Abstract
There is a need for new effective antivirals, particularly in response to the development of antiviral drug resistance and emerging RNA viruses such as SARS‐CoV‐2. Plants are a significant source of structurally diverse bioactive compounds for drug discovery suggesting that plant‐derived natural products could be developed as antiviral agents. This article reviews the antiviral activity of plant‐derived natural products against RNA viruses, with a focus on compounds targeting specific stages of the viral life cycle. A range of plant extracts and compounds have been identified with antiviral activity, often against multiple virus families suggesting they may be useful as broad‐spectrum antiviral agents. The antiviral mechanism of action of many of these phytochemicals is not fully understood and there are limited studies and clinical trials demonstrating their efficacy and toxicity in vivo. Further research is needed to evaluate the therapeutic potential of plant‐derived natural products as antiviral agents.
Keywords: antiviral, drug discovery, mechanism of action, natural product, phytochemical, plant, RNA virus
Significance and Impact of the Study: The COVID‐19 pandemic has emphasised the need for new antiviral agents, including broad‐spectrum drugs that can be rapidly deployed against emerging RNA viruses. Plant‐derived natural products act on a diverse range of targets at different stages of the RNA virus life cycle suggesting they may be useful for the development of novel antiviral drugs. However, this review demonstrates that the modes of action of antiviral phytochemicals are often not well understood, and there are limited studies pertaining to in vivo efficacy, pharmacokinetic profile and toxicity, indicating that further research is required to evaluate the therapeutic potential of these compounds.
Introduction
Viral infections present a significant global health and socioeconomic burden (Gasparini et al. 2012; Dye 2014; Klepser 2014). This is exemplified by the emergence of the Coronavirus Disease 2019 (COVID‐19) pandemic, which has caused >180 million infections and 4 million deaths (World Health Organization 2021) and is the largest pandemic since the 1918 influenza outbreak (Cascella et al. 2021). RNA viruses are the most common source of emerging human infectious diseases, with notable zoonotic viral infections including coronaviruses, Zika, Ebola, chikungunya and dengue virus (DENV) (Carrasco‐Hernandez et al. 2017). RNA viruses typically adapt more rapidly to new hosts owing to their relatively high mutation rates, given the error‐prone nature of the viral RNA dependent RNA polymerase (RdRp) and frequent recombination events (Holmes 2009). Understanding the threat of RNA viruses is key to limit and manage future global pandemics (Carrasco‐Hernandez et al. 2017). There are 214 RNA virus species that cause infection in humans (Woolhouse and Brierley 2018), ranging in severity from mild to fatal in addition to causing severe chronic infections such as human immunodeficiency virus (HIV) and hepatitis C virus (HCV). There are approximately 38 million people living with HIV globally (UNAIDS 2020), and 71 million people with HCV, which causes severe liver cirrhosis in 10% of cases (World Health Organization 2020).
The current COVID‐19 pandemic has emphasized a need for antiviral therapies that can be rapidly deployed against emerging viruses, particularly in the absence of an effective vaccine. Antiviral drugs have traditionally been developed as single spectrum agents and there is a lack of broad‐spectrum antivirals for treating emerging viral diseases (Vigant et al. 2015; Pattnaik and Chakraborty 2020). There has been increasing interest in repurposing of approved antiviral drugs for the treatment of new viral diseases and the discovery of broad‐spectrum antiviral agents (Vigant et al. 2015; Harrison 2020; Pattnaik and Chakraborty 2020). The corticosteroid dexamethasone was the first approved treatment for hospitalized COVID‐19 patients (Department of Health and Social Care 2020a) and the broad‐spectrum antiviral remdesivir was also approved in the European Union (European Medicines Agency 2021), United Kingdom (Department of Health and Social Care 2020b) and United States (Rubin et al. 2020), while a range of other drugs are undergoing clinical trials (de Clercq and Li 2016; Normand 2020). The majority of currently licensed antivirals are antiretroviral drugs for HIV, or those that target chronic and latent diseases such as HCV (de Clercq and Li 2016), yet due to the rapid evolutionary rates of RNA viruses, antiviral resistance is a significant problem. For example, drug‐resistant HIV has increased globally, and pan‐resistant isolates have been reported (Puertas et al. 2020). A potential strategy to limit the development of viral escape is to target host proteins used for viral replication rather than virus itself (Müller et al. 2012). Broad‐spectrum antivirals targeting conserved proteins are also hypothesized to have a higher threshold to resistance development; viral escape mutations in the coronavirus murine hepatitis virus (MHV) in the presence of remdesivir, which targets RdRp, had a significant cost to infection in a mouse model, suggesting conserved proteins are a promising target for broad‐spectrum antivirals (Agostini et al. 2018).
Plants have an extensive history of use as traditional medicines, both for communicable and non‐communicable diseases. Medicinal plants have been investigated as a source of novel therapeutic agents since the advent of modern drug discovery techniques (Beutler 2019). Plants produce an array of structurally diverse and often complex secondary metabolites with a range of functions; such compounds evolved for a particular biological function within plants, and they frequently exhibit biologically relevant privileged molecular scaffolds, which lends to their use as a reservoir for drug discovery (Rodrigues et al. 2016; Grigalunas et al. 2020). There has been a shift away from natural product research and towards high throughput screening of synthetic compounds for drug discovery over recent decades (Wright 2019). This can be attributed to the time‐consuming nature of identifying bioactive constituents, difficulties with synthesis and derivatization of natural product compounds, incompatibility with high throughput screening assays and the need for dereplication to remove previously characterized compounds (Atanasov et al. 2015; Beutler 2019; Wright 2019). A further challenge is that many phytochemicals exhibit poor solubility and bioavailability, requiring drug delivery strategies for their successful application as pharmaceutical products. The use of drug delivery strategies to improve the delivery of antiviral phytochemicals has been recently reviewed by Ben‐Shabat et al. (2020). Conversely, natural products are still a significant source of new drugs, as an estimated 34% of approved new chemical entities between 2000 and 2014 were derived from natural products of plants and other organisms (Newman and Cragg 2016). Plants could therefore be a reservoir of novel antiviral agents, and there is a large body of literature reporting the antiviral activity of plants and their constituents. Many natural products are also known to exhibit antitumor activity which could be used to prevent or treat cancers associated with viral infections such as HCV and HIV (Fatima et al. 2019; Akhtar et al. 2020).
Biologically active plant‐derived natural products are traditionally discovered using bioactivity guided fractionation. Crude extracts are first screened for their pharmacological activity, and fractionation steps are iteratively performed until the bioactive compounds are isolated and identified (Atanasov et al. 2021). There are a large number of published studies on the antiviral activity of plant crude extracts and their constituents, often based on plants used in traditional medicine. For example, bioactivity‐guided fractionation was recently employed to determine the antiviral activity of 26 Nigerian medicinal plants against echoviruses E7, E13 and E19, with 10 plant extracts demonstrating antiviral activity. Macaranga barteri methanolic extract was the most potent antiviral, with a 50% effective dose (EC50) of 0·0017–00·028 ng ml−1, of which the dichloromethane fraction (containing flavanoids) displayed the greatest activity with EC50 values of 0·0075–0·0175 ng ml−1 (Ogbole et al. 2018). Such studies often do not identify the active constituents of the crude extract. Plant extracts can be difficult to standardize, with variation in composition based on the growth conditions and extraction method (Wei et al. 2019), highlighting the importance of identifying active constituents. Moreover, a number of studies do not investigate beyond the initial determination of antiviral activity of crude extracts or their components, leaving the mechanism of action and molecular target undetermined. Advances in in silico approaches have enabled computational screening of natural product libraries for compounds that potentially bind to specific viral targets, which can then be subject to in vitro assays (Romano and Tatonetti 2019). Recently, computational methods have been employed extensively for natural products against potential severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) targets in response to the COVID‐19 pandemic, however the antiviral activity of identified compounds in vitro are not reported in many of these studies (Kandeel et al. 2020; Tahir ul Qamar et al. 2020; Gyebi et al. 2021). There is a need for further in‐depth studies to be performed against many of the antiviral plant products reported in the published literature to assess their importance in the development of antiviral agents. Despite this, the mode of action has been elucidated for a range of plant extracts with promising antiviral activity, targeting different steps throughout the viral life cycle and there are several examples of novel targets being identified, indicating that this is an under‐researched area for further development. The aim of this article was therefore to discuss the antiviral activity of natural product compounds known to target specific stages of the life cycle of RNA viruses and their potential utility for the development of antiviral agents.
Viral life cycle
Viruses hijack the host cell and induce an array of changes to enable replication and production of progeny viruses. Briefly, the viral life cycle can be divided into three major stages: entry, replication, and assembly and exit (Fig. 1). During entry, virions first attach to a specific host receptor and enter by endocytosis or fusion with the plasma membrane. The virus coat or capsid is then shed, either in the cytoplasm or in acidified endosomes, and the viral genome is released (Ryu 2017). The viral genome is then used for replication and transcription. The Baltimore system classifies viruses into seven genome types: double‐stranded (ds) DNA, single‐stranded (ss) DNA, dsRNA, positive sense (+) ssRNA, negative sense (–) ssRNA and +ssRNA viruses with a DNA intermediate (Flint et al. 2015; Aiewsakun and Simmonds 2018). The details of replication can vary significantly between RNA viruses depending on the genome type. In dsRNA viruses, the strand is transcribed into mRNA using virally encoded RdRp, and subsequently the mRNA is used to produce new copies of the genome. For +ssRNA viruses, the genome is directly translated into proteins, whereas in –ssRNA viruses require RdRp to transcribe the genome into mRNA. Finally, retroviruses use virally encoded reverse transcriptase (RT) to copy their +ssRNA genome into dsDNA, which is subsequently transcribed by host cell enzymes (Flint et al. 2015).
Figure 1.
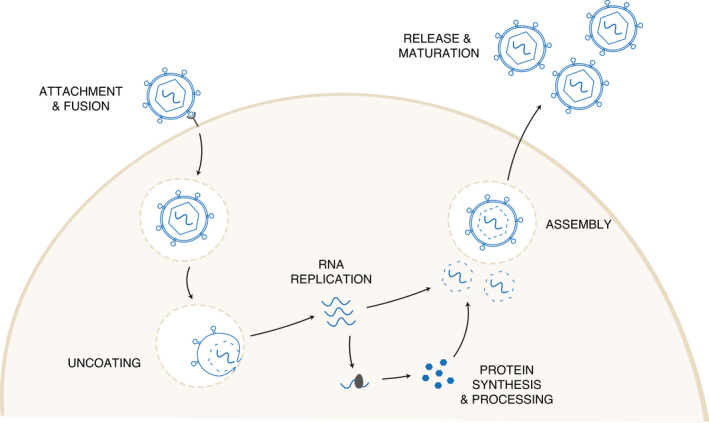
Stages of the viral life cycle.
Replication can occur in different sites within the host cell, such as in the nucleus for influenza A virus (IAV) or HIV (Samji 2009), and in the cytoplasm in replication complexes for HCV and DENV (Ci and Shi 2021). Protein translation relies on host cell processes, with viruses possessing various strategies to shut down host protein translation to increase efficiency of viral protein translation (Walsh and Mohr 2011). Many viruses also encode highly conserved proteases that cleave the polyprotein and play a crucial role in replication (Sharma and Gupta 2017). Newly synthesized proteins are trafficked through the endoplasmic reticulum and Golgi apparatus where capsid assembly and genome packaging occurs to produce progeny viruses. Capsid assembly and genome packaging can be a sequential process, for example in picornaviruses, or occur simultaneously such as in adenoviruses (Ryu 2017). Progeny are subsequently released from the host cell, either by cell lysis (non‐enveloped viruses) or budding (enveloped viruses), where the host cell lipid bilayer and transmembrane viral proteins surround the capsid as it is released from the plasma membrane (Willey et al. 2011). Finally, maturation occurs, where structural changes are induced to allow the virion to become infectious; for example, in HIV, the conformation of the viral capsid is altered by viral protease cleaving the Gag precursor protein into mature Gag proteins (Freed 2015).
