Abstract
African swine fever is an acute, haemorrhagic fever and contagious disease of pigs caused by African swine fever virus (ASFV), which has a great impact on the pig farming industry and related international trade. Understanding the response processes of various tissues in pigs after ASFV infection may help to address current major concerns, such as the exploration of key genes for vaccine development, the cooperative mechanism of the host response and the possibility of establishing active herd immunity. ASFV is able to infect core tissues and is associated with acute death. RNA and protein samples were obtained and verified from five tissues, including the lung, spleen, liver, kidney and lymph nodes. Multiple duplicate samples were quantitatively analyzed by corresponding transcriptomic and proteomic comparison. The results showed that different tissues cooperated in responses to ASFV infection and coordinated the defence against ASFV in the form of an inflammatory cytokine storm and interferon activation. The lung and spleen were mainly involved (dominant) in the innate immune response pathway; the liver and kidney were involved in the metabolic regulatory pathway and the inflammatory response; and the lymph nodes cooperated with the liver to complete energy metabolism regulation. The key pathways and responsive genes in each tissue of the contracted pigs were comprehensively mapped by infectomics, providing further evidence to investigate the complicated tie between ASFV and host cells.
Keywords: African swine fever virus, different tissues, proteomics, synergistic effect, transcriptomics
1. INTRODUCTION
African swine fever is an expanding and devastating viral infectious disease that currently threatens pig industry worldwide. Currently, vaccines and treatments are unavailable, and the disease has potentially devastating consequences for the availability of affordable protein‐production livelihood and trade (Brown et al., 2018). African swine fever virus (ASFV) is a bi‐enveloped, icosahedral double‐stranded DNA virus with a large genome, which has hindered efforts to rapidly obtain a full‐genome sequence. The genome of the virus replicates mainly in the cytoplasm of infected cells. ASFV can enter pigs through the mouth and upper respiratory system and infect the nasopharynx or tonsils, with close to 90% or higher mortality (Dixon et al., 2019; Molini et al., 2020). According to its virulence, ASFV is classified as high‐, moderate‐ or low‐virulence strain. Clinical manifestations range from acute death within 7 days of infection to chronic infection lasting several weeks or months. Infection caused by high virulence is usually followed by acute death in pigs of all ages. Clinical pathogen detection showed that blood, lymph nodes, spleen, bone marrow and lung are the best collection for dead pigs (Gómez‐Villamandos et al., 1995; Li et al., 2020).
Antiviral innate immunity is critical to the host response to viral infection. It has been reported that in ASFV infection, the spleen contains more macrophages, which are more prominently active and have greater differential expression than lymph nodes (Zhao et al., 2019). Other studies have shown that the host response to ASFV infection is rapid and intense (Takamatsu et al., 2013). Understanding the coordination between innate protective responses in different tissues and immune pathology might contribute to the development of strategies against ASFV. Nevertheless, these early records raise immediate questions, which tissues of pigs are involved in the battle against the virus, how do the tissues coordinate and cooperate to respond to virus invasion and what is the division of labour between the different tissues? The question is directly linked to several major concerns, namely, the best way to achieve efficacy of vaccines and subsequently herd immunity.
Comparative analysis of genome‐wide expression profiles and proteomics is increasingly being used to study specific animal virus–host interactions. Aiming to answer the above questions and respond to the concerns, we conducted a retrospective study on samples collected from different tissues of diseased pigs on a farm to accurately measure the intensity of up‐regulation or down‐regulation of genes by using infectomic analysis, including transcriptomics and proteomics. Based on this investigation, we illustrate the genes and protein profiles of different tissues specific to the virus and reveal the role and mechanism of the host viral cell response to ASFV infection. We believe the information will aid in vaccine development and prudent use of drugs, as well as other control strategies.
2. MATERIALS AND METHODS
2.1. Ethics statement
Samples were collected for African swine fever testing and surveillance under the agreement between the Ministry of Agriculture and Rural Affairs of the Chinese Government and farm owners by Centre for Animal Disease Control and Prevention of Guangdong Province with the protocols established by the World Organization for Animal Health. The protocol was approved by the ethics committee of the Military Veterinary Research Institute, Academy of Military Medical Sciences. The viruses were inactivated in the BSL‐3 laboratory, and the inactivated samples were transferred to a BSL‐2 level laboratory for genomic DNA extraction and detection. The analytical samples and protocols used in this study were approved by the Institute of Microbiology, Chinese Academy of Sciences, Research Ethics Committee (license number: PZIMCAS2019002).
2.2. Sample collection and preparation
Spleen, lung, lymph node, liver and kidney tissue samples were collected separately from three ASFV‐positive pigs in the field and one ASFV‐negative pig as a control. Tissue samples weighing 0.3 g were placed into sterile tubes to generate tissue homogenates for total RNA extraction using TRIzol reagent (Life Technologies, USA) according to the manufacturer's protocol. The quantity and quality of different total RNA samples were evaluated using NanoDrop ND‐1000 (Wilmington, USA). The integrity of RNA samples was estimated by the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Another set of tissue sample were ground into a fine power in liquid nitrogen. Subsequently, the power was suspended in lysis buffer (1% sodium deoxycholate (SDS), 8 M urea) which included appropriate protease inhibitor to inhibit protease activity. The mixture was allowed to settle at 4°C for 30 min during which the sample were vortexed at every 5 min, and treated by ultrasound at 40 kHz and 40 W for 2 min. After centrifugation at 16,000×g at 4°C for 30 min, the concentration of protein supernatant was determined by bicinchoninic acid (BCA) method by BCA Protein Assay Kit (Thermo, USA). Protein quantification was performed according to the kit protocol. The viruses were stored at the Military Veterinary Research Institute, Academy of Military Medical Sciences.
2.3. Quantitative real‐time PCR assay
Tissue samples were ground and lysed before viral nucleic acid extraction. ASFV genomic DNA was extracted from the tissue using GenElute Mammalian Genomic DNA Miniprep Kits (Sigma–Aldrich, USA). ASFV genomic DNA was quantified with TB Green Advantage qPCR Premix (Takara) on an Applied Biosystems 7500 Real Time Detection System (Roche, Germany) to determine the gene expression levels using the β‐actin gene promoter as a control. Each assay was performed in triplicate, and the level was calculated by the 2−∆∆ct method. The quantitative real‐time PCR (qPCR) results were consistent with the sequencing results.
