Abstract
Background and purpose
The antisense oligonucleotide nusinersen (Spinraza) regulates splicing of the survival motor neuron 2 (SMN2) messenger RNA to increase SMN protein expression. Nusinersen has improved ventilator‐free survival and motor function outcomes in infantile onset forms of spinal muscular atrophy (SMA), treated early in the course of the disease. However, the response in later onset forms of SMA is highly variable and dependent on symptom severity and disease duration at treatment initiation. Therefore, we aimed to identify novel noninvasive biomarkers that could predict the response to nusinersen in type II and III SMA patients.
Methods
Thirty‐four SMA patients were included. We applied next generation sequencing to identify microRNAs in the cerebrospinal fluid (CSF) as candidate biomarkers predicting response to nusinersen. Hammersmith Functional Motor Scale Expanded (HFMSE) was conducted at baseline and 6 months after initiation of nusinersen therapy to assess motor function. Patients changing by ≥3 or ≤0 points in the HFMSE total score were considered to be responders or nonresponders, respectively.
Results
Lower baseline levels of two muscle microRNAs (miR‐206 and miR‐133a‐3p), alone or in combination, predicted the clinical response to nusinersen after 6 months of therapy. Moreover, miR‐206 levels were inversely correlated with the HFMSE score.
Conclusions
Lower miR‐206 and miR‐133a‐3p in the CSF predict more robust clinical response to nusinersen treatment in later onset SMA patients. These novel findings have high clinical relevance for identifying early treatment response to nusinersen in later onset SMA patients and call for testing the ability of miRNAs to predict more sustained long‐term benefit.
Keywords: Biomarkers, Cerebrospinal Fluid, miR‐206, miR‐133, Muscle microRNAs, Nusinersen, Response predictors, Spinal muscular atrophy
INTRODUCTION
Spinal muscular atrophy (SMA) is a genetic disease with an incidence of ~1/11,000 live births, and a carrier frequency of ~1/40 [1, 2]. SMA is characterized by muscle weakness and atrophy, resulting from progressive degeneration of lower motor neurons in the spinal cord and the brain stem nuclei. Historically, SMA subtype classification is based on age at onset and the maximum motor abilities achieved. SMA type I patients have onset in early infancy and never sit. SMA type II patients have later infantile onset but never walk. SMA type III patients have more variable childhood onset, and achieve the ability to walk [3]. SMA IV patients typically present with milder muscle weakness in the second or third decade of life [4, 5, 6, 7]. The cause of SMA is homozygous deletion or compound heterozygous mutation involving exon 7 of the survival motor neuron 1 (SMN1) gene. However, SMN1 has a paralogous gene named SMN2, which undergoes alternative splicing, including the removal of exon 7, and produces ~10% functional SMN protein [8, 9]. Therefore, the number of SMN2 copies correlates with phenotypic severity and is the main genetic disease modifier [10, 11, 12].
Nusinersen was the first drug approved by the US Food and Drug Administration (FDA) to treat SMA. The drug is a synthetic antisense oligonucleotide that modulates pre‐messenger RNA splicing of the SMN2 gene. Despite the advance of novel molecular and gene therapies for SMA, nusinersen remains to this day the most widely used and available SMA disease‐modifying therapy. Nusinersen has been proven efficient in young type I SMA patients, and it is dramatically changing the natural history of this disease [13, 14]. However, the response in subjects with later onset forms of SMA (type II and III) is more variable, with 30%–40% presenting clinically meaningful improvement [15, 16]. Therefore, better understanding of the variable treatment response in this population is important, and novel biomarkers to predict treatment response to nusinersen in type II or type III SMA subjects are critical.
A main interest of our group has been to investigate the profile of microRNAs (miRNAs) in the disease context, as several endogenous noncoding RNAs are highly expressed in both neuronal and muscular tissues. Previous evidence indicates that some miRNAs are essential for motor neuron survival, and low expression levels have been demonstrated in motor neurons from an SMA mouse model and in postmortem neuronal tissues from patients with another motor neuron disease, amyotrophic lateral sclerosis (ALS) [17, 18, 19].
Moreover, miRNAs miR‐1/133a/133b/206, are mainly expressed in the skeletal muscle, where they are thought to play an important role in myoblast proliferation and differentiation [20, 21]. These are also known as myomiRs. Recent studies have demonstrated that the expression of myomiRs can be detected in biofluids from ALS patients [22, 23, 24], suggesting a potential role of these molecules in early diagnosis of the disease. In addition, serum myomiRs are in correlation with the response to ongoing therapy in SMA type II and III [25]. However, a prospective study that identifies responders to nusinersen therapy, based on a basal, pretreatment, molecular profile, was not yet reported.
On the basis of understanding the miRNA role in motor neuron diseases [17, 18, 19] and potential discovery of novel biomarkers [26, 27, 28], we sought to test the utility of specific miRNAs in the cerebrospinal fluid (CSF) as candidate molecules to predict the clinical response to nusinersen in type II/III SMA patients. We applied for the first time an unbiased next generation sequencing to investigate the potential of cell‐free microRNAs in the CSF of SMA patients receiving nusinersen therapy. Our novel findings indicate that the muscle miRNAs miR‐206 and miR‐133 predict the response to nusinersen treatment and, therefore, have the potential to help predict, when combined with other indicators, whether a given SMA patient is more or less likely to show an early response.
MATERIALS AND METHODS
Subjects, ethics, and motor function
This study includes a cohort of 45 type II or type III SMA patients, who were recruited between November 2016 and July 2019 at three different medical centers: Schneider Children's Medical Center of Israel (Israel, n = 13), Dana‐Dwek Children's Hospital, Tel Aviv Medical Center (Israel, n = 20), and Massachusetts General Hospital (USA, n = 12). Approval for the study was provided by local ethical committees (Schneider Medical Center, RMC‐0060‐18; Tel Aviv Medical Center, 0347‐18‐TLV; Massachusetts General Hospital, MGH #2016P000469), and it was conducted in accordance with the International Conference on Harmonization guidelines for Good Clinical Practice and the World Medical Association Declaration of Helsinki. Written informed parental consent was obtained from all participants and was the only inclusion criterion for treatment initiation. Nusinersen was administered on Days 1, 15, 29, and 64 for the loading phase, followed by an additional maintenance dose after 4 additional months from the loading phase. Baseline demographic and clinical data were collected. Motor function was assessed at each visit by a physical therapist with SMA clinical trial training [29] using the Hammersmith Functional Motor Scale Expanded (HFMSE) [30]. Scoliosis was evaluated on every visit by physical examination and through spine/chest radiograph. Bilevel positive airway pressure (BIPAP) machine usage data and subjective improvement in energy were collected from the medical records.
