Abstract
Aims
Agar art bridges the gap between science and art using microbes instead of paint. Afterwards, the art can change in response to microbial fluctuation, meaning preservation of the original art is essential. Here, formaldehyde and glutaraldehyde were investigated as preservatives, involving techniques used in healthcare settings to preserve samples.
Methods and Results
Formaldehyde was tested at 1.0%, 2.0% and 3.7%, w/v, whereas glutaraldehyde was tested at 1% and 2.5%, w/v. Both compounds and respective concentrations were tested for different time periods. Escherichia coli, Serratia marcescens, Staphlococcus aureus and Micrococcus luteus were used as bacteria for “drawing” the works of art. The effectiveness of fixation was determined using integrated densities and visual assessment. Initially, both compounds showed potential promise, albeit with a loss of bacteria. Ser. marcescens was prone to colour changes and glutaraldehyde caused discolouration of agar and bacteria. These could be caused by a pH decrease in the agar, due to residual free aldehyde groups. Reduction of this was tested using 300 mM sodium metabisulfite to neutralize excess aldehydes. This initially led to reduced bacterial loss and avoided colour changes, however measurements 24 h post‐fixation showed colour loss to some bacterial clusters.
Conclusions
Here, at least 2% formaldehyde for a short fixation period, typically 1 min, depending on the species, was most promising for the preservation of art. Given the success of this with different bacteria, it would make a good starting combination for anyone trying to fix agar art, although methodology refinement may be needed for optimisation depending on the bacterial species used.
Significance and Impact of Study
This study shows, for the first time, successful fixation and preservation of different bacterial species on agar. The impact of this is to preserve agar art while making it safe and non‐infective to those in contact with the microbial art.
Keywords: agar art, fixatives, microbes, microbial art, preservation
INTRODUCTION
Agar art is a form of bio‐art which involves producing images by growing various species and/or strains of bacteria or fungi on solid growth media. Effectively the agar artist is using microbes instead of paint, and agar instead of a canvas. Within the last decade, this area of microbiology has increased in popularity, possibly due to the American Society for Microbiology (ASM) launching a contest in 2015 which encouraged microbiologists from around the world to submit their agar artworks (ASM, 2019). The success of this competition, in terms of entries submitted each year has led to it becoming an annual event.
Although the competition for works of agar art is relatively new, the concept of using microbes to produce art is not new. The first attempt at producing agar art is unknown, but it has been reported that around a century ago Alexander Fleming used the different colours expressed by different bacteria to produce early examples of agar art (Adams & Hendry, 2002). However, microbes giving rise to works of art are almost certainly even older, albeit having happened inadvertently, with an example being the Bradshaw Rock art in Australia which remains vibrant despite none of the original paint remaining. This is due to microbes growing in place of the paint, thereby maintaining the outlines of the etchings, and creating a microenvironment allowing microbial growth (Pettigrew et al., 2010).
In addition, microbes are known to have other effects on historical works of art. For example many of the works of art from periods such as The Renaissance have been shown to have been historically influenced by microbes. One such is work which has analysed the microbiome of drawings by Leonardo da Vinci and shown the presence of microbes which are known to be degrader organisms (Piñar et al., 2020). In turn, this sort of information has been proposed as a means of helping to identify possible forgeries (Torralba et al., 2021), thereby demonstrating that the inter‐disciplinary nature of microbiology and art is greater than was originally first realized.
In terms of bio‐art, two principal different approaches can be taken by the artist; production of a finished piece of work following growth of the microbes over a defined period of time (e.g. after 24 h growth in an incubator) or a temporal change in the microbial pattern and colouration as different microbes dominate depending on the availability of nutrients and changes to the micro‐niche of the agar plate (e.g. changes in the factors such as osmotic levels).
Part of the problem with producing agar art over a defined time period as the final product is that it involves using living organisms. Therefore, the microbes present are at risk of dying as the nutritional resources are used up, thereby leading to a loss of at least part, if not all, of the original work. This means that the only way to preserve the original image and to have a record of the work, is to take a photograph of it. Unfortunately, if there has been any attempt to incorporate a three‐dimensional component to the work, this will be lost in a photograph. As a means of trying to maintain as much of the original form of artwork as possible, some form of biological fixation of material would be required. However, there is very little work which has been performed on fixing cells which have been grown on agar, as most preservation work involves either fixing tissue sections or cells which have been grown on a surface as part of tissue culture work (Thavarajah et al., 2012). Nevertheless, there have been a few examples of this type of work. One such example is Microbiology Infects Art (AIM) which is coordinated by Norbert Hoft at Weihenstephan‐Triesdorf University of Applied Sciences (https://www.hswt.de/person/norbert‐w‐hopf/mikrobiology‐infects‐art.html) with attempts to fix material on both agar plates and on nitro‐cellulose (Raups, 2015). Another involved looking at the vibrancy of microbes which have been grown (Schopf, 2011).
