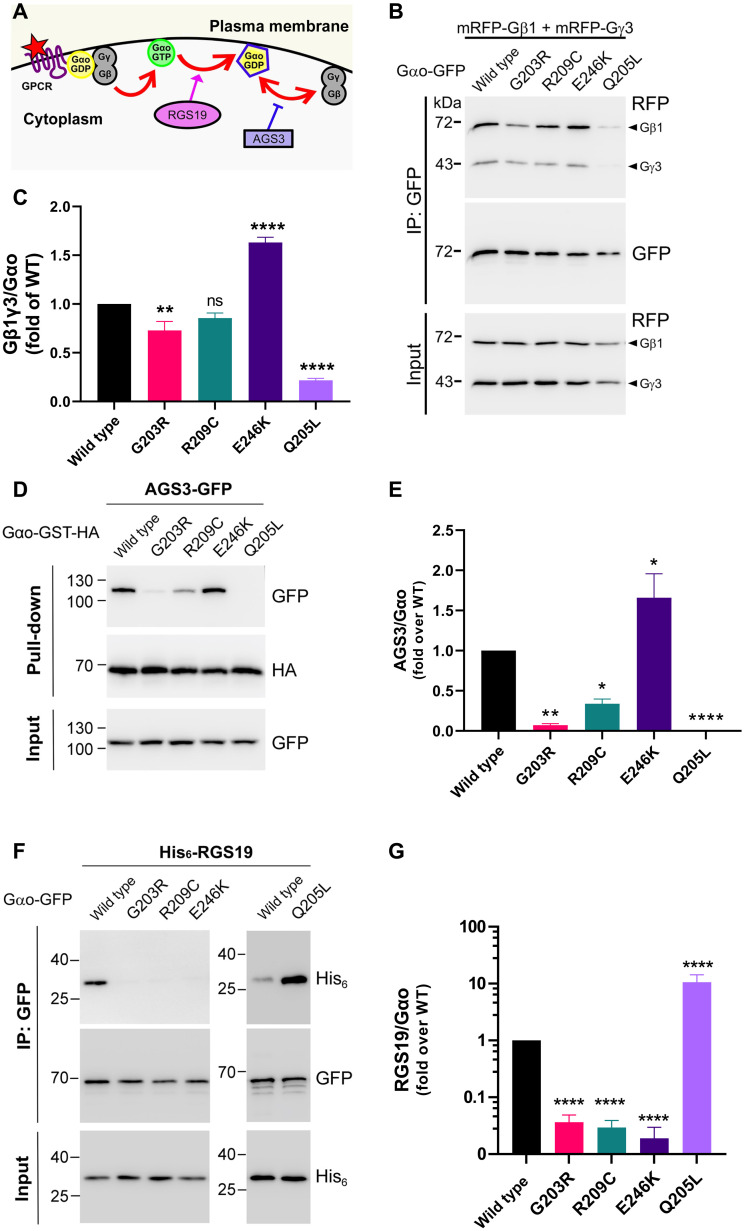
The distorted proportion of the GTP/GDP-state residence of the GNAO1 encephalopathy mutants inferred from the biochemical experiments must have consequences at the cellular level. We thus moved to express the mutants in mouse neuroblastoma Neuro-2a (N2a) cells, frequently used to study the Gαo function (16). In this, as other cell lines, wild-type Gαo shows dual localization at the plasma membrane and the Golgi apparatus, as previously reported by us (16, 19). The G203R, R209C, and E246K mutants show a similar dual localization (fig. S1, A to D), indicative of the normal posttranslational modifications of the mutants (20). Quantification of fluorescence intensities at the two compartments confirmed the near-equal distribution of wild-type Gαo and the three pathologic mutants (fig. S1, E and F). We next tested the interactions of the three pathologic Gαo mutants, along with the wild-type and the classical constitutively activated, disease-unrelated point mutant Gαo[Q205L], with a set of intracellular binding partners of Gαo. The following interaction/signaling partners were selected: RGS19 that preferentially binds the GTP-loaded form (4), Gβγ that preferentially interacts with the GDP-loaded form of Gαo (21), and AGS3 that also preferentially binds the GDP-loaded form (Fig. 2A) (17). We additionally tested the Golgi partners of Gαo: adenosine diphosphate ribosylation factor 1 (Arf1), Rab3a, and KDEL receptor (KDELR) (fig. S2A) (16). This broad assessment reveals a complicated nature of distortions of cellular interactions of the three pathologic Gαo mutants. First, we found that interactions with the Golgi partners were near normal for the pathologic mutations (fig. S2, B to G). Second, we found that interactions with Gβγ were differentially affected by the three pathologic mutations: decreased for G203R, insignificantly affected for R209C, and unexpectedly increased for E246K (Fig. 2, B and C). Third, another GDP-preferring partner of Gαo, AGS3, revealed the same tendency with even more pronounced effects: decreased binding to Gαo[G203R] and Gαo[G209C] and enhanced to Gαo[E246K] (Fig. 2, D and E).
Fig. 2. Aberrant cellular interactions of pathologic Gαo mutants.
(A) Analysis of plasma membrane/cytoplasmic partners of Gαo: Gβγ forming, with GDP-loaded Gαo, the heterotrimeric complex, competent to interact with a GPCR and respond to its activation by the GDP-GTP exchange and heterotrimer dissociation, RGS19 that speeds up GTP hydrolysis on Gαo, and AGS3 that competes with Gβγ for the GDP-loaded Gαo. Illustration modified from (16). (B and C) N2a cells were cotransfected with mRFP-Gβ1, mRFP-Gγ3, and Gαo-GFP (internally tagged) WT, G203R, R209C, E246K, or the activated Q205L mutant as control. Immunoprecipitation (IP) of Gαo was done using a nanobody against GFP, and the coprecipitation of Gβ1γ3 was analyzed by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot (B). Antibody (Ab) against GFP was used for detection of Gαo and against mRFP for Gβ1γ3 (arrowheads). Quantification of the coimmunoprecipitation (co-IP) of Gβ1γ3 by the different Gαo constructs (C). (D and E) After cotransfection with AGS3-GFP and Gαo-GST-HA (C-terminally tagged) variants, pull-down of Gαo was done using glutathione beads and samples were analyzed by SDS-PAGE and Western blot (D). Ab against hemagglutinin (HA) was used for detection of Gαo and against GFP for AGS3. Quantification of the coprecipitation of AGS3-GFP by Gαo-GST-HA WT or mutants (E). (F and G) After cotransfection with His6-RGS19 and Gαo-GFP (internally tagged) variants, IP of Gαo was done using a nanobody against GFP, and the coprecipitation of RGS19 was analyzed by SDS-PAGE and Western blot. Ab against GFP was used for detection of Gαo and against His6 for RGS19 (F). Quantification of the co-IP of RGS19 by the different Gαo constructs (G). Data in (C), (E), and (G) are means of ≥4 biological replicates ± SEM. Statistical significance: One-way ANOVA followed by Dunnett’s multiple comparisons test; *P < 0.05, **P < 0.01, and ****P < 0.0001.
However, the most notable and unexpected was the near-complete loss of the pathologic mutants’ interaction with RGS19, opposite to that seen for the constitutively active Q205L (Fig. 2, F and G). In these pull-downs, we used an internal green fluorescent protein (GFP) fusion [that does not impede the proper localization nor protein-protein interactions (14)] of the Gαo variants and a nanobody against GFP. To confirm that this placement of the GFP fusion has no decisive role in the markedly reduced interaction of the pathologic mutants with RGS19, we repeated the pull-downs with the C-terminally GFP-tagged Gαo variants, again revealing that the GNAO1 encephalopathy mutants lose the interactions with RGS19 (fig. S3, A and B).
Our findings may suggest that the pathologic Gαo mutants fail to adopt the proper conformation upon nucleotide binding (see Discussion). Gαi1 mutated in the amino acids equivalent to Gαo’s Arg209 and Glu246, as well as at Gly204 neighboring Gly203, has similarly been proposed to fail to adopt the activated conformation and to dissociate from Gβγ upon GTP binding (12). Failure to adopt the properly activated conformation upon GTP binding by a Gα protein has also been associated with the reduced ability to hold the GTP nucleotide after binding (12). To assess this feature, we prebound Gαo (wild type, G203R, R209C, and E246K or the constitutively active Q205L mutant) with BODIPY-GTPγS and then added excess of GDP, following (4, 12, 17). While wild-type and Q205L Gαo retain a substantial portion of the prebound BODIPY-GTPγS (fig. S4, A and B), the three pathologic mutants lose most of it (fig. S4, C to F). Furthermore, the rate of dissociation of BODIPY-GTPγS from the three pathologic mutants is strongly increased as compared with the wild type or Gαo[Q205L] (fig. S4, A to E and G). Importantly, this rate of BODIPY-GTPγS dissociation significantly exceeds the rate of BODIPY-GTPγS uptake by the three pathologic mutants, the feature not seen for the wild-type and Q205L Gαo (fig. S4G). These observations cumulatively argue that the three pathologic mutant forms of Gαo do not acquire the proper conformation upon GTP binding and potentially upon nucleotide binding in general.
Defective cellular GPCR signaling by the GNAO1 encephalopathy mutants
Aberrant cellular interactions of the G203R, R209C, and E246K mutants of Gαo described in the previous section suggest that some signaling routes mediated by the G protein could become aberrant in GNAO1 encephalopathies caused by these mutations to different extents: potentially more at the plasma membrane and potentially less in the Golgi. We thus next aimed at investigation of how efficiently different neuronal GPCRs known to couple to Gαo could activate signaling through these pathologic mutant variants.
To this end, we used a bioluminescence resonance energy transfer (BRET) system in human embryonic kidney (HEK) 293 cells. Cell transfection with Gαo (wild type or mutant) with the nanoluciferase tag, Gβ3, and Gγ9 with the Venus tag (22) results in a measurable BRET signal, indicative of the efficiency of the Gαo-βγ complex formation. Agreeing with the pull-down data presented above (Fig. 2, B and C), Gαo[E246K] demonstrates significantly higher BRET signal and, thus, Gαo-βγ complex formation than the wild type or the other two mutants (Fig. 3, A and B).
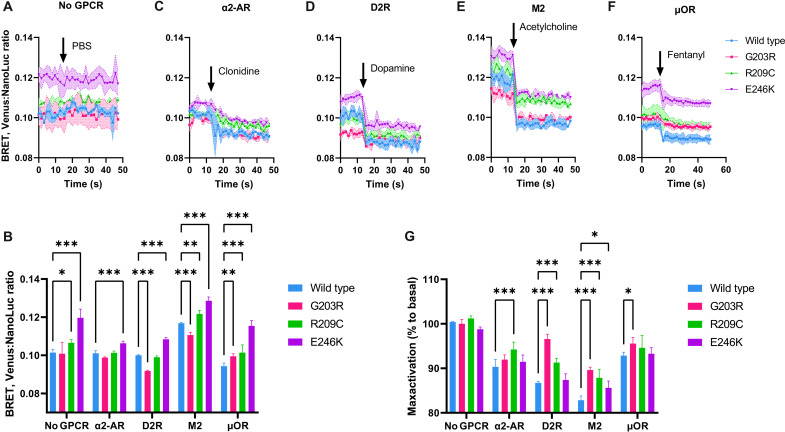
Fig. 3. Aberrant signaling from neuronal GPCRs by pathologic Gαo mutants.
(A) BRET ratio between Gαo (WT or mutant) tagged with nanoluciferase (NanoLuc) and Gβγ (Gβ3 and Gγ9 with the Venus tag) demonstrates different extent of association with Gβγ. Without cotransfection with a GPCR and upon a mock stimulation with phosphate-buffered saline (PBS), no change in the BRET signal is observed. (B) Quantification of the basal, nonstimulated levels of BRET (average of first 10 readings before agonist injection) for different Gαo variants in different receptor cotransfection settings. (C to F) When the proper neuronal GPCR is cotransfected, the addition of the respective agonist (arrow) induces a robust decrease in the BRET signal, indicative of the dissociation of the Gαo-βγ heterotrimer, reduced to a different extent for different GPCRs. The agonist concentrations were 10 μM clonidine for the α2-adrenergic receptor (α2-AR) (C), 10 μM dopamine for the D2 dopamine receptor (D2R) (D), 10 μM acetylcholine for the M2 muscarinic receptor (M2) (E), and 75 nM fentanyl for the μ-opioid receptor (μOR) (F). (G) Quantification of the plateau levels of the BRET decrease upon GPCR stimulation (ratio of 10 readings at the end of stimulation to 10 readings before stimulation). Data on all graphs are presented as a means from N = 4 independent repeats; SEMs are represented by colored zone in (A) and (C) to (F) and by bars in (B) and (G). Statistical analysis is done by two-way ANOVA with Sidak corrections for multiple comparisons; *P < 0.05, **P < 0.01, and ***P < 0.001.