These assessments were all performed prior to RNA sequencing, and therefore the clinical assessor was blinded to patients' miRNA profile. CSF samples were collected on Days 1 and 183 immediately before the nusinersen injection on each day and were stored frozen at −80°C.
Inclusion and exclusion criteria
All 45 patients had a confirmed genetic diagnosis of SMA, and were followed up to this point. Inclusion criteria required nonhemolyzed CSF and full documentation of HFMSE score. Therefore, all type II/III SMA patients who signed informed consent for downstream analysis were enrolled without any preselection. Thus, the cohort was unbiased by any selective/representative inclusion/exclusion criteria.
An improvement at the HFMSE final score of ≥3 points was considered clinically significant [31, 32] and patients with such an improvement were considered to be "responders", whereas those with a change of HFMSE final score of ≤0 were considered to be "non‐responders." Samples from 11 patients with an intermediate clinical response to nusinersen (HFMSE improvement of 1 or 2 points) were excluded from main analyses, but were included in supplementary analyses (Figures S2, S5, and S9, Table S2). Based on power analysis calculations, we found that the remaining 34 patients are sufficient to obtain an effect size of 3 for change in miRNA level with a power of 95% and a p‐value of 0.01.
Small RNA next generation sequencing
Total RNA was extracted from CSF using the miRNeasy Micro Kit (Qiagen) and quantified with a Qubit fluorometer using the RNA Broad‐Range Assay Kit (Thermo Fisher Scientific). For small RNA next generation sequencing (RNA‐seq), libraries were prepared from 7.5 ng of total RNA using the QIAseq miRNA Library Kit and QIAseq miRNA NGS 48 Index IL (Qiagen), by an experimenter who was blinded to the identity of patients from whom the samples were collected, as well as to their HFMSE scores. Samples were randomly allocated to library preparation and sequencing in batches. Precise linear quantification of miRNA was achieved by using unique molecular identifiers (UMIs), of random 12‐nucleotide after 3′ and 5′ adapter ligation, within the reverse transcription primers [26]. cDNA libraries were amplified by polymerase chain reaction for 23 cycles, with a 3′ primer that includes a six‐nucleotide unique index, followed by on‐bead size selection and cleaning. Library concentration was determined with a Qubit fluorometer (dsDNA High Sensitivity Assay Kit, Thermo Fisher Scientific) and library size with Tapestation D1000 (Agilent). Libraries with different indices were multiplexed and sequenced on NextSeq 500/550 v2 flow cell or Novaseq SP100 (Illumina), with 75‐bp single read and 6‐bp index read. Fastq files were demultiplexed using the User‐Friendly Transcriptome Analysis Pipeline [33]. Human miRNAs, as defined by miRBase [34], were mapped using Geneglobe Data Analysis Center (Qiagen).
Statistical analysis
As many as 2530 individual miRNA species were aligned to the human genome (GRCh37/hg19) across all samples. miRNAs with ≤100 UMIs on average across all samples were excluded from analysis. Sequencing data were normalized with the DESeq2 package [35] under the assumption that miRNA counts followed negative binomial distribution, and data were corrected for the library preparation batch to reduce its potential bias. Fold‐change values in miRNA abundance between responders and nonresponders were calculated as the ratio of normalized counts in responders (i.e., patients exhibiting meaningful clinical improvement) to the normalized counts in the nonresponders (patients exhibiting lack of clinical improvement), and transformed to log base 2. Statistical significance was determined using the Wald test.
Feature scaling to the range of 0–1 was further done for DESeq2 normalized counts of miR‐133a‐3p and miR‐206 by applying min–max scaling, whereby the scaled value (x′) is calculated by subtracting the minimum value of each feature, min (x), from each individual observation (x), and further dividing by the difference between the maximum and the minimum value, max (x) – min (x), as shown below:
Scaled values were then summed to generate a devised feature termed miR‐133a/206.
Correlation between miRNA levels and the clinical improvement in HFMSE score (HFMSE score post treatment – HFMSE score pretreatment) was calculated by both linear regression, wherein the improvement is a numerical value, and logistic regression, wherein the improvement is binarized into “failure” (HFMSE score change of ≤0 between post‐ and pretreatment) and “success” (HFMSE score change of ≥3 between post‐ and pretreatment). Logistic regression was performed with the aod and ggplot2 packages in R [36], either for a single explanatory variable (either one miRNA gene or a feature combining two miRNAs) or as multinomial regression with multiple explanatory variables (a combination of different miRNAs or of miRNAs and clinical features). The fitted logistic regression model is an exponential model from which predicted probabilities of “success” can be calculated based on values of the explanatory variable(s). Exponential equations for all models are available in Tables S1 and S2. Two values are derived from the logistic regression model: chi‐squared (χ 2), which expresses the goodness of fit for the whole model, and Akaike information criterion (AIC) [37], an estimation of the model prediction error. Statistical significance was determined for Spearman rho and χ 2 values, with p‐values <0.05 being considered statistically significant.
Leave‐one‐out cross‐validation (LOOCV) was performed on the logistic regression model, whereby model learning was done in n−1 samples out of the total n, and tested on the leave‐one‐out sample, yielding a predicted success probability for each sample. This procedure was repeated n times. Receiver operating characteristic (ROC) curves were generated by plotting true positive rates (sensitivity), that is, percentage of patients classified as responders, out of the total number of responders, against false positive rates (100% − specificity), that is, percentage of patients classified as responders, out of the total number of non‐responders, when each predicted success probability value is taken as a cutoff for binary classification. Area under the curve (AUC) and its respective p‐value were calculated for ROC curves, given null hypothesis of AUC = 0.5. Graphs were generated with Prism 5 (GraphPad Software).
RESULTS
Next generation sequencing analysis of CSF of SMA patients treated with nusinersen
We sought to explore miRNAs expressed in the CSF as potential biomarkers for monitoring response to nusinersen and to help determine, prior to treatment initiation, whether a patient was more or less likely to respond to the therapy. We used next generation sequencing to investigate, without an a priori bias, the comprehensive landscape of CSF miRNAs in 34 type II and type III SMA patients with documented demographic and clinical information available (Table 1). We analyzed RNA‐seq data from CSF samples collected before treatment and analyzed these data in the context of clinical response 6 months after initiation of nusinersen treatment (see study outline in Figure 1). Data from eleven additional patients with HFMSE change of a single or two units [1,2] were excluded.
TABLE 1.
