Abstract
Renal stones are a common urological disease with high prevalence and recurrence rates. Characterizing gut microbiome profiles of first‐onset renal calculi patients, both before and after surgery, may provide valuable insights and identify potential biomarkers for the disease. In this study, we explored the associations between the gut microbiome and renal stone formation using 16S ribosomal RNA (rRNA) gene sequencing. In brief, 20 patients were recruited, and information on health and eating habits within the previous 1–3 months was collected upon admission. A total of 493 operational taxonomic units (OTUs) were detected in 40 specimens, with an average of 67,888 ± 827 reads per sample. The results of OTU‐based partial least squares discriminant analysis (PLS‐DA) analysis showed differences between RS1 (fecal specimen before surgery) and RS2 (one month later after surgery) groups, with a significantly higher level of OTU7 in the RS2 group. Taxonomy‑based comparisons of the gut microbiome showed differences in the flora composition, with the prevalence of Enterobacteriales, Enterobacteriaceae, Gammaproteobacteria and Escherichia being higher in the RS2 group and the prevalence of Pseudomonadaceae, Pseudomonadales and Pseudomonas being higher in the RS1 group. Correlation analysis showed that an increased prevalence of Enterobacteriaceae, Gammaproteobacteria and Escherichia associated with a decreased level of urea, and a decreased creatinine level was correlated with an increased prevalence of Escherichia. These data strongly suggest that the gut microbiome plays an important role in kidney stone formation, and these findings may provide new insights for the prevention, diagnosis, and treatment of renal stones.
Keywords: 16s rRNA, gut microbiota, kidney stones
Significance and Impact of Study: 1. Faecal samples before surgery (RS1) and one month after surgery (RS2) showed difference in flora composition, with the prevalence of Enterobacteriales, Enterobacteriaceae, Gammaproteobacteria and Escherichia being higher in the RS2 group and the prevalence of Pseudomonadaceae, Pseudomonadales and Pseudomonas being higher in the RS1 group. 2. The increased prevalence of Enterobacteriaceae, Gammaproteobacteria and Escherichia were associated with a decreased level of urea, and a decreased creatinine level was correlated with an increased prevalence of Escherichia.

Background
Renal stones (RS) are a common condition affecting approximately 5 to 8·8% of the global population. A recent epidemiological survey showed that the prevalence of kidney stones was 6·4% in China (Zeng et al. 2017). Approximately 75% of kidney stones are primarily comprised of calcium oxalate, and urinary oxalate is considered a risk factor for the disease. Twin studies have revealed a 56% heritability risk for stones, whereas the other implicated factors are poor diet, lack of exercise, work environment and geographical region (Mehta et al. 2016).
In recent years, micro‐organisms have been reported to play a role in the pathogenesis and the prevention of kidney stones, and the involvement of the intestinal microbiome in renal disease has been an area of interest. The role of the intestinal microbiome in influencing the composition of the urine has been explored, which generated data suggesting that it affects the incidence of kidney stone formation (Mehta et al. 2016). Stern and his collaborator identified distinct differences in the gut microbiome of kidney stone patients compared to control subjects (Stern et al. 2016). A study conducted by Tang et al. (2018) using 16S rRNA gene sequencing revealed an altered composition of gut microbiota in individuals with kidney stones, and the gut microbial composition and diversity in patients with multiple kidney stones were different from those in healthy subjects. Suryavanshi et al. reported the dysbiosis of gut microbial communities in recurrent oxalate kidney stone sufferers using high throughput DNA sequencing and rationalized the first in‐depth surveillance of oxalate metabolizing bacterial species in the human gut and their association with hyperoxaluria (Suryavanshi et al. 2016). These data strongly suggest that the gut microbiome affects kidney stone formation.
However, most of these studies were focusing on the differences of gut microbiome between the health controls and RS patients; little literature was reported based on changes of gut microbiome on the RS patients, which may reduce the individual differences. In the present study, we performed 16S rRNA gene sequencing to assess differences between the composition and the abundance of the gut microbiome in first‐onset renal calculi patients, both before and after surgery.
Results and Discussions
Richness and diversity analysis
As a huge bacterial network, the interaction of gut microbes is complex. Intestinal microbes may play a role in the pathogenesis and the prevention of kidney stones. The gut‐kidney axis was first proposed by Meijers and Evenepoel (2011) based on the findings that abnormal intestinal metabolites were identified in patients with chronic renal diseases (Wu et al. 2011). This finding shows a close physiological and pathological relationship between the kidney and the intestine, and reveals the important role of intestinal flora dysbiosis in the progression of chronic renal disease.
In this study, we compared the gut microbiome profiles of patients with renal stones, both before and after the removal of the stones. In total, 2 715 523 usable raw reads were obtained from 40 specimens, with an average of 67 888 ± 827 reads per sample. The total number of operational taxonomic units (OTUs) at 97% similarity level was 493. The alpha diversity indices, including observed species, ACE index, Simpson’s diversity index and Good’s coverage index, of the gut microbiota of the RS1 group (before surgery) were all lower than those of the RS2 group (one month after surgery), but there was no statistical significance of the gut microbiota between RS1 and RS2 groups (Fig. S1).
