Abstract
Transcription factor PrfA controls the expression of virulence genes essential for Listeria monocytogenes pathogenesis. To gain insight into the structure-function relationship of PrfA, we devised a positive-selection system to isolate mutations reducing or abolishing transcriptional activity. The system is based on the observation that the listerial iap gene, encoding the p60 protein, is lethal if overexpressed in Bacillus subtilis. A plasmid in which the iap gene is placed under the control of the PrfA-dependent hly promoter was constructed and introduced into B. subtilis. This strain was rapidly killed when expression of iap was induced by introduction of a second plasmid carrying prfA. Two classes of B. subtilis survivor mutants were identified: one carried mutations in iap, and the second carried mutations in prfA. Sequence analysis of the defective prfA genes identified mutations in three regions of the PrfA protein: region A, between amino acids 58 and 67 in the β-roll domain of PrfA; region B, between amino acids 169 and 193, which corresponds to the DNA-binding helix-turn-helix motif; and region C, comprising the 38 C-terminal amino acids of PrfA, which form a leucine zipper-like structure. PrfA proteins with mutations in regions B and C were unable to bind to the PrfA-binding site in the target DNA, while mutations in region A resulted in a protein still binding the target DNA but unable to form a stable complex with RNA polymerase and initiate transcription in vitro.
The PrfA protein controls the expression of a regulon comprising essential virulence genes required by facultative intracellular pathogen Listeria monocytogenes to colonize its vertebrate hosts (17, 26, 31, 51). The occurrence of the prfA gene is not restricted to food-borne human pathogen L. monocytogenes (15, 17, 33); it is also present in animal pathogen Listeria ivanovii (32). In these two pathogenic Listeria species the prfA gene is part of a chromosomal island clustering several virulence genes, all regulated by PrfA, including prfA itself (25, 50). Recent studies have shown that other virulence-associated genes located outside of this gene cluster (including especially internalin [inl] genes) are also regulated by PrfA (12, 13, 14, 34, 37, 41, 43).
PrfA belongs to the large cyclic AMP (cAMP) receptor protein (CRP)/Fnr family of bacterial transcription factors, which are predominantly found in gram-negative bacteria. The amino acid sequence of PrfA shows substantial similarity to that of CRP of Escherichia coli (31), a key mediator of catabolite repression in conjunction with activator cofactor cAMP (8, 23). Recent studies indicate that structural conservation between CRP and PrfA is also functionally relevant, as deduced from mutational analysis of the helix-turn-helix domain interacting with DNA (46). Also, mutations in amino acids known to lead to cAMP-independent, constitutively active CRP proteins lead in the listerial regulator to PrfA protein forms which strongly activate PrfA-dependent expression in vivo (42) and which bind more strongly to PrfA-binding sites in vitro than wild-type PrfA (52). cAMP is not known to be produced by gram-positive bacteria, and thus it is not surprising that the cAMP-binding site of CRP is not well conserved in PrfA (52). There are, however, interesting differences between these two regulatory proteins, such as a second putative helix-turn-helix motif from amino acids 7 to 30 present in PrfA but absent in CRP and especially the additional 30-amino-acid C terminus of PrfA, which shows a leucine zipper-like motif (31).
To approach the structure-function analysis of PrfA, we developed a positive-selection system for the isolation of mutant PrfA proteins with reduced or lost activity. In our selection procedure, we used the listerial iap gene, encoding the extensively studied p60 protein, one of the major secreted proteins and a major protective antigen of L. monocytogenes (6, 22, 38). This protein possesses peptidoglycan hydrolase activity but seems to enhance also the uptake of L. monocytogenes by mammalian host cells (19, 27) especially by fibroblasts (28). Proteins related to p60 of L. monocytogenes were also found in all other listerial species (7). They all contain peptidoglycan hydrolase activity but differ from the L. monocytogenes p60 protein by the absence of a repeat sequence which is exclusively found in the central part of the L. monocytogenes protein (7) and which may be involved in the above-described enhanced internalization of L. monocytogenes. The isolated p60 protein lyses gram-positive bacteria including Bacillus subtilis, and the p60-like protein of Listeria grayi seems to exert the most efficient lytic activity on B. subtilis. In contrast to that of most virulence genes of L. monocytogenes, the expression of the iap gene is independent of PrfA (48) and appears to occur constitutively under all conditions studied (53).
We reasoned that overexpression of the iap gene in B. subtilis by a PrfA-dependent promoter in the presence of PrfA would kill the bacteria unless either the iap gene or the prfA gene was inactivated by mutations in functionally important domains of the two encoded proteins, p60 and PrfA. In this paper we show that this approach is feasible and leads to the expected mutant proteins. We present data on the prfA mutant alleles we generated with this strategy; these alleles encode a variety of PrfA− protein variants affected in obviously important functional domains of the listerial virulence gene regulator.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
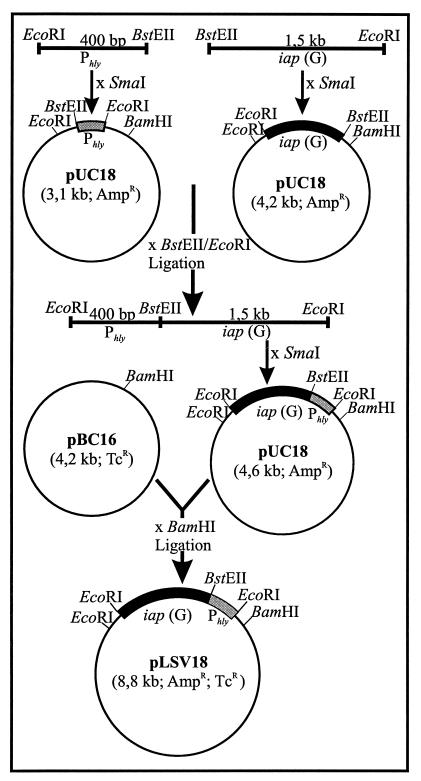
L. monocytogenes serovar 1/2a EGD was obtained from S. H. E. Kaufmann (Max-Planck-Institut für Infektionsbiologie, Berlin, Germany). Construction of the isogenic prfA deletion mutant has been described previously (47). The L. monocytogenes rough mutant RIII is derived from a smooth strain of serovar 1/2a (39) and was obtained from J. Potel (Institute for Medical Academy, Hannover, Germany). L. grayi was obtained from the Special Listeria Culture Collection (Institute for Hygiene and Microbiology, University of Würzburg, Würzburg, Germany). B. subtilis DB104 (hisH nprR2 nprE18 ΔaprE) was derived from R. Doi (University of California, Davis, Davis). This strain carries lesions in the structural genes for extracellular neutral (nprE) and serine (aprA) proteases (21). Plasmid pUC18 (57) was used for cloning experiments with E. coli JM109. Construction and use of plasmid shuttle vector pERL3502 carrying an erythromycin resistance gene and the prfA and plcA genes of L. monocytogenes have been previously described (33, 48). Plasmid pLSV 18 consists of gram-positive plasmid pBC16-1 (24), carrying a tetracycline resistance gene, and pUC18. Further details about its construction are given in Fig. 1.