Baseline characteristics at treatment onset: sex, spinal muscular atrophy type, number of sitters/walkers, scoliosis, BIPAP use, age, disease duration from symptom onset, and baseline HFMSE for each cohort
| Cohort | n (% females) | Type, II/III | Sitters/walkers | Scoliosis, no/yes | BIPAP use, no/yes | Age, years, median (range); adults, n | Disease duration, years, median (range) | Baseline HFMSE, median (range) |
|---|---|---|---|---|---|---|---|---|
| Before exclusion | ||||||||
| Schneider Medical Center | 13 (69%) | 7/6 | 10/3 | 3/10 | 12/1 | 11.2 (2.3–19.3); 1 | 10.4 (1.5–16.7) | 26 (1–52) |
| Tel Aviv Medical Center | 20 (55%) | 9/11 | 11/9 | 7/13 | 9/11 | 11.7 (1.7–24.4); 5 | 10.6 (0.2–21.8) | 11.5 (1–52) |
| MGH | 12 (42%) | 5/7 | 6/6 | 6/6 | 8/4 | 7.5 (2.1–56.6); 4 | 6.9 (1.9–48.6) | 41 (12–59) |
| Total | 45 (55%) | 21/24 | 27/18 | 16/29 | 29/16 | 11.0 (1.7–6.6); 10 | 10.1 (0.2–48.6) | 28 (1–59) |
| After exclusion | ||||||||
| Schneider Medical Center | 7 (100%) | 4/3 | 5/2 | 2/5 | 0/7 | 6.4 (2.8–17.3); 0 | 4.4 (1.8–15.3) | 21 (3–52) |
| Tel Aviv Medical Center | 17 (59%) | 8/9 | 8/9 | 7/10 | 8/9 | 11.1 (1.7–24.4); 3 | 10.1 (0.2–21.8) | 12 (2–50) |
| MGH | 10 (40%) | 4/6 | 5/5 | 5/5 | 7/3 | 11.1 (2.1–56.6); 4 | 9.1 (1.9–48.6) | 41 (12–59) |
| Total | 34 (53%) | 16/18 | 18/16 | 14/20 | 15/19 | 11.0 (1.7–56.6); 7 | 9.1 (0.2–48.6) | 27 (2–59) |
Data are presented for the total number of patients 45 and after exclusion of 11 patients (six from Schneider, three from Tel Aviv, and two from MGH) whose change in HFMSE score after 6 months of treatment was 1 or 2. These patients were excluded from the main analysis, but are included in supplementary analyses.
Abbreviations: BIPAP, bilevel positive airway pressure; HFMSE, Hammersmith Functional Motor Scale Expanded; MGH, Massachusetts General Hospital.
FIGURE 1.
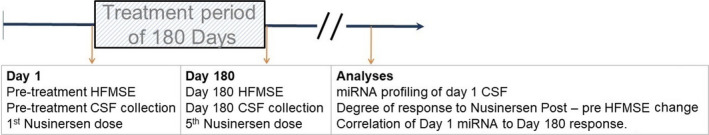
Summary of study outline. Patients were treated with nusinersen for 180 days. Behavioral assessment and miRNA quantification in cerebrospinal fluid (CSF) were conducted prior to treatment onset and prior to the last dose, and miRNA levels prior to treatment onset were compared between responders and non‐responders as defined by Hammersmith Functional Motor Scale Expanded (HFMSE) change post‐treatment versus pre‐treatment [Colour figure can be viewed at wileyonlinelibrary.com]
After next generation sequencing, 2530 miRNAs were annotated to the genome. Only 68 miRNA species exceeded a cutoff of ≥100 UMI counts per sample, averaged on all samples. Five of the 68 miRNAs changed posttreatment relative to pretreatment, with a fold change of ≥1.2 and p‐value of <0.05 by Wald test (Figure S1a). Analysis of these patients by stratified response to therapy (≥3 or ≤0 HFMSE points for responders or nonresponders, respectively) did not change the miRNA profile substantially (Figure S1b,c).
These data suggest that only minor effects were observed on miRNA profile after 6 months of nusinersen therapy. Therefore, measurements of miRNAs in the CSF after treatment were not a main exploratory effort and did not significantly contribute to the study's conclusions.
Baseline miR‐206/miR‐103 levels predict beneficial response to nusinersen therapy
The clinical response to nusinersen therapy is heterogeneous [38]. We tested whether the miRNA profile at baseline would be different between patients who clinically responded to nusinersen therapy (an increase of ≥3 points in the HFMSE total score [32]) and those who responded poorly (HFMSE score change of ≤0 points).
In responders' CSF, two miRNAs differed in a significant manner relative to nonresponders; miR‐103b increased by 2.6‐fold (Wald test: p = 0.002; Figure 2a), and miR‐206 decreased by 1.8‐fold (p = 0.048). Responders did not differ from nonresponders in their clinical characteristics (Figure 2b). During these analyses, we noticed that similar to the downregulated miR‐206 levels, other miRNAs that are known to be expressed mainly in the skeletal muscle (i.e., myomiRs) were relatively low in CSF of responders, including miR‐1‐3p, miR‐133a‐3p, and miR‐133b, at 1.4‐fold, 1.6‐fold, and 2.2‐fold, respectively. Our conclusion remains unchanged also when 11 additional patients with intermediate change in the HFMSE scores, which reflect modest response to therapy, were included as responders to therapy (Figure S2). We also note that miR‐1180‐5p and miR‐6849 were downregulated at baseline in patients that self‐reported energy improvement after therapy versus those who did not report improvement (p < 0.05, Wald test; Figure S3). Therefore, miR‐206/miR‐103 levels predict beneficial objective response to nusinersen therapy.
FIGURE 2.
