Abstract
In this study, we determined the boundaries of a 99-kb deletable element of Shigella flexneri 2a strain YSH6000. The element, designated the multiple-antibiotic resistance deletable element (MRDE), had recently been found to contain a 66-kb pathogenicity island (PAI)-like element (designated the SRL PAI) which carries the Shigella resistance locus (SRL), encoding resistance determinants to streptomycin, ampicillin, chloramphenicol, and tetracycline. The YSH6000 MRDE was found to be flanked by two identical IS91 elements present at the S. flexneri homologs of the Escherichia coli genes putA and mdoA on NotI fragment D. Sequence data from two YSH6000-derived MRDE deletants, YSH6000T and S2430, revealed that deletion of the MRDE occurred between the two flanking IS91 elements, resulting in a single IS91 element spanning the two original IS91 loci. Selection for the loss of tetracycline resistance confirmed that the MRDE deletion occurred reproducibly from the same chromosomal site and also showed that the SRL PAI and the SRL itself were capable of independent deletion from the chromosome, thus revealing a unique set of nested deletions. The excision frequency of the SRL PAI was estimated to be 10−5 per cell in the wild type, and mutation of a P4-like integrase gene (int) at the left end of the SRL PAI revealed that int mediates precise deletion of the PAI.
Shigella spp., the causative agents of bacillary dysentery, are responsible for the deaths of more than 1.1 million people every year (28). Infections are transmitted via the fecal-oral route either as a result of person-to-person contact or through ingestion of contaminated food or water, and result in watery diarrhea which may progress to the bloody mucoid stools typical of bacillary dysentery (12). In developing countries individuals affected by acute diarrhea are commonly treated by oral rehydration and antimicrobial therapy. However, rehydration therapy alone provides little benefit to patients with dysentery caused by invasive enteropathogens such as Shigella, and the global importance of dysentery in developing countries has increased as a result of ineffective treatment (26). In addition, resistance to antibiotics such as tetracycline and ampicillin, which were once highly efficacious in treatment of shigellosis, has grown considerably in the past few decades (45). In many cases, resistance genes are found to reside on easily transferable R plasmids. However, chromosomally borne resistance genes have recently been identified in a number of studies (10, 15, 29, 41, 42), although the basis of chromosomal resistance has not been widely investigated.
Rajakumar et al. (42) have described a spontaneous 99-kb chromosomal deletion that results in multi-antibiotic susceptibility in Shigella flexneri 2a YSH6000. The resistance locus carried on the 99-kb element was found to encode resistance determinants to streptomycin, ampicillin, chloramphenicol, and tetracycline. These determinants have since been collectively designated the SRL, for Shigella resistance locus. The SRL exhibits similarity in sequence and organization to the antibiotic resistance loci of NR-1, an R plasmid commonly found in Shigella, and transposon Tn2603 (41). Although the nature and exact location of the 99-kb multiple-antibiotic resistance deletable element (MRDE) harboring the SRL have not been determined, it has been mapped to a region of the chromosome on NotI fragment D bounded by the S. flexneri homologs of the Escherichia coli genes ompA and pyrC (42). The instability of the MRDE is reminiscent of the behavior of other large chromosomal regions in E. coli and Yersinia pestis, referred to as pathogenicity islands (PAIs) (7), suggesting that the MRDE may also be a PAI-like element.
The term “pathogenicity island” was first used to describe large unstable DNA regions in uropathogenic E. coli (UPEC) (7). However, the term is now used more generally to refer to sections of chromosome throughout a number of species that are often unstable and that frequently carry virulence genes (18). Since the introduction of the term PAI, islands have been identified in many species, including E. coli, Yersinia spp., Helicobacter pylori, Salmonella spp. and S. flexneri (20). In addition to virulence genes, PAIs also encode mobility elements such as integrases and insertion elements (IS elements) and commonly integrate into, or adjacent to, tRNA genes. These characteristics, which show remarkable similarity to those of bacteriophages, in conjunction with a G+C content that often differs from that of the host chromosome, have led to the suggestion that PAIs are acquired from different species via phage-mediated horizontal transfer (19). Indeed, it has recently been reported that the Vibrio cholerae PAI (VPI), which plays a role in the emergence of epidemic and pandemic cholera, is the genome of a prophage, and the VPIφ has been shown to transfer to one VPIφ-negative V. cholerae strain (24, 25).
In most cases, the instability of PAIs is due to their precise excision from the chromosome via recombination between identical sequences situated on either side of the element. PAIs are commonly arranged with short flanking direct repeats (DRs) of 9 to 20 bp which are analogous to phage att sites. These repeats are often identical to the 3′ sequence of the target tRNA gene, and upon PAI deletion only one copy of the DR remains on the chromosome (11). IS elements have also been found in the flanking region of some PAIs (19), and recombination between two flanking IS100 elements has been shown to occur upon deletion of the high-pathogenicity island (HPI) from Y. pestis (5, 13).
In this study, we investigated the various types of deletion events leading to loss of multiple-antibiotic resistance in S. flexneri 2a strain YSH6000. We report here that the antibiotic resistance genes of the SRL are lost following at least three distinct events including the precise deletion of the MRDE and the independent deletions of the SRL and of a PAI-like element termed the SRL PAI, both of which are entirely contained within the larger MRDE.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Strains were grown routinely at 37°C in Luria-Bertani (LB) medium (4) with the addition of ampicillin (100 μg/ml), kanamycin (50 μg/ml), trimethoprim (50 μg/ml), chloramphenicol (40 μg/ml), or tetracycline (10 μg/ml) when necessary.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| S. flexneri | ||
| YSH6000 | Wild-type S. flexneri 2a Japanese isolate; Smr Apr Cmr Tcr | 46 |
| YSH6000T | MRDE deletant of YSH6000; Sms Aps Cms Tcs | 38 |
| S2430 | MRDE deletant of YSH6000; Sms Aps Cms Tcs | 39 |
| SBA1304 | Wild-type S. dysenteriae 3 Japanese isolate | This study |
| SBA1363 | Spontaneous SRL PAI deletant of YSH6000; Sms Aps Cms Tcs | This study |
| SBA1365 | Spontaneous MRDE deletant of YSH6000; Sms Aps Cms Tcs | This study |
| SBA1366 | Spontaneous SRL deletant of YSH6000; Sms Aps Cms Tcs | This study |
| SBA1367 | Spontaneous SRL PAI deletant of YSH6000; Sms Aps Cms Tcs | This study |
| SBA1368 | Spontaneous SRL deletant of YSH6000; Sms Aps Cms Tcs | This study |
| SBA1369 | Spontaneous MRDE deletant of YSH6000; Sms Aps Cms Tcs | This study |
| AL11 | YSH6000 insertional int mutant; Smr Apr Cmr Tcr Knr | This study |
| AL108 | YSH6000 harboring pBRTprΔ; Smr Apr Cmr Tcr Tpr | This study |
| AL109 | AL11 harboring pBRTprΔ; Smr Apr Cmr Tcr Knr Tpr | This study |
| AL110 | AL11 harboring pAL66; Smr Apr Cmr Tcr Knr Tpr | This study |
| E. coli | ||
| DH5α | F−φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17 Deorthi-1 sunE441 gyrA96 relA1 | Bethesda Research Laboratories |
| S17-1(λpir) | RP4-2(Tcs::Mu)(Kns::Tn7) recA λpir Smr Tpr | 36 |
| JM109(λpir) | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) λpir | 36 |
| Plasmids | ||
| pWSK29 | pSC101-based low-copy-number vector; ΔlacZ Apr | 47 |
| pWSK129 | pSC101-based low-copy-number vector; ΔlacZ Knr | 47 |
| pBRTprΔ | 4.8-kb pBR322-based cloning vector; Tpr Tcr | C. Sasakawa |
| pJP5603 | R6K-based suicide vector; Knr | 40 |
| pSBA348 | pWSK29 harboring 23-kb BamHI fragment containing solA-mdoA region of YSH6000 | K. Rajakumar |
| pSBA509 | pWSK129 harboring part of SRL PAI and flanking regions | S. N. Luck |
| pSBA533 | pWSK129 harboring 14-kb EcoRI subclone from pSBA509 | S. N. Luck |
| pSBA574 | pWSK29 harboring SBA1366 SRL deletion region; Apr | This study |
| pSBA575 | pWSK29 harboring SBA1368 SRL deletion region; Apr | This study |
| pAL11 | pJP5603 harboring 666-bp internal SRL PAI integrase gene fragment | This study |
| pAL64 | pWSK29 harboring 1.3-kb PCR fragment containing int | This study |
| pAL66 | pBRTprΔ harboring 1.3-kb int fragment | This study |
Molecular biological techniques.