We next cotransfected the cells with neuronal GPCRs (D2 dopamine, μ-opioid, M2 muscarinic, or α2-adrenergic). Cell activation with respective GPCR agonists (dopamine, fentanyl, clonidine, or acetylcholine) results in a rapid decrease in the BRET, indicative of the GPCR-induced Gαo-βγ heterotrimer dissociation as the first step in activation of the corresponding signaling pathways (Fig. 3, C to F). Mock stimulations do not elicit any change in the BRET signal (Fig. 3A). As another control, we show that stimulation with the agonists without GPCR transfection does not elicit any signaling (fig. S5A). We also show that the expression levels of the tested Gαo variants harboring the nanoluciferase tag are similar within each experiment (fig. S5B). Last, we also found that addition of the excess of antagonist restores the BRET signal back to the basal levels (fig. S5C).
With these analyses, we show that the G203R, R209C, and E246K mutants have reduced, and, importantly, varying from receptor to receptor, ability to respond to a GPCR activation. Specifically, we find that the G203R mutant displays significantly reduced capacity to transmit the D2 dopamine and M2 muscarinic signals, while signaling from the α2-adrenergic and μ-opioid receptors is comparable to that mediated by wild-type Gαo (Fig. 3G). Similarly, R209C displays reduced signaling from the α2-adrenergic, D2 dopamine, and M2 muscarinic receptors and normal signaling from the μ-opioid receptors (Fig. 3G). Last, E246K is compromised in the M2 muscarinic signal transduction but not in signaling from the other three neuronal GPCRs receptors (Fig. 3G).
These findings double the number of GPCRs whose coupling to GNAO1 mutants has been studied [previously, the coupling to D2 dopamine and α2-adrenergic receptors was studied with different methods (11, 23)] and demonstrate that the three pathologic mutants are not completely incapable of activation of signaling by neuronal GPCRs. While all the three mutants display reduced (but not completely abrogated) responsiveness to the M2 muscarinic receptor activation, their signaling from the α2-adrenergic and μ-opioid receptors is only marginally or not at all reduced as compared to wild-type Gαo. These findings illustrate that varying degrees of defectiveness (from none to significant) in signaling from different neuronal GPCRs are attributable to different pathologic mutants. We next aimed at understanding the structural defects that underlie the distorted nucleotide handling, cellular interactions, and signaling capacities of the G203R, R209C, and E246K encephalopathy mutants.
Displacement of the GTPase catalytic residue in GNAO1 encephalopathy mutants in homology modeling and dynamic simulations
To gain insights into the possible structural deficiency in the G203R, R209C, and E246K encephalopathy mutants that lead to the deficient GTPase reaction and aberrant interactions with signaling partners, we performed homology modeling followed by molecular dynamics simulations of the GTP-loaded Gαo wild type, and the mutants, based on the Gαi1-GTPγS structure (24). Structural analysis of the energy-minimized state reveals that R209C and E246K, mutations of the amino acids normally forming a salt bridge to fasten switch II and the α3 helix to lock Gα in the active conformation upon the GTP binding (12, 25), result in a significant displacement of the Gln205 residue, the key to the catalytic GTPase reaction (24, 26), away from the γ-phosphate (fig. S6, A, B, and D). Through molecular dynamics simulations (100 ns), we further found a significant global destabilization of Gαo[E246K], in agreement with a similar analysis of the Gαi1 structure (12), but not of the G203R or R209C mutants (fig. S7A and movie S1). Furthermore, analysis of the energy-minimized model of Gαo[G203R] reveals a somewhat different arrangement as compared to E246K and R209C mutants: Instead of acting through switch II, the substituting Arg residue engages in a direct interaction with the catalytic Gln205, displacing the latter from the γ-phosphate and occupying the space normally used by the hydrolysis water molecule and thus inducing its displacement (fig. S6, C and D). Despite the limitations of the modeling and molecular dynamics approaches as compared to direct structural analysis, these findings suggest a common mechanism of the loss of the GTPase reaction in the three GNAO1 encephalopathy mutants as the displacement of the catalytic Gln205. The accompanying changes in Gαo structure likely lead to the failure of the G protein to adopt the fully activated conformation and thus make it poorly recognizable by RGS19. Another amino acid residue, T182 playing an important role in the Gα-GTP interaction with RGS proteins (27, 28), does not show a significant dislocation in the three pathologic Gαo mutants (fig. S7, C to F).
High-throughput assay aiming at recovering the GTPase activity of Gαo[E246K] identified zinc pyrithione as a drug specifically acting on the mutant but not wild-type proteins
We next argued that since the inability of the three Gαo encephalopathy mutant proteins to hydrolyze GTP represents an easily measurable biochemical characteristic, one could design an assay to screen for molecules potentially capable of correcting this deficiency. To build such a high-throughput screening (HTS) platform, we used Gαo[E246K] and BODIPY-GTP to monitor the GTP uptake and hydrolysis by the mutant, as described in the first section, and screened a library of 2736 U.S. Food and Drug Administration (FDA)–approved and pharmacopeial drugs (fig. S8A). While wild-type Gαo hydrolyzes all BODIPY-GTP provided to it in this biochemical setting within 10 min, Gαo[E246K] fails to do so, resulting in stable BODIPY fluorescence (see Fig. 1D). Thus, the initial screening was based on the identification of drugs capable of inducing a drop in fluorescence by the 10-min incubation of Gαo[E246K] with BODIPY-GTP. The screening was followed by hit validation, which resulted in three hit compounds: sennoside A, sennoside B, and zinc pyrithione (ZPT; Fig. 4, A and B, and fig. S8, B to E). Of those, sennosides A and B were found to decrease GTP uptake by Gαo[E246K] rather than GTP hydrolysis by it (fig. S8, D and E). In contrast, ZPT partially restored the GTP hydrolysis (Fig. 4B). When retesting the drugs on wild-type Gαo, sennosides were found to equally affect the GTP uptake by it, as seen by a decrease in the peak of BODIPY fluorescence upon treatment of wild-type Gαo with the compounds (fig. S8, F and G). In contrast, ZPT produced no effect on the GTP uptake and hydrolysis by the wild-type Gαo (Fig. 4C), revealing the specificity toward Gαo[E246K] and immediately raising our interest to this molecule.
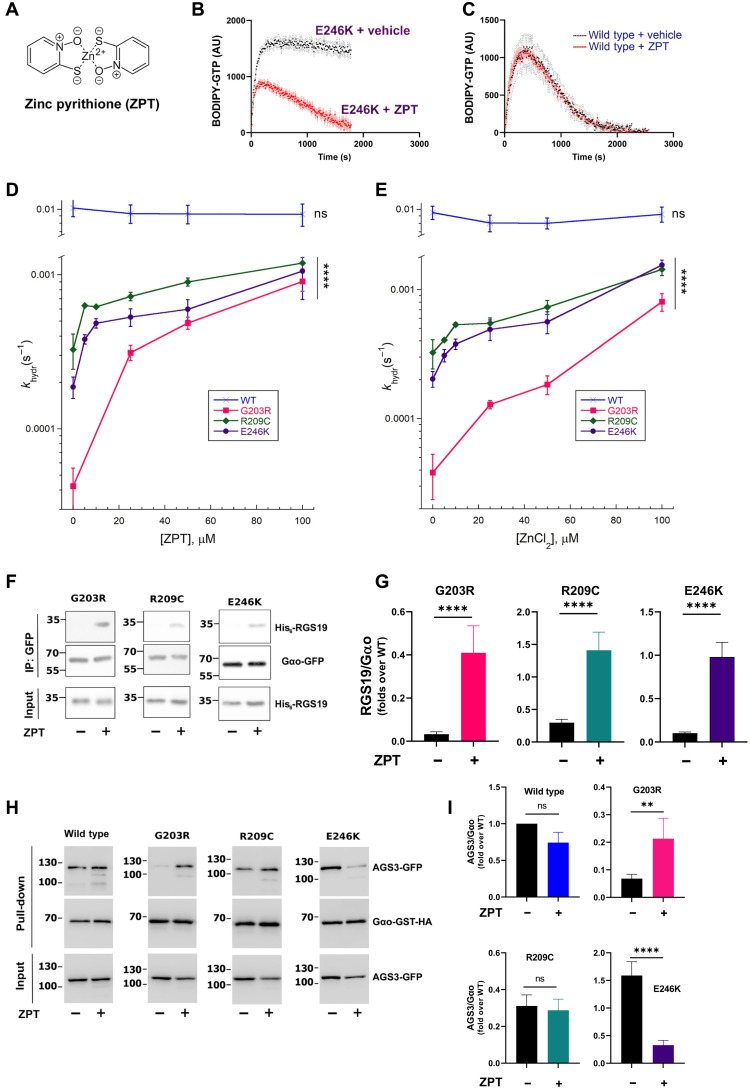
Fig. 4. ZPT and Zn2+ restore GTPase activity and cellular interactions of pathologic Gαo mutants.
(A to C) ZPT (A) restores GTPase activity of Gαo[E246K] (B), not affecting GTP binding and hydrolysis of WT Gαo (C). Representative curves of BODIPY-labeled GTP binding and hydrolysis are provided. (D and E) Quantification of khydr of Gαo WT, Gαo[G203R], Gαo[R209C], and Gαo[E246K] treated with ZPT (D) and ZnCl2 (E). Note the log scale in y axes. (F and G) N2a cells were cotransfected with His6-RGS19 and Gαo-GFP (C-terminally tagged) variants. The next day, cells were treated with dimethyl sulfoxide (DMSO) or 1 μM ZPT for 3 hours before IP of Gαo with nanobody against GFP; coprecipitation of RGS19 was analyzed by SDS-PAGE and Western blot. Abs against GFP and His6-tag were used to detect Gαo and RGS19, respectively (F). Quantification of co-IP of His6-RGS19 by Gαo variants, normalized to the binding levels of WT Gαo (G). (H and I) Cotransfection with AGS3-GFP and Gαo-GST-HA (C-terminally tagged) variants was similarly followed by treatment with DMSO or 1 μM ZPT before Gαo pull-down with glutathione beads. Ab against HA was used to detect Gαo and against GFP to detect AGS3 (H). Quantification of the coprecipitation of AGS3-GFP by Gαo-GST-HA variants normalized to the binding levels of WT Gαo (I). Data in (B) to (E), (G), and (I) are means of ≥3 biological replicates ± SEM; the means in (B) and (C) are shown as dots, and SEM deviations are shown as the error bars (black for no drug; red for ZPT). (D and E) ****P < 0.0005; ns, not significant by one-way ANOVA to verify the significance of the concentration dependence (as significance of the mean change upon changing concentrations of ZPT/ZnCl2). (G and I) **P < 0.01; ****P < 0.0001 on log-transformed data by two-way ANOVA with Sidak corrections for multiple comparisons.