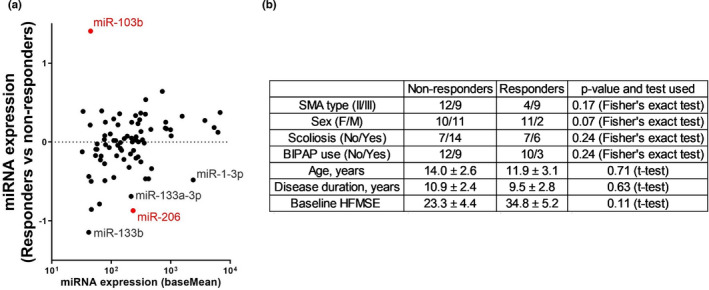
Pre‐treatment miRNA signature predicts the response to nusinersen treatment. (a) MA plot of differential miRNA expression in responders (n = 13, patients with Hammersmith Functional Motor Scale Expanded [HFMSE] change ≥ 3 after 6 months of treatment) versus non‐responders (n = 21, HFMSE change ≤ 0). Data from an additional 11 patients with HFMSE change of a single or two units [1,2] were excluded. Log 2 transformed fold change (y‐axis) is shown against mean miRNA abundance (x‐axis). Red indicates significantly changed miRNAs (p < 0.05, Wald test). Higher miR‐103b levels and lower muscle enriched miRNA miR‐1‐3p, miR‐133a/b, and miR‐206 levels are observed in prospective nusinersen responders. (b) A summary of the clinical data in non‐responders and responders, showing that they are not significantly different between the groups. Numerical data are presented as mean ± SEM. Categorical data were analyzed for significant differences in sex, type, scoliosis, and bilevel positive airway pressure (BIPAP) use distribution by Fisher exact test, whereas continuous data were analyzed for differences by Student t‐test. Notably, baseline HFMSE did not significantly differ between nonresponders and responders (p = 0.11). F, female; M, male; SMA, spinal muscular atrophy [Colour figure can be viewed at wileyonlinelibrary.com]
The levels of miR‐206 were negative predictors of positive treatment response, quantified as the change in the HFMSE final score (Spearman rho = −0.43, 95% confidence interval [CI] = −0.67 to −0.09, p = 0.01; Figure 3a), but were not correlated with baseline HFMSE score (Figure 3b).In addition, an orthogonal logistic regression model supported that miR‐206 levels correlated with clinical response to nusinersen therapy (χ 2 = 7, p = 0.008, AIC = 42.2; Figure 3c). We then tested whether miR‐206 levels can predict differential response, without considering clinical information about disease duration and SMA type (II/III). We found that miR‐206 is predictive of differential response to nusinersen independent of other baseline characteristics (Figure 3d,e). Therefore, miR‐206 is able to predict differential response between two patients with similar baseline characteristics.
FIGURE 3.
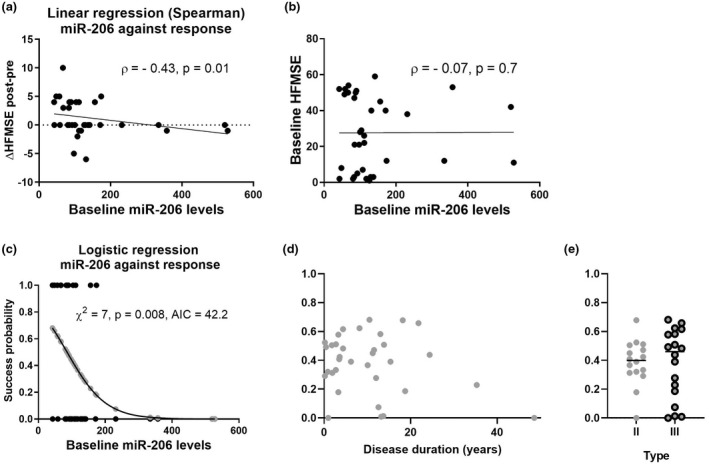
miR‐206 levels predict the clinical improvement of spinal muscular atrophy (SMA) patients following 6‐month nusinersen treatment. (a) Negative correlation of miR‐206 levels with response to nusinersen therapy (as posttreatment – pre‐treatment Hammersmith Functional Motor Scale Expanded [HFMSE] score difference, Spearman rho = −0.43, p = 0.01). (b) No correlation between miR‐206 levels and HFMSE score at baseline. (c) Logistic regression of baseline miR‐206 levels on clinical dichotomized response to therapy: responders (n = 13, HFMSE change ≥ 3 after 6 months of treatment) versus non‐responders (n = 21, HFMSE change ≤ 0). X‐axis represents miR‐206 levels. Y‐axis represents probability of successful response predicted by a logistic regression model (depicted by gray dots on the curve). Black dots represent dichotomized clinical response (non‐responders, −0; responders, −1). Statistical significance of the logistic regression model goodness of fit was assessed by the chi‐squared test. AIC, Akaike information criterion. (d, e) Success probabilities (y‐values the same as for gray dots in panel c) predicted by miR‐206 (d) plotted against disease duration at treatment onset or (e) stratified by SMA type
We next sought to validate the model on newly introduced data. We trained a machine learning procedure on 33 samples and tested the prediction on the single sample that was left out. The LOOCV procedure was reiterated 34 times. This analysis reveals that miR‐206 predicts response more than can be expected at random (ROC AUC = 0.7, 95% CI = 0.51–0.88, p = 0.06; Figure S4). As may be expected, the inclusion of 11 patients with intermediate clinical change decreased the association between miR‐206 levels and response to nusinersen, and reduced the accuracy of response prediction (Figure S5). None of the other myomiRs (miR‐1‐3p, miR‐133a‐3p, or miR‐133b) displayed a significant correlation with response to therapy (Figure S6) or with baseline HFMSE (not shown). However, miR‐103b levels correlated with the change in HFMSE score after therapy (Figure S7). Taken together, these data suggest that miR‐206 levels at baseline may predict the response to the nusinersen therapy.
Multifeature classifiers of response to nusinersen therapy
We next considered simultaneously more than a single miRNA. The correlation of miR‐206 and miR‐133a‐3p to clinical improvement and classification, when both are considered explanatory variables with weighted contributions to the model (multinomial logistic regression, equation available in Table S1), was higher than when miR‐206 was considered as a single feature (χ 2 = 9, p = 0.01, AIC = 42.1; ROC AUC = 0.74, 95% CI = 0.55–0.92, p = 0.022; Figure 4a). None of the other miRNAs (miR‐1‐3p, miR‐103b, or miR‐133b) displayed an improved prediction capacity when added to miR‐206 (Figure S8a–c). Thus, a feature composed of miR‐206 and miR‐133a‐3p levels has an improved prediction capacity for nusinersen therapy response. Adding SMA type (III vs. II) improved the correlation even further (<miR‐206 + miR‐133a‐3p + SMA type>, χ 2 = 12, p = 0.007, AIC = 41.2). We preserved the prediction capacity also by performing LOOCV (AUC = 0.73, 95 % CI = 0.55‐0.92, p = 0.024; Figure 4b). Input about patient sex further improved prediction (<miR‐206 + miR‐133a‐3p + SMA type + sex>, χ 2 = 14.5, p = 0.006, AIC = 40.7; ROC AUC = 0.76, 95 CI% = 0.56–0.95, p = 0.01; Figure 4c), with males being less likely to respond to therapy. Patient age at baseline did not improve prediction (χ 2 = 14.9, p = 0.01, AIC = 42.4; ROC AUC = 0.7, 95% CI = 0.475–0.925, p > 0.05; not shown), and was correlated with neither miR‐133a‐3p (Spearman ρ = −0.01, p = 0.96) nor miR‐206 levels (Spearman ρ = 0.175, p = 0.32), suggesting that miRNA levels predict response independently of age.