Genomic DNA was isolated by small-scale preparation as described previously (4). Plasmid DNA was isolated by a modification of the alkaline lysis method (37). Standard cloning procedures using the vector pWSK29, pJP5603, or pBRTprΔ were employed. E. coli DH5α was transformed using the rubidium chloride method (16). DNAs from both plasmids and PCR products were prepared for sequence analysis using the PRISM Ready Reaction Dye Deoxy Terminator Cycle, and chromatograms were produced on an Applied Biosystems model 373A DNA sequencing system.
Selection of tetracycline-sensitive derivatives of YSH6000.
Tetracycline-sensitive derivatives of YSH6000 were selected by plating dilutions of YSH6000 (grown in LB broth to a density of approximately 109 cells ml−1) onto LB agar supplemented with fusaric acid (12 μg/ml), chlortetracycline (50 μg/ml), and 0.1 mM ZnCl2 (31). Plates were incubated for 24 to 40 h at 37°C.
Southern hybridization.
After electrophoresis, DNA was transferred to a charged nylon membrane (Roche) using a vacuum blotting apparatus (TE80 Transvac; Hoefer) or by capillary transfer in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (pH 7.0). Overnight hybridization and subsequent washings were performed under high-stringency conditions at 68°C as recommended in the Roche digoxigenin labeling and detection kit instructions. Probes were labeled by PCR amplification with digoxigenin as specified by Boehringer Mannheim. The 0.5-kb phoH probe was amplified using primers BAP507 (5′-AATAAACCCTTCCCGCTTCC-3′) and T3 (5′-AATTAACCCTCACTAAAGGG-3′) from a pSBA509 template. The 2.0-kb csg probe was amplified using primers csgA forward (5′-AAAGAATTCGCTCTGGCAGGTGTTGTTCC-3′) and csgA reverse (5′-AAAAAGTCGACTTAACCAAAGCCAACCTGAGTCACG-3′) from an SBA1304 template. The 1.0-kb fec probe was amplified using primers BAP914 (5′-GCTCCCATTTCGCTCGGC-3′) and BAP935 (5′-GTTGTCGTCATAAGAGCGG-3′) with a YSH6000 template. The 1.0-kb int probe was amplified using BAP1005 (5′-GCTGGATTGGGAACTTACC-3′) and T7 (5′-GTAATACGACTCACTATAGGGC-3′) from a pSBA533 template.
PCR amplification of deletion point junctions.
Amplifications were performed on chromosomal DNA with the following oligonucleotide primers: for mapping the MRDE deletion endpoint by inverse PCR, BAP499 (5′-CGGGAAGAATACCTGTTGATG-3′) and BAP531 (5′-TATTTGATGCTATGAAGAAGGGGG-3′); for the MRDE deletion region, BAP649 (5′-AGCGGCAGCGGTATTCAC-3′) and BAP530 (5′-TTAATCTCTTCTTCACTTCGCCCC-3′) or 470 (5′-TCCAGCCACCTTTAGCGG-3′); for the SRL deletion region, BAP1249 (5′-TATCCCGCTTGCCGTCGC-3′) and BAP694 (5′-AGCGGCAGCGGTATTCAC-3′); and for the PAI deletion region, BAP679 (5′-GTGCTGCTTTCGGTGTGC-3′) and BAP1157 (5′-GCCAGCATTTCAACAGGAGG-3′).
Mutant construction.
Primers BAP1354 (5′-GCGGATTCCCCTGGCTTCGC-3′) and BAP1355 (5′-TTGGATTCAGGGGCGGGGGAAATGGG-3′) were used to amplify a 666-bp internal fragment of the integrase gene (bp 714 to 1380; GenBank accession no. AF326777), which was then ligated into the T-tailed HincII site of pJP5603. The recombinant plasmid carrying the int fragment, pAL11, isolated in JM109(λpir), was transferred by conjugation into YSH6000 using the mobilizing strain S17-1(λpir). Exconjugants carrying a single-crossover mutation of int were obtained by selection on kanamycin-ampicillin LB plates and confirmed by PCR and Southern hybridization.
Mutant complementation
A 1,344-bp product (bp 586 to 1930; GenBank accession no. AF326777) containing the entire int open reading frame (ORF) was amplified by PCR with primers BAP1636 (5′-TGGATGGGATCCCAGAGTGACGGGAATTAGC-3′) and BAP1637 (5′-ATGCCAGGATCCCATTACGAACTGGCATTG-3′). Both primers included BamHI sites at the 5′ ends, and this product was cloned into the T-tailed EcoRV site of pWSK29, designated pAL64, and sequenced to confirm that no errors were incorporated. The 1.3-kb BamHI fragment from pAL64 containing the int fragment was then cloned into the BamHI site of pBRTprΔ, and clones were selected by sensitivity to tetracycline. The orientation of the int fragment was confirmed by restriction enzyme digestion to be the same as that of the interrupted Tcr gene.
Computer analysis.
Sequencing chromatograms were analyzed using the Sequencher program (GeneCodes Corporation, Ann Arbor, Mich.). Nucleotide sequence similarity searches of the databases were performed using the BlastN or BlastX program (2). Protein sequence alignments were performed in eclustalw, available on the Australian National Genomic Information Server (ANGIS [http://www.angis.org.au]).
