Summary
Mosquito‐borne diseases remain a major cause of morbidity and mortality. Population replacement strategies involving the wMel strain of Wolbachia are being used widely to control mosquito‐borne diseases. However, these strategies may be influenced by temperature because wMel is vulnerable to heat. wMel infections in Drosophila melanogaster are genetically diverse, but few transinfections of wMel variants have been generated in Aedes aegypti. Here, we successfully transferred a wMel variant (termed wMelM) originating from a field‐collected D. melanogaster into Ae. aegypti. The new wMelM variant (clade I) is genetically distinct from the original wMel transinfection (clade III), and there are no genomic differences between wMelM in its original and transinfected host. We compared wMelM with wMel in its effects on host fitness, temperature tolerance, Wolbachia density, vector competence, cytoplasmic incompatibility and maternal transmission under heat stress in a controlled background. wMelM showed a higher heat tolerance than wMel, likely due to higher overall densities within the mosquito. Both wMel variants had minimal host fitness costs, complete cytoplasmic incompatibility and maternal transmission, and dengue virus blocking under laboratory conditions. Our results highlight phenotypic differences between Wolbachia variants and wMelM shows potential as an alternative strain in areas with strong seasonal temperature fluctuations.
Introduction
Aedes aegypti mosquitoes transmit some of the most important arboviral diseases such as dengue, which remain a major cause of morbidity and mortality across tropical regions (Kyle and Harris, 2008; Guzman et al., 2010). A promising approach to reduce mosquito‐borne disease involves the release of Ae. aegypti infected with the bacterium Wolbachia into wild populations (Hoffmann et al., 2011; Garcia et al., 2016; Indriani et al., 2020; Ahmad et al., 2021; Wang et al., 2021). Wolbachia are common intracellular bacteria that are transmitted maternally and have a range of effects on their insect hosts (Hoffmann and Turelli, 1997). Wolbachia often affect the reproduction of their hosts, particularly by causing cytoplasmic incompatibility (Hoffmann and Turelli, 1997), a phenomenon that results in sterility when a Wolbachia‐infected male mates with an uninfected female or a female carrying a different infection (Caspari and Watson, 1959; Hoffmann and Turelli, 1997). Cytoplasmic incompatibility can be applied directly to suppress field mosquito populations through male‐only releases (Zheng et al., 2019; Crawford et al., 2020; Beebe et al., 2021; Ng and Project Wolbachia‐Singapore Consortium, 2021) or replace populations with mosquitoes carrying Wolbachia (Hoffmann et al., 2011; Yen and Failloux, 2020). Replacement strategies are undertaken because Wolbachia reduce the transmission of dengue and other arboviruses by Ae. aegypti (Moreira et al., 2009; van den Hurk et al., 2012). Effective virus suppression is related to high Wolbachia infection frequencies in populations and high densities within individual mosquitoes (Lu et al., 2012; Pinto et al., 2021).
Through embryo microinjection, several Wolbachia infections including wMel, wMelPop, wMelCS and wAlbB have been established in Ae. aegypti following interspecific transfers from Drosophila, Culex and Aedes (Xi et al., 2005; McMeniman et al., 2009; Walker et al., 2011; Fraser et al., 2017). Releases to replace existing populations with those carrying Wolbachia have been achieved with both the wMel and wAlbB strains (Hoffmann et al., 2011; Nazni et al., 2019; Tantowijoyo et al., 2020). wMel originates from D. melanogaster (Hoffmann, 1988; Walker et al., 2011) and is now widely used for population replacement in Ae. aegypti, with releases undertaken in at least 10 countries including Australia, Brazil, Vietnam and Indonesia (Hoffmann et al., 2011; Garcia et al., 2019; Tantowijoyo et al., 2020; Hien et al., 2021). Mosquitoes with the wAlbB infection have been released in Malaysia, Australia, United States and Singapore for either population replacement or suppression (Nazni et al., 2019; Staunton et al., 2019; Crawford et al., 2020; Beebe et al., 2021; Ng and Project Wolbachia‐Singapore Consortium, 2021). There is now evidence of reduced dengue transmission after Wolbachia releases following replacement with both wMel and wAlbB (Nazni et al., 2019; Ryan et al., 2019; Indriani et al., 2020; Utarini et al., 2021).
The long‐term success of Wolbachia population replacement strategies will depend on the stability of Wolbachia infections in populations and their phenotypic effects (Bull and Turelli, 2013; Hoffmann et al., 2015; Ross et al., 2019a). One of these factors is the ability to maintain Wolbachia effects under fluctuating temperatures. Compared to the range that their hosts can tolerate, Wolbachia infections may be more vulnerable to temperature extremes (Corbin et al., 2017; Ross et al., 2017). However, Wolbachia strains in Ae. aegypti respond differently to heat stress, with wAlbB‐infected mosquitoes maintaining stronger cytoplasmic incompatibility, maternal transmission and Wolbachia density under heat stress compared to wMel (Ross et al., 2017). In addition, the lack of selection response in wMel for increased heat resistance (Ross and Hoffmann, 2018) could limit the suitability of this strain in locations with extreme temperature fluctuations. Cold temperatures and long‐term quiescence of eggs can also reduce Wolbachia densities, which may lead to reduced virus blockage or cytoplasmic incompatibility following a dry season (Lau et al., 2020; Lau et al., 2021).
The wMel strain is genetically diverse in natural D. melanogaster populations, with six major intraspecific clades (Richardson et al., 2012; Early and Clark, 2013; Ilinsky, 2013). Patterns of wMel clades have shifted in recent decades, with the global spread of the wMel variant (clade III) replacing many wMel‐CS‐like (clade VI) variants (Riegler et al., 2005). wMel variants have divergent phenotypic effects on hosts, inducing different levels of protection against viruses (Chrostek et al., 2013), expression of cytoplasmic incompatibility (Veneti et al., 2003) and fitness costs (Chrostek et al., 2013). Wolbachia titres also vary across lines and populations, which may relate to wMel variants (Chrostek et al., 2013; Early and Clark, 2013). The incidence of these variants in D. melanogaster is also affected by environmental conditions, with lower Wolbachia infection frequencies in cooler temperate regions (Hoffmann, 1988; Kriesner et al., 2016). Temperature influences the phenotypic effects of wMel, including levels of virus protection (Chrostek et al., 2021) and female fertility (Kriesner et al., 2016). However, these different environmental responses may vary among wMel and wMel‐like variants as indicated by impacts of variants on maternal transmission under cool temperatures (Hague et al., 2022), host temperature preference (Truitt et al., 2019; Hague et al., 2020) and survival following heat stress (Gruntenko et al., 2017; Burdina et al., 2021).
As wMel that has so far been transfected into Ae. aegypti is vulnerable to heat stress, and wMel variants in D. melanogaster have different thermal responses as noted earlier, we were motivated to generate new wMel variants in Ae. aegypti which could show greater environmental stability. Here, we report the generation and characterization of an Ae. aegypti line (termed wMelM) infected with a wMel variant from D. melanogaster collected near Melbourne, Australia. D. melanogaster invaded Australia fairly recently around 100 years ago (Hoffmann and Weeks, 2007) so our expectation is that the wMel strain present in Melbourne is associated with a robust invasive genotype. We compared wMelM to the original wMel strain (generated by Walker et al., 2011) in terms of life history, quiescent egg viability, Wolbachia density, vector competence, cytoplasmic incompatibility and maternal transmission under cold and heat stress. We found that wMelM had higher heat tolerance without an obvious fitness cost while maintaining dengue blocking, providing a new strain that may be useful for release in different contexts.
Methods
Insect strains and colony maintenance
Aedes aegypti mosquitoes were reared in temperature‐controlled insectaries at 26 ± 1°C with a 12 h photoperiod following Ross et al. (2017). Larvae were reared in trays filled with 4 L of reverse osmosis (RO) water and provided with fish food (Hikari tropical sinking wafers, Kyorin food, Himeji, Japan) ad libitum throughout their development. Four to five hundred adults were maintained in BugDorm‐1 (27 L) cages (Megaview Science C., Taichung, Taiwan). Four‐to‐six‐day‐old female mosquitoes were starved for 24 h and then fed on the forearm of a human volunteer. Blood feeding of female mosquitoes on human volunteers was approved by the University of Melbourne Human Ethics Committee (approval 0723847). All adult subjects provided informed written consent (no children were involved).
Three Ae. aegypti populations were used in this study, with an additional population (wAlbB) used in DENV vector competence experiments. wMel‐infected mosquitoes were collected from Cairns, Australia in 2013 from areas where wMel had established in the wild population (Hoffmann et al., 2011) following release of the wMel strain developed by Walker et al. (2011). The strain was developed with the shell vial technique, which was used to infect the RML‐12 cell line with wMel Wolbachia from D. melanogaster embryos from a laboratory stock (yw 67C23 ) and maintained by continuous serial passage. Wolbachia was then purified from this cell line and injected into Ae. aegypti embryos to transfer the wMel infection (Walker et al., 2011). In contrast, the wMelM‐infected mosquitoes in our study were generated by transferring cytoplasm directly from field‐collected D. melanogaster through microinjection (see embryonic microinjection below). wMel.tet mosquitoes were cured of their wMel infection by treating adults with 2 mg mL−1 tetracycline hydrochloride in a 10% sucrose solution across two consecutive generations. wAlbB‐infected mosquitoes were generated through microinjection as described previously (Ross et al., 2021). Before experiments commenced, all populations were backcrossed to an uninfected population collected from Cairns, Australia for three consecutive generations to control for genetic background.
To generate a line for source material for microinjection for the current study, D. melanogaster were collected from the Yarra Valley, Victoria, Australia in April 2019. Isofemale lines were established and their progeny were screened for Wolbachia infection (see Wolbachia detection and density), with only eggs from Wolbachia‐infected lines used in experiments. Flies were maintained on cornmeal media 19 ± 1°C with a 12 h photoperiod (Hoffmann et al., 1986).
Embryonic microinjection
wMelM from D. melanogaster was introduced to Ae. aegypti through embryonic microinjection. Cytoplasm was removed from approximately 1 h old donor eggs and injected directly into the recipient embryo according to Xi et al. (2005). Recipient eggs were collected by placing cups filled with larval rearing water and lined with filter paper (diameter 90 mm) into cages of mosquitoes blood fed >4 days prior. Eggs (<1.5 h old, light grey in colour) were lined up on filter paper and transferred to a cover slip with double‐sided tape. Eggs were covered in 100% halocarbon oil and injected using a MINJ‐1000 microinjection system (Tritech Research, Los Angeles, CA, USA). Eggs were gently removed from the oil after 10 min using a fine paintbrush and placed on a moist piece of filter paper. Eggs were hatched 3 days post‐injection by submerging filter papers in containers filled with RO water, a few grains of yeast and half tablet of fish food. Hatching larvae were reared to adulthood and F0 females were crossed to uninfected males, blood fed and isolated for oviposition. We screened the females for wMelM infection after producing viable offspring using loop‐mediated isothermal amplification (LAMP) assay (Goncalves et al., 2019; Jasper et al., 2019). This crossing process was repeated for three further generations, with only progeny from Wolbachia‐positive females contributing to the next generation. We stopped the backcrossing and the detection once the wMelM infection reached fixation. We initially maintained the population as five independent lines derived from different F1 females (but the same F0 female). Three isofemale lines were eventually pooled to generate the wMelM line, which was used in experiments. All comparisons of treatments and lines were undertaken as fully randomized designs where comparisons were done at the same time. There were no repeats (blocks) in the experiments.