FIGURE 4.
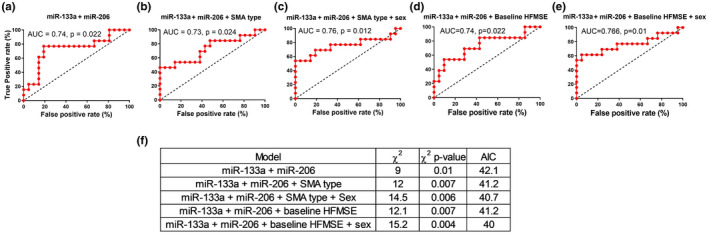
A predictor, based on miR‐133a‐3p and miR‐206, predicts response to nusinersen. (a–e) Receiver operating characteristic curves with leave‐one‐out cross‐validation (LOOCV) for (a) miR‐133a‐3p + miR‐206, (b) miR‐133a‐3p + miR‐206 + spinal muscular atrophy (SMA) type, (c) miR‐133a‐3p + miR‐206 + SMA type + sex, (d) miR‐133a‐3p + miR‐206 + baseline Hammersmith Functional Motor Scale Expanded (HFMSE) score, and (e) miR‐133a‐3p + miR‐206 + baseline HFMSE score + sex. (f) A summary of logistic regression metrics for five multiple feature models. AIC, Akaike information criterion; AUC, area under the curve. For the sake of brevity, miR‐133a‐3p is referred to in this figure and the next figure as "miR‐133a" [Colour figure can be viewed at wileyonlinelibrary.com]
Considering baseline HFMSE as a feature, together with miRNAs (miR‐206 and miR‐133a‐3p), yielded comparable or slightly superior predictive capacities (without sex: χ 2 = 12.1, p = 0.007, AIC = 41.2; ROC AUC = 0.74, 95% CI = 0.55–0.92, p = 0.022, Figure 4d; with sex: χ 2 = 15.2, p = 0.004, AIC = 40; ROC AUC = 0.766, 95% CI = 0.58–0.95, p = 0.01; Figure 4e,f).
In addition to the multinomial regression of miR‐133a‐3p and miR‐206, we also devised a feature by normalization of the miRNA values by min–max scaling and summation of the scaled values. The new predictor, miR‐133a/206, displays a χ 2 of 9 (p = 0.003; Figure 5a), with a drop in predicted model errors relative to when miR‐133a‐3p and miR‐206 are independently considered (AIC = 40.2 vs. 42.1, respectively). The single standardized feature, miR‐133a/206, displays an ROC AUC of 0.74 (95% CI = 0.55–0.92, p = 0.02; Figure 5b), and information about SMA type (II or III based on clinical manifestation) and sex further improved correlation to clinical response and predictive capacity (χ 2 = 14.5, p = 0.002, AIC = 38.8; ROC AUC = 0.784, 95% CI = 0.6–0.97, p = 0.006; Figure 5c). A model that includes baseline HFMSE performs comparably (χ 2 = 15.2, p = 0.0017, AIC = 38.0; ROC AUC = 0.78, 95% CI = 0.6–0.96, p = 0.007; Figure 5d,e).
FIGURE 5.

A predictor, based on a summation of the scaled values of miR‐133a and miR‐206, predicts response to nusinersen. (a) Logistic regression analysis of miR‐133a/206 at baseline and clinical response to therapy after 6 months of nusinersen treatment. When the extreme sample whose miR‐133a/206 score was 2 is excluded, chi‐squared slightly decreases (from 9 to 8) and the p‐value marginally increases (from 0.003 to 0.0046), suggesting the overall data and interpretation remain unchanged. (b–d) Receiver operating characteristic curves based on leave‐one‐out cross‐validation (LOOCV) for (b) miR‐133a/206, (c) miR‐133a/206 with spinal muscular atrophy (SMA) type and sex, and (d) miR‐133a/206 with baseline Hammersmith Functional Motor Scale Expanded (HFMSE) and sex. (e) A summary of logistic regression metrics for three models. AIC, Akaike information criterion; AUC, area under the curve [Colour figure can be viewed at wileyonlinelibrary.com]
Given inclusion of eleven additional patients with intermediate degree of clinical response to nusinersen therapy, miR‐133a‐3p and miR‐206 remained valuable predictors alone or in combination, with a notable reduction in accuracy (Figure S9).
Data were available for only 26 patients at a longer follow‐up period of 12 months. Logistic regression analysis on miR‐206 levels or on scaled levels of miR‐133a/miR‐206 is consistent with earlier time point analysis indicating that extremely high levels of myomiRs predict poor response to nusinersen (Figure S10). In summary, two miRNAs in the CSF are able to predict response to nusinersen therapy in SMA type II and III patients and may be useful for pre‐treatment clinical decisions.
DISCUSSION
Given the expanding treatment options available for motor neuron diseases, novel biomarkers to help track disease progression and therapeutic response are urgently needed. Here, we report novel findings derived from the first screening for small RNAs in the CSF of type II/III SMA patients treated with nusinersen. These data demonstrate that baseline levels of the myomiRs miR‐133a‐3p and miR‐206 predict the response to nusinersen therapy, suggesting that these myomiRs could play an important role in the clinical setting to help identify those patients most likely to demonstrate a more robust response to nusinersen therapy. In presenting these data, we hope to encourage incorporation of examination of CSF levels of miR‐133a‐3p and miR‐206 as well as other myomiRs in longitudinal follow‐up studies, across cohorts of SMA patients receiving nusinersen or other novel therapies, to help validate the current findings.
Identifying biomarkers to track progression of motor neruon diseases and the response to molecular therapies is an important focus of our group and others. We have previously demonstrated the usefulness of miR‐181 in prediction of progression in ALS [28] and of electrophysiologic measures of peripheral denervation in tracking peripheral motor unit integrity [39, 40]. Circulating SMN protein levels are a candidate biomarker for monitoring SMA disease progression, although changes in peripheral SMN levels are not expected with at least two of the three currently FDA‐approved therapies [41]. CSF and plasma neurofilaments are markers of neuronal damage and have been identified as a potential treatment response biomarker in pediatric SMA and other neurodegenerative diseases, but baseline CSF or circulating neurofilament levels do not seem to necessarily predict treatment response in pediatric SMA patients [42, 43, 44, 45]. With three FDA‐approved therapies and the anticipation of additional emerging treatments for SMA, comprehensive longitudinal cohort studies remain critically necessary to identify useful biomarkers that could predict the response to individual therapies.