The two groups shared 415 OTUs, with 40 and 38 OTUs unique to the RS1 group and RS2 group respectively (Fig. 1a). The result of data analysis showed that OTU7, the taxonomy of which was constituted by Bacteria, Proteobacteria, Gammaproteobacteria, Enterobacteriales, Enterobacteriaceae, Escherichia and Escherichia_coli, was significantly higher in the RS2 group than in the RS1 group (Fig. 1b). The results of OTU‐based PLS‐DA analysis showed differences between the two groups (Fig. 1c), RS1 group and RS2 group samples are separated in the direction of the horizontal axis. The OTU accumulation curves using the R package showed the sufficiency of sample quantity and species richness (Fig. 1d).
Figure 1.
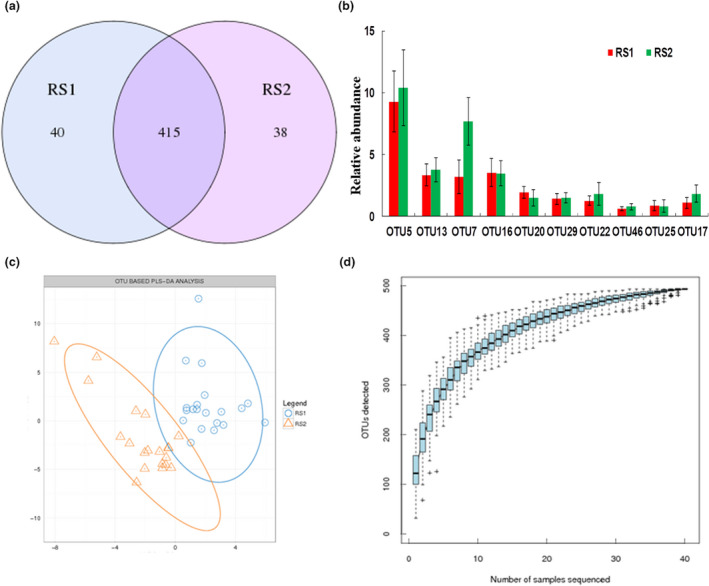
OTU clustering and analysis. (a) OTU Venn diagram, blue circle indicated the number of OTUs unique to RS1, purple circle indicated the number of OTUs unique to RS2, the overlap part showed the number of OTUs shared by the two groups. (b) OTU abundance analysis; Data were expressed by Mean ± SEM. One‐way ANOVA test showed OTU7 was significantly different between the two groups (P < 0·05). (c) OUT‐based PLS‐DA analysis; RS1 group and RS2 group samples are separated in the direction of the horizontal axis. (d) species accumulation curves, reflects the impact of the numbers of samples sequenced on species diversity.
Taxonomy‑based comparisons of gut microbiota
After comparing with the database, OTU species were classified. Taxonomy‐based comparisons of gut microbiota in the two groups showed differences in the abundance of OTU species. The results of species richness analysis showed that there were differences in class (Fig. 2a), genus (Fig. 2b), family (Fig. 2c), phylum (Fig. 2d), order (Fig. 3e) and species (Fig. 2f).
Figure 2.
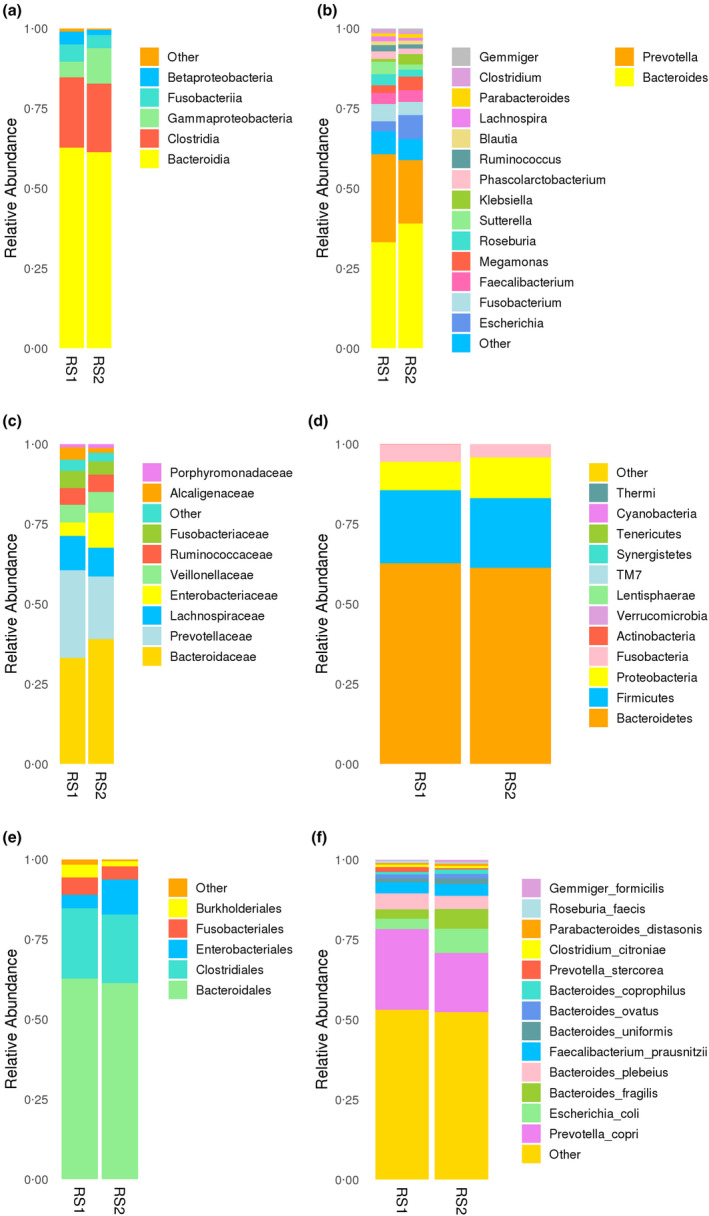
OTU species abundance analysis. All species with an abundance below 0·5% were merged into others at other levels. (a) class; (b) genus; (c) family; (d) phylum; (e) order; (f) species.