RESULTS
Mapping the MRDE deletion point.
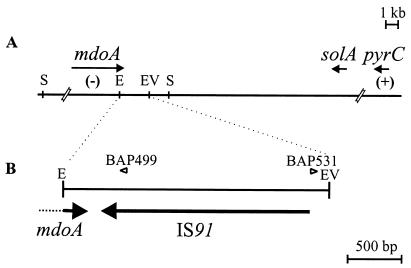
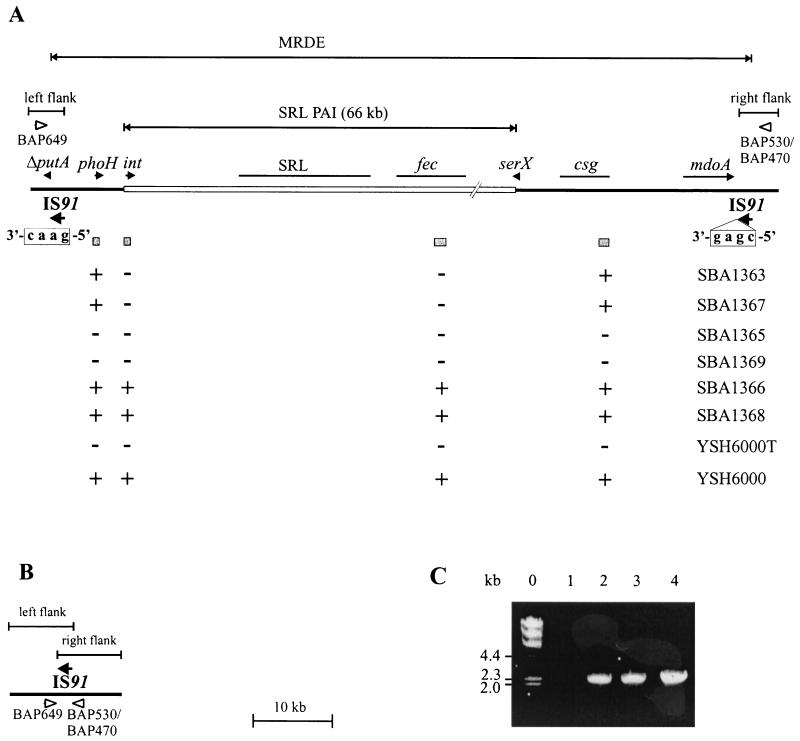
The MRDE of S. flexneri 2a YSH6000 had previously been mapped to chromosomal NotI fragment D, between the ompA and pyrC genes (42). Southern analysis of the wild-type strain, YSH6000, and the spontaneous MRDE deletion strains, YSH6000T and S2430, with a series of probes derived from the ompA-pyrC region showed that the right MRDE endpoint lies within a 2.4-kb EcoRI-EcoRV fragment between mdoA and solA (Fig. 1A). Sequencing of this fragment revealed that this region contained the 3′ end of the mdoA locus and a 1,829-bp element showing 93% similarity at the nucleotide level to IS91 of E. coli (Fig. 1B). Due to the high number of IS elements in Shigella, Southern hybridization could not be used to determine the deletion endpoint within this region. As the position of the left endpoint of the MRDE was unknown, inverse PCR with primers situated near the right deletion endpoint was employed to amplify a product which traversed the deletion site in strain YSH6000T. A PCR product of approximately 6 kb was amplified from SalI-digested and religated YSH6000T genomic DNA using primers BAP499 and BAP531 (Fig. 1B). As the positions of these primers allowed the amplification of a maximum of 2 kb of right flanking region, this suggested that the amplified inverse PCR product extended across the MRDE deletion point and 4 kb into the left flanking region. Sequence analysis revealed that this fragment contained the right end of an IS91 element followed by a sequence exhibiting high-level identity to the 3′ region of the E. coli putA gene, which is situated approximately 31 kb upstream of mdoA in E. coli. These data showed that a single copy of the IS91 element spanned the deletion region in YSH6000T and suggested two possible explanations. There may have been a single IS91 element adjacent to the mdoA locus that, after the deletion event, spanned the region between this locus and the putA sequence. Alternatively, an IS91 element may have been present at each locus in YSH6000, and after the deletion event, only one copy of the element remained on the chromosome. To decide between these two alternatives, direct PCR and sequence analysis of both the left flanking region of the MRDE in YSH6000 and the deletion regions of YSH6000T and S2430 were carried out. Identical intact IS91 elements were found at both flanks of the MRDE in YSH6000, confirming that two distinct IS91 elements were present, one downstream of the mdoA locus and the other interrupting the putA sequence. The data also showed that an identical intact IS91 was present at the deletion point in both S2430 and YSH6000T. The organizational similarity between E. coli and S. flexneri, together with the known sequence of the SRL PAI, which is located within this region (S. N. Luck, S. A. Turner, K. Rajakumar, H. Sakellaris, and B. Adler, submitted for publication), allowed us to propose a structure for the MRDE in YSH6000 and for the corresponding regions bearing the deletion points in YSH6000T and S2430 (Fig. 2A). This organization suggests that the MRDE was not acquired or did not evolve as a unit but is composed of a PAI that is situated within a distinct deletable region of the chromosome defined by two IS91 elements.
FIG. 1.
Map of the right flank of the YSH6000 MRDE. (A) Schematic representation of the YSH6000 mdoA-pyrC region. (B) The 2.4-kb EcoRI/EcoRV fragment within pSBA348 bearing the right deletion endpoint of the MRDE. Chromosomal DNA is represented as a thin black line, and genes are represented as arrows. The presence (+) or absence (−) of regions as tested by Southern hybridization in MRDE deletants YSH6000T and S2430 is also shown. Open arrowheads indicate the positions of primers BAP499 and BAP531, which were used to amplify across the MRDE deletion region in YSH6000T by inverse PCR of SalI-digested and religated genomic DNA. Abbreviations for restriction sites: E, EcoRI; EV, EcoRV; S, SalI.
FIG. 2.
Deletion of the YSH6000 MRDE. (A) Schematic representation of the genetic organization of the wild-type YSH6000 MRDE. The boundaries of the MRDE are defined by the two IS91 elements, indicated by arrows. The SRL PAI is represented as an open rectangle. Genes and IS elements are represented as arrows, with truncations indicated by a capital delta. The fec, csg, and SRL loci are shown as thin black lines. The tetramers immediately downstream of the IS91 elements at the left and right flanks of the MRDE are shown boxed below each element. Shaded boxes indicate the positions of probes used to determine the presence (+) or absence (−) of these regions within six streptomycin-, ampicillin-, chloramphenicol-, and tetracycline-sensitive strains, YSH6000T, and YSH6000. Open arrowheads represent primers BAP649 and BAP530/BAP470, which were used to amplify across the deletion endpoint. The physical distance between putA and csg is based on PCR and/or sequence analysis, while the distance between csg and mdoA was calculated based on the original sizing of the MRDE at 99 kb (42). Regions designated the MRDE left and right flanks, common to both the wild type and MRDE deletants, are indicated above the diagram. (B) Schematic representation of the resultant structure across the deletion point following loss of the MRDE element in YSH6000T, S2430, SBA1365, and SBA1369. (C) Detection of the MRDE deletion point. Shown are PCR products amplified using primers BAP649 and BAP470 (panels A and B). Lanes: 0, HindIII-digested λ DNA size markers; 1, YSH6000; 2, YSH6000T; 3, SBA1365; 4, SBA1369.