Plant‐derived natural products targeting specific steps in the viral life cycle
Viral entry
Attachment and fusion
There are several examples of plant‐derived natural products from a range of compound classes that inhibit the interactions between viral ligands and host cell receptors, suggesting they could potentially be developed as antiviral agents (Tables 1 and 2). Lectins are a structurally diverse group of non‐enzymatic proteins that bind to carbohydrates (Mitchell et al. 2017). Binding to carbohydrates occurs via hydrogen bonds between polar residues and the hydroxyl groups of carbohydrates (Bellande et al. 2017). Lectins derived from plants have been extensively investigated as potential entry inhibitors (Table 1), primarily focusing on their applications as topical prophylactics against sexually transmitted diseases including HIV (Mitchell et al. 2017). BanLec, a lectin isolated from banana fruits, exhibited antiviral activity against a range of HIV‐1 and HIV‐2 isolates at an EC50 ranging 0·48–2·06 nmol l−1 (Swanson et al. 2010); the mechanism of action was attributed to BanLec binding to high mannose type glycans present on the viral envelope glycoprotein gp120, subsequently blocking glycan‐mediated attachment to the host cell receptor, CD4, and/or co‐receptors CCR5 and CXCR4 (Swanson et al. 2010; Hopper et al. 2017). Further development of BanLec was subsequently performed to ameliorate potential mitogenic effects of this compound, which could lead to side effects in clinical applications; a BanLec mutant (H84T) substituting a histidine residue for threonine was significantly less mitogenic while retaining a similar EC50 against a range of HIV isolates (0·33–4·10 nmol l−1) as wild type BanLec (0·87–14·00 nmol l−1). Prophylactic vaginal application of H84T BanLec prevented HIV infection in a bone‐marrow‐liver‐thymus humanized mouse model, with no detectable infections in the sample group, compared to infection in 50% of the control group (Table 3). BanLec also demonstrated antiviral activity against HCV and H1N1, H3N2 and H5N1 strains of IAV, which also possess high mannose type glycans on their surface, suggesting that this compound has promise as a broad‐spectrum antiviral (Swanson et al. 2015). Griffithsin is another well‐characterized mannose‐specific lectin, derived from red marine algae (Griffithisia sp.), with significant antiviral activity against HIV (Emau et al. 2007). Griffithsin also inhibits coronaviruses; the infectivity of Middle East respiratory syndrome coronavirus (MERS‐CoV) was reduced by over 90% in MRC‐5, Vero and Huh‐7 cells by 2 μg ml−1 griffithsin (Millet et al. 2016), while severe acute respiratory syndrome coronavirus (SARS‐CoV) was inhibited by griffithsin at an EC50 of 0·61‐1·19 µg ml−1 (O’Keefe et al. 2010). Griffithsin binds to the spike glycoprotein of coronaviruses, and while binding to the host cell receptor human angiotensin‐converting enzyme 2 (ACE2) still occurs, viral entry is inhibited (O’Keefe et al. 2010). O’Keefe et al. (2010) also reported that griffithsin (10 mg kg−1 day−1) significantly increased the survival of SARS‐CoV infected mice to 100% compared to 30% in the control group (Table 3). These studies suggest that plant lectins could be useful for the development of broad‐spectrum antiviral agents.
Table 1.
Antiviral activity (EC50 or percentage inhibition) and cytotoxicity (CC50 or percentage inhibition) of plant derived natural products targeting different stages within the viral life cycle (– Not defined; * Extract contains mixture of compounds)
| Life cycle stage | Extract/compound | Compound type | Structure | Virus | Target | EC50/inhibition (%) | CC50/inhibition (%) | Reference |
|---|---|---|---|---|---|---|---|---|
| Attachment/Fusion | BanLec | Lectin |
PDB ID: 7KMU (Coves‐Datson et al. 2021; Meagher and Stuckey 2021)
|
HIV | Surface glycoprotein high mannose type glycans | 0·48–2·06 nmol l−1 | – | Swanson et al. (2010, 2015) |
| HCV | Surface glycoprotein high mannose type glycans | 9·5–20·8 nmol l−1 | – | Swanson et al. (2015) | ||||
| IAV | Surface glycoprotein high mannose type glycans | 0·06–11 µg ml−1 | – | Swanson et al. (2015) | ||||
| H84T BanLec | Lectin | HIV | Surface glycoprotein high mannose type glycans | 0·33–4·10 nmol l−1 | – | Swanson et al. (2015) | ||
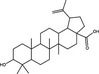
| Betulinic acid | Triterpenoid |
|
HCV | – | 4·2%, 1 µmol l−1 | ~75%, 50 µmol l−1 | Xiao et al. (2014) | |
| β‐cyclodextrin‐betulinic acid conjugates | Triterpenoid | HCV | – | 37·1–88·2%, 1 µmol l−1 | ~15–85%, 50 µmol l−1 | Xiao et al. (2014) | ||
| Betulinic acid derivatives | Triterpenoid | HIV | gp120 | 0·31 to >40 µmol l−1 | – | Sun et al. (2002) | ||
| Echinocystic acid | Triterpenoid |
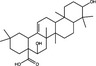
|
HCV | HCV envelope glycoprotein 2 | 90%, 10 µmol l−1 | >50 µmol l−1 | Yu et al. (2013) | |
| HCV | HCV envelope glycoprotein 2 | 27·5%, 1 µmol l−1 | 0%, 50 µmol l−1 | Xiao et al. (2014) | ||||
| β‐cyclodextrin‐echinocystic acid conjugates | Triterpenoid | HCV | – | −4·0 to 86·5%, 1 µmol l−1 | ~1–90%, 50 µmol l−1 | Xiao et al. (2014) | ||
| Echinocystic acid‐galactose conjugate | Triterpenoid | IAV | HA | 5 µmol l−1 | >200 µmol l−1 | Yu et al. (2014) | ||
| Fortunella margarita EO | – | * | IAV | – | 6·77 µg ml−1 | 239·54 µg ml−1 | Ibrahim et al. (2015) | |
| Griffithsin | Lectin |
PDB ID: 2GTY (Ziolkowska and Wlodawer 2006; Ziolkowska et al. 2006)
|
HIV | Surface glycoprotein high mannose type glycans | 0·023–0·21 nmol l−1 | >500 nmol l−1 | Emau et al. (2007) | |
| MERS‐CoV | Surface glycoprotein high mannose type glycans | 44·7–63·2%, 0·125 µg ml−1 | >2 µg ml−1 | Millet et al. (2016) | ||||
| SARS‐CoV | Surface glycoprotein high mannose type glycans | 0·61–1·19 µg ml−1 | >100 µg ml−1 | O’Keefe et al. (2010) | ||||
| Isorhamnetin | Flavonoid |
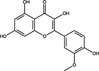
|
SARS‐CoV‐2 | ACE2 | 47·7%, 50 µmol l−1 | >200 µmol l−1 | Zhan et al. (2021) | |
| Oleanolic acid | Triterpenoid |
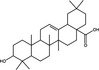
|
HCV | HCV envelope glycoprotein 2 | 60%, 10 µmol l−1 | >50 µmol l−1 | Yu et al. (2013) | |
| HCV | HCV envelope glycoprotein 2 | 22·4%, 1 µmol l−1 | ~5%, 50 µmol l−1 | Xiao et al. (2014) | ||||
| β‐cyclodextrin‐ oleanolic acid conjugates | Triterpenoid | HCV | – | 25·2–82·7%, 1 µmol l−1 | ~0–90%, 50 µmol l−1 | Xiao et al. (2014) | ||
| Quercetin | Flavonoid |
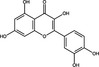
|
IAV | HA2 subunit | 1·93–7·76 µg ml−1 | >250 µg ml−1 | Wu et al. (2015) | |
| Ursolic acid | Triterpenoid |

|
HCV | – | 13·8%, 1 µmol l−1 | ~90%, 50 µmol l−1 | Xiao et al. (2014) | |
| β‐cyclodextrin‐ursolic acid acid conjugates | Triterpenoid | HCV | – | 13·6–62·6%, 1 µmol l−1 | ~1–90%, 50 µmol l−1 | Xiao et al. (2014) | ||
| Uncoating | Meliacine | Cyclic peptide | Cyclic peptide with aliphatic amino acids, MW 2·2–2·3 kDa | Foot and mouth disease virus | Lysosome acidification | 0·5 µg ml−1 | >100 µg ml−1 | Wachsman et al. (1998) |
| Tea tree EO | – | * | IAV | Lysosome acidification | 0·0006% (v/v) | 0·025% (v/v) | Garozzo et al. (2011) | |
| Terpinen‐4‐ol | Monoterpenoid |
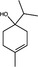
|
IAV | Lysosome acidification | 0·002% (v/v) | – | Garozzo et al. (2011) | |
| Polyprotein processing | Caffeic acid | Hydroxycinnamic Acid |
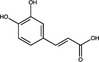
|
HIV | HIV Protease | 90·2%, 1 mg ml−1 | – | Wang et al. (2019) |
| Ethyl caffeate | Hydroxycinnamic Acid |
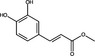
|
HIV | HIV Protease | 100%, 1 mg ml−1 | – | Wang et al. (2019) | |
| Isovanillin | Phenolic Aldehyde |
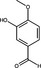
|
HIV | HIV Protease | 61·2%, 1 mg ml−1 | – | Wang et al. (2019) | |
| Corilagin | Polyphenol |
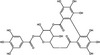
|
HCV | NS3 protease | 13·59 µmol l−1 | 96·65 µmol l−1 | Liu et al. (2012) | |
| Excoecariphenol D | Polyphenol |
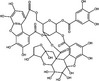
|
HCV | NS3 protease | 12·61 µmol l−1 | 56·25 µmol l−1 | Li et al. (2012) | |
| Shuanghuanglian | – | * | SARS‐CoV‐2 | 3CLPro | 0·93–1·20 µl ml−1 | >12·50 µl ml−1 | Su et al. (2020) | |
| Baicalin | Flavonoid |
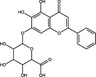
|
SARS‐CoV‐2 | 3CLPro | 27·87 µmol l−1 | >200 µmol l−1 | Su et al. (2020) | |
| Baicalein | Flavonoid |
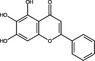
|
SARS‐CoV‐2 | 3CLPro | 2·94 µmol l−1 | >200 µmol l−1 | Su et al. (2020) | |
| Apigenin | Flavonoid |
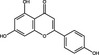
|
HCV | RdRp NS5B | 50 µmol l−1 | 75%, 100 µmol l−1 | Manvar et al. (2012) | |
| RNA Replication | Calanolide A | Coumarin |
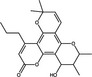
|
HIV | RT | 0·08–0·50 µmol l−1 | 7·3‐>10·0 µmol l−1 | Buckheit et al. (1999) |
| Eclipra alba aqueous extract | – | * | HCV | RdRp NS5B | 95%, 130 µg ml−1 | 40%, 100 µg ml−1 | Manvar et al. (2012) | |
| Wedelolactone | Coumestan |
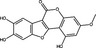
|
HCV | RdRp NS5B | 80%, 50 µmol ml−1 | 79%, 100 µmol l−1 | Manvar et al. (2012) | |
| Luteolin | Flavonoid |
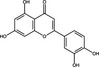
|
HCV | RdRp NS5B | 50 µmol l−1 | 88%, 100 µmol l−1 | Manvar et al. (2012) | |
| Green tea extract | – | * | SARS‐CoV‐2 | NSp15 endo‐ribonuclease | 0·24 µg ml−1 | – | Hong et al. (2021) | |
| Epigallocatechin gallate | Polyphenol |
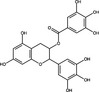
|
SARS‐CoV‐2 | NSp15 endo‐ribonuclease | 0·092 µg ml−1 | – | Hong et al. (2021) | |
| Protein Synthesis | Aeginetia indica aqueous extract | – | * | HCV | NS5A phosphorylation | 90%, 8 mg ml−1 | 14 mg ml−1 | Lin et al. (2018) |
| Camptothecin | Alkaloid |
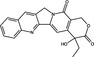
|
Echovirus 71 | Topoisomerase 1 | 90%, 10 µmol l−1 | >100 µmol l−1 | Wu and Chu (2017) | |
| Castanospermine | Iminosugar |
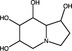
|
Zika virus | α‐glucosidases | ~60–90%, 1 µmol l−1 | >100 μmol l−1 | Bhushan et al. (2020) | |
| Curcumin | Polyphenol |
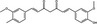
|