Figure 3.
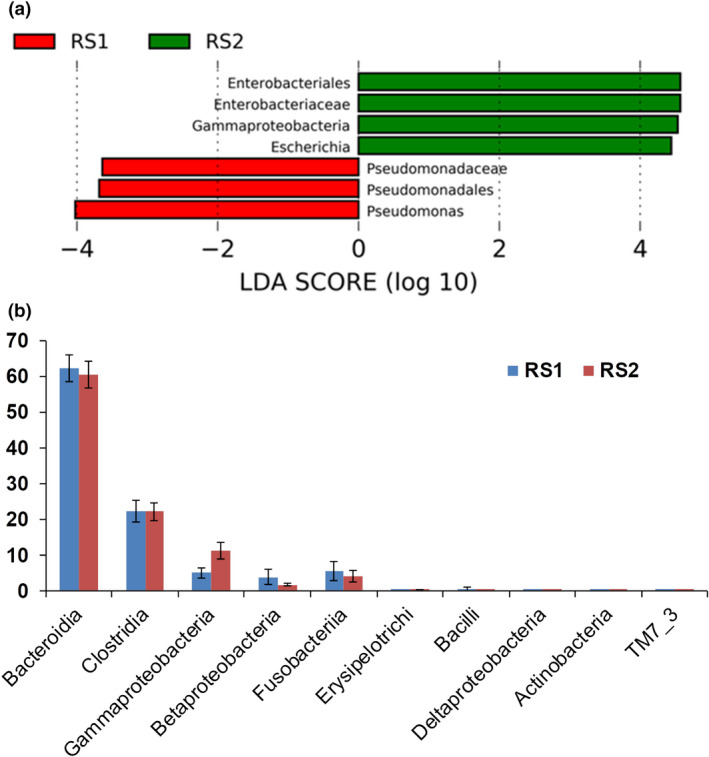
Species difference analysis. (a) LDA diagram of LEfSe analysis; the absolute value of LDA was set at 2·0. The red histogram represents RS1 group, and green histogram represents RS2 group. The length of the histogram represents LDA score, which is the significant difference between different groups. (b) Comparison of key class differences. Top ten classes with an average relative abundance of each group and the significance of difference was displayed. Data were expressed by Mean ± SEM. One‐way ANOVA test showed Grammaproteobcateria was significantly different between the two groups (P < 0·05).
According to the analysis, the prevalence of Enterobacteriales, Enterobacteriaceae, Gammaproteobacteria (Fig. 3b) and Escherichia was higher in the RS2 group, whereas the prevalence of Pseudomonadaceae, Pseudomonadales and Pseudomonas was higher in the RS1 group (Fig. 3a).
Gammaproteobacteria is closely correlated with the expression of antibiotic resistance genes and also directly associated with host locomotion and oxidative stress levels (Zhang et al. 2021). Renal stones caused by Escherichia coli infection have been reported in numerous studies (Amimanan et al. 2017; Tavichakorntrakool et al. 2017; Chen et al. 2019; An et al. 2021), and E. coli is one of the most commonly identified organisms in stone cultures (Parkhomenko et al. 2017). E. coli is associated with kidney stone disease, either as a cause or an effect (secondary or recurrent urinary tract infection, UTI) (Tavichakorntrakool et al. 2017). E. coli is the most common bacterium isolated from urine and stone matrices of calcium oxalate (CaOx) from individuals with kidney stone (Amimanan et al. 2017). Recent evidence has suggested that E. coli is not only involved in recurrent UTI following kidney stone disease, but it is also a microbe that can cause kidney stone formation (Tavichakorntrakool et al. 2012). Interestingly, previous in vitro and in vivo studies have demonstrated that E. coli can promote CaOx crystal growth and aggregation (Chutipongtanate et al. 2013; Barr‐Beare et al. 2015), both of which are critical for kidney stone formation.