Although the S. flexneri IS91 showed considerable similarity to the E. coli IS91, some differences were noted. IS91 is known to show absolute insertion specificity for the tetramer GAAC or CAAG, always inserting such that the right inverted repeat (IRR) is adjacent to either of these target sequences (34). The left flanking IS91 in YSH6000 also exhibits this specificity, but the right flanking IS91 is inserted adjacent to the sequence CGAG (Fig. 2A), indicating that this element may have an insertion specificity different from that of the E. coli IS91. Additionally, the sequences showing similarity to the two major ORFs of IS91, ORF121, which is implicated in insertion specificity (6) and tnpA, the transposase ORF (33, 35), are shortened by 12 and 31 amino acids, respectively, at the carboxy termini compared with their E. coli homologs.
Spontaneous deletion of the MRDE.
In order to assess whether the MRDE deletes reproducibly from the same point in the chromosome, we selected for the loss of tetracycline resistance by growth of YSH6000 on LB medium supplemented with fusaric acid. Colonies that grew on fusaric acid medium were confirmed for sensitivity to tetracycline and further tested for susceptibility to the antibiotics streptomycin, ampicillin, and chloramphenicol. Using this method, six Sms, Aps, Cms, and Tcs strains were identified: SBA1363 and SBA1365 through SBA1369. PCR using the inwardly directed primers BAP649 and BAP470 was employed to confirm MRDE deletions, as the intact deletion region would be amplified only in MRDE− strains. Products were amplified in strains SBA1365 and SBA1369 (Fig. 2C, lanes 3 and 4); sequencing revealed that these strains each carried a single IS91 element at the deletion point, confirming that the MRDE deletions in these strains were identical to those in YSH6000T and S2430 (Fig. 2B).
Novel deletions leading to loss of antibiotic resistance.
As typical MRDE deletions could be confirmed by PCR in only two of the six multi-antibiotic-sensitive strains, the basis for antibiotic sensitivity in the remaining strains was investigated further. Pulsed-field gel electrophoresis of NotI-digested chromosomal DNA showed that in both SBA1365 and SBA1369, NotI fragment D carrying the MRDE underwent a deletion of the same size as those observed in YSH6000T and S2430 (data not shown). However, in the remaining strains a variety of smaller deletions were observed (data not shown). To localize these deletions more precisely, strains were analyzed by Southern hybridization with a series of probes corresponding to four regions within the MRDE (Fig. 2A). With the exception of SBA1365 and SBA1369, which had undergone deletion events identical to that in YSH6000T, the deletions were confined to the region between phoH and csg (Fig. 2A).
Deletion of the SRL.
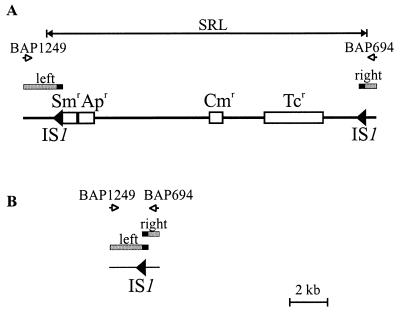
Sequence analysis of the region responsible for antibiotic resistance in S. flexneri YSH6000 revealed that the MRDE harbors a cluster of resistance genes, now called the SRL (Fig. 2A). The SRL shows significant similarity to the resistance region in the Shigella R plasmid NR-1 but also includes an oxaI cassette, encoding β-lactamase (41), therefore conferring resistance to the antibiotics streptomycin, ampicillin, chloramphenicol, and tetracycline. The SRL is 16.7 kb in length, including the two flanking 768-bp IS1 elements (Fig. 3A). Each IS1 contains an intact insAB′ and ΔinsA-B′-insB ORF, both of which have been implicated in transposition (30, 32), suggesting that they are still functional. However, the IS1 elements were not identical; the left and right IS1 elements showed 99 and 97% identity, respectively, to the E. coli IS1 nucleotide sequence.
FIG. 3.
Deletion of the SRL. (A) Diagram of the SRL in YSH6000. (B) Schematic representation of the structure across the deletion point following loss of the SRL element in SBA1366 and SBA1368. IS elements are represented as arrows, and chromosomal DNA is represented by thin black lines. Determinants encoding resistance to streptomycin, ampicillin, chloramphenicol, and tetracycline are represented as open boxes labeled Smr, Apr, Cmr, and Tcr, respectively. Left and right flanking regions common to both the wild type and SRL deletants are indicated by shaded boxes, with the solid part of each box corresponding to the identical 487 bp of each IS1.
Although the profiles of SBA1366 and SBA1368 indicated that all probed areas were present (Fig. 2A), these strains were susceptible to all four antibiotics to which resistance was encoded by the SRL. For this reason it was considered that these two strains might harbor deletions of the resistance locus itself. Inward-facing primers, BAP1249 and BAP694, situated on either side of the SRL were used to amplify this region. These primers are separated by 18.5 kb in the wild type and thus do not result in the amplification of a product (Fig. 3A), but amplification of a 2.6-kb product occurred in both SBA1366 and SBA1368. The PCR products were cloned into EcoRV-digested, T-tailed pWSK29 and designated pSBA574 and pSBA575, respectively. Sequence analysis revealed that these fragments contained sequences identical to that flanking the left end of the SRL, a stretch of sequence identical to the left IS1 element, followed directly by sequences identical to that flanking the right end of the SRL (Fig. 3B). These data indicate that the SRL was able to delete from the chromosome independently of the MRDE and that upon deletion, a single IS1 element identical to the left IS1 of the SRL spanned the deleted area. However, it is not possible to determine whether the IS1 element spanning the deletion point has its origin entirely from the left IS1 of the SRL or whether it is a composite of the left and right IS1 elements, which share an identical sequence throughout bp 1 to 487 (Fig. 3).
Deletion of the SRL PAI.