EV71 | Protein kinase Cδ | ~50%, 10 µmol l−1 | 40–50%, 50 µmol l−1 | Huang et al. (2018) | |
| Celgosivir | Iminosugar |
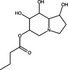
|
DENV | α‐glucosidases | 5·17 μmol l−1 | >100 μmol l−1 | Sayce et al. (2016) | |
| Zika virus | α‐glucosidases | ~44–97%, 1 µmol l−1 | >100 μmol l−1 | Bhushan et al. (2020) | ||||
| Deoxy‐nojirimycin | Iminosugar |
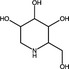
|
Zika virus | α‐glucosidases | ~50–99%, 1 µmol l−1 | >100 μmol l−1 | Bhushan et al. (2020) | |
| Luteolin | Flavonoid |
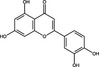
|
Coxsackie A16 | – | 7·4 µmol l−1 | 148·02 µmol l−1 | Xu et al. (2014) | |
| EV71 | 10·31 µmol l−1 | 148·02 µmol l−1 | Xu et al. (2014) | |||||
| Silvestrol | Flavagline |
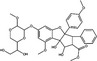
|
Ebola virus | eIF4a | 0·8 nmol l−1 | >10 nmol l−1 | Müller et al. (2018) | |
| Human coronavirus 229E | eIF4a | 3 nmol l−1 | >10 nmol l−1 | Müller et al. (2018) | ||||
| MERS‐CoV | eIF4a | 1·3 nmol l−1 | >10 nmol l−1 | Müller et al. (2018) | ||||
| Human rhinovirus A1 | eIF4a | 100 nmol l−1 | >10 nmol l−1 | Müller et al. (2018) | ||||
| Poliovirus | eIF4a | 20 nmol l−1 | >10 nmol l−1 | Müller et al. (2018) | ||||
| Zika virus | eIF4a | ~45%, 10 nmol l−1 | ~70%, 50 nmol l−1 | Elgner et al. (2018) | ||||
| Assembly | Berberine | Alkaloid |
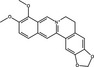
|
IAV | MAPK/ERK pathway (ribo‐nucleoprotein export) | 52 μmol l−1 | 1035 μmol l−1 | Botwina et al. (2020) |
| Hemanthamine | Alkaloid |
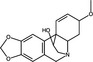
|
IAV | Ribo‐nucleoprotein export | 1·48 μmol l−1 | 50 μmol l−1 | He et al. (2013) | |
| Lycorine | Alkaloid |
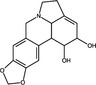
|
IAV | Ribo‐nucleoprotein export | <0·46 μmol l−1 | 20·9 μmol l−1 | He et al. (2013) | |
| Maturation | Bevirimat | Triterpenoid |
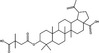
|
HIV | Gag polyprotein | 0·065 μmol l−1 | >2 μmol l−1 | Zhao et al. (2021) |
| Luteolin | Flavonoid |
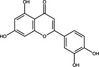
|
DENV | Furin protease | 4·36–8·38 μmol l−1 | 45·89 μmol l−1 | Peng et al. (2017) |
Table 2.
Inhibition (IC50 or percentage inhibition) of target viral or host proteins by plant‐derived natural products. (– Not defined; *Extract contains mixture of compounds)
| Life cycle stage | Extract/compound | Compound type | Structure | Target | IC50/inhibition (%) | Reference |
|---|---|---|---|---|---|---|
| Attachment/Fusion | Geranium EO | – | * | ACE‐2 | 10·63%, 50 μg ml−1 | Senthil Kumar et al. (2020) |
| Lemon EO | – | * | ACE‐2 | 24·79%, 25 μg ml−1 | Senthil Kumar et al. (2020) | |
| Citronellol | Monoterpenoid |
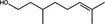
|
ACE‐2 | 57·39%, 50 μmol ml−1 | Senthil Kumar et al. (2020) | |
| Geraniol | Monoterpenoid |
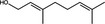
|
ACE‐2 | 42·00%, 50 μmol ml−1 | Senthil Kumar et al. (2020) | |
| Limonene | Monoterpene |
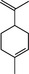
|
ACE‐2 | 28·22%, 50 μmol ml−1 | Senthil Kumar et al. (2020) | |
| Neryl acetate | Monoterpenoid |
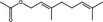
|
ACE‐2 | 7·61%, 50 μmol ml−1 | Senthil Kumar et al. (2020) | |
| Quercetin | Flavonoid |
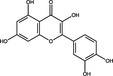
|
ACE2 | 4·48 μmol l−1 | Liu et al. (2020) | |
| Polyprotein Processing | Caffeic acid | Hydroxycinnamic acid |
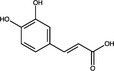
|
HIV Protease | 1·5 μmol l−1 | Wang et al. (2019) |
| Ethyl Caffeate | Hydroxycinnamic acid |
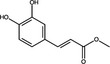
|
HIV Protease | 1 μmol l−1 | Wang et al. (2019) | |
| Isovanillin | Phenolic aldehyde |
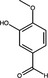
|
HIV Protease | 3·5 μmol l−1 | Wang et al. (2019) | |
| Corilagin | Polyphenol |
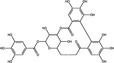
|
HCV NS3 Protease | 3·45 μmol l−1 | Li et al. (2012) | |
| Excoecariphenol D | Polyphenol |
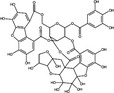
|
HCV NS3 Protease | 6·93 μmol l−1 | Li et al. (2012) | |
| Shuanghuanglian | – | * | SARS‐CoV‐2 3CLPro | 0·01–0·09 μl ml−1 | Su et al. (2020) | |
| Baicalin | Flavonoid |
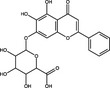
|
SARS‐CoV‐2 3CLPro | 6·41 μmol l−1 | Su et al. (2020) | |
| Baicalein | Flavonoid |
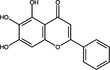
|
SARS‐CoV‐2 3CLPro | 0·94 μmol l−1 | Su et al. (2020) | |
| RNA replication | Eclipta alba aqueous extract | – | * | HCV NS5B | 11 μg ml−1 | Manvar et al. (2012) |
| Apigenin | Flavonoid |
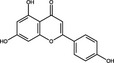
|
HCV NS5B | 175·5 μg ml−1 | Manvar et al. (2012) | |
| Luteolin | Flavonoid |
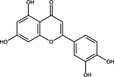
|
HCV NS5B | 11·3 μg ml−1 | Manvar et al. (2012) | |
| Wedolactone | Coumestan |
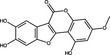
|
HCV NS5B | 7·7 μg ml−1 | Manvar et al. (2012) | |
| Green tea extract | – | * | SARS‐CoV‐2 NSp15 | 2·54 μg ml−1 | Hong et al. (2021) | |
| Epigallocatechin gallate | Polyphenol |
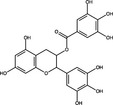
|
SARS‐CoV‐2 NSp15 | 0·74 μg ml−1 | Hong et al. (2021) | |
| Peptide AIHIILI | Peptide | 7 amino acid peptide | HIV RT | 274 nmol l−1 | Seetaha et al. (2021) | |
| Peptide LIAVSTNIIFIVV | Peptide | 13 amino acid peptide | HIV RT | 236·4 nmol l−1 | Seetaha et al. (2021) |
Table 3.
In vivo efficacy of plant‐derived natural products that target specific phases of the viral life cycle
| Life cycle stage | Extract/compound | Virus | Model | Results | Reference |
|---|---|---|---|---|---|
| Attachment/Fusion | BanLec H84T (Lectin) | HIV | Bone‐marrow‐liver‐thymus humanized mouse model; 75 μg, prophylactic vaginal administration | Prophylactic treatment with H84T significantly (P ≤ 0·05) reduced the incidence of infection, with no HIV infection detected in the H84T treatment group compared to 50% of the control group | Swanson et al. (2015) |
| Griffithsin (Lectin) | SARS‐CoV | Mouse, 10 mg kg−1 day−1, intranasal administration | All mice in the griffithsin treatment group survived SARS‐CoV infection compared to 30% survival in the no treatment group. Lung viral titres and weight loss were also significantly (P ≤ 0·05) reduced by griffithsin treatment | O’Keefe et al. (2010) | |
| RNA replication | Calanolide A (Coumarin) | HIV | Human, phase 1 clinical trial of healthy volunteers (n = 47); escalating doses for 5 days | Transient mild to moderate adverse effects were observed. Adverse effects and laboratory abnormalities were non‐dose dependent. Absorption profiles and plasma levels of calanolide A varied significantly | Eiznhamer et al. (2002) |
| Protein synthesis | Celgosivir (Iminosugar) | DENV | Mouse; 33 mg kg−1 three times daily, oral gavage | Significant reduction (P ≤ 0·05) in viral RNA load. Marginal but non‐significant (P > 0·05) reduction in infectious viral load | Sayce et al. (2016) |
| DENV | Human phase 1b trial of dengue fever patients (n = 50); initial dose 400 mg, followed by 200 mg every 12 h for 4·5 days | Celgosivir was generally well tolerated. Treatment marginally but non‐significantly (P > 0·05) reduced viral load compared to the control (−1·86 vs −1·64 virological log10 reduction). No significant differences (P > 0·05) in fever burden or haematological and biochemical markers were observed | Low et al. (2014) | ||
| UV‐4B (Iminosugar) | IAV | Mouse; single dose 100–1000 mg kg−1, oral gavage | Significant dose‐dependent reduction in mortality at doses; >250 mg kg−1 produced a 30–70% survival compared to 10% in the control group | Warfield et al. (2020) | |
| DENV | Mouse; single dose 250–1000 mg kg−1, oral gavage | Significant dose‐dependent reduction in mortality at doses, with 25–70% survival compared to 10% in the control group | Warfield et al. (2020) | ||
| Maturation | Luteolin (Flavonoid) | DENV | Mouse; 100 mg kg−1, oral, four times per day | A significant (P ≤ 0·05; twofold) reduction in viraemia compared to the control was observed 3 days post‐infection | Peng et al. (2017) |
| Bevirimat (Triterpenoid) | HIV | Human phase I and II clinical trial of HIV patients (n = 24); single dose of 75, 150 or 250 mg, oral administration | Significant reductions in HIV viral RNA load were observed in 150 and 250 mg groups (0·46–0·47 log10 reductions) compared to the placebo (0·15 log10 reduction). No significant adverse effects were reported | Smith et al. (2007) |
Pentacyclic triterpenoids are a large class of plant secondary metabolite reported to possess a range of biological activities (Bachořík and Urban 2021). Pentacyclic triterpenoids (Table 1) are hydrophobic isoprenoid compounds derived from oxidized squalene precursors (Cárdenas et al. 2019; Li et al. 2020). Echinocystic acid and oleanolic acid are members of the oleanane group of pentacyclic triterpenoids, which are derived from β‐amyrin (Thimmappa et al. 2014), and have been reported to block viral attachment (Yu et al. 2013; Li et al. 2020). Yu et al. (2013) demonstrated that echinocystic acid and oleanolic acid inhibited the entry of HCV into Huh‐7 cells by targeting HCV envelope glycoprotein 2, with respective EC50 values of 1·4 and ~10 µmol l−1 and 50% cytotoxic concentration (CC50) values of >50 µmol l−1 (Yu et al. 2013). An echinocystic acid‐galactose conjugate also inhibited IAV entry into MDCK cells at an EC50 of 5 µmol l−1 by binding to haemagglutinin (HA), thus disrupting its interaction with sialic acid on host cells (Yu et al. 2014). The insolubility of triterpenoids in water is a significant barrier to their potential use as antiviral agents and chemical modifications have been attempted to improve the bioavailability of triterpenoids. A range of triazole‐bridged β‐cyclodextrin (CD)–pentacyclic triterpene (betulinic acid, oleanolic acid, ursolic acid and echinocystic acid) conjugates inhibited the entry of HCV into Huh‐7 cells and demonstrated improved water solubility and reduced hemolytic activity (Xiao et al. 2014). Derivates of betulinic acid, a lupane‐type pentacyclic triterpene (Bachořík and Urban 2021) have been shown to act as fusion inhibitors against HIV by targeting gp120 (Sun et al. 2002; Aiken and Chen 2005).