2.4. Transmission electron microscopy
Ultrastructural examination was done on different infected tissues and selected lung, spleen, kidney, liver from ASFV‐infected pigs. Tissues are centrifuged and fixed in buffered 2.5% glutaraldehyde for 24 h. Specimens are postfixed in 1% osmium tetroxide, washed, dehydrated through a graded series of alcohol and acetone, and embedded in a mixture of epoxy resin (Salguero et al., 2004). Then, the thin sections are stained with 2% uranyl acetate and lead citrate and examined with an electron microscope (JEM‐1200EXII; Japan).
2.5. Library construction and sequencing of transcriptome
Paired‐end libraries for RNA sequencing were constructed using a TruSeq™ RNA Sample Prep Kit (Illumina) according to the manufacturer's instructions. Library preparation began with the extraction of poly‐A mRNA from total RNA using oligo‐(dT)‐coupled magnetic beads. Fragmentation buffer was added to randomly break mRNA into small fragments of approximately 300 bp. cDNA was synthesized with integrated DNase treatment, followed by end repair. The libraries were amplified using 15 cycles and purified. After quantification with TBS380 (PicoGreen), the libraries were pooled in equimolar concentrations and sequenced on the Illumina HiSeq Xten platform by Majorbio Bio‐Pharm Technology Co., Ltd. using 2 × 150 bp reads.
2.6. Library construction and sequencing of proteome
To reveal the changes in protein expression related to ASFV infection, we selected S2 samples of the treatment pigs and a control pig for tandem mass tag (TMT)‐based proteomic analysis. Ten purified tissue proteins were obtained and protein quantification was performed using a Thermo Fisher Scientific Pierce BCA Protein Assay Kit, and SDS‐PAGE was also applied to determine protein purity. One hundred micrograms of protein from each tissue were dissolved in 100 mM TEAB (Sigma). The protein solution was reduced with 10 mM TCEP (Thermo Scientific) for 1 h at 37°C and alkylated with 40 mM iodoacetamide (Sigma) for 40 min at room temperature in darkness. Cold acetone (Sinopharm Chemical) was added at a 6:1 acetone‐to‐protein mass ratio for a 4 h precipitation at −20°C, and the precipitate was obtained by centrifugation at 10,000×g for 20 min. For trypsin digestion, the precipitate was dissolved by adding 50 mM TEAB, and trypsin (Promega) was added at a 1:50 trypsin‐to‐protein mass ratio for digestion overnight. After trypsin digestion, one unit of TMT reagent (defined as the amount of reagent required to label 100 μg of protein; Thermo Fisher) was thawed and reconstituted in acetonitrile (Fisher Chemical). The peptide mixtures were then incubated for 2 h at room temperature, pooled, desalted and dried by vacuum centrifugation. The samples were fractionated by high‐pH reverse‐phase HPLC using an ACQUITY UPLC BEH C18 column (1.7 μm, 2.1 mm × 250 mm; Waters Corporation, USA). LC–MS/MS analysis was then performed as described previously (García‐Hernández et al., 2018).
2.7. Transcriptomics data analysis
After the raw RNA‐seq data were trimmed for low‐quality reads/adapters and filtered by FASTX‐Toolkit (version 0.0.14), the sequencing reads of the clean data were aligned to the Sus scrofa genome (version Sscrofa11.1) with the use of HISAT2 (version 2.1.0) (Kim et al., 2015). The output SAM files were sorted and converted to BAM files using SAMtools (version 1.9) (Li et al., 2009), and count files were generated with featureCounts (version 1.6.4) (Liao et al., 2014). Differentially expressed genes (DEGs) between groups were identified using the EdgeR package (version 3.26.8) and limma package (version 3.40.6) in R (Ritchie et al., 2015; Robinson et al., 2010). The threshold for DEGs was set at log2(fold change (FC)) ≤−1 or ≥1 and false discovery rate (FDR) < 0.05. It should be noted that only DEGs with the same expression trend across the three samples were used for subsequent functional annotation. Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed using clusterProfiler (version 3.12.0) in R (Yu et al., 2012). Correction of multiple hypothesis testing was carried out using standard FDR control methods. GO term or KEGG pathway with a p value < .05 after a two‐tailed Fisher's exact test was considered significantly enriched.
2.8. Proteomic data analysis
Raw mass spectrometry (MS) data were processed using the Mascot search engine (Version 2.3; Matrix Science) embedded into Proteome Discoverer 2.2 (Thermo Fisher Scientific). Tandem mass spectra were searched against the UniProt Sus_scrofa_Sscrofa11.1 database. A peptide mass tolerance of 20 ppm, fragment mass tolerance of 0.02 Da and maximum missed cleavages of two were allowed. The modification set was as follows: static modification: carbamidomethyl (C), TMT 6plex (K), and TMT 6plex (N‐terminus); and dynamic modification: oxidation (M) and acetylation (protein N‐terminus). The stringent FDR was set to 0.01 for peptide and protein identification. Proteins with a FC ≥ 1.4 and FDR < 0.05 in two comparable groups (treatment versus control) were considered differentially expressed proteins (DEPs).
For functional annotation of these DEPs, GO enrichment analysis and KEGG pathway analysis were performed. Correction of multiple hypothesis testing was carried out using standard FDR control methods. A GO term or KEGG pathway with a p value < .05 after a two‐tailed Fisher's exact test was considered significantly enriched.
3. RESULTS
3.1. The virus detected in core tissues of pigs infected with ASFV
Autopsies are performed on death of pigs and showed pulmonary congestion, pneumonia with caseous necrosis (sometimes accompanied by local calcification), foaming of the trachea and bronchus, and severe alveolar and interstitial pulmonary oedema (Figure 1(a)). Notably, the most obvious necropsy lesions were enlargement and friability of the spleen with dark red or even black round edges and petechiation on the capsule of the kidney. There were also ecchymoses in the kidneys (cortexes and pelves), hepatic congestion, gallbladder bleeding, lymph node enlargement and severe bleeding. The ultramicroscopy results in Figure 1(b) also are used to confirm what the macroscopic pathological lesions suggest, and the sequencing data showed that multiple tissues were infected by ASFV. All the pigs in the study were infected with the SY‐18 strain identified by qPCR.