Whole Wolbachia genome sequencing
We sequenced the whole Wolbachia genome of the wMelM transinfection generated in this study at F3 post‐transinfection. We also sequenced populations from three lines at F7, which were derived from three single F1 females. Genomic DNA was extracted from a pooled sample of five individuals using a DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany). Extracted DNA was randomly fragmented to a size of 350bp then end‐polished, A‐tailed, and ligated with Illumina sequencing adapters using a NEBNext® Ultra™ DNA Library Prep Kit (New England Biolabs, Ipswich MA, USA), and further PCR enriched with primers of P5 and P7 oligos. The PCR products, which comprised the final libraries, were purified (AMPure XP system, Beckman Coulter Life Sciences, Indianapolis IN, USA) and subjected to quality control tests that included size distribution by Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA), and molarity measurement using real‐time PCR. The libraries were then pooled and sequenced on a NovaSeq 6000 (Illumina) using 2 × 150 bp chemistry by Novogene HK Company Limited, Hong Kong.
Genome assembly
Quality filtering of raw sequencing reads was performed with Trimmomatic (Bolger et al., 2014), using the following settings: leading = 20, trailing = 20, sliding window = 4:20, minlen = 70. Adapter sequences were removed using the ILLUMINACLIP function, with maximum seed mismatches = 2 and the palindrome clip threshold = 30. Filtered reads were mapped to a wMel reference genome (GenBank accession: NC_002978.6) and an Ae. aegypti mitochondrial reference genome (GenBank accession: NC_035159.1) using the Burrows–Wheeler Aligner (BWA), with the bwa mem algorithm and default parameters (Li, 2013). SAMtools and BCFtools were used to perform the quality filtering of alignments and variant calling (Li et al., 2009; Danecek et al., 2021). PCR duplicates were excluded from downstream analyses by soft masking. Reads with a MAPQ score < 25 were removed from alignments, except for those with MAPQ = 0, which were retained to allow for mapping to repetitive regions. A maximum of 2000 reads per position were used to calculate genomic likelihoods. Ploidy was set to haploid for variant calling. The variant call output was used to create a consensus genome sequence for each sample, wherein low coverage positions (depth < 5) were masked as ‘N’. For Wolbachia genomes, Kraken2 (Wood et al., 2019) was used to search for sequence contamination with the Standard‐8 precompiled reference database (https://benlangmead.github.io/aws-indexes/k2; downloaded 17 September 2021). The sequencing reads mapped by bwa to the wMel reference genome were filtered to remove reads matching taxa other than Wolbachia, and genome assemblies were then repeated with the filtered datasets, using the above pipeline. Genome sequences were inspected and aligned with Geneious v 9.1.8 (https://www.geneious.com). Read mapping densities were analysed with CNVpytor (Suvakov et al., 2021) using the read depth approach, in order to search for putative deletions and duplications within the wMel genomes. Bin sizes of 100 and 500 were tested for the histogram, partition and CNV call steps.
Phylogenetic analysis
A phylogenetic tree was constructed using the wMel genomes from the present study and two large genome‐wide single nucleotide polymorphism (SNP) datasets that include representatives of the different wMel clades, obtained from D. melanogaster sampled from various international locations (Richardson et al., 2012; Chrostek et al., 2013; Early and Clark, 2013). Polymorphic sites that were common to both datasets were combined into a single SNP matrix, with sites that contained ambiguous base calls in any sample excluded from the analysis. A total of 58 polymorphic loci were retained. Maximum likelihood trees were constructed with RAxML‐HPC v8.2.12 (Stamatakis, 2014) on XSEDE, using a general time reversible model of nucleotide substitution under the gamma model of rate heterogeneity (GTR‐GAMMA model) with the Lewis method of ascertainment bias correction (Lewis, 2001), 44 alignment patterns and rapid bootstrapping (100 inferences). Bootstrap scores were plotted onto the best scoring ML tree. RAxML‐HPC was accessed through the CIPRES Science Gateway (Miller et al., 2010).
Life history parameters and cytoplasmic incompatibility
To compare the effects of wMelM and wMel infection on mosquito life history, we performed phenotypic assessments of the wMel, wMelM and wMel.tet populations. Fresh stored eggs (<1 week old) from each population were hatched in trays filled with 3 L of RO water, a few grains of yeast, and one tablet of fish food. One day after hatching, 100 larvae were counted into trays filled with 500 mL of RO water and provided with fish food ad libitum, with 12 replicate trays per population. To determine the average larval development time for each tray, pupae were counted and sexed every day in the morning and evening. Pupae from each population were pooled across replicate trays (but with sexes separate) and used in the following experiments. The remaining pupae were pooled across sexes and released into BugDorm‐1 cages for the quiescent egg viability experiment.
To determine adult longevity, 25 females and 25 males were placed in 3 L cages with cups of 10% sucrose and water, with eight replicate cages per population. Females were blood fed once per week. Dead mosquitoes were recorded and removed three times per week until all mosquitoes had died.
To determine female fertility and patterns of cytoplasmic incompatibility between wMel variants, we set up reciprocal crosses between wMel, wMelM and wMel.tet adults. Fifty females (1 day old) from the wMel, wMelM or wMel.tet populations were aspirated into 3 L cages containing 50 wMel, wMelM or wMel.tet males (1 day old), for a total of nine crosses. Five‐day‐old females were blood fed and 30 engorged females per cross were isolated for oviposition in 70 mL specimen cups with mesh lids that were filled with 15 mL of larval rearing water and lined with a strip of sandpaper (Norton Master Painters P80 sandpaper, Saint‐Gobain Abrasives Pty., Thomastown, Victoria, Australia). Eggs were collected, partially dried, and then hatched 3 days after collection. Female fecundity was determined by counting the total number of eggs on each sandpaper strip, while hatch proportions were determined by dividing the number of hatched eggs (with a clearly detached egg cap) by the total number of eggs per female. Females that did not lay eggs or died before laying eggs were excluded from the analysis. To evaluate the fertility across gonotrophic cycles, females from the within‐strain crosses (e.g. wMel × wMel) were blood fed again after laying eggs. Eggs were collected from four gonotrophic cycles in total, when females were 9, 14, 19 and 23 days old.
As an estimate of body size, one wing each from 20 males and 20 females per population was dissected and measured for its length (Ross et al., 2016). Damaged wings were excluded from the analysis.
Quiescent egg viability
To test quiescent egg viability which is an important ecological trait that can be affected by Wolbachia infections (Lau et al., 2021), cages of 5‐day‐old females from wMel, wMelM or wMel.tet populations were blood fed and six cups filled with larval rearing water and lined with sandpaper strips were placed inside in each cage. Eggs were collected 5 days after blood feeding, partially dried, then placed in a sealed chamber with an open container of saturated potassium chloride (KCl) solution to maintain a constant humidity of ~84%. When eggs were 1, 2, 4, 6, 8, 10, 12, 14, 16, 18, 20 and 22 weeks old, small sections of each sandpaper strip were removed and submerged in water with a few grains of yeast and fish food to hatch. Twelve replicate batches of eggs were hatched per population at each time point, with around 50–100 eggs per batch. Hatch proportions were determined 3 days after immersion in water by dividing the number of hatched eggs (with a clearly detached egg cap) by the total number of eggs in each batch.
Wolbachia density across life stages
We compared the Wolbachia density of wMelM and wMel during development by storing random subsets of first instar larvae, third instar larvae, female pupae, male pupae, female adults and male adults (within 24 h of emergence) in 100% ethanol. Fifteen individuals from each life stage, sex and wMel variant were then measured for Wolbachia density using qPCR (see section ‘Wolbachia detection and density’).
DENV‐2 virus challenge and quantification
We measured DENV‐2 virus titres and infection frequencies as a proxy for the vector competence of wMelM for DENV‐2 relative to two other Wolbachia variants (wMel and wAlbB) and a matched uninfected population (wMel.tet). Experiments were performed in a quarantine insectary with Ae. aegypti held in incubators (PG50 Plant Growth Chambers, Labec Laboratory Equipment, Marrickville, NSW, Australia) set to a constant 26°C with a 12:12 light: dark photoperiod. Eggs (10 days old) from the wMel, wMelM, wMel.tet and wAlbB populations were hatched and larvae were reared to adulthood in five replicate trays of 100 larvae per temperature treatment for each population. Adults were released into 13.8 Lcages (BugDorm‐4F2222, Megaview Science C, Taichung, Taiwan) and provided with 10% sucrose solution. Adults were starved for 24 h before virus challenge and experiments were performed with 6–7‐day‐old females.
We performed virus challenge experiments with a 1 × 107 TCID50 mL−1 dose of DENV‐2 in human blood. DENV‐2 (Cosmopolitan) provided by VDRL and isolated in Melbourne from a traveller in 2016 and was grown in C6/36 cells before use in experiments. Females were fed human blood sourced from the Red Cross under agreement number 16‐10VIC‐02 and spiked with DENV‐2. Blood was provided through 6 mL Hemotek membrane feeders (Hemotek, Blackburn Lancashire, Great Britain) which were placed on the top of each cage for 30 min and heated with pocket hand warmers (Kathmandu). Fully engorged mosquitoes were transferred to cages with sucrose solution and cups lined with sandpaper strips for oviposition, with non‐blood and partially fed mosquitoes discarded. Females were maintained at 26°C for 12 day before processing. Cages were chilled briefly, then heads were dissected from bodies on a cold plate and stored individually in 1.7 mL tubes with 100 μL crushing media (DMEM with 1000 μ mL−1 streptomycin/penicillin and 2 μg mL−1 amphoterin B) and two 3 mm glass beads. Tubes were stored at −20°C for 48 h before removal from the quarantine insectary for long‐term storage at −80°C.
Virus titres were quantified from whole bodies (heads removed) from 30 individuals per population with TCID50 assays as described previously (Duchemin et al., 2017). Briefly, 30 μL of crushed mosquito were serially 10‐fold diluted in medium (DMEM containing 2% FBS). Using 96‐well plates, 50 μL of each dilution was sequentially placed in wells (six replicates). The 100 μL of fresh medium containing Vero cells (final cell confluency of 50%–60%) was then overlaid. The cells were incubated for 7 days before they were observed for cytopathic effect (CPE). The 50% endpoint was calculated using the Reed and Muench method (Reed and Muench, 1938). The infection rate was calculated as the proportion (percentage) of all experimentally infected mosquitoes (n = 30) in which DENV was detected.