Quercetin, a flavonoid compound extracted from a wide range of plants, has been known to possess antiviral activity for decades (Colunga Biancatelli et al. 2020). Quercetin belongs to the flavonol group of flavonoids (Tables 1 and 2), possessing a backbone of 3‐hydroxyflavone (D’Andrea 2015). There have been published reports of concentration‐dependent inhibition of poliovirus, parainfluenza virus and herpes simplex virus type 1 in 1985 (Kaul et al. 1985). Since then, there has been a continued interest in the antiviral activity of quercetin; Wu et al. (2015) demonstrated that quercetin inhibited the infection of HCDK cells with IAV at an EC50 ranging 1·93–7·76 μg ml−1. Time of addition assays demonstrated that quercetin inhibited the infectivity of IAV when used as a virus pre‐treatment or co‐treatment, suggesting that quercetin inhibits viral entry; in accordance expression of viral HA was limited when quercetin was added to infected cells up to 2 h post‐infection. Quercetin also binds to the HA2 subunit of IAV, and inhibits HA‐mediated haemolysis of erythrocytes, suggesting that quercetin may inhibit membrane fusion (Wu et al. 2015). More recently quercetin and related flavonoids have been investigated as a potential antiviral agent against SARS‐CoV‐2 using in silico approaches or in vitro protein assays (Derosa et al. 2021). Quercetin inhibited recombinant ACE2 at a 50% inhibitory concentration (IC50) of 4·48 μmol l−1 (Table 2). ACE2 acts as a receptor for SARS‐CoV‐2 spike protein and inhibition of ACE2 activity was proposed as a potential strategy to block binding and entry of SARS‐CoV‐2 (Liu et al. 2020). There is limited published research on the inhibition of SARS‐CoV‐2 infection by quercetin and other flavonoids, which is required to evaluate their potential utility as lead antiviral compounds. Zhan et al. (2021) reported that the flavonol isorhamnetin (50 μmol l−1) reduced entry of a SARS‐CoV‐2 spike protein pseudotyped virus into ACE2‐overexpressing HEK293 cells by 47·7%, indicating that isorhamnetin could inhibit ACE2‐mediated entry; in contrast, quercetin did not significantly (P > 0·05) affect viral entry. Isorhamnetin did not display significant cytotoxicity up to 200 μmol l−1, suggesting that further investigation of its antiviral activity may be warranted (Zhan et al. 2021).
Essential oils (EOs) are volatile crude extracts of plants, predominantly containing terpenes, phenylpropanoids and related compounds, with well‐established in vitro antibacterial and antifungal properties (Tariq et al. 2019). Research on the antiviral activity of EOs and their components appears to mainly comprise of preliminary antiviral screening assays. EOs derived from the fruits of Fortunella margarita inhibited H5N1 IAV at an EC50 of 6·77 μg ml−1, compared to a CC50 of 239·54 μg ml−1 (Ibrahim et al. 2015); the active compounds and mechanism of action of this effect were not established. Recently EOs have been reported to inhibit ACE2. EOs derived from geranium and lemon (25–50 μg ml−1) inhibited ACE2 protein concentrations (10·63–24·79% compared to control) and downregulated expression of ACE2 in HT‐29 cells. Further screening of the major components of these EOs demonstrated that citronellol (50 µmol l−1), geraniol (50 µmol l−1), limonene (50 µmol l−1), and neryl acetate (50 µmol l−1) significantly reduced ACE2 levels by 7·61–57·39% compared to the control and did not exhibit cytotoxic activity at 100 µmol l−1 (Senthil Kumar et al. 2020). The effect of these compounds against SARS‐CoV‐2 infectivity was not established, which is required to understand the significance of their ACE2 inhibitory activity.
Uncoating
There are few reports of plant‐derived natural products that target uncoating (Table 1). Tea tree (Melaleuca alternifolia) EO and its terpenoid constituent terpinen‐4‐ol inhibited IAV, with respective EC50 values of 0·0006 and 0·002% (v/v), compared to a CC50 of 0·025% (v/v) against MDCK cells. Time‐of‐addition assays demonstrated that tea tree oil and terpinen‐4‐ol were active against IAV 1 h after viral adsorption and inhibited the acidification of MDCK lysosomes (Garozzo et al. 2011). Acidification of lysosomes is a key stage in the uncoating of IAV by prompting the fusion of the viral envelope and the endosomal membrane, thereby releasing the nucleocapsid into the cytoplasm (Guinea and Carrasco 1995; Garozzo et al. 2011). Similarly, meliacine, a cyclic peptide isolated from Melia azedarach, blocked lysosomal acidification in BHK‐21 cells, resulting in the inhibition of foot and mouth disease virus uncoating (Wachsman et al. 1998), although the intracellular target that blocked acidification was not established.
Genome replication
Polyprotein processing
Upon entry into the host cell, the incoming RNA is translated and polyproteins processed to synthesize viral proteins involved in RNA replication. There are some examples of plant natural products that may inhibit proteases in the published literature (Table 1). A well‐characterized target for coronaviruses is the main protease 3‐Chymotrypsin‐like protease (3CLPro) (Zhang et al. 2020), which cleaves nascent polypeptides into non‐structural proteins required for genome replication (Mody et al. 2021). 3CLPro inhibitors have been of significant interest for the development of antivirals for SARS‐CoV‐2, and as 3CLPro is highly conserved across coronaviruses, could also be active against SARS‐CoV and MERS‐CoV (Rathnayake et al. 2020). Shuanghuanglian is a Chinese traditional medicine extracted from the herbs Lonicera japonica, Scutellariia baicalensis and Forsythia suspense and used as a remedy for respiratory tract infections. Both oral liquid and lyophilized powder preparations of Shuanghuanglian were recently demonstrated to inhibit SARS‐CoV‐2 infection in Vero cells (EC50 = 0·93–1·20 μl ml−1) with activity being attributed to inhibition of 3CLPro. Investigation of the constituents of Shuanghuanglian identified that the two major flavonoid components baicalin and baicalein also showed 3CLPro‐inhibitory activity (IC50 = 6·41 and 0·94 μmol l−1 respectively; Table 2) and respective EC50 values against SARS‐CoV‐2 replication of 27·87 and 2·94 μmol l−1; the CC50 were >200 μmol l−1 (Su et al. 2020). Molecular docking analysis suggested that two alkaloid compounds (10‐hydroxyusambarensine and cryptoquindoline) and two pentacyclic triterpenoids (6‐oxoisoiguesterin and 22‐hydroxyhopan‐3‐one) derived from an African plant library bind to SARS‐CoV‐2 3CLPro, suggesting that they could possess antiviral activity against SARS‐CoV‐2 by inhibiting viral protein cleavage (Gyebi et al. 2021). Quercetin was also identified as a potential inhibitor of SARS‐CoV‐2 3CLPro by molecular docking and in vitro enzyme inhibition assays (Abian et al. 2020). A range of plant compounds inhibit HIV protease (Ribeiro Filho et al. 2010) including the hydroxycinnamic acids ethyl caffeate and caffeic acid and isovanillin, a phenolic aldehyde compound at IC50 values of 1·0–3·5 μmol l−1 (Table 2) (Wang et al. 2019). Further examples of plant‐derived protease inhibitors include the polyphenols corilagin and excoecariphenol D, which inhibited HCV NS3 protease (IC50 = 3·45 and 6·93 μmol l−1, respectively; Table 2) and RNA production in Huh‐7.5 cells with respective EC50 values of 13·59 and 12·61 μmol l−1 (Li et al. 2012).
RNA replication
Retroviruses undergo reverse transcription of their RNA genome, catalysed by RT, before integrating into the host genome. RT is a key target for current anti‐HIV drugs and as such has been the subject of interest for natural product drug discovery (Table 1). The coumarin compound calanolide A, present in the Malaysian tree Calophyllum langigerum strongly inhibits RT (Currens et al. 1996; el Sayed 2000; Eiznhamer et al. 2002). Buckheit Jr et al. (1999) reported that calanolide A inhibited HIV‐1 with EC50 values ranging 0·08–0·50 µmol l−1; however, no antiviral activity was detected against HIV‐2 or simian immunodeficiency virus, with EC50 > 10 µmol l−1). Calanolide A has subsequently been the subject of clinical trials to assess its safety and pharmacokinetic profile as an anti‐HIV agent, showing a reasonable safety profile with mild to moderate side effects in 47 healthy HIV negative participants (el Sayed 2000; Eiznhamer et al. 2002). Other plant‐derived natural products that inhibit RT include two hydrolysed peptides AIHIILI and LIAVSTNIIFIVV from Quercus infectoria fruit peel, with IC50 values of 236–274 nmol ml−1 against RT (Table 2) (Seetaha et al. 2021).
The RNA genome of HCV is replicated by the RdRp non‐structural protein 5B (NS5B). NS5B is a key target for the development of anti‐HCV drugs, with several such drugs either approved for clinical use or under investigation (de Clercq and Li 2016). Eclipta alba has been used in traditional Indian medicine for the treatment of liver diseases. An aqueous extract of E. alba inhibited the replication of HCV in vitro, with a 90% reduction in viral RNA after treatment with 130 μg ml−1. NS5B was also directly inhibited with an IC50 of 11 μg ml−1 (Table 2). Bioactivity‐guided fractionation identified wedelolactone, luteolin and apigenin as active compounds, with respective IC50 values against NS5B of 7·7, 11·3 and 175·5 μmol ml−1 (Table 2). HCV replication was reduced by 80% upon treatment with 50 μmol ml−1 wedelolactone (coumestan) and by 50% with 50 μmol ml−1 luteolin (flavone) and apigenin (flavone). Cytotoxicity was observed at 50 μmol ml−1, suggesting that the activity and selectivity of these compounds are limited (Manvar et al. 2012).
The infectivity of enterovirus 71 (EV71) and coxsackievirus A16 was reduced by luteolin, a flavone compound. EC50 values of luteolin were 10·31 μmol l−1 against EV71 and 7·40 μmol l−1 against coxsackievirus A16, compared to CC50 values of 148·02 μmol l−1. Although the molecular target was not elucidated, mechanism of action studies suggested that luteolin targeted post‐attachment stages of infection, where viral RNA expression in infected cells was suppressed upon treatment with luteolin (Xu et al. 2014).