FIGURE 1.
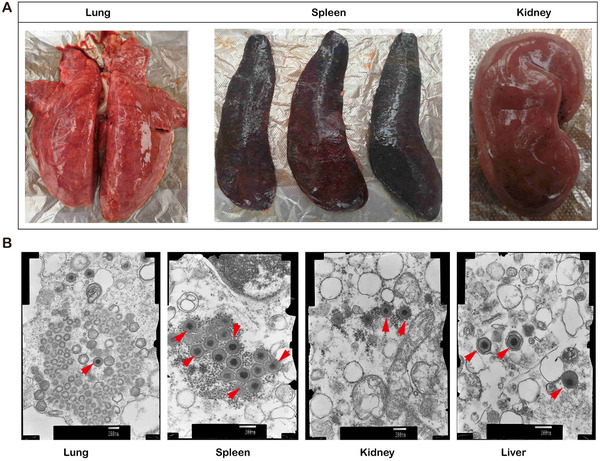
Clinical and pathological characteristics of pig tissues infected with African swine fever virus (ASFV). (a) Pulmonary haemorrhage, splenomegaly and renal congestion in acute African swine fever. (b) Transmission electron micrograph (TEM) showing different infected tissues (lung, spleen, kidney, liver) of the red arrow ASFV. TEM provides high‐resolution images of biological structures, including the types and arrangement of organelles within a cell and the locations of plasma membranes of complexly shaped cells. Mature virions, immature virions and midbody are visible
3.2. Summary of the DEGs in each tissue
We extracted the RNA and constructed a library for deep sequencing. Illumina‐based RNA‐seq was performed on the HiSeq 4000 platform in five groups (each group was repeated three times) of different tissue samples, namely, those from the lung, liver, lymph node, spleen and kidney. Each group had three biological replicates. The sequences were used for subsequent analysis. Finally, the total number of up‐regulated and down‐regulated genes in each tissue was obtained (Table 1).
TABLE 1.
Numbers of up‐regulated and down‐regulated differentially expressed genes at transcriptome level in different infected tissues
| Lung | Liver | Lymphaden | Spleen | Kidney | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | IN | S1 | S2 | S3 | IN | S1 | S2 | S3 | IN | S1 | S2 | S3 | IN | S1 | S2 | S3 | IN | |
| Up | 2246 | 745 | 1650 | 158 | 1903 | 750 | 1327 | 242 | 3058 | 3689 | 1637 | 929 | 835 | 2699 | 958 | 192 | 1177 | 2592 | 1089 | 145 |
| Down | 2084 | 1044 | 1057 | 227 | 1944 | 1159 | 1730 | 588 | 2867 | 2910 | 778 | 394 | 1184 | 1143 | 1972 | 363 | 2811 | 2121 | 1337 | 428 |
IN represents the common genes of DEGs of S1, S2 and S3.
As shown in Figure 2(a), all the promoted and suppressed DEGs were identified in tissues infected by ASFV. Notably, the DEGs of S2 samples were relatively different, especially in lymph nodes and spleen. Upon importing the dataset of genes with altered expression profiles obtained from the sequencing analysis to analyze GO pathway enrichment, we examined possible biological interactions among the DEGs and identified important functional networks. The results of the GO analysis of the common subclasses of cellular processes are presented in Figure S1. Furthermore, the DEGs were mapped to the KEGG pathway database to further explore the functional analysis (Figure S2). Hundreds of signaling pathways were found to be enriched in each tissue, among which the pathways were found to be significantly altered (Q value < .05 and p < .01) (Table 2).
FIGURE 2.
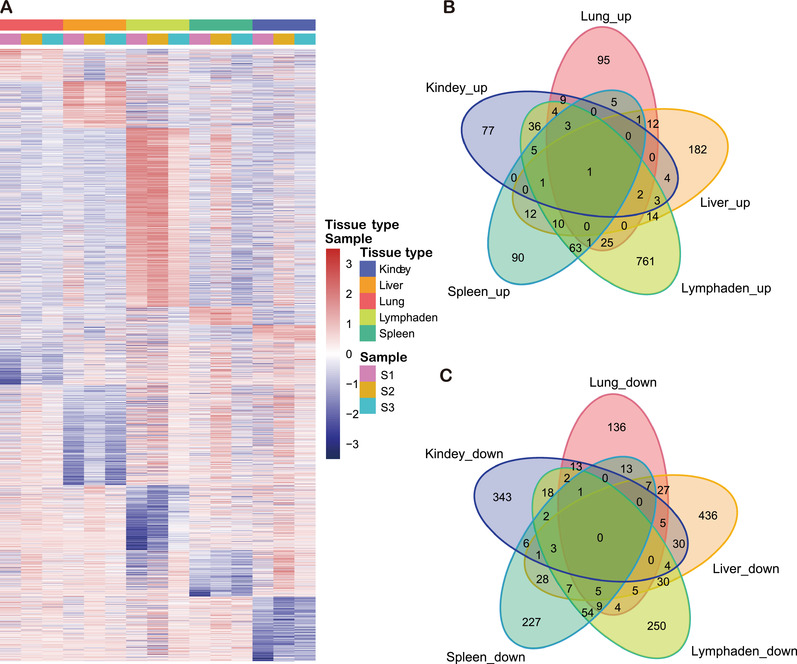
Summary of differentially expressed genes (DEGs) among infected kidney, liver, lung, lymph node and spleen tissues. (a) A hierarchical clustering heat map was used to classify gene expression patterns among each infected tissue. Genes with an FDR‐corrected p value ≤ .05 and a fold change value ≥2 were considered DEGs. Each column represents a sample, and each row represents a gene. The intensity of colour indicates gene expression levels normalized according to log10 (TPM+1) values. Red indicates a high expression level and blue indicates a low expression level. Panels (b) and (c) were Venn diagram analysis of the common up‐regulated and down‐regulated DEGs in different ASFV‐infected tissues
TABLE 2.