Cytoplasmic incompatibility and Wolbachia density following cold and heat stress
We measured cytoplasmic incompatibility and Wolbachia density in adults after being exposed to cyclical cold or heat stress during the egg stage for 1 week. Eggs were collected on sandpaper from both Wolbachia‐infected populations. Four days after collection, batches of 40–60 eggs were brushed off the sandpaper and tipped into 0.2 ml PCR tubes (12 replicate tubes per population) and exposed to cyclical temperatures of 3–13°C, 4–14°C, 6–16°C, 27–37°C, 28–38°C or 29–39°C for 7 days in Biometra TProfessional TRIO 48 thermocyclers (Biometra, Göttingen, Germany) according to Kong et al. (2016) and Ross et al. (2019b). Eggs of the same age from each population, as well as wMel.tet eggs, were kept at 26°C. Eggs from all treatments were brought to 26°C and hatched synchronously. Larvae were reared at a controlled density (up to 100 larvae per tray of 500 ml water). Pupae were sexed and 8–30 adults per population were stored in absolute ethanol within 24 h of emergence for Wolbachia density measurements (see ‘Wolbachia detection and density’ section). The remaining pupae were left to emerge into 3 L cages (with each sex, temperature treatment and population held in separate cages) for cytoplasmic incompatibility crosses. Due to low survivorship of eggs held at 3–13°C and 4–14°C, individuals from these treatments were not used in crossing experiments.
We established two sets of crosses to test cytoplasmic incompatibility induced by Wolbachia‐infected males and test the ability of Wolbachia‐infected females to restore compatibility with Wolbachia‐infected males. In the first set, untreated wMel.tet females were crossed with Wolbachia‐infected males from each temperature treatment. In the second set, Wolbachia‐infected females from each temperature treatment were crossed with untreated Wolbachia‐infected males. Five‐day‐old females were blood‐fed and 20 females per cross were isolated for oviposition. Hatch proportions per female were determined according to previous experiments (see above).
Maternal transmission of Wolbachia
We compared the ability of wMel and wMelM females to transmit Wolbachia infections to their offspring after parental eggs were exposed to cyclical heat stress (28–38°C) for 1 week, or held at 26°C. Eggs were returned to 26°C and reared to adulthood, and wMel and wMelM females from each temperature treatment were crossed to wMel.tet males. Twenty females per cross were isolated for oviposition after blood feeding. We scored 10 offspring for their Wolbachia infection status from 10 females per infection type at each temperature to assess the loss of Wolbachia in next generation.
Wolbachia detection and density
We used LAMP assays for rapid detection of wMel during microinjection experiments according to Jasper et al. (2019) using wMel‐specific primer sets (Goncalves et al., 2019). qPCR assays were used to confirm the presence or absence of Wolbachia infection and measure relative density in all other experiments (Lee et al., 2012; Axford et al., 2016). DNA was extracted using 250 μL of 5% Chelex 100 resin (Bio‐Rad oratories, Hercules, CA) according to methods described previously (Hoffmann et al., 2014). Wolbachia infections were detected using a LightCycler® 480 High Resolution Melting Master (HRMM) kit (Roche; Cat. No. 04909631001, Roche Diagnostics Australia Pty. Ltd., Castle Hill New South Wales, Australia) and IMMOLASETM DNA polymerase (5 U μL−1) (Bioline; Cat. No. BIO‐21047) as described by Lee et al. (2012). Three primer sets were used to amplify markers specific to mosquitoes (mRpS6_F 5′AGTTGAACGTATCGTTTCCCGCTAC3′ and mRpS6_R 5′ GAAGTGACGCAGCTTGTGGTCGTCC3′), Ae. aegypti (aRpS6_F 5′ATCAAGAAGCGCCGTGTCG3′ and aRpS6_R 5′CAGGTGCAGGATCTTCATGTATTCG3′), and wMel (w1_F 5′AAAATCTTTGTGAAGAGGTGATCTGC3′ and w1_R 5′ GCACTGGGATGACAGGAAAAGG3′). Relative Wolbachia densities were determined by subtracting the Cp value of the Wolbachia‐specific marker from the Cp value of the mosquito‐specific marker. Differences in Cp were averaged across two to three consistent replicate runs, then transformed by 2 n .
Statistical analysis
All analyses were conducted using SPSS statistics version 26.0 for Mac (SPSS, Chicago, IL). Egg hatch proportions were characterized per female (i.e. individual eggs were not treated as independent data points) and this trait as well as longevity were not normally distributed according to Shapiro–Wilk tests; therefore, we analysed these data with nonparametric log‐rank tests for longevity and Kruskal–Wallis for egg hatch proportions. The development time and wing length for females and males were measured by one‐way ANOVAs. Fecundity and Wolbachia density under heat and cold were analysed by two‐way ANOVAs. Wolbachia density for different life stages was compared with a two‐way ANOVA and t test. DENV‐2 TCID50 comparisons across strains were undertaken using a Mann‐Whitney test and differences in the infection rate were evaluated using two‐tailed Fisher's tests. Because we undertook treatment/line comparisons at the same time in randomized designs, there were no experimental block/repeat effects in the analysis.
Results
Genomic analyses of the wMelM transinfection
The genome of the original wMel transinfection (Walker et al., 2011) was very similar to both the wMel reference genome (Wu et al., 2004) and the wMel genome reported by Chrostek et al. (2013), differing from each by five and four polymorphic loci, respectively, consistent with the three strains having originated from same Drosophila stock (yw 67C23 ) less than 5000 generations ago (Supporting Information Table S2). Phylogenetic analysis placed these genomes in a single monophyletic cluster together with the other members of wMel clade III (Fig. 1).
Fig. 1.
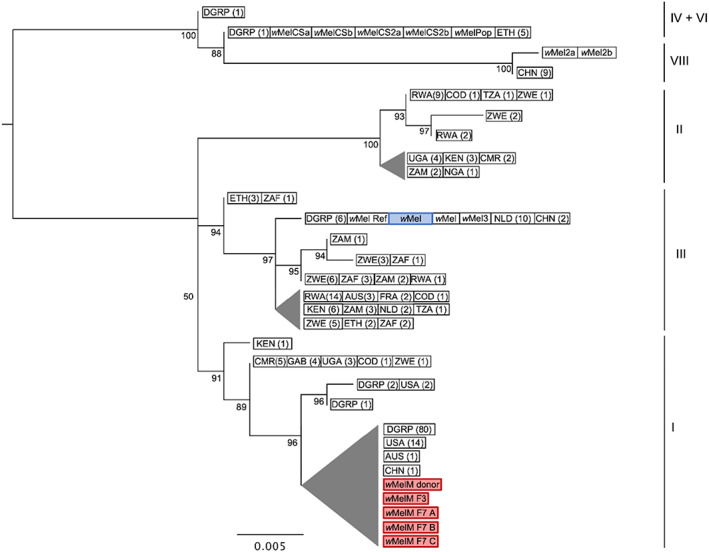
Genome‐wide SNP phylogenetic analysis of wMel variants. SNP data from the six genomes in this study and 253 previously published wMel genomes were analysed (Richardson et al., 2012; Chrostek et al., 2013; Early and Clark, 2013). The wMel variant highlighted in blue is the original wMel transinfection (Walker et al., 2011) used in phenotypic comparisons with wMelM. wMelM sequences are highlighted in red. Maximum likelihood trees were constructed with RAxML‐HPC using 58 SNP loci and ascertainment bias correction; scale bar = number of substitutions per SNP matrix site. Nodes with bootstrap values less than 50% have been collapsed; triangle height = length of longest branch within node. Wolbachia wMel clades are shown on the right. In cases where multiple samples had identical SNP haplotypes, one representative sequence was used for tree construction. The number of sequences corresponding to each entry is shown in parentheses. For wild populations, the location of sampling is shown (refer to Table S1 for sample names). AUS = Australia; CHN = China; CMR = Cameroon; COD = Democratic Republic of the Congo; ETH = Ethiopia; FRA = France; GAB = Gabon; GIN = Guinea; KEN = Kenya; NGA = Nigeria; NLD = Netherlands; RWA = Rwanda; TZA = Tanzania; UGA = Uganda; USA = USA; ZAF = South Africa; ZMB = Zambia; ZWE = Zimbabwe; DGRP = Drosophila Genetic Reference Panel (originally sampled from Raleigh NC, USA).
Clear differences were observed between the genome sequence of the original wMel transinfection strain and the wMelM variant. Phylogenetic analysis placed wMelM within wMel clade I, with greatest similarity to wMel variants from Ithaca NY, USA and Tasmania, Australia (Fig. 1). Thirty‐six SNPs and small indels were identified, with approximately half predicted to cause a change to an amino acid sequence (Supporting Information Table S2). Several of these changes were located within proteins with functions including transportation and secretion; amino acid, porphyrin, and nucleotide metabolism; and DNA replication and repair, while many others were located within hypothetical proteins of unknown function. Most SNPs identified were the same SNPs identified by Hague et al. (2022) in their comparison of wMel genomes from temperate (clade III) and tropical (clade I) D. melanogaster, with five of these being significantly associated with temperature according to their analysis (P < 0.05; see Hague et al., 2022). Analysis of read mapping densities, performed with CNVpytor, did not identify any regions of significant copy number variation within the wMelM genome, when compared with the wMel reference genome. We did, however, identify two large deletions through standard genome assembly – one within a gene encoding a S49 family peptidase and one within an ankyrin repeat containing gene. Examination of allele frequencies indicated that in addition to wMelM, a wMel variant with high identity to the wMel reference sequence is likely to have been present within the Drosophila wMelM donor population, but only wMelM appears to have been retained within the transinfected mosquito populations. The wMelM genome sequences in Ae. aegypti at F3 and three isofemale lines at F7 were identical, suggesting no genetic changes across several generations following transinfection. Relative to the Ae. aegypti wMelM genomes, the Drosophila wMelM consensus sequence contained eight SNPs and lacked the two large deletions mentioned above. It is probable that these differences are artefacts introduced by the presence of the other wMel variant in this sample.
The mitochondrial genome sequences of the five mosquito populations included in the study were very similar, with only five SNPs observed between them, four of which were located outside of known coding regions. Most of the SNPs were associated with anomalously low read depths, relative to their surrounding positions, making it likely that they represent sequencing artefacts.
Limited effects of wMelM on life history traits
We compared the effects of wMel and wMelM infection on Ae. aegypti life history traits. Overall, wMelM infection had no effect, or slightly increased mosquito fitness compared to uninfected (wMel.tet) mosquitoes (Fig. 2). Wolbachia infection had no clear effect on development time (one‐way ANOVA: P = 0.124 for females and 0.194 for males; Fig. 2A and B). Both wMel variants increased adult female longevity (Log‐rank test: wMel vs wMel.tet, P = 0.011, χ2 = 6.436, df = 1, wMelM vs wMel.tet: P = 0.019, χ2 = 5.501, df = 1; Fig. 2C) relative to wMel.tet, but male longevity was unaffected by Wolbachia infection status (Log‐rank test: wMel vs wMel.tet, P = 0.251, χ2 = 1.32, df = 1, wMelM vs wMel.tet: P = 0.091, χ2 = 2.849, df = 1; Fig. 2D). Fecundity was influenced by Wolbachia infection type (two‐way ANOVA: F 2,326 = 3.753, P = 0.024), with wMelM females consistently laying more eggs than the wMel and wMel.tet females (Fig. 2E). Median egg hatch proportions were close to 100% across all gonotrophic cycles, regardless of Wolbachia infection type (Fig. 2F). Both female (one‐way ANOVA: P = 0.124) and male (one‐way ANOVA: P = 0.194) wing length were unaffected by Wolbachia infection type (Fig. 2G and H).
