Abstract
Aims
The DNA marker HF183 is a partial 16S rRNA gene sequence highly specific to human‐associated Bacteroides including Bacteroides dorei. While HF183 is used to assess human faecal contamination in aquatic environments worldwide, little is known about the existence of HF183 and B. dorei in human microbiomes outside of the human gastrointestinal tract and faeces.
Methods and Results
Previously published human skin and urine microbiome data sets from five independent human body skin studies, the Human Microbiome Project (HMP) and three independent human urine studies were analysed. The HF183 gene sequence was detected in all skin data sets, with the ratios of positive samples ranging from 0.5% to 36.3%. Popliteal fossa (knee), volar forearm and inguinal (groin) creases were identified as hot spots. HF183 was detected in two of three urine data sets, with ratios of positive samples ranging from 0% to 37.5%. All HF183‐containing sequences from these data sets were classified as associated with B. dorei.
Conclusions
HF183 is widespread on human skin and present in urine.
Significance and Impact of Study
Skin and urine microbiomes could be sources of HF183 to environmental waters. Such non‐faecal sources of HF183 might explain low concentrations of HF183 in recreational waters when swimmers are present.
Keywords: Bacteroides dorei, HF183, human faecal contamination, human microbiomes, skin, urine
INTRODUCTION
Faecal contamination of water bodies is a serious public health problem due to the introduction of waterborne human pathogens. Monitoring environmental microbiological water quality by standard culturing and isolation of human pathogens is impractical, given resources required to surveil a diversity of waterborne human pathogens (Harwood et al., 2014). High throughput sequencing techniques used in metagenomics have been validated and applied in simultaneous detection of diverse waterborne human pathogens including viruses (Bibby et al., 2011; Escobedo‐Hinojosa & Pardo‐López, 2017), yet such approaches do not address infectivity and are still comparatively unaffordable for routine use in monitoring. Faecal indicator bacteria (FIB) including total coliforms, faecal coliforms, Escherichia coli and enterococci have been used as indicators of microbiological quality of environmental waters for over a century due to their high abundance in faeces and easy detection. However, FIB are found broadly within faeces of warm‐blooded animals, and so do not specifically reveal human faecal contamination which is of greatest public health concern due to host specificity of pathogens. Further, as FIB can persist and multiply in environments such as sediments, soils and aquatic vegetation (Field & Samadpour, 2007; Harwood et al., 2014), FIB may not indicate recent faecal pollution. These issues can result in elevated FIB levels being associated with relatively low human health risks (Ferguson et al., 1996). To assess microbial risk and implement appropriate water quality management practices, faecal contamination sources should be accurately assigned and located via microbial source tracking (MST).
In MST, host‐specific faecal DNA markers from human and animal‐associated micro‐organisms such as Bacteroidales – using amplification by polymerase chain reaction (PCR), quantitative PCR (qPCR) and droplet digital PCR (ddPCR) – have been widely applied (Boehm et al., 2013; Field & Samadpour, 2007; Harwood et al., 2014; Meays et al., 2004; Reischer et al., 2013; Scott et al., 2002; Stoeckel & Harwood, 2007). Numerous human‐associated markers are used to track human faecal contamination (Harwood et al., 2014; Hughes et al., 2017; Roslev & Bukh, 2011). Based on its occurrence in human faecal samples, HF183 – termed for a particular partial 16S rRNA gene sequence highly specific to human‐associated Bacteroides including Bacteroides dorei – has been used to detect and quantify human faecal contamination worldwide (Bakir et al., 2006; Bernhard & Field, 2000a; Bernhard & Field, 2000b; Haugland et al., 2010; Kirs et al., 2016). Diverse PCR and qPCR‐based methods use the HF183 gene sequence as the basis for the forward primer, paired with reverse primers and probes targeting broader phylogenetic groups such as Bac708R, BFDRev and BFDFAM – all targeting the entire Bacteroidales family (Bernhard & Field, 2000b; Haugland et al., 2010), as well as BacR287 and BacP234MGB designed based on an alignment of Bacteroidetes 16S rRNA gene sequences containing HF183 (Green et al., 2014). High specificity and sensitivity of HF183 to human faeces compared to other DNA markers have defined HF183 as one of the best performing bacterial MST markers (Boehm et al., 2013). Human faecal contamination based on HF183 detection has been widely documented in various environmental waters including treated wastewater, urban stormwater, rivers, streams, lakes, marine waters and groundwater (Bradshaw et al., 2016; Diston et al., 2015; Jardé et al., 2018; Mayer et al., 2015; Molina et al., 2014; Sidhu et al., 2013; Staley et al., 2016; Staley et al., 2018). HF183 was also consistently observed in beach waters along the California coast in many previous studies associated with sampled drainage, creek and river waters (Cao et al., 2017; McQuaig et al., 2012; Russell et al., 2013; Sercu et al., 2009). In recent studies of several beaches, swimmers were found to be sources of chronic, low concentration, HF183 markers in beach surf zone waters (Li, Van De Werfhorst, Steets, Ervin, Murray, Blackwell, et al., 2021; Li, van De Werfhorst, Steets, Ervin, Murray, Devarajan, & Holden, 2021; Toubiana et al., 2021). These results raised the question: could aquatic HF183 detections originate from other than human faecal releases or faecal shedding into the environment?