GO and KEGG pathway analysis results at transcriptome level in different infected tissues
| GO Terms | ||||
|---|---|---|---|---|
| BP | CC | MF | Pathway | |
| Lung | Regulation of lipase activity, plasma lipoprotein particle assembly, protein–lipid complex assembly, positive regulation of lipase activity, regulation of sterol transport, regulation of cholesterol transport, regulation of plasma lipoprotein particle levels, triglyceride catabolic process, cholesterol efflux, phospholipid transport and more (71) | Extracellular space, high‐density lipoprotein particle, protein–lipid complex, plasma lipoprotein particle, lipoprotein particle | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen, iron ion binding, enzyme regulator activity, monooxygenase activity, enzyme inhibitor activity, transition metal ion binding, serine‐type endopeptidase inhibitor activity, peptidase inhibitor activity, purinergic receptor activity, heme binding and more (24) | Complement and coagulation cascades, chemical carcinogenesis, linoleic acid metabolism, retinol metabolism, metabolism of xenobiotics by cytochrome P450, cholesterol metabolism, arachidonic acid metabolism |
| Liver | Immune response, locomotion, cell migration, cell motility, localization of cell, chemotaxis, taxis, regulation of cell migration, regulation of cellular component movement, regulation of cell motility and more (51) | Extracellular space, MHC protein complex, cell surface, external side of plasma membrane, protein–lipid complex, plasma lipoprotein particle, lipoprotein particle, collagen‐containing extracellular matrix, high‐density lipoprotein particle, collagen trimer | Tetrapyrrole binding, monooxygenase activity, heme binding, iron ion binding, oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, purinergic receptor activity, G protein‐coupled receptor binding, calcium ion binding, cofactor binding, oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen and more (15) | Staphylococcus aureus infection, cytokine–cytokine receptor interaction, intestinal immune network for IgA production, rheumatoid arthritis, Rap1 signaling pathway, chemokine signaling pathway, retinol metabolism, leishmaniasis, cell adhesion molecules (CAMs), platelet activation and more (26) |
| Lymphaden | Angiogenesis, cardiovascular system development, blood vessel development, blood vessel morphogenesis, vasculature development, regulation of anatomical structure morphogenesis, cell migration, locomotion, anatomical structure formation involved in morphogenesis, circulatory system development and more (26) | Extracellular matrix, collagen‐containing extracellular matrix, membrane raft, membrane microdomain, apical part of cell, membrane region, apical plasma membrane, lipid droplet, caveola | Chemokine activity, chemokine receptor binding, receptor ligand activity, G protein‐coupled receptor binding, receptor regulator activity, glycosaminoglycan binding, cytokine activity, heparin binding, oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor, sulphur compound binding and more (2) | Viral protein interaction with cytokine and cytokine receptor, cytokine–cytokine receptor interaction, PPAR signaling pathway, regulation of lipolysis in adipocytes, chemokine signaling pathway, Herpes simplex virus 1 infection, amoebiasis, retinol metabolism |
| Spleen | Immune response, leukocyte migration, inflammatory response, cell migration, locomotion, defence response, cell motility, localization of cell, respiratory electron transport chain, drug metabolic process and more (50) | Extracellular space, respiratory chain, sarcolemma, respiratory chain complex, extracellular matrix | NADH dehydrogenase (ubiquinone) activity, NADH dehydrogenase (quinone) activity, monocarboxylic acid binding, NADH dehydrogenase activity, oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor, transition metal ion binding, serine hydrolase activity, serine‐type endopeptidase activity, serine‐type peptidase activity, carboxylic acid binding and more (18) | T cell receptor signaling pathway, chemical carcinogenesis, primary immunodeficiency, haematopoietic cell lineage, Th1 and Th2 cell differentiation, Parkinson disease, retinol metabolism, natural killer cell mediated cytotoxicity, cell adhesion molecules (CAMs), steroid hormone biosynthesis and more (14) |
| Kidney | Ion transport, anion transport, organic anion transport, sulphur compound metabolic process, sulphur amino acid metabolic process, transmembrane transport | Apical part of cell, apical plasma membrane, integral component of plasma membrane, intrinsic component of plasma membrane, extracellular space, plasma membrane region | Heme binding, tetrapyrrole binding, oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen, ion transmembrane transporter activity, ammonium transmembrane transporter activity, monooxygenase activity, transmembrane transporter activity, cation transmembrane transporter activity, transition metal ion binding, transporter activity and more (12) | Retinol metabolism, steroid hormone biosynthesis, fatty acid degradation, chemical carcinogenesis, metabolism of xenobiotics by cytochrome P450, PPAR signaling pathway |
The numbers in brackets were total pathway number in the term, and bold were immune‐related pathways.
According to the results of Venn diagram analysis of up‐regulated DEGs, there were no identical genes with major crossover in each tissue. One of the up‐regulated DEGs was transforming acid‐coiled‐containing protein 1 (TACC1), which participates in the biological process of pigs and is involved in mitotic spindle formation (Figure 2(b)). It was also found that TACC1 is involved in nuclear receptor‐induced transcriptional regulation, including that of the T3 thyroid hormone and all‐trans retinoic acid pathways, which may promote receptor nuclear localization and promote cell division prior to the formation of differentiated tissues. In contrast, there was no intersection among the down‐regulated DEGs of the five tissues (Figure 2(c)).
3.3. Different tissues collaborate against ASFV infection
The S2 samples were selected for proteomic sequencing, and the proteomics and transcriptomics data were matched to find the common genes in each tissue. GO and KEGG analyses were also employed to reveal the possible functions of the common gene transcripts in samples. Table S1 shows the biological processes and KEGG functional annotations of the intersecting DEGs between the transcriptome and proteome in S2 samples. With examination of the common genes related to cellular functions and metabolic processes in tissues, we found that the most enriched biological processes mainly included defence response, immune system process regulation and inflammatory response in the lung and spleen (Figures 3(a) and 3(b)), with antigen processing and presentation and complement and coagulation cascades as the most significant molecular functions (Figures S3 and S4). The results showed that pig tissues cooperated in response to ASFV infection and coordinated defence against the virus in the form of an inflammatory cytokine storm and interferon (IFN) activation.