Methods for fixation are broadly classified into four groups and involve the use of one of the following groups of chemicals: aldehydes; alcohols; oxidizing agents; or metallic agents. Unfortunately, no single fixative can give the desired effect on all samples so different fixatives are used for different samples (Hobro & Smith, 2017), with there is a necessity to check the efficiency of different fixatives for different purposes (Lawrence & Singer, 1985) and even the order of work after fixing (McEwan, 1998). The temperature at which material is fixed is also critical, as elevating it can ensure morphological preservation by increasing the rate at which the chemical reaction occurs, although this is offset by the fact that it can also speed up the degeneration of any unfixed tissues (Grizzle & Fredenburgh, 2001, 2005; Rolls, 2011). Another important variable in the fixation process may be the time left to fix and allow cross‐linking, as this can continue to occur weeks after the fixative has been applied (Suvarna et al., 2018). While fixation is an attempt to prevent deterioration of the tissue during short and long‐term storage from microorganisms (Suvarna et al., 2018), there is also an acceptance that most fixation methods will give some change to tissue such as shrinkage or hardening. For this reason, it is important to investigate different factors which may influence the effectiveness and rate of a fixative.
The current work aimed to provide an evaluation and comparison of two of the more common chemicals used for fixation and different times of fixation as a means of preserving works of agar art.
MATERIALS AND METHODS
Chemicals and media
Formaldehyde (37% v/v) and sodium metabisulfite (300 mM) were purchased from Fischer Chemicals and glutaraldehyde (25% v/v) was obtained from Sigma‐Aldrich. Nutrient broth (Cat# CM0001), nutrient agar (Cat# CM003) and phosphate‐buffered saline (PBS) tablets were all purchased from Oxoid. Agar and nutrient broth were prepared following the manufacturer's instructions, with each batch being autoclaved using an ASV490 autoclave, Astell Scientific (Sidcup, England) at 121°C for 15 min at 15 psi, 103 kPa prior to use. Nutrient broth was then stored at 4°C and nutrient agar at 55°C for a maximum of 7 days before use.
Bacterial species and bacterial growth
Bacteria used were Escherichia coli 4174, Staphlococcus aureus 6571, Serratia marcescens 1377 and Micrococcus luteus 2665, and were all supplied by NCIMB (National Collection of Industrial, Food and Marine Bacteria, Aberdeen, Scotland). Aliquots of stock cultures were grown overnight in nutrient broth with gentle shaking (180 rpm) on an orbital shaker at 37°C. All agar art was carried out using 9 cm petri dishes with 1.5% (w/v) nutrient agar as a growth medium. A template, photocopied onto a piece of paper, was attached to the underside of the petri dish to provide uniformity of images being drawn. A dissection needle, which was sterilized in a Bunsen burner flame between manipulations, was used to draw images by dipping the needle into a flask of the relevant bacterium and then scoring the appropriate area of the agar as depicted by the template on the base of the plate. A maximum of one or two different bacteria were used per plate and each area was scored once at each position within the plate. Plates with M. luteus were grown for 48 h. All other combinations were grown for 24 h. All plates were set up in triplicate in anticipation of the number of fixation conditions to be used. For example, if only two conditions were being compared then six plates would be set up; where two fixative at two concentrations were used 12 plates would be prepared, etc. Likewise, the same template was used for all microbes grown at the same time to give uniformity of results.
Fixing bacterial cells and image capturing
Prior to any fixation step, images were captured of the bacteria which had grown on the agar on the petri dishes by scanning with a ProtoCol 3 (Synbiosis).
Bacteria cells on agar plates were then fixed for varying times and with varying concentrations of two different fixatives (5 ml) (see Table 1 for details). Immediately after fixing, plates were rinsed three times with 10 ml of PBS. Image capturing was performed as above immediately after the third wash and plates were returned to the incubator for a further 24 h to allow penetration of the fixative, after which image capturing of plates was repeated once more.