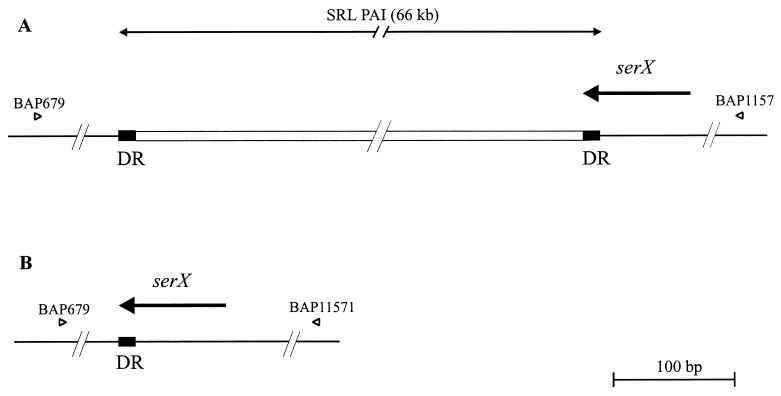
We have recently sequenced the region surrounding the SRL, revealing the presence of a 66-kb element which displays characteristics of a PAI. This element, designated the SRL PAI, is contained completely within the MRDE (Fig. 2A). The SRL PAI carries an integrase-like gene at the left boundary, contains numerous IS elements and phage-related sequences, and also encodes a ferric dicitrate transport system (fec) (Luck et al., submitted). The genetic organization in the wild-type strain YSH6000 showed the SRL PAI to be flanked by 14-bp DRs which correspond to the 3′ terminus of the tRNA gene serX (5′-GGGGGAGTGGCGGT-3′). The right flank of the PAI harbored an intact serX, but similarity to the E. coli sequence ended directly downstream of the 14-bp sequence. At the left end of the SRL PAI, the 14-bp repeat was followed by a stretch of sequence similar to that found downstream of the serX tRNA gene in E. coli, indicating that the PAI had inserted into the 3′ end of the serX gene (Fig. 4A). Although deletion of this element had not been previously reported, the Southern hybridization profiles of SBA1363 and SBA1367 suggested that the PAI might be capable of excision (Fig. 2A). Inward-facing primers situated on either side of serX were used to amplify a product spanning the right and left flanking regions of the PAI. A product of approximately 1.1 kb was amplified in both SBA1363 and SBA1367, and sequence analysis of this fragment revealed the presence of an intact serX gene, with only a single copy of the 14-bp repeat present. Upstream and downstream regions were identical to the left and right flanking regions of the PAI, demonstrating that the SRL PAI itself is capable of precise excision from the chromosome, leaving behind an intact serX gene (Fig. 4B).
FIG. 4.
Deletion of the SRL PAI. (A) Diagram of the genetic organization of the left and right SRL PAI flanking regions in YSH6000. (B) Schematic representation of the structure across the deletion point following loss of the SRL PAI in SBA1363 and SBA1367. Thin black line, chromosomal DNA; open rectangle, SRL PAI; arrows, genes; solid boxes, 14-bp DRs. Also shown are the locations of primers BAP1157 and BAP679, used to amplify across the SRL PAI deletion point.
Role of integrase in excision of the SRL PAI.
Hacker et al. (18) have suggested that flanking repeats may act as targets for site-specific recombinases, facilitating integration and/or excision of PAIs. It was thought that the 14-bp repeat may therefore represent the core sequence of the SRL PAI attachment (att) site, acting in a manner similar to that of the att sites of site-specific bacteriophages (9). The presence of mobility genes encoding determinants, such as integrases, on PAIs had been noted previously (18, 19), but with the exception of the PAI-like V. cholerae SXT element (21), the role of integrases in the excision of integrated PAIs had not been demonstrated. The SRL PAI encodes an ORF at its left boundary that shows similarity at the amino acid level to several integrase proteins from the P4 prophage Int family (Luck et al., submitted). This ORF, designated int, encodes a putative protein of 405 amino acids and contains the highly conserved HXXR, Y motif necessary for integrase function (3). As the model of PAI deletion described above suggests the involvement of a site-specific recombinase, the role of the SRL PAI int in deletion of the element was investigated.
An insertion mutation in the int gene was constructed in S. flexneri 2a strain YSH6000 (see Materials and Methods), and the frequencies of spontaneous SRL PAI excision in YSH6000 and the int mutant strain, AL11, were compared. Spontaneous SRL PAI excisants were isolated by taking advantage of the selective properties of fusaric acid against tetracycline resistance encoded by the SRL PAI (see Materials and Methods). Tetracycline-sensitive derivative strains were further tested for susceptibility to the antibiotics streptomycin, ampicillin, and chloramphenicol, and PAI deletions were confirmed by PCR using primers to amplify a 1.1-kb product across the intact serX gene, as performed previously for SBA1363 and SBA1367 (Fig. 4). By comparing the number of PAI excisants to the total cell count, the PAI deletion frequency was estimated to be approximately 10−5 per cell in the wild-type strain YSH6000. However, precise excision of the SRL PAI was not detected at all in the mutant strain AL11 (detection limit = 1.1 × 10−7 per cell). Since deletion of the MRDE occurs at a rate of 10−6 per cell in both the wild type and AL11 (data not shown), the int mutation was responsible for at least a 10-fold decrease in the SRL PAI excision rate, suggesting that the integrase gene is essential for precise excision of the PAI.
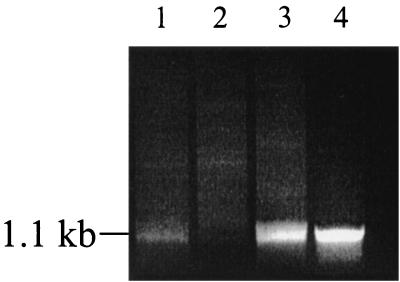
To confirm that the loss of PAI excision in this strain was due to inactivation of the integrase, AL11 was complemented with pBRTprΔ, carrying an intact int gene (pAL66). The resultant strain (designated AL110), YSH6000/pBRTprΔ (AL108), and AL11/pBRTprΔ (AL109) were tested for spontaneous SRL PAI excision using a PCR assay. Genomic DNA extracted from AL108, AL109, and AL110 was standardized for concentration and assayed for excision of the PAI using the inward-facing primers BAP1157 and BAP679 to amplify serX as described previously. Although PAI excision is an infrequent event, a PCR product could be detected using wild-type AL108 DNA (Fig. 5, lane 1), but as expected, no PCR product was detected when AL109 DNA was used as the template (Fig. 5, lane 2), confirming the previous findings that strains lacking a functional int gene are unable to undergo PAI deletion. An amplification product across the PAI deletion point was again detected using the int-complemented strain AL110, confirming that int was required for SRL PAI excision (Fig. 5, lane 3).
FIG. 5.
Complementation of the int mutation. PCR products spanning the SRL PAI excision site were obtained by amplification using primers BAP1157 and BAP679 (Fig. 4). From 100-μl PCR mixtures, all was loaded for lanes 1 and 2, 75 μl was loaded for lane 3, and 25 μl was loaded for lane 4. Lanes: 1, AL108 (YSH6000/pBRTprΔ); 2, AL109 (AL11/pBRTprΔ); 3, AL110 (AL11/pAL66); 4, SBA1363 (YSH6000-derived spontaneous PAI deletant).