FIG. 1.
Construction of the Listeria-E. coli shuttle vector pLSV18 carrying iap(G) under the control of the hly promoter, Phly. The DNA sequences of all primers used for amplification of cloned fragments are given in Materials and Methods.
Media and reagents.
L. monocytogenes and B. subtilis strains were grown in brain heart infusion (BHI) broth (Difco Laboratories, Detroit, Mich.). E. coli strains were cultured in Luria-Bertani media. Erythromycin (5 μg/ml for Listeria or Bacillus strains; 300 μg/ml for E. coli strains), 7.5 μg of tetracycline/ml, or 50 μg of ampicillin/ml (E. coli) was added to broth or agar when required. Electroporation and protoplast transformation methods were used as previously described (55, 56). DNA-modifying enzymes were purchased from Roche (Mannheim, Germany), Pharmacia (Uppsala, Sweden), and Eurogentec (Brussels, Belgium). [γ-32P]ATP (3,000 Ci/mmol) was purchased from Amersham (Braunschweig, Germany).
Amplification procedures, DNA manipulations, and DNA sequencing.
For amplification of the iap gene under the control of the Phly promoter, primer pairs 5′-ATGTCGAATTCCATGGTCATCGTATCATGTGTACCTG-3′ (EcoRI) and 5′-TCATGGGTTACCCTCTCCTTCTAC-3′ (BstEII), amplifying the Phly region, and 5′-GAGAGGGTTACCATGAATATGAAAAAAGCAACGATCG-3′ (BstEII) and 5′-AAGTGAATTCTTATACGCGACCGAAGCCAA-3′ (EcoRI), for the iap open reading frame, were used. For cloning experiments, restriction sites (underlined; the associated restriction endonucleases are in parentheses) were introduced into the primers. The amplification of DNA sequences by PCR was performed in a 100-μl reaction volume in accordance with protocols by Innis et al. (20). DNA sequencing was performed with the T7 sequencing kit (Pharmacia) and various internal primers from the known iap and hly gene sequences.
The procedures for isolation of plasmid DNA from E. coli and for recombinant DNA techniques were according to standard protocols (45).
Isolation of plasmid DNA from B. subtilis and L. monocytogenes was performed using the Nucleobond kit (Macherey-Nagel). The manufacturer‘s protocol was modified by incubation of the bacteria in buffer S1 with 10 mg of lysozyme/ml for 5 min at 37°C.
For PCR amplifications, chromosomal DNA of L. monocytogenes was obtained by incubating bacteria in 1× PCR buffer (Eurogentec) containing 2.4 mg of lysozyme (Sigma, St. Louis, Mo.)/ml for 15 min at 37°C and then incubating them for 15 min at 56°C with proteinase K (final concentration, 0.24 mg/ml; Sigma). The lysis procedure was completed by incubating bacteria for 5 min at 110°C and transferring them immediately on ice. For PCR, 1 to 2 μl from this crude lysate was used.
Preparation of the anti-L. grayi p60 antiserum.
To generate an antiserum specific for the L. grayi p60, synthetic peptide KQLNKLDSDRIVPG (positions 54 to 67 from the N terminus) was derived from the known amino acid sequence (7). The peptide was produced as a multiple-antigen peptide immobilized via a lysine branch to a polyethylene glycol resin (10) in a peptide sequencer (Applied Biosystems, Foster City, Calif.) with 9-fluorenylmethoxy carbonyl chemistry (49). By using this type of peptide immobilization, no subsequent coupling to a carrier protein is necessary. A New Zealand White rabbit (Charles River, Kisslegg, Germany) was subcutaneously injected with 750 μg of the peptide emulsified in oil adjuvant for primary immunization, boosted three times with 750 μg of the peptide, also emulsified in oil adjuvant, on days 14, 21, and 28, and then bled after 38 days (18). The serum was stored in aliquots at −20°C. The anti-PrfA antiserum, generated by immunization of a guinea pig with a recombinant PrfA carrying a histidine tag, was described recently (2). Preparation of the rabbit anti-L. monocytogenes p60 antiserum has been described earlier (22).
Protein preparations, SDS-PAGE, and immunoblotting.
Supernatant proteins were precipitated with trichloroacetic acid (7% [vol/vol]), washed with acetone, and solubilized in Laemmli sample buffer (22, 30). Total cellular proteins from 2 ml of a Bacillus or Listeria culture grown to logarithmic phase were obtained by incubating bacteria for 15 min at 37°C in 20 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA; pH 8.0) containing 20 mg of lysozyme (Sigma)/ml. After 5 μl of fivefold-concentrated Laemmli sample buffer was added, bacteria were heated for 5 min at 95°C. Nucleic acids were then degraded by incubating the samples for 10 min at 37°C in the presence of 11 U of Benzonase (Sigma). All samples were heated at 95°C for 5 min before electrophoresis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 12.5% polyacrylamide gels was used for protein separations as previously described (30). For immunoblotting, proteins were transferred from SDS-polyacrylamide gels by semidry electrotransfer (29). Nitrocellulose filters were blocked and incubated in either anti-p60(M) (L. monocytogenes) (22) or anti-p60(G) (L. grayi) or anti-PrfA antiserum and horseradish peroxidase-conjugated anti-rabbit or anti-guinea pig immunoglobulins (DAKO, Hamburg, Germany). The blot was developed with 4-chloro-1-naphthol (0.5 mg/ml) and hydrogen peroxide (0.025%). All comparative immunoblotting was performed three times.
Purification of overexpressed His-tagged proteins.
The mutant prfA genes were cloned into pQE30 expression vectors (Qiagen, Hilden, Germany). The six-His-tagged PrfA proteins were purified by chromatography by Ni-nitrilotriacetic acid affinity chromatography (Qiagen) as previously described (2). Protein purity was analyzed by Coomassie staining of SDS-polyacrylamide gels as described by Laemmli (30).
EMSA.
Electrophoretic mobility shift assays (EMSA) with protein extracts were performed as described by Böckmann et al. (2). EMSA with the purified mutant PrfA proteins were performed with the 109-bp DNA probe of the hly promoter region as described previously (11). For CI complex formation, partially purified RNA polymerase (RNAP) from L. monocytogenes, cultivated in BHI medium and shifted to minimal medium for 30 min (RNAPMEM) (M. Lalic-Muelthaler, J. Bohne, and W. Goebel, submitted for publication), was added to the binding assay mixtures containing PrfA bound to the DNA. These were further incubated for 5 min at 37°C and 10 min on ice.