FIGURE 3.
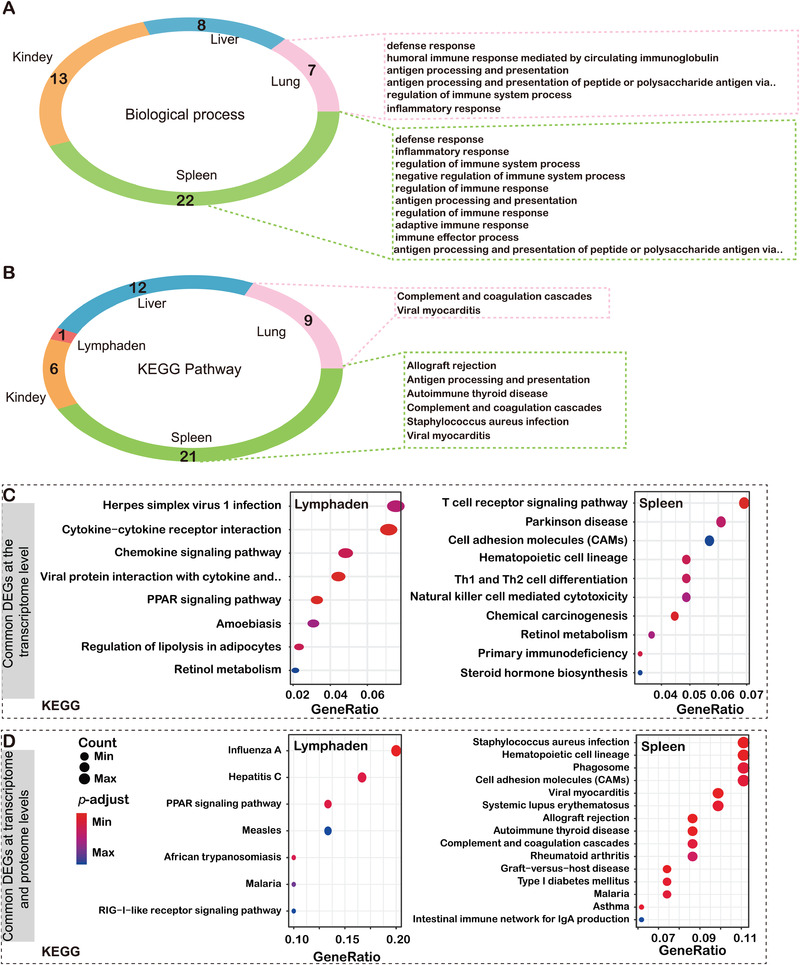
Common pathway based on biological process and KEGG analysis at the RNA and protein levels. (a) Biological process and (b) KEGG pathway. Different colours represent different tissues of pigs, and numbers indicate the number of biological processes and KEGG pathways. Those shown in the dotted box are immune‐related pathways. Common DEGs at transcriptome level (c) and proteome level (d) in lymph nodes and spleen
ASFV mainly replicates in monocytes and macrophages of lymph nodes adjacent to the site of virus invasion, and then transfers through blood or lymph nodes to the sites (e.g., spleen, lung, liver, kidney, etc.) of secondary replication. In this regard, it is particularly important concerning the response of lymph nodes and spleen after virus infection, which mainly focused on the innate immune response pathway and energy metabolism regulation (Figures 3(c) and 3(d)). By contrast, the liver and kidney were focused on the metabolic regulatory pathway and the inflammatory response, and the lung cooperated with the lymph nodes and liver to complete the transfer of energy.
4. DISCUSSION
Highly virulent ASFV can cause acute infection in domestic pigs, which are lethal to infected pigs, especially with poor ventilation breeding conditions, high stocking densities and under stress (Dixon et al., 2019; Malogolovkin & Kolbasov, 2019). The clinical presentation of hosts infected with ASFV depend on the complex interaction between the viruses and host (Galindo & Alonso, 2017). In addition to the virus itself, the active response of different tissues and host restriction proteins play an irreplaceable role in viral replication (Dixon et al., 2013). In our study, core tissues were collected from pigs that died due to ASFV infection. Transcriptomic and proteomic analyses showed that a systemic immune response occurs in the body, which is embodied in each tissue that together play an antiviral role (Forth et al., 2020). Our research systematically demonstrated the functional characteristics of each tissue after infection, including the key pathways and genes related to the innate immunity to ASFV. With the increasing understanding of the interaction between virus and hosts at the molecular, cellular and animal levels, new insights have been provided into different symptoms of ASFV‐infected hosts, which promote the further development of ASFV prevention and treatment approaches, including vaccine development.
It is widely known that the core tissues involved in innate immunity are the lungs and spleen, with the coordination of lymph nodes. Previous studies have shown that the target cells of ASFV are monocytes/macrophages, which have different contents in each tissue. Then, the release of effect factors by infected cells results in different effects on the cells in different target tissues. As a result, the spleen and lymph nodes have the most active viral replication and the highest viral content (Carrillo et al., 1994; Sánchez‐Torres et al., 2003). GO enrichment analysis of DEGs showed that ASFV infection can affect host signal transduction, activate leukocytes/lymphocytes and mediate immunity, which is consistent with the suppression of host immune system activation and evasion of the host natural immunity after viral infection. ASFV infection also affects the nucleation and nuclear localization of host proteins and cellular component migration, which may be related to the need to recruit specific host proteins or organelle components during viral replication. In addition, ASFV infection also affects bone marrow cell differentiation and haematopoietic function, the motor system, and cell adhesion. The fluctuation of ATP8 gene expression is involved in intracellular oxidative phosphorylation and is a major participant in energy production and transformation. The up‐regulation of ATP8 expression during ASFV infection may support the massive replication of the virus. In addition, ASFV mainly infects the mononuclear macrophage system during invasion, and the spleen and lymph nodes have the most mononuclear macrophages. One of the characteristics of acute ASFV infection is apoptosis of a large number of uninfected B and T cells in the lymph nodes, but the exact mechanism is not clear. It is speculated that the virus and its infected cells induce apoptosis of neighbouring uninfected cells by secreting or presenting certain molecules on the cell surface (Ramiro‐Ibáñez et al., 1996). Extensive destruction of lymphocytes caused by apoptosis may also induce immunosuppression.