A recent in silico study investigated potential inhibitors of SARS‐CoV‐2 RdRp from a library of 1664 chemicals previously approved for use in humans by the United States Food and Drug Administration. Although the in vitro antiviral activities of the compounds were not established, there were eight natural products identified with high molecular docking scores, including three glycosides (aloin, digoxin and sennoside B), two triterpenes (asiaticoside and glycyrrhizin), two flavonoids (quercetin and taxifolin) and one flavanone (neohesperidin diydrochalcone). These results provide potential candidates for repurposing non‐toxic and well‐characterized compounds as SARS‐CoV‐2 replication inhibitors (Kandeel et al. 2020).
A natural product library of 28 compounds was screened for inhibitors of SARS‐CoV‐2 non‐structural protein 15 (NSp15) endoribonuclease, an enzyme involved in the processing of viral RNA during replication. Green tea extract and its major constituent epigallocatechin gallate inhibited NSp15 with respective IC50 values of 2·54 and 0·74 μg ml−1 (Table 2), and inhibited SARS‐CoV‐2 infectivity against Vero cells at EC50 values of 0·24 and 0·092 μg ml−1 respectively (Hong et al. 2021).
Protein synthesis
There are several examples demonstrating the inhibition of various stages of protein synthesis (Table 1). Aeginetia indica is a parasitic plant found in Asia and historically used in folk medicine for liver diseases. An aqueous extract of A. indica was reported to possess antiviral activity against HCV; a dose of 8 mg ml−1 reduced HCV infection of Huh‐7.5.1 cells by 90% compared to a CC50 of 14 mg ml−1. It was concluded that A. indca targeted the translation and replication of HCV, as the expression of RNA and non‐structural protein 3 (NS3) were reduced, and internal ribosome entry site‐mediated translation activity was inhibited. Phosphorylation of NS5A protein at serine 235, a key post‐translational event in the replication of HCV was significantly reduced, which was posited as the mechanism of action (Lin et al. 2018). The composition of the crude extract and the active compound were not elucidated, necessitating further study.
Another strategy for inhibiting viral protein synthesis is to target the host cell. Host cell proteins are less likely to mutate and lead to viral escape, compared to viral protein targets (Müller et al. 2012); conversely, there may be an increased risk of side effects from non‐specific interactions when targeting host cells (Biedenkopf et al. 2017). Curcumin a polyphenolic compound extracted from turmeric (Curcuma longa) (Sharifi et al. 2020), was reported to inhibit HCV infection of Huh‐7.5 cells (10–20 μmol l−1) by upregulating the expression of heme oxygenase‐1 and inhibiting PI3K‐AKT signaling, thus inhibiting HCV protein expression (Chen et al. 2012). Curcumin (10 μmol l−1) also inhibited EV71; pre‐treatment of HCT29 cells with 10 μmol l−1 curcumin resulted in a significant (P ≤ 0·05) decrease in viral titre and RNA expression at 6 to 9 h post‐infection, whilst host cell viability was significantly (P ≤ 0·05) increased. The expression of phosphorylated protein kinase Cδ was upregulated in EV71‐infected HT29 cells, yet suppressed up to 6 h post‐infection upon treatment with curcumin (Huang et al. 2018). Protein kinase Cδ is involved in a range of cellular processes including cell proliferation and has previously been shown to play a role in virus replication (Zhao et al. 2014). It was hypothesized that suppression protein kinase Cδ activation inhibits EV71 protein expression (Huang et al. 2018). Curcumin was significantly (P ≤ 0·05) cytotoxic above 20 μmol l−1 in both studies (Chen et al. 2012; Huang et al. 2018), suggesting that its activity is not selective which may limit its utility. Indeed, curcumin interacts with a wide range of cellular targets including, for example, cytochrome P450 and NF‐κB (Huang et al. 2018).
Camptothecin, a quinoline alkaloid compound derived from the Camptotheca acuminata tree, has been extensively studied as an anticancer agent because of its topoisomerase 1 inhibitory activity (Li et al. 2017) but has more recently been investigated as a potential antiviral agent. Wu and Chu (2017) reported that of camptothecin inhibited EV71; protein expression was suppressed in both replication‐incompetent and replication‐competent EV71, suggesting that both the transcription and translation of viral RNA were blocked by camptothecin. It was noted that the antiviral efficacy of camptothecin was relatively limited, with a 90% reduction in infectivity at a concentration of 10 µmol l−1; investigations of its mechanism of action may enhance understanding of host‐virus interactions and in turn could potentially lead to the development of novel antiviral agents for enteroviruses (Wu and Chu 2017).
Silvestrol is a flavagline compound isolated from Aglaia sp. trees that was first investigated as a potential anticancer agent but has more recently been demonstrated to possess antiviral activity (Oran Schulz et al. 2021). Silvestrol inhibits the ATP‐dependent RNA helicase eukaryotic initiation factor 4a (eIF4a) by increasing its binding affinity to mRNA, thereby preventing protein translation (Biedenkopf et al. 2017; Oran Schulz et al. 2021). Silvestrol was demonstrated to inhibit Ebola virus at an EC50 of 0·8 nmol l−1 in Huh‐7 cells and exhibited antiviral activity when added both before and after infection; no cytotoxicity was demonstrated up to 10 nmol l−1. Promising antiviral activity was also reported against human coronavirus 229E, MERS‐CoV, human rhinovirus A1 and poliovirus in MRC‐5 cells, with respective EC50 values of 3, 1·3, 100 and 20 nmol l−1 and strong selectivity indices (SI) ranging >100 to 3330 (Müller et al. 2018). The infectivity of Zika virus was also significantly reduced (Elgner et al. 2018). These results suggest that silvestrol may be a candidate for the development of antiviral agents against a range of RNA viruses (Müller et al. 2018).
Iminosugars are a class of sugar analogues possessing a nitrogen atom in substitution of cyclic oxygen (Bhushan et al. 2020) that occur in plants and bacteria. Iminosugars competitively inhibit ⍺‐glucosidases, thereby inhibiting viruses that rely on the endoplasmic reticulum glycosylation pathway for correct folding of glycoproteins (Evans DeWald et al., 2020). Iminosugars and their derivatives reportedly inhibit enveloped viruses, for instance celgosivir, a prodrug of the plant‐derived iminosugar castanospermine, was demonstrated to inhibit DENV in vitro (EC50 = 5·17 μmol l−1) and in vivo, where oral treatment significantly reduced viral load in a mouse model (Table 3) (Sayce et al. 2016). However, in phase 1b clinical trials celgosivir showed no significant reductions in viral load or fever in dengue fever patients compared to the placebo (Low et al. 2014). Celgosivir, castanospermine and deoxynojirimycin also significantly inhibited Zika virus infection in vitro at 1 μmol l−1 (Bhushan et al. 2020). A single oral dose of N‐(9′‐methoxynonyl)‐1‐deoxynojirimycin hydrochloride salt UV‐4B, a derivative of deoxynojirimycin, also significantly reduced mortality of influenza and DENV‐infected mice (Table 3) (Warfield et al. 2020).
Assembly
Natural products targeting viral particle assembly have been less commonly reported in the published literature (Table 1). Two alkaloid compounds (lycorine and hemanthamine) derived from Lycoris radiata, a plant used in traditional Chinese medicine, block assembly of H5N1 IAV (He et al. 2013). Respective EC50 values of lycorine and hemanthamine were <0·46 and 1·48 μmol ml−1, while the CC50 values against MDCK cells were 20·9 and 50 μmol ml−1. The SI of these compounds was therefore moderate at 34 to >45, (where 2–10 was considered a weak SI, 10–50 moderate and >50 as a strong SI). Time‐of‐addition assays demonstrated that these compounds exhibited an antiviral effect when added post‐infection rather than during pre‐treatment or co‐treatment. It was subsequently demonstrated that lycorine and hemanthamine blocked the export of viral ribonucleoprotein from the nucleus into the cytoplasm (He et al. 2013), where virion assembly occurs (Samji 2009). Recently, the alkaloid berberine was demonstrated to possess antiviral activity against IAV by impairing the export of ribonucleotides from the nucleus into the cytoplasm; this was attributed to the suppression of virus‐induced phosphorylation of the mitogen‐activated protein kinase/extracellular signal‐related kinase 1 (MAPK/ERK1) signalling pathway (Botwina et al. 2020).
Maturation
Natural products may also target the maturation of virions (Table 1). The flavonoid luteolin was reported to inhibit DENV infection by blocking viral maturation. The infectivity of all DENV serotypes (DENV1‐DENV4) was reduced against Huh‐7 cells, where EC50 values were 4·36–8·38 μmol ml−1 and the CC50 was 45·89 μmol ml−1, giving an SI of 8·84 (Peng et al. 2017). Free virions isolated from the supernatant of infected cells contained elevated precursor membrane (prM) protein levels after treatment with luteolin, suggesting that the virions were immature. DENV prM is a structural protein that, along with the Envelope (E) protein, encapsulate immature virions. Immature virions are trafficked to the Golgi apparatus where furin protease cleaves prM, producing a mature, infectious virion (Screaton et al. 2015; Peng et al. 2017). Further investigation showed that luteolin was a non‐competitive inhibitor of furin protease (inhibitory constant of 58·6 μmol ml−1). Luteolin exhibited a weak efficacy in vivo using a mouse model, where luteolin (100 mg kg−1 oral administration, four times per day) reduced viral titre in blood samples by twofold compared to the control (Table 3). This could potentially be attributed to the limited bioavailability of luteolin or low potency of the compound and suggests that compound optimization or drug delivery strategies are required (Peng et al. 2017).
In addition to fusion inhibition, betulinic acid derivatives block HIV maturation. Bevirimat (3‐O‐(3′,3′‐dimethylsuccinyl)betulinic acid) is a first‐in‐class HIV maturation inhibitor derived from betulinic acid (Zhao et al. 2021). This compound binds to Gag polyprotein, inhibiting the cleavage of the mature capsid protein p24 from capsid spacer protein‐1 (p25), thereby preventing the development of mature infectious virions (Li et al. 2003; Martin et al. 2008). Phase I and II clinical trials were conducted in HIV patients (n = 24), suggesting that bevirimat (150–250 mg) significantly (P ≤ 0·05) reduced HIV viral RNA load and did not produce significant adverse effects (Smith et al. 2007), however drug resistance was reported in up to 50% of patients due to the presence of polymorphisms in Gag at the capsid spacer 1 region in later studies (Adamson et al. 2010; Zhao et al. 2021). Investigation of alternative betulinic acid derivatives as potential HIV maturation inhibitors is ongoing (Zhao et al. 2021).
Conclusion and future perspectives
A wide range of plant‐derived natural products have antiviral activity and act on a diverse range of targets at different stages of the virus life cycle suggesting they may be useful for the development of novel antiviral drugs (Table 1). Several plant extracts and compounds have demonstrated broad‐spectrum antiviral activity in vitro. Future research needs to further investigate the antiviral efficacy of these natural products against a wider range of viruses and establish a fuller understanding of their mechanisms of action, including the molecular target of natural products, which will aid in the design of more potent derivatives (Aiken and Chen 2005). Computational methods have also revealed an abundance of compounds that interact with viral protein targets, and recently in silico studies have been performed against SARS‐CoV‐2 (Kandeel et al. 2020; Tahir ul Qamar et al. 2020; Gyebi et al. 2021). The in vitro activity of compounds identified by in silico techniques are often not reported, however in order for natural products to be developed as antiviral drugs this efficacy data is paramount. Additionally, there are a number of in vitro studies that only report the antiviral efficacy of crude extracts without elucidating active constituents or mechanism of action which require more in‐depth study. Natural products known to possess other bioactive properties have also seen increasing interest for their antiviral activity, including the repurposing of approved compounds which could be a strategy to accelerate the development of new antivirals (Kandeel et al. 2020). Another area of interest for future studies includes combinations of plant‐derived natural products that exhibit synergistic interactions by blocking different stages of the viral life cycle, which could potentially enhance the potency of natural product compounds and limit the emergence of resistant isolates (Morán‐Santibañez et al. 2018).