Zn2+ is the active component of ZPT restoring GTPase activity of the three encephalopathy Gαo mutants in vitro
We next found that ZPT revealed a concentration-dependent restoration of the GTPase activity of all the three encephalopathy Gαo mutants that we studied: G203R, R209C, and E246K (Fig. 4D and fig. S9, A to C). ZPT is a coordinated complex of pyrithione, a membrane-permeable ionophore (29), and Zn2+ (Fig. 4A) and has the primary indication to treat dandruff and seborrheic dermatitis (30); other biological activities of ZPT including antiviral and anticancer have also been reported (31, 32). Zn2+ ions are poorly penetrant through cellular membranes (33), and the pyrithione moiety of the drug serves to deliver the ions inside cells (34). To test whether Zn2+ ions are the active component of ZPT in restoring the GTPase activity, we applied 1 mM EGTA, an efficient chelator of Zn2+ but not Mg2+ (35), the latter present in our experiments at the concentration of 10 mM (see Materials and Methods). As shown in fig. S9 (D and D′), zinc chelation by EGTA abolishes the restoration of the GTPase activity on Gαo[G203R]. We next directly tested the two components of ZPT, Zn2+ and pyrithione, for their ability to restore the GTPase reaction, finding that ZnCl2, in an EGTA-sensitive manner, was active in restoring the GTPase activity unlike the “empty” ionophore (fig. S9, D, D′, E, and E′). We further tested several other metal ions, revealing that none of them recapitulated the effect of Zn2+: Co2+, Fe2+, Ni2+, Mn2+, and Li+ were inactive in restoring the GTPase reaction, while Cu2+ at 100 μM appeared to completely inactivate the G protein (fig. S9, F and F′). Last, concentration dependence analysis showed that ZnCl2 was similar to ZPT in restoration of the GTPase activity in the three encephalopathy Gαo mutants (Fig. 4E and fig. S9, G to I). In contrast, the effect of ZPT or ZnCl2 on wild-type Gαo was confirmed to be insignificant (Fig. 4, D and E).
Potential mechanism of action of Zn2+ in restoring the GTPase activity of GNAO1 encephalopathy mutants suggested by homology modeling and dynamic simulations
We applied structural modeling and molecular dynamics simulations to gain insights into the potential mechanism of action of Zn2+ in the restoration of the GTPase activity of the three Gαo mutants. We argued that Zn2+ can replace Mg2+ in the Gαo’s active center upon the interaction with GTP: It is well known that Mg2+-binding sites of proteins can generally be substituted by a broad range of other divalent metal ions, with Zn2+ being one of the most potent substitutes (36). We used the CHARMM36m field in dynamic simulations that provides distinct parameters for the Mg2+ and Zn2+ ions (see Materials and Methods), permitting comparative assessment of their effects. Our analysis shows that the substitution of Zn2+ for Mg2+ in the active center does not affect the global structure in the energy-minimized state of wild-type Gαo (fig. S7B and movie S2). The global rearrangement observed in Gαo[E246K] was, to a certain degree, further aggravated in the Zn2+-bound protein; no global effect of Zn2+, however, was seen for the G203R and R209C mutants (fig. S7B and movie S2), suggesting that it is unlikely to represent the general or main mechanism of Zn2+ action to restore the GTPase activity.
We thus next paid special attention to the position and flexibility of the catalytic Gln205 that we found to be displaced from the GTP’s γ-phosphate in the pathologic mutants (see fig. S6). Our molecular dynamics simulations reveal that, for all three mutant variants, distance between the γ-N atom of Gln205 and the γ-P atom of GTP (increased as compared to the wild-type Gαo) is reduced back to the wild-type levels by Zn2+ (Fig. 5). Note that in the wild-type protein, Zn2+ also decreases the distance between Q205 and γ-phosphate but to a considerably smaller extent that has no further influence on the catalytic activity (Fig. 5, A to D). This effect is particularly evident for the Gαo[E246K] and Gαo[R209C] mutants (Fig. 5, A, B, and D to F), agreeing well with the fact that the residues E246 and R209 normally form the salt bridge to fasten the active conformation upon the GTP binding (12, 25). The effect of Zn2+ on the distance between Gln205 of Gαo[G203R] and the γ-phosphate was also clear (Fig. 5, C, D, and G). We also measured the dynamics of the bond, as the distance between the γ-N atom of Gln205 and the ζ-atom of Arg203, in the Mg2+-bound versus Zn2+-bound conformations. This new Arg in position 203 forms a hydrogen bond with Gln205, and this might contribute to the loss of the GTPase activity in the mutant [see fig. S6 (C and D)]. Unexpectedly, we found that Zn2+, if anything, stabilized the Arg203-Gln205 interaction (fig. S7, G and H).
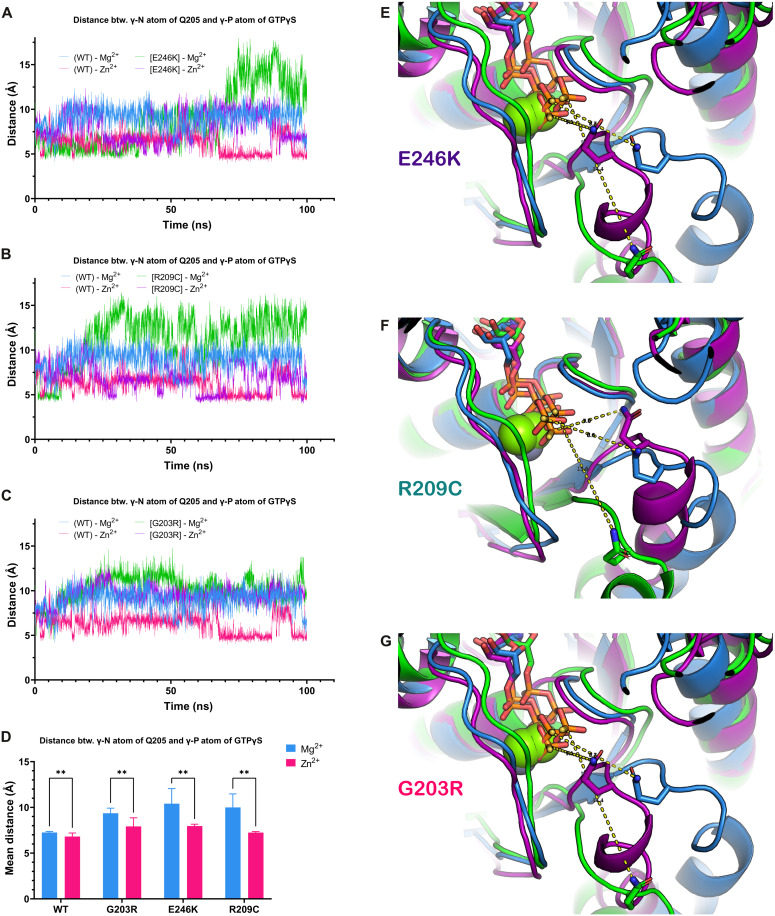
Fig. 5. Zn2+ restores the defects induced to the catalytic Q205 by the pathologic Gαo mutations.
(A to D) Analysis of the distance between the γ-N atom of Q205 residue in Gαo and the γ-P atom of GTP in WT and mutant proteins over 100 ns of molecular dynamics simulation. The panels show the plots of the measured distance between these atoms in WT protein containing Mg2+ or Zn2+ and the same in the [E246K] mutant (A), [R209C] mutant (B), and the [G203R] mutant (C). The graphs demonstrate that, over the major or significant part of the simulation trajectories in all three mutants, the Q205 residue is further away from the active site than in WT protein, which is rescued by the presence of Zn2+ in the active site. (A to C) Individual MD simulations. (D) Summary of the last 50 ns of three independent MD simulations presented as means ± SD. Statistical analysis: Two-way ANOVA followed by Fisher’s least significant difference multiple comparisons test; **P < 0.01. (E to G) Three-dimensional structures of the representative states with the maximal removal of Q205 from the active site are shown for the WT in the Mg2+-bound conformations versus the mutants in the Mg2+- and Zn2+-bound conformations: [E246K] (E), [R209C] (F), and [G203R] (G). It is evident that, upon Zn2+ binding, the mutants’ Q205 is brought back to the γ-P atom of GTP (the hydrogen bonds are indicated). Color coding as in the respective (A) to (C) panels.
Overall, our structural modeling and molecular dynamics analysis suggests the atomistic mechanism for the action of Zn2+ on the restoration of the GTPase activity of the E246K, R209C, and G203R mutants as the bringing back of the catalytic Gln205 to the γ-phosphate of GTP, otherwise swayed away by the mutations. More complex modeling (e.g., through quantum mechanics or hybrid quantum/molecular mechanics, taking into consideration differences in coordination sphere configurations of Zn2+ and Mg2+) and biophysical measurements will further advance these initial findings.
ZPT restores cellular RGS19 interactions of GNAO1 encephalopathy mutants
Our in vitro data show that ZPT and its ion component, Zn2+, are able to restore the GTPase activity of the pathologic Gαo. We next wondered whether such a restoration could be seen in cells and be reflected in restored interactions with Gαo partners. We first tested whether the mutants’ interaction with RGS19 can be restored in the N2a cells. It is however known that, acutely, Zn2+ can cross the cellular membranes either by active transporters (33) or with the help of ionophores such as pyrithione (34). On the other hand, a significant neuronal and other cell toxicity of ZPT has been reported, mainly because of its effectiveness in bringing large concentrations of zinc inside the cells (32, 37, 38). Thus, we first investigated the cytotoxicity of ZPT, pyrithione, and ZnCl2 in N2a cells. This analysis confirms the neurotoxicity of ZPT with the half-maximal inhibitory concentration of ca. 5 μM; ZnCl2, in contrast, was not toxic up to the concentrations of 100 μM (fig. S10A), as, presumably, it failed to penetrate the cells in this acute setting. For the subsequent experiments on the restoration of the Gαo-RGS19 interactions in N2a cells, we took the highest tested agents’ concentrations that did not display any cytotoxicity: 1 μM for ZPT and 100 μM for ZnCl2.
Using these concentrations, we found that ZPT could recover the ability of the G203R, R209C, and E246K mutants to interact with RGS19 (Fig. 4, F and G); ZnCl2, in contrast, was ineffective (fig. S10, B and C). The interaction of wild-type Gαo with RGS19 appeared to increase by ZPT ca. twofold (fig. S10, B and C), a modest increase as compared to the effect of ZPT on the mutant Gαo versions: ca. 6-fold for Gαo[R209C], ca. 12-fold for Gαo[E246K], and 20-fold for Gαo[G203R], recovering the interactions with RGS19 to levels of the wild-type protein (Fig. 4, F and G, and fig. S10, B and C).
We similarly tested whether ZPT could correct the aberrant cellular interactions of the Gαo mutants with another partner, AGS3. As we showed above (Fig. 2, D and E), Gαo[G203R] and Gαo[R209C] demonstrate decreased interactions and Gαo[E246K] increased interactions with AGS3 as compared to the wild-type protein. We found that cell treatment with ZPT tends to normalize these interactions, increasing the binding of AGS3 to the G203R mutant and decreasing to the E246K mutant (Fig. 4, H and I). Last, we tested the effect of ZPT on the most aberrant, among the three pathologic Gαo mutants, interaction with Gβγ, the twofold increased binding by Gαo[E246K] (Fig. 2C), finding a correction to the wild-type levels of the Gαo[E246K]-Gβγ binding by the drug (fig. S10, D and E).
Thus, we conclude that cell supplementation with Zn2+ ions restores the normal conformation on the three pathologic mutants of Gαo, bringing to the wild-type level the G protein interactions with RGS19, the partner preferring the GTP-bound form of Gαo, as well as with AGS3 and even Gβγ preferring the GDP-bound state of Gαo. These recovery effectiveness in cells prompted us to next ask whether zinc can be effective at a higher level of complexity, i.e., in an organism model of GNAO1 encephalopathy.
Zn2+ dietary supplementation rescues the Drosophila model of GNAO1 encephalopathy
Animal models have the instrumental role in deciphering the human disease mechanisms and in identifying/validating the treatment routes. The fruit fly Drosophila melanogaster represents an excellent model organism for studies in various fields of biology (39), and our recent work highlights the power of Drosophila as the host to model GNAO1 encephalopathy (40). To establish a GNAO1 encephalopathy model in the fruit fly, we applied the CRISPR-Cas9 mutagenesis together with phiC31-mediated recombinase-mediated cassette exchange (RMCE) (41) to introduce the pathogenic G203R mutation into the Drosophila Gαo (see Materials and Methods and fig. S11). We found that the resultant [G203R]/[G203R] flies are second larval stage lethal [as compared to the late embryonic lethality of homozygous Gαo null mutants (42, 43)], while [G203R]/+ flies are viable and fertile yet reveal a number of deficiencies. Specifically, the heterozygous mutant flies manifest a significant motor dysfunction, measured in the negative geotaxis assay as a reduced capacity to climb up the wall (Fig. 6, A and B), reminiscent of the motor dysfunction in the human patients. Furthermore, the encephalopathy mutant Drosophila displays a twofold reduction in the life span (Fig. 6C). Regional brain atrophy, sometimes progressive, has been described in GNAO1 patients with the G203R mutation and may be the outcome of epileptic onsets (6, 44–46). Analysis of 35-day-old [G203R]/+ flies revealed limited yet significant brain degeneration (fig. S12). However, this degeneration was very modest as compared to that observed, e.g., in Drosophila models of Alzheimer’s disease (47), the finding that may be aligned with the fact that the G203R mutant flies did not display any signs of spontaneous epilepsy. Given the neonatal lethality of GNAO1[G203R]/+ mice (9), we thus establish the first viable animal model of G203R encephalopathy with this Drosophila line capable of recapitulating some of the clinical manifestations of the disease.