In contrast to the functions of the lung and spleen, the liver and kidney are mainly responsible for the regulation of metabolism. In the liver and kidney following ASFV infection, the pathways related to energy metabolism were abnormally excited and sharply up‐regulated. KEGG enrichment analysis of the DEGs showed that it was mainly concentrated in the Th cell differentiation, antigen presentation processing, T/B cell receptor signaling and chemokine signaling pathways. Takamatsu et al., 2013 summed that it may be increase in the activity of natural killer cells after ASFV infection, and the cytotoxic activity of specific subsets of CD8+ T lymphocytes may also play an important role in this process (Takamatsu et al., 2013). It was supported by DNA immunization studies, demonstrating a correlation between the protection afforded against lethal challenge and the detection of a large number of vaccine‐induced antigen‐specific CD8+ T lymphocytes (Hühr et al., 2020). Interestingly, IgA‐mediated intestinal immune pathways were enriched in both the kidneys and liver, suggesting that intestinal mucosal immunity may play an important role in the process of ASFV infection. In addition, cytochrome P450 (CYP450) and the peroxisome proliferator‐activated receptor (PPAR) signaling pathway have also attracted our attention. Obviously, the activity of these two pathways is caused by the massive activation of energy metabolism (Lamb et al., 2019; Thulasi Raman et al., 2020). In other words, in pigs with acute ASFV infection, the kidneys and livers struggle to provide energy for the antiviral effect. However, exactly how these key metabolites work is still unknown and should be the focus of our attention.
Tissues produce inflammatory factor storms and IFN activation as a synergistic defence against ASFV infection. Previous studies have demonstrated that the IFN system plays an important role in controlling ASFV replication and inducing protective immune responses in infected pigs. Studies have shown that ASFV can inhibit the expression of type I and II IFNs (Esparza et al., 1988). However, previous progress revealed that IFN‐α and IFN‐β can be detected in the serum of animals infected with highly virulent viruses (Karalyan et al., 2012). The results of our analysis showed that the expression of type I and III IFNs was also up‐regulated to varying degrees, which was consistent with previous studies. In fact, ASFV is a DNA virus that replicates primarily in the cytoplasm. The DNA receptor signaling pathway plays a key role in ASFV recognition and type I IFN expression induction. However, viral DNA can also be transcribed into dsRNA by host RNA polymerase III or viral RNA polymerase. Therefore, RNA receptors may also play an important role in ASFV recognition. NF‐κB not only activates the type I IFN promoter but also promotes the transcriptional activation of a large number of genes involved in the inflammatory response (Zhuo et al., 2020). Those associated with PA238L and A179L were expressed in the early stage of viral infection according to our data. Previous studies have shown that PA238L shuttles between the cytoplasm and nucleus; it inhibits not only the production of TNF‐α but also the downstream signaling pathways activated by TNF‐α, inducible nitric oxide synthase and cyclooxygenase (Castelló et al., 2009; Vallée et al., 2001). The high expression of A179L in the early and late stages of ASFV infection indicates that A179L inhibits apoptosis and inflammation during the process of ASFV infection but does not participate in the entry of the core virus particles into the cytoplasm.
Our results provide a reliable method to study the interaction of ASFV with host cells. Cackett et al., 2020 reported that the majority of ASFV genes show some degree of differential expression from early to late infection. Indeed, the choice of exact time point is of great relevance to the conclusion (Cackett et al., 2020). Our sampling time was in the late period of infection, due to the pigs were close to death and all core tissues had undergone pathological phenomena, which belonged to the late period of virus infecting the host. The results of the analysis of the infection group revealed the genes and pathways that are responsive to ASFV infection in pigs. We believe that infectomic maps of ASFV infection provide a foundation for investigating the invasion mechanism, vaccine targets and prevention and control strategies of ASFV infection. At the same time, these results also raised many follow‐up scientific problems for further study. For example, how do DCs play a role in the immune response to ASFV infection? MGF360 and MGF505 of the strains can un‐inhibit the production and response of type I IFN, however, the mechanism of action is still unknown (Sánchez‐Cordón et al., 2018). In addition, the mechanisms of PA238L and A179L expression pattern/phase in infected cells and in vivo remain to be further studied.
CONFLICTS OF INTEREST
The authors declare no conflict of interests.
AUTHOR CONTRIBUTIONS
J. L. and R. H. conceived and designed the project. W. F., P. J., Y. C., P. Y. and H. Z. performed data analyses. T. C., X. Z. and Y. Q. performed the viral replication ability tests and sample testing. J. L. and W. F. prepared the manuscript and completed its revision. L. S., D. L. and W. L. made many suggestions in this study. All authors read and approved the final manuscript.
Supporting information
Supplementary Figure 1. GO analysis results based on common DEGs of three samples in different infected tissues at transcriptome level.
Supplementary Figure 2. KEGG analysis results based on common DEGs of three samples in different infected tissues (Lung, Liver, and Kidney) at transcriptome level.
Supplementary Figure 3. GO analysis results based on common DEGs of three samples in different infected tissues at transcriptome and proteome level.
Supplementary Figure 4. KEGG analysis results based on common DEGs of three samples in different infected tissues (Lung, Liver, and Kidney) at transcriptome and proteome level.
Supplementary Table 1. Common pathway annotations based on Biological process and KEGG analysis at transcriptome and proteome level.
ACKNOWLEDGEMENTS
We thank Dr. Nan Li for the help in TEM assay and all colleagues of BSL‐3 in the Institute of Military Veterinary Medicine, Academy of Military Medical Science for their support during this study. This work was supported by grants from the National Key R&D Program of China (2018YFC0840402), the National Natural Science Foundation of China (31941015), and the Research Project of African Swine Fever of Chinese Academy of Sciences (KJZD‐SW‐L06). J. L. is supported by the Youth Innovation Promotion Association of CAS (2019091).