TABLE 1.
Fixatives used, their working concentrations, and the fixation times used prior to washing with PBS
| Fixative | Concentration | Fixation times used |
|---|---|---|
| Formaldehyde | 1.0%; 2.0%; 3.7% | 1 min; 5 min; 10 min |
| Glutaraldehyde | 1%; 2.5% | 2 min; 5 min |
Abbreviation: PBS, phosphate‐buffered saline.
ImageJ software (developed by the National Institutes of Health and Laboratory for Optical and Computational Instrumentation) was used to quantify images at all stages. The software was set to an area of 248,296 data points for each plate and the background (no bacterial growth) was subtracted to obtain the integrated density values at each point. All values were converted to percentages of the values for the plate prior to fixation for comparisons between fixation methods. After the final measurements were recorded, all plates were sealed with Parafilm to reduce levels of drying out of agar and the plates were stored at 4°C in a fridge. Plates were checked occasionally (approximately every 2–3 weeks) over a period of 3 months to determine if there had been a noticeable change in the appearance of pattern on the plates.
To determine if residual free aldehydes played a role in the level of colouration 300 mM sodium metabisulfite was added to reduce the abundance of any free aldehydes which might remain.
Statistical analysis
Plates which were prepared on the same day and had the same organisms grown on them were treated as single batches. Mean values and standard deviations were calculated for all triplicates.
Three sets of density measurements were determined for all plates; prior to the fixation step; immediately after the third post‐fixation wash step; and 24 h after fixation step. The post‐wash sample and the 24‐h sample were each expressed as a percentage of the pre‐fixation value.
Differences between samples on different groups of triplicate plates were compared by two‐way analysis of variance (ANOVA) with replication for percentage density values. Calculations were also taken for percentage density measurements immediately after the third PBS wash, and a second round of ANOVA calculations were performed 24 h later after storage at 4°C. At all points, the three replica plates had data recorded individually. Comparisons between density scores for different groups of triplicate plates were performed at 24‐h intervals using paired t‐tests. In all cases, a value of p < 0.05 was accepted as showing statistically significant differences.
RESULTS
An example of the images obtained is shown in Figure 1, with an example of a plate where Ser. marcescens and M. luteus were used to create the image. In the case of the red colouration, this was produced in the areas where Ser. marcescens was used.
FIGURE 1.
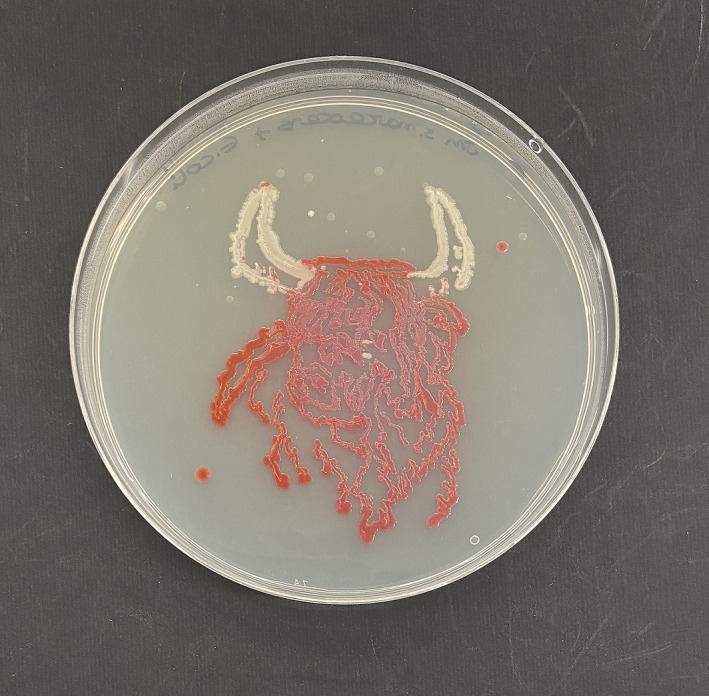
Example of agar art produced using Serratia marcescens (areas in red) and Micrococcus luteus. Images were repeated in triplicate
Values for plates with a combination of E. coli and Staph. aureus which were fixed with formaldehyde are shown in Table 2. Statistically significant (p < 0.001) differences were detected based on the formaldehyde concentration and the time of fixation, as well as the concentration × time interaction. Moreover, most samples showed a statistically significant temporal reduction in density values after 24 h, with three exceptions (3.7% formaldehyde for 5 min, 3.7% formaldehyde for 10 min and 2.0% formaldehyde for 1 min).