DISCUSSION
In this study we demonstrated three independent mechanisms for the deletion of the resistance locus of S. flexneri 2a YSH6000: deletion of the MRDE involving IS91 elements, deletion of the SRL involving IS1 elements, and deletion of the SRL PAI occurring via int-mediated recombination of 14-bp DRs located at each extremity of the element. In many ways, the different deletion events involving the SRL PAI resemble the variety of deletion events that the HPI undergoes in different species of Yersinia. Like the SRL PAI, in Yersinia pseudotuberculosis IP32637, the HPI deletes via recombination between flanking 17-bp DRs (8). However, like MRDE deletion, in Y. pestis or Yersinia enterocolitica Ye8081 deletion of the HPI is associated with loss of flanking chromosome either by homologous recombination of flanking IS100 elements or by as-yet-undefined mechanisms, respectively (5, 13). To the best of our knowledge, the SRL PAI of S. flexneri 2a YSH6000 is the first PAI that has been observed to undergo both integrase-mediated and non-integrase-mediated excision in the same strain. It is only the second such element to be described which carries multi-antibiotic resistance determinants (the first was the SXT element of V. cholerae [20]).
The structuring of the three nested elements is itself unique, and although the MRDE was the first element to be described as carrying the SRL (41, 42), it seems unlikely that the entire 99 kb inserted en bloc into the Shigella flexneri 2a YSH6000 chromosome. Rather, it would appear that insertion of the IS91 elements and insertion of the SRL PAI were distinct events. This hypothesis is supported by the occurrence of independent deletions involving the PAI and MRDE, and it also explains the remarkable sequence and organizational conservation of the regions surrounding the PAI and the corresponding region in the E. coli chromosome. Additionally, had the entire MRDE inserted into the S. flexneri chromosome, this would presumably have led to duplication of the regions between the mdoA locus and putA, a phenomenon which was not observed in YSH6000 (S. A. Turner, unpublished data).
We have shown that the int gene was not required for MRDE deletion, and the loss of one of the flanking IS91 elements after deletion suggested that this element itself may be involved in excision of the MRDE. IS91 is the prototype of a small, unique family of IS elements that are believed to propagate by a rolling-circle replication mechanism (30). IS91 was originally isolated from a hemolysin-encoding plasmid of E. coli and has since been implicated in the spread of hemolysin (hly) genes (49, 50). hly genes have been reported on several PAIs in E. coli (17), and although no hly genes were discovered on the SRL PAI, it does carry an ORF showing similarity to a hemolysin expression-modulating protein of E. coli (Hha) (Luck et al., submitted). It is an intriguing possibility that hly determinants may have originally been present on the SRL PAI.
IS element-mediated deletion of adjacent DNA has been demonstrated for some IS elements (14). However, it has been reported that IS91 is unable to cause deletions of adjacent DNA (6); thus, the deletion of the MRDE is probably not mediated by transposition of the elements themselves. It is likely that MRDE deletion is the result of recA-mediated homologous recombination between the two flanking elements, which would cause a looping out of the intervening region, resulting in a single residual copy of the IS91 element on the chromosome. Such RecA-dependent “adjacent deletions” have been shown to occur for other IS elements (27), and work is currently under way to determine if the MRDE deletion is the result of RecA-dependent homologous recombination.
Deletion of the SRL also appears to involve IS elements. Two potentially functional IS1 elements flank the SRL, although upon deletion, only a single IS1 element remains on the chromosome. IS1 is known to mediate deletions of adjacent DNA that end precisely at one boundary of the element (14). The loss of the SRL may be an example of such a deletion.
Alternatively, the SRL may delete via homologous recombination between the flanking IS1 elements. As described previously, the YSH6000 SRL shares many similarities with the resistance determinant (r-det) of NR1, an archetypal resistance plasmid of Shigella (41). The r-det of NR1, which has a size of 23.3 kb and is flanked by DRs of IS1, is also a transposable unit (48). Using phage P1 as a carrier, the IS1-flanked r-det (Tn2671) was shown to move both to the site of the resident IS1 in the P1 genome and to another region of the P1 genome (22). It was proposed that the former mechanism would involve an intermediate circular form of the r-det carrying a single copy of the IS1 that excises from NR1 via homologous recombination between the flanking IS1 elements (22). The organization of the YSH6000 SRL deletants suggests that a similar recombination event may have taken place between the two flanking SRL IS1 elements, thus leaving a single copy of an IS1 element on the chromosome. Interestingly, after mobilization to P1, Tn2671 was subsequently mobilized to E. coli recipients and then to the genome of phage P7, demonstrating the existence of a natural mechanism for spread of antibiotic resistance genes (23). This highlights the potential of the SRL to spread to other genomes, independent of mechanisms involving the MRDE or SRL PAI deletions.
In this study we have demonstrated that the SRL PAI of S. flexneri 2a strain YSH6000, which is flanked by 14-bp DRs, excises from the 3′ end of the serX tRNA gene via mechanisms that resemble the site-specific recombination exhibited by some prophages. The absence of detectable precise deletion of the SRL PAI in the int mutant suggests that recombination between the flanking DRs is int-mediated site-specific recombination, a mechanism similar to that observed for some phages. Phage transduction has previously been implicated in the mobility of some PAIs (e.g., the V. cholerae VPI and the staphylococcal PAI SaPI), and bacteriophage proteins encoded on PAIs are often assumed to have played a role in the original mobilization of the elements (20). However, with the exception of the SXT element of V. cholerae (21), a role for phage-like integrases in the excision of integrated PAIs has not been demonstrated. Here, the S. flexneri 2a strain YSH6000 SRL PAI integrase was shown to be required for the precise excision of the element, confirming that the integrase is functional and plays an important role in the deletion of the SRL PAI.
The SRL PAI is one of a growing number of PAIs that appear to delete precisely through site-specific recombination of short flanking DRs. It is probable that, like phages, these PAIs originally integrated into the 3′ termini of tRNA genes, with the DR sequence acting in a manner similar to the core of attB sites during phage integration (9). Other elements in this category may include PAI I536 and II536 of UPEC, with 16- and 18-bp DRs, respectively (7), the she PAI of S. flexneri with 22-bp DRs (1), and the HPI of Y. pseudotuberculosis with 17-bp DRs (8). Additionally, these five PAIs also carry sequences near one end of the element exhibiting similarity to the P4 family of phage-like integrases (1, 8, 17). Indeed, many other PAIs have been found to possess sequences with similarity to integrases and other phage-related ORFs. Interestingly, the insertion site of the HPI has been found to contain a region showing partial similarity to the P4 attB site (8). The 3′ end of serX near the SRL PAI integration site also possesses a sequence that matches 12 of the 20 bp of the P4 attB (Luck et al., submitted). The she PAI in S. flexneri and the HPI of Y. pseudotuberculosis have been found inserted in at least two different phe and asn tRNA genes possessing the target DR of each, respectively (8; K. Al-Hasani, B. Adler, K. Rajakumar, and H. Sakellaris, submitted for publication). Similarly, the SRL PAI has been found inserted in both serX and a paralog of this gene, serW, in Shigella (S. A. Turner, unpublished data), indicating that, like the chromosomal integration of phages, there is a high level of insertion specificity for these PAIs.