Fig. 2.
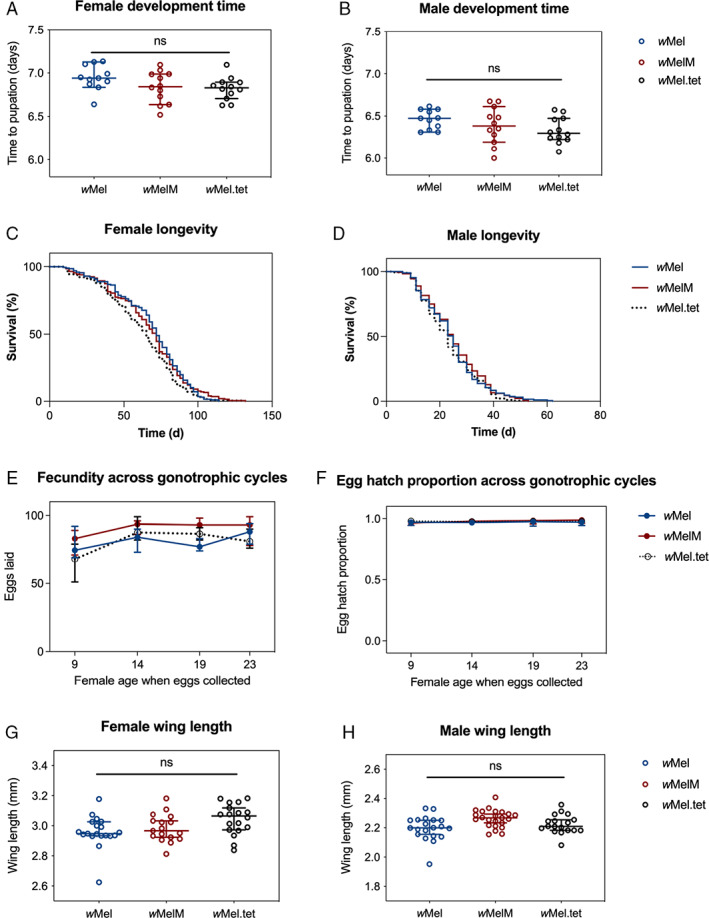
Life history parameters of backcrossed wMel, wMelM and wMel.tet Aedes aegypti populations. The wMel (blue), wMelM (red) and wMel.tet (black) populations were evaluated for the following traits: larval development time for (A) females and (B) males, longevity of (C) females and (D) males, (E) female fecundity across gonotrophic cycles, (F) egg hatch proportion across gonotrophic cycles, and wing length of (G) females and (H) males. Each point represents data averaged across a replicate container of 100 individuals (A and B) or data from individual mosquitoes (panels E–H). For each treatment/strain, we measured 12 replicate trays for development time (A–B), 8 replicate cages of 25 females and 25 males for longevity (C and D), 50 individual females for fecundity and egg hatch (E and F) and 20 individuals for wing length (G and H). Medians and 95% confidence intervals are shown in lines and error bars.
wMel and wMelM induce complete cytoplasmic incompatibility
We tested the ability of wMel and wMelM‐infected males to induce cytoplasmic incompatibility with wMel.tet females. These females produced no viable offspring when crossed to wMel and wMelM males, indicating that both variants induce complete cytoplasmic incompatibility (Table 1). Reciprocal crosses between wMel and wMelM variants resulted in high egg hatch proportions (>90%), indicating that the two variants are bidirectionally compatible (Table 1).
Table 1.
Cytoplasmic incompatibility between wMel, wMelM and wMel.tet Aedes aegypti populations.
| Female | ||||
|---|---|---|---|---|
| wMel.tet | wMel | wMelM | ||
| Male | wMel.tet | 0.980 (0.967, 1) | 0.930 (0.867, 0.969) | 0.952 (0.904, 0.990) |
| wMel | 0 (0, 0) | 0.971 (0.945, 0.983) | 0.981 (0.955, 0.990) | |
| wMelM | 0 (0, 0) | 0.963 (0.850, 0.986) | 0.962 (0.942, 0.986) | |
Median egg hatch proportions are shown with 95% confidence intervals (lower, upper).
wMel variants decrease quiescent egg viability
Stored eggs from each population were hatched every 2 weeks to determine quiescent egg viability. Egg viability decreased across time for all three infection types, but hatch proportions for both wMel variants were lower than wMel.tet from week 2 onwards (Kruskal–Wallis test, week 2: H = 12.434, df = 2, P = 0.002; Fig. 3). wMel and wMelM did not differ significantly in hatch proportion across all weeks combined (statistical test P = 0.114), suggesting that the two variants have similar quiescent egg viability.
Fig. 3.
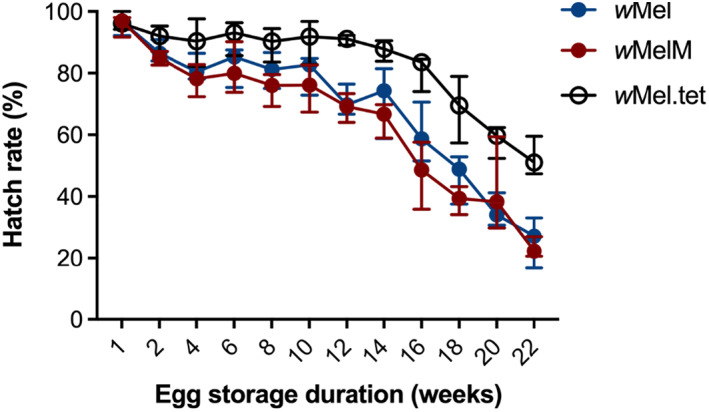
Quiescent egg viability of backcrossed wMel, wMelM and wMel.tet Aedes aegypti populations across 22 weeks. Twelve replicate batches of eggs produced by 5 day‐old females from wMel, wMelM or wMel.tet populations were hatched per population at each time point, with around 50–100 eggs per batch. Symbols show median egg hatch proportions while error bars show 95% confidence intervals.
wMel variants have similar Wolbachia density across life stages
Wolbachia density in whole individuals differed across life stages (two‐way ANOVA: F 5,177 = 75.85, P < 0.001), with low densities observed in third instar larvae (Fig. 4). We did not find differences in pairwise comparisons between the wMel and wMelM strains (t‐test: all P > 0.05) except for male pupae (P = 0.011), where wMelM male pupae had higher Wolbachia densities than wMel male pupae (Fig. 4). Overall, the Wolbachia density of wMelM was slightly higher than wMel across all life stages, but this effect was not significant (two‐way ANOVA: F 1,177 = 2.636, P = 0.106).
Fig. 4.
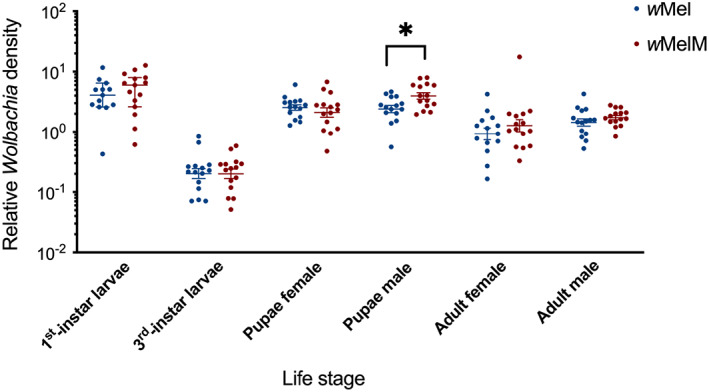
Wolbachia density of different life stages in backcrossed wMel and wMelM Aedes aegypti populations. Each point represents the relative density for an individual averaged across two to three technical replicates. We measured fifteen individuals from each life stage, sex and wMel variant. We used mRpS6 as the reference gene to calculate relative density. Medians and 95% confidence intervals are shown in lines and error bars.
wMelM infection reduces DENV‐2 viral titre and infection prevalence in mosquitoes
We estimated the vector competence of wMelM‐infected Ae. aegypti by feeding mosquitoes an infectious blood meal and comparing DENV‐2 prevalence and titres against wMel, wAlbB and wMel.tet populations. DENV‐2 titres (P = 0.016) and the proportion of DENV‐2‐infected females (P = 0.033) were significantly lower in wMelM compared with wMel.tet (Fig. 5). There was no significant difference in DENV‐2 titres between wMel and wMel.tet (P = 0.114), but the proportion of DENV‐2‐infected females (P = 0.017) was significantly lower in wMel (Fig. 5). There was no detectable difference between wMelM blockage and that of the other two Wolbachia strains, wMel and wAlbB, as measured by both DENV‐2 replication and infection proportion (all P > 0.05).
Fig. 5.
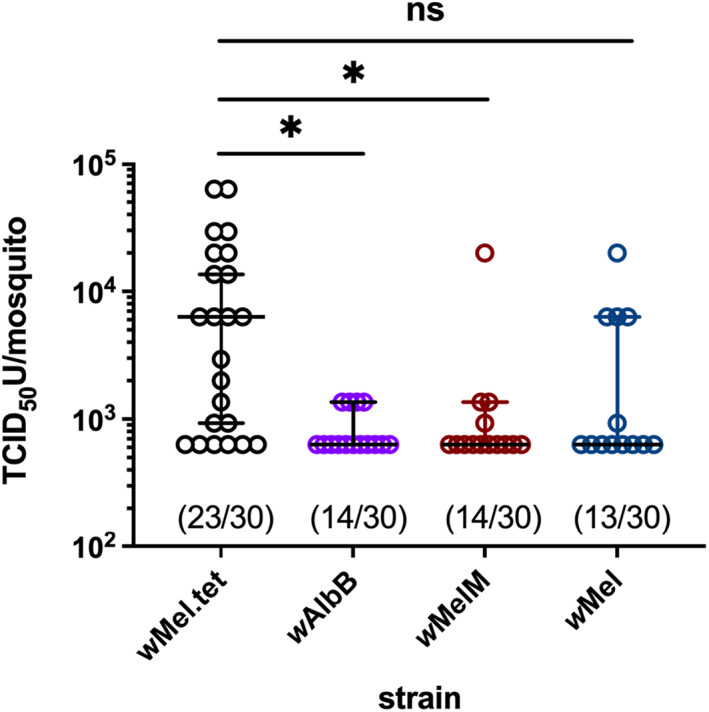
wMelM infection reduces DENV‐2 titre and prevalence in virus‐fed Aedes aegypti. Virus titres were quantified from whole bodies (heads removed) from 30 individuals per population. DENV‐2 titres (P = 0.016) and the proportion of DENV‐2 infected females (P = 0.033) were significantly lower in wMelM compared to wMel.tet. Statistical comparisons of DENV‐2 titre were compared using a Mann‐Whitney test where * indicates P < 0.05. Each point on the plot represents an individual infected mosquito (with negative mosquitoes excluded), and lines and error bars denote medians ± 95% CIs. The proportion of DENV‐2 positive mosquitoes out of the total tested (proportion infected) is indicated below each plot.
wMelM induces stronger cytoplasmic incompatibility than wMel under cyclical heat stress
We tested the ability of wMel and wMelM‐infected males to induce cytoplasmic incompatibility when eggs were exposed to cold or heat stress. Males from both wMel and wMelM lines induced complete cytoplasmic incompatibility with wMel.tet females at 6–16°C and 26°C (Fig. 6A). Incomplete cytoplasmic incompatibility was observed at 27–37°C and above, with an increasing proportion of viable offspring produced as the temperature increased (Fig. 6A). However, wMelM retained stronger cytoplasmic incompatibility than wMel at high temperatures, with significantly lower egg hatch proportions when males were exposed to 28–38°C (Kruskal–Wallis test: H = 6.743, df = 1, P = 0.009) and 29–39°C (H = 5.344, df = 1, P = 0.021).