A study by Lobel et al. illustrated a clear and robust signal of Enterobacteriaceae enrichment in patients with chronic kidney disease (Lobel et al. 2020), and Enterobacteriaceae was reported to cause urinary tract infections (Milliner et al. 2018; Lee and Yeo 2020; Simoes et al. 2020). Pseudomonas is a common, Gram‐negative bacterium. It can be a significant pathogenic factor of severe infections in humans (Mielko et al. 2019), and end‐stage kidney disease patients undergoing peritoneal dialysis therapy presented lower species richness, with dominance by Pseudomonadaceae and Prevotelaceae families (Simoes‐Silva et al. 2020). The results of correlation analysis found strong associations of the clinical features with the microbiome.
Function analysis and correlation analysis
The prediction of the function showed enrichment in (1) amino acid degradation; (2) nucleoside and nucleotide biosynthesis; (3) cofactor, prosthetic group, electron carrier, and vitamin biosynthesis; (4) fatty acid and lipid biosynthesis; and (5) carbohydrate biosynthesis (Fig. 4a). Functional difference analysis showed the RS1 and RS2 groups shared significant differences in amino acid biosynthesis, alcohol degradation, aldehyde degradation, and fatty acid and lipid degradation (Fig. 4b).
Figure 4.
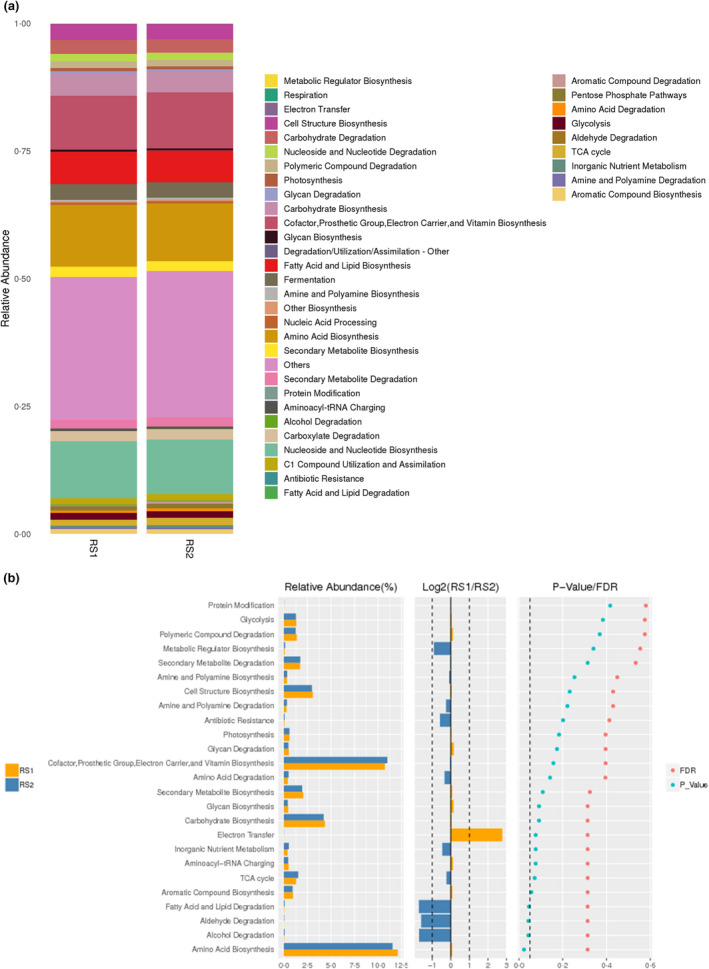
The prediction of function. (a) MetaCyc classification; the Metacyc pathway in bacterial community were obtained by PICRUST 2. (b) Functional difference analysis. The left histogram showed relative abundance of species in each pathway, the middle represents the log2 value of the average relative abundance ratio of the same species in the two groups, and the right figure shows the P value and FDR values obtained by Wilcox test.
The clinical data of the patients before surgery and one month after surgery are listed in Tables S1–3. A routine urine test showed more epithelial cells, small round cells, cast and small red blood cells in the RS2 group (P < 0·05), whereas the RS1 group had higher levels of creatinine (P = 0·009), urea (P = 0·008), total bile acid (P = 0·019) and calcium (P = 0·027), but lower HCO3 (P = 0·045) in electrolyte, liver function and kidney function tests. In the routine blood test, the RS1 group showed more had higher white blood cells, neutrophils, as well as a higher neutrophils percentage (P < 0·05), whereas the RS2 group had more lymphocytes and a higher of lymphocyte percentage (P < 0·05). We combined the clinical data and 16S RNA sequencing results, and the correlation network is shown in Fig. 5. The correlations between microbes and clinical features were based on Pearson correlation analysis. Correlations with R‐value greater than 0·4 and the P value less than 0·05 were shown in the figure.
Figure 5.