In this study, the SRL PAI was shown to have a deletion rate of approximately 10−5, which is consistent with those of PAI I536 and II536 (10−4 to 10−5) (18), the HPI (10−4) (8), and the she PAI (10−5 to 10−6) (43). Other similarities include flanking DRs, the presence of integrases, and evidence of site-specific deletion. In light of evidence presented here that the int gene is required for precise excision of the SRL PAI, the shared features of these PAIs provide a strong argument that integrase-dependent, site-specific recombination is likely to be a common mechanism of excision among these elements. Indeed, the P4-like integrase from the Y. pestis HPI has been shown to act in a site-specific manner (44), while in the HPI of Y. enterocolitica, interruption of the integrase gene and loss of conservation in the flanking 17-bp DRs are thought to be responsible for the “stabilization” of this element in the chromosome. In the HPI of Y. pseudotuberculosis, the integrase gene and DRs are intact, probably explaining why this element undergoes precise excision from the chromosome (5). It is noteworthy that although the int sequences of both PAI I536 and II536 of UPEC are thought to be nonfunctional, these PAIs delete from the chromosome at a rate similar to that of the SRL PAI (17, 18), implying that other recombinases may mediate the excision of these elements.
Further evidence for the importance of integrases is found in the V. cholerae SXT element, which exhibits many similarities to PAIs, including mobility, insertion into a specific chromosomal site, and the presence of 17-bp DRs flanking the inserted element. The int gene of the SXT element is necessary for excision and/or production of an extrachromosomal circular form of the element, which is required for the transfer of SXT to both V. cholerae and E. coli recipients (21). Recently Rakin et al. provided evidence that the integrase from Y. pestis HPI promotes both excision and integration of a minimal integrative HPI module into the asn tRNA target site (44). These findings demonstrate that the int of the SRL PAI and other PAIs may play equally important roles not only in excision but also in acquisition and dissemination of the PAI-like elements on which they reside.
Importantly, from a clinical perspective, these elements pose a substantial risk given their ability to alter dramatically both the virulence and the antibiotic susceptibility profile of a pathogen.
ACKNOWLEDGMENTS
We acknowledge the expert technical assistance of Ian McPherson and Vicki Vallance.
This work was supported by the National Health and Medical Research Council, Canberra, Australia.
REFERENCES
- 1.Al-Hasani K, Rajakumar K, Bulach D, Robins-Browne R, Adler B, Sakellaris H. Genetic organization of the she pathogenicity island in Shigella flexneri 2a. Microb Pathog. 2001;30:1–8. doi: 10.1006/mpat.2000.0404. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argos P, Landy A, Abremski K, Egan J B, Haggard-Ljungquist E, Heoss R H, Kahn M L, Kalionis B, Narayana S V L, Pierson III L S, Sternberg N, Leong J M. The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986;5:433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley Interscience; 1993. [Google Scholar]
- 5.Bach S, Buchrieser C, Prentice M, Guiyoule A, Msadek T, Carniel E. The high-pathogenicity island of Yersinia enterocolitica Ye8081 undergoes low-frequency deletion but not precise excision, suggesting recent stabilization in the genome. Infect Immun. 1999;67:5091–5099. doi: 10.1128/iai.67.10.5091-5099.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernales I, Mendiola M V, de la Cruz F. Intramolecular transposition of insertion sequence IS91 results in second-site simple insertions. Mol Microbiol. 1999;33:223–234. doi: 10.1046/j.1365-2958.1999.01432.x. [DOI] [PubMed] [Google Scholar]
- 7.Blum G, Ott M, Lischewski A, Ritter A, Imrich H, Tschape H, Hacker J. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect Immun. 1994;62:606–614. doi: 10.1128/iai.62.2.606-614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchrieser C, Brosch R, Bach S, Guiyoule A, Carniel E. The high-pathogenicity island of Yersinia pseudotuberculosis can be inserted into any of the three chromosomal asn tRNA genes. Mol Microbiol. 1998b;30:965–978. doi: 10.1046/j.1365-2958.1998.01124.x. [DOI] [PubMed] [Google Scholar]
- 9.Campbell A M. Chromosomal insertion sites for phages and plasmids. J Bacteriol. 1992;174:7495–7499. doi: 10.1128/jb.174.23.7495-7499.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casalino M, Nicoletti M, Salvia A, Colonna B, Pazzani C, Calconi A, Mohamud K A, Maimone F. Characterization of endemic Shigella flexneri strains in Somalia: antimicrobial resistance, plasmid profiles, and serotype correlation. J Clin Microbiol. 1994;32:1179–1183. doi: 10.1128/jcm.32.5.1179-1183.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dozois C M, Curtiss R., III Pathogenic diversity of Escherichia coli and the emergence of “exotic” islands in the gene stream. Vet Res. 1999;30:157–179. [PubMed] [Google Scholar]
- 12.DuPont H L, Levine M M, Hornick R B, Formal S B. Inoculum size in shigellosis and implications for expected mode of transmission. J Infect Dis. 1989;159:1126–1128. doi: 10.1093/infdis/159.6.1126. [DOI] [PubMed] [Google Scholar]
- 13.Fetherston J D, Schuetze P, Perry R D. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol Microbiol. 1992;6:2693–2704. doi: 10.1111/j.1365-2958.1992.tb01446.x. [DOI] [PubMed] [Google Scholar]
- 14.Galas D, Chandler M. Bacterial insertion sequences. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 109–162. [Google Scholar]
- 15.Gebre-Yohannes A, Drasar B S. Plasmid profiles of antibiotic-resistance Shigella dysenteriae types 2, 3, 4, 6, and 7 isolated in Ethiopia during 1976–85. Epidemiol Infect. 1990;105:65–72. doi: 10.1017/s0950268800047658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glover D M, editor. DNA cloning: a practical approach. Vol. 1. Oxford, United Kingdom: IRL Press Ltd.; 1985. [Google Scholar]
- 17.Hacker J, Blum-Oehler G, Janke B, Nagy G, Goebel W. Pathogenicity islands of extraintestinal Escherichia coli. In: Kaper J B, Hacker J, editors. Pathogenicity islands and other mobile virulence elements. Washington, D.C.: American Society for Microbiology; 1999. pp. 59–76. [Google Scholar]
- 18.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 19.Hacker J, Kaper J B. The concept of pathogenicity islands. In: Kaper J B, Hacker J, editors. Pathogenicity islands and other mobile virulence elements. Washington, D.C.: American Society for Microbiology; 1999. pp. 1–12. [Google Scholar]