Fig. 6.
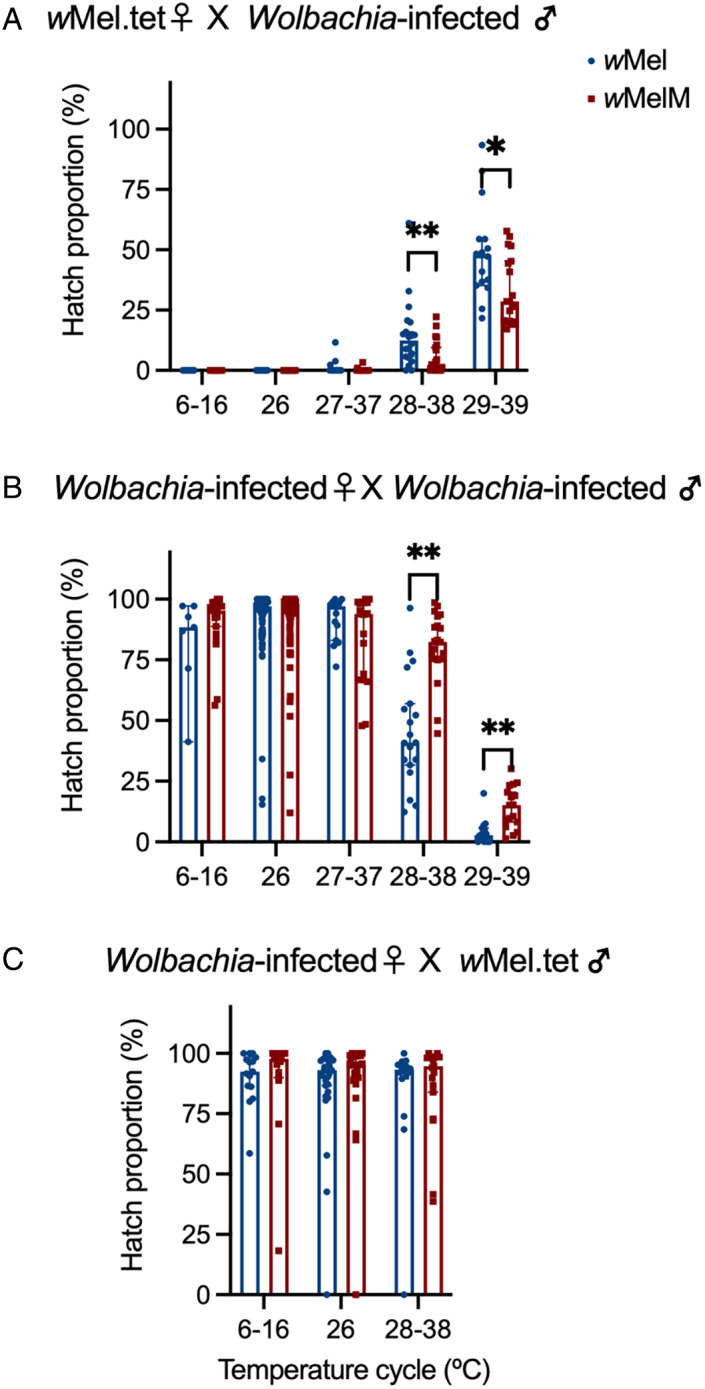
Egg hatch proportions in crosses with wMel/wMelM variants and wMel.tet under cold and heat stress. We performed crosses to test the ability of (A) Wolbachia‐infected males to induce cytoplasmic incompatibility with wMel.tet females and (B) Wolbachia‐infected females to restore compatibility with Wolbachia‐infected males. (C) Egg hatch proportions from crosses between Wolbachia‐infected females and wMel.tet males used in the maternal transmission experiment. In each cross, males (A) or females (B, C) were exposed to cyclical temperatures for 7 day or held at 26°C during the egg stage. Each point represents the hatch proportion of eggs from a single female, with up to 20 females measured per cross. Bars show medians while error bars show 95% confidence intervals.
Wolbachia‐infected females exposed to cyclical temperatures as eggs were crossed to Wolbachia‐infected males reared at 26°C to test the ability of females to restore compatibility (Fig. 6B). Compared to wMelM, wMel had a slightly decreased hatch rate under cold stress, but treatments did not differ significantly (Kruskal–Wallis test: H = 1.838, df = 1, P = 0.175). After the exposure to cyclical temperatures of 27–37°C for 7 days, hatch rate reduced. Hatch rates in both strains decreased with temperature, but wMelM had a higher hatch rate under cyclical temperatures of 28–38°C (Kruskal–Wallis test: H = 18.000, df = 1, P < 0.001) and 29–39°C (H = 13.766, df = 1, P < 0.001).
wMelM has increased maternal transmission fidelity compared to wMel under cyclical heat stress
We compared the ability of wMel and wMelM females to transmit Wolbachia to their offspring when they were exposed to 28–38°C as eggs for 7 days or reared at a constant 26°C (Fig. 6C). wMel and wMelM females transmitted the infection to all their offspring at 26°C. In contrast, wMel and wMelM‐infected females failed to transmit the infection to some of their progeny at 28–38°C (Fig. 7). More progeny from wMelM mothers had Wolbachia than from wMel mothers (66.7% ± 6.87%) at a cycling 28–38°C (Kruskal–Wallis test: H = 3.891, df = 1, P = 0.048; Fig. 7), suggesting that the transmission of wMelM may be more stable at high temperatures.
Fig. 7.
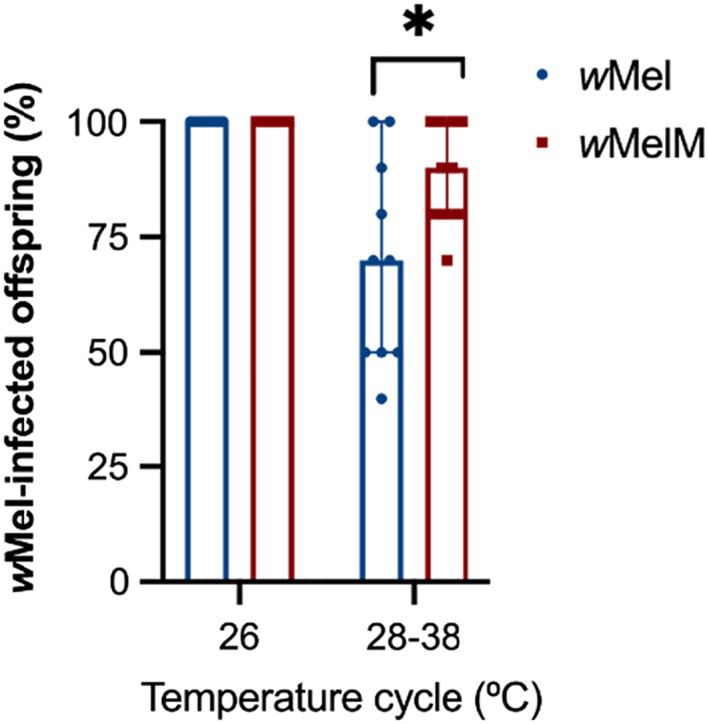
Maternal transmission of wMel/wMelM variants following maternal heat stress exposure. Wolbachia‐infected females were exposed to cyclical temperatures of 28–38°C for 7 days or held at 26°C during the egg stage, then crossed to wMel.tet males. We scored 10 offspring for their Wolbachia infection status from 10 females per infection type at each temperature. Each point represents the percent of Wolbachia‐positive offspring from a single female. Medians and 95% confidence intervals are shown in lines.
wMelM has a higher density than wMel under heat and cold stress
We measured Wolbachia density when eggs were exposed to cold or heat stress. We found no significant effect of sex on Wolbachia density (P > 0.05), so data from males and females were pooled for the following analysis. In the two‐way ANOVA, Wolbachia density was influenced both by the wMel variant (F 1,142 = 26.425, P < 0.001) and temperature (F 2,142 = 247.264, P < 0.001). At high temperatures, the density of wMel and wMelM decreased sharply (Fig. 8B). For the lower temperature, the Wolbachia density was relatively stable but was still influenced by wMel variant (F 1,99 = 12.658, P = 0.001) and temperature (F 2,99 = 18.323, P < 0.001) (Fig. 8A). Although wMel and wMelM had similar density at the adult stage when reared at constant 26°C (Fig. 4), we did find that wMelM had a higher Wolbachia density under all the cycling heat and cold stress regimes (Fig. 8). Compared to wMel, the density of wMelM was around two times higher overall in this experiment.
Fig. 8.
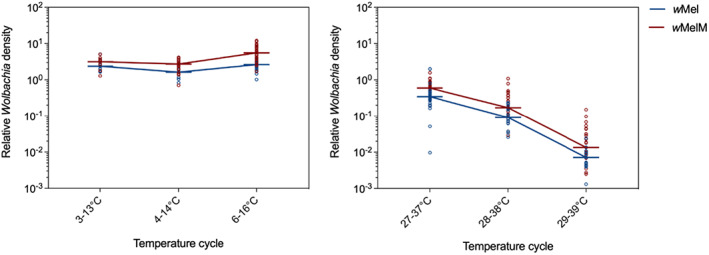
Wolbachia density in adult mosquitoes following exposure to cyclical (A) cold and (B) heat stress during the egg stage. Eggs were exposed to cyclical temperatures for 7 days. Each point represents the relative density for an individual averaged across two to three technical replicates, pooled across females and males (with up to 15 individuals tested per sex). Medians are shown as short horizontal lines and lines join medians across different temperature cycles.