Runoff in vitro transcription assays.
In vitro transcription was performed in runoff experiments as previously described (1; Lalic-Muelthaler et al., submitted) using the probe of the plcA promoter region (1), resulting in a 109-base transcript and the same RNAP as that described above (see “EMSA”).
RESULTS
Overexpression of iap(G) under the control of the hly promoter in B. subtilis is lethal in the presence of functional PrfA.
Plasmid pLSV18 was constructed as shown in Fig. 1. It carries the iap gene of L. grayi, iap(G), under the control of the L. monocytogenes PrfA-dependent hly promoter, Phly, instead of the L. monocytogenes iap gene, iap(M), since previous studies have shown that p60(G), the product of the iap(G) gene, exhibits the highest bacteriolytic activity of all listerial p60 proteins (A. Bubert, unpublished data).
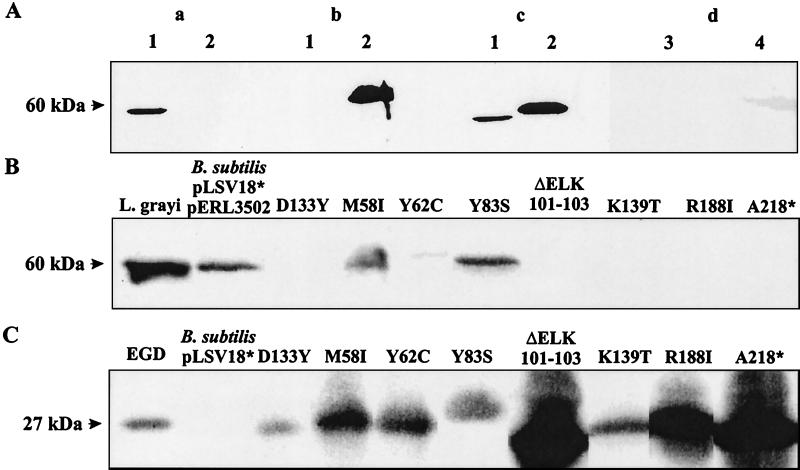
To test whether induced expression of iap(G) in the presence of PrfA may be lethal for B. subtilis, we transformed B. subtilis DB104 (21) with pLSV18 and pERL3502. The latter plasmid carries the prfA gene under the control of PrfA-specific promoters P1 and P2 and the PrfA-dependent plcA promoter (16), resulting in efficient synthesis of PrfA (5). In the transformation protocol we first introduced pLSV18 into DB104 and monitored the expression of p60(G) with an antiserum which was raised against a p60(G)-specific peptide (7). As shown in Fig. 2A, this antiserum specifically recognized p60(G) in the supernatant of L. grayi. When the supernatant of B. subtilis DB104(pLSV18) transformants were analyzed, only small amounts of p60(G) were detected (Fig. 2A, lane d4), suggesting that expression of Phly-iap(G) in B. subtilis is very low in the absence of PrfA.
FIG. 2.
Specific recognition of p60(G) with anti-L. grayi p60 antiserum and determination of extracellular p60(G) produced by group II transformants of B. subtilis carrying pLSV18 and pERL3205. (A) Immunoblotting of culture supernatant proteins (1 ml) was performed with p60(G) antiserum (a), L. monocytogenes p60 antiserum (b), or a cocktail of both antisera (same dilutions) (c). Lanes: 1, supernatant of L. grayi; 2, supernatant of L. monocytogenes EGD. (d) Supernatants of B. subtilis DB104 (lane 3) and B. subtilis carrying pLSV18 and pERL3502 (lane 4). Proteins from supernatants (2 ml) were separated by SDS-PAGE and immunostained with the anti-p60 antisera (1:400 dilution). (B) Supernatant from the p60(G) protein of group II transformants of B. subtilis carrying pLSV18 and pERL3502. Immunoblotting of proteins from culture supernatants (2 ml) was performed with anti-p60(G) antiserum (1:400 dilution). As controls equal volumes of supernatant from L. grayi and B. subtilis pLSV18* and pERL3502 were used. pLSV18* produces an inactive p60 protein. (C) PrfA protein of the same transformants of B. subtilis carrying pLSV18 and pERL3502. Immunoblotting of bacterial lysates, separated by SDS-PAGE, was performed with PrfA antiserum (1:500 dilution). Controls are lysates from L. monocytogenes EGD (positive) and B. subtilis pLSV18* without pERL3502 (negative). The data shown are derived from differently developed immunoblots of various gel runs, which explains the different intensities of the PrfA protein bands.
DB104(pLSV18) was then transformed with pERL3502. The expected double transformants were selected on agar plates containing tetracycline (pLSV18 carries a tetracycline resistance gene) and erythromycin (resistance to this antibiotic is carried by pERL3502). Only a few colonies were repeatedly obtained in these transformation assays (about 10 per assay, while more than 104 transformants were routinely obtained when pERL3502 or pLSV18 was separately transformed into DB104). The resistance to both antibiotics suggested the presence of both plasmids in these transformants.
We next analyzed the presence of p60(G) in the culture supernatants of these transformants with p60(G) antiserum. Based on the amount of produced p60(G), the obtained transformants fell into three different groups: group I transformants did not produce any detectable p60(G), group II transformants secreted amounts of p60(G) as small as that secreted by the B. subtilis strain without pERL3502, and group III transformants secreted large amounts of p60(G). Further analysis of the iap(G) and prfA genes by PCR with iap- or prfA-specific primers revealed that group I transformants carried an intact prfA gene but that the iap(G) gene was deleted. Group III transformants carried an intact prfA gene, which led to the observed induced production of p60(G), but the p60(G) proteins produced were functionally inactive, as revealed by a previously described chain disruption assay using L. monocytogenes mutant RIII (5, 22) (data not shown).
Group II transformants, which did not produce large amounts of p60 (Fig. 2B), were tested for the presence of PrfA by immunoblotting with anti-PrfA antiserum. Some of these group II transformants did not produce any PrfA, and PCR analysis revealed the complete loss of the prfA gene in these transformants. Other transformants of this group synthesized PrfA proteins whose sizes were the same as or smaller than that of the wild-type PrfA protein (Fig. 2C). These PrfA proteins were apparently strongly impaired in their ability to activate expression of Phly-iap(G), suggesting that they carried mutations causing the loss or reduction of their transcriptional activity.
Sequence analysis of the prfA genes of group II transformants.