Fan, W. , Cao, Y. , Jiao, P. , Yu, P. , Zhang, H. , Chen, T. , Zhou, X. , Qi, Y. , Sun, L. , Liu, D. , Zhu, H. , Liu, W. , Hu, R. , & Li, J. (2022). Synergistic effect of the responses of different tissues against African swine fever virus. Transboundary and Emerging Diseases, 69, e204–e215. 10.1111/tbed.14283
Contributor Information
Rongliang Hu, Email: ronglianghu@hotmail.com.
Jing Li, Email: lj418@163.com.
DATA AVAILABILITY STATEMENT
The raw bulk RNA‐seq data have been deposited to Sequence Read Archive (SRA) (https://submit.ncbi.nlm.nih.gov) accession to cite for these SRA data: PRJNA719293. The MS proteomics data have been deposited at the iProX (http://www.iprox.org) repository with the dataset identifier Project ID IPX0002956000.
REFERENCES
- Brown, A. A. , Penrith, M. L. , Fasina, F. O. , & Beltran‐Alcrudo, D. (2018). The African swine fever epidemic in West Africa, 1996–2002. Transboundary and Emerging Diseases, 65, 64–76. 10.1111/tbed.12673 [DOI] [PubMed] [Google Scholar]
- Cackett, G. , Matelska, D. , Sýkora, M. , Portugal, R. , Malecki, M. , Bähler, J. , Dixon, L. , & Werner, F. (2020). The African swine fever virus transcriptome. Journal of Virology, 94. 10.1128/JVI.00119-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo, C. , Borca, M. V. , Afonso, C. L. , Onisk, D. V. , & Rock, D. L. (1994). Long‐term persistent infection of swine monocytes/macrophages with African swine fever virus. Journal of Virology, 68, 580–583. 10.1128/jvi.68.1.580-583.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelló, A. , Quintas, A. , Sánchez, E. G. , Sabina, P. , Nogal, M. , Carrasco, L. , & Revilla, Y. (2009). Regulation of host translational machinery by African swine fever virus. PLoS Pathogens, 5, e1000562. 10.1371/journal.ppat.1000562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, L. K. , Chapman, D. A. , Netherton, C. L. , & Upton, C. . (2013). African swine fever virus replication and genomics. Virus Research, 173, 3–14. 10.1016/j.virusres.2012.10.020 [DOI] [PubMed] [Google Scholar]
- Dixon, L. K. , Sun, H. , & Roberts, H. (2019). African swine fever. Antiviral Research, 165, 34–41. 10.1016/j.antiviral.2019.02.018 [DOI] [PubMed] [Google Scholar]
- Esparza, I. , González, J. C. , & Viñuela, E. (1988). Effect of interferon‐alpha, interferon‐gamma and tumour necrosis factor on African swine fever virus replication in porcine monocytes and macrophages. Journal of General Virology, 69(Pt 12), 2973–2980. 10.1099/0022-1317-69-12-2973 [DOI] [PubMed] [Google Scholar]
- Forth, J. H. , Forth, L. F. , Václavek, P. , Barták, P. , Höper, D. , Beer, M. , & Blome, S. (2020). Whole‐genome sequence of an African swine fever virus isolate from the Czech Republic. Microbiology Resource Announcements, 9. 10.1128/MRA.00948-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo, I. , & Alonso, C. (2017). African swine fever virus: A review. Viruses, 9, 103. 10.3390/v9050103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Hernández, V. , Sánchez‐Bernal, C. , Schvartz, D. , Calvo, J. J. , Sanchez, J. C. , & Sánchez‐Yagüe, J. (2018). A tandem mass tag (TMT) proteomic analysis during the early phase of experimental pancreatitis reveals new insights in the disease pathogenesis. Journal of Proteomics, 181, 190–200. 10.1016/j.jprot.2018.04.018 [DOI] [PubMed] [Google Scholar]
- Gómez‐Villamandos, J. C. , Hervás, J. , Méndez, A. , Carrasco, L. , Martín de las Mulas, J. , Villeda, C. J. , Wilkinson, P. J. , & Sierra, M. A. (1995). Experimental African swine fever: Apoptosis of lymphocytes and virus replication in other cells. Journal of General Virology, 76(Pt 9), 2399–2405. 10.1099/0022-1317-76-9-2399 [DOI] [PubMed] [Google Scholar]
- Hühr, J. , Schäfer, A. , Schwaiger, T. , Zani, L. , Sehl, J. , Mettenleiter, T. C. , Blome, S. , & Blohm, U. (2020). Impaired T‐cell responses in domestic pigs and wild boar upon infection with a highly virulent African swine fever virus strain. Transboundary and Emerging Diseases, 67, 3016–3032. 10.1111/tbed.13678 [DOI] [PubMed] [Google Scholar]
- Karalyan, Z. , Zakaryan, H. , Sargsyan, K. , Voskanyan, H. , Arzumanyan, H. , Avagyan, H. , & Karalova, E. (2012). Interferon status and white blood cells during infection with African swine fever virus in vivo. Veterinary Immunology and Immunopathology, 145, 551–555. 10.1016/j.vetimm.2011.12.013 [DOI] [PubMed] [Google Scholar]
- Kim, D. , Langmead, B. , & Salzberg, S. L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nature Methods, 12, 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, D. C. , Follmer, A. H. , Goldstone, J. V. , Nelson, D. R. , Warrilow, A. G. , Price, C. L. , True, M. Y. , Kelly, S. L. , Poulos, T. L. , & Stegeman, J. J. (2019). On the occurrence of cytochrome P450 in viruses. Proceedings of the National Academy of Sciences of the United States of America, 116, 12343–12352. 10.1073/pnas.1901080116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , He, X. , Yang, Y. , Gong, W. , Huang, K. , Zhang, Y. , Yang, Y. , Sun, X. , Ren, W. , Zhang, Q. , Wu, X. , Zou, Z. , & Jin, M. (2020). Rapid and visual detection of African swine fever virus antibody by using fluorescent immunochromatography test strip. Talanta, 219, 121284. 10.1016/j.talanta.2020.121284 [DOI] [PubMed] [Google Scholar]