Fig. 6. Dietary Zn2+ rescues motor dysfunction and life span in Drosophila model of GNAO1 encephalopathy.
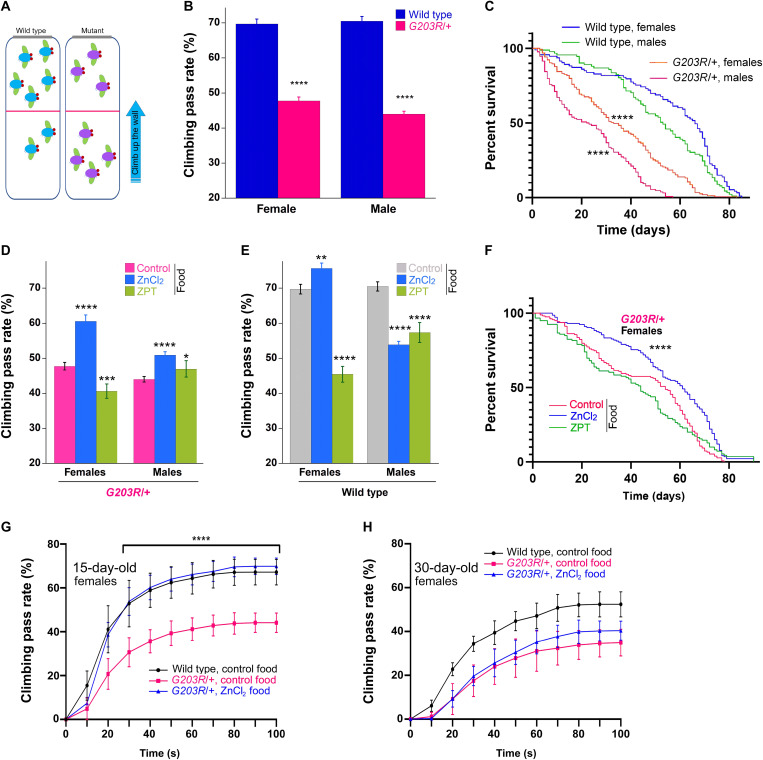
(A) Negative geotaxis: After tapping down, flies climb up the vial (blue arrow). The climbing pass rate is the percent of flies passing 10 cm from the bottom (red line) in 10 s. (B) Negative geotaxis of 100 to 110 male and female flies (in groups of ca. 15 flies). Data are means ± SEM, n = 10. Two-way ANOVA with Sidak corrections for multiple comparisons shows significant differences between WT and G203R/+ flies, P < 0.0001. (C) Life span of 250 flies (110 males and 140 females) determined for 85 days. Mantel-Cox and Gehan-Breslow-Wilcoxon tests both show significant (P < 0.0001) drop in survival of G203R/+ flies. Intersex life-span differences were also significant (P = 0.0124 for WT and P < 0.0001 for G203R/+ flies, Mantel-Cox test). (D) Locomotion dysfunction of G203R/+ flies rescued by dietary ZnCl2; ZPT shows no consistent effect. (E) Effects of dietary ZnCl2/ZPT on locomotion of WT flies. Data presentation and analysis in (D) and (E) as in (B); *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (F) Drop in life span of female G203R/+ flies rescued by dietary ZnCl2. Data presentation and analysis as in (C). (G and H) WT and G203R/+ female flies, 15 (G) and 30 (H) days old, tested in video-recorded quantitative negative geotaxis assay. Younger G203R/+ flies show complete locomotion recovery if raised on ZnCl2-supplemented food (t test, P < 0.0001 for the time points starting from 30 s for the difference between control food and ZnCl2 supplementation). Older flies reveal only a tendency of recovery. Data are means ± SD, n = 7 to 10. Age-dependent decrease in the climbing rate for WT [black curves in (G) and (H)] and G203R/+ flies [red curves in (G) and (H)] is highly significant (P < 0.0001 for both WT and mutant flies).
Dietary zinc supplementation has been applied to treat various human health conditions, including the neurological ones such as depression (48), epilepsy (49), psychiatric and neurodegenerative diseases (50), or sleep disorders (51), as well as to support normal neonatal development (52). We thus wondered whether dietary supplementation of zinc in the Drosophila model of GNAO1 encephalopathy may reveal any beneficial effects. Although, in the acute setting, we could not observe an effect of ZnCl2 to N2a cells (fig. S10), we argued that the continuous supplementation through diet could make a difference. Food supplementation of ZnCl2 to the final concentration of 200 μM has been previously shown to rescue the survival of Drosophila mutant for dZip1 and dZip2, the gut zinc transporters (53), providing us a guideline. We also tested 10 μM ZPT food supplementation, following the application of up to 15 μM ZPT in rats (54). Drosophila lines, G203R/+ and the wild-type control (see Materials and Methods and fig. S11), were raised from the egg at the standard food or that supplemented with ZnCl2 or ZPT. The climbing capacities of the resultant populations were compared, along with their longevities.
We found that the ZnCl2-containing food significantly improves the motor function of the G203R/+ flies; ZPT did not show consistent effects (Fig. 6D). The effect of ZnCl2 was particularly strong for female Drosophila, bringing the climbing capacity toward the wild-type levels. The control wild-type flies showed sex-variable effects: slight improvement for females and decrease for males (Fig. 6E); ZPT was decreasing the climbing rate for both sexes, likely reflecting its toxicity upon systemic administration (54). ZnCl2 food supplementation also rescued the reduced life span of female G203R/+ flies (Fig. 6F); no effect could however be seen for ZPT or for male flies (Fig. 6F and fig. S13A).
To gain more insights into the motor dysfunction of the G203R/+ flies and its restoration by the dietary zinc, placed in the context of the animal aging, we applied a modified, more detailed negative geotaxis assay (55) that permits to quantify the locomotion over different distances and time frames (see movie S3). With this analysis, comparing wild-type and G203R/+ flies of the age of 15 and 30 days, we see a progressive age-dependent decline in the climbing capacity of both genotypes (Fig. 6, G and H). Younger female G203R/+ flies reveal a complete recovery of their climbing capacity upon the ZnCl2 dietary supplementation (Fig. 6G and movie S3), while ZPT was ineffective. Less pronounced but significant effects were also seen in males, where dietary ZPT had a similar effect (fig. S13B). The ability of dietary ZnCl2 to restore the mutant fly locomotion declined with age (Fig. 6H and fig. S13, C and D).
Overall, these observations reveal strong improvements of the behavioral and life span conditions in the Drosophila model of GNAO1 encephalopathy by dietary zinc supplementation. Given the large body of clinical evidence on the dietary zinc supplements for human patients with diverse neurological conditions, our findings may speak for the applicability of such supplementation for the patients with pediatric GNAO1 encephalopathy. These issues and the sex- and age-sensitive effects of the dietary zinc supplementation are discussed in the next section in the context of the probable need for the continuous dietary zinc supplementation to produce therapeutic effects.
DISCUSSION
GNAO1-dependent pediatric encephalopathy is a recently diagnosed rare yet devastating neurological disease. The number of found different, mostly missense point, mutations in GNAO1 causing this malady steadily increases every year since the first 2013 report (5). However, despite some insights (5, 9, 19, 23, 56, 57), the understanding of the molecular etiology underlying the pathological developments has been largely missing. This delay in the understanding blocks development of therapies to treat the patients. Being mostly unresponsive to the conventional anti-epileptic treatments, the sick children have so far demonstrated the best, albeit partial, response to the symptomatic, highly invasive and poorly accessible therapy: deep-brain stimulation (13).
The molecular dysfunction that we have “diagnosed” here for the three most common GNAO1 encephalopathy mutations is the constitutive GTP-binding state of the mutant Gαo proteins, resulting from a strongly increased rate of GTP uptake concomitant with a markedly reduced rate of GTP hydrolysis. Molecular dynamics and structural modeling provide us with the likely unifying mechanism of this dysfunction: Each of the three mutations induces a displacement of the catalytic Gln205; this change is also likely the reason for the defective interaction of the three mutants with RGS19. We have further observed abnormal cellular interactions with some other Gαo partners, such as Gβγ and AGS3, which interact less with Gαo[G203R] and Gαo[R209C] but unexpectedly more with Gαo[E246K] than with wild-type Gαo. In contrast, interactions with the Golgi partners of Gαo are near normal in the pathologic mutants: the findings that, together with the decreased response of the mutants to neuronal GPCRs, suggest that the three encephalopathy mutations that we studied here affect rather the plasma membrane than the Golgi pools of Gαo-mediated activities (16, 19).
The following considerations are needed before interpreting the complex cellular interaction aberrations of the pathologic Gαo variants. Long-lived pools of monomeric GDP-loaded Gαo are induced by GPCRs (58), and many signaling partners of Gαo do not discriminate between GTP- and GDP-bound states of the protein (16, 59–61). In contrast, regulators of Gαo activities “care” about the nucleotide state of the G protein: Gβγ and AGS3 act as guanine nucleotide dissociation inhibitor proteins that bind the GDP form of Gαo (2, 17, 21, 62), and RGS19 acts as a GTPase activating protein that recognizes the GTP form (3, 4). Put together with our biochemical and cellular characterizations of the pathologic Gαo mutants, these considerations may speak for the following. First, given the lack of cellular interaction with the RGS protein whose function is to speed up GTP hydrolysis (3, 4), the cellular GTP residence of the mutants is expected to be even more pronounced, which could be one possible explanation for the reduced interactions of Gαo[G203R] and [R209C] (but not of [E246K]) with Gβγ and AGS3. Second, despite the inferred increased residence in the GTP-bound state, the mutant Gαo proteins likely do not adopt the truly activated conformation and hence fail to interact with RGS19. Third, the abnormal interactions with Gβγ and AGS3 (with the paradoxical increase of this interaction for Gαo[E246K]) might indicate that the pathological mutants have aberrated conformations not only in their GTP-bound state but also when GDP-bound. As Mg2+ was found to occupy the same location in the catalytic site of Gαi1 in both nucleotide states (63), its substitution with Zn2+ could be expected to influence both GTP and GDP forms of Gαo. In agreement, we find that cell treatment with ZPT not only recovers the RGS19 interactions of the three pathologic mutants but also tends to correct their interactions with AGS3: up for the G203R and down for the E246K. The aberrant E246K-Gβγ interactions are also corrected by ZPT. More work may be needed to gain structural insights into the GDP-GTP conformations and dynamics of the Gαo mutants and the effect of zinc ions on them.
The constitutive GTP binding by the G203R, R209C, and E246K mutant forms of Gαo suggests the gain-of-function (GOF) nature of these mutations: hypermorph in the classical Muller classification of gene mutations (64). This observation is consistent with the findings that Gαo[R209C] induced higher levels of Akt and S6 kinase phosphorylation than the wild-type protein in white blood cells, stimulating cell proliferation and neoplastic transformation, ultimately contributing to childhood acute lymphoblastic leukemia (65). Similarly, Gαo[G203R] and Gαo[E246K] have been delineated as GOF mutants on the basis of their ability to decrease the median effective concentration values for α2A adrenergic receptor-mediated inhibition of cyclic adenosine monophosphate as compared to wild-type Gαo (11).
However, we find that, despite GTP binding, the three mutants fail to adopt the proper activated conformation, resulting in the aberrant interaction with Gαo partner proteins. Furthermore, we find that G203R, R209C, and E246K have a reduced potential to respond to a panel of neuronal GPCRs. These features rather speak for a loss-of-function (LOF) or a partial LOF nature of the mutants, amorph and hypomorph, respectively (64), which is the conclusion that agrees with some other studies on these GNAO1 mutations (23, 56). Some features of the Drosophila model of the disease, namely, the delayed lethality of the Gαo[G203R]/Gαo[G203R] mutant flies as compared to homozygous Gαo null mutants (42, 43), also speak in favor of the hypomorph nature of the mutation. Last, from human patients to the Drosophila Gαo[G203R]/+ model, agreeing also with mouse (9, 23) and nematode (56, 57) models of the disease, and further agreeing with some in vitro observations (23), the encephalopathy GNAO1 mutations have been described in many instances as dominant negative or antimorph in the Muller classification (64).