HF183 and human‐associated Bacteroides including B. dorei were previously identified and isolated from human faeces (Bakir et al., 2006; Bernhard & Field, 2000a), and HF183 sequences were occasionally detected in chicken and dog faeces in low quantities (Feng & McLellan, 2019; Harwood et al., 2014). However, little is known about the distribution of HF183 and B. dorei beyond the human gastrointestinal tract and faeces. In the last decade, numerous studies have been performed to reveal human skin and urine microbiomes (Fouts et al., 2012; Grice et al., 2009; Lehtimäki et al., 2017; Lewis et al., 2013; Meisel et al., 2016; Oh et al., 2014; Siddiqui et al., 2011; Turnbaugh et al., 2007; van Rensburg et al., 2015). Considering the possibilities for contamination of human body skin and the urinary tract by HF183 and human‐associated B. dorei, as well as the potential for any HF183 populations on humans to be sources to environmental waters during bather shedding (Li, Van De Werfhorst, Steets, Ervin, Murray, Blackwell, et al., 2021; Li, van De Werfhorst, Steets, Ervin, Murray, Devarajan, & Holden, 2021; Toubiana et al., 2021), it is worth determining whether HF183 and human‐associated B. dorei are within sequences associated with human skin and urine microbiomes. In this study, we examine this issue based on secondary examination of previously published human skin and urine microbiome gene sequence data. The results reveal previously undiscussed sources of HF183 and human‐associated Bacteroides in human microbiomes outside of faeces, and add new understanding useful to interpreting HF183 detections in water bodies such as recreational waters.
MATERIALS AND METHODS
Human body skin and urine microbiome data sets
The HF183 marker is located in the V2 region of the 16S rRNA gene sequence, thus only published studies utilizing primers of nearly full length (27F and 1492R) or the V1‐V3 region (27F and 534R) of the 16S rRNA gene for sequencing were selected. Five independent previously published studies (S1–S5) focusing on human body skin microbiomes and three studies (U1–U3) focusing on human urine microbiomes were selected for downloading sequencing data sets (Fouts et al., 2012; Grice et al., 2009; Lehtimäki et al., 2017; Lewis et al., 2013; Meisel et al., 2016; Oh et al., 2014; Siddiqui et al., 2011; van Rensburg et al., 2015). A part of the sequencing data sets for human skin microbiomes was also downloaded from the HMP project (https://hmpdacc.org/). Based on the sampling information provided in these studies, all of the human skin samples were collected by study personnel or nurses. An exception may be the HMP project for which the sampling information is unavailable. All of the urine samples were collected following a similar protocol, by which either volunteers were instructed to provide a clean‐catch, mid‐stream voided, urine sample into a sterile container or urine was obtained directly via a catheter in subjects utilizing sterile catheters. The details of all nine data sets and the sampling locations on human skin or in urine are listed in Table S1. These nine data sets were sequenced by Sanger, 454 or Illumina sequencing. In brief, 71.3 Gb of data representing 2943 skin samples from more than 15 human body sites were acquired, and 1.5 Gb of data from 77 urine samples were downloaded. Due to the remarkable variation in the amount of sequencing data obtained for each sample across the nine data sets, the sequencing data were not normalized in this study.
Bioinformatic analyses
To confirm the source of HF183 as B. dorei, the human faeces specific sequence HF183 (5’‐ATCATGAGTTCACATGTCCG‐3′) used as the forward primer in HF183 assays (Table S2) was aligned against the NCBI NT and SILVA SSU r132 RefNR database (https://www.arb‐silva.de/) using blastn as well as the RDP database using Probe Match (https://rdp.cme.msu.edu/). HF183 was then searched in all six data sets from the human skin microbiome studies and three data sets from human urine microbiome studies using blastn with criteria of 100% match and 100% coverage. The HF183 containing sequences from skin and urine microbiome data sets were further used to generate a phylogenetic tree together with all B. dorei reference 16S rRNA sequences as well as all Bacteroides genus type strain 16S rRNA sequences archived in the RDP database. The sequences were aligned using MAFFT (Katoh et al., 2002), and the phylogenetic tree was constructed using MEGA X by the neighbour‐joining algorithm and the Jukes–Cantor distance estimation method with bootstrap analyses for 1000 replicates (shown in Figure 1) (Kumar et al., 2018). Additionally, all reverse primers and probes (Table S2) were aligned against the HF183 containing sequences with nearly full length (using primers 27F and 1492R) using blastn with criteria of 100% match and 100% coverage. The nearly full‐length sequences were selected considering the variable loci of reverse primers and probes.
FIGURE 1.