One striking finding from our observations here is that myomiRs were detected in the CSF and that CSF myomiR levels were lower in SMA patients who responded well to nusinersen, when compared to those who presented a limited response. Although the myomiRs discussed here were identified via an unbiased screening approach in CSF, our data corroborate recent findings demonstrating that serum myomiR levels are readouts of nusinersen response in SMA [25].
We discuss two potential models that may explain these observations. First, the presence of myomiRs in SMA CSF could be an indication of blood–CSF or blood–spinal cord barrier disruption [46, 47], which might enable the entrance of miRNAs that are derived from breakdown of muscle cells in the setting of acute denervation [22, 23, 24]. Thus, lower myomiR levels prior to treatment with nusinersen, which predicts those with a better response potential, could reflect reduced muscle cell breakdown, presumably associated with higher muscle mass and higher numbers of functional motor units. Moreover, disease‐associated damage to CSF barrier integrity might allow miRNA spilled from muscle to arrive at the CSF.
In addition, there is evidence for expression of miR‐133 [48] in the brain and for upregulation of miR‐206 in the brains in Alzheimer disease subjects [49]. Therefore, miR‐133/miR‐206 may be expressed in the brain, and their levels may be further changed in SMA.
Together, the presence of muscle‐specific miRNAs in CSF of patients with SMA can be due to change in the transcription of these miRNAs, which are commonly thought to be muscle‐specific but are expressed also in neurons [48, 49], or to genuine spilling of dying myotubes along with pathological breakdown of the blood–CSF barrier.
We previously demonstrated that serum creatinine, a product of skeletal muscle creatine metabolism, was also identified as a candidate biomarker to track SMA disease progression [50], and that creatinine may be a sensitive predictor for the onset of denervation in infants with three SMN2 copies [50]. Similarly, circulating creatine kinase levels correlated with SMA severity and decreased during nusinersen therapy [51]. Therefore, in the future, the capacity to predict response to nusinersen therapy may be tested by protein biomarkers in combination with miRNAs such as miR‐206/133a‐3p.
Limitations of our study include a relatively small cohort and a limited follow‐up period. When larger cohorts are studied, over longer follow‐up periods, the use of Revised Upper Limb Module (RULM) and 6‐min walk test clinical scores in addition to HFMSE may enable better clinical estimation. Work is required also for creating means for quantification of absolute miRNA concentrations before these findings can be applied to the clinic.
In summary, the current study demonstrates that the levels of muscle miRNAs miR‐206 and miR‐133a‐3p in CSF can predict the clinical response to nusinersen treatment in SMA patients. These novel findings have high clinical relevance and may prove useful in helping to design appropriate protocols intended to refine and improve treatment outcomes. Targeted approaches for quantification of miR‐133a‐3p and miR‐206 may in the future increase the accessibility to miRNA measurements and as a result, CSF miRNAs may be used as a means to determine whether to initiate treatment. However, to get to that point, future studies should be performed in bigger, ethnically diverse groups of patients, and ethical issues should be considered.
AUTHOR CONTRIBUTIONS
Iddo Magen: Conceptualization (lead), data curation (lead), formal analysis (lead), investigation (lead), methodology (lead), project administration (lead), resources (lead), software (lead), supervision (lead), validation (lead), visualization (lead), writing–original draft (lead), writing–review & editing (lead). Sharon Aharoni: Conceptualization (equal), data curation (lead), formal analysis (supporting), methodology (lead), project administration (supporting), resources (lead), writing–original draft (equal), writing–review & editing (equal). Nancy Sarah Yacovzada: Formal analysis (supporting), methodology (supporting), software (supporting), writing–original draft (supporting), writing–review & editing (supporting). Itay Tokatly Latzer: Data curation (lead), investigation (lead), resources (lead), writing–original draft (supporting), writing–review & editing (supporting). Christiano R. R. Alves: Data curation (lead), investigation (lead), resources (lead), writing–original draft (supporting), writing–review & editing (supporting). Liora Sagi: Investigation (lead). Aviva Fattal‐Valevski: Data curation (lead), investigation (lead), resources (lead), writing–original draft (supporting), writing–review & editing (supporting). Kathryn J. Swoboda: Data curation (lead), investigation (lead), resources (lead), writing–original draft (supporting), writing–review & editing (supporting). Jacob Katz: Data curation (supporting). Elchanan Bruckheimer: Data curation (supporting). Yoram Nevo: Conceptualization (equal), data curation (lead), formal analysis (supporting), methodology (lead), project administration (supporting), resources (lead), writing–original draft (equal), writing–review & editing (equal). Eran Hornstein: Conceptualization (lead), formal analysis (lead), funding acquisition (lead), methodology (lead), project administration (lead), resources (lead), supervision (lead), validation (lead), visualization (lead), writing–original draft (lead), writing–review & editing (lead).
CONFLICT OF INTEREST
C.R.R.A. and K.J.S. are inventors on a patent filed by Mass General Brigham that describes genome‐engineering technologies to treat SMA. K.J.S. has received clinical trial funding from AveXis/Novartis and Biogen. She has received speaker fees from Biogen and has served on scientific advisory boards for AveXis, Biogen, and Roche/Genentech. S.A. has received clinical trial funding from AveXis/Novartis Gene Therapies and Biogen and has served on scientific advisory boards for AveXis/Novartis Gene Therapies. None of the other authors has any conflict of interest to disclose.