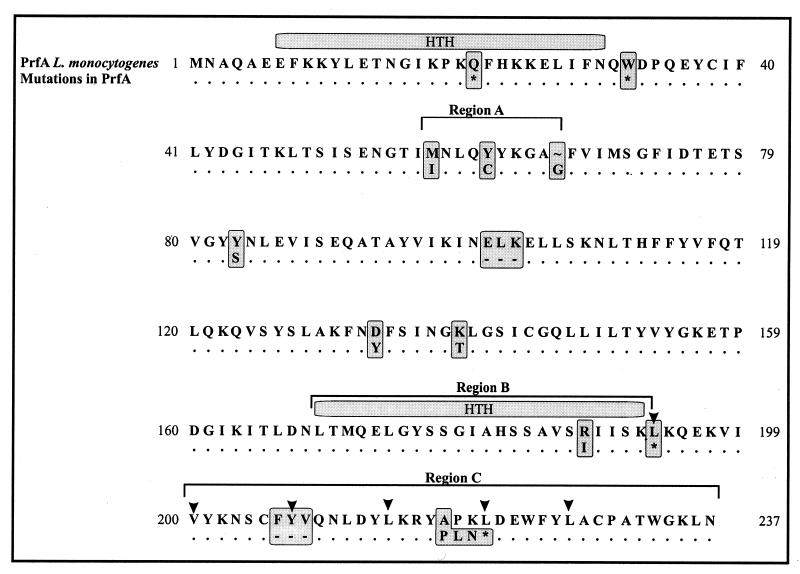
We sequenced all prfA genes of type II transformants that showed normal-size prfA-specific PCR products and some prfA genes of reduced size. Figure 3 summarizes the results of this sequence analysis. As expected, all prfA genes which yielded PCR products with normal sizes carried either missense or nonsense point mutations, leading to PrfA proteins with single amino acid exchanges or to shorter PrfA products. Two of the mutant prfA genes carried in-frame deletions leading to PrfA products which had lost three amino acids. Interestingly, most of the missense point mutations were clustered in three regions of the prfA gene, corresponding to amino acids 58 to 67 (region A), 169 to 193 (region B), and the C-terminal 38 amino acids (region C). In the last region we observed an interesting mutation resulting in loss of most of the C-terminal extension of PrfA, which is missing in the related CRP of E. coli. This last deletion and some of the described missense mutations (especially mutation R188I) were obtained several times in independent mutagenesis experiments with B. subtilis, suggesting that our collection of prfA mutations probably includes some affecting amino acid positions critical for PrfA function. This procedure will not identify mutations in the promoter(s) responsible for the expression of prfA in pERL3502 since these would result in the loss of prfA transcription.
FIG. 3.
Amino acid exchanges in the mutant PrfA proteins obtained from the group II transformants of B. subtilis DB104 carrying pLSV18 and pERL3502. Upper lane, amino acid sequence encoded by wild-type prfA; the amino acids below indicate the altered positions in the various group II mutants. HTH, helix-turn-helix motif; arrowheads, putative leucine zipper motifs; asterisk, stop codon; -, deleted amino acids. Each marked amino acid alteration belongs to an individual mutant.
Binding of mutant PrfA proteins to the PrfA-binding site of the hly/plcA promoter region.
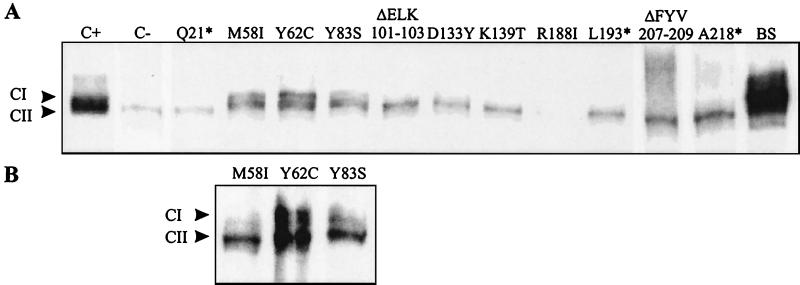
To test whether the loss of or reduction in transcriptional activation of the PrfA-dependent hly promoter by the mutant PrfA proteins observed in B. subtilis is caused by an impaired capacity to bind to the target DNA sequence (the PrfA box) or to other factors required for transcription initiation of PrfA-dependent genes (2, 11, 40), we prepared bacterial extracts of the B. subtilis transformants carrying these mutant prfA genes. These bacterial extracts were tested by EMSA using a P32-labeled DNA fragment carrying the PrfA box shared by the hly and plcA genes. As shown in Fig. 4A, extracts containing mutant PrfA proteins were either unable to form a CI complex (RNAP plus PrfA bound to the promoter [11]) with this DNA fragment (Q21*, ΔELK101-103, D133Y, K139T, R188I, L193*, DFYV207-209, and A218*) or showed strongly reduced CI formation (M58I, Y62C, and Y83S). Extracts of a prfA-deleted L. monocytogenes strain expressing the last three mutant PrfA proteins also showed reduced CI formation (Fig. 4B), while this L. monocytogenes strain, carrying the other prfA mutants, did not yield any CI complex (data not shown).
FIG. 4.
(A) EMSA with a 32P-labeled hly promoter DNA fragment and protein extracts of B. subtilis DB104 transformants carrying the various mutated prfA genes (see Fig. 3). As controls protein extracts of L. monocytogenes EGD (C+), L. monocytogenes prfA mutant Δ42 (C−), and B. subtilis DB104 carrying pERL3502 (BS) were used. (B) EMSA with protein extracts from L. monocytogenes Δ42 with mutant PrfA proteins M58I, Y62C, and Y83S. CI and CII, complexes of target DNA with PrfA and RNAP (CI) or with target DNA and RNAP alone (CII) (11).
We further studied the functional defect in these PrfA mutants using purified proteins. For this analysis we selected mutants M58I (region A), R188I (region B), and A218* (region C) (Fig. 3). As shown previously (2), PrfA tagged with six His’s at the N terminus retains full activity in vitro and in vivo. The binding of these proteins to the hly promoter region was assessed by an EMSA as described before using a double-stranded oligonucleotide carrying the PrfA box and the −10 motif (35).
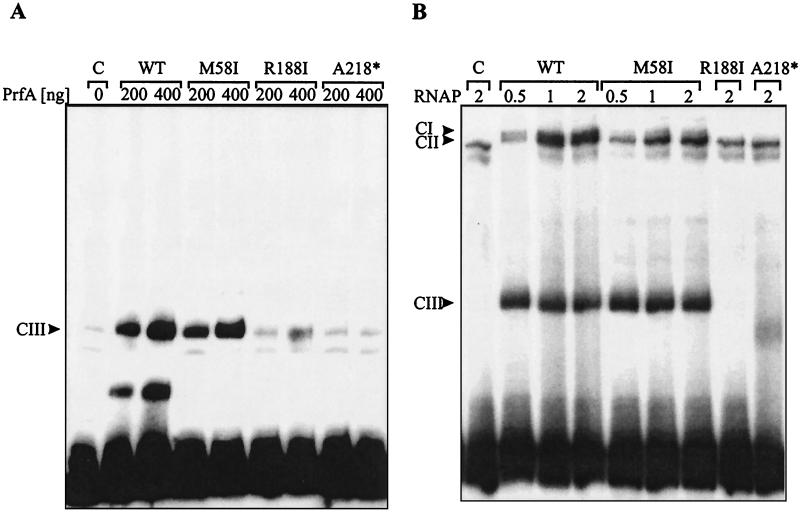
As shown in Fig. 5A, the wild-type PrfA protein led to the formation of the CIII complex in a dose-dependent fashion, as previously described (11). The PrfA protein with a mutation in region A (mutant M58I) also yielded a CIII complex, albeit at a reduced efficiency (about 60% compared to that for the wild-type PrfA protein). In contrast, the PrfA proteins with mutations in region B and C did not yield CIII complexes or yielded them in very small amounts.