This multitude of ways to genetically ascribe the nature of the encephalopathy GNAO1 mutations has created a significant confusion and debate in the field. Individual amino acid changes may have counteracting effects on different biochemical properties of the G protein (nucleotide binding and hydrolysis, interaction with regulators and effectors, structure of the active state, etc.), resulting in the emergence of the pathogenic mutations as hypermorph, amorph/hypomorph, or antimorph depending on the specific biological property that is considered. Alternatively, these mutations might be described using the only remaining type within the Muller classification, the neomorph. As described in Muller’s classical genetics work (64), neomorph represents a “change in the nature of the gene at the original locus, giving an effect not produced, or at least not produced to an appreciable extent, by the original normal gene.” In cancer, a large number of oncogenic mutations in different genes, previously considered GOF or LOF, now emerge as neomorphs (66). The recognition of a given disease-causing mutation as neomorphic imposes important restrains on the drug discovery efforts, as mere increase in the normal (nonmutated) gene function or pharmacological modulation of the function of the wild-type allele cannot be efficient in counterbalancing the neomorphic pathological function (64, 66). Instead, drug discovery efforts should be dedicated directly to the aberrant neomorphic activity, ideally (for the sake of minimizing side effects) not affecting the wild-type protein.
It is in this paradigm that we designed our drug discovery aiming at recovering the pathologic, biochemically measurable function of mutant Gαo, in the counter screen testing the drug candidates against wild-type Gαo. It is remarkable that a simple ion, Zn2+ emerging from our screening of drug candidates to recover the GTPase deficiency of the mutant Gαo, is efficient in restoring the structural rearrangements induced by the pathologic mutations with a minimal action on the wild-type protein. Our initial molecular mechanics modeling, albeit simplistic, suggested that, replacing Mg2+ from the GTP pocket of Gαo, Zn2+ has a tendency to bring back the catalytic Gln205 to the vicinity of the γ-phosphate of GTP. This trend in the structural rearrangements likely reflects the mechanism behind the ability of Zn2+ to recover the GTPase reaction of the three pathologic mutants, their cellular interactions with RGS19 and AGS3, and ultimately provide motor activity and longevity ameliorations in the Drosophila model of GNAO1 encephalopathy upon dietary zinc supplementation. In the future, more quantitative and in-depth insights into the effects of Mg2+ substitution by Zn2+, by modeling and experiments, will deepen the molecular understanding of this phenomenon.
Dietary zinc supplements have found numerous applications in human health. Being safe, they have been shown to improve neonatal brain development (52) and sleep quality in adults (51) and to ameliorate health conditions in depression (48), epilepsy (49), and a set psychiatric and neurodegenerative states (50). For example, daily 25 mg of Zn2+ applied for 6 weeks in one study (67), as well as daily 220 mg of zinc sulfate (providing 50 mg of zinc) applied for 12 days in another (68), was found to positively act on patients with depression. The upper limits of dietary zinc with no observed adverse effects, as set by the World Health Organization, are 13 mg/day for the age of 7 to 12 months, 23 mg/day for the age of 1 to 6 years, and 45 mg/day for adults (69). In this regard, dietary zinc supplementation might be considered as a potential treatment option to improve the conditions of patients with GNAO1 encephalopathy, at least those carrying the G203R, R209C, and E246K mutations. More studies will show how applicable is the Zn2+-restorable GTPase deficiency mechanism to the other GNAO1 encephalopathy mutants.
Despite decades of diverse applications of zinc supplements in human health, the details of bioavailability, pharmacokinetics, pharmacodynamics, and potential toxicities of the dietary zinc are still controversial (50, 70). Multiple factors confound the efficiency of zinc absorption from the gut, its penetration through the blood-brain barrier, and the ultimate entry and activities within neuronal cells (50, 71, 72). For example, the different efficiency of dietary zinc supplementation in rescuing the motor dysfunction and reduced life span in female versus male fruit flies might be related to different expression of certain zinc transporters in the two sexes (73). Alternatively, higher food consumption by female Drosophila [by ca. threefold over that in males given the need of egg production (74, 75)] possibly results in higher accumulation of dietary zinc, thus contributing to the better responses. We find using the Drosophila model that the rescuing capacity of the dietary zinc decreases with age. Adult Drosophila consumes much less food than larvae, and a further marked decline in food consumption in adults after the age of 2 weeks has been observed (74), thus possibly contributing to the decreased performance of aging G203R/+ flies despite the availability of the zinc-supplemented diet. Collectively, these observations might speak for necessity of continuous dietary supplementation to maintain the healing effects in patients.
Pyrithione is a chelator bringing zinc ions across cell membranes (29, 34), hence the ability of ZPT to restore the mutant Gαo functions in cell cultures. However, given the toxicity of ZPT upon systemic administration (54) also seen in our Drosophila experiments, its indications as a drug are restricted to topical applications to treat dandruff and seborrheic dermatitis (30). Diverse approaches to enhance and control safe zinc uptake and delivery are being developed, from nutritional chelators and nanoparticle carriers to intravenous, cerebrospinal, or intrabrain injections (50, 72, 76), and might be considered in the future prospective applications to patients with GNAO1 encephalopathy. As a possible alternative to the dietary supplementation, intravenous zinc (as sulfate, chloride, or gluconate salts) is part of the parenteral nutrition protocols, with the dosages of 400 μg/kg per day recommended for premature infants, 250 μg/kg per day for term infants below 3 months, and 100 μg/kg per day for children above 3 months of age; adult dosages vary from 2.5 to 30 mg/day (77).
To sum up, the study presented here sweeps from the understanding, at the molecular and even atomistic level, of the core biochemical dysfunctions seen in the three most frequent GNAO1 encephalopathy mutations to assay establishment and screening for drug candidates to rescue this dysfunction, followed by the candidate validation in biochemical and cellular models. Last, we establish an animal model of GNAO1 encephalopathy and show that a dietary supplementation of Zn2+, the active component of the treatment, provides a significant rescue of the movement disorder and life-span shortening of the mutant animals. Our work sheds light on the basic functions of the major neuronal G protein and on the molecular etiology of GNAO1 encephalopathy and might serve as grounds for recommendation of the dietary zinc supplementation as a treatment option for patients with GNAO1 encephalopathy.
MATERIALS AND METHODS
Plasmids and molecular cloning
The plasmids for the Gαo-GFP (C-terminally and internally tagged), Gαo-GST-HA (C-terminally tagged), monomeric RFP (mRFP)–Gβ1, mRFP-Gγ3, His6-RGS19, GFP-Rab3a, and AGS3-GFP were previously described (4, 14, 16, 78, 79). Arf1-mRFP was generated by replacing the Age I/Not I GFP sequence of Arf1-GFP (16) with the corresponding mRFP sequence from the mRFP-N1 plasmid (16). KDELR-HA (hemagglutinin) was obtained by replacing the Age I/Not I GFP sequence of KDELR-GFP (16) with three consecutive HA tags generated by aligned oligonucleotides (table S1). The Gαo mutants were obtained by site-directed mutagenesis in the pcDNA3.1 plasmid (4) using the primers as listed in table S1. The plasmid pET-23b encoding wild-type N-terminally tagged 6xHis-Gαo (4) was used to create E246K, R209C, and G203R mutants through subcloning using restriction sites Sph I and Eco RI from the constructs in pcDNA3.1.
Protein production and purification
The Rosetta-gami Escherichia coli strain was transformed with pET23b-Gαo wild type, pET23b-Gαo[G203R], pET23b-Gαo[R209C], or pET23b-Gαo[E246K] and grown at 37°C to an optical density at 600 nm of 0.6 before induction with 1 mM isopropyl-β-d-thiogalactopyranoside and additional growth overnight at 18°C. Cells then were harvested by centrifugation 3500g at 4°C and resuspended in tris-buffered saline (TBS) [20 mM tris-HCl (pH 7.5) and 150 mM NaCl] supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF) and 30 mM imidazole. Cells were disrupted with a high-pressure cell press homogenizer; the debris was removed by centrifugation at 15,000g for 15 min at 4°C. The supernatant was applied to the Ni2+ resin (QIAGEN) overnight in a rotary shaker at 4°C. The Ni2+ resin was washed twice with 10 resin volumes of TBS supplemented with 10 mM imidazole. On the third wash, the washing buffer was supplemented with 3% glycerol, 10 mM MgCl2, 0.1 mM dithiothreitol, and 200 μM GDP. The Ni2+ resin was washed two more times with 10 resin volumes of the washing buffer. Proteins were then eluted with TBS containing 300 mM imidazole. To subsequently remove imidazole, the protein buffer was exchanged into TBS using Vivaspin concentrator. Protein concentration was measured using the Bradford assay, and the purity was analyzed using SDS–polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie staining. Gαo[Q205L] was purified in parallel as described (4).
GTP-binding and hydrolysis assay
The GTP-binding and hydrolysis assay using BODIPY-GTP (Invitrogen) or BODIPY-GTPγS (Invitrogen) was performed as described (4). Gαo was diluted to 1 μM in the reaction buffer [TBS supplemented with 10 mM MgCl2 and 0.5% bovine serum albumin (BSA)]. The mixture was then pipetted into black 384-well plates (Greiner), and BODIPY-GTP or BODIPY-GTPγS (1 μM; Invitrogen) was added into the wells. Fluorescence measurements were performed with a Tecan Infinite M200 PRO plate reader with excitation at 485 nm and emission at 530 nm at 28°C. To trace the fast kinetics of the Gαo[G203R] and Gαo[R209C], BODIPY-GTPγS was added using the injector unit followed by immediate measurement. The GTP-binding and hydrolysis data of Gαo–wild type were fit to obtain the kbind and khydr rate constants as previously described (4), where the end point was set as the baseline. As Gαo[G203R], Gαo[R209C], and Gαo[E246K] were incapable of GTP hydrolysis, the BODIPY-GTP curve was fit with the initial fluorescence value as baseline, and khydr was calculated using this curve. For the BODIPY-GTPγS displacement analysis, 5 μl of 50 μM GDP solution in TBS was injected in 20 μl of the BODIPY-GTPγS–loaded Gαo solution at the indicated time to the final concentration of 10 μM without interruption of measurements.
Homology modeling and molecular dynamics analysis
The structure of wild-type GTPγS-bound Gαo was homology modeled using the Protein Data Bank (PDB) 1GIA structure (24) on the SWISS-MODEL server with the user template setting (80). This structure was used as a base to generate amino acid substitutions in the PyMOL 2.4.0 software and metal ion substitutions using Check My Metal web interface (81). The resulting draft PDB models of Gαo mutants bound to Mg2+ or Zn2+ were directly used in the GROMACS 2021.2 software (82, 83) to generate both the energy-minimized models and the molecular dynamics runs. To this end, the CHARMM36m all-atom force field was used (mackerell.umaryland.edu/charmm_ff.shtml#gromacs). The structures were solvated in a cubic box with 1-nm distance from protein edges; the phosphate group charge was neutralized by Na+ ions. Subsequently, energy minimization and temperature and pressure equilibration were performed using typical parameters (duration, 50 ps; step, 2 fs). A 100-ns production run was performed on high-performance computation cluster of University of Geneva with 2-fs step and leap-frog integrator and with 1-nm cutoffs for van der Waals and electrostatic cutoffs. Subsequent analysis of the trajectories and structures was performed using both built-in functions of GROMACS package and PyMOL using custom scripts.
High-throughput screening
HTS for mutant Gαo modulators was performed using the Gαo[E246K] protein and FDA Approved & Pharmacopeial Drug Library (HY-L066, MedChemExpress). Dimethyl sulfoxide (DMSO) or compounds in DMSO (12.5 μM) were mixed with Gαo[E246K] at 1 μM in a reaction buffer and BODIPY-GTP at 1 μM as described in the “GTP-binding and hydrolysis assay” section above. Reaction was carried out for 10 min.