Discussion
We successfully used microinjection to transfer Wolbachia directly from D. melanogaster to Ae. aegypti to generate the wMelM line. wMelM differs from the original wMel transinfection (Walker et al., 2011) both in terms of the source of the wMel material (laboratory vs field‐derived D. melanogaster) and initial passage through a cell line. Although different methods were used to develop the wMel and wMelM transinfection, the wMelM variant like the original wMel has minimal effects on mosquito fitness. The wMelM variant was associated with a somewhat higher host fecundity compared to the wMel strain and a longer host life span when compared to wMel.tet mosquitoes, traits that would be likely to improve the success of release strategies in a field setting aimed at replacement. Apart from these fitness effects, we found minor differences in the density of Wolbachia, which may be correlated with the degree of virus blocking (Lu et al., 2012; Osborne et al., 2012; Fraser et al., 2017). For wMelM, Wolbachia density was higher but only under cycling heat and cold conditions; this leads to the expectation that virus blockage should be similar in the two strains, consistent with the similar virus blocking ability for wMelM, wMel and wAlbB observed at 26°C. The high level of transmission and incompatibility generated by wMelM under the varied thermal conditions tested here may facilitate its invasion into natural field populations and persistence at a high level.
In native Drosophila hosts, wMel Wolbachia density can vary substantially (Early and Clark, 2013); for instance, individuals from Ithaca NY, USA, most of which were infected with wMel from clade I, had higher Wolbachia titres when reared at room temperature than individuals from Tasmania, Australia, Zimbabwe and the Netherlands, most of which were infected with wMel from clade III. It is possible that a common mechanism underlies this finding and our observation of higher densities of wMelM (clade I) in temperature‐stressed Ae. aegypti relative to the original wMel transinfection strain (clade III). However, the cause of these differences in Wolbachia density remains to be determined. In more distantly related wMelCS‐like variants (clade VI), high Wolbachia titres are associated with both the loss of and the amplification of the Octomom genome region (Chrostek et al., 2013; Chrostek and Teixeira, 2015; Duarte et al., 2021). In our case, we did not detect any alteration of Octomom copy number in wMelM. However, we did observe non‐silent changes in genes encoding a Wolbachia surface protein (wspB), a member of the type IV secretion system, and proteins involved in amino acid metabolism, all functions that are likely to be important in interactions between endosymbiont and host (Pichon et al., 2009; Caragata et al., 2014; Rice et al., 2017; Jimenez et al., 2019; Epis et al., 2020).
Previous studies have shown that strong geographic structuring exists among global wMel populations. While multiple wMel variants can often be found within a single location or region, usually one variant predominates at any given location, suggestive of adaptation to local environmental conditions (Richardson et al., 2012; Early and Clark, 2013). There have been very few changes in the original wMel strain since transinfection (Huang et al., 2020), but this strain is different genomically from some wMel strains present in D. melanogaster from Australia (which provided the donor for wMelM). Phylogenetic analysis placed the original wMel transinfection (Walker et al., 2011) within clade III, which contains 13 of the 14 other Australian (Tasmanian) samples included in the tree, and wMelM within clade I, which contains a single Tasmanian sample. This pattern raises the possibility that clade III wMel variants may have a competitive advantage over clade I variants in some parts of southern Australia, such as Tasmania, which lies at the southern boundary of the cline of decreasing Wolbachia infection incidence (Kriesner et al., 2016). Most of the SNPs in the wMelM genome were identified in a study of wMel SNPs that distinguished different geographic climate zones, with five of these shown to be significantly associated with temperature (Hague et al., 2022). In our analysis, we did find a SNP in the gene encoding the Wolbachia outer membrane protein B (wspB, WD0009), which results in a premature stop codon and is likely a major determinant of Wolbachia thermal sensitivity (Hague et al., 2022). It may be that the polymorphisms in the wMelM genome and the higher densities observed for wMelM under heat and cold stress are selected against in regions with lower yearly minimum temperatures (Kriesner et al., 2016). It would be interesting to determine the frequency of different wMel variants in natural D. melanogaster populations across Australia, especially in more northerly regions, where Ae. aegypti population replacement programmes are being conducted.
Phenotypic differences between wMel and wMelM may relate to the donor line of natural Drosophila used in the transinfection as well as the difference in methodology related to mosquito cell passaging rather than direct transfer. In eastern Australian D. melanogaster, Wolbachia were first identified from incompatibility generated through crossing experiments between populations from along a latitudinal cline (Hoffmann, 1988) and wMel shows a stable latitudinal pattern from being near 100% in incidence to being at a very low frequency (Kriesner et al., 2016). This pattern suggests environmental effects on Wolbachia dynamics and/or differences in the Wolbachia strain/host backgrounds affecting these dynamics. Transplant experiments followed by semi‐field cage experiments point to a complex pattern of Wolbachia and host associated fitness effects at ends of the cline (Olsen et al., 2001) while experimental studies implicate environmental effects interacting with Wolbachia fitness (Kriesner et al., 2016). Other data also point to different wMel variants changing over time at different temperatures (Versace et al., 2014). Furthermore, environmental and genetic difference sometimes contribute together to Wolbachia transmission and frequency variation expected to influence the Wolbachia spread (Hague et al., 2020).
For Wolbachia infections generally, high temperatures can have an impact on release success by weakening the reproductive effects induced by Wolbachia (Trpis et al., 1981; Johanowicz and Hoy, 1998; Ross et al., 2017) and Wolbachia can even be eliminated under sufficient thermal stress (Trpis et al., 1981; Stouthamer et al., 1990). In the case of Ae. aegypti, high temperature effects depend on the Wolbachia strain, with wAlbB performing better than wMel (Ross et al., 2017). The higher Wolbachia density of wAlbB under heat may explain the fact that its phenotypic effects are not altered much by high temperatures. This also appears to be the case for the wMelM variant. When eggs were exposed to high temperatures, Ae. aegypti infected with wMelM had a higher Wolbachia density, stronger cytoplasmic incompatibility and an increased fidelity of maternal transmission compared to wMel. While these patterns were consistent across different egg heat treatments, differences may depend on the timing of heat treatment given that some life stages are more sensitive to heat and Wolbachia density can recover (Ulrich et al., 2016; Ross et al., 2020; Mancini et al., 2021). Differences in tissue distribution could also help to explain the increased stability of wMelM, as for wMel variants in Drosophila at cold temperatures (Hague et al., 2022), but this requires further investigation.
Given the impact of high temperature on the dynamics of Wolbachia‐infected mosquitoes and the benefits of Wolbachia releases targeting Ae. aegypti in several countries from different climate zones where dengue is common (Bhatt et al., 2013; Ritchie, 2018), it may be prudent to consider multiple strains in releases in different environments (Ross et al., 2019a). The wMelM variant generated here has minimal fitness effects but a higher phenotypic stability at high temperatures compared to wMel. Our results suggest that this new transinfected strain may be useful in environments where temperatures are variable. Given that temperature conditions also change seasonally, the new strain may allow for releases in spring when temperatures are often variable. It is also worth considering other strains from field populations for transfections.
Data availability
The wMelM Aedes aegypti strain is available from the authors on request. Sequence data for the samples included in this study are available from NCBI GenBank and have the following accession codes: BioProject – PRJNA805284, PRJNA791959; BioSample – SAMN24793266 (raw data for wMel genome), SAMN25851851 – SAMN25851855 (raw data for wMelM), SAMN24371057 (Wolbachia genomes for wMel), SAMN25853239 – SAMN25853242 (Wolbachia genomes for wMelM) and SAMN25853243 (Wolbachia genomes for donor flies Drosophila melanogaster).
Supporting information
Table S1. Sample information for wMel variants included in the phylogentic analysis. For references refer to main text.
Table S1. SNP matrix used for phylogenetic analysis
Table S1. SNP haplotype sequences (from SNP matrix) and wMel clade information.
Table S2. Genome‐wide polymorphisms, relative to the reference genome, are shown for each sample. The number of high quality bases and the allele frequency are shown in parentheses. Green = change present in consensus genome sequence, beige = no change, blue = low coverage, red text = same SNP identified in Hague et al. (2022), bold text = SNP related to temperature (P < 0.05, Hague et al., (2022)).
Table S2. KEGG gene ontology information is provided for loci where available.
Table S2. Summary of functional category information. The number of loci corresponding to each category is shown.
Acknowledgements
The authors thank Kelly Richardson for collecting the Drosophila line used for microinjection, Marianne Coquilleau for the assistance in the experiments and Moshe Jasper for support with LAMP assays. A.A.H. was supported by the National Health and Medical Research Council (1132412, 1118640, www.nhmrc.gov.au). P.A.R. was supported by a University of Melbourne Early Career Research Grant. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Open access publishing facilitated by The University of Melbourne, as part of the Wiley ‐ The University of Melbourne agreement via the Council of Australian University Librarians.