Plant natural products are often limited by poor solubility and bioavailability, and there are limited studies on the pharmacokinetic profile and in vivo toxicity of antiviral plant natural products. Derivatization has proven successful for increasing solubility (Xiao et al. 2014) and reducing toxicity (Swanson et al. 2015), while drug delivery systems have also been explored to improve the bioavailability of natural products (Wei et al. 2014; Ben‐Shabat et al. 2020). Similar modifications may be an area of future study for recently identified antiviral plant compounds.
There are a limited number of in vivo studies (Table 3) on the antiviral activity of plant‐derived natural products which are required to further understand the efficacy of compounds with promising in vitro activity (Tables 1 and 2). Few clinical trials related to antiviral natural products have been performed, and several recent systematic reviews have concluded that there is weak evidence that traditional herbal medicines are effective for viral diseases such as HIV, the common cold and SARS, due to the small number of studies and sample sizes, in addition to inappropriate methodologies for the majority of studies (Liu et al. 2005, 2012; Wu et al. 2007). As a result, there is a need for progression from in vitro efficacy tests and target elucidation towards in vivo studies and robust clinical trials for natural products to be developed as antiviral agents.
Author contributions
LO contributed to conceptualization, investigation and writing—original draft preparation. KL contributed to conceptualization and writing—review and editing. MS contributed to conceptualization, investigation and writing—review and editing.
Conflict of Interest
The authors declare that there is no conflict of interest.
References
- Abian, O. , Ortega‐Alarcon, D. , Jimenez‐Alesanco, A. , Ceballos‐Laita, L. , Vega, S. , Reyburn, H.T. , Rizzuti, B. and Velazquez‐Campoy, A. (2020) Structural stability of SARS‐CoV‐2 3CLpro and identification of quercetin as an inhibitor by experimental screening. Int J Biol Macromol 164, 1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson, C.S. , Sakalian, M. , Salzwedel, K. and Freed, E.O. (2010) Polymorphisms in Gag spacer peptide 1 confer varying levels of resistance to the HIV‐1 maturation inhibitor bevirimat. Retrovirology 7, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini, M.L. , Andres, E.L. , Sims, A.C. , Graham, R.L. , Sheahan, T.P. , Lu, X. , Smith, E.C. , Case, J.B. et al. (2018) Coronavirus susceptibility to the antiviral remdesivir (GS‐5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio 9, e00221‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiewsakun, P. and Simmonds, P. (2018) The genomic underpinnings of eukaryotic virus taxonomy: creating a sequence‐based framework for family‐level virus classification. Microbiome 6, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiken, C. and Chen, C.H. (2005) Betulinic acid derivatives as HIV‐1 antivirals. Trends Mol Med 11, 31–36. [DOI] [PubMed] [Google Scholar]
- Akhtar, M.F. , Saleem, A. , Rasul, A. , Baig, M.M.F.A. , Bin‐Jumah, M. and Daim, M.M.A. (2020) Anticancer natural medicines: an overview of cell signaling and other targets of anticancer phytochemicals. Eur J Pharmacol 888, 173488. [DOI] [PubMed] [Google Scholar]
- Atanasov, A.G. , Waltenberger, B. , Pferschy‐Wenzig, E.M. , Linder, T. , Wawrosch, C. , Uhrin, P. , Temml, V. , Wang, L. et al. (2015) Discovery and resupply of pharmacologically active plant‐derived natural products: a review. Biotechnol Adv 33, 1582–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasov, A.G. , Zotchev, S.B. , Dirsch, V.M. and Supuran, C.T. (2021) Natural products in drug discovery: advances and opportunities. Nat Rev Drug Discov 20, 200–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachořík, J. and Urban, M. (2021) Biocatalysis in the chemistry of lupane triterpenoids. Molecules 26, 2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellande, K. , Bono, J.J. , Savelli, B. , Jamet, E. and Canut, H. (2017) Plant lectins and lectin receptor‐like kinases: how do they sense the outside? Int J Mol Sci 18, 1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben‐Shabat, S. , Yarmolinsky, L. , Porat, D. and Dahan, A. (2020) Antiviral effect of phytochemicals from medicinal plants: applications and drug delivery strategies. Drug Deliv Transl Res 10, 354–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler, J.A. (2019) Natural products as a foundation for drug discovery. Curr Protoc Pharmacol 86, e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan, G. , Lim, L. , Bird, I. , Chothe, S.K. , Nissly, R.H. , and Kuchipudi, S.V. (2020) Iminosugars with endoplasmic reticulum α‐glucosidase inhibitor activity inhibit zikv replication and reverse cytopathogenicity in vitro . Front Microbiol 11, 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedenkopf, N. , Lange‐Grünweller, K. , Schulte, F.W. , Weißer, A. , Müller, C. , Becker, D. , Becker, S. , Hartmann, R.K. et al. (2017) The natural compound silvestrol is a potent inhibitor of Ebola virus replication. Antiviral Res 137, 76–81. [DOI] [PubMed] [Google Scholar]
- Botwina, P. , Owczarek, K. , Rajfur, Z. , Ochman, M. , Urlik, M. , Nowakowska, M. , Szczubiałka, K. and Pyrc, K. (2020) Berberine hampers influenza A replication through inhibition of MAPK/ERK pathway. Viruses 12, 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckheit, R.W. Jr , White, E.L. , Fliakas‐Boltz, V. , Russell, J. , Stup, T.L. , Kinjerski, T.L. , Osterling, M.C. , Weigand, A. et al. (1999) Unique anti‐human immunodeficiency virus activities of the nonnucleoside reverse transcriptase inhibitors calanolide A, costatolide, and dihydrocostatolide. Antimicrob Agents Chemother 43, 1827–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas, P.D. , Almeida, A. and Bak, S. (2019) Evolution of structural diversity of triterpenoids. Front Plant Sci 10, 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco‐Hernandez, R. , Jácome, R. , López Vidal, Y. and Ponce de León, S. (2017) Are RNA viruses candidate agents for the next global pandemic? A review. ILAR J 58, 343–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella, M. , Rajnik, M. , Aleem, A. , Dulebohn, S.C. and di Napoli, R. (2021) Features, evaluation, and treatment of coronavirus (COVID‐19). StatPearls [internet]. Treasure Island: StatPearls Publishing. [PubMed] [Google Scholar]
- Chen, M.‐H. , Lee, M.‐Y. , Chuang, J.‐J. , Li, Y.‐Z. , Ning, S.‐T. , Chen, J.‐C. and Liu, Y.‐W. (2012) Curcumin inhibits HCV replication by induction of heme oxygenase‐1 and suppression of AKT. Int J Mol Med 30, 1021–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ci, Y. and Shi, L. (2021) Compartmentalized replication organelle of flavivirus at the ER and the factors involved. Cell Mol Life Sci 78, 4939–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Clercq, E. and Li, G. (2016) Approved antiviral drugs over the past 50 years. Clin Microbiol Rev 29, 695–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colunga Biancatelli, R.M.L. , Berrill, M. , Catravas, J.D. and Marik, P.E. (2020) Quercetin and vitamin C: an experimental, synergistic therapy for the prevention and treatment of SARS‐CoV‐2 related disease (COVID‐19). Front Immunol 11, 1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coves‐Datson, E.M. , King, S.R. , Legendre, M. , Swanson, M.D. , Gupta, A. , Claes, S. , Meagher, J.L. , Boonen, A. et al. (2021) Targeted disruption of pi‐pi stacking in Malaysian banana lectin reduces mitogenicity while preserving antiviral activity. Sci Rep 11, 656–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currens, M.J. , Gulakowski, R.J. , Mariner, J.M. , Moran, R.A. , Buckheit, R.W. , Gustafson, K.R. , McMahon, J.B. and Boyd, M.R. (1996) Antiviral activity and mechanism of action of calanolide A against the human immunodeficiency virus type‐1. J Pharmacol Exp Ther 279, 645–651. [PubMed] [Google Scholar]
- D'Andrea, G. (2015) Quercetin: a flavonol with multifaceted therapeutic applications? Fitoterapia 106, 256–271. [DOI] [PubMed] [Google Scholar]
- Department of Health and Social Care (2020) World first coronavirus treatment approved for NHS use by government [Online]. https://www.gov.uk/government/news/world‐first‐coronavirus‐treatment‐approved‐for‐nhs‐use‐by‐government Accessed 19/07/2021. [Google Scholar]
- Department of Health and Social Care (2020) Selected NHS patients to access coronavirus treatment remdesivir [Online]. https://www.gov.uk/government/news/selected‐nhs‐patients‐to‐access‐coronavirus‐treatment‐remdesivir Accessed 19/07/2021. [Google Scholar]
- Derosa, G. , Maffioli, P. , D’Angelo, A. and di Pierro, F. (2021) A role for quercetin in coronavirus disease 2019 (COVID‐19). Phytother Res 35, 1230–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye, C. (2014) After 2015: infectious diseases in a new era of health and development. Philos Trans R Soc Lond B Biol Sci 369, 20130426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiznhamer, D.A. , Creagh, T. , Ruckle, J.L. , Tolbert, D.T. , Giltner, J. , Dutta, B. , Flavin, M.T. , Jenta, T. et al. (2002) Safety and pharmacokinetic profile of multiple escalating doses of (+)‐calanolide A, a naturally occurring nonnucleoside reverse transcriptase inhibitor, in healthy HIV‐negative volunteers. HIV Clin Trials 3, 435–450. [DOI] [PubMed] [Google Scholar]
- Elgner, F. , Sabino, C. , Basic, M. , Ploen, D. , Grünweller, A. and Hildt, E. (2018) Inhibition of Zika virus replication by silvestrol. Viruses 10, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emau, P. , Tian, B. , O’Keefe, B.R. , Mori, T. , McMahon, J.B. , Palmer, K.E. , Jiang, Y. , Bekele, G. et al. (2007) Griffithsin, a potent HIV entry inhibitor, is an excellent candidate for anti‐HIV microbicide. J Med Primatol 36, 244–253. [DOI] [PubMed] [Google Scholar]
- European Medicines Agency (2021) Veklury|European Medicines Agency [Online]. https://www.ema.europa.eu/en/medicines/human/EPAR/veklury Accessed 19/07/2021. [Google Scholar]
- Evans DeWald, L. , Starr, C. , Butters, T. , Treston, A. and Warfield, K.L. (2020) Iminosugars: a host‐targeted approach to combat Flaviviridae infections. Antiviral Res 184, 104881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatima, I. , Kanwal, S. and Mahmood, T. (2019) Natural products mediated targeting of virally infected cancer. Dose Response 17, 1559325818813227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint, J. , Racaniello, V.R. , Rall, G.F. , Skalka, A.M. and Enquist, L.W. (2015) Principles of Virology. Volume I: Molecular Biology, 4th edn. Washington, DC: ASM Press. [Google Scholar]