FIG. 5.
EMSA with 32P-labeled hly promoter DNA fragment and purified wild-type (WT) and mutant PrfA proteins M58I, R188I, and A218* (see Fig. 3). (A) Formation of CIII complex with 200 or 400 ng of PrfA and target DNA. Arrowhead, position of the CIII complex. For a discussion of the other bands in the control (C) and the EMSA with the wild-type and mutant PrfA proteins see text. (B) Formation of CI complex with increasing amounts of partially purified RNAP (0.5, 1.0, and 2.0 ng of RNAPMEM) and 200 ng of PrfA per lane. CI, CII, and CIII, positions of the various formed complexes (see text for details). Control (C) contains the 32P-labeled promoter probe but no PrfA protein. The band seen below the CIII complexes and in the control is of unknown origin and was seen with this promoter probe but not with other preparations. Note again the complete absence of the CIII complex in the EMSA with the mutant PrfA proteins R188I and A218*.
The minor band migrating at a position similar to that for the CIII complex, seen in the shift experiments with PrfA mutant proteins R188I and A218*, seems to derive mainly from the oligonucleotide probe as this band is present in a similar concentration in the control lane (C), which contains the same amount of the radioactively labeled oligonucleotide used in the shift experiments but no PrfA protein. The band migrating below the CIII complex in the EMSA with wild-type PrfA is of unknown nature but is often observed at higher PrfA concentrations and may represent a complex of monomeric PrfA bound to the oligonucleotide probe.
The data suggest that the region A PrfA mutant protein retained the capacity to bind to its target DNA while the PrfA proteins with mutations in regions B and C lost this binding capacity.
Previous studies have shown that the CIII complex is shifted to the CI complex by adding protein extract from a prfA deletion mutant (11) or partially purified listerial RNAP. This shift was readily obtained in the control with wild-type PrfA (Fig. 5B), whereas with PrfA from mutant M58I mainly the CII complex (RNAP bound to the promoter [11]) and little CI complex (about 5 to 10% of the amount of CII complex observed with the wild-type PrfA) was obtained. This inability of M58I to shift complex CIII to CI is not due to a reduced amount of CIII complex as the levels of CIII complexes for the wild-type PrfA and the M58I mutant PrfA proteins are similar (Fig. 5B). These results suggest that mutant M58I PrfA protein can still interact with its target site but is strongly impaired in its ability to bind RNAP. As expected no CI complex formation was observed with PrfA mutant proteins R188I and A218* (Fig. 5B).
Activity of the PrfA− mutant protein in an in vitro transcription assay.
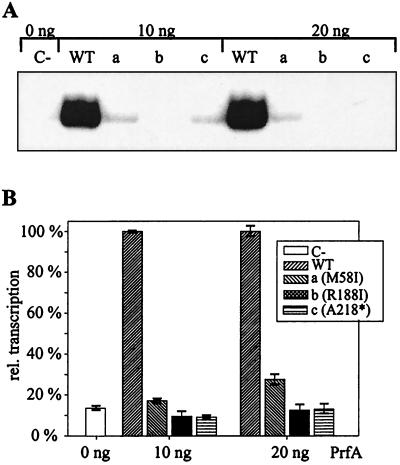
The lack of CI complex formation also suggested that the three mutant PrfA proteins may not be able to initiate transcription starting at PrfA-dependent promoters. This was directly tested in a recently developed in vitro transcription assay using partially purified RNAP (1; Lalic-Muelthaler et al., submitted). For the runoff assay we applied a DNA template which initiates transcription at the PrfA-dependent plcA promoter (PplcA) and RNAP from L. monocytogenes (RNAPMEM) cultivated in BHI medium and shifted into minimal medium for 30 min (Lalic-Muelthaler et al., submitted). As shown in Fig. 6, transcription starting from PplcA was obtained in the presence of the wild-type PrfA protein but not with the mutant PrfA proteins (R188I and A218*). Transcriptional activation of the mutant M58I is strongly reduced compared to that of the wild-type PrfA protein (about 5% residual activity with 20 ng of mutant PrfA protein), which is in accord with the reduction in CI complex formation shown in Fig. 5B.
FIG. 6.
(A) In vitro transcription starting from the plcA promoter with RNAPMEM (0.25 ng) and the purified wild-type (WT) or mutant PrfA proteins (a to c) or without PrfA (C-). Purified PrfA proteins (10 and 20 ng) were applied. Runoff assays were carried out as described previously (1). (B) Densitometric analysis of the autoradiograms shown in panel A. Transcription efficiency with wild-type PrfA was set to 100%.
DISCUSSION
We report here the identification of mutant proteins which are impaired in the transcriptional activation of PrfA by taking advantage of a system which selects for mutations in the prfA gene that lead to the loss or reduction in the activity of the L. monocytogenes virulence gene regulator. The system is based on the strong bacteriolytic activity of the p60 protein of L. grayi for B. subtilis (5). The p60 protein of L. monocytogenes and the p60-related proteins produced by all listerial species (7) possess peptidoglycan hydrolase activity, which appears to act in a late step of cell wall synthesis of Listeria and which is preferentially secreted into the supernatant during the stationary phase (44, 55). In recent studies it has been shown (A. Bubert, unpublished data) that p60(G) exerts the highest lytic activity on B. subtilis and other gram-positive bacteria, while B. subtilis is able to tolerate a significantly higher concentration of the L. monocytogenes p60 (55). The iap gene of L. grayi encoding the p60(G) protein is not expressed in B. subtilis when put under the control of the promoter of the PrfA-dependent listeriolysin gene (hly) of L. monocytogenes in the absence of functional PrfA. However, introduction of the prfA gene activates this promoter and leads only to the production of p60(G), which kills most bacilli. Under these conditions, B. subtilis survives if mutations have inactivated either the iap or the prfA gene. Such mutants are obtained at a frequency of about 10−8 after transformation of B. subtilis with plasmid pERL3502, which carries the prfA gene under the control of the PrfA-dependent plcA promoter, leading to efficient synthesis of PrfA (35).