TABLE 2.
Mean and standard deviation (in parenthesis) ProtoCol 3 density values from plates with Escherichia coli and Staphylococcus aureus which were fixed with different concentrations of formaldehyde for either 1, 5 or 10 min
| Formaldehyde concentration (%) | Fixation time (min) | p‐values | ||||
|---|---|---|---|---|---|---|
| 1 | 5 | 10 | ||||
| Post PBS wash | 3.7 | 81 (2) | 70 (5) | 70 (2) | Concentration | <0.001 |
| 2.0 | 82 (4) | 71 (5) | 80 (3) | Fixation time | <0.001 | |
| 1.0 | 87 (6) | 81 (11) | 90 (2) | Interaction | <0.001 | |
| After 24 h | 3.7 | 72 (2) | 66 (2) | 68 (6) | Concentration | <0.001 |
| 2.0 | 87 (7) | 64 (4) | 72 (1) | Fixation time | <0.001 | |
| 1.0 | 73 (7) | 73 (8) | 80 (7) | Interaction | <0.001 | |
Note: Measurements were taken immediately after the third PBS wash and after 24 h. All values were then expressed as a percentage of the pre‐fixation density, n = 3. Statistical comparisons were made using two‐way ANOVA with replication.
Abbreviations: ANOVA, analysis of variance; PBS, phosphate‐buffered saline.
Values for plates with a combination of Ser. marcescens and M. luteus which were fixed with formaldehyde are shown in Table 3. Statistically significant differences (p < 0.05) were detected based on the formaldehyde concentration and the time of fixation, as well as the concentration × time interaction. Moreover, with the exception of the fixation with 3.7% formaldehyde for 1 min, all samples showed a statistically significant reduction in density values after 24 h. Images of agar art are shown in Figure 2 highlighting the effects of addition of 2% formaldehyde for 5 min. Images were taken before fixation, following fixation and 24 h after fixation.
TABLE 3.
Mean and standard deviation (in parenthesis) ProtoCol 3 density values from plates with Serratia marcescens and Micrococcus luteus which were fixed with different concentrations of formaldehyde for either 1, 5 or 10 min
| Formaldehyde concentration (%) | Fixation time (min) | p‐values | ||||
|---|---|---|---|---|---|---|
| 1 | 5 | 10 | ||||
| Post PBS wash | 3.7 | 85 (7) | 79 (6) | 87 (8) | Concentration | <0.001 |
| 2.0 | 92 (12) | 88 (7) | 87 (7) | Fixation time | <0.001 | |
| 1.0 | 96 (8) | 89 (8) | 84 (2) | Interaction | <0.01 | |
| After 24 h | 3.7 | 85 (10) | 63 (15) | 84 (8) | Concentration | <0.05 |
| 2.0 | 82 (9) | 78 (9) | 80 (7) | Fixation time | <0.001 | |
| 1.0 | 81 (9) | 73 (8) | 75 (4) | Interaction | <0.001 | |
Note: Measurements were taken immediately after the third PBS wash and after 24 h. All values were then expressed as a percentage of the pre‐fixation density, n = 3. Statistical comparisons were made using two‐way ANOVA with replication.
Abbreviations: ANOVA, analysis of variance; PBS, phosphate‐buffered saline.
FIGURE 2.
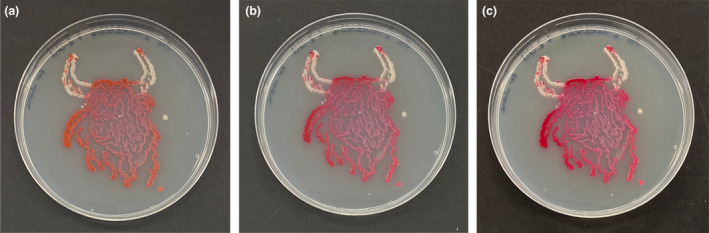
(a) Serratia marcescens and Micrococcus luteus agar art before fixation. (b) Following fixation of 2% formaldehyde for 5 min. (c) Preserved agar art 24 h after fixation
Values for plates with E. coli and S.aureus which were fixed with glutaraldehyde are shown in Table 4. No statistically significant differences were detected based on the glutaraldehyde concentration and the time of fixation. However, for the 24‐h sample only, there was a difference in terms of the concentration × time interaction. All samples showed a statistically significant (p < 0.001) reduction in density values after 24 h.