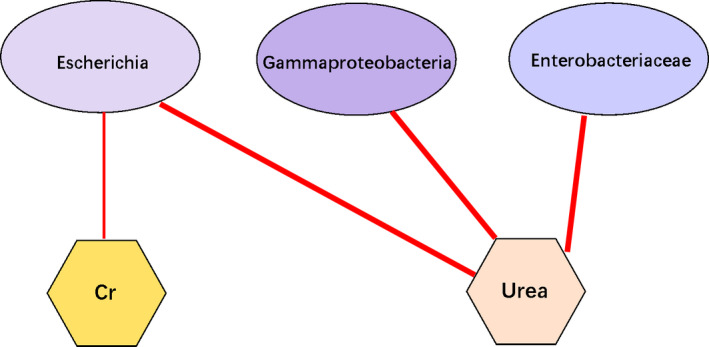
Clinical‐microbiome correlation network. Cr, creatinine. Red line, negative correlations. Bold line, P < 0·01; fine line, P < 0·05.
The results of correlation analysis showed that an increased prevalence of Enterobacteriaceae, Gammaproteobacteria and Escherichia associated with a decreased level of urea (RS1: 4·611 ± 1·043 mmol l−1 vs RS2: 3·950 ± 1·045 mmol l−1), and a decreased creatinine level was correlated with an increased prevalence of Escherichia. Aberrant gut microbiota alters host metabolome and impacts renal failure in humans and rodents (Wang et al. 2020). Intestinal barrier is often impaired in patient with chronic kidney disease, promoting the penetrance of toxins into the circulation system (Vaziri et al. 2016), aggravating the development of kidney disease and its complications.
Rat studies have demonstrated that O. formigenes stimulates oxalate secretion from the plasma to the gut (Hatch et al. 2006; Arvans et al. 2017), which may attenuate urinary oxalate excretion and limit kidney stone formation risk. Lifestyle and diet changes are the key approach for the prevention of kidney stone recurrences (D'Alessandro et al. 2019). Differences in human population, inclusion criteria, and detection methods may lead to differences in results. It is noteworthy that several risk factors for kidney stone formation are modifiable and related to lifestyle and dietary habits (Ferraro et al. 2017). Dietary intervention (Oliveira et al. 2014) aims to correct urinary abnormalities known to induce lithogenesis, but also to prevent weight gain, hypertension, diabetes, or obesity. Based on the patients’ descriptions, the medical and health questionnaire in our study found that most of the patients with renal stones showed no significant differences in alcohol and tobacco use, defecation, exercise and eating habits, and mood and sleep states.
An increasing body mass index (BMI) was associated with an increasing urine sodium level and a decreasing pH in men and increasing urine uric acid and sodium levels, and a decreasing urine citrate level in women (Shavit et al. 2015). Similar to obese individuals with kidney stones, overweight individuals with kidney stone show alterations in metabolic urinary profiles that are associated with an increased overall risk of stone formation (Shavit et al. 2015). An analysis of our unpublished data shows that the BMI values of kidney stone formers (27·59 ± 1·08, n = 24) were higher than those of normal controls (23·54 ± 0·94, n = 16; P = 0·018).
It was proposed that a precise diagnosis of the kidney stone composition and the urinary risk factors is the basis for successful treatment and prevention, and this should be done for all kidney stone patients. The bacteria with a lower content may be more susceptible to the environment and diet. The urinary excretion of oxalate in patients with idiopathic calcium‐oxalate urolithiasis can be greatly reduced with treatment using a high concentration of freeze‐dried lactic acid bacteria (Campieri et al. 2001).
However, this study had several limitations. First, due to the population mobility and COVID19 epidemic situation, the number of the patients enrolled was limited. Second, matched healthy controls were missed; family member or colleagues may be a good choice. Third, the biological functions of some related species should be explored and verified in renal stone rat models.
In this study, we performed 16S rRNA gene sequencing to assess differences between the composition and the abundance of the gut microbiome in 20 first‐onset renal calculi patients, both before and after surgery. Taxonomy‑based comparisons of the gut microbiome showed differences in the flora composition, with the prevalence of Enterobacteriales, Enterobacteriaceae, Gammaproteobacteria and Escherichia being higher in the RS2 group and the prevalence of Pseudomonadaceae, Pseudomonadales and Pseudomonas being higher in the RS1 group. The increased prevalence of Enterobacteriaceae, Gammaproteobacteria and Escherichia were associated with a decreased level of urea, and a decreased creatinine level was correlated with an increased prevalence of Escherichia. These data strongly suggest that the gut microbiome plays an important role in kidney stone formation, and these findings may provide new insights for the prevention, diagnosis, and treatment of renal stones.
Materials and Methods
Sampling
The study participants were local residents of Shenzhen city for more than 1 year. The 20 male patients with nephrolithiasis, aged 26–48 years (36·45 ± 6·50 years, mean ± STD), were from The People’s Hospital of Longhua District (Shenzhen, China). All patients were first diagnosed with positive calculi by X‐ray plain film radiography, intravenous pyelography or CT. All participants had no history of surgery or extracorporeal shock wave lithotripsy. No patient was on any medication before sample collection. All participants provided written consent for inclusion before participation, and the study was reviewed and approved by the Ethics Committee of The People’s Hospital of Longhua, Shenzhen (No. KY2019021). The information on health and eating habits within the previous 1–3 months was collected upon admission. The first faecal specimen was collected from each patient before surgery, and the second one was collected one month later when the patients returned to the hospital for the removal of the double J stent. Faecal samples were frozen immediately after sampling and stored at −80°C.