Supporting information
App S1
ACKNOWLEDGMENTS
E.H. is the Mondry Family Professorial Chair and Head of the Andi and Larry Wolfe Center for Neuroimmunology and Neuromodulation. Research at the Hornstein laboratory is supported by the CReATe consortium and ALSA (program: "Prognostic Value of miRNAs in Biofluids From ALS Patients"); the RADALA Foundation; AFM Telethon (20576); the Weizmann–Brazil Center for Research on Neurodegeneration at Weizmann Institute of Science; the Minerva Foundation, with funding from the Federal German Ministry for Education and Research; the ISF Legacy Heritage Fund (828/17); the Israel Science Foundation (135/16, 3497/21, 424/22, 425/22); Target ALS (118945); the Thierry Latran Foundation for ALS Research; the European Research Council under the European Union's Seventh Framework Program ([FP7/2007–2013]/ERC grant agreement number 617351); ERA‐Net for Research Programs on Rare Diseases (eRARE FP7) via the Israel Ministry of Health; Dr Sydney Brenner and friends; Edward and Janie Moravitz; A. Alfred Taubman through IsrALS; Yeda‐Sela; Yeda‐CEO; the Israel Ministry of Trade and Industry; the Y. Leon Benoziyo Institute for Molecular Medicine; the Nella and Leon Benoziyo Center for Neurological Disease; the Kekst Family Institute for Medical Genetics; the David and Fela Shapell Family Center for Genetic Disorders Research; the Crown Human Genome Center; the Nathan, Shirley, Philip, and Charlene Vener New Scientist Fund; the Julius and Ray Charlestein Foundation; the Fraida Foundation; the Wolfson Family Charitable Trust; the Adelis Foundation; Merck (UK); M. Halphen; the estates of F. Sherr, L. Asseof, and L. Fulop; the Goldhirsh‐Yellin Foundation; the Redhill Foundation–Sam and Jean Rothberg Charitable Trust; Dr. Dvora and Haim Teitelbaum Endowment Fund. N.S.Y. was supported by the Israeli Council for Higher Education via the Weizmann Data Science Research Center. I.M. was supported by Teva Pharmaceutical Industries as part of the Israeli National Network of Excellence in Neuroscience (fellowship 117941). C.R.R.A. was funded by MGH Executive Committee on Research. K.J.S. was funded by the National Institutes of Health (NIH) National Institute of Child Health and Human Development (R01HD054599), NIH National Institute of Neurological Disorders and Stroke (R21NS108015), Biogen, AveXis, and Cure SMA. We are grateful to all the patients and families who participated in this study.
Magen I, Aharoni S, Yacovzada NS, et al. Muscle microRNAs in the cerebrospinal fluid predict clinical response to nusinersen therapy in type II and type III spinal muscular atrophy patients. Eur J Neurol. 2022;29:2420–2430. doi: 10.1111/ene.15382
Iddo Magen and Sharon Aharoni contributed equally.
Contributor Information
Yoram Nevo, Email: yoramne@clalit.org.il.
Eran Hornstein, Email: eran.hornstein@weizmann.ac.il.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Ogino S, Leonard DG, Rennert H, Wilson RB. Spinal muscular atrophy genetic testing experience at an academic medical center. J Mol Diagn. 2002;4(1):53‐58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Verhaart IEC, Robertson A, Wilson IJ, et al. Prevalence, incidence and carrier frequency of 5q‐linked spinal muscular atrophy ‐ a literature review. Orphanet J Rare Dis. 2017;12(1):124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Prior TW, Leach ME, Finanger E. Spinal Muscular Atrophy. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Mirzaa G, Gripp KW, Mirzaa GM, Amemiya A, editors. GeneReviews(R) . University of Washington, Seattle; 1993. [PubMed] [Google Scholar]
- 4. Brahe C, Servidei S, Zappata S, Ricci E, Tonali P, Neri G. Genetic homogeneity between childhood‐onset and adult‐onset autosomal recessive spinal muscular atrophy. Lancet. 1995;346(8977):741‐742. [DOI] [PubMed] [Google Scholar]
- 5. Clermont O, Burlet P, Lefebvre S, Burglen L, Munnich A, Melki J. SMN gene deletions in adult‐onset spinal muscular atrophy. Lancet. 1995;346(8991‐8992):1712‐1713. [DOI] [PubMed] [Google Scholar]
- 6. Wadman RI, Stam M, Gijzen M, et al. Association of motor milestones, SMN2 copy and outcome in spinal muscular atrophy types 0–4. J Neurol Neurosurg Psychiatry. 2017;88(4):365‐367. [DOI] [PubMed] [Google Scholar]
- 7. Zerres K, Rudnik‐Schoneborn S, Forrest E, Lusakowska A, Borkowska J, Hausmanowa‐Petrusewicz I. A collaborative study on the natural history of childhood and juvenile onset proximal spinal muscular atrophy (type II and III SMA): 569 patients. J Neurol Sci. 1997;146(1):67‐72. [DOI] [PubMed] [Google Scholar]
- 8. Groen EJN, Talbot K, Gillingwater TH. Advances in therapy for spinal muscular atrophy: promises and challenges. Nat Rev Neurol. 2018;14(4):214‐224. [DOI] [PubMed] [Google Scholar]
- 9. Faravelli I, Nizzardo M, Comi GP, Corti S. Spinal muscular atrophy–recent therapeutic advances for an old challenge. Nat Rev Neurol. 2015;11(6):351‐359. [DOI] [PubMed] [Google Scholar]
- 10. Mailman MD, Heinz JW, Papp AC, et al. Molecular analysis of spinal muscular atrophy and modification of the phenotype by SMN2. Genet Med. 2002;4(1):20‐26. [DOI] [PubMed] [Google Scholar]
- 11. Feldkotter M, Schwarzer V, Wirth R, Wienker TF, Wirth B. Quantitative analyses of SMN1 and SMN2 based on real‐time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet. 2002;70(2):358‐368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Crawford TO, Paushkin SV, Kobayashi DT, et al. Evaluation of SMN protein, transcript, and copy number in the biomarkers for spinal muscular atrophy (BforSMA) clinical study. PLoS One. 2012;7(4):e33572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aartsma‐Rus A. FDA approval of nusinersen for spinal muscular atrophy makes 2016 the year of splice modulating oligonucleotides. Nucleic Acid Ther. 2017;27(2):67‐69. [DOI] [PubMed] [Google Scholar]
- 14. Messina S. New directions for SMA therapy. J Clin Med. 2018;7(9):251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hagenacker T, Wurster CD, Gunther R, et al. Nusinersen in adults with 5q spinal muscular atrophy: a non‐interventional, multicentre, observational cohort study. Lancet Neurol. 2020;19(4):317‐325. [DOI] [PubMed] [Google Scholar]