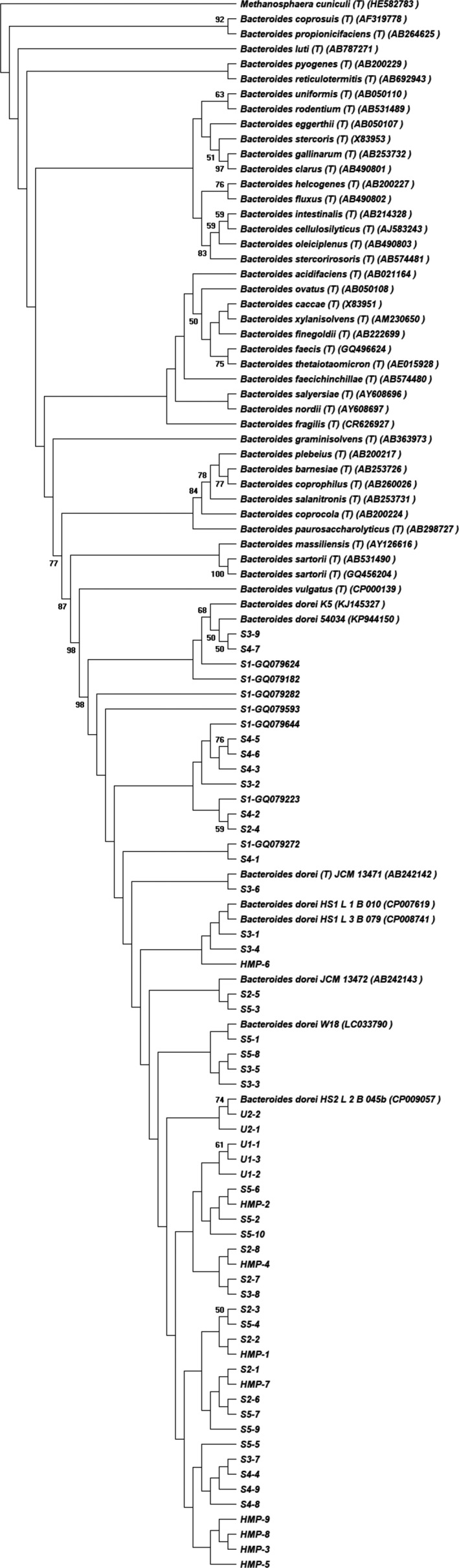
Phylogenetic relationships of representative HF183 containing sequences from skin microbiome data sets S1–S5, HMP project, urine microbiome data sets U1–U3, together with all B. dorei reference 16S rRNA sequences as well as all Bacteroides genus type stain 16S rRNA sequences archived in RDP database determined by the neighbour‐joining method. Bootstrap values of >50% (obtained with 1000 resamplings) are shown at nodes. Methanosphaera cuniculi type strain DSM 4103 (HE582783) was used as an outgroup. GenBank accession numbers are in parentheses
RESULTS
By aligning against the RDP, SILVA and NCBI NT databases, HF183 was only matched to sequences affiliated with B. dorei (29, 18 and 15 sequences respectively) and all other matches belonged to uncultured bacteria without definite species names (Table S3). The matching results of HF183 to human skin and urine microbiome data sets are listed in Table S1 and summarized in Table 1. HF183 was detected in all six skin data sets, and the ratios of positive samples ranged from 0.5% to 36.3%. Study S1 collected skin microbiome samples from healthy adults (Grice et al., 2009) and showed the lowest detection of HF183, possibly due to less sequencing depth (less than 1000 sequences per sample) obtained by cloning and sequencing compared to the other studies which utilized high‐throughput sequencing methods. Study S5 investigated the microbiome on volar forearm of 275 children of ages ranging from 2 months to 14 years (Lehtimäki et al., 2017) and showed the highest overall ratio of HF183 detection as 36.3%. This value was significantly higher than the ratios of other studies, suggesting that children might have more HF183 associated bacteria in their skin microbiome compared with adults. Studies S2 (Meisel et al., 2016) and HMP both characterized the microbiome in healthy adults and have similar overall HF183 detection ratios (4.7% and 3.1% respectively). The HF183 detection level was also similar (4.7%) in study S3 which analysed the skin microbiome before, during and after experimental inoculation of the arm with Haemophilus ducreyi in adult volunteers who subsequently resolved the infection or formed abscesses (van Rensburg et al., 2015). HF183‐associated bacteria on skin were not obviously influenced by infection (data not shown) (van Rensburg et al., 2015). Study S4 has the largest number of samples (1542 samples) obtained from healthy volunteers of 23 to 39 years of age without any chronic skin diseases (Oh et al., 2014). For that study, the highest ratio of HF183 positive samples came from popliteal fossa (37.6%), following by inguinal crease (18.8%), volar forearm (13.3%) and back (12.5%).
TABLE 1.