DNA extraction and 16S rRNA gene amplicon sequencing
Genomic DNA was extracted from each fecal sample using the QIAamp Fast DNA Stool Mini Kit (QIAGEN, Germany). The amount of DNA was determined, and the integrity and size of DNA were assessed by 1·0% (W/V) agarose gel electrophoresis. All DNA samples were stored at −20°C before further processing. The 16S rRNA gene was amplified, and PCR amplicons were analysed by gel electrophoresis and purified using the QIAquick PCR Purification Kit (QIAGEN). The amplicon libraries were constructed using the TruSeq DNA PCR‐Free Sample Preparation Kit (Illumina, USA), and library sequencing was performed on the Illumina HiSeq 2500 platform (Illumina).
Bioinformatics analysis
Raw reads were filtered to remove adaptors and low‐quality and ambiguous bases, and paired‐end reads were added to tags by the Fast Length Adjustment of Short Reads Program (FLASH, v1.2.11) (Magoc and Salzberg 2011) (http://www.cbcb.umd.edu/software/flash). The tags were clustered into OTUs with a cutoff value of 97% using UPARSE Software (v7.0.1090) (Edgar 2013) (http://drive5.com/uparse/), and chimeric sequences were compared with the Gold Database using UCHIME (v4.2.40)(Edgar et al. 2011) (http://drive5.com/uchime). Subsequently, OTU representative sequences were taxonomically classified using the Ribosomal Database Project Classifier (v.2.2) with a minimum confidence threshold of 0·6 and trained on the Greengenes Database (v201305) by QIIME (v1.8.0) (Caporaso et al. 2010) (http://qiime.sourceforge.net/). The USEARCH Global Program (Edgar 2010) (http://www.drive5.com/usearch) was used to compare all tags back to OTUs to generate the OTU abundance statistics table for each sample.
Alpha and beta diversity were estimated by MOTHUR (v1.31.2)(Schloss et al. 2009) (https://www.mothur.org) and QIIME (Caporaso et al. 2010) at the OTU level respectively. Sample clustering was conducted by QIIME (Caporaso et al. 2010) based on UPGMA. KEGG and COG functions were predicted using PICRUSt Software (Wilkinson et al. 2018) (http://picrust.github.io/picrust/). Bar plots and heatmaps of different classification levels were plotted with R package (v3.4.1) and R package “gplots” respectively.
Statistical analysis
Statistical analysis was performed using Sigmaplot 16.0 Software (Systat Software, Inc.). Statistical significance was evaluated by one‐way ANOVA followed by Fisher's protected least‐significant difference post hoc test, unless otherwise specified. P < 0·05 was considered a statistically significant difference.
Author Contributions
All of the listed authors participated in conducting the experiments, data analysis, and manuscript preparing. Q.D. received grants, and H.L. reviewed the manuscript.
Funding information
This study was supported by research grants from Natural Science Foundation (31800984), The Science and Technology Planning Project of Guangdong Province (2018A0303130337, B2022065), Shenzhen Fundamental Research Project (JCYJ20180228164047023, JCYJ20190808095407464) and Longhua Science and Technology Planning Project (11501A20190513BA7B6B, 2020006, 2020026).
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethics approval
The study was approved by the ethics committee of the People’s Hospital of Longhua, Shenzhen (Approval no. KY2019021).
Consent to participate
Written consents were obtained from the patients.
Consent for publication
Written consents were obtained from the patients.
Supporting information
Figure S1. Alpha diversity box chart. The differences between the two groups were determined by Wilcox Test. (a) Observed species; (b) Chao; (c) Ace; (d) Shannon; (e) Simpson; (f) Coverage.
Table S1. Urine routine.
Table S2. Electrolyte, liver function and kidney function.
Table S3. Routine blood test.
Acknowledgement
The authors thank Zi Liu and Qiu’e Shao from The People’s Hospital of Longhua for sampling, investigation on health and eating habits and secretarial assistance. The authors also thank Wei Zhao, Dabin Zhong, Xiaoshuang Dai, PhD, and Bingfeng Leng, PhD (BGI Genomics, Shenzhen) for the quality control of the clinical samples and data analysis.