TABLE 4.
Mean and standard deviation (in parenthesis) ProtoCol 3 density values from plates with Escherichia coli and Staphylococcus aureus which were fixed with different concentration of glutaraldehyde for either 2 or 5 min
| Glutaraldehyde concentration (%) | Fixation time (min) | p‐values | |||
|---|---|---|---|---|---|
| 2 | 5 | ||||
| Post PBS wash | 2.5 | 81 (2) | 79 (1) | Concentration | 0.325 |
| 1.0 | 80 (5) | 79 (3) | Fixation time | 0.325 | |
| Interaction | 0.999 | ||||
| After 24 h | 2.5 | 38 (2) | 29 (4) | Concentration | 0.380 |
| 1.0 | 38 (2) | 31 (6) | Fixation time | <0.001 | |
| Interaction | 0.380 | ||||
Note: Measurements were taken immediately after the third PBS wash and after 24 h. All values were then expressed as a percentage of the pre‐fixation density, n = 3. Statistical comparisons were made using two‐way ANOVA with replication.
Abbreviations: ANOVA, analysis of variance; PBS, phosphate‐buffered saline.
Values for plates with Ser. marcescens and M. luteus which were fixed with glutaraldehyde are shown in Table 5. After the fixation and washing process, the density values were generally higher than those on the plate's pre‐fixation. No statistical differences were detected based on either glutaraldehyde concentration or fixation time, although the concentration × time interaction was significantly different (p < 0.001) for measurements taken immediately after the third PBS wash. However, when measurements were repeated 24 h later, significant differences were observed for both the glutaraldehyde concentration and time spent fixing, but no interaction differences were observed. Most samples showed a statistically significant reduction in density values after 24 h, with the exception of 2.5% glutaraldehyde for 5 min. It should also be noted that following the fixation with glutaraldehyde, there was an obvious yellowing of the surface of the plate which was visible to the eye. Even the use of 300 mM sodium metabisulfite to quench free aldehydes did not significantly change the reading values (data not shown). Images of agar art are shown in Figure 3 highlighting the effects of addition of 2.5% glutaraldehyde for 5 min. Images were taken before fixation, following fixation and 24 h after fixation and finally 24 h after fixation with the addition of 300 mM sodium metabisulfite.
TABLE 5.
Mean and standard deviation (in parenthesis) ProtoCol 3 density values from plates with Serratia marcescens and Micrococcus luteus which were fixed with different concentration of glutaraldehyde for either 2 or 5 min
| Glutaraldehyde concentration (%) | Fixation time (min) | p‐values | |||
|---|---|---|---|---|---|
| 2 | 5 | ||||
| Post PBS wash | 2.5 % | 117 (3) | 104 (10) | Concentration | 0.387 |
| 1.0 % | 100 (5) | 117 (8) | Fixation time | 0.397 | |
| Interaction | <0.001 | ||||
| After 24 h | 2.5 % | 92 (4) | 102 (7) | Concentration | <0.001 |
| 1.0 | Fixation time | <0.001 | |||
| 1.0 % | 83 (3) | 102 (8) | Interaction | <0.05 | |
Note: Measurements were taken immediately after the third PBS wash and after 24 h. All values were then expressed as a percentage of the pre‐fixation density, n = 3. Statistical comparisons were made using two‐way ANOVA with replication.
Abbreviations: ANOVA, analysis of variance; PBS, phosphate‐buffered saline.
FIGURE 3.