The ratios of HF183 positive human skin and urine microbiome samples in five independent studies S1–S5, HMP project and three independent studies U1–U3 a
| Site | S1 b | S2 c | S3 d | S4 e | S5 f | HMP g | U1 h | U2 i | U3 j |
|---|---|---|---|---|---|---|---|---|---|
| Back | 0% (0/10) | 12.5% (2/16) | |||||||
| Cubital fossa | 0% (0/10) | 11.1% (2/18) | 10.9% (43/395) | 5.2% (5/96) | |||||
| Forehead | 11.1% (2/18) | ||||||||
| Inguinal crease | 0% (0/10) | 18.8% (6/32) | |||||||
| Manubrium | 0% (0/10) | 11.8% (2/17) | |||||||
| Nare | 0% (0/10) | 12.2% (32/263) | |||||||
| Occiput | 10% (1/10) | 0% (0/16) | 0% (0/17) | ||||||
| Palm | 0% (0/10) | 11.1% (2/18) | 6.1% (2/33) | ||||||
| Plantar heel | 0% (0/10) | 0% (0/36) | |||||||
| Popliteal fossa | 0% (0/10) | 37.6% (38/101) | |||||||
| Retroauricular crease | 0% (0/10) | 0% (0/18) | 8.1% (15/186) | 3.1% (4/128) | |||||
| Toe web | 0% (0/10) | 5.6% (1/18) | 3.0% (1/33) | ||||||
| Umbilicus | 0% (0/10) | 0% (0/18) | |||||||
| Upper arm | 4.7% (9/191) | ||||||||
| Volar forearm | 0% (0/10) | 13.3% (41/308) | 36.3% (207/571) | ||||||
| Other | 0% (0/70) | 0% (0/26) | 0% (0/105) | 0% (0/65) | |||||
| Urine | 37.5% (3/8) | 2.0% (1/51) | 0% (0/16) | ||||||
| Total | 0.5% (1/200) | 4.7% (7/150) | 4.7% (9/191) | 11.8% (182/1542) | 36.3% (207/571) | 3.1% (9/289) | 37.5% (3/8) | 2.0% (1/51) | 0% (0/16) |
The number of positive samples and the total number of samples characterized at each skin site or in urine are shown in parentheses. The sites not characterized in each study are left as blank. Data sets in studies S1–S5 and U1–U3 were fully downloaded and analysed, while 100 sequencing files of human skin microbiome in the HMP project were randomly selected and analysed in this study. The amount of sequencing data for each study is shown in Table S1.
Reference Grice et al. (2009) using Sanger sequencing for 16S rRNA clone library.
Reference Meisel et al. (2016) using Illumina sequencing.
Reference van Rensburg et al. (2015) using 454 sequencing.
Reference Oh et al. (2014) using 454 sequencing.
Reference Lehtimäki et al. (2017) using Illumina sequencing.
The HMP project (https://hmpdacc.org/) with the sequencing techniques unavailable.
Reference Siddiqui et al. (2011) using 454 sequencing.
Reference Fouts et al. (2012) using 454 sequencing.
Reference Lewis et al. (2013) using 454 sequencing.
Not all body skin sites were characterized in the six skin microbiome data sets, but there were several skin sites where HF183 seemed to be consistently present in the different studies, such as cubital fossa, volar forearm, palm, retroauricular crease and toe web (Table 1). Comparing to the other skin sites, popliteal fossa, volar forearm and inguinal crease were hot spots for HF183 sequences, which might be from faecal contamination during showering, or defecation. Meanwhile, HF183 was also detected at some unique body sites such as nare and retroauricular creases where there would seem to be less likelihood of contamination from faecal sources. The detection ratios still ranged from 3.1% to 12.2%, indicating that the presence of HF183 at these unique sites was not occasional.
Human faecal marker HF183 was also detected in 2 of 3 human urine microbiomes. The ratios of positive samples ranged from 0% to 37.5%. No HF183 was present in any microbiome of voided urine from asymptomatic adults in study U3. However, HF183 was detected in three of eight healthy adult female urine microbiomes in study U1 and present in a urine sample of one female with neuropathic bladder due to spinal cord injury in study U2, with all urine specimens culturing negative.
As shown in Figure 1, all HF183 containing sequences obtained from nine data sets used in this study were grouped with B. dorei reference 16S rRNA sequences and affiliated with B. dorei, which confirmed the HF183‐associated bacteria as B. dorei. Furthermore, the HF183 containing sequences with nearly full length (using primers 27F and 1492R) were selected to test the coverage of all reverse primers and probes (Table S2). In total, 22 full‐length sequences (longer than 1350 bp) matched HF183, among which 21 sequences (approximately 95.5%) matched reverse primers and probes Bac242R (Seurinck et al., 2005), BacR287 (Green et al., 2014) and BacP234MGB (Green et al., 2014), and all of these 22 sequences matched Bac708 (Bernhard and Field 2000), BFDRev (Haugland et al., 2010) and BFDFAM (Haugland et al., 2010).