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
References
- Amimanan, P. , Tavichakorntrakool, R. , Fong‐ngern, K. , Sribenjalux, P. , Lulitanond, A. , Prasongwatana, V. , Wongkham, C. , Boonsiri, P. et al. (2017) Elongation factor Tu on Escherichia coli isolated from urine of kidney stone patients promotes calcium oxalate crystal growth and aggregation. Sci Rep 7, 2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, L. , Wu, W. , Li, S. , Lai, Y. , Chen, D. , He, Z. , Chang, Z. , Xu, P. et al. (2021) Escherichia coli aggravates calcium oxalate stone formation via PPK1/Flagellin‐mediated renal oxidative injury and inflammation. Oxid Med Cell Longev 2021, 9949697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvans, D. , Jung, Y.C. , Antonopoulos, D. , Koval, J. , Granja, I. , Bashir, M. , Karrar, E. , Roy‐Chowdhury, J. et al. (2017) Oxalobacter formigenes‐derived bioactive factors stimulate oxalate transport by intestinal epithelial cells. J Am Soc Nephrol 28, 876–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr‐Beare, E. , Saxena, V. , Hilt, E.E. , Thomas‐White, K. , Schober, M. , Li, B. , Becknell, B. , Hains, D.S. et al. (2015) The interaction between Enterobacteriaceae and calcium oxalate deposits. PLoS One 10, e0139575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campieri, C. , Campieri, M. , Bertuzzi, V. , Swennen, E. , Matteuzzi, D. , Stefoni, S. , Pirovano, F. , Centi, C. et al. (2001) Reduction of oxaluria after an oral course of lactic acid bacteria at high concentration. Kidney Int 60, 1097–1105. [DOI] [PubMed] [Google Scholar]
- Caporaso, J.G. , Kuczynski, J. , Stombaugh, J. , Bittinger, K. , Bushman, F.D. , Costello, E.K. , Fierer, N. , Peña, A.G. et al. (2010) QIIME allows analysis of high‐throughput community sequencing data. Nat Methods 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D. , Jiang, C. , Liang, X. , Zhong, F. , Huang, J. , Lin, Y. , Zhao, Z. , Duan, X. et al. (2019) Early and rapid prediction of postoperative infections following percutaneous nephrolithotomy in patients with complex kidney stones. BJU Int 123, 1041–1047. [DOI] [PubMed] [Google Scholar]
- Chutipongtanate, S. , Sutthimethakorn, S. , Chiangjong, W. and Thongboonkerd, V. (2013) Bacteria can promote calcium oxalate crystal growth and aggregation. J Biol Inorg Chem 18, 299–308. [DOI] [PubMed] [Google Scholar]
- D'Alessandro, C. , Ferraro, P.M. , Cianchi, C. , Barsotti, M. , Gambaro, G. and Cupisti, A. (2019) Which diet for calcium stone patients: a real‐world approach to preventive care. Nutrients 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R.C. (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. [DOI] [PubMed] [Google Scholar]
- Edgar, R.C. (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10, 996–998. [DOI] [PubMed] [Google Scholar]
- Edgar, R.C. , Haas, B.J. , Clemente, J.C. , Quince, C. and Knight, R. (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro, P.M. , Taylor, E.N. , Gambaro, G. and Curhan, G.C. (2017) Dietary and lifestyle risk factors associated with incident kidney stones in men and women. J Urol 198, 858–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch, M. , Cornelius, J. , Allison, M. , Sidhu, H. , Peck, A. and Freel, R.W. (2006) Oxalobacter sp. reduces urinary oxalate excretion by promoting enteric oxalate secretion. Kidney Int 69, 691–698. [DOI] [PubMed] [Google Scholar]
- Lee, Y.R. and Yeo, S. (2020) Cefiderocol, a new siderophore cephalosporin for the treatment of complicated urinary tract infections caused by multidrug‐resistant pathogens: preclinical and clinical pharmacokinetics, pharmacodynamics, efficacy and safety. Clin Drug Investig 40, 901–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobel, L. , Cao, Y.G. , Fenn, K. , Glickman, J.N. and Garrett, W.S. (2020) Diet posttranslationally modifies the mouse gut microbial proteome to modulate renal function. Science 369, 1518–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc, T. and Salzberg, S.L. (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, M. , Goldfarb, D.S. and Nazzal, L. (2016) The role of the microbiome in kidney stone formation. Int J Surg 36(Pt D), 607–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijers, B.K. and Evenepoel, P. (2011) The gut‐kidney axis: indoxyl sulfate, p‐cresyl sulfate and CKD progression. Nephrol Dial Transplant 26, 759–761. [DOI] [PubMed] [Google Scholar]
- Mielko, K.A. , Jablonski, S.J. , Milczewska, J. , Sands, D. , Lukaszewicz, M. and Mlynarz, P. (2019) Metabolomic studies of Pseudomonas aeruginosa . World J Microbiol Biotechnol 35, 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milliner, D. , Hoppe, B. and Groothoff, J. (2018) A randomised Phase II/III study to evaluate the efficacy and safety of orally administered Oxalobacter formigenes to treat primary hyperoxaluria. Urolithiasis 46, 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira, L.M. , Hauschild, D.B. , Leite Cde, M. , Baptista, D.R. , Carvalho, M. (2014) Adequate dietary intake and nutritional status in patients with nephrolithiasis: new targets and objectives. J Ren Nutr 24, 417–422. [DOI] [PubMed] [Google Scholar]