- Freed, E.O. (2015) HIV‐1 assembly, release and maturation. Nat Rev Microbiol 13, 484–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garozzo, A. , Timpanaro, R. , Stivala, A. , Bisignano, G. and Castro, A. (2011) Activity of Melaleuca alternifolia (tea tree) oil on influenza virus A/PR/8: study on the mechanism of action. Antiviral Res 89, 83–88. [DOI] [PubMed] [Google Scholar]
- Gasparini, R. , Amicizia, D. , Lai, P.L. and Panatto, D. (2012) Clinical and socioeconomic impact of seasonal and pandemic influenza in adults and the elderly. Hum Vaccin Immunother 8, 21–28. [DOI] [PubMed] [Google Scholar]
- Grigalunas, M. , Burhop, A. , Christoforow, A. and Waldmann, H. (2020) Pseudo‐natural products and natural product‐inspired methods in chemical biology and drug discovery. Curr Opin Chem Biol 56, 111–118. [DOI] [PubMed] [Google Scholar]
- Guinea, R. and Carrasco, L. (1995) Requirement for vacuolar proton‐ATPase activity during entry of influenza virus into cells. J Virol 69, 2306–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyebi, G.A. , Ogunro, O.B. , Adegunloye, A.P. , Ogunyemi, O.M. and Afolabi, S.O. (2021) Potential inhibitors of coronavirus 3‐chymotrypsin‐like protease (3CLpro): an in silico screening of alkaloids and terpenoids from African medicinal plants. J Biomol Struct Dyn 39, 3396–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, C. (2020) Coronavirus puts drug repurposing on the fast track. Nat Biotechnol 38, 379–381. [DOI] [PubMed] [Google Scholar]
- He, J. , Qi, W.B. , Wang, L. , Tian, J. , Jiao, P.R. , Liu, G.Q. , Ye, W.C. and Liao, M. (2013) Amaryllidaceae alkaloids inhibit nuclear‐to‐cytoplasmic export of ribonucleoprotein (RNP) complex of highly pathogenic avian influenza virus H5N1. Influenza Other Respir Viruses 7, 922–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, E.C. (2009) The evolutionary genetics of emerging viruses. Ann Rev Ecol Syst 40, 353–372. [Google Scholar]
- Hong, S. , Seo, S.H. , Woo, S.‐J. , Kwon, Y. , Song, M. and Ha, N.‐C. (2021) Epigallocatechin gallate inhibits the uridylate‐specific endoribonuclease Nsp15 and efficiently neutralizes the SARS‐CoV‐2 strain. J Agri Food Chem 69, 5948–5954. [DOI] [PubMed] [Google Scholar]
- Hopper, J.T.S. , Ambrose, S. , Grant, O.C. , Krumm, S.A. , Allison, T.M. , Degiacomi, M.T. , Tully, M.D. , Pritchard, L.K. et al. (2017) The tetrameric plant lectin Banlec neutralizes HIV through bidentate binding to specific viral glycans. Structure 25, 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H.I. , Chio, C.C. and Lin, J.Y. (2018) Inhibition of EV71 by curcumin in intestinal epithelial cells. PLoS One 13, e0191617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim, N.A. , El‐Hawary, S.S. , Mohammed, M.M.D. , Farid, M.A. , Abdel‐Wahed, N.A.M. , Ali, M.A. and El‐Abd, E.A.W. (2015) Chemical composition, antiviral against avian influenza (H5N1) virus and antimicrobial activities of the essential oils of the leaves and fruits of Fortunella margarita, lour. swingle, growing in Egypt. J Appl Pharm Sci 5. [Google Scholar]
- Kandeel, M. , Kitade, Y. and Almubarak, A. (2020) Repurposing FDA‐approved phytomedicines, natural products, antivirals and cell protectives against SARS‐CoV‐2 (COVID‐19) RNA‐dependent RNA polymerase. PeerJ 8, e10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul, T.N. , Middleton, E. and Ogra, P.L. (1985) Antiviral effect of flavonoids on human viruses. J Med Virol 15, 71–79. [DOI] [PubMed] [Google Scholar]
- Klepser, M.E. (2014) Socioeconomic impact of seasonal (epidemic) influenza and the role of over‐the‐counter medicines. Drugs 74, 1467–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F. , Goila‐Gaur, R. , Salzwedel, K. , Kilgore, N.R. , Reddick, M. , Matallana, C. , Castillo, A. , Zoumplis, D. et al. (2003) PA‐457: A potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc Natl Sci Acad USA 100, 13555–13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F. , Jiang, T. , Li, Q. and Ling, X. (2017) Camptothecin (CPT) and its derivatives are known to target topoisomerase I (Top1) as their mechanism of action: did we miss something in CPT analogue molecular targets for treating human disease such as cancer? Am J Cancer Res 7, 2350–2394. [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Sun, J. , Xiao, S. , Zhang, L. and Zhou, D. (2020) Triterpenoid‐mediated inhibition of virus−host interaction: is now the time for discovering viral entry/release inhibitors from nature? J Med Chem 63, 15371–15388. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Yu, S. , Liu, D. , Proksch, P. and Lin, W. (2012) Inhibitory effects of polyphenols toward HCV from the mangrove plant Excoecaria agallocha L. Bioorg Med Chem Lett 22, 1099–1102. [DOI] [PubMed] [Google Scholar]
- Lin, C.W. , Lo, C.W. , Tsai, C.N. , Pan, T.C. , Chen, P.Y. and Yu, M.J. (2018) Aeginetia indica decoction inhibits hepatitis C virus life cycle. Int J Mol Sci 19, 208. 10.3390/ijms19010208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J.P. , Manheimer, E. and Yang, M. (2005) Herbal medicines for treating HIV infection and AIDS. Cochrane Database Syst Rev 20, CD003937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Raghuvanshi, R. , Ceylan, F.D. and Bolling, B.W. (2020) Quercetin and its metabolites inhibit recombinant human angiotensin‐converting enzyme 2 (ACE2) activity. J Agric Food Chem 68, 13982–13989. [DOI] [PubMed] [Google Scholar]
- Liu, X. , Zhang, M. , He, L. and Li, Y. (2012) Chinese herbs combined with Western medicine for severe acute respiratory syndrome (SARS). Cochrane Database Syst Rev 10, CD004882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low, J.G. , Sung, C. , Wijaya, L. , Wei, Y. , Rathore, A.P.S. , Watanabe, S. , Tan, B.H. , Toh, L. et al. (2014) Efficacy and safety of celgosivir in patients with dengue fever (CELADEN): a phase 1b, randomised, double‐blind, placebo‐controlled, proof‐of‐concept trial. Lancet Infect Dis 14, 706–715. [DOI] [PubMed] [Google Scholar]
- Manvar, D. , Mishra, M. , Kumar, S. and Pandey, V.N. (2012) Identification and evaluation of anti hepatitis C virus phytochemicals from Eclipta alba . J Ethnopharmacol 144, 545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, D.E. , Salzwedel, K. and Allaway, G.P. (2008) Bevirimat: a novel maturation inhibitor for the treatment of HIV‐1 infection. Antiviral Chem Chemother 19, 107–113. [DOI] [PubMed] [Google Scholar]
- Meagher, J.L. and Stuckey, J.A. (2021) Structure of WT Malaysian banana lectin. 10.2210/pdb7KMU/pdb [DOI]
- Millet, J.K. , Séron, K. , Labitt, R.N. , Danneels, A. , Palmer, K.E. , Whittaker, G.R. , Dubuisson, J. and Belouzard, S. (2016) Middle East respiratory syndrome coronavirus infection is inhibited by griffithsin. Antiviral Res 133, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, C.A. , Ramessar, K. and O’Keefe, B.R. (2017) Antiviral lectins: selective inhibitors of viral entry. Antiviral Res 142, 37–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mody, V. , Ho, J. , Wills, S. , Mawri, A. , Lawson, L. , Ebert, M.C.C.J.C. , Fortin, G.M. , Rayalam, S. et al. (2021) Identification of 3‐chymotrypsin like protease (3CLPro) inhibitors as potential anti‐SARS‐CoV‐2 agents. Commun Biol 4, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morán‐Santibañez, K. , Peña‐Hernández, M.A. , Cruz‐Suárez, L.E. , Ricque‐Marie, D. , Skouta, R. , Vasquez, A.H. , Rodríguez‐Padilla, C. & Trejo‐Avila, L.M. (2018) Virucidal and synergistic activity of polyphenol‐rich extracts of seaweeds against measles virus. Viruses, 10, 465. 10.3390/v10090465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, C. , Schulte, F.W. , Lange‐Grünweller, K. , Obermann, W. , Madhugiri, R. , Pleschka, S. , Ziebuhr, J. , Hartmann, R.K. et al. (2018) Broad‐spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona‐ and picornaviruses. Antiviral Res 150, 123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, K.H. , Kakkola, L. , Nagaraj, A.S. , Cheltsov, A.V. , Anastasina, M. and Kainov, D.E. (2012) Emerging cellular targets for influenza antiviral agents. Trends Pharmacol Sci 33, 89–99. [DOI] [PubMed] [Google Scholar]
- Newman, D.J. and Cragg, G.M. (2016) Natural products as sources of new drugs from 1981 to 2014. J Nat Prod 79, 629–661. [DOI] [PubMed] [Google Scholar]
- Normand, S.‐L.‐T. (2020) The RECOVERY platform. N Engl J Med 384, 757–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe, B.R. , Giomarelli, B. , Barnard, D.L. , Shenoy, S.R. , Chan, P.K.S. , McMahon, J.B. , Palmer, K.E. , Barnett, B.W. et al. (2010) Broad‐spectrum in vitro activity and in vivo efficacy of the antiviral protein griffithsin against emerging viruses of the family Coronaviridae. J Virol 84, 2511–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogbole, O.O. , Akinleye, T.E. , Segun, P.A. , Faleye, T.C. and Adeniji, A.J. (2018) In vitro antiviral activity of twenty‐seven medicinal plant extracts from Southwest Nigeria against three serotypes of echoviruses. Virol J 15, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oran Schulz, G. , Victoria, C. , Kirschning, A. and Steinmann, E. (2021) Rocaglamide and silvestrol: a long story from anti‐tumor to anti‐coronavirus compounds. Nat Prod Rep 38, 18–23. [DOI] [PubMed] [Google Scholar]
- Pattnaik, G.P. and Chakraborty, H. (2020) Entry inhibitors: efficient means to block viral infection. J Membr Biol 253, 425–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, M. , Watanabe, S. , Chan, K.W.K. , He, Q. , Zhao, Y. , Zhang, Z. , Lai, X. , Luo, D. et al. (2017) Luteolin restricts dengue virus replication through inhibition of the proprotein convertase furin. Antiviral Res 143, 176–185. [DOI] [PubMed] [Google Scholar]
- Puertas, M.C. , Ploumidis, G. , Ploumidis, M. , Fumero, E. , Clotet, B. , Walworth, C.M. , Petropoulos, C.J. and Martinez‐Picado, J. (2020) Pan‐resistant HIV‐1 emergence in the era of integrase strand‐transfer inhibitors: a case report. Lancet Microbe 1, e130–e135. [DOI] [PubMed] [Google Scholar]
- Rathnayake, A.D. , Zheng, J. , Kim, Y. , Perera, K.D. , Mackin, S. , Meyerholz, D.K. , Kashipathy, M.M. , Battaile, K.P. et al. (2020) 3C‐like protease inhibitors block coronavirus replication in vitro and improve survival in MERS‐CoV–infected mice. Sci Transl Med 12, 5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro Filho, J. , de Sousa Falcao, H. , Maria Batista, L. , Maria Barbosa Filho, J. and Regina Piuvezam, M. (2010) Effects of plant extracts on HIV‐1 protease. Curr HIV Res 8, 531–544. [DOI] [PubMed] [Google Scholar]