With this selection system we have obtained two classes of mutations; in one class iap deletion or point mutations within the iap gene apparently eliminated or inactivated the p60 protein, while in the second class mutations within prfA led to the inactivation of essential functions of the PrfA protein. Some of the prfA point mutations generate stop codons leading to rather short PrfA peptides, but most others represent missense mutations leading to the synthesis of mutant PrfA proteins with single amino acid exchanges. The latter mutations and also some of the shorter in-frame deletions are clustered in three regions of PrfA, designated A, B, and C.
Mutations in region B affect the helix-turn-helix domain of PrfA, which is highly homologous to the corresponding DNA binding domain of the related regulatory CRP of E. coli (31). Recent studies (46) have indeed shown that this PrfA domain is responsible for the interaction of PrfA with its specific binding site, i.e., the PrfA box, a sequence of dyad symmetry comprising 14 nucleotides (16, 36). Within PrfA region B, the R188I exchange was obtained six times in independent mutagenesis experiments, suggesting that this amino acid position is very critical for the function of the helix-turn-helix domain. Not surprisingly, this amino acid exchange in PrfA leads to a mutant protein which is strongly impaired in its ability to bind to the PrfA box of Phly/PplcA, used as specific PrfA target site in our binding assays.
More interesting are the mutations in prfA affecting the C-terminal part of PrfA or region C. This 38-amino-acid C-terminal sequence of PrfA exhibits a leucine zipper-like motif and is missing in the related CRP and other regulatory proteins belonging to the CRP/Fnr family of regulatory proteins (31). Two of these mutant PrfA proteins, one of which carries a 3-amino-acid deletion in this region of PrfA while the other has lost the last 17 amino acids, were further analyzed. Both mutants, similar to region B mutants, are completely unable to bind to the PrfA box. Preliminary results indicate that these mutant PrfA proteins are unable to dimerize, at least under in vitro conditions, suggesting that the leucine zipper-like domain of this sequence may be involved in the dimerization of PrfA, as already shown for other proteins carrying similar leucine zipper domains (31). Nevertheless, our results do not exclude the possibility that the loss of this C-terminal region alters the overall conformation of the PrfA protein, thus prohibiting the proper folding of the helix-turn-helix DNA binding domain.
Region A of PrfA is located between amino acids 58 and 67. The corresponding region in CRP represents a potential activator loop that might directly contact RNAP (54). It has been shown that this loop may be involved in transcriptional activation of class II promoters by CRP; such promoters carry the CRP binding site at position −42 (8, 9, 23), i.e., similar to the position of the PrfA box in PrfA-dependent listerial promoters.
In contrast to the mutant PrfA proteins of the other two classes, purified PrfA protein from region A mutants binds to the PrfA box of the hly/plcA promoter region with an efficiency of about 60% of that of wild-type PrfA. These data suggest that the binding of this mutant PrfA protein to its DNA target site is essentially intact. However, the binding of this PrfA protein to its specific target site does not permit the subsequent association of RNAP with this complex, which readily occurs with the wild-type PrfA protein. In addition, in vitro transcription starting from the PrfA-dependent PplcA promoter is not activated in the presence of this mutant PrfA protein. These results suggest that region A of PrfA represents or at least includes the site of interaction between PrfA and RNAP.
In summary, we have presented a collection of mutations that impair the function of PrfA. These mutations fall essentially in three regions, which may represent functional domains in this transcription factor. One should, however, keep in mind that the described positive selection procedure for prfA mutations is performed in B. subtilis, a heterologous bacterial system which does not necessarily reproduce all the characteristics and subtleties of PrfA-dependent regulation in L. monocytogenes (3, 4). There is evidence that an additional factor(s) is needed for this differential regulation by PrfA (1, 3, 11, 40, 52) and that this factor(s) may not be present in B. subtilis. In addition, the culture conditions used (growth in BHI medium, a rich medium) may not necessarily select for the interaction of PrfA with such a factor(s), and hence possible regions in PrfA essential for the interaction of PrfA with this factor(s), possibly relevant to the differential regulation of PrfA-dependent genes, are therefore not expected to be identified with this selection procedure. Anyhow, the fact that we were able to repeatedly identify with our selection system the same mutations in several independent experiments provides at least circumstantial evidence that the three regions of PrfA identified may represent critical functional domains. These are presumably essential for binding to RNAP (region A), for binding to its target DNA site (region B), and, possibly, for dimerization (region C). The amino acids affected by the point mutations may possibly play a key role in these functions. The PrfA mutants described here, together with the elucidation of the three-dimensional structure of the PrfA protein, may help in the understanding of the structure and function of this listerial key regulator.
ACKNOWLEDGMENTS
M. Herler and A. Bubert contributed equally to this work.
We thank D. Palm (University of Würzburg) for assistance in the preparation of the anti-L. grayi p60 antiserum. H. Kestler is thanked for construction of plasmids used in this study.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 479-B1), the Fonds der Chemischen Industrie, the European Union (grant BMH4-CT-96-0659), and the Spanish Ministry for Science and Technology (grant BMC2000-0553).