References
- Ahmad, N.A. , Mancini, M.V. , Ant, T.H. , Martinez, J. , Kamarul, G.M.R. , Nazni, W.A. , et al. (2021) Wolbachia strain wAlbB maintains high density and dengue inhibition following introduction into a field population of Aedes aegypti . Philos Trans R Soc Lond B Biol Sci 376: 20190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axford, J.K. , Ross, P.A. , Yeap, H.L. , Callahan, A.G. , and Hoffmann, A.A. (2016) Fitness of wAlbB Wolbachia infection in Aedes aegypti: parameter estimates in an outcrossed background and potential for population invasion. Am J Trop Med Hyg 94: 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe, N.W. , Pagendam, D. , Trewin, B.J. , Boomer, A. , Bradford, M. , Ford, A. , et al. (2021) Releasing incompatible males drives strong suppression across populations of wild and Wolbachia‐carrying Aedes aegypti in Australia. Proc Natl Acad Sci USA 118: e2106828118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt, S. , Gething, P.W. , Brady, O.J. , Messina, J.P. , Farlow, A.W. , Moyes, C.L. , et al. (2013) The global distribution and burden of dengue. Nature 496: 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger, A.M. , Lohse, M. , and Usadel, B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull, J.J. , and Turelli, M. (2013) Wolbachia versus dengue: evolutionary forecasts. Evol Med Public Health 2013: 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdina, E.V. , Bykov, R.A. , Menshanov, P.N. , Ilinsky, Y.Y. , and Gruntenko, N.E. (2021) Unique Wolbachia strain wMelPlus increases heat stress resistance in Drosophila melanogaster . Arch Insect Biochem 106: e21776. [DOI] [PubMed] [Google Scholar]
- Caragata, E.P. , Rances, E. , O'Neill, S.L. , and McGraw, E.A. (2014) Competition for amino acids between Wolbachia and the mosquito host, Aedes aegypti . Microb Ecol 67: 205–218. [DOI] [PubMed] [Google Scholar]
- Caspari, E. , and Watson, G.S. (1959) On the evolutionary importance of cytoplasmic sterility in mosquitoes. Evolution 13: 568–570. [Google Scholar]
- Chrostek, E. , Marialva, M.S.P. , Esteves, S.S. , Weinert, L.A. , Martinez, J. , Jiggins, F.M. , and Teixeira, L. (2013) Wolbachia variants induce differential protection to viruses in Drosophila melanogaster: a phenotypic and phylogenomic analysis. PLoS Genet 9: e1003896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrostek, E. , Martins, N.E. , Marialva, M.S. , and Teixeira, L. (2021) Wolbachia‐conferred antiviral protection is determined by developmental temperature. mBio 12: e02923–e02920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrostek, E. , and Teixeira, L. (2015) Mutualism breakdown by amplification of Wolbachia genes. PLoS Biol 13: e1002065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin, C. , Heyworth, E.R. , Ferrari, J. , and Hurst, G.D.D. (2017) Heritable symbionts in a world of varying temperature. Heredity 118: 10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, J.E. , Clarke, D.W. , Criswell, V. , Desnoyer, M. , Cornel, D. , Deegan, B. , et al. (2020) Efficient production of male Wolbachia‐infected Aedes aegypti mosquitoes enables large‐scale suppression of wild populations. Nat Biotechnol 38: 482–492. [DOI] [PubMed] [Google Scholar]
- Danecek, P. , Bonfield, J.K. , Liddle, J. , Marshall, J. , Ohan, V. , Pollard, M.O. , et al. (2021) Twelve years of SAMtools and BCFtools. Gigascience 10: giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte, E.H. , Carvalho, A. , Lopez‐Madrigal, S. , Costa, J. , and Teixeira, L. (2021) Forward genetics in Wolbachia: regulation of Wolbachia proliferation by the amplification and deletion of an addictive genomic island. PLoS Genet 17: e1009612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchemin, J.B. , Mee, P.T. , Lynch, S.E. , Vedururu, R. , Trinidad, L. , and Paradkar, P. (2017) Zika vector transmission risk in temperate Australia: a vector competence study. Virol J 14: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early, A.M. , and Clark, A.G. (2013) Monophyly of Wolbachia pipientis genomes within Drosophila melanogaster: geographic structuring, titre variation and host effects across five populations. Mol Ecol 22: 5765–5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epis, S. , Varotto‐Boccazzi, I. , Crotti, E. , Damiani, C. , Giovati, L. , Mandrioli, M. , et al. (2020) Chimeric symbionts expressing a Wolbachia protein stimulate mosquito immunity and inhibit filarial parasite development. Commun Biol 3: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, J.E. , De Bruyne, J.T. , Iturbe‐Ormaetxe, I. , Stepnell, J. , Burns, R.L. , Flores, H.A. , and O'Neill, S.L. (2017) Novel Wolbachia‐transinfected Aedes aegypti mosquitoes possess diverse fitness and vector competence phenotypes. PLoS Pathog 13: e1006751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, G.D. , dos Santos, L.M.B. , Villela, D.A.M. , and Maciel‐de‐Freitas, R. (2016) Using Wolbachia releases to estimate Aedes aegypti (Diptera: Culicidae) population size and survival. PLoS One 11: e0160196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, G.D. , Sylvestre, G. , Aguiar, R. , da Costa, G.B. , Martins, A.J. , Lima, J.B.P. , et al. (2019) Matching the genetics of released and local Aedes aegypti populations is critical to assure Wolbachia invasion. PLoS Neglect Trop D 13: e0007023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves, D.D. , Hooker, D.J. , Dong, Y. , Baran, N. , Kyrylos, P. , Iturbe‐Ormaetxe, I. , et al. (2019) Detecting wMel Wolbachia in field‐collected Aedes aegypti mosquitoes using loop‐mediated isothermal amplification (LAMP). Parasit Vector 12: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruntenko, N.E. , Ilinsky, Y.Y. , Adonyeva, N.V. , Burdina, E.V. , Bykov, R.A. , Menshanov, P.N. , and Rauschenbach, I.Y. (2017) Various Wolbachia genotypes differently influence host Drosophila dopamine metabolism and survival under heat stress conditions. BMC Evol Biol 17: 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman, M.G. , Halstead, S.B. , Artsob, H. , Buchy, P. , Jeremy, F. , Gubler, D.J. , et al. (2010) Dengue: a continuing global threat. Nat Rev Microbiol 8: S7–S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hague, M.T. , Shropshire, J.D. , Caldwell, C.N. , Statz, J.P. , Stanek, K.A. , Conner, W.R. , and Cooper, B.S. (2022) Temperature effects on cellular host‐microbe interactions explain continent‐wide endosymbiont prevalence. Curr Biol 32: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hague, M.T.J. , Mavengere, H. , Matute, D.R. , and Cooper, B.S. (2020) Environmental and genetic contributions to imperfect wMel‐Like Wolbachia transmission and frequency variation. Genetics 215: 1117–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hien, N.T. , Anh, D.D. , Le, N.H. , Yen, N.T. , Phong, T.V. , Nam, V.S. , et al. (2021) Environmental factors influence the local establishment of Wolbachia in Aedes aegypti mosquitoes in two small communities in central Vietnam. Gates Open Res 5: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, A.A. (1988) Partial cytoplasmic incompatibility between two Australian populations of Drosophila melanogaster . Entomol Exp Appl 48: 61–67. [Google Scholar]
- Hoffmann, A.A. , Iturbe‐Ormaetxe, I. , Callahan, A.G. , Phillips, B. , Billington, K. , Axford, J.K. , et al. (2014) Stability of the wMel Wolbachia infection following invasion into Aedes aegypti populations. PLoS Neglect Trop D 8: e3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, A.A. , Montgomery, B.L. , Popovici, J. , Iturbe‐Ormaetxe, I. , Johnson, P.H. , Muzzi, F. , et al. (2011) Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 476: 454–457. [DOI] [PubMed] [Google Scholar]
- Hoffmann, A.A. , Ross, P.A. , and Rasic, G. (2015) Wolbachia strains for disease control: ecological and evolutionary considerations. Evol Appl 8: 751–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, A.A. , and Turelli, M. (1997) Cytoplasmic incompatibility in insects. In Influential Passengers: Inherited Microorganisms and Arthropod Reproduction, O'Neill, S.L. , Hoffmann, A.A. , and Werren, J.H. (eds), pp. 42–80. Oxford: Oxford Univ. Press. [Google Scholar]
- Hoffmann, A.A. , Turelli, M. , and Simmons, G.M. (1986) Unidirectional incompatibility between populations of Drosophila simulans . Evolution 40: 692–701. [DOI] [PubMed] [Google Scholar]
- Hoffmann, A.A. , and Weeks, A.R. (2007) Climatic selection on genes and traits after a 100 year‐old invasion: a critical look at the temperate‐tropical clines in Drosophila melanogaster from eastern Australia. Genetica 129: 133–147. [DOI] [PubMed] [Google Scholar]
- Huang, B.X. , Yang, Q. , Hoffmann, A.A. , Ritchie, S.A. , van den Hurk, A.F. , and Warrilow, D. (2020) Wolbachia genome stability and mtDNA variants in Aedes aegypti field populations eight years after release. Iscience 23: 101572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilinsky, Y. (2013) Coevolution of Drosophila melanogaster mtDNA and Wolbachia Genotypes. PLoS One 8: e54373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indriani, C. , Tantowijoyo, W. , Rances, E. , Andari, B. , Prabowo, E. , Yusdi, D. , et al. (2020) Reduced dengue incidence following deployments of Wolbachia‐infected Aedes aegypti in Yogyakarta, Indonesia: a quasi‐experimental trial using controlled interrupted time series analysis. Gates Open Res 4: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasper, M.E. , Yang, Q. , Ross, P.A. , Endersby‐Harshman, N. , Bell, N. , and Hoffmann, A.A. (2019) A LAMP assay for the rapid and robust assessment of Wolbachia infection in Aedes aegypti under field and laboratory conditions. PLoS One 14: e0225321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez, N.E. , Gerdtzen, Z.P. , Olivera‐Nappa, A. , Salgado, J.C. , and Conca, C. (2019) A systems biology approach for studying Wolbachia metabolism reveals points of interaction with its host in the context of arboviral infection. PLoS Neglect Trop D 13: e0007678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanowicz, D.L. , and Hoy, M.A. (1998) Experimental induction and termination of non‐reciprocal reproductive incompatibilities in a parahaploid mite. Entomol Exp Appl 87: 51–58. [Google Scholar]
- Kong, J.D. , Axford, J.K. , Hoffmann, A.A. , and Kearney, M.R. (2016) Novel applications of thermocyclers for phenotyping invertebrate thermal responses. Methods Ecol Evol 7: 1201–1208. [Google Scholar]
- Kriesner, P. , Conner, W.R. , Weeks, A.R. , Turelli, M. , and Hoffmann, A.A. (2016) Persistence of a Wolbachia infection frequency cline in Drosophila melanogaster and the possible role of reproductive dormancy. Evolution 70: 979–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyle, J.L. , and Harris, E. (2008) Global spread and persistence of dengue. Annu Rev Microbiol 62: 71–92. [DOI] [PubMed] [Google Scholar]
- Lau, M.J. , Ross, P.A. , Endersby‐Harshman, N.M. , and Hoffmann, A.A. (2020) Impacts of low temperatures on Wolbachia (Rickettsiales: Rickettsiaceae)‐infected Aedes aegypti (Diptera: Culicidae). J Med Entomol 57: 1567–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, M.J. , Ross, P.A. , and Hoffmann, A.A. (2021) Infertility and fecundity loss of Wolbachia‐infected Aedes aegypti hatched from quiescent eggs is expected to alter invasion dynamics. PLoS Neglect Trop D 15: e0009179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.F. , White, V.L. , Weeks, A.R. , Hoffmann, A.A. , and Endersby, N.M. (2012) High‐throughput PCR assays to monitor Wolbachia infection in the dengue mosquito (Aedes aegypti) and Drosophila simulans . Appl Environ Microb 78: 4740–4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, P.O. (2001) A likelihood approach to estimating phylogeny from discrete morphological character data. Syst Biol 50: 913–925. [DOI] [PubMed] [Google Scholar]
- Li, H. (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA‐MEM. arXiv: 1303.3997. [Google Scholar]
- Li, H. , Handsaker, B. , Wysoker, A. , Fennell, T. , Ruan, J. , Homer, N. , et al. (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, P. , Bian, G.W. , Pan, X.L. , and Xi, Z.Y. (2012) Wolbachia induces density‐dependent inhibition to dengue virus in mosquito cells. PLoS Neglect Trop D 6: e1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini, M.V. , Ant, T.H. , Herd, C.S. , Martinez, J. , Murdochy, S.M. , Gingell, D.D. , et al. (2021) High temperature cycles result in maternal transmission and dengue infection differences between Wolbachia strains in Aedes aegypti . Mbio 12: e00250–e00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMeniman, C.J. , Lane, R.V. , Cass, B.N. , Fong, A.W.C. , Sidhu, M. , Wang, Y.F. , and O'Neill, S.L. (2009) Stable introduction of a life‐shortening Wolbachia infection into the mosquito Aedes aegypti . Science 323: 141–144. [DOI] [PubMed] [Google Scholar]
- Miller, M.A. , Pfeiffer, W. , and Schwartz, T. (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees" in proceedings of the gateway computing environments workshop (GCE), 14 Nov. New Orleans LA: 1‐8.