DISCUSSION
Although it is possible that the HF183 marker sequence could match sequences that are not related to Bacteroides spp. at all, the HF183 containing sequences found in the microbiome data sets of this study were all affiliated with B. dorei as shown in Figure 1 by aligning with all B. dorei reference 16S rRNA sequences as well as all Bacteroides genus type strain 16S rRNA sequences archived in the RDP database. This confirmed the high specificity in this study of HF183 marker sequence to B. dorei. Multiple HF183 detection assays have been established (Bernhard and Field 2000; Seurinck et al., 2005; Haugland et al., 2010; Green et al., 2014). All of these assays use the same HF183 marker sequence as forward primer, but use different reverse primers and probes with variable specificities either to the whole Bacteroidales family including B. dorei (Bernhard & Field, 2000b; Haugland et al., 2010), or some Bacteroidetes sequences containing HF183 (Green et al., 2014). Thus, these reverse primers and probes are less specific when comparing to the HF183 specific sequence, and the specificities of these HF183 detection assays are determined by the HF183 marker sequence as the forward primer. To validate this, almost full length 16S rRNA gene sequences obtained using 27F and 1492R primers in the human microbiomes of this study were selected and aligned by HF183 sequence and all reverse primers and probes. All reverse primers and probes match almost all 22 sequences containing HF183. Although only one sequence did not match the reverse primers/probes Bac242R (Seurinck et al., 2005), BacR287 and BacP234MGB (Green et al., 2014), this sequence still belonged to B. dorei as confirmed by Figure 1, and matched other reverse primers/probes Bac708 (Bernhard and Field 2000), BFDRev (Haugland et al., 2010) and BFDFAM (Haugland et al., 2010) which were all designed to match the whole Bacteroidales family. The BacR287 primer and BacP234MGB probe in the Green et al. (2014) method were designed based on an alignment of available Bacteroidetes 16S rRNA gene sequences with 100% similarity to the HF183 primer. Thus, it is not surprising that the BacR287 primer and BacP234MGB probe do not match all of the sequences containing HF183 sequence in this study. However, in real applications of MST, the less specific reverse primers and probes could result in more nonspecific amplifications compared to more specific primers and probes such as the BacR287 primer and BacP234MGB probe (Green et al., 2014).
The results of this study indicated that HF183 was widespread on human skin based on the results of the six data sets although these data sets were obtained by different sequencing techniques including Sanger, 454 and Illumina. Compared to Sanger sequencing, 454 and Illumina sequencing can generate much larger amounts of sequence data which clearly increased the chance of finding sequences containing HF183 (Table S2). However, to avoid biasing the findings of this study to certain sequencing techniques, the data sets obtained using different sequencing techniques were retained in this study but not normalized due to the high variance of the sequencing depth. Overall, the results of this study still demonstrated that HF183 was widespread on human skin regardless of sequencing technique. The presence of HF183 was further observed on not only some hot spots but also some unique body sites such as nare and retroauricular creases. While the source of HF183 in these locations is unclear and was not studied here, toilet flushing is known to aerosolize human faecal bacteria (Abney et al., 2021) which could include B. dorei. Human urine within the urinary tract has in general been considered sterile, based on a lack of culturable micro‐organisms present in urine specimens obtained by the clean‐catch method and by catheterization (Fouts et al., 2012; Lewis et al., 2013; Siddiqui et al., 2011). However, advances in sequencing techniques have proven that a diverse spectrum of bacterial profiles is associated with healthy, culture negative human urine (Fouts et al., 2012; Lewis et al., 2013; Siddiqui et al., 2011). The results of this study demonstrated the occasional presence of HF183 in human urine microbiomes. It should be noted that self‐collected urine samples are prone to faecal contamination when proper collection procedures are not followed (Franz & Hörl, 1999), although a sterile sampling protocol was followed based on the sampling description of the studies used here (Fouts et al., 2012; Lewis et al., 2013; Siddiqui et al., 2011). Although B. dorei has been described as strictly anaerobic (Bakir et al., 2006), other obligate anaerobic Bacteroides species such as B. vulgatus have also been found in human skin microbiomes in abundance (Meisel et al., 2016). Considering that B. dorei was widespread on human skin and frequently present in human urine according to the data sets of this study, the human body skin and urinary track might be not only contaminated by human faeces but also be potential niches for B. dorei outside of faeces.
Since the widespread application of HF183 in MST, particularly using qPCR and ddPCR (Steele et al., 2018), human faecal contamination has been documented worldwide in diverse environmental waters (Bradshaw et al., 2016; Diston et al., 2015; Jardé et al., 2018; Mayer et al., 2015; Molina et al., 2014; Sidhu et al., 2013; Staley et al., 2016; Staley et al., 2018). The results of this study suggest possible sources of HF183 in environmental waters could be associated with human skin and urine microbiomes, although HF183 on human skin or in urine might still originate from human faecal contamination. HF183 has been consistently detected in beach waters along California coast (Cao et al., 2017; McQuaig et al., 2012; Russell et al., 2013; Sercu et al., 2009), and public health risks have been estimated based on quantitative microbial risk assessment (QMRA) of HF183 detection levels (Boehm et al., 2015; Brown et al., 2017). Correlations between swimmers and the presence of low levels of HF183 have been reported, suggesting bather shedding as a source of HF183 to recreational waters (Li, Van De Werfhorst, Steets, Ervin, Murray, Blackwell, et al., 2021; Li, van De Werfhorst, Steets, Ervin, Murray, Devarajan, & Holden, 2021; Toubiana et al., 2021). The results of this study may inform such correlations by suggesting that B. dorei in skin or urine microbiomes of bathers might contribute to the observed low levels of HF183 in environmental waters.