REFERENCES
- 1.Böckmann R, Dickneite C, Goebel W, Bohne J. PrfA mediates specific binding of RNA polymerase of Listeria monocytogenes to PrfA-dependent virulence gene promoters resulting in a transcriptionally active complex. Mol Microbiol. 2000;36:487–497. doi: 10.1046/j.1365-2958.2000.01868.x. [DOI] [PubMed] [Google Scholar]
- 2.Böckmann R, Dickneite C, Middendorf B, Goebel W, Sokolovic Z. Specific binding of the Listeria monocytogenes transcriptional regulator PrfA to target sequences requires additional factor(s) and is influenced by iron. Mol Microbiol. 1996;22:643–653. doi: 10.1046/j.1365-2958.1996.d01-1722.x. [DOI] [PubMed] [Google Scholar]
- 3.Bohne J, Kestler H, Uebele C, Sokolovic Z, Goebel W. Differential regulation of the virulence genes of Listeria monocytogenes by the transcriptional activator PrfA. Mol Microbiol. 1996;20:1189–1198. doi: 10.1111/j.1365-2958.1996.tb02639.x. [DOI] [PubMed] [Google Scholar]
- 4.Bubert A, Sokolovic Z, Chun S K, Papatheodorou L, Simm A, Goebel W. Differential expression of Listeria monocytogenes virulence genes in mammalian host cells. Mol Gen Genet. 1999;261:323–336. doi: 10.1007/pl00008633. [DOI] [PubMed] [Google Scholar]
- 5.Bubert A, Kestler H, Götz M, Böckmann R, Goebel W. The Listeria monocytogenes iap gene as an indicator gene for the study of PrfA-dependent regulation. Mol Gen Genet. 1997;256:54–62. doi: 10.1007/s004380050545. [DOI] [PubMed] [Google Scholar]
- 6.Bubert A, Schubert A P, Köhler S, Frank S, Goebel W. Synthetic peptides derived from the Listeria monocytogenes p60 protein as antigens for the generation of polyclonal antibodies specific for secreted cell-free L. monocytogenes p60 proteins. Appl Environ Microbiol. 1994;60:3120–3127. doi: 10.1128/aem.60.9.3120-3127.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bubert A, Kuhn M, Goebel W, Köhler S. Structural and functional properties of the p60 proteins from different Listeria species. J Bacteriol. 1992;174:8166–8171. doi: 10.1128/jb.174.24.8166-8171.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Busby S, Ebright R H. Transcription activation by catabolite activator protein (CAP) J Mol Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 9.Busby S, Ebright R H. Transcription activation at class II CAP-dependent promoters. Mol Microbiol. 1997;23:853–859. doi: 10.1046/j.1365-2958.1997.2771641.x. [DOI] [PubMed] [Google Scholar]
- 10.Butz S, Rawer S, Rapp W, Birsner U. Immunization and affinity purification of antibodies using resin-immobilized lysine-branched synthetic peptides. Pept Res. 1994;7:20–23. [PubMed] [Google Scholar]
- 11.Dickneite C, Boeckmann R, Spory A, Goebel W, Sokolovic Z. Differential interaction of the transcription factor PrfA and the PrfA-activating factor (Paf) of Listeria monocytogenes with target sequences. Mol Microbiol. 1998;27:915–928. doi: 10.1046/j.1365-2958.1998.00736.x. [DOI] [PubMed] [Google Scholar]
- 12.Dramsi S, Kocks C, Forestier C, Cossart P. Internalin-mediated invasion of epithelial cells by Listeria monocytogenes is regulated by the bacterial growth state, temperature and the pleiotropic activator prfA. Mol Microbiol. 1993;9:931–941. doi: 10.1111/j.1365-2958.1993.tb01223.x. [DOI] [PubMed] [Google Scholar]
- 13.Engelbrecht F, Dickneite C, Lampidis R, Goetz M, DasGupta U, Goebel W. Sequence comparison of the chromosomal regions encompassing the internalin C genes (inlC) of Listeria monocytogenes and L. ivanovii. Mol Gen Genet. 1998;257:186–197. doi: 10.1007/s004380050638. [DOI] [PubMed] [Google Scholar]
- 14.Engelbrecht F, Chun S K, Ochs C, Hess J, Lottspeich F, Goebel W, Sokolovic Z. A new PrfA-regulated gene of Listeria monocytogenes encoding a small, secreted protein which belongs to the family of internalins. Mol Microbiol. 1996;21:823–837. doi: 10.1046/j.1365-2958.1996.541414.x. [DOI] [PubMed] [Google Scholar]
- 15.Farber J M, Peterkin P I. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev. 1991;55:476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freitag N E, Rong L, Portnoy D A. Regulation of the prfA transcriptional activator of Listeria monocytogenes: multiple promoter elements contribute to intracellular growth and cell-to-cell spread. Infect Immun. 1993;61:2537–2544. doi: 10.1128/iai.61.6.2537-2544.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goebel W, Kreft J, Böckmann R. Regulation of virulence genes in pathogenic Listeria. In: Fischetti V A, Novick R P, Ferretti J J, Portnoy D A, Rood J I, editors. Gram-positive pathogens. Washington, D.C.: American Society for Microbiology; 2000. pp. 499–506. [Google Scholar]
- 18.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 19.Hess J, Gentschev I, Szalay G, Ladel C, Bubert A, Goebel W, Kaufmann S H. Listeria monocytogenes p60 supports host cell invasion by and in vivo survival of attenuated Salmonella typhimurium. Infect Immun. 1995;63:2047–2053. doi: 10.1128/iai.63.5.2047-2053.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Innis M A, Gelfand D H, Sninsky J J. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press; 1990. [Google Scholar]
- 21.Kawamura F, Doi R H. Construction of a Bacillus subtilis double mutant deficient in extracellular alkaline and neutral proteases. J Bacteriol. 1984;160:442–444. doi: 10.1128/jb.160.1.442-444.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Köhler S, Leimeister-Wächter M, Chakraborty T, Lottspeich F, Goebel W. The gene coding for protein p60 of Listeria monocytogenes and its use as a specific probe for Listeria monocytogenes. Infect Immun. 1990;58:1943–1950. doi: 10.1128/iai.58.6.1943-1950.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolb A, Busby S, Buc H, Garges S, Adhya S. Transcriptional regulation by cAMP and its receptor protein. Annu Rev Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- 24.Kreft J, Bernhard K, Goebel W. Recombinant plasmids capable of replication in B. subtilis and E. coli. Mol Gen Genet. 1978;162:59–67. doi: 10.1007/BF00333851. [DOI] [PubMed] [Google Scholar]
- 25.Kreft J, Vazquez-Boland J-A, Ng E, Goebel W. Virulence gene clusters and putative pathogenicity islands in Listeria. In: Kaper J, Hacker J, editors. Pathogenicity islands and other mobile genetic elements. Washington, D.C.: American Society for Microbiology; 1999. pp. 219–232. [Google Scholar]
- 26.Kuhn M, Goebel W. Molecular studies on the virulence of Listeria monocytogenes. Genet Eng. 1995;17:31–51. [PubMed] [Google Scholar]
- 27.Kuhn M, Goebel W. Identification of an extracellular protein of Listeria monocytogenes possibly involved in intracellular uptake by mammalian cells. Infect Immun. 1989;57:55–61. doi: 10.1128/iai.57.1.55-61.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuhn M, Kathariou S, Goebel W. Hemolysin supports survival but not entry of the intracellular bacterium Listeria monocytogenes. Infect Immun. 1988;56:79–82. doi: 10.1128/iai.56.1.79-82.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kyhse-Andersen J. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J Biochem Biophys Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Lampidis R, Gross R, Sokolovic Z, Goebel W, Kreft J. The virulence regulator protein of Listeria ivanovii is highly homologous to PrfA from Listeria monocytogenes and both belong to the Crp-Fnr family of transcription regulators. Mol Microbiol. 1994;13:141–151. doi: 10.1111/j.1365-2958.1994.tb00409.x. [DOI] [PubMed] [Google Scholar]