- Rodrigues, T. , Reker, D. , Schneider, P. and Schneider, G. (2016) Counting on natural products for drug design. Nat Chem 8, 531–541. [DOI] [PubMed] [Google Scholar]
- Romano, J.D. and Tatonetti, N.P. (2019) Informatics and computational methods in natural product drug discovery: A review and perspectives. Front Genet 30, 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, D. , Chan‐Tack, K. , Farley, J. and Sherwat, A. (2020) FDA approval of Remdesivir – a step in the right direction. N Engl J Med 383, 2598–2600. [DOI] [PubMed] [Google Scholar]
- Ryu, W.‐S. (2017) Virus life cycle. Mol Virol Human Pathog Viruses 2017, 31–45. [Google Scholar]
- Samji, T. (2009) Influenza A: understanding the viral life cycle. Yale J Biol Med 82, 153–159. [PMC free article] [PubMed] [Google Scholar]
- Sayce, A.C. , Alonzi, D.S. , Killingbeck, S.S. , Tyrrell, B.E. , Hill, M.L. , Caputo, A.T. , Iwaki, R. , Kinami, K. et al. (2016) Iminosugars inhibit dengue virus production via inhibition of ER alpha‐glucosidases—not glycolipid processing enzymes. PLOS Negl Trop Dis 10, e0004524. 10.1371/journal.pntd.0004524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Sayed, K.A. (2000) Natural products as antiviral agents. In Studies in Natural Products Chemistry ed. Rahman, A.U. pp. 473–572. Amsterdam: Elsevier. [Google Scholar]
- Screaton, G. , Mongkolsapaya, J. , Yacoub, S. and Roberts, C. (2015) New insights into the immunopathology and control of dengue virus infection. Nat Rev Immunol 15, 745–759. [DOI] [PubMed] [Google Scholar]
- Seetaha, S. , Hannongbua, S. , Rattanasrisomporn, J. and Choowongkomon, K. (2021) Novel peptides with HIV‐1 reverse transcriptase inhibitory activity derived from the fruits of Quercus infectoria . Chem Biol Drug Des 97, 157–166. [DOI] [PubMed] [Google Scholar]
- Senthil Kumar, K.J. , Vani, M.G. , Wang, C.S. , Chen, C.C. , Chen, Y.C. , Lu, L.P. , Huang, C.H. , Lai, C.S. et al. (2020) Geranium and lemon essential oils and their active compounds downregulate angiotensin‐converting enzyme 2 (ACE2), a SARS‐CoV‐2 spike receptor‐binding domain, in epithelial cells. Plants 9, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi, S. , Fathi, N. , Memar, M.Y. , Hosseiniyan Khatibi, S.M. , Khalilov, R. , Negahdari, R. , Zununi Vahed, S. and Maleki Dizaj, S. (2020) Anti‐microbial activity of curcumin nanoformulations: new trends and future perspectives. Phytother Res 34, 1926–1946. [DOI] [PubMed] [Google Scholar]
- Sharma, A. and Gupta, S.P. (2017) Fundamentals of viruses and their proteases. In Viral Proteases and Their Inhibitors ed. Gupta, S.P. pp. 1–24. London: Academic Press. [Google Scholar]
- Smith, P.F. , Ogundele, A. , Forrest, A. , Wilton, J. , Salzwedel, K. , Doto, J. , Allaway, G.P. and Martin, D.E. (2007) Phase I and II study of the safety, virologic effect, and pharmacokinetics/pharmacodynamics of single‐dose 3‐o‐(3′, 3′‐dimethylsuccinyl) betulinic acid (bevirimat) against human immunodeficiency virus infection. Antimicrob Agents Chemother 51, 3574–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, H.‐X. , Yao, S. , Zhao, W.‐F. , Li, M.‐J. , Liu, J. , Shang, W.‐J. , Xie, H. , Ke, C.‐Q. et al. (2020) Anti‐SARS‐CoV‐2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol Sin 41, 1167–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, I.C. , Chen, C.H. , Kashiwada, Y. , Wu, J.H. , Wang, H.K. and Lee, K.H. (2002) Anti‐AIDS agents 49. Synthesis, anti‐HIV, and anti‐fusion activities of IC9564 analogues based on betulinic acid. J Med Chem 45, 4271–4275. [DOI] [PubMed] [Google Scholar]
- Swanson, M. , Boudreaux, D. , Salmon, L. , Chugh, J. , Winter, H. , Meagher, J. , André, S. , Murphy, P. et al. (2015) Engineering a therapeutic lectin by uncoupling mitogenicity from antiviral activity. Cell 163, 746–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, M.D. , Winter, H.C. , Goldstein, I.J. and Markovitz, D.M. (2010) A lectin isolated from bananas is a potent inhibitor of HIV replication. J Biol Chem 285, 8646–8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahir ul Qamar, M. , Alqahtani, S.M. , Alamri, M.A. and Chen, L.‐L. (2020) Structural basis of SARS‐CoV‐2 3CLpro and anti‐COVID‐19 drug discovery from medicinal plants. J Pharm Anal 10, 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq, S. , Wani, S. , Rasool, W. , Shafi, K. , Bhat, M.A. , Prabhakar, A. , Shalla, A.H. & Rather, M.A. (2019) A comprehensive review of the antibacterial, antifungal and antiviral potential of essential oils and their chemical constituents against drug‐resistant microbial pathogens. Microb Pathog, 134, 103580. 10.1016/j.micpath.2019.103580 [DOI] [PubMed] [Google Scholar]
- Thimmappa, R. , Geisler, K. , Louveau, T. , O'Maille, P. and Osbourn, A. (2014) Triterpene biosynthesis in plants. Ann Rev Plant Biol 65, 225–257. [DOI] [PubMed] [Google Scholar]
- UNAIDS (2020) UNAIDS Data 2020. Geneva: UNAIDS. [Google Scholar]
- Vigant, F. , Santos, N.C. and Lee, B. (2015) Broad‐spectrum antivirals against viral fusion. Nat Rev Microbiol 13, 426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachsman, M.B. , Castilla, V. and Coto, C.E. (1998) Inhibition of foot and mouth disease virus (FMDV) uncoating by a plant‐derived peptide isolated from Melia azedarach L leaves. Arch Virol 143, 581–590. [DOI] [PubMed] [Google Scholar]
- Walsh, D. and Mohr, I. (2011) Viral subversion of the host protein synthesis machinery. Nat Rev Microbiol 9, 860–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Wei, Y. , Tian, W.‐Y. , Sakharkar, M.K. , Liu, Q. , Yang, X. , Zhou, Y.‐Z. , Mou, C.‐L. et al. (2019) Characterization of nine compounds isolated from the acid hydrolysate of Lonicera fulvotomentosa Hsu et S. C. Cheng and evaluation of their in vitro activity towards HIV protease. Molecules 24, 4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfield, K.L. , Alonzi, D.S. , Hill, J.C. , Caputo, A.T. , Roversi, P. , Kiappes, J.L. , Sheets, N. , Duchars, M. et al. (2020) Targeting endoplasmic reticulum α‐glucosidase I with a single‐dose iminosugar treatment protects against lethal influenza and dengue virus infections. J Med Chem 63, 4205–4214. [DOI] [PubMed] [Google Scholar]
- Wei, J.N. , Liu, Z.H. , Zhao, Y.P. , Zhao, L.L. , Xue, T.K. and Lan, Q.K. (2019) Phytochemical and bioactive profile of Coriandrum sativum L. Food Chem 286, 260–267. [DOI] [PubMed] [Google Scholar]
- Wei, Y. , Guo, J. , Zheng, X. , Wu, J. , Zhou, Y. , Yu, Y. , Ye, Y. , Zhang, L. et al. (2014) Preparation, pharmacokinetics and biodistribution of baicalin‐loaded liposomes. Int J Nanomed 9, 3623. 10.2147/IJN.S66312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey, J.M. , Sherwood, L.M. and Woolverton, C.J. (2011) Prescott’s Microbiology, 8th edn. New York: McGraw‐Hill. [Google Scholar]
- Woolhouse, M.E.J. and Brierley, L. (2018) Epidemiological characteristics of human‐infective RNA viruses. Sci Data 5, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2020) Hepatitis C [Online]. https://www.who.int/news‐room/fact‐sheets/detail/hepatitis‐c. Acccessed 18/07/2021. [Google Scholar]
- World Health Organization (2021) WHO Coronavirus (COVID‐19) Dashboard|WHO Coronavirus (COVID‐19) Dashboard with Vaccination Data [Online]. https://covid19.who.int/. Accessed 18/07/2021. [Google Scholar]
- Wright, G.D. (2019) Unlocking the potential of natural products in drug discovery. Microb Biotechnol 12, 55–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, K.X. and Chu, J.J.H. (2017) Antiviral screen identifies EV71 inhibitors and reveals camptothecin‐target, DNA topoisomerase 1 as a novel EV71 host factor. Antiviral Res 143, 122–133. [DOI] [PubMed] [Google Scholar]
- Wu, W. , Li, R. , Li, X. , He, J. , Jiang, S. , Liu, S. and Yang, J. (2015) Quercetin as an antiviral agent inhibits influenza a virus (IAV) entry. Viruses 8, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, S. , Wang, Q. , Si, L. , Shi, Y. , Wang, H. , Yu, F. , Zhang, Y. , Li, Y. et al. (2014) Synthesis and anti‐HCV entry activity studies of β‐cyclodextrin‐pentacyclic triterpene conjugates. ChemMedChem 9, 1060–1070. [DOI] [PubMed] [Google Scholar]
- Xu, L. , Su, W. , Jin, J. , Chen, J. , Li, X. , Zhang, X. , Sun, M. , Sun, S. et al. (2014) Identification of luteolin as enterovirus 71 and coxsackievirus A16 inhibitors through reporter viruses and cell viability‐based screening. Viruses 6, 2778–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F. , Wang, Q.I. , Zhang, Z. , Peng, Y. , Qiu, Y. , Shi, Y. , Zheng, Y. , Xiao, S. et al. (2013) Development of oleanane‐type triterpenes as a new class of HCV entry inhibitors. J Med Chem 56, 4300–4319. 10.1021/jm301910a [DOI] [PubMed] [Google Scholar]
- Yu, M. , Si, L. , Wang, Y. , Wu, Y. , Yu, F. , Jiao, P. , Shi, Y. , Wang, H. et al. (2014) Discovery of pentacyclic triterpenoids as potential entry inhibitors of influenza viruses. J Med Chem 57, 10058–10071. [DOI] [PubMed] [Google Scholar]
- Zhan, Y. , Ta, W. , Tang, W. , Hua, R. , Wang, J. , Wang, C. and Lu, W. (2021) Potential antiviral activity of isorhamnetin against SARS‐CoV‐2 spike pseudotyped virus in vitro . Drug Dev Res 13, 1124–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Lin, D. , Sun, X. , Curth, U. , Drosten, C. , Sauerhering, L. , Becker, S. , Rox, K. et al. (2020) Crystal structure of SARS‐CoV‐2 main protease provides a basis for design of improved a‐ketoamide inhibitors. Science 368, 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Wu, T. , Zhang, J. , Yan, Q. , Xie, L. and Liu, G.J. (2007) Chinese medicinal herbs for the common cold. Cochrane Database Syst Rev 24, CD004782. 10.1002/14651858.CD004782.pub2. [DOI] [PubMed] [Google Scholar]
- Zhao, H. , Guo, X.K. , Bi, Y. , Zhu, Y. and Feng, W.H. (2014) PKCδ is required for porcine reproductive and respiratory syndrome virus replication. Virology 468–470, 96–103. [DOI] [PubMed] [Google Scholar]
- Zhao, Y. , Chen, C.H. , Morris‐Natschke, S.L. and Lee, K.H. (2021) Design, synthesis, and structure activity relationship analysis of new betulinic acid derivatives as potent HIV inhibitors. Eur J Med Chem 215, 113287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziolkowska, N.E. , O’Keefe, B.R. , Mori, T. , Zhu, C. , Giomarelli, B. , Voidani, F. , Palmer, K.E. , McMahon, J.B. et al. (2006) Domain‐swapped structure of the potent antiviral protein griffithsin and its mode of carbohydrate binding. Structure 14, 1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziolkowska, N.E. and Wlodawer, A. (2006) Crystal structure of unliganded griffithsin. 10.2210/pdb2GTY/pdb. [DOI]