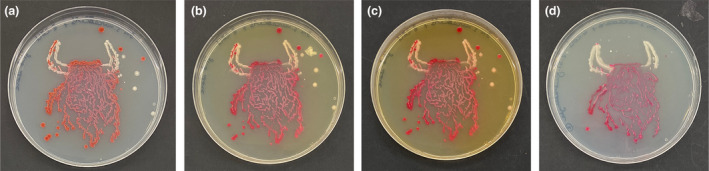
(a) Serratia marcescens and Micrococcus luteus agar art before fixation. (b) Following fixation of 2.5% glutaraldehyde for 5 min. (c) Preserved agar art 24 h after fixation with glutaraldehyde. (d) Preserved agar art 24 h after fixation with glutaraldehyde and addition of sodium metabisulfite
Following the measurements at 24 h, the agar plates were placed in a fridge at 4°C. Over the course of the next 3 months, agar plates were inspected by eye, although quantitative measurements were not repeated. During this time most plates did not show any obvious signs of change, and despite the thickness of the agar reducing, the impact of drying out generally did not impact on the works of bio‐art.
DISCUSSION
The data presented here demonstrate that, as with most requirements for the fixation of biological material, there is no single solution to the problem. However, the work does show that there is the potential for some level of fixation to take place.
Two different fixatives were used in this work; formaldehyde and glutaraldehyde. They are both aldehyde molecules and react with amines, but with different modes of action. Formaldehyde works by forming covalent bonds between proteins; specifically, it reacts with an available free amine on a protein, producing a Schiff base, followed by reacting with another free amine leading to a covalent bond being created. This then maintains the integrity of the lipid membrane and thus preserves the bacteria (Chao & Zhang, 2011). Glutaraldehyde is a carbonyl reagent which condenses amines through Mannich reactions and/or reductive amination.
In addition to having different chemical properties as fixatives, the two are also different in size, with glutaraldehyde molecules being considerably larger (molecular weight of 100.11 Da), than the formaldehyde molecules (molecular weight of 30.026 Da) and so are considered to need longer times to penetrate the material, which leads to longer fixation times (Rolls, 2011). Moreover, there are differences in terms of concentrations which are routinely used for fixation, for example 2.5% glutaraldehyde as opposed to 2.0% for formaldehyde (Chao & Zhang, 2011).
It should be noted that generally after the fixation process, followed by the PBS washes, the reading values generally drop below that originally recorded prior to fixation. The one exception to this pattern was seen in the case of plates where Ser. marcescens and M. luteus had been grown and then fixed with glutaraldehyde (Table 5). Even after 24 h, these values had not dropped for fixation times of 5 min with the two concentrations of glutaraldehyde (1% and 2.5%). However, although this provided some degree of encouragement for glutaraldehyde as a fixative with agar art works, this was not the case when E. coli and Staph. aureus had been fixed with it, with a loss of density values in samples immediately after the fixation and PBS wash, and even more so when measurements were repeated 24 h later (Table 4). This loss after 24 h typically resulted in values of less than 40% of the original pre‐fixation value. Moreover, many of the agar plates had a yellowish appearance to them after fixation with glutaraldehyde (Figure 2c), and therefore had an impact on the physical appearance of the agar art, which defeated the purpose of the fixation process. Taking these factors into consideration, it was decided that although the use of glutaraldehyde may be useful for fixation of some bacteria grown on agar plates, it was generally unreliable and prone to a change in physical appearance of the agar plates within the areas where no bacteria were streaked and so had no bacterial growth.
The other example of a chemical routinely used for fixation which was tested was formaldehyde. This routinely gave a slightly lower reading after the post‐fixation wash measurement, and a further drop after the measurement 24 h later. However, there was never the same level of loss of measurement seen with the glutaraldehyde, and neither was the extensive discolouration observed after 24 h. As with the use of glutaraldehyde, the plates which had a fixation with formaldehyde had a lower set of measurements with the E. coli and Staph. aureus plates (Table 2 vs. Table 3) but never had values dropping down to the same extent as was seen with the equivalent plates fixed with glutaraldehyde (Table 4).
Although statistically significant differences were seen for formaldehyde concentration, fixation time, and the interaction between the concentration and time, there was no obvious pattern emerging in terms of a recommended combination of formaldehyde concentration for a specified fixation time as, again, there were differences seen depending on the species of organism which had been used to generate the piece of agar art.
It is also worth noting that following casual inspection on a regular basis for around 3 months after the quantitative measurements had been completed, there was no obvious deterioration in terms of the image. Although the depth of the agar reduced over time, presumably due to some level of drying out, although as mentioned already this did not obviously impact on the appearance of the bio‐art.
The use of biological fixation has the additional bonus of enhancing biological safety aspects as the fixation process should kill all of the microbes being used in the production of the art. In our work, we only used strains of species which required low‐level containment facilities, but in the event of someone wanting to use a slightly higher risk organism, the fixation process should reduce the risk of having this work on public display.