- Moreira, L.A. , Iturbe‐Ormaetxe, I. , Jeffery, J.A. , Lu, G.J. , Pyke, A.T. , Hedges, L.M. , et al. (2009) A Wolbachia symbiont in Aedes aegypti limits infection with dengue, chikungunya, and plasmodium. Cell 139: 1268–1278. [DOI] [PubMed] [Google Scholar]
- Nazni, W.A. , Hoffmann, A.A. , NoorAfizah, A. , Cheong, Y.L. , Mancini, M.V. , Golding, N. , et al. (2019) Establishment of Wolbachia strain wAlbB in Malaysian populations of Aedes aegypti for dengue control. Curr Biol 29: 4241–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, L.C. , and Project Wolbachia‐Singapore Consortium . (2021) Wolbachia‐mediated sterility suppresses Aedes aegypti populations in the urban tropics. medRxiv 2021: 21257922. [Google Scholar]
- Olsen, K. , Reynolds, K.T. , and Hoffmann, A.A. (2001) A field cage test of the effects of the endosymbiont Wolbachia on Drosophila melanogaster . Heredity 86: 731–737. [DOI] [PubMed] [Google Scholar]
- Osborne, S.E. , Iturbe‐Ormaetxe, I. , Brownlie, J.C. , O'Neill, S.L. , and Johnson, K.N. (2012) Antiviral protection and the importance of Wolbachia density and tissue tropism in Drosophila simulans . Appl Environ Microb 78: 6922–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon, S. , Bouchon, D. , Cordaux, R. , Chen, L.M. , Garrett, R.A. , and Greve, P. (2009) Conservation of the type IV secretion system throughout Wolbachia evolution. Biochem Bioph Res Co 385: 557–562. [DOI] [PubMed] [Google Scholar]
- Pinto, S.B. , Riback, T.I.S. , Sylvestre, G. , Costa, G. , Peixoto, J. , Dias, F.B.S. , et al. (2021) Effectiveness of Wolbachia‐infected mosquito deployments in reducing the incidence of dengue and other Aedes‐borne diseases in Niteroi, Brazil: A quasi‐experimental study. PLoS Neglect Trop D 15: e0009556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, L.J. , and Muench, H. (1938) A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27: 493–497. [Google Scholar]
- Rice, D.W. , Sheehan, K.B. , and Newton, I.L.G. (2017) Large‐scale identification of Wolbachia pipientis effectors. Genome Biol Evol 9: 1925–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, M.F. , Weinert, L.A. , Welch, J.J. , Linheiro, R.S. , Magwire, M.M. , Jiggins, F.M. , and Bergman, C.M. (2012) Population genomics of the Wolbachia endosymbiont in Drosophila melanogaster . PLoS Genet 8: e1003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegler, M. , Sidhu, M. , Miller, W.J. , and O'Neill, S.L. (2005) Evidence for a global Wolbachia replacement in Drosophila melanogaster . Curr Biol 15: 1428–1433. [DOI] [PubMed] [Google Scholar]
- Ritchie, S.A. (2018) Wolbachia and the near cessation of dengue outbreaks in Northern Australia despite continued dengue importations via travellers. J Travel Med 25: tay084. [DOI] [PubMed] [Google Scholar]
- Ross, P.A. , Axford, J.K. , Yang, Q. , Staunton, K.M. , Ritchie, S.A. , Richardson, K.M. , and Hoffmann, A.A. (2020) Heatwaves cause fluctuations in wMel Wolbachia densities and frequencies in Aedes aegypti . PLoS Neglect Trop D 14: e0007958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, P.A. , Endersby, N.M. , and Hoffmann, A.A. (2016) Costs of three Wolbachia infections on the survival of Aedes aegypti larvae under starvation conditions. PLoS Neglect Trop D 10: e0004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, P.A. , Gu, X.Y. , Robinson, K.L. , Yang, Q. , Cottingham, E. , Zhang, Y.F. , et al. (2021) A wAlbB Wolbachia transinfection displays stable phenotypic effects across divergent Aedes aegypti mosquito backgrounds. Appl Environ Microb 87: e01264–e01221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, P.A. , and Hoffmann, A.A. (2018) Continued susceptibility of the wMel Wolbachia infection in Aedes aegypti to heat stress following field deployment and selection. Insects 9: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, P.A. , Ritchie, S.A. , Axford, J.K. , and Hoffmann, A.A. (2019b) Loss of cytoplasmic incompatibility in Wolbachia‐infected Aedes aegypti under field conditions. PLoS Negl Trop Dis 13: e0007357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, P.A. , Turelli, M. , and Hoffmann, A.A. (2019a) Evolutionary ecology of Wolbachia releases for disease control. Annu Rev Genet 53: 93–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, P.A. , Wiwatanaratanabutr, I. , Axford, J.K. , White, V.L. , Endersby‐Harshman, N.M. , and Hoffmann, A.A. (2017) Wolbachia infections in Aedes aegypti differ markedly in their response to cyclical heat stress. PLoS Pathog 13: e1006006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, P.A. , Turley, A.P. , Wilson, G. , Hurst, T.P. , Retzki, K. , Brown‐Kenyon, J. , et al. (2019) Establishment of wMel Wolbachia in Aedes aegypti mosquitoes and reduction of local dengue transmission in Cairns and surrounding locations in northern Queensland, Australia. Gates Open Res 3: 1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis, A. (2014) RAxML version 8: a tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staunton, K.M. , Yeeles, P. , Townsend, M. , Nowrouzi, S. , Paton, C.J. , Trewin, B. , et al. (2019) Trap location and premises condition influences on Aedes aegypti (Diptera: Culicidae) catches using biogents sentinel traps during a 'Rear and Release' program: implications for designing surveillance programs. J Med Entomol 56: 1102–1111. [DOI] [PubMed] [Google Scholar]
- Stouthamer, R. , Luck, R.F. , and Hamilton, W.D. (1990) Antibiotics cause parthenogenetic Trichogramma (Hymenoptera, Trichogrammatidae) to revert to sex. Proc Natl Acad Sci USA 87: 2424–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvakov, M. , Panda, A. , Diesh, C. , Holmes, I. , and Abyzov, A. (2021) CNVpytor: a tool for copy number variation detection and analysis from read depth and allele imbalance in whole‐genome sequencing. Gigascience 10: giab074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantowijoyo, W. , Andari, B. , Arguni, E. , Budiwati, N. , Nurhayati, I. , Fitriana, I. , et al. (2020) Stable establishment of wMel Wolbachia in Aedes aegypti populations in Yogyakarta, Indonesia. PLoS Neglect Trop D 14: e0008157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trpis, M. , Perrone, J.B. , Reissig, M. , and Parker, K.L. (1981) Control of cytoplasmic incompatibility in the Aedes scutellaris complex. J Hered 72: 313–317. [Google Scholar]
- Truitt, A.M. , Kapun, M. , Kaur, R. , and Miller, W.J. (2019) Wolbachia modifies thermal preference in Drosophila melanogaster . Environ Microbiol 21: 3259–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich, J.N. , Beier, J.C. , Devine, G.J. , and Hugo, L.E. (2016) Heat sensitivity of wMel Wolbachia during Aedes aegypti development. PLoS Neglect Trop D 10: e0004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utarini, A. , Indriani, C. , Ahmad, R.A. , Tantowijoyo, W. , Arguni, E. , Ansari, M.R. , et al. (2021) Efficacy of Wolbachia‐infected mosquito deployments for the control of dengue. New Engl J Med 384: 2177–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hurk, A.F. , Hall‐Mendelin, S. , Pyke, A.T. , Frentiu, F.D. , McElroy, K. , Day, A. , et al. (2012) Impact of Wolbachia on infection with chikungunya and yellow fever viruses in the mosquito vector Aedes aegypti . PLoS Negl Trop Dis 6: e1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veneti, Z. , Clark, M.E. , Zabalou, S. , Karr, T.L. , Savakis, C. , and Bourtzis, K. (2003) Cytoplasmic incompatibility and sperm cyst infection in different Drosophila‐Wolbachia associations. Genetics 164: 545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versace, E. , Nolte, V. , Pandey, R.V. , Tobler, R. , and Schlotterer, C. (2014) Experimental evolution reveals habitat‐specific fitness dynamics among Wolbachia clades in Drosophila melanogaster . Mol Ecol 23: 802–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, T. , Johnson, P.H. , Moreira, L.A. , Iturbe‐Ormaetxe, I. , Frentiu, F.D. , McMeniman, C.J. , et al. (2011) The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 476: 450–453. [DOI] [PubMed] [Google Scholar]
- Wang, G.H. , Gamez, S. , Raban, R.R. , Marshall, J.M. , Alphey, L. , Li, M. , et al. (2021) Combating mosquito‐borne diseases using genetic control technologies. Nat Commun 12: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, D.E. , Lu, J. , and Langmead, B. (2019) Improved metagenomic analysis with Kraken 2. Genome Biol 20: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, M. , Sun, L.V. , Vamathevan, J. , Riegler, M. , Deboy, R. , Brownlie, J.C. , et al. (2004) Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol 2: 327–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi, Z.Y. , Khoo, C.C.H. , and Dobson, S.L. (2005) Wolbachia establishment and invasion in an Aedes aegypti laboratory population. Science 310: 326–328. [DOI] [PubMed] [Google Scholar]
- Yen, P.S. , and Failloux, A.B. (2020) A Review: Wolbachia‐based population replacement for mosquito control shares common points with genetically modified control approaches. Pathogens 9: 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X.Y. , Zhang, D.J. , Li, Y.J. , Yang, C. , Wu, Y. , Liang, X. , et al. (2019) Incompatible and sterile insect techniques combined eliminate mosquitoes. Nature 572: 56–61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Sample information for wMel variants included in the phylogentic analysis. For references refer to main text.
Table S1. SNP matrix used for phylogenetic analysis
Table S1. SNP haplotype sequences (from SNP matrix) and wMel clade information.
Table S2. Genome‐wide polymorphisms, relative to the reference genome, are shown for each sample. The number of high quality bases and the allele frequency are shown in parentheses. Green = change present in consensus genome sequence, beige = no change, blue = low coverage, red text = same SNP identified in Hague et al. (2022), bold text = SNP related to temperature (P < 0.05, Hague et al., (2022)).
Table S2. KEGG gene ontology information is provided for loci where available.
Table S2. Summary of functional category information. The number of loci corresponding to each category is shown.
Data Availability Statement
The wMelM Aedes aegypti strain is available from the authors on request. Sequence data for the samples included in this study are available from NCBI GenBank and have the following accession codes: BioProject – PRJNA805284, PRJNA791959; BioSample – SAMN24793266 (raw data for wMel genome), SAMN25851851 – SAMN25851855 (raw data for wMelM), SAMN24371057 (Wolbachia genomes for wMel), SAMN25853239 – SAMN25853242 (Wolbachia genomes for wMelM) and SAMN25853243 (Wolbachia genomes for donor flies Drosophila melanogaster).