CONFLICT OF INTEREST
No conflict of interest declared.
Supporting information
Tables S1
Tables S2‐S3
ACKNOWLEDGEMENTS
This study was funded by the State of California Clean Beach Initiative, using funds from Proposition 84, and from funds provided by Mr. Henry (Sam) Wheeler.
Li, D. , Van De Werfhorst, L.C. & Holden, P.A. (2022) Genetic sequence data evidence that human faecal‐associated HF183 sequences are on human skin and in urine. Journal of Applied Microbiology, 133, 232–240. Available from: 10.1111/jam.15577
REFERENCES
- Abney, S.E. , Bright, K.R. , McKinney, J. , Ijaz, M.K. & Gerba, C.P. (2021) Toilet hygiene‐review and research needs. Journal of Applied Microbiology, 131, 2705–2714. 10.1111/jam.15121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakir, M.A. , Sakamoto, M. , Kitahara, M. , Matsumoto, M. & Benno, Y. (2006) Bacteroides dorei sp. nov., isolated from human faeces. The International Journal of Systematic and Evolutionary Microbiology, 56, 1639–1643. [DOI] [PubMed] [Google Scholar]
- Bernhard, A.E. & Field, K.G. (2000a) Identification of nonpoint sources of fecal pollution in coastal waters by using host‐specific 16S ribosomal DNA genetic markers from fecal anaerobes. Applied and Environmental Microbiology, 66, 1587–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard, A.E. & Field, K.G. (2000b) A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides‐Prevotella genes encoding 16S rRNA. Applied and Environmental Microbiology, 66, 4571–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby, K. , Viau, E. & Peccia, J. (2011) Viral metagenome analysis to guide human pathogen monitoring in environmental samples. Letters in Applied Microbiology, 52, 386–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm, A.B. , Soller, J.A. & Shanks, O.C. (2015) Human‐associated fecal quantitative polymerase chain reaction measurements and simulated risk of gastrointestinal illness in recreational waters contaminated with raw sewage. Environmental Science and Technology Letters, 2, 270–275. [Google Scholar]
- Boehm, A.B. , Van De Werfhorst, L.C. , Griffith, J.F. , Holden, P.A. , Jay, J.A. , Shanks, O.C. et al. (2013) Performance of forty‐one microbial source tracking methods: a twenty‐seven lab evaluation study. Water Research, 47, 6812–6828. [DOI] [PubMed] [Google Scholar]
- Bradshaw, J.K. , Snyder, B.J. , Oladeinde, A. , Spidle, D. , Berrang, M.E. , Meinersmann, R.J. et al. (2016) Characterizing relationships among fecal indicator bacteria, microbial source tracking markers, and associated waterborne pathogen occurrence in stream water and sediments in a mixed land use watershed. Water Research, 101, 498–509. [DOI] [PubMed] [Google Scholar]
- Brown, K.I. , Graham, K.E. , Soller, J.A. & Boehm, A.B. (2017) Estimating the probability of illness due to swimming in recreational water with a mixture of human‐ and gull‐associated microbial source tracking markers. Environmental Science Processes & Impacts, 19, 1528–1541. [DOI] [PubMed] [Google Scholar]
- Cao, Y. , Raith, M.R. , Smith, P.D. , Griffith, J.F. , Weisberg, S.B. , Schriewer, A. et al. (2017) Regional assessment of human fecal contamination in southern California coastal drainages. International Journal of Environmental Research and Public Health, 14, 874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diston, D. , Sinreich, M. , Zimmermann, S. , Baumgartner, A. & Felleisen, R. (2015) Evaluation of molecular‐ and culture‐dependent MST markers to detect fecal contamination and indicate viral presence in good quality groundwater. Environmental Science & Technology, 49, 7142–7151. [DOI] [PubMed] [Google Scholar]
- Escobedo‐Hinojosa, W. and Pardo‐López, L. (2017) Analysis of bacterial metagenomes from the southwestern Gulf of Mexico for pathogens detection. Pathogens and Disease 75, ftx058. [DOI] [PubMed] [Google Scholar]
- Feng, S. & McLellan, S.L. (2019) Highly specific sewage‐derived Bacteroides quantitative PCR assays target sewage‐polluted waters. Applied and Environmental Microbiology, 85, e02696–e02618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, C.M. , Coote, B.G. , Ashbolt, N.J. & Stevenson, I.M. (1996) Relationships between indicators, pathogens and water quality in an estuarine system. Water Research, 30, 2045–2054. [Google Scholar]
- Field, K.G. & Samadpour, M. (2007) Fecal source tracking, the indicator paradigm, and managing water quality. Water Research, 41, 3517–3538. [DOI] [PubMed] [Google Scholar]
- Fouts, D.E. , Pieper, R. , Szpakowski, S. , Pohl, H. , Knoblach, S. , Suh, M.J. et al. (2012) Integrated next‐generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. Journal of Translational Medicine, 10, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz, M. & Hörl, W.H. (1999) Common errors in diagnosis and management of urinary tract infection. I: pathophysiology and diagnostic techniques. Nephrology, Dialysis, Transplantation, 14, 2746–2753. [DOI] [PubMed] [Google Scholar]