- 32.Landschulz W H, Johnson P F, McKnight S L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 33.Leimeister-Wächter M, Haffner C, Domann E, Goebel W, Chakraborty T. Identification of a gene that positively regulates expression of listeriolysin, the major virulence factor of Listeria monocytogenes. Proc Natl Acad Sci USA. 1990;87:8336–8340. doi: 10.1073/pnas.87.21.8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lingnau A, Domann E, Hudel M, Bock M, Nichterlein T, Wehland J, Chakraborty T. Expression of the Listeria monocytogenes EGD inlA and inlB genes, whose products mediate bacterial entry into tissue culture cell lines, by PrfA-dependent and -independent mechanisms. Infect Immun. 1995;63:3896–3903. doi: 10.1128/iai.63.10.3896-3903.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mengaud J, Dramsi S, Gouin E, Vazquez-Boland J-A, Milon G, Cossart P. Pleiotropic control of Listeria monocytogenes virulence factors by a gene that is autoregulated. Mol Microbiol. 1991;5:2273–2283. doi: 10.1111/j.1365-2958.1991.tb02158.x. [DOI] [PubMed] [Google Scholar]
- 36.Mengaud J, Vicente M F, Cossart P. Transcriptional mapping and nucleotide sequence of the Listeria monocytogenes hlyA region reveal structural features that may be involved in regulation. Infect Immun. 1989;57:3695–3701. doi: 10.1128/iai.57.12.3695-3701.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michel E, Mengaud J, Galsworthy S, Cossart P. Characterization of a large motility gene cluster containing the cheR, motAB genes of Listeria monocytogenes and evidence that PrfA downregulates motility genes. FEMS Microbiol Lett. 1998;169:341–347. doi: 10.1111/j.1574-6968.1998.tb13338.x. [DOI] [PubMed] [Google Scholar]
- 38.Pamer E G. Direct sequence identification and kinetic analysis of an MHC class I-restricted Listeria monocytogenes CTL epitope. J Immunol. 1994;152:686–694. [PubMed] [Google Scholar]
- 39.Potel J, Schulze-Lammers J. Listeria monocytogenes-vaccine: production and control. Zentbl Bakteriol Mikrobiol Hyg A. 1985;259:331–340. doi: 10.1016/s0176-6724(85)80035-3. [DOI] [PubMed] [Google Scholar]
- 40.Renzoni A, Klarsfeld A, Dramsi S, Cossart P. Evidence that PrfA, the pleiotropic activator of virulence genes in Listeria monocytogenes, can be present but inactive. Infect Immun. 1997;65:1515–1518. doi: 10.1128/iai.65.4.1515-1518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ripio M T, Vazquez-Boland J-A, Vega Y, Nair S, Berche P. Evidence for expressional crosstalk between the central virulence regulator PrfA and the stress response mediator ClpC in Listeria monocytogenes. FEMS Microbiol Lett. 1998;158:45–50. doi: 10.1111/j.1574-6968.1998.tb12798.x. [DOI] [PubMed] [Google Scholar]
- 42.Ripio M T, Dominguez-Bernal G, Lara M, Suarez M, Vazquez-Boland J-A. A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. J Bacteriol. 1997;179:1533–1540. doi: 10.1128/jb.179.5.1533-1540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ripio M T, Brehm K, Lara M, Suarez M, Vazquez-Boland J-A. Glucose-1-phosphate utilization by Listeria monocytogenes is PrfA dependent and coordinately expressed with virulence factors. J Bacteriol. 1997;179:7174–7180. doi: 10.1128/jb.179.22.7174-7180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruhland G J, Hellwig M, Wanner G, Fiedler F. Cell-surface location of Listeria-specific protein p60—detection of Listeria cells by indirect immunofluorescence. J Gen Microbiol. 1993;139:609–616. doi: 10.1099/00221287-139-3-609. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1998. [Google Scholar]
- 46.Sheehan B, Klarsfeld A, Ebright R, Cossart P. A single substitution in the putative helix-turn-helix motif of the pleiotropic activator PrfA attenuates Listeria monocytogenes virulence. Mol Microbiol. 1996;20:785–797. doi: 10.1111/j.1365-2958.1996.tb02517.x. [DOI] [PubMed] [Google Scholar]
- 47.Sheehan B, Klarsfeld A, Msadek T, Cossart P. Differential activation of virulence gene expression by PrfA, the Listeria monocytogenes virulence regulator. J Bacteriol. 1995;177:6469–6476. doi: 10.1128/jb.177.22.6469-6476.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sokolovic Z, Riedel J, Wuenscher M, Goebel W. Surface-associated, PrfA-regulated proteins of Listeria monocytogenes synthesized under stress conditions. Mol Microbiol. 1993;8:219–227. doi: 10.1111/j.1365-2958.1993.tb01566.x. [DOI] [PubMed] [Google Scholar]
- 49.Van Regenmortel M H, Daney de Marcillac G. An assessment of prediction methods for locating continuous epitopes in proteins. Immunol Lett. 1988;17:95–107. doi: 10.1016/0165-2478(88)90076-4. [DOI] [PubMed] [Google Scholar]
- 50.Vázquez-Boland, J.-A., G. Domínguez-Bernal, B. González-Zorn, W. Goebel, and J. Kreft. Pathogenicity islands and virulence evolution in Listeria. Microb. Infect., in press. [DOI] [PubMed]
- 51.Vázquez-Boland J-A, Kuhn M, Berche P, Chakraborty T, Domínguez-Bernal G, Goebel W, Gonzalez-Zorn B, Wehland J, Kreft J. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev. 2001;14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vega Y, Dickneite C, Ripio M T, Böckmann R, Gonzalez-Zorn B, Novella S, Dominguez-Bernal G, Goebel W, Vazquez-Boland J-A. Functional similarities between the Listeria monocytogenes virulence regulator PrfA and cyclic AMP receptor protein: the PrfA* (Gly145Ser) mutation increases binding affinity for target DNA. J Bacteriol. 1998;180:6655–6660. doi: 10.1128/jb.180.24.6655-6660.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wagner M, Schmid M, Juretschko S, Trebesius K H, Bubert A, Goebel W, Schleifer K H. In situ detection of a virulence factor mRNA and 16S rRNA in Listeria monocytogenes. FEMS Microbiol Lett. 1998;160:159–168. doi: 10.1111/j.1574-6968.1998.tb12906.x. [DOI] [PubMed] [Google Scholar]
- 54.Williams R, Bell A, Sims G, Busby S. The role of two surface exposed loops in transcription activation by the Escherichia coli CRP and FNR proteins. Nucleic Acids Res. 1991;19:6705–6712. doi: 10.1093/nar/19.24.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wuenscher M D, Köhler S, Bubert A, Gerike U, Goebel W. The iap gene of Listeria monocytogenes is essential for cell viability, and its gene product, p60, has bacteriolytic activity. J Bacteriol. 1993;175:3491–3501. doi: 10.1128/jb.175.11.3491-3501.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wuenscher M D, Köhler S, Goebel W, Chakraborty T. Gene disruption by plasmid integration in Listeria monocytogenes: insertional inactivation of the listeriolysin determinant lisA. Mol Gen Genet. 1991;228:177–182. doi: 10.1007/BF00282463. [DOI] [PubMed] [Google Scholar]
- 57.Yannisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]