- 20.Hacker J, Kaper J B. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol. 2000;54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- 21.Hochhut B, Waldor M K. Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol Microbiol. 1999;32:99–110. doi: 10.1046/j.1365-2958.1999.01330.x. [DOI] [PubMed] [Google Scholar]
- 22.Iida S, Arber W. On the role of IS1 in the formation of hybrids between the bacteriophage P1 and the R plasmid NR1. Mol Gen Genet. 1980;177:261–270. doi: 10.1007/BF00267437. [DOI] [PubMed] [Google Scholar]
- 23.Iida S, Hanni C, Echarti C, Arber W. Is the IS1-flanked r-determinant of the R plasmid NR1 a transposon? J Gen Microbiol. 1981;126:413–425. doi: 10.1099/00221287-126-2-413. [DOI] [PubMed] [Google Scholar]
- 24.Karaolis D K R, Kaper J B. Pathogenicity islands and other mobile virulence elements of Vibrio cholerae. In: Kaper J B, Hacker J, editors. Pathogenicity islands and other mobile virulence elements. Washington, D.C.: American Society for Microbiology; 1999. pp. 167–187. [Google Scholar]
- 25.Karaolis D K R, Somara S, Maneval D R, Jr, Johnson J A, Kaper J B. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature. 1999;399:375–379. doi: 10.1038/20715. [DOI] [PubMed] [Google Scholar]
- 26.Khan M U. Fourteen years of shigellosis in Dhaka: an epidemiological analysis. Int J Epidemiol. 1985;14:607–613. doi: 10.1093/ije/14.4.607. [DOI] [PubMed] [Google Scholar]
- 27.Kleckner N. Transposon Tn10. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 227–268. [Google Scholar]
- 28.Kotloff K L, Winickoff J P, Ivanoff B, Clemens J D, Swerdlow D L, Sansonetti P J, Adak G K, Levine M M. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull W H O. 1999;77:651–665. [PMC free article] [PubMed] [Google Scholar]
- 29.Ling J M, Shaw P C, Kam K M, Cheng A F, French G L. Molecular studies of plasmids of multiply-resistant Shigella spp. in Hong Kong. Epidemiol Infect. 1993;110:437–446. doi: 10.1017/s095026880005086x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maloy S R, Nunn W D. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol. 1981;145:1110–1112. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsutani S. Genetic evidence for IS1 transposition regulated by InsA and the δInsA-B′-InsB species, which is generated by translation from two alternative internal initiation sites and frameshifting. J Mol Biol. 1994;240:52–65. doi: 10.1006/jmbi.1994.1417. [DOI] [PubMed] [Google Scholar]
- 33.Mendiola M V, de la Cruz F. IS91 transposase is related to the rolling-circle-type replication proteins of the pUB110 family of plasmids. Nucleic Acids Res. 1992;20:3521. doi: 10.1093/nar/20.13.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mendiola M V, de la Cruz F. Specificity of insertion of IS91, an insertion sequence present in alpha-haemolysin plasmids of Escherichia coli. Mol Microbiol. 1989;3:979–984. doi: 10.1111/j.1365-2958.1989.tb00247.x. [DOI] [PubMed] [Google Scholar]
- 35.Mendiola M V, Jubete Y, de la Cruz F. DNA sequence of IS91 and identification of the transposase gene. J Bacteriol. 1992;174:1345–1351. doi: 10.1128/jb.174.4.1345-1351.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller V C, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morelle G. A plasmid extraction procedure on a miniprep scale. Focus. 1989;11:7–8. [Google Scholar]
- 38.Nakata N, Sasakawa C, Okada N, Tobe T, Fukuda I, Suzuki T, Komatsu K, Yoshikawa M. Identification and characterization of virK, a virulence-associated large plasmid gene essential for intercellular spreading of Shigella flexneri. Mol Microbiol. 1992;6:2387–2395. doi: 10.1111/j.1365-2958.1992.tb01413.x. [DOI] [PubMed] [Google Scholar]
- 39.Okada N, Sasakawa C, Tobe T, Yamada M, Nagai S, Talukder K A, Komatsu K, Kanegasaki S, Yoshikawa M. Virulence-associated chromosomal loci of Shigella flexneri identified by random Tn5 insertion mutagenesis. Mol Microbiol. 1991;5:187–195. doi: 10.1111/j.1365-2958.1991.tb01839.x. [DOI] [PubMed] [Google Scholar]
- 40.Penfold R J, Pemberton J M. An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene. 1992;118:145–146. doi: 10.1016/0378-1119(92)90263-o. [DOI] [PubMed] [Google Scholar]
- 41.Rajakumar K, Bulach D, Davies J, Ambrose L, Sasakawa C, Adler B. Identification of a chromosomal Shigella flexneri multi-antibiotic resistance locus which shares sequence and organizational similarity with the resistance region of the plasmid NR1. Plasmid. 1997;37:159–168. doi: 10.1006/plas.1997.1280. [DOI] [PubMed] [Google Scholar]
- 42.Rajakumar K, Sasakawa C, Adler B. A spontaneous 99-kb chromosomal deletion results in multi-antibiotic susceptibility and an attenuation of contact haemolysis in Shigella flexneri 2a. J Med Microbiol. 1996;45:64–75. doi: 10.1099/00222615-45-1-64. [DOI] [PubMed] [Google Scholar]
- 43.Rajakumar K, Sasakawa C, Adler B. Use of a novel approach, termed island probing, identifies the Shigella flexneri she pathogenicity island, which encodes a homolog of the immunoglobulin A protease-like family of proteins. Infect Immun. 1997;65:4606–4614. doi: 10.1128/iai.65.11.4606-4614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rakin A, Noelting C, Schropp P, Heesemann J. Integrative module of the high-pathogenicity island of Yersinia. Mol Microbiol. 2001;39:407–415. doi: 10.1046/j.1365-2958.2001.02227.x. [DOI] [PubMed] [Google Scholar]
- 45.Sack R B, Rahman M, Yunus M, Khan E H. Antimicrobial resistance in organisms causing diarrheal disease. Clin Infect Dis. 1997;24(Suppl. 1):S102–S105. doi: 10.1093/clinids/24.supplement_1.s102. [DOI] [PubMed] [Google Scholar]
- 46.Sasakawa C, Kamata K, Sakai T, Murayama S Y, Makino S, Yoshikawa M. Molecular alteration of the 140-megadalton plasmid associated with loss of virulence and Congo red binding activity in Shigella flexneri. Infect Immun. 1986;51:470–475. doi: 10.1128/iai.51.2.470-475.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 48.Womble D D, Rownd R H. Genetic and physical map of plasmid NR1: comparison with other IncFII antibiotic resistance plasmids. Microbiol Rev. 1988;52:433–451. doi: 10.1128/mr.52.4.433-451.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zabala J C, de la Cruz F, Ortiz J M. Several copies of the same insertion sequence are present in alpha-hemolytic plasmids belonging to four different incompatibility groups. J Bacteriol. 1982;151:472–476. doi: 10.1128/jb.151.1.472-476.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zabala J C, Garcia-Lobo J M, Diaz-Aroca E, de la Cruz F, Ortiz J M. Escherichia coli alpha-haemolysin synthesis and export genes are flanked by a direct repetition of IS91-like elements. Mol Gen Genet. 1984;197:90–97. doi: 10.1007/BF00327927. [DOI] [PubMed] [Google Scholar]