- Li, H. , Handsaker, B. , Wysoker, A. , Fennell, T. , Ruan, J. , Homer, N. , Marth, G. , Abecasis, G. , & Durbin, R. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25, 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y. , Smyth, G. K. , & Shi, W. (2014). featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30, 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Malogolovkin, A. , & Kolbasov, D. (2019). Genetic and antigenic diversity of African swine fever virus. Virus Research, 271, 197673. 10.1016/j.virusres.2019.197673 [DOI] [PubMed] [Google Scholar]
- Molini, U. , Mushonga, B. , Settypalli, T. B. K. , Dundon, W. G. , Khaiseb, S. , Jago, M. , Cattoli, G. , & Lamien, C. E. (2020). Molecular characterization of African swine fever virus from outbreaks in Namibia in 2018. Transboundary and Emerging Diseases, 67, 1008–1014. 10.1111/tbed.13399 [DOI] [PubMed] [Google Scholar]
- Ramiro‐Ibáñez, F. , Ortega, A. , Brun, A. , Escribano, J. M. , & Alonso, C. (1996). Apoptosis: A mechanism of cell killing and lymphoid organ impairment during acute African swine fever virus infection. Journal of General Virology, 77(Pt 9), 2209–2219. 10.1099/0022-1317-77-9-2209 [DOI] [PubMed] [Google Scholar]
- Ritchie, M. E. , Phipson, B. , Wu, D. , Hu, Y. , Law, C. W. , Shi, W. , & Smyth, G. K. (2015). limma powers differential expression analyses for RNA‐sequencing and microarray studies. Nucleic Acids Research, 43, e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M. D. , McCarthy, D. J. , & Smyth, G. K. (2010). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salguero, F. J. , Sánchez‐Cordón, P. J. , Sierra, M. A. , Jover, A. , Núñez, A. , & Gómez‐Villamandos, J. C. (2004). Apoptosis of thymocytes in experimental African Swine Fever virus infection. Histology and Histopathology, 19, 77–84. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Cordón, P. J. , Jabbar, T. , Berrezaie, M. , Chapman, D. , Reis, A. , Sastre, P. , Rueda, P. , Goatley, L. , & Dixon, L. K. (2018). Evaluation of protection induced by immunisation of domestic pigs with deletion mutant African swine fever virus BeninΔMGF by different doses and routes. Vaccine, 36, 707–715. 10.1016/j.vaccine.2017.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez‐Torres, C. , Gómez‐Puertas, P. , Gómez‐del‐Moral, M. , Alonso, F. , Escribano, J. M. , Ezquerra, A. , & Domínguez, J. (2003). Expression of porcine CD163 on monocytes/macrophages correlates with permissiveness to African swine fever infection. Archives of Virology, 148, 2307–2323. 10.1007/s00705-003-0188-4 [DOI] [PubMed] [Google Scholar]
- Takamatsu, H. H. , Denyer, M. S. , Lacasta, A. , Stirling, C. M. , Argilaguet, J. M. , Netherton, C. L. , Oura, C. A. , Martins, C. , & Rodríguez, F. (2013). Cellular immunity in ASFV responses. Virus Research, 173, 110–121. 10.1016/j.virusres.2012.11.009 [DOI] [PubMed] [Google Scholar]
- Thulasi Raman, S. N. , Latreille, E. , Gao, J. , Zhang, W. , Wu, J. , Russell, M. S. , Walrond, L. , Cyr, T. , Lavoie, J. R. , Safronetz, D. , Cao, J. , Sauve, S. , Farnsworth, A. , Chen, W. , Shi, P. Y. , Wang, Y. , Wang, L. , Rosu‐Myles, M. , & Li, X. (2020). Dysregulation of Ephrin receptor and PPAR signaling pathways in neural progenitor cells infected by Zika virus. Emerging Microbes and Infections, 9, 2046–2060. 10.1080/22221751.2020.1818631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallée, I. , Tait, S. W. , & Powell, P. P. (2001). African swine fever virus infection of porcine aortic endothelial cells leads to inhibition of inflammatory responses, activation of the thrombotic state, and apoptosis. Journal of Virology, 75, 10372–10382. 10.1128/JVI.75.21.10372-10382.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G. , Wang, L. G. , Han, Y. , & He, Q. Y. (2012). clusterProfiler: An R package for comparing biological themes among gene clusters. Omics, 16, 284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, D. , Liu, R. , Zhang, X. , Li, F. , Wang, J. , Zhang, J. , Liu, X. , Wang, L. , Zhang, J. , Wu, X. , Guan, Y. , Chen, W. , Wang, X. , He, X. , & Bu, Z. (2019). Replication and virulence in pigs of the first African swine fever virus isolated in China. Emerging Microbes and Infections, 8, 438–447. 10.1080/22221751.2019.1590128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo, Y. , Guo, Z. , Ba, T. , Zhang, C. , He, L. , Zeng, C. , & Dai, H. (2020). African swine fever virus MGF360‐12L inhibits type I interferon production by blocking the interaction of importin α and NF‐κB signaling pathway. Virologica Sinica, 10.1007/s12250-020-00304-4:1-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. GO analysis results based on common DEGs of three samples in different infected tissues at transcriptome level.
Supplementary Figure 2. KEGG analysis results based on common DEGs of three samples in different infected tissues (Lung, Liver, and Kidney) at transcriptome level.
Supplementary Figure 3. GO analysis results based on common DEGs of three samples in different infected tissues at transcriptome and proteome level.
Supplementary Figure 4. KEGG analysis results based on common DEGs of three samples in different infected tissues (Lung, Liver, and Kidney) at transcriptome and proteome level.
Supplementary Table 1. Common pathway annotations based on Biological process and KEGG analysis at transcriptome and proteome level.
Data Availability Statement
The raw bulk RNA‐seq data have been deposited to Sequence Read Archive (SRA) (https://submit.ncbi.nlm.nih.gov) accession to cite for these SRA data: PRJNA719293. The MS proteomics data have been deposited at the iProX (http://www.iprox.org) repository with the dataset identifier Project ID IPX0002956000.