To analyze the data generated by the HTS, two parameters were calculated: (i) binding constant (kbind) and (ii) maximal GTP uptake. For candidates affecting the kbind, the hits were picked if the compound modulated kbind by ≥2 SD of DMSO-treated wells. For candidates affecting the maximal BODIPY-GTP uptake, the hits were picked if the compound modulated the maximal GTP uptake by ≥3 SD of DMSO-treated wells. The hits were subsequently validated by performing the GTP-binding assay at 50 μM of compounds using both Gαo wild type and Gαo[E246K]. Validations were performed using commercially available sennosides (USP), ZPT (Sigma-Aldrich), ZnCl2 (Sigma-Aldrich), and pyrithione (Sigma-Aldrich).
Cell lines and culture conditions
Male mouse neuroblastoma N2a [American Type Culture Collection (ATCC), CCL-131] cells were maintained in minimum essential medium (MEM; Thermo Fisher Scientific), supplemented with 10% fetal calf serum, 2 mM l-glutamine, 1 mM pyruvate, and 1% penicillin-streptomycin at 37°C and 5% CO2. Human HEK293T (ATCC, CRL-3216) cells grew in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific), supplemented as above and under the same culture conditions.
Immunoprecipitation and pull-down
Immunoprecipitation and glutathione S-transferase (GST)–based pull-down of Gαo-GFP and Gαo-GST-HA constructs was performed as previously described (16, 20). Briefly, N2a cells were transfected with the constructs indicated in the corresponding figures, and after 24 hours, cells were directly harvested or incubated with fresh media supplemented with 1 μM ZPT, 100 μM ZnCl2, or DMSO for 3 hours at normal culture conditions. Cells were harvested with ice-cold GST lysis buffer [20 mM tris-HCl (pH 8.0), 1% Triton X-100, and 10% glycerol in phosphate-buffered saline (PBS)] supplemented with a protease inhibitor cocktail (Roche). Cell lysates were cleared by centrifugation at 16,000g for 15 min at 4°C. For GST-based pull-downs, supernatants were directly incubated with 20 μl of a 50% slurry of Glutathione Sepharose 4B beads (GE Healthcare) overnight on a rotary shaker at 4°C. For immunoprecipitation, cleared supernatants were incubated with 2 μg of nanobody against GFP (84) on ice for 30 min; then, 20 μl of Glutathione Sepharose 4B beads was added, and samples were incubated as above. Beads were repeatedly washed with lysis buffer, and bound proteins were eluted by boiling the beads with SDS-PAGE sample buffer. Samples were lastly analyzed by SDS-PAGE followed by Western blot using antibodies against GFP (dilution, 1:2000; GeneTex, GTX113617), His6-tag (dilution, 1:2000; QIAGEN, 34650), mRFP (dilution, 1:250; Santa Cruz Biotechnology, sc-101526), and HA-tag (dilution, 1:2000; Roche, 3F10). Peroxidase-conjugated antibodies were from Jackson ImmunoResearch (dilution, 1:20,000; 115-035-062 and 111-035-144). Quantification of blots was done using ImageJ.
Immunofluorescence and microscopy
For microscopy, N2a cells were transfected for 7 hours, trypsinized, and seeded on poly-l-lysine–coated coverslips in complete MEM for an additional 15 hours before fixation. Cells were fixed for 20 min with 4% paraformaldehyde in PBS, were permeabilized for 1 min using ice-cold PBS supplemented with 0.1% Triton X-100, blocked for 30 min with PBS supplemented with 1% BSA, incubated with the primary antibody against GM130 (dilution, 1:500; BD Biosciences, 610823) in blocking buffer for 2 hours at room temperature, washed, and subsequently incubated with the secondary antibody and 4′,6-diamidino-2-phenylindole (DAPI) in blocking buffer for 2 hours at room temperature. The Cy3-labeled secondary antibody was from Jackson ImmunoResearch (dilution, 1:1000; 115-165-146). Coverslips were lastly mounted with VECTASHIELD on microscope slides. Cells were recorded with a Plan-Apochromat 63×/1.4 oil objective on a LSM 800 confocal microscope and further processed using the ZEN Blue software (all Zeiss). Quantification of relative localization of different variants of Gαo at plasma membrane and Golgi was performed as in (20).
MTT assay
N2a cells (3000 cells per well) were distributed into a transparent 384-well plate. The medium of each well was replaced by 50 μl of fresh medium the next day containing the indicated concentrations of ZPT, ZnCl2, or pyrithione. After incubation for 3 hours, the medium in each well was replaced by 50 μl of Thiazolyl blue (0.5 mg/ml; Carl Roth) solution in 1× PBS. The plates were incubated for 3 hours at 37°C. Then, the solution was removed, and 30 μl of DMSO was added into each well. Absorbance at 570 nm was measured in a Tecan Infinite M200 PRO plate reader.
Analysis of the heterotrimeric G protein complex formation and dissociation by BRET
The plasmid Go1-CASE encoding nanoluciferase-tagged Gαo, Gβ3, and Gγ9 with the Venus tag was provided by G. Schulte (22). The pathologic mutations were introduced by site-directed mutagenesis using the same primers as described above to introduce corresponding mutations and the following two flanking primers 5′-AATCCAAGAGTGCTTCAACCGGTC and 5′-ATATTAACGCTTACAATTTACGCC to generate two overlapping polymerase chain reaction (PCR) fragments containing mutation for subsequent Gibson assembly in Af lII/Xma I–linearized Go1-CASE. These plasmids were cotransfected at 1:1 ratio in HEK293T cells with pcDNA3.1 as a control or the following GPCR-encoding plasmids: dopamine D2 receptor (85), ɑ2-adrenergic receptor, TANGO-tagged (86), M2 muscarinic receptor (cDNA Resource Center, #MAR0200000), and μ-opioid receptor (87). Twelve hours after transfection, the cells were seeded at 6000 cells per well in the transparent-bottom black 384-well plates. After an additional 24 hours, the medium was replaced by 10 μl of PBS. Furimazine was injected to 10 μM immediately before measurement, and agonist and antagonist solutions were injected sequentially at the indicated times to indicated concentrations in PBS. Reading was performed with a Tecan Infinite plate reader.
Plasmids for Drosophila dGαo editing
Donor plasmid pGao47-LattP-pBacDsRed-attPR for the CRISPR-Cas9 step of transgenesis
Plasmid pHD-ScarlessDsRed (Drosophila Genomics Resource Center, Bloomington, USA, stock #1364) was modified by adding LoxP sequences after DsRed coding region, for which the annealed complementary oligonucleotides loxPfw and loxPrev (table S1) were cloned into the Not I site of this plasmid. The resultant plasmid (pScarlessDsRed-lox) was digested with Aar I and Sap I and assembled with two 110–base pair (bp) attP sequences, using the NEBuilder HiFi DNA Assembly Cloning Kit (New England Biolabs, catalog no. E5520S). Plasmid pTA-attP (Addgene, #18930) was used as a template for PCR amplifications of attP, which were performed with the primer sets attPfwRI/attPrevHpaI and attPfwKpnI/attPrevHpaI (see the list of primers below). The resultant pattP-pBacDsRed-lox-attP plasmid contains the pBac transposon with the fluorescent DsRed marker flanked with two inverted attP sequences. The left homologous arm (LHA) was PCR-amplified with the dGao47Lfw and dGao47Lrev primers from Drosophila genomic DNA, producing the 815-bp PCR product, which was treated with Eco RI and further cloned into the pattP-pBacDsRed-lox-attP plasmid by the Eco RI site producing the construct pLattP-pBacDsRed-lox-attP. The right homologous arm (RHA) was PCR-amplified with the dGao47Rfw and dGao47Rrev primers, and the resulting 500-bp PCR product was treated with Kpn I and then cloned into the plasmid pLattP-pBacDsRed-lox-attP digested with Kpn I and Sma I. The resultant donor plasmid pLattP-pBacDsRed-attPR contains the DsRed marker flanked with inverted attPs and the 815-bp-long LHA and 500-bp-long RHA. All PCRs were performed using Phusion High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA, USA, catalog no. M0530S).
Plasmids providing expression of guide RNAs under the control of the Drosophila U6:2 promoter
CRISPR targets sites were identified using Target Finder (88) (targetfinder.flycrispr.neuro.brown.edu/). Four targets sites (two upstream of the fourth coding exon and two downstream of the seventh exon of dGαo) were selected. Complimentary oligonucleotides gRNAR2fw and gRNAR2rev were annealed and cloned into pENTR1A-DUAL-CRISPR (provided by A. Glotov, Umea University, Sweden), which was digested with Bbs I and Sap I (New England Biolabs, catalog nos. R0539S and R0569S). The resultant plasmid pDUAL-L1R2 contains two guide RNAs (gRNAs) for induction of double-strand breaks upstream of exon 4 and downstream of exon 7 of dGαo. The same approach was performed to construct pDUAL-R1L2 using the gRNAL2fw and gRNAL2rev; gRNAR1fw and gRNAR1rev oligonucleotides. The plasmids pDUAL-L1R2 and pDUAL-R1L2 combined with the donor plasmid pGao47-LattP-pBacDsRed-attPR were used for CRISPR-Cas9 step of transgenesis (fig. S11).
Donor plasmids for RMCE
Plasmid piB-GFP (Addgene, #13844) containing two attB sequences was used as a template for PCR amplification of the 3100-bp fragment (plasmid body flanked by attB sequences) with the primer attBcircle. Drosophila genomic DNA was used as a template for PCR amplification of three fragments with the primer sets attBNco_Gao47L/attBNco_Gao47R (1190 bp), attBNco_Gao47L/G203Rrev (367 bp), and G203Rfw/attBNco_Gao47R (725 bp). The amplified fragments 3100 and 1190 bp were mixed and circulated using the NEBuilder HiFi DNA Assembly Cloning Kit (p2xattB-dGaoWT). The amplified fragments 3100, 367, and 725 bp were mixed and treated identically (p2xattB-dGaoG203R). The resultant plasmid p2xattB-dGaoWT has two attB sites, which flank sequences between exons 4 and 7 of dGαo with about 100 bp of adjacent noncoding regions. p2xattB-dGaoG203R has the identical structure but bears G203R mutation in exon 5. Both plasmids were used as donor plasmids for RMCE step of transgenesis (fig. S11).
Drosophila lines and germline transformation
Flies were maintained at 25°C on the standard medium. For dietary experiment, the food was supplemented with 200 μM ZnCl2 or 10 μM ZPT. The line y[1], sc[*], v[1], sev[21]; P{y[+t7.7] v[+t1.8]=nos-Cas9.R}attP2 expressing Cas9 in the germline under the control of the nos promoter [Bloomington Drosophila Stock Center (BDSC), Bloomington USA, stock #78782] was used for germline transformation in the CRISPR-Cas9 step of transgenesis (fig. S11). Transgenic flies were selected by red fluorescence in eyes provided by the expression of DsRed under the eye-specific 3xP3 promoter. The resultant fly stock dGαo[47dsRed] was combined with P{y[+t7.7]=nos-phiC31\int.NLS}X (BDSC, stock #34770) and balanced over CyO. This stock was used for the RMCE step of transgenesis (fig. S11). Transgenic flies in this case were selected by the absence of red fluorescence in eyes. The resultant alleles and dGαo[WT-control] were balanced over CyO. The stock dGαo[G203R]/CyO is maintained in the heterozygous state as the G203R mutation leads lethality in homozygous. The stock dGαo[WT-control] is viable and fertile in the homozygous state. This allele having the identical background to that of the mutant one was used as the wild-type control in the negative geotaxis assay and for calculation of the longevity. Germline transformation was performed as described previously (40). A fluorescence stereomicroscope [Zeiss SteREO Discovery.V8, Carl Zeiss, Jena Germany with the Filter Set 43 HE for the DsRed fluorescent dye detection (excitation BP 550/25; emission BP 605/70)] was used for selection of transgenic flies with/without fluorescence in the eyes.
Molecular analysis of established fly stocks
Genomic DNA was isolated from individual flies of different genotypes as described previously (89). The established transgenic fly strains were verified by PCR with the primers annealing to the neighboring genomic sequences outside the homologous arms and inside the donor plasmids. PCR analysis was carried out with different primer sets: dGao47Test3 (1)/dGao47Rnew1RevLong (2), dGao47Test3 (1)/pBacWTlong (3) and pBac_rev (4)/dGao47Rnew1RevLong (2) for dGαo[47dsRed]; dGao47Test3 (1)/dGao47Rnew1RevLong (2) and dGao47Test3 (1)/G203Rrev (5) for dGαo[WT-control] and dGαo[G203R]/CyO (fig. S11 and table S1). All PCRs were performed using Phusion High-Fidelity DNA Polymerase (New England Biolabs) following the manufacturer’s instructions. Intactness of inserted sequences via RMCE (stocks dGαo[WT-control] and dGαo[G203R]/CyO) was verified by sequencing. Proper expression of these alleles was verified by sequencing their cDNA performed as described previously (40).