- Parkhomenko, E. , De Fazio, A. , Tran, T. , Thai, J. , Blum, K. and Gupta, M. (2017) A multi‐institutional study of struvite stones: patterns of infection and colonization. J Endourol 31, 533–537. [DOI] [PubMed] [Google Scholar]
- Schloss, P.D. , Westcott, S.L. , Ryabin, T. , Hall, J.R. , Hartmann, M. , Hollister, E.B. , Lesniewski, R.A. , Oakley, B.B. et al. (2009) Introducing mothur: open‐source, platform‐independent, community‐supported software for describing and comparing microbial communities. Appl Environ Microbiol 75, 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shavit, L. , Ferraro, P.M. , Johri, N. , Robertson, W. , Walsh, S.B. , Moochhala, S. and Unwin, R. (2015) Effect of being overweight on urinary metabolic risk factors for kidney stone formation. Nephrol Dial Transplant 30, 607–613. [DOI] [PubMed] [Google Scholar]
- Simoes, E.S.A.C. , Oliveira, E.A. and Mak, R.H. (2020) Urinary tract infection in pediatrics: an overview. J Pediatr (Rio J) 96(Suppl 1), 65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes‐Silva, L. , Araujo, R. , Pestana, M. , Soares‐Silva, I. and Sampaio‐Maia, B. (2020) Peritoneal microbiome in end‐stage renal disease patients and the impact of peritoneal dialysis therapy. Microorganisms 8, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, J.M. , Moazami, S. , Qiu, Y. , Kurland, I. , Chen, Z. , Agalliu, I. , Burk, R. and Davies, K.P. (2016) Evidence for a distinct gut microbiome in kidney stone formers compared to non‐stone formers. Urolithiasis 44, 399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryavanshi, M.V. , Bhute, S.S. , Jadhav, S.D. , Bhatia, M.S. , Gune, R.P. and Shouche, Y.S. (2016) Hyperoxaluria leads to dysbiosis and drives selective enrichment of oxalate metabolizing bacterial species in recurrent kidney stone endures. Sci Rep 6, 34712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, R. , Jiang, Y. , Tan, A. , Ye, J. , Xian, X. , Xie, Y. , Wang, Q. , Yao, Z. et al. (2018) 16S rRNA gene sequencing reveals altered composition of gut microbiota in individuals with kidney stones. Urolithiasis 46, 503–514. [DOI] [PubMed] [Google Scholar]
- Tavichakorntrakool, R. , Boonsiri, P. , Prasongwatana, V. , Lulitanond, A. , Wongkham, C. and Thongboonkerd, V. (2017) Differential colony size, cell length, and cellular proteome of Escherichia coli isolated from urine vs. stone nidus of kidney stone patients. Clin Chim Acta 466, 112–119. [DOI] [PubMed] [Google Scholar]
- Tavichakorntrakool, R. , Prasongwattana, V. , Sungkeeree, S. , Saisud, P. , Sribenjalux, P. , Pimratana, C. , Bovornpadungkitti, S. , Sriboonlue, P. et al. (2012) Extensive characterizations of bacteria isolated from catheterized urine and stone matrices in patients with nephrolithiasis. Nephrol Dial Transplant 27, 4125–4130. [DOI] [PubMed] [Google Scholar]
- Vaziri, N.D. , Zhao, Y.Y. and Pahl, M.V. (2016) Altered intestinal microbial flora and impaired epithelial barrier structure and function in CKD: the nature, mechanisms, consequences and potential treatment. Nephrol Dial Transplant 31, 737–746. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Yang, S. , Li, S. , Zhao, L. , Hao, Y. , Qin, J. , Zhang, L. , Zhang, C. et al. (2020) Aberrant gut microbiota alters host metabolome and impacts renal failure in humans and rodents. Gut 69, 2131–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, T.J. , Huws, S.A. , Edwards, J.E. , Kingston‐Smith, A.H. , Siu‐Ting, K. , Hughes, M. , Rubino, F. , Friedersdorff, M. et al. (2018) CowPI: a rumen microbiome focussed version of the PICRUSt functional inference software. Front Microbiol 9, 1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, I.‐W. , Hsu, K.‐H. , Lee, C.‐C. , Sun, C.‐Y. , Hsu, H.‐J. , Tsai, C.‐J. , Tzen, C.‐Y. , Wang, Y.‐C. et al. (2011) p‐Cresyl sulphate and indoxyl sulphate predict progression of chronic kidney disease. Nephrol Dial Transplant 26, 938–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, G. , Mai, Z. , Xia, S. , Wang, Z. , Zhang, K. , Wang, L.I. , Long, Y. , Ma, J. et al. (2017) Prevalence of kidney stones in China: an ultrasonography based cross‐sectional study. BJU Int 120, 109–116. [DOI] [PubMed] [Google Scholar]
- Zhang, Q. , Zhang, Z. , Lu, T. , Yu, Y. , Penuelas, J. , Zhu, Y.G. and Qian, H. (2021) Gammaproteobacteria, a core taxon in the guts of soil fauna, are potential responders to environmental concentrations of soil pollutants. Microbiome 9, 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Alpha diversity box chart. The differences between the two groups were determined by Wilcox Test. (a) Observed species; (b) Chao; (c) Ace; (d) Shannon; (e) Simpson; (f) Coverage.
Table S1. Urine routine.
Table S2. Electrolyte, liver function and kidney function.
Table S3. Routine blood test.
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.