- 16. Walter MC, Wenninger S, Thiele S, et al. Safety and treatment effects of nusinersen in longstanding adult 5q‐SMA type 3 ‐ a prospective observational study. J Neuromuscul Dis. 2019;6(4):453‐465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Emde A, Eitan C, Liou LL, et al. Dysregulated miRNA biogenesis downstream of cellular stress and ALS‐causing mutations: a new mechanism for ALS. EMBO J. 2015;34(21):2633‐2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reichenstein I, Eitan C, Diaz‐Garcia S, et al. Human genetics and neuropathology suggest a link between miR‐218 and amyotrophic lateral sclerosis pathophysiology. Sci Transl Med. 2019;11(523):eaav5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haramati S, Chapnik E, Sztainberg Y, et al. miRNA malfunction causes spinal motor neuron disease. Proc Natl Acad Sci USA. 2010;107(29):13111‐13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen J‐F, Mandel EM, Thomson JM, et al. The role of microRNA‐1 and microRNA‐133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38(2):228‐233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen J‐F, Tao Y, Li J, et al. microRNA‐1 and microRNA‐206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J Cell Biol. 2010;190(5):867‐879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. de Andrade HM, de Albuquerque M, Avansini SH, et al. MicroRNAs‐424 and 206 are potential prognostic markers in spinal onset amyotrophic lateral sclerosis. J Neurol Sci. 2016;368:19‐24. [DOI] [PubMed] [Google Scholar]
- 23. Toivonen JM, Manzano R, Olivan S, Zaragoza P, Garcia‐Redondo A, Osta R. MicroRNA‐206: a potential circulating biomarker candidate for amyotrophic lateral sclerosis. PLoS One. 2014;9(2):e89065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Waller R, Goodall EF, Milo M, et al. Serum miRNAs miR‐206, 143–3p and 374b–5p as potential biomarkers for amyotrophic lateral sclerosis (ALS). Neurobiol Aging. 2017;55:123‐131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bonanno S, Marcuzzo S, Malacarne C, et al. Circulating MyomiRs as potential biomarkers to monitor response to nusinersen in pediatric SMA patients. Biomedicines. 2020;8(2):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Coenen‐Stass AML, Magen I, Brooks T, et al. Evaluation of methodologies for microRNA biomarker detection by next generation sequencing. RNA Biol. 2018;15(8):1133‐1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Magen I, Yacovzada N‐S, Warren JD, et al. miRNA biomarkers for diagnosis of ALS and FTD, developed by a nonlinear machine learning approach. medRxiv. 2021:2020.01.22.20018408. [Google Scholar]
- 28. Magen I, Yacovzada NS, Yanowski E, et al. Circulating miR‐181 is a prognostic biomarker for amyotrophic lateral sclerosis. Nat Neurosci. 2021;24(11):1534‐1541. [DOI] [PubMed] [Google Scholar]
- 29. Glanzman AM, Mazzone ES, Young SD, et al. Evaluator training and reliability for SMA global nusinersen trials1. J Neuromuscul Dis. 2018;5(2):159‐166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. O'Hagen JM, Glanzman AM, McDermott MP, et al. An expanded version of the Hammersmith functional motor scale for SMA II and III patients. Neuromuscul Disord. 2007;17(9‐10):693‐697. [DOI] [PubMed] [Google Scholar]
- 31. Mercuri E, Darras BT, Chiriboga CA, et al. Nusinersen versus Sham control in later‐onset spinal muscular atrophy. N Engl J Med. 2018;378(7):625‐635. [DOI] [PubMed] [Google Scholar]
- 32. Swoboda KJ, Scott CB, Reyna SP, et al. Phase II open label study of valproic acid in spinal muscular atrophy. PLoS One. 2009;4(5):e5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kohen R, Barlev J, Hornung G, et al. UTAP: user‐friendly transcriptome analysis pipeline. BMC Bioinformatics. 2019;24:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kozomara A, Griffiths‐Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68‐D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Multiple Logistic Regression. Applied Logistic Regression; 2000;31‐46.
- 37. Akaike H. Information theory and an extension of the maximum likelihood principle. In: Parzen E, Tanabe K, Kitagawa G, eds. Selected Papers of Hirotugu Akaike. Springer, New York; 1998:199‐213. [Google Scholar]
- 38. Finkel RS, Mercuri E, Darras BT, et al. Nusinersen versus sham control in infantile‐onset spinal muscular atrophy. N Engl J Med. 2017;377(18):1723‐1732. [DOI] [PubMed] [Google Scholar]
- 39. Bromberg MB, Swoboda KJ. Motor unit number estimation in infants and children with spinal muscular atrophy. Muscle Nerve. 2002;25(3):445‐447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lewelt A, Krosschell KJ, Scott C, et al. Compound muscle action potential and motor function in children with spinal muscular atrophy. Muscle Nerve. 2010;42(5):703‐708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Alves CRR, Zhang R, Johnstone AJ, et al. Whole blood survival motor neuron protein levels correlate with severity of denervation in spinal muscular atrophy. Muscle Nerve. 2020;62(3):351‐357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Darras BT, Crawford TO, Finkel RS, et al. Neurofilament as a potential biomarker for spinal muscular atrophy. Ann Clin Transl Neurol. 2019;6(5):932‐944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eichelberger EJ, Alves CRR, Zhang R, et al. Increased systemic HSP70B levels in spinal muscular atrophy infants. Ann Clin Transl Neurol. 2021;8:1495‐1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. De Vivo DC, Bertini E, Swoboda KJ, et al. Nusinersen initiated in infants during the presymptomatic stage of spinal muscular atrophy: interim efficacy and safety results from the phase 2 NURTURE study. Neuromuscul Disord. 2019;29(11):842‐856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Winter B, Guenther R, Ludolph AC, Hermann A, Otto M, Wurster CD. Neurofilaments and tau in CSF in an infant with SMA type 1 treated with nusinersen. J Neurol Neurosurg Psychiatry. 2019;90(9):1068‐1069. [DOI] [PubMed] [Google Scholar]
- 46. Müschen LH, Osmanovic A, Binz C, et al. Cerebrospinal fluid parameters in antisense oligonucleotide‐treated adult 5q‐spinal muscular atrophy patients. Brain Sci. 2021;11(3):296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Somers E, Lees RD, Hoban K, et al. Vascular defects and spinal cord hypoxia in spinal muscular atrophy. Ann Neurol. 2016;79(2):217‐230. [DOI] [PubMed] [Google Scholar]
- 48. Kim J, Inoue K, Ishii J, et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317(5842):1220‐1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee ST, Chu K, Jung KH, et al. miR‐206 regulates brain‐derived neurotrophic factor in Alzheimer disease model. Ann Neurol. 2012;72(2):269‐277. [DOI] [PubMed] [Google Scholar]
- 50. Alves CRR, Zhang R, Johnstone AJ, et al. Serum creatinine is a biomarker of progressive denervation in spinal muscular atrophy. Neurology. 2020;94(9):e921‐e931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Freigang M, Wurster CD, Hagenacker T, et al. Serum creatine kinase and creatinine in adult spinal muscular atrophy under nusinersen treatment. Ann Clin Transl Neurol. 2021;8(5):1049‐1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
App S1
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