- Green, H.C. , Haugland, R.A. , Varma, M. , Millen, H.T. , Borchardt, M.A. , Field, K.G. et al. (2014) Improved HF183 quantitative real‐time PCR assay for characterization of human fecal pollution in ambient surface water samples. Applied and Environmental Microbiology, 80, 3086–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice, E.A. , Kong, H.H. , Conlan, S. , Deming, C.B. , Davis, J. , Young, A.C. et al. (2009) Topographical and temporal diversity of the human skin microbiome. Science, 324, 1190–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood, V.J. , Staley, C. , Badgley, B.D. , Borges, K. & Korajkic, A. (2014) Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiology Reviews, 38, 1–40. [DOI] [PubMed] [Google Scholar]
- Haugland, R.A. , Varma, M. , Kelty, C.A. , Peed, L. , Sivaganesan, M. & Shanks, O.C. (2010) Evaluation of genetic markers from the 16S rRNA gene V2 region for use in quantitative detection of selected Bacteroidales species and human fecal waste by real‐time PCR. Systematic and Applied Microbiology, 33, 348–357. [DOI] [PubMed] [Google Scholar]
- Hughes, B. , Beale, D.J. , Dennis, P.G. , Cook, S. & Ahmed, W. (2017) Cross‐comparison of human wastewater‐associated molecular markers in relation to fecal indicator bacteria and enteric viruses in recreational beach waters. Applied and Environmental Microbiology, 83, e00028–e00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardé, E. , Jeanneau, L. , Harrault, L. , Quenot, E. , Solecki, O. , Petitjean, P. et al. (2018) Application of a microbial source tracking based on bacterial and chemical markers in headwater and coastal catchments. Science of the Total Environment, 610–611, 55–63. [DOI] [PubMed] [Google Scholar]
- Katoh, K. , Misawa, K. , Kuma, K. & Miyata, T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research, 30, 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirs, M. , Caffaro‐Filho, R.A. , Wong, M. , Harwood, V.J. , Moravcik, P. & Fujioka, R.S. (2016) Human‐associated Bacteroides spp. and human polyomaviruses as microbial source tracking markers in Hawaii. Applied and Environmental Microbiology, 82, 6757–6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. , Li, M. , Knyaz, C. & Tamura, K. (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35, 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtimäki, J. , Karkman, A. , Laatikainen, T. , Paalanen, L. , von Hertzen, L. , Haahtela, T. et al. (2017) Patterns in the skin microbiota differ in children and teenagers between rural and urban environments. Scientific Reports, 7, 45651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, D.A. , Brown, R. , Williams, J. , White, P. , Jacobson, S.K. , Marchesi, J.R. et al. (2013) The human urinary microbiome, bacterial DNA in voided urine of asymptomatic adults. Frontiers in Cellular and Infection Microbiology, 3, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, D. , Van De Werfhorst, L.C. , Steets, B. , Ervin, J. , Murray, J.L.S. , Blackwell, A. et al. (2021) Sources of low level human fecal markers in recreational waters of two Santa Barbara, CA beaches: roles of WWTP outfalls and swimmers. Water Research, 202, 117378. [DOI] [PubMed] [Google Scholar]
- Li, D. , van De Werfhorst, L.C. , Steets, B. , Ervin, J. , Murray, J.L.S. , Devarajan, N. et al. (2021) Bather shedding as a source of human fecal markers to a recreational beach. Frontiers in Microbiology, 12, 673,190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, R.E. , Vierheilig, J. , Egle, L. , Reischer, G.H. , Saracevic, E. , Mach, R.L. et al. (2015) Automated sampling procedures supported by high persistence of bacterial fecal indicators and Bacteroidetes genetic microbial source tracking markers in municipal wastewater during short‐term storage at 5°C. Applied and Environmental Microbiology, 81, 5134–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuaig, S. , Griffith, J. & Harwood, V.J. (2012) Association of fecal indicator bacteria with human viruses and microbial source tracking markers at coastal beaches impacted by nonpoint source pollution. Applied and Environmental Microbiology, 78, 6423–6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meays, C.L. , Broersma, K. , Nordin, R. & Mazumder, A. (2004) Source tracking fecal bacteria in water: a critical review of current methods. Journal of Environmental Management, 73, 71–79. [DOI] [PubMed] [Google Scholar]
- Meisel, J.S. , Hannigan, G.D. , Tyldsley, A.S. , SanMiguel, A.J. , Hodkinson, B.P. , Zheng, Q. et al. (2016) Skin microbiome surveys are strongly influenced by experimental design. The Journal of Investigative Dermatology, 136, 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina, M. , Hunter, S. , Cyterski, M. , Peed, L.A. , Kelty, C.A. , Sivaganesan, M. et al. (2014) Factors affecting the presence of human‐associated and fecal indicator real‐time quantitative PCR genetic markers in urban‐impacted recreational beaches. Water Research, 64, 196–208. [DOI] [PubMed] [Google Scholar]