In conclusion, for anyone trying to fix a piece of agar art using a biological fixative, we would recommend using formaldehyde as the fixative of choice, to minimize the potential for damage to the colour of the agar outwith the areas which have been drawn upon. However, the selection of formaldehyde concentration and time of fixation used will probably have to be developed depending on the choice of bacterial species used for producing the agar art.
CONFLICT OF INTEREST
No conflict of interest declared.
Wilson, S. , Law, S.P. , McEwan, N.R. , Wright, R. & Macaskill, J.S. (2022) Development of a fixative protocol using formaldehyde and gluteraldehyde for preservation of microbial art on agar plates. Journal of Applied Microbiology, 133, 665–672. Available from: 10.1111/jam.15597
Funding information
Partially funded by Society for Applied Microbiology and partially by departmental research fund.
REFERENCES
- Adams, M. & Hendry, P. (2002) The lost art of bacteriology. The Microbiologist, 14–15. [Google Scholar]
- American Society for Microbiology (2019) What is ‘agar art’? Available at: https://asm.org/Events/2019‐ASM‐Agar‐Art‐Contest/Home Accessed March 14, 2020.
- Chao, Y. & Zhang, T. (2011) Optimisation of fixation methods for observation of bacterial cell morphology and surface ultrastructures by atomic force microscopy. Applied Microbiology and Biotechnology, 92(2), 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grizzle, W.E. & Fredenburgh, J. (2001) Avoiding biohazards in medical, veterinary and research labs. Biotechnic & Histochemistry, 76, 183–206. [PubMed] [Google Scholar]
- Grizzle, W.E. & Fredenburgh, J. (2005) Safety in biomedical and other laboratories. Molecular Diagnostics, 33, 421–428. [Google Scholar]
- Hobro, A.J. & Smith, N.I. (2017) An evaluation of fixation methods: spatial and compositional cellular changes observed by Raman imaging. Vibrational Spectroscopy, 91, 31–45. [Google Scholar]
- Lawrence, J.B. & Singer, R.H. (1985) Quantitative analysis of in situ hybridization methods for the detection of actin gene expression. Nucleic Acids Research, 13, 1777–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NR, M.E. (1998) Immunocytochemistry and in situ hybridization ‐ is the order important? The World Wide Web Journal of Biology, 3, 3. [Google Scholar]
- Pettigrew, J. , Callistemon, C. , Weiler, A. , Gorbushina, A. , Krumbein, W. & Weiler, R. (2010) Living pigments in Australian Bradshaw rock art. Antiquity, 326, f1–f6. [Google Scholar]
- Piñar, G. , Sclocchi, M.C. , Pinzari, F. , Colaizzi, P. , Graf, A. , Sebastiani, M.L. et al. (2020) The microbiome of Leonardo da Vinci's drawings: a bio‐archive of their history. Frontiers in Microbiology, 11, 593401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raups S. Microbiology infects art Konservierung der Kunst . Available at: https://www.hswt.de/fileadmin/_processed_/csm_Konservierung_der_Kunst_711039dce6.jpg Accessed April 14, 2022
- Rolls, G. (2011) Fixation and fixatives (2) – factors influencing chemical fixation, formaldehyde and glutaraldehyde. Deer Park, IL: Leica Biosystems Publications. [Google Scholar]
- Schopf, E. (2011) Eine Symbiose aus Kunst und Wissenschaft: Malen mit Bakterien. Labor&more, 1.11, 22–31. [Google Scholar]
- Suvarna, S.K. , Layton, C. & Bancroft, J.D. (Eds.) (2018) Chapter 4: Fixation of tissues. In: Bancroft's theory and practice of histological techniques, 8th edition, pp. 40–63. London, UK: Elsevier. [Google Scholar]
- Thavarajah, R. , Mudimbaimannar, V.K. , Elizabeth, J. , Rao, U.K. & Ranganathan, K. (2012) Chemical and physical basics of routine formaldehyde fixation. Journal of Oral & Maxillofacial Pathology, 16, 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torralba, M.G. , Kuelbs, C. , Moncera, K.J. , Roby, R. & Nelson, K.E. (2021) Characterizing microbial signatures on sculptures and paintings of similar provenance. Microbial Ecology., 81, 1098–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]