Locomotion, life span, and neurodegeneration in Drosophila
The negative geotaxis assay was performed as described previously (40) using 5- to 6-day-old flies. Further quantitative locomotion analyses were performed as described (55) with 15- and 30-day-old flies. Measurement of the life span was performed as described (90); A total of 110 males and 140 females of each genotype and on each supplemented food were monitored. Adult brain degeneration was assessed following (47) using costaining with Alexa Fluor 488 phalloidin (1:100; Thermo Fisher Scientific) and DAPI (1:1000) after fixation in 4% paraformaldehyde/0.5% Triton X-100 in PBS. Fluorescent images (Z-stacks) were acquired with a Zeiss LSM 800 Airyscan confocal microscope.
Acknowledgments
We thank B. Ben-Zeev and D. Silachev for critically reading the manuscript, S. Troccaz for the technical assistance, and the HumanaFly facility of the Faculty of Medicine, University of Geneva for hosting the Drosophila experiments.
Funding: This work was supported by the Swiss National Science Foundation grant #31003A_175658 to V.L.K. J.V. was also supported by a research fellowship from the Bow Foundation.
Author contributions: Y.A.L. and A.K. performed biochemical experiments and HTS. A.K. performed structural modeling and molecular dynamics. M.S. generated and performed experiments with Drosophila models. G.P.S., Y.A.L., and J.V. performed cellular experiments. V.L.K. designed and supervised the study and wrote the manuscript. All authors contributed to revising the manuscript.
Competing interests: The HTS and initial hit validation phases of this work were, in part, financed by a private donation from the family of a GNAO1-encephalopath child. The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S13
Table S1
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S3
REFERENCES AND NOTES
- 1.Hauser A. S., Attwood M. M., Rask-Andersen M., Schiöth H. B., Gloriam D. E., Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 16, 829–842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oldham W. M., Hamm H. E., Heterotrimeric G protein activation by G-protein-coupled receptors. Nat. Rev. Mol. Cell Biol. 9, 60–71 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Ross E. M., Wilkie T. M., GTPase-activating proteins for heterotrimeric G proteins: Regulators of G protein signaling (RGS) and RGS-like proteins. Annu. Rev. Biochem. 69, 795–827 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Lin C., Koval A., Tishchenko S., Gabdulkhakov A., Tin U., Solis G. P., Katanaev V. L., Double suppression of the Gα protein activity by RGS proteins. Mol. Cell 53, 663–671 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Nakamura K., Kodera H., Akita T., Shiina M., Kato M., Hoshino H., Terashima H., Osaka H., Nakamura S., Tohyama J., Kumada T., Furukawa T., Iwata S., Shiihara T., Kubota M., Miyatake S., Koshimizu E., Nishiyama K., Nakashima M., Tsurusaki Y., Miyake N., Hayasaka K., Ogata K., Fukuda A., Matsumoto N., Saitsu H., De novo mutations in GNAO1, encoding a Gαo subunit of heterotrimeric G proteins, cause epileptic encephalopathy. Am. J. Hum. Genet. 93, 496–505 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schirinzi T., Garone G., Travaglini L., Vasco G., Galosi S., Rios L., Castiglioni C., Barassi C., Battaglia D., Gambardella M. L., Cantonetti L., Graziola F., Marras C. E., Castelli E., Bertini E., Capuano A., Leuzzi V., Phenomenology and clinical course of movement disorder in GNAO1 variants: Results from an analytical review. Parkinsonism Relat. Disord. 61, 19–25 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Kelly M., Park M., Mihalek I., Rochtus A., Gramm M., Pérez-Palma E., Axeen E. T., Hung C. Y., Olson H., Swanson L., Anselm I., Briere L. C., High F. A., Sweetser D. A.; Undiagnosed Diseases Network, Kayani S., Snyder M., Calvert S., Scheffer I. E., Yang E., Waugh J. L., Lal D., Bodamer O., Poduri A., Spectrum of neurodevelopmental disease associated with the GNAO1 guanosine triphosphate-binding region. Epilepsia 60, 406–418 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Axeen E., Bell E., Robichaux Viehoever A., Schreiber J. M., Sidiropoulos C., Goodkin H. P., Results of the first GNAO1-related neurodevelopmental disorders caregiver survey. Pediatr. Neurol. 121, 28–32 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Silachev D., Koval A., Savitsky M., Padmasola G., Quairiaux C., Thorel F., Katanaev V. L., Mouse models characterize GNAO1 encephalopathy as a neurodevelopmental disorder leading to motor anomalies: From a severe G203R to a milder C215Y mutation. Acta Neuropathol. Commun. 10, 9 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akamine S., Okuzono S., Yamamoto H., Setoyama D., Sagata N., Ohgidani M., Kato T. A., Ishitani T., Kato H., Masuda K., Matsushita Y., Ono H., Ishizaki Y., Sanefuji M., Saitsu H., Matsumoto N., Kang D., Kanba S., Nakabeppu Y., Sakai Y., Ohga S., GNAO1 organizes the cytoskeletal remodeling and firing of developing neurons. FASEB J. 34, 16601–16621 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Feng H., Sjögren B., Karaj B., Shaw V., Gezer A., Neubig R. R., Movement disorder in GNAO1 encephalopathy associated with gain-of-function mutations. Neurology 89, 762–770 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knight K. M., Ghosh S., Campbell S. L., Lefevre T. J., Olsen R. H. J., Smrcka A. V., Valentin N. H., Yin G., Vaidehi N., Dohlman H. G., A universal allosteric mechanism for G protein activation. Mol. Cell 81, 1384–1396.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benato A., Carecchio M., Burlina A., Paoloni F., Sartori S., Nosadini M., d’Avella D., Landi A., Antonini A., Long-term effect of subthalamic and pallidal deep brain stimulation for status dystonicus in children with methylmalonic acidemia and GNAO1 mutation. J. Neural Transm. (Vienna) 126, 739–757 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Solis G. P., Kozhanova T. V., Koval A., Zhilina S. S., Mescheryakova T. I., Abramov A. A., Ishmuratov E. V., Bolshakova E. S., Osipova K. V., Ayvazyan S. O., Lebon S., Kanivets I. V., Pyankov D. V., Troccaz S., Silachev D. N., Zavadenko N. N., Prityko A. G., Katanaev V. L., Pediatric encephalopathy: Clinical, biochemical and cellular insights into the role of Gln52 of GNAO1 and GNAI1 for the dominant disease. Cell 10, 2749 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larrivee C. L., Feng H., Quinn J. A., Shaw V. S., Leipprandt J. R., Demireva E. Y., Xie H., Neubig R. R., Mice with GNAO1 R209H movement disorder variant display hyperlocomotion alleviated by risperidone. J. Pharmacol. Exp. Ther. 373, 24–33 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Solis G. P., Bilousov O., Koval A., Lüchtenborg A.-M., Lin C., Katanaev V. L., Golgi-resident Gαo promotes protrusive membrane dynamics. Cell 170, 939–955.e24 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Kopein D., Katanaev V. L., Drosophila GoLoco-protein pins is a target of Galpha(o)-mediated G protein-coupled receptor signaling. Mol. Biol. Cell 20, 3865–3877 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egger-Adam D., Katanaev V. L., The trimeric G protein Go inflicts a double impact on axin in the Wnt/frizzled signaling pathway. Dev. Dyn. 239, 168–183 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Solis G. P., Katanaev V. L., Gαo (GNAO1) encephalopathies: Plasma membrane vs. Golgi functions. Oncotarget 9, 23846–23847 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solis G. P., Kazemzadeh A., Abrami L., Valnohova J., Alvarez C., van der Goot F. G., Katanaev V. L., Local and substrate-specific S-palmitoylation determines subcellular localization of Gαo. Nat. Commun. 13, 2072 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katanayeva N., Kopein D., Portmann R., Hess D., Katanaev V. L., Competing activities of heterotrimeric G proteins in Drosophila wing maturation. PLOS ONE 5, e12331 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schihada H., Shekhani R., Schulte G., Quantitative assessment of constitutive G protein-coupled receptor activity with BRET-based G protein biosensors. Sci. Signal. 14, eabf1653 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Muntean B. S., Masuho I., Dao M., Sutton L. P., Zucca S., Iwamoto H., Patil D. N., Wang D., Birnbaumer L., Blakely R. D., Grill B., Martemyanov K. A., Gαo is a major determinant of cAMP signaling in the pathophysiology of movement disorders. Cell Rep. 34, 108718 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coleman D. E., Berghuis A. M., Lee E., Linder M. E., Gilman A. G., Sprang S. R., Structures of active conformations of Gi alpha 1 and the mechanism of GTP hydrolysis. Science 265, 1405–1412 (1994). [DOI] [PubMed] [Google Scholar]
- 25.Iiri T., Farfel Z., Bourne H. R., Conditional activation defect of a human Gsalpha mutant. Proc. Natl. Acad. Sci. U.S.A. 94, 5656–5661 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sondek J., Lambright D. G., Noel J. P., Hamm H. E., Sigler P. B., GTPase mechanism of Gproteins from the 1.7-A crystal structure of transducin alpha-GDP-AIF-4. Nature 372, 276–279 (1994). [DOI] [PubMed] [Google Scholar]
- 27.Kimple A. J., Bosch D. E., Giguere P. M., Siderovski D. P., Regulators of G-protein signaling and their Gα substrates: Promises and challenges in their use as drug discovery targets. Pharmacol. Rev. 63, 728–749 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slep K. C., Kercher M. A., Wieland T., Chen C. K., Simon M. I., Sigler P. B., Molecular architecture of Galphao and the structural basis for RGS16-mediated deactivation. Proc. Natl. Acad. Sci. U.S.A. 105, 6243–6248 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barnett B. L., Kretschmar H. C., Hartman F. A., Structural characterization of bis(N-oxopyridine-2-thionato)zinc(II). Inorg. Chem. 16, 1834–1838 (1977). [Google Scholar]
- 30.Faergemann J., Management of seborrheic dermatitis and pityriasis versicolor. Am. J. Clin. Dermatol. 1, 75–80 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Qiu M., Chen Y., Chu Y., Song S., Yang N., Gao J., Wu Z., Zinc ionophores pyrithione inhibits herpes simplex virus replication through interfering with proteasome function and NF-κB activation. Antiviral Res. 100, 44–53 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Chen M., Ding Y., Ke Y., Zeng Y., Liu N., Zhong Y., Hua X., Li Z., Xiong Y., Wu C., Yu H., Anti-tumour activity of zinc ionophore pyrithione in human ovarian cancer cells through inhibition of proliferation and migration and promotion of lysosome-mitochondrial apoptosis. Artif. Cells Nanomed. Biotechnol. 48, 824–833 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Sekler I., Sensi S. L., Hershfinkel M., Silverman W. F., Mechanism and regulation of cellular zinc transport. Mol. Med. 13, 337–343 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding W.-Q., Lind S. E., Metal ionophores—An emerging class of anticancer drugs. IUBMB Life 61, 1013–1018 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Anderegg G., Komplexone XXXVI. Reakinsenthalpie und -entropie bei der bildung der metallkomplexe der höheren EDTA-homologen. Helvetica Chim. Acta 47, 1801–1814 (1964). [Google Scholar]
- 36.Dudev T., Lim C., Principles governing Mg, Ca, and Zn binding and selectivity in proteins. Chem. Rev. 103, 773–788 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Carraway R. E., Dobner P. R., Zinc pyrithione induces ERK- and PKC-dependent necrosis distinct from TPEN-induced apoptosis in prostate cancer cells. Biochim. Biophys. Acta Mol. Cell Res. 1823, 544–557 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Koh J. Y., Choi D. W., Zinc toxicity on cultured cortical neurons: Involvement of N-methyl-d-aspartate receptors. Neuroscience 60, 1049–1057 (1994). [DOI] [PubMed] [Google Scholar]
- 39.S. E. Mohr, in First in Fly: Drosophila Research and Biological Discovery, (Harvard Univ. Press, 2018), p. xii. [Google Scholar]