- Oh, J. , Byrd, A.L. , Deming, C. , Conlan, S. , NISC Comparative Sequencing Program , Kong, H.H. et al. (2014) Biogeography and individuality shape function in the human skin metagenome. Nature, 514, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischer, G.H. , Ebdon, J.E. , Bauer, J.M. , Schuster, N. , Ahmed, W. , Aström, J. et al. (2013) Performance characteristics of qPCR assays targeting human‐ and ruminant‐associated bacteroidetes for microbial source tracking across sixteen countries on six continents. Environmental Science & Technology, 47, 8548–8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roslev, P. & Bukh, A.S. (2011) State of the art molecular markers for fecal pollution source tracking in water. Applied Microbiology and Biotechnology, 89, 1341–1355. [DOI] [PubMed] [Google Scholar]
- Russell, T.L. , Sassoubre, L.M. , Wang, D. , Masuda, S. , Chen, H. , Soetjipto, C. et al. (2013) A coupled modeling and molecular biology approach to microbial source tracking at Cowell Beach, Santa Cruz, CA, United States. Environmental Science & Technology, 47, 10,231–10,239. [DOI] [PubMed] [Google Scholar]
- Scott, T.M. , Rose, J.B. , Jenkins, T.M. , Farrah, S.R. & Lukasik, J. (2002) Microbial source tracking: current methodology and future directions. Applied and Environmental Microbiology, 68, 5796–5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sercu, B. , Van De Werfhorst, L.C. , Murray, J. & Holden, P.A. (2009) Storm drains are sources of human fecal pollution during dry weather in three urban southern California watersheds. Environmental Science & Technology, 43, 293–298. [DOI] [PubMed] [Google Scholar]
- Seurinck, S. , Defoirdt, T. , Verstraete, W. & Siciliano, S.D. (2005) Detection and quantification of the human‐specific HF183 Bacteroides 16S rRNA genetic marker with real‐time PCR for assessment of human faecal pollution in freshwater. Environmental Microbiology, 7, 249–259. [DOI] [PubMed] [Google Scholar]
- Siddiqui, H. , Nederbragt, A.J. , Lagesen, K. , Jeansson, S.L. & Jakobsen, K.S. (2011) Assessing diversity of the female urine microbiota by high throughput sequencing of 16S rDNA amplicons. BMC Microbiology, 11, 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu, J.P. , Ahmed, W. , Gernjak, W. , Aryal, R. , McCarthy, D. , Palmer, A. et al. (2013) Sewage pollution in urban stormwater runoff as evident from the widespread presence of multiple microbial and chemical source tracking markers. Sci Total Environ, 463–464, 488–496. [DOI] [PubMed] [Google Scholar]
- Staley, Z.R. , Boyd, R.J. , Shum, P. & Edge, T.A. (2018) Microbial source tracking using quantitative and digital PCR to identify sources of fecal contamination in stormwater, river water, and beach water in a Great Lakes area of concern. Applied and Environmental Microbiology, 84, e01634–e01618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley, Z.R. , Grabuski, J. , Sverko, E. & Edge, T.A. (2016) Comparison of microbial and chemical source tracking markers to identify fecal contamination sources in the Humber River (Toronto, Ontario, Canada) and associated storm water outfalls. Applied and Environmental Microbiology, 82, 6357–6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele, J.A. , Blackwood, A.D. , Griffith, J.F. , Noble, R.T. & Schiff, K.C. (2018) Quantification of pathogens and markers of fecal contamination during storm events along popular surfing beaches in San Diego, California. Water Research, 136, 137–149. [DOI] [PubMed] [Google Scholar]
- Stoeckel, D.M. & Harwood, V.J. (2007) Performance, design, and analysis in microbial source tracking studies. Applied and Environmental Microbiology, 73, 2405–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toubiana, M. , Salles, C. , Tournoud, M.G. , Licznar‐Fajardo, P. , Zorgniotti, I. , Trémélo, M.L. et al. (2021) Monitoring urban beach quality on a summer day: determination of the origin of fecal indicator bacteria and antimicrobial resistance at Prophète Beach, Marseille (France). Frontiers in Microbiology, 12, 710346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh, P.J. , Ley, R.E. , Hamady, M. , Fraser‐Liggett, C. , Knight, R. & Gordon, J.I. (2007) The human microbiome project: exploring the microbial part of ourselves in a changing world. Nature, 449, 804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rensburg, J.J. , Lin, H. , Gao, X. , Toh, E. , Fortney, K.R. , Ellinger, S. et al. (2015) The human skin microbiome associates with the outcome of and is influenced by bacterial infection. MBio, 6, e01315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1
Tables S2‐S3