- 40.Savitsky M., Solis G. P., Kryuchkov M., Katanaev V. L., Humanization of drosophila Gαo to model GNAO1 paediatric encephalopathies. Biomedicine 8, 395 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bateman J. R., Lee A. M., Wu C.-t., Site-specific transformation of Drosophila via ϕC31 integrase-mediated cassette exchange. Genetics 173, 769–777 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frémion F., Astier M., Zaffran S., Guillèn A., Homburger V., Sémériva M., The heterotrimeric protein Go is required for the formation of heart epithelium in Drosophila. J. Cell Biol. 145, 1063–1076 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katanaev V. L., Ponzielli R., Semeriva M., Tomlinson A., Trimeric G protein-dependent frizzled signaling in Drosophila. Cell 120, 111–122 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Saitsu H., Fukai R., Ben-Zeev B., Sakai Y., Mimaki M., Okamoto N., Suzuki Y., Monden Y., Saito H., Tziperman B., Torio M., Akamine S., Takahashi N., Osaka H., Yamagata T., Nakamura K., Tsurusaki Y., Nakashima M., Miyake N., Shiina M., Ogata K., Matsumoto N., Phenotypic spectrum of GNAO1 variants: Epileptic encephalopathy to involuntary movements with severe developmental delay. Eur. J. Hum. Genet. 24, 129–134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arya R., Spaeth C., Gilbert D. L., Leach J. L., Holland K. D., GNAO1-associated epileptic encephalopathy and movement disorders: c.607G>A variant represents a probable mutation hotspot with a distinct phenotype. Epileptic Disord. 19, 67–75 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Schorling D. C., Dietel T., Evers C., Hinderhofer K., Korinthenberg R., Ezzo D., Bönnemann C. G., Kirschner J., Expanding phenotype of de novo mutations in GNAO1: Four new cases and review of literature. Neuropediatrics 48, 371–377 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Behnke J. A., Ye C., Moberg K. H., Zheng J. Q., A protocol to detect neurodegeneration in Drosophila melanogaster whole-brain mounts using advanced microscopy. STAR Protoc. 2, 100689 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.da Silva L. E. M., de Santana M. L. P., Costa P. R. F., Pereira E. M., Nepomuceno C. M. M., de Oliveira Queiroz V. A., de Oliveira L. P. M., da Conceição- Machado M. E. P., de Sena E. P., Zinc supplementation combined with antidepressant drugs for treatment of patients with depression: A systematic review and meta-analysis. Nutr. Rev. 79, 1–12 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Doboszewska U., Młyniec K., Wlaź A., Poleszak E., Nowak G., Wlaź P., Zinc signaling and epilepsy. Pharmacol. Ther. 193, 156–177 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Grabrucker A. M., Rowan M., Garner C. C., Brain-delivery of zinc-ions as potential treatment for neurological diseases: Mini review. Drug Deliv. Lett. 1, 13–23 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cherasse Y., Urade Y., Dietary zinc acts as a sleep modulator. Int. J. Mol. Sci. 18, 2334 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brion L. P., Heyne R., Lair C. S., Role of zinc in neonatal growth and brain growth: Review and scoping review. Pediatr. Res. 89, 1627–1640 (2021). [DOI] [PubMed] [Google Scholar]
- 53.Qin Q., Wang X., Zhou B., Functional studies of Drosophilazinc transporters reveal the mechanism for dietary zinc absorption and regulation. BMC Biol. 11, 101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.H. Fliss, Zinc ionophores as anti-stress agent. Patent number: PCT/CA02/00458. Date of filling: 4 April 2002; issued date of publication: 17 October 2002.
- 55.Madabattula S. T., Strautman J. C., Bysice A. M., O’Sullivan J. A., Androschuk A., Rosenfelt C., Doucet K., Rouleau G., Bolduc F., Quantitative analysis of climbing defects in a drosophila model of neurodegenerative disorders. J. Vis. Exp. , 52741 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang D., Dao M., Muntean B. S., Giles A. C., Martemyanov K. A., Grill B., Genetic modeling of GNAO1 disorder delineates mechanisms of Gαo dysfunction. Hum. Mol. Genet. 31, 510–522 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Di Rocco M., Galosi S., Lanza E., Tosato F., Caprini D., Folli V., Friedman J., Bocchinfuso G., Martire A., Schiavi E. D., Leuzzi V., Martinelli S., Caenorhabditis elegans provides an efficient drug screening platform for GNAO1-related disorders and highlights the potential role of caffeine in controlling dyskinesia. Hum. Mol. Genet. 31, 929–941 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katanaev V. L., Chornomorets M., Kinetic diversity in G-protein-coupled receptor signalling. Biochem. J. 401, 485–495 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luchtenborg A. M., Solis G. P., Egger-Adam D., Koval A., Lin C., Blanchard M. G., Kellenberger S., Katanaev V. L., Heterotrimeric Go protein links Wnt-Frizzled signaling with ankyrins to regulate the neuronal microtubule cytoskeleton. Development 141, 3399–3409 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katanaev V. L., The Wnt/Frizzled GPCR signaling pathway. Biochemistry (Mosc.) 75, 1428–1434 (2010). [DOI] [PubMed] [Google Scholar]
- 61.Lin C., Katanaev V. L., Kermit interacts with Gαo, vang, and motor proteins in Drosophila planar cell polarity. PLOS ONE 8, e76885 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Willard F. S., Kimple R. J., Siderovski D. P., Return of the GDI: The GoLoco motif in cell division. Annu. Rev. Biochem. 73, 925–951 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Coleman D. E., Sprang S. R., Crystal structures of the G protein Gi alpha 1 complexed with GDP and Mg2+: A crystallographic titration experiment. Biochemistry 37, 14376–14385 (1998). [DOI] [PubMed] [Google Scholar]
- 64.Muller H. J., Further studies on the nature and causes of gene mutations. Int. Congr. Genet. 6 1, 213–255 (1932). [Google Scholar]
- 65.Song L., Yu B., Yang Y., Liang J., Zhang Y., Ding L., Wang T., Wan X., Yang X., Tang J., Wang S., Li B., Li Y., Feng H., Identification of functional cooperative mutations of GNAO1 in human acute lymphoblastic leukemia. Blood 137, 1181–1191 (2021). [DOI] [PubMed] [Google Scholar]
- 66.Takiar V., Ip C. K., Gao M., Mills G. B., Cheung L. W., Neomorphic mutations create therapeutic challenges in cancer. Oncogene 36, 1607–1618 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nowak G., Siwek M., Dudek D., Zieba A., Pilc A., Effect of zinc supplementation on antidepressant therapy in unipolar depression: A preliminary placebo-controlled study. Pol. J. Pharmacol. 55, 1143–1147 (2003). [PubMed] [Google Scholar]
- 68.Salari S., Khomand P., Arasteh M., Yousefzamani B., Hassanzadeh K., Zinc sulphate: A reasonable choice for depression management in patients with multiple sclerosis: A randomized, double-blind, placebo-controlled clinical trial. Pharmacol. Rep. 67, 606–609 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Gibson R. S., King J. C., Lowe N., A review of dietary zinc recommendations. Food Nutr. Bull. 37, 443–460 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Duan M., Li T., Liu B., Yin S., Zang J., Lv C., Zhao G., Zhang T., Zinc nutrition and dietary zinc supplements. Crit. Rev. Food Sci. Nutr., 1–16 (2021). [DOI] [PubMed] [Google Scholar]
- 71.Frederickson C. J., Koh J. Y., Bush A. I., The neurobiology of zinc in health and disease. Nat. Rev. Neurosci. 6, 449–462 (2005). [DOI] [PubMed] [Google Scholar]
- 72.Udechukwu M. C., Collins S. A., Udenigwe C. C., Prospects of enhancing dietary zinc bioavailability with food-derived zinc-chelating peptides. Food Funct. 7, 4137–4144 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Lye J. C., Richards C. D., Dechen K., Paterson D., de Jonge M. D., Howard D. L., Warr C. G., Burke R., Systematic functional characterization of putative zinc transport genes and identification of zinc toxicosis phenotypes in Drosophila melanogaster. J. Exp. Biol. 215, 3254–3265 (2012). [DOI] [PubMed] [Google Scholar]
- 74.Wong R., Piper M. D., Wertheim B., Partridge L., Quantification of food intake in Drosophila. PLOS ONE 4, e6063 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Deshpande S. A., Carvalho G. B., Amador A., Phillips A. M., Hoxha S., Lizotte K. J., Ja W. W., Quantifying Drosophila food intake: Comparative analysis of current methodology. Nat. Methods 11, 535–540 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Abdelnour S. A., Alagawany M., Hashem N. M., Farag M. R., Alghamdi E. S., Hassan F. U., Bilal R. M., Elnesr S. S., Dawood M. A. O., Nagadi S. A., Elwan H. A. M., ALmasoudi A. G., Attia Y. A., Nanominerals: Fabrication methods, benefits and hazards, and their applications in ruminants with special reference to selenium and zinc nanoparticles. Animals 11, 1916 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hardy G., Menendez A. M., Manzanares W., Trace element supplementation in parenteral nutrition: Pharmacy, posology, and monitoring guidance. Nutrition 25, 1073–1084 (2009). [DOI] [PubMed] [Google Scholar]
- 78.Vural A., Fadillioglu E., Kelesoglu F., Ma D., Lanier S. M., Role of G-proteins and phosphorylation in the distribution of AGS3 to cell puncta. J. Cell Sci. 131, jcs216507 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miete C., Solis G. P., Koval A., Brückner M., Katanaev V. L., Behrens J., Bernkopf D. B., Gαi2-induced conductin/axin2 condensates inhibit Wnt/β-catenin signaling and suppress cancer growth. Nat. Commun. 13, 674 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F. T., de Beer T. A. P., Rempfer C., Bordoli L., Lepore R., Schwede T., SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Handing K. B., Niedzialkowska E., Shabalin I. G., Kuhn M. L., Zheng H., Minor W., Characterizing metal-binding sites in proteins with x-ray crystallography. Nat. Protoc. 13, 1062–1090 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., Lindahl E., GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19–25 (2015). [Google Scholar]
- 83.S. Páll, M. J. Abraham, C. Kutzner, B. Hess, E. Lindahl, Tackling exascale software challenges in molecular dynamics simulations with GROMACS. In Solving Software Challenges for Exascale, S. Markidis, E. Laure, Eds. (International Conference on Exascale Applications and Software, EASC 2014, Stockholm, Sweden, April 2-3, 2014; Lecture Notes in Computer Science, vol 8759. Springer, 2015), pp. 3–27. [Google Scholar]
- 84.Katoh Y., Nozaki S., Hartanto D., Miyano R., Nakayama K., Architectures of multisubunit complexes revealed by a visible immunoprecipitation assay using fluorescent fusion proteins. J. Cell Sci. 128, 2351–2362 (2015). [DOI] [PubMed] [Google Scholar]
- 85.Laschet C., Dupuis N., Hanson J., A dynamic and screening-compatible nanoluciferase-based complementation assay enables profiling of individual GPCR-G protein interactions. J. Biol. Chem. 294, 4079–4090 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kroeze W. K., Sassano M. F., Huang X. P., Lansu K., McCorvy J. D., Giguère P. M., Sciaky N., Roth B. L., PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat. Struct. Mol. Biol. 22, 362–369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stoeber M., Jullié D., Lobingier B. T., Laeremans T., Steyaert J., Schiller P. W., Manglik A., von Zastrow M., A genetically encoded biosensor reveals location bias of opioid drug action. Neuron 98, 963–976.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gratz S. J., Ukken F. P., Rubinstein C. D., Thiede G., Donohue L. K., Cummings A. M., O’Connor-Giles K. M., Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gloor G. B., Preston C. R., Johnson-Schlitz D. M., Nassif N. A., Phillis R. W., Benz W. K., Robertson H. M., Engels W. R., Type I repressors of P element mobility. Genetics 135, 81–95 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.N. J. Linford, C. Bilgir, J. Ro, S. D. Pletcher, Measurement of lifespan in Drosophila melanogaster. J. Vis. Exp., 50068 (2013). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S13
Table S1
Movies S1 to S3